Abstract
Dietary rice bran (RB) has shown capacity to influence metabolism by modulation of gut microbiota in individuals at risk for colorectal cancer (CRC), which warranted attention for delineating mechanisms for bi-directional influences and cross-feeding between the host and RB-modified gut microbiota to reduce CRC. Accordingly, in the present study, fermented rice bran (FRB, fermented with a rice bran responsive microbe Bifidobacterium longum), and non-fermented rice bran (RB) were fed as 10% w/w (diet) to gut microbiota-intactspf or germ-free micegf to investigate comparative efficacy against inflammation-associated azoxymethane /dextran sodium sulfate (AOM/DSS)-induced CRC. Results indicated both microbiota-dependent and independent mechanisms for RB meditated protective efficacy against CRC that was associated with reduced neoplastic lesion size and local-mucosal/systemic inflammation, and restoration of colonic epithelial integrity. Enrichment of beneficial commensals (such as, Clostridiales, Blautia, Roseburia), phenolic metabolites (benzoate and catechol metabolism), and dietary components (ferulic acid-4 sulfate, trigonelline, and salicylate) were correlated with anti-CRC efficacy. Germ-free studies revealed gender-specific physiological variables could differentially impact CRC growth and progression. In the germ-free females, the RB dietary treatment showed a ~72% reduction in the incidence of colonic epithelial erosion when compared to the ~40% reduction in FRB-fed micegf. Ex vivo fermentation of RB did not parallel the localized-protective benefits of gut microbial metabolism by RB in damaged colonic tissues. Findings from this study suggest potential needs for safety considerations of fermented fiber rich foods as dietary strategies against severe inflammation-associated colon tumorigenesis (particularly with severe damage to the colonic epithelium).
Keywords: colon carcinogenesis, cancer intervention, azoxymethane, germ free mice, rice bran, microbiome, metabolomics, probiotics, fermentation
1. INTRODUCTION
Colorectal cancer (CRC) is the 2nd leading cause of cancer-related deaths in US 1 and develops as a result of complex interactions between microbiota, dietary components, and gut mucosal immune/ inflammatory mediators 2-4. Multiple research efforts have associated the differential impact of human gut microbiota and microbiota-associated metabolism changes with CRC 2,3,5-9. Given the high CRC incidence and mortality statistics, host and environmental factors that affect CRC, alongside novel agents that modulate host and gut microbial metabolism merit attention to reduce CRC. Thus, practical dietary interventions that modulate these interactions and in-turn reduce incidence of adenomatous polyps and/or prevent their progression to CRC are urgently needed.
Whole grain, brown rice and rice bran components with bioactivity have been reported for exhibiting protection against CRC 10-12. Rice bran is an agricultural co-product of rice processing that has shown preventive efficacy against chronic colitis and CRC in numerous pre-clinical studies 10,11,13-17. The probiotic metabolism of rice bran 11,18,19 and completed human studies provide compelling support for rice bran metabolism by colonic microbiota in favor of reducing CRC 20-25. Stool metabolite analysis from healthy adults and CRC survivors showed changes after 4 weeks of 30g/day rice bran consumption; these clinical results were corroborated in another clinical trial in CRC survivors following 24 weeks of 30g/day rice bran feeding 20-22,24. Some of the identified rice bran-modified gut microbiota (e.g. Bifidobacterium spp.) are highlighted for CRC chemoprevention 26. Notably, germ-free mice receiving human fecal transplantation with rice-bran modified stool microbes showed significantly less colon cancerous lesions compared to mice humanized by stool microbes from control fed adults (without rice-bran intervention) 27.
Taken together, these studies provided convincing rationale for testing the hypothesis that rice bran and Bifidobacterium-fermented rice bran can re-structure the ‘high risk’ or dysbiotic gut microbiota towards the colonization of microbial communities that protect against CRC growth and progression. However, the microbiome-mediated and microbiota-independent colonic tissue/tumor effects and metabolic pathways impacted following dietary rice bran or fermented rice bran remain largely unknown. This gap in literature needs to be addressed because establishing anti-CRC actions as dependent and/or independent of an individual’s gut microbiota composition can guide future precision nutrition based approaches for CRC control and prevention. In addition, we also hypothesized that some phytochemicals in rice bran serve as gut microbial substrates and are metabolized into bioactive metabolites that impact the colonic microenvironment for enhanced efficacy against CRC. Accordingly, Bifidobacterium longum fermented rice bran and non-fermented dietary rice bran were fed to gut microbiota-intactspf or germ-free micegf and evaluated for comparative efficacy to a control diet against azoxymethane /dextran sodium sulfate (AOM/DSS)-induced CRC (a pre-clinical animal model with inflammation-associated colon tumorigenesis 28.
2. MATERIALS AND METHODS
2.1. Antibodies and reagents
Azoxymethane (AOM, #A5486) was from Millipore Sigma (Temecula, CA, USA) and Dextran sodium sulfate (DSS, mol.wt. 36,000–50,000, #02160110-CF) was from MP Biomedicals (Santa Ana, CA, USA). Antibodies for Ki67 (#ab16667, RRID:AB_302459), CD44 (#ab157107, RRID:AB_2847859), NF-κB (p65) (#ab16502, RRID:AB_443394), ZO-1 (#ab96587, RRID:AB_10680012), Claudin-2 (#ab53032, RRID:AB_869174) and Alcian Blue PAS Stain Kit (#ab245876) were from Abcam (Cambridge, MA). Antibody for β-catenin (#sc-59737, RRID:AB_781850) and Cox-2 (#sc-376861, RRID:AB_2722522) were from Santa Cruz Biotechnology (Dallas, TX, USA). Biotin conjugated secondary anti-rabbit (#E0432, RRID:AB_2313609) and anti-mouse (#E0433, RRID:AB_2687905) antibodies were from Dako/Agilent technologies (West Cedar Creek, TX, USA) and Streptavidin-HRP (#434323, RRID:AB_2619743) and CAS-Block (#008120) were from ThermoFisher Scientific (Waltham, MA, USA). The diaminobenzidine (DAB) kit (#SK4100, RRID:AB_2336382) was from Vector Laboratories (Burlingame, CA, USA). The Proteome Profiler Mouse XL Cytokine Array kit (#ARY028) was purchased from R&D systems (Minneapolis, MN, USA).
2.2. Experimental study-diet preparation
Ri300 heat-stabilized rice bran (110 °C for 30 min) was purchased from Rice Bran Technologies (Sacramento, CA, USA). Bifidobacterium longum (ATCC-55813; American Type Culture Collection, Manassas, VA, USA) fermented rice bran was harvested and lyophilized for animal study as described previously 18. 4% Corn oil diet (AIN-93M diet, Envigo, Madison, WI, USA) was used as the control diet. Heat-stabilized rice bran and the B. longum-fermented rice bran were incorporated at 10% w/w into the diets (Envigo); nutrient ingredients were adjusted in the supplemented diets and the diets were all gamma-irradiated (sterilization process) and tested for absence of microbial contamination.
2.3. Animal study design, necropsy, and sample collection/processing
Six-week old male Balb/c micespf (Charles River Laboratories, Wilmington, MA, USA) were housed in specific pathogen free animal facility at UC Denver-AMC, and fed a control AIN-93M pellet diet for one-week (acclimatization period) and then administered a single intraperitoneal (i.p) injection of AOM 10 mg/kg body weight in saline followed by exposure to 2% DSS in drinking water for five days [as per modified protocol of Suzuki et al 28] with one water change once after 2 days providing fresh DSS. After completion of DSS exposure, mice were switched to supplemented diets with rice bran (RB, n=20), B. longum-fermented rice bran (FRB, n=20) or maintained on a control AIN-93M diet (n=20) for 15 weeks. Non-AOM/DSS exposed mice (on control AIN-93M diet) served as overall negative controls (NC, n=5) in the study; non-AOM/DSS exposed but fed-with RB (n=5) or FRB (n=4) diets were also included as negative controls. An overview of the study design is depicted in Supplementary Figure S1. Weekly body weight, diet consumption, and general health (stool consistency, incidents of bloody diarrhea/loose stools, and rectal-prolapse were monitored closely for confirmation of colitis events). Aseptic condition was maintained to avoid cross contamination of microbiota between different groups throughout the study. At the end of 15-week feeding phase, all groups were euthanized via CO2 asphyxiation followed by exsanguination. Details about necropsy procedure, quantification of lesions, collection of blood, intestinal tissue/contents and microbiome/metabolomics procedures evaluations are all detailed in Supplementary Methods section.
C57BL/6 germ free micegf breeding colony and germ-free litters were housed in sterile vinyl isolators in the gnotobiotic facility at UC Denver-AMC. Micegf were maintained in isolators and provided with ad libitum access to autoclaved water and gamma-irradiated AIN-93M pellet diet. All micegf were exposed to the AOM/DSS procedure as detailed above. This was then followed by 14 days of normal drinking water (resting phase). This cycle (DSS exposure + resting phase) was repeated twice to yield a total of three DSS cycles as the C57BL/6 strain is more resistant to AOM/DSS induced pathological manifestations. Micegf (male or female) were randomized into three groups according to their diet: Controlgf (M = 10, F = 3); RBgf (M = 8, F = 7) and FRBgf (M = 10, F = 5) mice. After 1st cycle of DSS exposure, mice were switched to RB, FRB or maintained on control diets for 18 weeks and then euthanized and tissue/samples were collected as detailed above. An overview of the study design is depicted in Supplementary Figure S3.
2.4. Ethics statement.
All animal studies were performed under Institutional guidelines using approved Institutional Animal Care and Use Committee (IACUC) protocol [(B- 57916(04)1E] in the specific pathogen free facility and gnotobiotic core facility at UC Denver-AMC.
2.5. Microbiome and Metabolomic analysis.
Briefly, 16s RNA sequencing was performed on the cecum and colon tissue contents as described previously 18. Clean (devoid of any colon fecal contents) proximal and distal colon tissue slivers (50 mg/sample) and plasma (1 mL/sample) from mice were sent to Metabolon Inc., (Durham, NC) for metabolite extraction and identification as described previously 18,27. Procedures, evaluations and statistical analysis are all detailed in Supplementary Methods section.
2.6. Immunohistochemical (IHC), and cytokine analysis.
Paraffin-embedded sections (5 μm thick) were deparaffinized and stained using specific primary antibodies followed by DAB staining, as described previously 29. Primary antibody dilutions used were Ki-67 (1:25), β-catenin (1:25), Cox-2 (1:25), NF-κB (p65)(1:250), CD44 (1:250), Claudin-2 (1:50) and ZO-1(1:100). Rabbit anti-mouse IgG (Dako) and goat anti-rabbit IgG (Dako) were used as a biotinylated secondary antibody. % Positive cells were quantified as the number of brown-stained cells x100 per total number of cells counted under ×400 magnification in 5-8 randomly selected fields in each sample. Immunoreactivity based on intensity of brown staining) was scored as 0 (no staining), +1 (very weak), +2 (weak), +3 (moderate) and +4 (strong). Alcian blue and Periodic acid Schiff (PAS) stains were used for simultaneous differentiation of acidic and neutral mucins in colon tissue as described previously 30. The expression of cytokines and chemokines in mouse plasma were determined using commercially available Proteome Profiler Mouse XL Cytokine Array (#ARY028, R&D Systems, Minneapolis, MN, USA). Membrane dot blots were quantified using Image Studio Lite software.
2.7. Statistical and microscopic analyses.
All pathology and IHC based statistical analyses were carried out with GraphPad Prism software version-5 and two-sided P values <0.05 were considered significant. Fisher’s Exact test was used to compare incidence of epithelial erosion, and grades of dysplasia. Unpaired two-tailed Student’s t-test was employed as needed. Difference between different groups was determined by one-way ANOVA followed by Tukey-test for multiple comparisons. All microscopic analyses were done by Zeiss Axio Imager.A1 microscope (Carl Zeiss, Inc., Jena, Germany) and photomicrographs captured by AxioCam MrC5 camera (Carl Zeiss).
3. RESULTS
3.1. Differential effect of RB and FRB intake on inflammation-associated colon tumorigenesis in gut microbiota-intactspf (specific pathogen-free) mice.
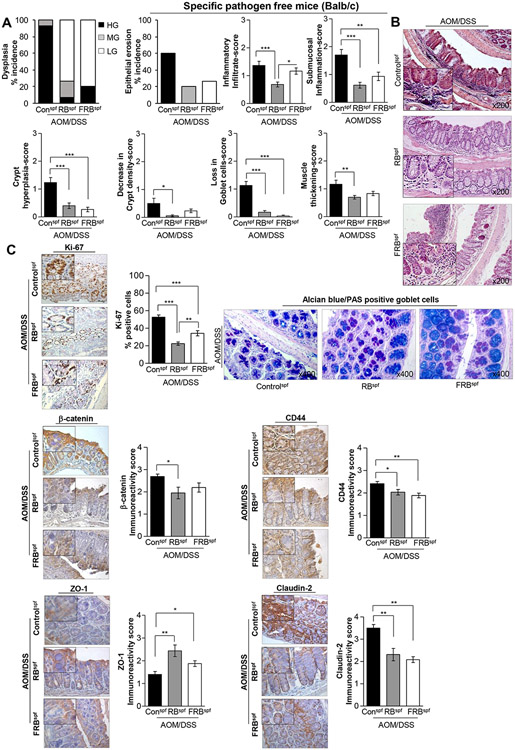
Dietary intervention with RB and FRB feeding for 15 weeks (post-AOM/DSS induction) in a gut microbiota-intact micespf did not result in changes to food consumption and no significant differences were observed in body weight between FRB and RB supplemented diet-fed mice and AOM/DSS controlsspf (data not shown). At necropsy, it was observed that both RB and FRB-fed groups had relatively increased colon length, and less colonic (proximal, middle, distal regions) macro-tumor lesions (though statistically not significant) which were significantly smaller in size compared to AOM/DSS controlsspf (Supplementary Fig.S1). Given that these gross morphological observations did not reveal differences in the relative efficacy of RB vs. FRB against AOM/DSS associated colon tumorigenesis, H&E-stained colon sections were microscopically examined for histopathological changes and scored as detailed in Supplementary Table 1 [according to published criteria with slight modifications 31,32] . Histopathological evaluation (Fig. 1A-B) revealed that colon tissue from untreated and treated AOM/DSS micespf had either dysplastic and/or adenoma/adenocarcinoma characteristics, with no evidence of invasive adenocarcinoma. Briefly, observed dysplastic lesions and other histological manifestations were classified into (a) low grade [LG], (b) moderate grade [MG], (c) high grade [HG] dysplasia. In AOM/DSS controlsspf, ~93% of mice showed HG and ~7% showed MG dysplasia characteristics (Fig. 1A). In comparison, there was only ~7% incidence of HG and ~73% incidence of LG dysplasia (including ~20% incidence of MG) in RBspf (AOM/DSS) group (Fig. 1A). On the other hand, compared to RB-fed group, there was a relatively higher incidence of HG dysplasia in FRBspf (AOM/DSS) group; specifically, there was ~20% incidence of HG and ~80% incidence of LG dysplasia in FRB-fed group (Fig. 1A). The photomicrographs, representative of a treatment group, are shown in Fig. 1B.
Fig 1. Effect of RB and FRB-intake on AOM/DSS-induced colon tumorigenesis in gut microbiota-intactspf (specific pathogen-free) Balb/c mice.
(A) Histopathological grading of AOM/DSS micespf colon tissues (with and without RB and FRB-supplemented diets); a score ranging from 0 for normal (or lower) to 3 (extreme events) was assigned. (B) Representative pictographs of distal colon (H&E images) showing histopathological changes; images were acquired at x200 with digitally magnified insets. (C, upper left-and lower-panel) Pictorial and data representation for % Ki-67 positive cells, and immunoreactivity scores for β-catenin, CD44, ZO-1, and claudin-2 expression levels based on immunohistochemical staining. Images were acquired at x400 with digitally magnified insets. % Positive cells were quantified as the number of brown-stained cells x100 per total number of cells counted under ×400 magnification in 5-8 randomly selected fields in each sample. Immunoreactivity (represented by intensity of brown staining) was scored as 0 (no staining), +1 (weak), +2 (moderate), +3 (strong) and +4 (very strong). (C, upper right-panel) Representative pictograph (x400 magnification) of alcian blue-PAS staining in the distal colon tissue depicting changes in mucin composition and distribution. Quantified data is represented as columns (mean for each group); bars represent SEM. ***P ≤0 .001, **P ≤ 0.01, and *P ≤ 0.05. RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; HG, high grade; MG, moderate grade; LG, low grade dysplasia; PAS, Periodic acid–Schiff; micespf, specific pathogen-free mice.
Apart from assessing the severity of the colonic lesions, tissue sections were further analyzed for the signs of epithelial erosion and muscular thickening (if any), presence of inflammatory infiltration (including extent of submucosal inflammation), crypt hyperplasia, decrease in crypt density and associated loss/and or type of goblet cells (Fig. 1A-C). For each of these pathological manifestations, a score ranging from 0 for normal (or lower) to 3 (extreme events) was assigned as per published criteria 31,32 (Supplementary Table S1). Moderate colonic epithelial erosion events were observed in 60% of AOM/DSS controlsspf, while the RBspf and FRBspf (AOM/DSS) groups displayed only mild erosion of epithelium in certain regions, and the incidence was also reduced to ~20-27%, respectively. There was increased presence of inflammatory cells in the mucosa/lamina propria and clusters of infiltrating cells were observed in the sub-mucosa of AOM/DSS controlsspf. Notably, there was an overall ~50% decrease (P<0.001) in the presence of mucosal inflammatory cells in the RBspf (AOM/DSS) group compared to the AOM/DSS controlsspf; sub-mucosal inflammation (if any) was minimal (only individual cells), and no clusters were observed. On the other hand, there was only slight decrease (which was non-significant) in the mucosal inflammatory cells of the FRBspf (AOM/DSS) group; however, the extent of sub-mucosal infiltration was markedly reduced (45%, P<0.01) compared to AOM/DSS controlsspf. Increase in crypt lengths (slight crypt hyperplasia), as observed in AOM/DSS controlsspf, was not very evident (P<0.001) in RB and FRB-fed groups. Only slight decrease (<10%) in crypt density was observed in FRBspf (AOM/DSS) and AOM/DSS controlsspf, such changes were negligent in the RB-fed group. Moreover, thickness of the colon muscularis externa was slightly increased across all groups.
For AOM/DSS controlsspf, a marked decrease was observed in mucin secreting goblet cells. The goblet cells were smaller in size and appeared irregularly scattered/or absent in the regions of high proliferative activity and dysplastic zones (Fig. 1C). However, there was a significant increase in both goblet cell number and size in RBspf and FRBspf (AOM/DSS) groups compared to AOM/DSS controlsspf; though, RB-fed group displayed a relatively higher number than FRB-fed group. Dual staining with alcian blue (AB) and periodic acid schiffs (PAS) stain to detect acidic and neutral mucins 30, respectively, indicated a shift towards the presence of more acidic mucins with RB and FRB feeding (Fig. 1C). The RB-fed group displayed a mixed combination of ‘dense’ blue stained/acidic mucin rich goblet cells and dual stained (violet) goblet cells of moderately uniform size, whereas the FRB-fed group had relatively larger sized but less dense blue stained/acidic mucin rich goblet cells as well as dual stained-but smaller sized goblet cells.
Next, IHC analysis of colon tissues was performed to determine the effect on expression pattern of CRC associated molecular markers. Quantification of Ki-67 staining showed a decrease in proliferation indices by ~57% and ~35% (P<0.001, for both) in RBspf and FRBspf (AOM/DSS) groups, respectively (Fig. 1C). β-catenin expression was decreased by ~28% (P<0.05) in RBspf (AOM/DSS) mice, but FRB-feeding had no effect (Fig. 1C); also, RB and FRB feeding decreased CD44 expression by ~16% (P<0.05) and ~22% (P<0.01), respectively, in AOM/DSS micespf mice (Fig. 1C). With regards to the impact on tight cellular junction complexes that regulate permeability of mucosal barrier such as occludins and claudins 33, both RB and FRB feeding in AOM/DSS micespf (Fig. 1C) strongly decreased claudin-2 expression (a marker of leaky epithelia) by ~34% and ~41% (P<0.01, for both), respectively; notably, the expression of scaffold -forming intracellular zonula occludin (ZO-1) that has been shown to decrease with CRC progression, was significantly increased ~1.7 folds (P<0.01) and ~1.3 folds (P<0.05) by RB and FRB feeding, respectively.
3.2. Effect of RB and FRB-intake on gut microbiota composition during inflammation-associated colon tumorigenesis in gut microbiota-intactspf mice.
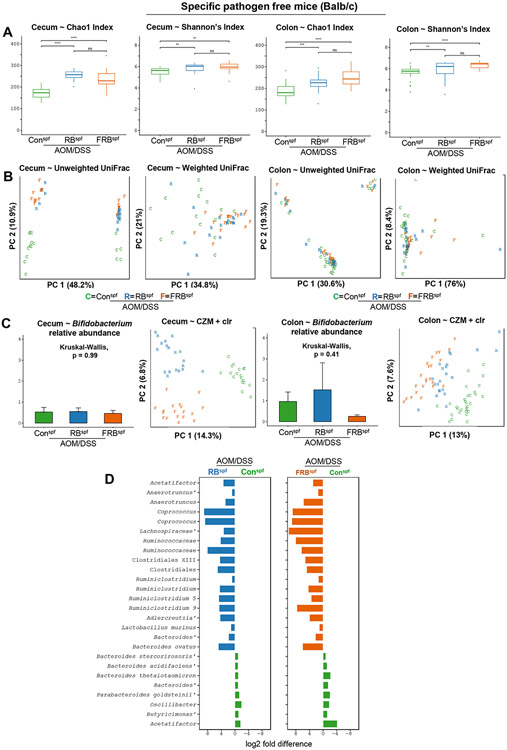
To assess the impact of rice bran supplemented diets (with and without fermentation) during AOM/DSS facilitated colon tumorigenesis on species richness and species diversity of cecum and colon microbiota, in RBspf and FRBspf-AOM/DSS groups, we performed pairwise Kruskal-Wallis testing across each diet group for two non-phylogenetic alpha diversity metrics: Chao1 index (richness) and Shannon’s Index (diversity) (Fig.2A). We detected significant differences in both richness and diversity of microbiota between AOM/DSS controlsspf vs. RBspf and FRBspf (AOM/DSS) groups (Fig.2A). However, no differences for either metric were detected between RBspf vs. FRBspf (AOM/DSS) groups. We next compared phylogenetic-based beta diversity of cecum and colon microbiota in all groups through principal coordinates analysis (PCoA) of unweighted and weighted UniFrac distances. Ordination demonstrated clear separation of cecum microbiota by diet group for both UniFrac metrics; however, colon microbiota showed relative similarity across each diet group and metric (Fig. 2B). These observations were corroborated through pairwise PERMANOVA comparisons where statistically significant differences were observed in cecum microbiota in all three groups for both metrics. Additionally, in unweighted UniFrac, colon microbiota also exhibited statistically significant differences between AOM/DSS controlsspf vs. RBspf and FRBspf (AOM/DSS) groups; however, no differences were observed in colon microbiota for any of the pairwise diet comparisons of weighted UniFrac distance.
Fig 2. Effect of RB and FRB-intake on gut microbiome (caecum and colon) composition during inflammation associated colon tumorigenesis in gut microbiota-intactspf mice.
A) species richness and diversity were analyzed by Shannon/Chao indices, B) clear separation in community composition based upon sample type and diet group by Principal components analysis (PCA) of centred log-ratio transformed abundances for all amplicon sequence variants. Percentage values along each axis indicate the amount of variation explained by each of the first two principal components C) Bifidobacterium longum abundance was not changed between dietary groups in caecum, and had greater expression in RB diet supplemented mice. Letters and colors denote study diet group for each sample type (caecum or distal colon). D) Bar charts of log2 fold differences for 26 differentially abundant (FDR-P<0.1) amplicon sequence variants when comparing rice bran or B. longum-fermented rice bran diets compared to control.
Next, Kruskal-Wallis testing of the per-sample relative abundance of 11 amplicon sequence variants (ASVs) assigned to the genus Bifidobacterium was performed to determine whether an enrichment of the fermentation strain occurred in the gut microbiomes of FRBspf (AOM/DSS) mice (which were consuming the B. longum fermented rice bran). Results indicated lack of enrichment of the fermenting strain in these mice (non-significant p-values for both cecum and colon samples, Fig. 2C); these results are consistent with our previous findings in healthy mice fed a similar FRB diet 18.
In recognition of the inherent compositional nature of microbiota datasets acquired from next-generation sequencing platforms, we also employed a compositional analysis framework that used the count zero multiplicative (CZM) method to replace zeros in the ASV absolute abundance table followed by transformation using the centered log-ratio (clr). Principal components analysis (PCA) of CZM-adjusted clr abundances displayed striking separation by diet groups for both cecum and colon microbiota (Fig. 2C). To explore specific ASVs that may be driving differences between diet groups, we utilized ALDEx2 for differential abundance testing in pairwise fashion for all diet combinations. This analysis identified 72 ASVs in cecum samples of AOM/DSS controlsspf and RBspf (AOM/DSS) mice; specifically, 17 enriched in AOM/DSS controlsspf and 55 enriched in RBspf (AOM/DSS) group. Furthermore, this analysis identified 69 ASVs in cecum samples of AOM/DSS controlsspf and FRBspf (AOM/DSS) groups; specifically, 25 enriched in AOM/DSS controlsspf and 44 enriched in FRBspf (AOM/DSS) group. Notably, 24 ASVs were identified in cecum samples of RBspf and FRBspf (AOM/DSS) groups; specifically, 9 enriched in FRBspf (AOM/DSS) and 15 enriched in RBspf (AOM/DSS) group. For colon samples, analysis identified 52 ASVs in AOM/DSS controlsspf and RBspf (AOM/DSS) mice: 17 enriched in AOM/DSS controlsspf and 35 enriched in RBspf (AOM/DSS) group; 85 ASVs in colon samples for AOM/DSS controlsspf and FRBspf (AOM/DSS) groups: 16 enriched in AOM/DSS controlsspf and 69 enriched in FRBspf (AOM/DSS) groups. This analysis also identified 33 ASVs in colon samples for RBspf and FRBspf (AOM/DSS) groups: 27 enriched in FRBspf (AOM/DSS) and 6 enriched in RBspf (AOM/DSS) group. Importantly, a total of 18 ASVs were consistently enriched in both cecum and colon samples of either RBspf or FRBspf (AOM/DSS) groups compared to AOM/DSS controlsspf (Fig. 2D). ASVs affiliated with the genus Bacteroides dominated the microbiota of AOM/DSS controlsspf, while mice fed either of the supplemented diets showed enrichments for ASVs assigned as Clostridiales, Lachnospiraceae, Ruminiclostridium, and Coprococcus, among others (Fig. 2D). Notably, seven of these ASVs also exhibited further enrichment in FRBspf (AOM/DSS) mice compared to RBspf (AOM/DSS) group, indicating the potential of B. longum-fermented diet intake to cause enrichment of gut ASVs. (Fig. 2D). These additional ASVs in the FRBspf (AOM/DSS) mice included Blautia, Streptococcus, and an identical Roseburia-ASV previously identified in healthy mice fed a B. longum-fermented diet 18.
3.3. Effect of RB and FRB-intake on microbial metabolites in gut and systemic circulation after inflammation-associated colon tumorigenesis in gut microbiota-intactspf mice.
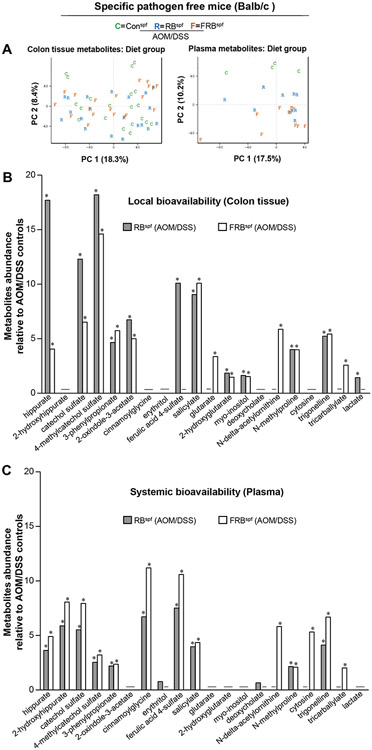
A species-rich and diverse gut ecosystem has the inherent potential and increased capacity for the fermentation of nondigestible and undigested substrates, which can result in the production of bioactive metabolites with either health benefits or adverse effects-impacting various physiological/metabolic processes and diseases, including CRC 3,34,35. Therefore, we next assessed the metabolic by-products generated after the intake of RB and FRB-supplemented diets in AOM/DSS micespf model. Specifically, the bioavailability of the metabolites generated in different groups was assessed locally (uptake in colon tissue) as well as systemically (presence in plasma). For this, a non-targeted metabolomics approach using ultra-performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS) was employed to assess relative abundance of metabolites in the distal colon tissue and plasma of AOM/DSS controlsspf, RBspf and FRBspf (AOM/DSS) mice. A total of 536 differentially abundant metabolites were identified in the colon and plasma metabolomes from AOM/DSS treated micespf; the metabolite profiles differed across control, RB and FRB diet fed-AOM/DSS groups. The PCA for the metabolite profile composition across all groups is depicted in Fig 3A. Non-targeted metabolomics revealed identity of 21 metabolites in plasma and colon tissue, whereby the levels were significantly different between experimental groups (Fig. 3B-C). Specifically, there was a ~4-8 fold increase (P<0.05) in the plasma levels of hippurate and catechol sulphate in both RBspf and FRBspf (AOM/DSS) mice; importantly, the levels increased by ~12-17 folds (P<0.05) in the colon tissue of RB fed-AOM/DSS micespf. The 2-hydroxyhippurate was not detected in the colon tissue of mice fed either of the supplemented diets, yet plasma levels were significantly increased (~6-8 folds, P<0.05) when compared to AOM/DSS controlsspf. The 4-methylcatechol sulphate levels were also significantly increased (~15-18 folds, P<0.05) in the colon tissue and plasma (~3 folds, P<0.05) of RBspf and FRBspf (AOM/DSS) mice. Interestingly, cinnamoylglycine was not detected in the colon tissue of RBspf and FRBspf (AOM/DSS) mice, but its plasma levels increased by ~6-11 folds (P<0.05) compared to AOM/DSS controlsspf. Cytosine was another compound that was not detected in the colon tissue, but its plasma level was increased by ~5 folds (P<0.05) in the plasma of FRBspf (AOM/DSS) mice. The metabolite, salicylate, showed a ~4-10 folds increase (P<0.05) in the plasma and colon tissue levels, respectively, of both RBspf and FRBspf (AOM/DSS) mice; on similar lines, N-methylproline and 3-phenylpropionate levels were also increased in the plasma (~2 folds) and colon tissue (~4-6 folds) of both these groups (P<0.05 for both) compared to the AOM/DSS controlsspf. Trigonelline levels were also increased in the plasma (~4-7 folds; P<0.05) and colon tissue (~5 folds; P<0.05) of both RBspf and FRBspf (AOM/DSS) mice. Notably, ferulic acid-4 sulphate levels were significantly increased (~8-10 folds, P<0.05) in the plasma and colon tissue of RBspf (AOM/DSS) mice, respectively; however, it was not detected in the colon tissue of FRB diet fed-AOM/DSS micespf (though there was ~10 folds increase in its plasma levels in this group). Tricarballylate had increased abundance (~2 fold; P<0.05) in the plasma and colon tissue of FRBspf (AOM/DSS) mice (and not the RB-fed) compared to the AOM/DSS controlsspf. Importantly, N-delta-acetylornithine levels were significantly increased (~5-6 folds; P<0.05) in both plasma and colon tissue of FRBspf (AOM/DSS) mice compared to RBspf (AOM/DSS) and AOM/DSS controlsspf.
Fig 3. Effect of RB and FRB-intake on microbial metabolites found locally in gut or systemic circulation following AOM/DSS-induced colon tumorigenesis in microbiota-intactspf mice.
(A) Principal components analysis (PCA) of colonic and plasma metabolomes from control, rice bran and fermented rice bran diet groups. (B-C) Median scaled relative abundance for selected metabolites in colon tissue and plasma for rice bran or B. longum-fermented rice bran diet groups compared to control. Differential metabolite abundance is reported as relative to values obtained in control diet fed AOM/DSS treated micespf. Quantified data is represented as Columns (relative fold change); *P ≤ 0.05. RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; micespf, specific pathogen-free mice.
3.4. Effect of RB and FRB-intake on the expression of inflammatory mediators during inflammation-associated colon tumorigenesis in gut microbiota-intactspf mice.
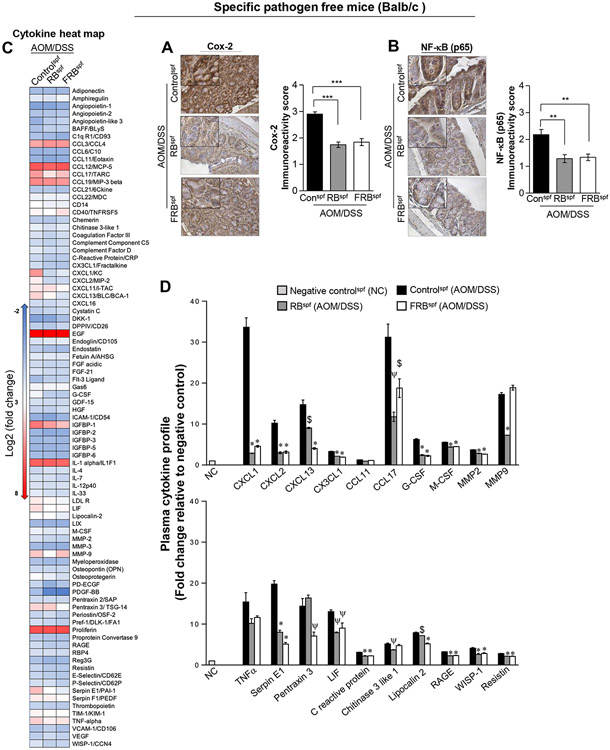
Given that in the H&E analysis we observed significant decrease in immune cell infiltration in the colonic tissues of RBspf and FRBspf (AOM/DSS) mice, we next assessed the expression of inflammation mediators Cox-2 and NF-kB/p65 molecular levels in the colonic tissues. The earlier pathological results were corroborated by analysis of the colonic mucosa which showed significant decrease in the expression of inflammation mediators Cox-2 (P < 0.001) and NF-kB/p65 (P < 0.02) in the colonic tissue of RBspf and FRBspf (AOM/DSS) mice (Fig. 4A-B). Next, we performed a proteome profiler array to detect effect of RB and FRB feeding on inflammation modulators-cytokine/chemokine profiles in the plasma of AOM/DSSspf mice (Fig. 4C-D). Our results indicated that various chemokines and cytokines implicated in cancer growth and progression are significantly increased in AOM/DSS controlsspf compared to negative controls (NC, no-AOM/DSS group) (Fig. 4C-D, Supplementary Fig. S2), and were significantly downregulated by dietary intervention. Notably, the expression of CXCL1 was decreased by ~90% and ~86% by RB and FRB-feeding (P<0.001, for both), respectively. On similar lines, RB and FRB feeding decreased CXCL2 (~70%), CXCL13 (~39-73%), and cytokine CCL17 (~62% and ~40%) plasma levels, respectively, in AOM/DSSspf mice. Plasma levels of G-CSF, M-CSF and MMP-2 were also decreased by the supplemented-diets; however, MMP9 levels were significantly decreased only by RB-feeding. Furthermore, expression of other inflammatory mediators such as Serpin E1, Pentraxin 3, c-reactive protein, Lipocalin-2, RAGE, WISP-1, and Resistin were also differentially modulated by RB and FRB feeding in AOM/DSSspf mice (Fig. 4C-D, Supplementary Fig. S2).
Fig 4. Effect of RB and FRB-intake on the expression of inflammatory mediators during AOM/DSS-induced colon tumorigenesis in gut microbiota-intactspf mice.
Pictorial and data representation for colonic expression of inflammation-associated molecules (A) Cox-2, and (B) NF-κB (p65). Images were acquired at x400 with digitally magnified insets. Immunoreactivity (represented by intensity of DAB-stained brown staining) was scored as 0 (no staining), +1 (weak), +2 (moderate), +3 (strong) and +4 (very strong). Quantified data is represented as Columns (mean for each group); bars represent SEM. ***P ≤0 .001, **P ≤ 0.01, and *P ≤ 0.05. (C) Heat map depicting relative changes in the plasma expression of various cytokine/chemokines and other inflammatory-mediators involved in immune responses, and CRC growth and progression. Plasma collected from untreated (negative controls) and AOM/DSS micespf (with and without RB and FRB-supplemented diets) was evaluated for the presence of various cytokines/chemokines using a mouse-Proteome profiler membrane array containing antibodies to 111 different cytokines/chemokines. Colors are assigned according to the relative scale of expression, ranging from ****−2 to 8, and represent fold change compared to negative controls. (D) Densitometric analysis of dot blots from pooled samples in each group are shown as fold changes relative to negative controls (NC). Quantified data is represented as Columns (mean for each group); bars represent SEM. $; p<0.05, ψ; p<0.01, *; p<0.001). RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; micespf, specific pathogen free mice.
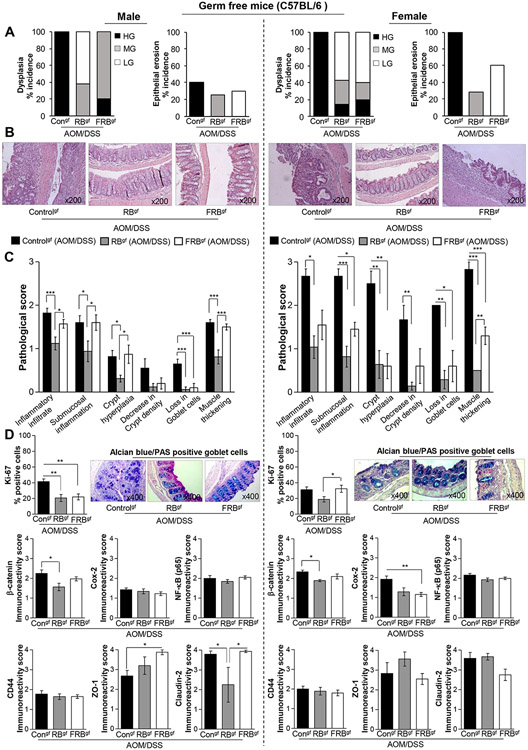
3.5. Gender specific effects of RB and FRB-intake on inflammation-associated colon tumorigenesis in germ-free micegf.
To determine whether the protective effects of non-fermented RB were dependent/independent of gut microbiota presence, we next assessed the RB and FRB intervention benefits against AOM/DSS carcinogenesis in a germ-free mice model. Given the varied susceptibility of the male and female germ-free mice to AOM/DSS-induced health impacts (weight loss) including colonic disease manifestations (in-house preliminary study observations) we performed the efficacy studies in both sexes. Note: both female and male germ-free mice were in the C57BL/6 background (given the limited availability of our in-house bred colony-strain) unlike the microbiota-intact mice (BALB/c strain); hence, to account for variability due to strain differences, the results of both studies were viewed individually. In the germ-free AOM/DSS control groups (AOM/DSS controlsgf) the females had relatively higher number of colonic macro-lesions compared to their male counterparts; notably, RB and FRB intervention decreased both number and size of these tumor lesions irrespective of the mice gender (Supplementary Fig. S3). Microscopically, though, both male and female AOM/DSS controlsgf also displayed 100% incidence of HG dysplasia characteristics, the female colonic tissue was marred by significant epithelial erosion (100% of mice showed these characteristics) Fig. 5A. In comparison, RB-intervention in the male mice protected against HG dysplasia and displayed signs of LG and MG dysplasia in the colonic tissue whereas FRB intervention resulted in a marked decrease in HG dysplasia together with presence of MG dysplasia Fig. 5A. In the female mice, both RB and FRB feeding resulted in significant decrease in HG dysplasia incidence (~14-20 %); however, in FRBgf (AOM/DSS) group, 60% of mice still had significant colonic epithelial erosion, while only ~28% of RB-fed mice displayed erosion characteristics. The photomicrographs, representative of a treatment group, are shown in Fig. 5B. Importantly, compared to male mice, female-AOM/DSS controlsgf showed higher pathological score for colonic inflammatory infiltrate, submucosal inflammation, crypt hyperplasia, decrease in crypt density, loss in goblet cells and muscle thickening (Fig. 5C); the inflammatory cellular infiltrates were more evident in the regions of eroded epithelium. While, RBgf (AOM/DSS) group showed overall low pathological scores irrespective of gender, FRB-feeding had differential effects based on gender of germ-free mice (Fig. 5C). In female-FRBgf (AOM/DSS) group, inflammation and crypt hyperplasia scores were significantly decreased compared to AOM/DSS controlsgf; however, this effect was not evident in the male mice.
Fig 5. Gender-specific effects of RB and FRB-intake on AOM/DSS-induced colon tumorigenesis in the absence of gut microbiota (germ-free micegf).
Histopathological analysis of AOM/DSS micegf colon tissues (with and without RB and FRB-supplemented diets) in male (left-panel) and female (right-panel) C57Bl/6gf mice; (A) % incidence of colonic dysplasia and epithelial erosion, (B) Representative pictographs of distal colon (H&E images) showing histopathological changes; images were acquired at x200, and (C) Pathological scores for inflammatory infiltrates, submucosal inflammation, crypt hyperplasia, decrease in crypt density and loss in goblet cells; a score ranging from 0 for normal (or lower) to 3 (extreme events) was assigned. (D, upper right-panel, male and female) Representative pictograph (x400 magnification) of alcian blue-PAS staining in the distal colon tissue depicting changes in mucin composition and distribution; (D, upper left-and lower-panel, male and female) Pictorial and data representation for % Ki-67 positive cells, and immunoreactivity scores for β-catenin, Cox-2, NF-κB (p65), ZO-1, and claudin-2 expression levels based on immunohistochemical staining. % Positive cells were quantified as the number of brown-stained cells x100 per total number of cells counted under ×400 magnifications in 5-8 randomly selected fields in each sample. Immunoreactivity (represented by intensity of brown staining) was scored as 0 (no staining), +1 (weak), +2 (moderate), +3 (strong) and +4 (very strong). Quantified data is represented as Columns (mean for each group); bars represent SEM. ***P ≤0 .001, **P ≤ 0.01, and *P ≤ 0.05. RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; HG, high grade; MG, moderate grade; LG, low grade dysplasia; PAS, Periodic acid-Schiff; micegf, germ free mice.
Also, in the male AOM/DSS controlsgf , the goblet cells (Fig. 5D, left panel) were fewer in number (compared to gut microbiota -intact mice), small-sized, irregular shaped and absent in many places; these were predominantly stained dark blue (acidic and neutral mucins) in the dorsal colonic region while the proximal regions had more neutral mucins (magenta colored). In the male- RBgf (AOM/DSS) mice, the goblets cells were bigger and more uniform in shape; also, the goblet cells in the lower part of the crypts were mostly alcian blue stained (acidic mucins) whereas the upper parts had more neutral mucins (PAS/magenta stained). On the other hand in the FRB group, the goblet cells were still bigger but less dense and stained mostly as acidic mucins. In the female AOM/DSS controlsgf (Fig. 5D, right panel) in the distal region most of the goblet cells were lost due to epithelial erosion, remnants of crypts had irregularly shaped deep blue goblet cells while proximal had mostly magenta stained neutral mucins. In the female- RBgf (AOM/DSS) mice the number of goblet cells were considerably increased which stained more deep blue than the controls; due to more epithelial erosion-the female FRB-fed mice also had fewer goblet cells, but whatever little were present were dispersed around (either containing magenta stained neutral mucins) or contained deep blue-stained mucins.
Tissue based molecular analysis (Fig. 5D) indicated that both RB and FRB-feeding could significantly decrease proliferating cells (% Ki-67 positive cells) in male-AOM/DSSgf mice (~51-54% decrease, P < 0.01 for both); however, FRB-effect was compromised in the female mice. On similar lines while RB-feeding decreased β-catenin expression (~20-31%, P < 0.05) in both male and female AOM/DSS micegf, FRB-feeding showed no effect. CD44 and NF-kB/p65 expression were unaltered by RB and FRB feeding in both genders; however, Cox-2 levels were decreased in RB (~33% decrease, though not significant) and FRB-fed females (40% decrease, P < 0.01) while Cox-2 levels in male mice were unaffected. Additionally, it was observed that under germ-free conditions RB and FRB feeding did not have any significant effect on the expression levels of tight junction proteins ZO-1 or claudin-2 in the female-AOM/DSS micegf. However, in the male-AOM/DSS micegf, while RB had no effect on ZO-1 levels, it was able to significantly decrease claudin-2 levels (P < 0.01); on the other hand, FRB-feeding significantly increased ZO-1 levels (~1.4 folds) and had no effect on claudin-2 levels.
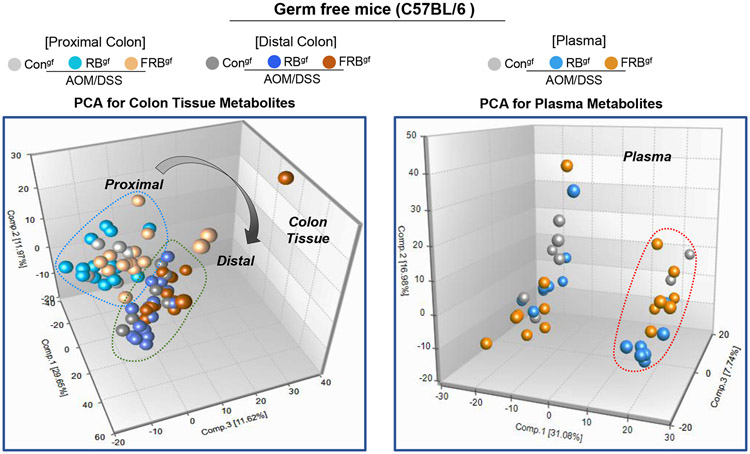
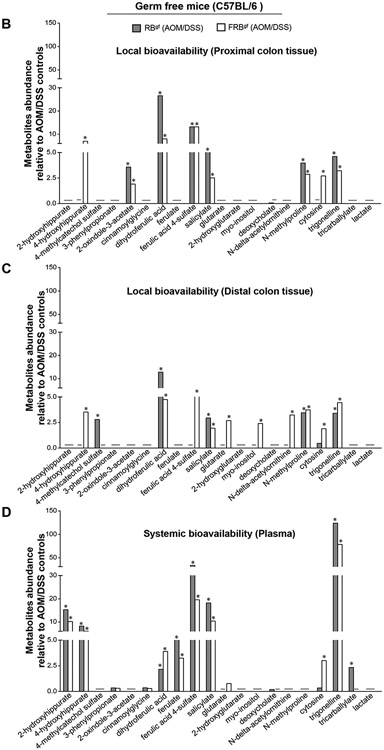
3.6. Effect of RB and FRB-intake on microbial metabolites in gut and systemic circulation after inflammation-associated colon tumorigenesis in germ-free micegf.
Next, we assessed the metabolic by-products generated after the intake of RB and FRB-supplemented diets in the absence of gut-microbiota in the AOM/DSS micegf model (Fig 6). Principal component analysis (PCA) of the metabolites composition across all groups indicated no gender specific-effects on the colonic metabolites generated after RB and FRB-intake (data not shown). Under germ-free conditions, both proximal and distal tissue regions were identified to be involved in AOM/DSS-induced neoplastic changes, and the local gut-directed bioavailability of metabolites was assessed for proximal and distal colonic tissues and in systemic circulation (Fig 6A-D). In the proximal colon tissue of FRB and RB-fed AOM/DSS micegf, levels of dihydroferulic acid (~8-27 folds), ferulic acid 4-sulfate (~13 folds), salicylate (~3-5 folds), N-methylproline (~3-4 folds), and trigonelline (~3-5 folds) were significantly increased (P<0.05, for all) compared to AOM/DSS controlsgf (Fig 6B). Importantly, 4-hydroxyhippurate levels were significantly increased (P<0.05) in the proximal (~7 folds) and distal colon (~4 folds) tissues of FRBgf (AOM/DSS) mice only. Ferulic acid 4-sulfate, glutarate, myo-inositol, N-delta-acetylornithine were other metabolites that showed significant expression in the distal colonic tissues of the FRBgf (AOM/DSS) mice and not in the RB-fed group (Fig 6C); also, cytosine was markedly increased in proximal and distal colonic tissues after FRB-feeding compared to RB-fed group. Notably, 2-hydroxyhippurate (~10-15 folds), 4-hydroxyhippurate (~6-8 folds), dihydroferulic acid (~2-4 folds), ferulate (~3-5 folds), ferulic acid 4-sulfate (~20-34 folds), salicylate (~10-18 folds), and trigonelline (~79-124 folds) were significantly increased (P<0.05, for all) in the plasma of RBgf and FRBgf (AOM/DSS) groups (Fig 6D). As in the case of colonic tissues, glutarate and cytosine levels were also increased (~1-3 folds P<0.05) in the plasma of FRBgf (AOM/DSS) groups. We did not observe any gender-specific impact on the type of RB and FRB metabolites present in the plasma; however, the relative abundance of most of the plasma metabolites in male mice were comparatively higher to female mice in the RB-fed micegf (Supplementary Figure S5).
Fig 6. Effect of RB and FRB-intake on colon and plasma metabolite profiles during AOM/DSS-induced colon tumorigenesis in germ-free micegf.
(A) Principal components analysis of colon (proximal and distal) and plasma metabolite profiles. Relative abundance of metabolites (B) in proximal colon tissue; (C) in distal colon tissue, and (D) in systemic circulation-plasma. Differential metabolite abundance is reported relative to values obtained in control diet fed AOM/DSS treated micegf. Quantified data is represented as Columns (relative fold change); *P ≤ 0.05. RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; micegf, germ free mice.
4. DISCUSSION
The capacity for rice bran and fermented rice bran to suppress AOM/DSS- inflammation induced CRC is novel and supports previously demonstrated cancer control and preventive properties of dietary rice bran against genetically susceptible and carcinogen induced murine models of CRC 10,15-17. In this study, we performed extensive pre-clinical investigations utilizing non-fermented and fermented diets to determine whether the anti-CRC efficacy of dietary rice bran is dependent or independent of gut microflora. Primary study outcomes support that both RB and FRB -intake demonstrated protective benefits against AOM/DSS induced colon tumorigenesis under the gut microbiota-intactspf state, yet with RB showing relatively better efficacy than FRB following in-depth histopathological and molecular analysis of the colonic tissues. Notably, the supplemented diets also reduced sub-mucosal inflammation, epithelial erosion, restored mucosal barrier integrity 33, and increased the presence (number and content) of acidic mucin secreting goblet cells [which is otherwise substantially decreased during colon tumorigenesis] 30. In the colon tissue microenvironment, there was a significant decrease in the expression of inflammatory mediators, Cox-2 and NF-κB, after RB and FRB-intervention compared to AOM/DSS controlsspf. In the absence of gut-microbiotagf, RB diet supplementation was able to manifest protective benefits against CRC growth and progression, whereas the efficacy of FRB-supplemented diets was markedly compromised compared to RB-intervention. Given the strain differences between the gut microbiota-intact and -absent studies, the outcomes were compared between the groups within a study and not compared to assess differences across the two studies. However, generalized inferences based on results from both studies are being discussed for a broader perspective.
In the microbiota-intactspf mice, the anti-CRC benefits of RB and FRB were associated with modulation of cecal and colonic microbiota according to increased species richness and diversity indices. Both the supplemented diets were able to increase colonization of native gut probiotics and showed enrichments in Clostridiales, Lachnospiraceae, Ruminiclostridium, and Coprococcus, among others; furthermore, in the FRBspf (AOM/DSS) mice, enrichments were also observed in Blautia, Streptococcus, and Roseburia. These enrichments indicate a shift towards increased beneficial commensals that are involved in maintaining intestinal homeostasis, attenuation of intestinal inflammation, strengthening/maintaining intestinal barrier 34-38; and thus, could play an important role in RB/FRB mediated anti-CRC effects. Over time, these diet induced-changes in gut microbiota composition showed functional differences to modulate microbial metabolism 3,9,39. Rice bran is a rich source of diverse substrates for gut microbiota, including but not limited to prebiotics, amino acids, lipids and phytochemicals 10,40. A significant accumulation of phenolic metabolites (derived from RB) such as hippurate, catechol sulfate, 4-methyl catechol sulfate, and 3-phenyl propionate were identified in the colonic tissue and plasma of RB and FRB -fed gut microbiota-intactspf mice. Tracking the relative abundance of these metabolites in local colonic tissue supported greater abundance of colonic metabolites after intake of RB-supplemented diets. Importantly, phenolic metabolites were either not present or present in low quantities in the colon tissues and plasma of germ-free AOM/DSSgf mice fed with either of the supplemented diets to support that they originated from RB, and that RB derived bioactives were a result of gut microbial transformation of various aromatic polyphenolic components [benzoate and catechol metabolism] from the diet. These gut microbial digested or transformed RB metabolites had contributed to colon cancer protective efficacy under the gut microbiota-intact state 3,39,41,42. Furthermore, high levels of another phenolic compound ferulic acid 4-sulfate, a derivative of ferulic acid [with known chemopreventive potential 43], was found increased in colonic tissue and plasma with RB intake. Notably, colonic tissues and plasma levels of salicylate, and nicotinic acid metabolite trigonelline [compounds with anti-cancer benefits in CRC and pancreatic cancer, respectively 44,45] were increased in RB and FRB-supplemented groups under both microbiota-intact and germ-free conditions.
Interestingly, the relative abundance of ferulic acid 4-sulfate, cinnamoylglycine [a cinnamic acid conjugate associated with health benefits of berries in CRC patients 46], and most of the phenolic metabolites (that were higher in colonic tissues of RB-fed mice compared to FRB-group) such as, hippurate, catechol sulfate, 4-methyl catechol sulfate, showed increased presence in the plasma of the FRB-fed micespf compared to the RB-fed micespf. Fermentation of RB had increased bioavailability of a number of bioactive food components; specifically, ex-vivo fermentation of RB by Bifidobacterium longum resulted in rapid absorption and systemic circulation uptake of hippurates and catechol sulfates. When the RB diet (as is) acts as a substrate for microbiota in the gut, there is higher production of abundant microbial metabolites, but it takes relatively more time to yield these microbial metabolites when compared to easily accessible microbial metabolites from the FRB diet. As such, the metabolites generated after gut microbiota-mediated metabolism of RB have a longer duration and possibly longer half-life in the colonic environment and become more enriched in the local colonic tissue. The systemic bioavailability of RB-metabolites is slower than when FRB-is fed to the micespf. These findings suggest a superior opportunity for native gut microbial metabolism of RB to exert direct-effects (via local tissue presence) against CRC growth and progression when compared to dietary FRB.
It is important to note that FRB-intake resulted in unique food-derived metabolites (N-delta-acetylornithine, and tricarballylate) in the plasma and colon tissues of FRB-fed micespf .We have previously reported that N-delta acetylornithine and tricarballylate moieties were significantly increased in colonic tissues of healthy mice after FRB-intervention 18. The nutritional and anti-cancer importance of higher levels of N-delta-acetylornithine is supported by healthy mucosal colonocytes differing in expression to CRC tumors 47, and tricarballylate modifies energy balance via aconitase inhibition in the tricarboxylic acid cycle [TCA] 48. Thus, the disruption of cancer cell metabolism by fermented diet derived metabolites could play an important role in FRB-mediated CRC effects. On the other hand, the plasma metabolites observed after RB and FRB intervention could exert indirect effects (via systemic circulation) against CRC growth and progression (as is evident by significant anti-inflammatory effects via modulation of plasma cytokines/chemokines). The RB/FRB supplemented diets were able to significantly decrease the expression of CXC chemokines (CXCL-1, 2, 13) that are known to play pivotal role in immune cell recruitment, inflammation-associated CRC growth and progression 49,50. Furthermore, other inflammatory mediators, which are recognized to be associated with tumor invasiveness and poor prognosis in various cancers, including CRC 49,50, such as, TNFα, Serpin E1, pentraxin3, c-reactive protein, etc., were also decreased by the RB/FRB dietary intervention. Taken together, the results from the present studies under gut microbiota-intactspf state corroborate our earlier scientific findings that the RB and FRB alter host metabolism and generated microbial metabolites which have predominately anti-inflammatory and anti-cancer potential 18.
Even in the absence of gut microbiota, the relative abundance and diversity of metabolites were comparatively higher after the intake of RB-supplemented diets compared to FRB-intake. Though, RB diets were never exposed to microbial fermentation events (ex-vivo or within the gut environment), the RB is rich in phenolic constituents that are subjected to diverse non-microbial metabolic mechanisms, which result in the production of relatively more abundant metabolites with anti-CRC benefits. However, gender-specific pathologies were observed in the AOM/DSS modelgf in the absence of gut microbiota which in turn could have resulted in differential effects of RB and FRB-supplemented diets on CRC growth and progression. In the AOM/DSS controlsgf , the females had relatively higher number of colonic macro-lesions/adenoma compared to their male counterparts. Notably, female-AOM/DSSgf mice were also marred by severe epithelial erosion in their colonic tissues; these pathological aberrations were comparatively less evident in the malegf mice. While RB-intake showed protective benefits against epithelial erosion, FRB-intake was not significantly effective in reducing epithelial erosion characteristics in the female micegf. Immune cell infiltrates including submucosal inflammation were also significantly higher in the female-AOM/DSSgf mice compared to male micegf. Whether, FRB-intake has the potential to alter the number and phenotype of these colonic immune/inflammatory cells to execute its anti-CRC effects warrants further investigation.
Gender -based differential effects of RB and FRB-supplemented diets in micegf highlight the importance of gender-specific physiological variables that could differentially impact CRC growth and progression in people. One of the study limitations herein is that gender-based effects under gut microbiota- intact conditions (as in the germ-free state) were not assessed and merit future attention. Another limitation is that fermentation of RB-diet was conducted with a single probiotic species and a microbial community cocktail for fermentation of RB (to yield a larger suite of bioactive molecules) was not compared. Further, a dietary dose response with the FRB-diet will be essential prior to investigations in a human feeding trial. Moreover, a longitudinal analysis of colon tissue profiles at multiple time points after AOM-DSS treatment could identify earlier windows of opportunity for dietary modulation and a pattern of metabolites according to tumorigenic specific stages during inflammation-associated carcinogenesis.
Conclusion
Rice bran is a nutrient and fiber rich food with functional phytochemicals that exert protective effects against colon tumorigenesis in the presence as well as absence of gut microbiota. Rice bran prebiotic properties and its constituents as microbial substrates promote healthy gut microbiota composition and function in the colon via production of novel, bioactive microbial metabolites that significantly modulate multiple pathways involved in colon carcinogenesis. Fermentation of rice bran did not substantially impact gut microbiota metabolism and did not show increased protective efficacy against CRC when compared to non-fermented rice bran. These findings demonstrate that caution is needed for the use of fermented rice bran food products during severe gut inflammation or colon tumorigenesis-associated with severe damage to the colonic epithelial lining. The inability of FRB to provide local-protective benefits to the damaged colonic tissues when compared to RB suggests further investigations are needed to establish comparative efficacy. The present study highlights the importance of gender-specific physiological variables that could differentially impact CRC growth and progression and influence the microbiome-dependent versus -independent actions of functional whole grains.
Supplementary Material
Supplementary Fig S1. Effect of RB and FRB-intake on AOM/DSS-induced colonic macroscopic lesions in gut microbiota-intactspf (specific pathogen-free) Balb/c mice. (A) Experimental study design; six-week old male Balb/c micespf fed a control AIN-93 pellet diet for one-week acclimatization period and then administered a single intraperitoneal (i.p) injection of AOM 10 mg/kg body weight in saline followed by exposure to 2% DSS in drinking water for five days. After DSS treatment mice were switched to rice bran (RB, n=20), B. longum-fermented rice bran (FRB, n=20) supplemented diets or maintained on a control AIN-93M diet (n=20) for 15 weeks and sacrificed to harvest tissues. Non-AOM/DSS exposed mice (on control AIN-93M diet) served as overall negative controls (NC, n=5) in the study; non-AOM/DSS exposed but fed-with RB (n=5) or FRB (n=4) diets were also included as negative controls. Effect of RB and FRB-supplemented diets on (B) colon length, (C) total macroscopic lesions/colon, and (D) size distribution of macroscopic lesions in proximal, middle, and distal colon (lesion size were categorized as <1mm, 1-2mm, >2-3mm, and >3mm). Quantified data is represented as columns (mean for each group); ***P ≤0 .001, **P ≤ 0.01, and *P ≤ 0.05. RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; micespf, specific pathogen-free mice.
Supplementary Fig S2. Proteome profiler-based plasma cytokine/chemokine dot-blot array depicting changes in their expression after intake of RB and FRB-supplemented diets during AOM/DSS-induced colon tumorigenesis in gut microbiota-intactspf mice. Plasma collected from untreated (negative controls) and AOM/DSS micespf (with and without RB and FRB-supplemented diets) was evaluated for the presence of various cytokines/chemokines and other inflammatory-mediators involved in immune responses, and CRC growth and progression. using a mouse-Proteome profiler membrane array containing antibodies to 111 different molecules. Densitometric analysis of dot blots from pooled samples in each group are shown as fold changes relative to negative controls (NC). Quantified data is represented as Columns (mean for each group); bars represent SEM. *P ≤ 0.05. RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; micespf, specific pathogen-free mice.
Supplementary Fig S3. Gender-specific effects of RB and FRB-intake on AOM/DSS-induced colonic macroscopic lesions in the absence of gut microbiota (germ-free micegf). (A) Experimental study design: germ-free micegf (C57Bl/6 mice-inbred in gnotobiotic facility) were administered a single intraperitoneal (i.p) injection of 10 mg/kg AOM in saline. Seven days after AOM injection, mice were exposed to 2% DSS in drinking water for five days; this was then followed by 14 days of normal drinking water (resting phase). This cycle (DSS exposure + resting phase) was repeated twice to yield a total of three DSS cycles. After 1st cycle of DSS exposure, Micegf (male or female) were randomized into three groups according to their diet: Controlgf (M = 10, F = 3); RBgf (M = 8, F = 7) and FRBgf (M = 10, F = 5) for 18 weeks and then were sacrificed to harvest tissues. Effect of RB and FRB-supplemented diets on (B) colon length, (C) total macroscopic lesions/colon, and (D) size distribution of macroscopic lesions in proximal, middle, and distal colon (lesion size were categorized as <1mm, 1-2mm, and >2-3mm). Quantified data is represented as columns (mean for each group).***P ≤0 .001, **P ≤ 0.01, and *P ≤ 0.05. RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; micegf, germ free mice
Supplementary Fig S4. Representative pictographs of immunohistochemical staining for the expression of Ki-67 positive cells, and immunoreactivity scores for β-catenin, Cox-2, NF-κB (p65), ZO-1, and claudin-2 expression levels during AOM/DSS-induced colon tumorigenesis in gut microbiota-intactspf mice. Gender specific effects are shown as male (left-panel), and female (right-panel). Images were acquired at x400 with digitally magnified insets. % Positive cells were quantified as the number of brown-stained cells x100 per total number of cells counted under ×400 magnification in 5-8 randomly selected fields in each sample. Immunoreactivity (represented by intensity of brown staining) was scored as 0 (no staining), +1 (weak), +2 (moderate), +3 (strong) and +4 (very strong). RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; micegf, germ free mice.
Supplementary Fig S5. Gender-specific effect of RB and FRB-intake on plasma metabolites, during AOM/DSS-induced colon tumorigenesis in the absence of gut microbiota (germ-free micegf). Relative abundance of metabolites in plasma of (A) male and female (RBspf-AOM/DSS) mice; (B) male and female (FRBspf-AOM/DSS) mice. Differential metabolite abundance is reported relative to values obtained in control diet fed AOM/DSS treated micegf. (C) Relative abundance of plasma metabolites (male vs. female) in RB and FRB-fed AOM/DSSgf mice. Quantified data is represented as Columns (relative fold change); *P ≤ 0.05. RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; micegf, germ free mice.
Funding:
National Institute of Health (NIH)/National Cancer Institute (NCI), Research grant R01 CA201112 to KR and EPR. Acknowledge the support of the Kevin and Lorie Haarberg Funds for Cancer Research to KR.
Abbreviations:
- AOM
azoxymethane
- DSS
dextran sodium sulfate
- CRC
colorectal cancer
- RB
rice bran
- FRB
fermented rice bran
- B. longum
Bifidobacterium longum
- micespf
specific pathogen -free mice
- RBspf
RB-fed specific pathogen-free mice
- FRBspf
FRB-fed specific pathogen-free mice
- controlsspf
control diet-fed specific pathogen-free mice
- micegf
germ-free mice
- RBgf
RB-fed germ-free mice
- FRBgf
FRB-fed germ-free mice
- controlsgf
control diet fed germ-free mice
- H&E
hematoxylin and eosin
- HG
high grade
- MG
moderate grade
- LG
low grade (dysplasia)
- DAB
3,3′-diaminobenzidine
- AB
alcian blue
- PAS
periodic acid-schiff
- PCoA
principal coordinates analysis
- ASVs
amplicon sequence variants
- CZM
count zero multiplicative
- clr
centered log-ratio
- PCA
principal components analysis
- UPLC-MS/MS
ultra-performance liquid chromatography tandem mass spectrometry
- IHC
immunohistochemical
- CD44
cluster of differentiation 44
- ZO-1
zonula occluding
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- Cox-2
Cyclooxygenase-2
- CXCL
chemokine (C-X-C motif) ligand
- CCL17
CC chemokine ligand 17
- G-CSF
Granulocyte-colony-stimulating factor
- M-CSF
macrophage-colony-stimulating factor
- MMP
matrix metalloproteinase
- RAGE
receptor for advanced glycation end-products
- WISP-1
WNT1-inducible-signaling pathway protein 1
Footnotes
Conflict of Interest Statement: The authors declare that there are no conflicts to disclose.
Institutional Review Board Statement: All animal studies were performed under Institutional guidelines using approved Institutional Animal Care and Use Committee (IACUC) protocol [(B- 57916(04)1E] in the specific pathogen free facility and gnotobiotic core facility at UC Denver-AMC.
Supplementary Data
Supplementary Material and Methods, Figures and Table are available as an addendum
Data Sharing and Data availability
To ensure transparency and complete data reproducibility, the amplicon sequence data supporting the conclusions of this manuscript are available via NCBI SRA BioProject Accession PRJNA516457. Sample data are available in Supplementary Metadata File S1. The R code to create the manifest for importing FASTQ files into QIIME 2 can also be found on this project’s GitHub repository located at github.com/kdprkr/MerlinsManuscript.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. [DOI] [PubMed] [Google Scholar]
- 2.Zackular JP, Baxter NT, Iverson KD, et al. The gut microbiome modulates colon tumorigenesis. mBio. 2013;4(6):e00692–00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins SL, Patterson AD. The gut microbiome: an orchestrator of xenobiotic metabolism. Acta Pharmaceutica Sinica B. 2020;10(1):19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potrykus M, Czaja-Stolc S, Stankiewicz M, Kaska Ł, Małgorzewicz S. Intestinal Microbiota as a Contributor to Chronic Inflammation and Its Potential Modifications. Nutrients. 2021;13(11):3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tjalsma H, Bolejia A, Marchesi JR, Dutilh BE A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10(8):575–582. [DOI] [PubMed] [Google Scholar]
- 6.Geng J, Fan H, Tang X, Zhai H, Zhang Z. Diversified pattern of the human colorectal cancer microbiome. Gut pathogens. 2013;5(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PloS one. 2013;8(8):e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16(11):690–704. [DOI] [PubMed] [Google Scholar]
- 9.Rowland I, Gibson G, Heinken A, et al. Gut microbiota functions: metabolism of nutrients and other food components. European journal of nutrition. 2018;57(1):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan EP. Bioactive food components and health properties of rice bran. Journal of the American Veterinary Medical Association. 2011;238(5):593–600. [DOI] [PubMed] [Google Scholar]
- 11.Henderson AJ, Ollila CA, Kumar A, et al. Chemopreventive properties of dietary rice bran: current status and future prospects. Adv Nutr. 2012;3(5):643–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norris L, Malkar A, Horner-Glister E, et al. Search for novel circulating cancer chemopreventive biomarkers of dietary rice bran intervention in Apc(Min) mice model of colorectal carcinogenesis, using proteomic and metabolic profiling strategies. Mol Nutr Food Res. 2015;59(9):1827–1836. [DOI] [PubMed] [Google Scholar]
- 13.Forster GM, Raina K, Kumar A, et al. Rice varietal differences in bioactive bran components for inhibition of colorectal cancer cell growth. Food Chem. 2013;141(2):1545–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agista AZ, Rusbana TB, Islam J, et al. Fermented rice bran supplementation prevents the development of intestinal fibrosis due to DSS-induced inflammation in mice. Nutrients. 2021;13(6):1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katyama M, Yoshimi N, Yamada Y, et al. Preventive effect of fermented brown rice and rice bran against colon carcinogenesis in male F344 rats. Oncology reports. 2002;9(4):817–822. [PubMed] [Google Scholar]
- 16.Tan BL, Norhaizan ME. Scientific evidence of rice by-products for cancer prevention: chemopreventive properties of waste products from rice milling on carcinogenesis in vitro and in vivo. BioMed research international. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verschoyle R, Greaves P, Cai H, Edwards R, Steward W, Gescher A. Evaluation of the cancer chemopreventive efficacy of rice bran in genetic mouse models of breast, prostate and intestinal carcinogenesis. British journal of cancer. 2007;96(2):248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nealon NJ, Parker KD, Lahaie P, et al. Bifidobacterium longum-fermented rice bran and rice bran supplementation affects the gut microbiome and metabolome. Benef Microbes. 2019;10(8):823–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibayama J, Kuda T, Shikano A, et al. Effects of rice bran and fermented rice bran suspensions on caecal microbiota in dextran sodium sulphate-induced inflammatory bowel disease model mice. Food Bioscience. 2018;25:8–14. [Google Scholar]
- 20.Sheflin AM, Borresen EC, Wdowik MJ, et al. Pilot dietary intervention with heat-stabilized rice bran modulates stool microbiota and metabolites in healthy adults. Nutrients. 2015. ;7(2):1282–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown DG, Borresen EC, Brown RJ, Ryan EP. Heat-stabilised rice bran consumption by colorectal cancer survivors modulates stool metabolite profiles and metabolic networks: a randomised controlled trial. Br J Nutr. 2017;117(9):1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheflin AM, Borresen EC, Kirkwood JS, et al. Dietary supplementation with rice bran or navy bean alters gut bacterial metabolism in colorectal cancer survivors. Mol Nutr Food Res. 2017;61(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.So WK, Law BM, Law PT, Chan CW, Chair SY. Current Hypothesis for the Relationship between Dietary Rice Bran Intake, the Intestinal Microbiota and Colorectal Cancer Prevention. Nutrients. 2016;8(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.So WKW, Chan JYW, Law BMH, et al. Effects of a Rice Bran Dietary Intervention on the Composition of the Intestinal Microbiota of Adults with a High Risk of Colorectal Cancer: A Pilot Randomised-Controlled Trial. Nutrients. 2021;13(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zarei I, Oppel RC, Borresen EC, Brown RJ, Ryan EP. Modulation of plasma and urine metabolome in colorectal cancer survivors consuming rice bran. Integr Food Nutr Metab. 2019;6(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh J, Rivenson A, Tomita M, Shimamura S, Ishibashi N, Reddy BS. Bifidobacterium longum, a lactic acid-producing intestinal bacterium inhibits colon cancer and modulates the intermediate biomarkers of colon carcinogenesis. Carcinogenesis. 1997;18(4):833–841. [DOI] [PubMed] [Google Scholar]
- 27.Parker KD, Maurya AK, Ibrahim H, et al. Dietary Rice Bran-Modified Human Gut Microbial Consortia Confers Protection against Colon Carcinogenesis Following Fecal Transfaunation. Biomedicines. 2021;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki R, Kohno H, Sugie S, Nakagama H, Tanaka T. Strain differences in the susceptibility to azoxymethane and dextran sodium sulfate-induced colon carcinogenesis in mice. Carcinogenesis. 2006;27(1):162–169. [DOI] [PubMed] [Google Scholar]
- 29.Derry MM, Raina K, Balaiya V, et al. Grape seed extract efficacy against azoxymethane-induced colon tumorigenesis in A/J mice: interlinking miRNA with cytokine signaling and inflammation. Cancer Prev Res (Phila). 2013;6(7):625–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasprzak A, Seraszek-Jaros A, Jagielska J, et al. The Histochemical Alterations of Mucin in Colorectal Carcinoma Quantified by Two Efficient Algorithms of Digital Image Analysis. International Journal of Molecular Sciences. 2019;20(18):4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koelink PJ, Wildenberg ME, Stitt LW, et al. Development of Reliable, Valid and Responsive Scoring Systems for Endoscopy and Histology in Animal Models for Inflammatory Bowel Disease. J Crohns Colitis. 2018;12(7):794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan ME, Zheng B, Koelink PJ, et al. New perspective on dextran sodium sulfate colitis: antigen-specific T cell development during intestinal inflammation. PloS one. 2013;8(7):e69936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salvador E, Burek M, Förster CY. Tight junctions and the tumor microenvironment. Current pathobiology reports. 2016;4(3):135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinninella E, Raoul P, Cintoni M, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. Bmj. 2018;361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo P, Zhang K, Ma X, He P. Clostridium species as probiotics: potentials and challenges. Journal of Animal Science and Biotechnology. 2020;11(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Mao B, Gu J, et al. Blautia—a new functional genus with potential probiotic properties? Gut microbes. 2021;13(1):1875796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rezasoltani S, Aghdaei HA, Dabiri H, Sepahi AA, Modarressi MH, Mojarad EN. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microbial pathogenesis. 2018;124:244–249. [DOI] [PubMed] [Google Scholar]
- 39.Mosele JI, Macià A, Motilva M-J. Metabolic and microbial modulation of the large intestine ecosystem by non-absorbed diet phenolic compounds: a review. Molecules. 2015;20(9):17429–17468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan EP, Heuberger AL, Weir TL, Barnett B, Broeckling CD, Prenni JE. Rice bran fermented with saccharomyces boulardii generates novel metabolite profiles with bioactivity. Journal of agricultural and food chemistry. 2011;59(5):1862–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cardona F, Andrés-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuño MI. Benefits of polyphenols on gut microbiota and implications in human health. The Journal of nutritional biochemistry. 2013;24(8):1415–1422. [DOI] [PubMed] [Google Scholar]
- 42.Corrêa TAF, Rogero MM, Hassimotto NMA, Lajolo FM. The two-way polyphenols-microbiota interactions and their effects on obesity and related metabolic diseases. Frontiers in nutrition. 2019:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Z, Moghadasian MH. Chemistry, natural sources, dietary intake and pharmacokinetic properties of ferulic acid: A review. Food Chemistry. 2008;109(4):691–702. [DOI] [PubMed] [Google Scholar]
- 44.Duthie GG, Wood AD. Natural salicylates: foods, functions and disease prevention. Food Funct. 2011;2(9):515–520. [DOI] [PubMed] [Google Scholar]
- 45.Arlt A, Sebens S, Krebs S, et al. Inhibition of the Nrf2 transcription factor by the alkaloid trigonelline renders pancreatic cancer cells more susceptible to apoptosis through decreased proteasomal gene expression and proteasome activity. Oncogene. 2013;32(40):4825–4835. [DOI] [PubMed] [Google Scholar]
- 46.Pan P, Skaer CW, Stirdivant SM, et al. Beneficial regulation of metabolic profiles by black raspberries in human colorectal cancer patients. Cancer Prevention Research. 2015;8(8):743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gómez de Cedrón M, Acin Perez R, Sánchez-Martínez R, et al. Micro RNA-661 modulates redox and metabolic homeostasis in colon cancer. Molecular Oncology. 2017;11(12):1768–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watson JA, Fang M, Lowenstein JM. Tricarballylate and hydroxycitrate: substrate and inhibitor of ATP: citrate oxaloacetate lyase. Arch Biochem Biophys. 1969;135(1):209–217. [DOI] [PubMed] [Google Scholar]
- 49.Cytokines Klampfer L., inflammation and colon cancer. Current cancer drug targets. 2011;11(4):451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuomisto AE, Mäkinen MJ, Väyrynen JP. Systemic inflammation in colorectal cancer: Underlying factors, effects, and prognostic significance. World journal of gastroenterology. 2019;25(31):4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig S1. Effect of RB and FRB-intake on AOM/DSS-induced colonic macroscopic lesions in gut microbiota-intactspf (specific pathogen-free) Balb/c mice. (A) Experimental study design; six-week old male Balb/c micespf fed a control AIN-93 pellet diet for one-week acclimatization period and then administered a single intraperitoneal (i.p) injection of AOM 10 mg/kg body weight in saline followed by exposure to 2% DSS in drinking water for five days. After DSS treatment mice were switched to rice bran (RB, n=20), B. longum-fermented rice bran (FRB, n=20) supplemented diets or maintained on a control AIN-93M diet (n=20) for 15 weeks and sacrificed to harvest tissues. Non-AOM/DSS exposed mice (on control AIN-93M diet) served as overall negative controls (NC, n=5) in the study; non-AOM/DSS exposed but fed-with RB (n=5) or FRB (n=4) diets were also included as negative controls. Effect of RB and FRB-supplemented diets on (B) colon length, (C) total macroscopic lesions/colon, and (D) size distribution of macroscopic lesions in proximal, middle, and distal colon (lesion size were categorized as <1mm, 1-2mm, >2-3mm, and >3mm). Quantified data is represented as columns (mean for each group); ***P ≤0 .001, **P ≤ 0.01, and *P ≤ 0.05. RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; micespf, specific pathogen-free mice.
Supplementary Fig S2. Proteome profiler-based plasma cytokine/chemokine dot-blot array depicting changes in their expression after intake of RB and FRB-supplemented diets during AOM/DSS-induced colon tumorigenesis in gut microbiota-intactspf mice. Plasma collected from untreated (negative controls) and AOM/DSS micespf (with and without RB and FRB-supplemented diets) was evaluated for the presence of various cytokines/chemokines and other inflammatory-mediators involved in immune responses, and CRC growth and progression. using a mouse-Proteome profiler membrane array containing antibodies to 111 different molecules. Densitometric analysis of dot blots from pooled samples in each group are shown as fold changes relative to negative controls (NC). Quantified data is represented as Columns (mean for each group); bars represent SEM. *P ≤ 0.05. RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; micespf, specific pathogen-free mice.
Supplementary Fig S3. Gender-specific effects of RB and FRB-intake on AOM/DSS-induced colonic macroscopic lesions in the absence of gut microbiota (germ-free micegf). (A) Experimental study design: germ-free micegf (C57Bl/6 mice-inbred in gnotobiotic facility) were administered a single intraperitoneal (i.p) injection of 10 mg/kg AOM in saline. Seven days after AOM injection, mice were exposed to 2% DSS in drinking water for five days; this was then followed by 14 days of normal drinking water (resting phase). This cycle (DSS exposure + resting phase) was repeated twice to yield a total of three DSS cycles. After 1st cycle of DSS exposure, Micegf (male or female) were randomized into three groups according to their diet: Controlgf (M = 10, F = 3); RBgf (M = 8, F = 7) and FRBgf (M = 10, F = 5) for 18 weeks and then were sacrificed to harvest tissues. Effect of RB and FRB-supplemented diets on (B) colon length, (C) total macroscopic lesions/colon, and (D) size distribution of macroscopic lesions in proximal, middle, and distal colon (lesion size were categorized as <1mm, 1-2mm, and >2-3mm). Quantified data is represented as columns (mean for each group).***P ≤0 .001, **P ≤ 0.01, and *P ≤ 0.05. RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; micegf, germ free mice
Supplementary Fig S4. Representative pictographs of immunohistochemical staining for the expression of Ki-67 positive cells, and immunoreactivity scores for β-catenin, Cox-2, NF-κB (p65), ZO-1, and claudin-2 expression levels during AOM/DSS-induced colon tumorigenesis in gut microbiota-intactspf mice. Gender specific effects are shown as male (left-panel), and female (right-panel). Images were acquired at x400 with digitally magnified insets. % Positive cells were quantified as the number of brown-stained cells x100 per total number of cells counted under ×400 magnification in 5-8 randomly selected fields in each sample. Immunoreactivity (represented by intensity of brown staining) was scored as 0 (no staining), +1 (weak), +2 (moderate), +3 (strong) and +4 (very strong). RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; micegf, germ free mice.
Supplementary Fig S5. Gender-specific effect of RB and FRB-intake on plasma metabolites, during AOM/DSS-induced colon tumorigenesis in the absence of gut microbiota (germ-free micegf). Relative abundance of metabolites in plasma of (A) male and female (RBspf-AOM/DSS) mice; (B) male and female (FRBspf-AOM/DSS) mice. Differential metabolite abundance is reported relative to values obtained in control diet fed AOM/DSS treated micegf. (C) Relative abundance of plasma metabolites (male vs. female) in RB and FRB-fed AOM/DSSgf mice. Quantified data is represented as Columns (relative fold change); *P ≤ 0.05. RB, rice bran; FRB, Bifidobacterium longum-fermented RB; AOM, azoxymethane; DSS, dextran sodium sulfate; micegf, germ free mice.
Data Availability Statement
To ensure transparency and complete data reproducibility, the amplicon sequence data supporting the conclusions of this manuscript are available via NCBI SRA BioProject Accession PRJNA516457. Sample data are available in Supplementary Metadata File S1. The R code to create the manifest for importing FASTQ files into QIIME 2 can also be found on this project’s GitHub repository located at github.com/kdprkr/MerlinsManuscript.