Abstract
Head and neck squamous cell carcinoma is highly innervated by peripheral sensory neurons. Local neurotransmitter release (e.g., calcitonin gene-related peptide (CGRP)) from sensory neurons innervating the cancer has been linked to tumorigenesis. CGRP-immunoreactive nerve presence comprised 9.53±1.9% of total nerve area across 11 HNSCC patients. A syngeneic tongue tumor transplant mouse model of oral cancer and a global Calca knockout mouse (CGRPKO) were used to investigate the impact of CGRP signaling on tumor growth and the associated immune response in vivo. In tumor-bearing CGRPKO mice, there was a significant reduction in tumor size over time compared to wildtype mice using two different mouse oral cancer cell lines. Furthermore, tumor tissue from CGRPKO mice had a significant increase in tumor infiltrating CD4+ T cells, cytotoxic CD8+ T cells and NK1.1+ NK cells compared to wildtype. Fluorescent-activated cell sorting and real-time qPCR were used to confirm that CD4+ T cells isolated from tumor-bearing wildtype mice contained high expression of Ramp1 compared to sham mice. These data suggest that sensory neurotransmitter CGRP may modulate oral cancer progression via tumor immunosurveillance. Understanding the relationship between sensory neurons and cancer will aid in repurposing clinically available nervous system drugs for the treatment of cancer.
Keywords: calcitonin gene-related peptide, oral, cancer, neurotransmission, immune
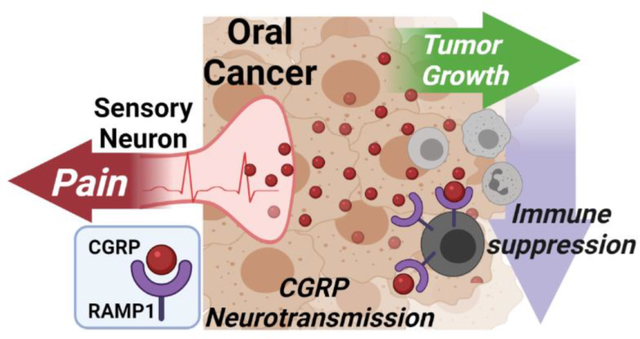
Graphical Abstract
Peripheral nerves can influence the tumor-associated microenvironment. In oral squamous cell carcinoma, pain is a primary feature and calcitonin gene-related peptide (CGRP)-releasing sensory nerves dominate tumor innervation. CGRP signaling drives a pro-tumor microenvironment by limiting immunosurveillance of cancer. Reducing CGRP release from tumor-innervating sensory nerves may be a viable therapeutic strategy to reduce pain and slow cancer progression.
1.0. Introduction
Head and neck cancer (HNC) affects approximately 36,000 people in the United States per year, with demographics shifting to a younger population.[1] The majority of these tumors arise from the epithelial cells of the oral cavity, oropharynx, larynx, or hypopharynx.[2, 3] Risk factors can be behavioral (i.e. tobacco and alcohol use) or infection-associated (i.e. human papillomavirus (HPV)), and these factors vary with geographic location.[4] Head and neck squamous cell carcinoma (SCC) constitutes 90% of cases of HNC and is associated with severe disease and high rates of recurrence despite advances in cancer treatment.[1]
The oral cavity is innervated by cranial nerves with a high density of sensory nerves, originating mainly from the trigeminal ganglia.[5] To date, histological patterns of cancer-nerve interaction, defined as perineural invasion (PNI), are identified as the presence of tumor cell clusters within the peripheral nerve sheath or infiltrating the nerves and/or tumor cells encircling one-third of the nerve circumference.[6] PNI was detected in up to 70% of oral SCC in the tongue and/or floor of the mouth and has been demonstrated as an independent predictor of poor prognosis and an indicator of aggressive tumor behavior.[7–10] Prior research by Rahima et al.[11] and Laske et al.[12] found that PNI in early stage oral and oropharyngeal carcinomas has a highly negative association with recurrence-free survival and tumor differentiation. Systemic analysis of the neural influences within the cancer microenvironment, including identification of how PNI and other nerve-cancer interactions generally relate to poor prognosis, is crucial given that ongoing research is focusing on neural invasion for targeted therapies of tumor regression.[13–18]
Calcitonin gene-related peptide (CGRP) is the most abundant neurotransmitter in trigeminal ganglia neurons (TGN) innervating the tongue[19, 20] and serves a prominent role in efferent signaling; upon activation of sensory nerve fibers, CGRP is released from peripheral nerve terminals and exerts paracrine effects on surrounding tissues[21], including tumor cells. Two isoforms of CGRP exist; αCGRP, derived from the CALCA gene, is the principal form found in the central and peripheral nervous system, whereas βCGRP, derived from the CALCB gene, is found mainly in the enteric nervous system.[21] In a rat oral cancer model, a high percentage of αCGRP-immunoreactive nerve sprouting was found in the tumor microenvironment and orofacial sensitization was accompanied by upregulated αCGRP expression in the maxillary and mandibular trigeminal branches.[22] High levels of CGRP have been identified in tumor tissues and serum of patients with medullary thyroid carcinoma[23], although the isoform was not specified. Both α and βCGRP in peripheral tissues act as potent vasodilators; however, the inflammatory role has been debated with evidence for both pro- and anti-inflammatory effects.[21] The CGRP receptor is a G-protein coupled receptor complex identified mainly by a single transmembrane-spanning protein designated receptor activity-modifying protein 1 (RAMP1), which is required for CGRP binding to the receptor complex.[24] RAMP1 signaling has been implicated in both innate and adaptive inflammation[25, 26], and CGRP has been shown to influence immune cell activity.[27] However, the role of CGRP signaling in tumor progression and the associated tumor immune response is currently unknown.
In the current study, we explored the mechanisms of cancer-sensory nerve interaction with the hypothesis that peripheral sensory neurotransmission in the oral cancer microenvironment modulates tumor growth and the tumor-associated immune response. We sought to (1) confirm the presence of αCGRP-expressing sensory nerves innervating tumor tissue from HNSCC patients and a syngeneic oral cancer mouse model, (2) determine the effect of sensory nerve signaling via αCGRP on oral cancer cell proliferation in vitro and tumor growth in vivo, and (3) determine the effect of αCGRP on the tumor-associated immune response via RAMP1 signaling.
2.0. Materials and Methods
2.1. Patient Data:
We quantified peripheral nerve presence in HNSCC patients who underwent surgical resection at the University of Pittsburgh Medical Center between 2015 and 2018. Eleven patients were collected for these analyses; all were >18 years, underwent surgical resection for squamous cell carcinoma of the oral cavity or oropharynx, and had perineural invasion (PNI) denoted in the pathology report by an oral and maxillofacial pathologist. Patients with recurrent disease or second primaries were excluded. Demographics and clinical characteristics were obtained from medical record review and included: age at diagnosis, sex, race, surgical procedure [i.e., oropharyngectomy (tonsil, base of tongue), mandibulectomy/composite resection, glossectomy/floor of mouth resection, laryngectomy/pharyngectomy], tumor size, nodal status, HPV status for oropharyngeal tumors, history of tobacco use, history of alcohol use, PNI (yes/no) and extracapsular spread (yes/no/not evaluated) (Table 1). Patient-reported pain was measured via the Functional Assessment of Cancer Therapy – Head and Neck (FACT-HN version 4). The FACT-HN consists of 37-items grouped in five subscales: 1) physical well-being, 2) social/family well-being, 3) emotional well-being, 4) functional well-being, and 5) a head and neck cancer-specific score. Each item is scored from 0–4 with higher scores representing worse pain. Head and neck-additional concerns question 12 (HN12: “I have pain in my mouth, throat or neck”) was used to assess the presence of pain prior to surgery. HNSCC tumor tissue from patients was acquired through the University of Pittsburgh Medical Center (UPMC) Hillman Cancer Center’s Head and Neck SPORE (P50CA097190; R.L. Ferris, PI) collection of head and neck cancer tissue specimens. All patients provided informed written consent; this study was approved by the Institutional Review Board associated with University of Pittsburgh Cancer Institute (STUDY20050058).
Table 1.
Demographic and Clinical Characteristics.
| Variables | M±SD | n(%) |
|---|---|---|
|
| ||
| Age, years | 59.09±7.56 | |
|
| ||
| Sex | ||
| Male | 8(72.73) | |
| Female | 3(27.27) | |
|
| ||
| Race | ||
| White | 9(81.82) | |
| Other | 2(18.18) | |
|
| ||
| Radiation | ||
| No | 2(18.18) | |
| Yes | 9(81.82) | |
|
| ||
| Chemotherapy | ||
| No | 5(45.45) | |
| Yes | 6(54.55) | |
|
| ||
| Immunotherapy | ||
| No | 10(90.91) | |
| Yes | 1(9.09) | |
|
| ||
| Nodal Stage | ||
| N0 | 1(9.09) | |
| N1 | 2(18.18) | |
| N2 | 8(72.73) | |
|
| ||
| Tumor Stage | ||
| T1 | 0(0) | |
| T2 | 5(45.45) | |
| T3 | 3(27.27) | |
| T4 | 3(27.27) | |
|
| ||
| Metastasis | ||
| M0 | 1(9.09) | |
| MX | 10(90.91) | |
|
| ||
| Site | ||
| Oropharynx | 4(36.36) | |
| HPV | ||
| Neg | 0(0) | |
| Pos | 4(100) | |
| Oral cavity | 7(63.64) | |
|
| ||
| Smoking | ||
| No | 4(36.36) | |
| Yes | 7(63.64) | |
|
| ||
| Alcohol | ||
| No | 3(27.27) | |
| Yes | 8(72.73) | |
|
| ||
| PNI | ||
| No | 0(0) | |
| Yes | 11(100.00) | |
|
| ||
| ECS | ||
| No | 6(54.55) | |
| Yes | 4(36.36) | |
| Not Evaluated | 1(9.09) | |
2.2. Animals:
Adult (10–12 weeks, 20–25 g) male and female C57Bl/6 (stock #000664; Jackson Labs, Bar Harbor, ME), athymic nude (stock #002019, Jackson Labs) and CalcaKO (stock #033168, Jackson Labs) mice were used for in vivo experiments. All mice were housed in a temperature-controlled room on a 12:12-hour light cycle (0700–1900 hours light), with unrestricted access to food and water. Researchers were trained under the Animal Welfare Assurance Program. All procedures were approved by the University of Pittsburgh Institutional Animal Care and Use Committees and performed in accordance with National Institutes of Health guidelines for the use of laboratory animals in research.
2.3. Retrograde tracer labeling:
At least 10 days prior to any manipulation (e.g. tumor cell inoculation, sham injection), retrograde tracer 1,19-dioctadecyl-3,3,39,39-tetramethylindocarbocyanine perchlorate (DiI; Invitrogen, Carlsbad, CA) was injected peripherally into the anterior lateral portion of the tongue to label tongue-innervating trigeminal ganglia sensory neurons. For oral cancer mouse model, mice were injected with DiI 10 days before inoculation of tumor cells or sham injection. The tracer was dissolved at 170 mg/mL in DMSO, diluted 1:10 in sterile saline, and injected bilaterally using a 30 g needle and a Hamilton syringe for a total volume of 5 to 7 μL per tongue under isoflurane anesthesia.
2.4. Cell Culture:
All cell lines were cultured in 10 cm diameter cell culture dishes at 37°C with 5% CO2. Human oral squamous cell carcinoma (SCC) cell line, HSC3 (Sekisui XenoTech, Kansas City, KS), non-tumorigenic cell line HaCaT (ThermoFisher), were cultured in DMEM (Gibco, Waltham, MA) supplemented with 10% fetal bovine serum (FBS; Corning) and penicillin/streptomycin (P/S, 50 U/mL, Corning; DMEM-COMP). SCC cell line SCC-4 (ATCC, Manassas, VA) was cultured in DMEM/F12 (Gibco) supplemented with 10% FBS and P/S. Dysplastic oral keratinocyte cell line DOK (Sigma-Aldrich, St. Louis, MO) was cultured in DMEM supplemented with 10% FBS, P/S, and 5μg/mL hydrocortisone (Sigma-Aldrich). Human cell line authenticity was independently verified by PCR using short tandem repeat profiles (ATCC). Mouse oral SCC cell lines MOC1 and MOC2 (Kerafast) were cultured in IMDM/F12 (2:1; Gibco) supplemented with 5% FBS, P/S, 5 μg/mL insulin (Sigma-Aldrich), 40 ng/mL hydrocortisone, and 5 ng/mL epidermal growth factor (EMD Millipore; MOC-COMP) Cell pellets for gene expression analysis from all cell lines were collected from a passage number less than 14.
2.5. Proliferation assay:
HaCaT and HSC3 cell lines were cultured in DMEM-COMP media; MOC1 was cultured in MOC-COMP media. Tissue culture-treated plates (48-well, Costar) were coated with 1 ug/mL Poly-L-ornithine solution (Sigma-Aldrich), allowed to dry, then rinsed and dried thoroughly prior to plating. HaCaT (passage 13), HSC3 (passage 8), and MOC1 (passage 9) cells were seeded at a density of 2 × 103 cells/well in complete media. Cells were incubated for 24 hours. Treatment dilutions in media were added to generate final concentrations of 0.03nM, 0.1nM, and 0.3nM αCGRP as well as 0.3nM αCGRP + 1nM of a CGRP receptor antagonist (i.e. AC-187, CGRP 8–37). Blockers were added 10–15 minutes prior to αCGRP. Cells were incubated for 48 hours. For quantification of cell proliferation, media was replaced with media containing NucBlue live nuclear stain (2 drops/mL; ThermoFisher), incubated 30 minutes, and each well was imaged on BZ-X810 fluorescent microscope at 10x magnification (Keyence, Japan). Well images were stitched using BZ-X Analyzer (Keyence), and the number of cells were counted using ImageJ software (NIH, Bethesda, MD). All images were taken, stitched, and analyzed under identical settings within each cell line.
2.6. Enzyme-linked immunosorbent assay (ELISA)
αCGRP protein concentration was measured in homogenized tongue tumor tissue by ELISA (Cayman Chemical Cat#589001). For mouse tumor tissue collection, sham mice and MOC1or MOC2 tumor-bearing mice were perfused with PBS and tongues were harvested and snap frozen in dry ice. Tumor tissue was then dissected and minced on dry ice and stored at −80°C until needed. Frozen minced tissue (20–40 mg) was homogenized in 500μl RIPA buffer (Pierce Biotechnology) containing 5μl HALT Protease Inhibitor Cocktail (Sigma) and agitated for an additional 15min at 4°C. Lysates were centrifuged at 16,000xg for 15 min. Supernatants were removed and used immediately. Total protein concentrations were determined for all samples using a Bradford Assay (Bio-Rad Laboratories, Inc.) and ELISA was run per the manufacturer’s instructions. The optical densities of the standards and samples were read at 405 nm using a GloMax Explorer GM3500 Microplate Reader (Promega). CGRP protein concentrations were calculated based on manufacturer’s instructions and normalized to total protein in the sample.
2.7. Immunohistochemistry:
Head and neck cancer tumor tissue and mouse tumor tissue staining:
Paraffin-embedded tumor tissue blocks from HNSCC patients were obtained from the Head and Neck Tissue Bank, and sectioned (5um) and stained by the Developmental Pathology Laboratory in the Department of Pathology. For tissue obtained from the mouse model, adult C57Bl/6 mice with MOC2 tumors (PID 14) under 3–5% isoflurane anesthesia were transcardially perfused with phosphate buffered saline (PBS) followed by 20mL cold 4% paraformaldehyde solution (PFA, Electron Microscopy Sciences). Tongue tissue was harvested, bisected along the sagittal plane and stored in 4% PFA at 4°C for up to 1 week. Tissue was then washed with PBS, paraffin-embedded and sectioned (5μm). For both human and mouse tissue, antigen retrieval was performed using a citrate buffer (Dako, Carpinteria, CA). The S100 antibody (rabbit polyclonal, Cat# IR50461–2 DAKO (Agilent), Carpenteria, CA) was applied using a 1:200 dilution at room temperature. The secondary antibody consisted of SignalStain Boost (HRP, Rabbit) (Cell Signaling, Danvers, MA). The substrate used was 3,3, Diaminobenzidine + (Dako). Lastly, the slides were counterstained with Hematoxylin (Dako). The sections were scanned at 10x, 20x, and 40x magnification and S100 labeling was used to identify all nerve bundles. Total S100 immunoreactive area was quantified using Aperio ImageScope software (Leica Biosystems). Within each nerve bundle, CGRP immunoreactive area was quantified and summed for total CGRP area, and the percent of CGRP+ area relative to total S100 area was then calculated.
Mouse sensory ganglia staining:
For tissue harvest, mice under 3–5% isoflurane anesthesia were transcardially perfused with PBS followed by 4% PFA. The trigeminal ganglia (TG) were dissected, postfixed for 1 hour in 4% PFA, and cryoprotected in 30% sucrose at 4°C overnight. TG were embedded in Tissue-Tek OCT compound (Sakura Finetek, Torrance, CA), sectioned (14μm), and mounted on Superfrost Plus slides (Fisher Scientific, Waltham, MA, USA). Slides were incubated in primary antibody rabbit anti-CGRP (1:500, Cell Signaling Technologies) in PBS containing 1% bovine serum albumin overnight at room temperature. Slides were extensively washed in PBS, incubated in goat anti-rabbit Alexa fluor 488 (1:250, Jackson ImmunoResearch) for 2.5 hours, extensively washed, and cover-slipped with Fluoro-Gel II mounting media containing DAPI (Electron Microscopy Sciences). Using a Leica DMi8 microscope with LAS software, TG sections were photographed at 20x magnification within the intersection of the mandibular and maxillary branch, where most retrograde labeled trigeminal tongue neurons reside. Trigeminal ganglia neurons (TGN) with distinct nuclei and at least 50% of the cell area labeled with DiI were counted in every fifth section (9 sections/mouse). ImageJ software (NIH, Bethesda, MD) was used to count retrograde labeled neurons which overlapped with anti-TH immunoreactivity per animal.
2.8. Oral cancer mouse model:
Mice were inoculated with either 5×105 MOC1 cells, 2×104 MOC2, or 1×105 HSC3 cells in 30 μL of 1:1 DMEM and Matrigel (Corning) into the anterior lateral portion of the tongue as previously described.[28] Tongue tumor size was quantified weekly by calipers and calculated using the volume of ellipsoid formula V = 4/3π x L x W x H. Tongue tumor area was quantified at PID 35 and PID 14 for MOC1 and MOC2 tumors respectively. At the end of the experiment, tongues were harvested, fixed in 10% neutral buffered formalin, bisected, paraffin-embedded, and sectioned at 5μm thickness through the entire block (about 50 sections). Average tumor area relative to total tongue area in the 1st, 10th, 20th, 30th and 40th section was quantified using hematoxylin and eosin (H&E) stain and ImageJ. The experimenters conducting tumor quantification were blinded to the treatment groups.
2.9. Quantitative real-time PCR:
Total RNA was isolated from pelleted cells (1–1.5×106) from cell lines, fluorescently sorted lymph node immune cells, and whole TG using the Qiagen RNeasy Plus Mini Kit (Qiagen Inc.). Reverse transcription was performed with Quantitect Reverse Transcription Kit (Qiagen Inc.) according to the manufacturer’s instructions. cDNA was diluted with nuclease free water to a 5 ng/μL concentration. For single cell analysis, DiI-positive single neurons were identified using fluorescence microscopy, picked up using glass capillaries (World Precision Instruments) held by micromanipulator (Sutter Instruments), headstage (EPC10 HEKA) and electrode holder under brightfield optics. Cell size was not considered during selection. Each cell was transferred into a 0.2 ml PCR tube containing 9μl of single cell lysis solution and Dnase I from Single Cell-to-CT™ Kit (ThermoFisher), incubated for 5 minutes and immediately stored at −80°C until further use. Cells were collected within 1 h of removal from the incubator and within 8 h of removal from the animals (n=4 C57Bl/6 mice, 4 MOC1 bearing mice, 4 MOC2 bearing mice). Reverse transcription, cDNA pre-amplification and Real-Time PCR were executed per manufacturer’s instructions. Relative expression levels of Calca and CGRP receptor gene components, RAMP1 and CALCR1, were assessed using TaqMan Gene Expression Assays and TaqMan Gene Expression Master Mix (ThermoFisher), using a 96 well Quantstudio 3 Real-Time PCR System (ThermoFisher). We used assays from Life Technologies for the following genes: Calca (Mm00801463_g1), Ramp1 (Mm00489796_m1), Calcr1 (Mm00516986_m1), RAMP1 (Hs00195288_m1), CALCR1 (Hs00907738_m1). GUSB and Gusb (Hs00939627_m1 and Mm01197698_m1) were used as the internal control gene for human and mouse respectively. Relative quantification analysis of gene expression data was calculated using the 2−ΔCt method. For single cell PCR, any cell with a Gusb Ct threshold of 28 or higher was excluded from further analysis. Genes were considered ‘not expressed’ if one sample either failed to detect expression or the Ct was above 38.
2.10. Single cell RNA sequencing and analyses:
For sourcing and processing of publicly available data, filtered single cell data including gene counts and sample annotations were procured and transformed as previously described[29], then further manipulations were conducted in R. Scaling normalization of raw gene counts was accomplished using the scran package’s computeSumFactors function.[30] Top variable genes were ascertained using the scran package’s modelGeneVar and getTopHVGs functions, then used as the subset of features for principal component analysis using the scater package’s runPCA and the base values for building a UMAP from runUMAP.[31] For comparison to the clusters given in the original dataset, new labels were obtained by clustering cells using the nearest neighbor algorithm in scran, buildSNNGraph, and clusters were identified using the igraph function cluster_louvain[32]. Ten different immune cell subtypes were obtained and parsed from the annotated genes which included CD19+ B cells, CD14+ monocytes, CD16+ myeloid derived suppressor cells, CD8+ cytotoxic T cells, dendritic cells, mast cells, cytotoxic NK cells, plasma dendritic cells, CD4+FOXP3– T conventional cells and CD4+FOXP3+ T regulatory cells. For differential gene expression analysis, pseudo-bulk RNAseq matrices were created from single cell data representing the entire dataset and each annotated cell type. Each attribute was then modeled as a design matrix in DESeq2, and contrasts were used to find the differential expression between conditions in the full matrix and within each cell type classification. A wald test between specific groups was used to target the specific comparisons.
2.11. Analytical Cytometry and Fluorescence-Activated Cell Sorting:
Analytical cytometry and fluorescence-activated cell sorting (FACS) were used to assess immune infiltrate in mouse tumors and bilateral submandibular and cervical lymph nodes and to collect subpopulations of immune cells from dissociated sham and MOC1- or MOC2-tumor tongue tissue. Mouse tongues were harvested and dissociated as previously described.[33] Briefly, tongue tissue was dissected and minced in DMEM with antibiotics, collagenase-H (0.5 mg/mL; Sigma-Aldrich, 34 units/mg), DNase (0.5 mg/mL) and 20 mM 4-(2-hydroxyethyl)-1-piperazine ethanesulfonic acid (HEPES, Fisher Scientific), and then incubated at 37°C for 1 h. The tissue was then mechanically dissociated using a fire-polished pipette, washed twice with fresh DMEM containing antibiotics and HEPES, and resuspended in CaCl2/MgCl2 free phosphate buffered saline (Sigma-Aldrich) containing 3% FBS, 1mM EDTA (Sigma-Aldrich), and 0.02% sodium azide (Sigma-Aldrich) and filtered through a 40 mm cell strainer (Falcon brand, Fisher Scientific). To isolate subpopulations, cells were stained with fluorescently conjugated rat anti-mouse mAbs. Leukocytes from the spleen were used for compensation controls (i.e. correction of a signal overlap between emission spectra of different fluorochromes used). Cell viability marker was used to exclude dead cells. Forward and side scatter parameters were used to confirm the size and granularity. Post-sort purity was >97%. FACS was performed on a five laser Beckman Coulter MoFlo Astrios High Speed Sorter. Tongue infiltrating immune cells were sorted into lysis buffer containing DNase for RNA purification and qPCR using the cell lysis solution and Dnase I from the Cell-to-CT™ Kit (ThermoFisher); Reverse transcription, cDNA pre-amplification and Real-Time PCR were executed per manufacturer’s instructions. Submandibular and cervical lymph node immune cells were sorted into sterile-filtered PBS, spun down at 300xg for 4 min, snap frozen, and stored at −80°C until needed.
2.12. Statistics:
Statistical significance was set at p < 0.05. All statistical analyses were performed using Prism (version 8) statistical software (Graphpad Software Inc., La Jolla, CA, USA). Results were presented as mean ± standard error of the mean. Box/scatter or violin/scatter configurations were used to show the biological variability when illustrative. Student’s t test was employed to evaluate the difference between two groups. Tissue and data from oral cancer mouse models were never directly compared; tumor-bearing mice were only compared to time-matched (i.e. equal time post inoculation) sham mice. Analysis of variance (ANOVA) was employed to evaluate the difference between groups regarding time and treatment. To adjust for multiple comparisons, the post-hoc Holm-Sidak test statistic was employed. Kuskal-Wallis test was used as a non-parametric test for statistical differences in relative gene expression distribution in sensory neurons between treatment groups.
3.0. Results
3.1. Human and mouse CGRP-expressing nerves innervate oral cancer
Alpha calcitonin gene-related peptide (αCGRP)-immunoreactivity (IR) was used to identify sensory CGRP-releasing neurons innervating the tumor microenvironment in HNSCC patients. Anti-S100, a Schwann cell marker useful for evaluating nerve sheath, was used to identify all peripheral nerve bundles within the tumor tissue. In serial sections, αCGRP-IR fibers were found within S100-denoted nerve bundles (Figure 1A). Next, we quantified the relative percent of αCGRP-IR nerve area relative to total S100-IR nerve area in HNSCC tumor tissue. We found that αCGRP-IR nerve fibers comprise 9.53±1.9% of total S100-IR nerve area on average across 11 HNSCC patients (Figure 1B). Given evidence that CGRP neuropeptide is a marker for nociceptive neurons, we sought to determine if patient-reported pain correlated with αCGRP-IR nerve presence in the tumor. We correlated the percentage of CGRP+ nerve area with patient-reported pain using the Functional Assessment of Cancer Therapy-Head and Neck (FACT H&N) questionnaire version 4, specifically Additional Concerns Question 12, “I have pain in my mouth, throat or neck”. The response to this question is rated on a scale of 0 (not at all) to 4 (very much). We found a positive correlation between patient-reported pain and CGRP-immunoreactive nerve presence in the tumor; however, this relationship was not significant (Pearson correlation, r2=0.352. p=0.054; Supplemental Figure 1).
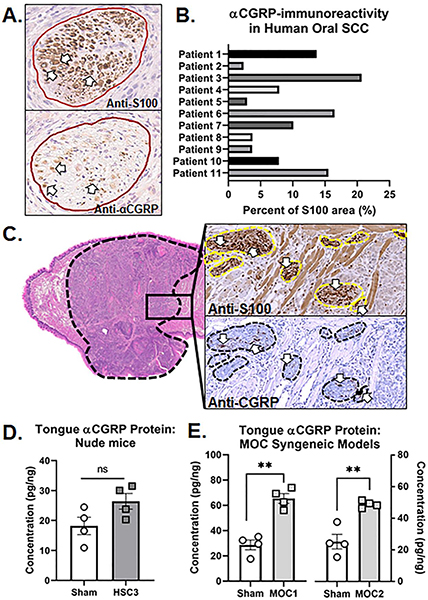
Figure 1. Anti-CGRP immunoreactivity (IR) in tissue from HNSCC patients and oral cancer mouse models.
A) Representative images of HNSCC tumor serial sections stained with either anti-S100 (top) or anti-CGRP (bottom). Nerve bundles are outline in red and positive stain is indicated by white arrows. Image magnification = 20x. B) Quantification of the percent of total αCGRP-IR nerve area relative to total S100-IR nerve area across tumor tissue sections from 11 HNSCC patients with PNI pathology reported by an oral and maxillofacial pathologist. Patient demographics are located in Table 1. C) Representative image of CGRP-IR in serial tumor sections stained with either hematoxylin and eosin (H&E, left, 4x magnification), S100 (right top), or CGRP (right bottom). Tumor is outlined in black and nerve bundles are outlined in yellow (top inset) and black (bottom inset). Positive stain is indicated by white arrows. Image magnification = 20x. αCGRP protein concentration in the tumor was measured in tongue tissue from (D) HSC3 tumor bearing athymic nude mice at post-inoculation day (PID) 28 compared to sham mice and in (E) MOC1 (PID 35) and MOC2 (PID 14) tumor bearing C57Bl/6 mice compared to sham mice by ELISA (n=4/group). αCGRP concentration was normalized to total protein isolated in the tongue tumor tissue for each sample. Student’s t test **p<0.01.
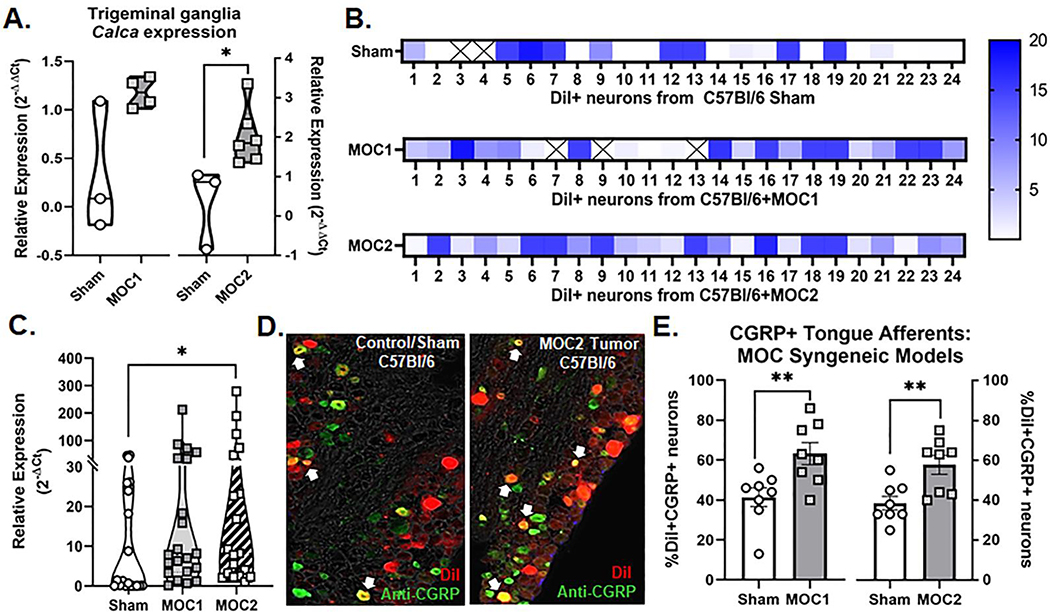
Next, utilizing a syngeneic transplant mouse model of tongue SCC in C57Bl/6 mice, we identified CGRP-IR nerves within S100-denoted nerve bundles (Figure 1C) innervating the mouse oral cancer (MOC1) tumor microenvironment. CGRP is reported to be the most prevalent neurotransmitter released by trigeminal sensory nerves innervating the oral cavity.[21, 34] We first sought to determine if CGRP is released into the tumor microenvironment during tumorigenesis. αCGRP protein concentration was quantified in homogenized tumor tissue from tumor-bearing mice compared to tongue tissue from sham mice as well as in MOC1 and MOC2 cancer cell pellets by ELISA and normalized to total protein isolated in the sample. Tissue from both the immunocompromised xenograft model (i.e. HSC3 transplant) and syngeneic allograft model (i.e. MOC1 or MOC2 transplant) were used. While there was no significant difference in mouse αCGRP protein concentration in HSC3-bearing mice compared to sham (t=2.092, p=0.082; Figure 1D), tongue tumor tissue from MOC1-bearing mice (65.49±3.9 pg/ng, n=4; t=6.720, p=0.001) and MOC2-bearing mice (48.86±1.08 pg/ng, n=4; t=4.998, p=0.003) contained significantly more mouse αCGRP compared to sham mice (n=4/time point; Figure 1E). We found no detectable αCGRP protein in MOC1 or MOC2 oral cancer cell pellets (1×106 cells per pellet). We next sought to determine if the trigeminal ganglia (TG), which hold the neuronal somas of the nerves innervating the tongue, express αCGRP and if this expression changes in the presence of oral cancer. Intact TG from sham C57Bl/6 mice and MOC1- and MOC2-bearing C57Bl/6 mice were probed for the Calca gene using qPCR; relative expression was normalized to sham mice for both groups. There was an average two-fold increase in relative Calca gene expression in TG from MOC2-bearing mice normalized to sham mice (t=2.846, p=0.0248; Figure 2A). However, ganglia are comprised of multiple cell types, including satellite glia and immune cells[35], as well as neurons that do not innervate the tongue[36] and consequently should not be affected by tumor growth. In order to assess changes in tongue-innervating neuron-specific expression, ganglia from retrograde labeled mice were dissociated, and individual neurons were collected for single cell analysis based on fluorescence; cell size was not considered. RNA from 24 individual, DiI-labeled neurons from each of the following groups was subjected to qPCR analysis: 2 sham C57Bl/6, 2 MOC1-bearing C57Bl/6, and 2 MOC2-bearing C57Bl/6 mice. The majority of neurons tested contained detectable Calca gene expression (22/24 sham, 21/24 MOC1, 24/24 MOC2) (Figure 2B) and there was a significant increase in the relative magnitude of Calca expression in neurons from MOC2-bearing mice compared to sham (H(2)=6.213, P=0.045) (Figure 2C). CGRP protein expression was also visualized in tongue-innervating neurons using retrograde labeling and immunohistochemistry in TG sections from sham C57Bl/6 mice and MOC1- and MOC2-bearing C57Bl/6 mice (Figure 2D). There was a significantly higher percentage of tongue-innervating (i.e. DiI+) CGRP-positive neurons in TG tissue from both MOC1 (t=3.050, p=0.009) and MOC2 (t=3.309, p=0.005) bearing mice compared to time-matched sham tissue (Figure 2E).
Figure 2. CGRP expression in tongue trigeminal ganglia neurons (TGN).
A) Quantification of Calca gene expression in whole trigeminal ganglia isolated from sham (n=3/time point), MOC1-tumor bearing mice (n=4, PID 35), and MOC2-tumor bearing mice (n=6, PID 14) using RT-PCR. Expression was normalized to housekeeping gene GusB and then to average expression in sham mice. Student’s t test *p<0.05. B) Calca gene expression from retrogradely labeled (DiI+) tongue-innervating TG neurons from sham C57Bl/6 mice (n=24, 2 mice), MOC1-tumor bearing mice (n=24, 2 mice PID 35), and MOC2-tumor bearing mice (n=24, 2 mice PID 14) that were manually picked to perform single-cell PCR. Gene expression was calculated for relative mRNA expression for each cell for each gene. Gusb was used as an internal control. Data are presented as heatmap for each gene for each cell. Boxes containing an X indicate no detectable expression (CT value > 38) for that cell. C) The relative expression distribution across treatment group was compared using Kruskal-Wallis test *p<0.05. D) Representative images of retrograde labeling (red) and anti-CGRP immunoreactivity (green) in the mandibular branch of the TG from a sham C57Bl/6 mouse and a MOC2-tumor bearing mouse PID 14. E) Quantitative analysis of DiI-positive tongue-innervating neurons with distinct visible nuclei with CGRP-IR in TG sections from sham (n=8/time point), MOC1-tumor bearing mice (n=8, PID 35), and MOC2-tumor bearing mice (n=8, PID 14). Student’s t test **p<0.01.
3.2. Transplant tumors grow more slowly in CGRPKO mice compared to wildtype control mice
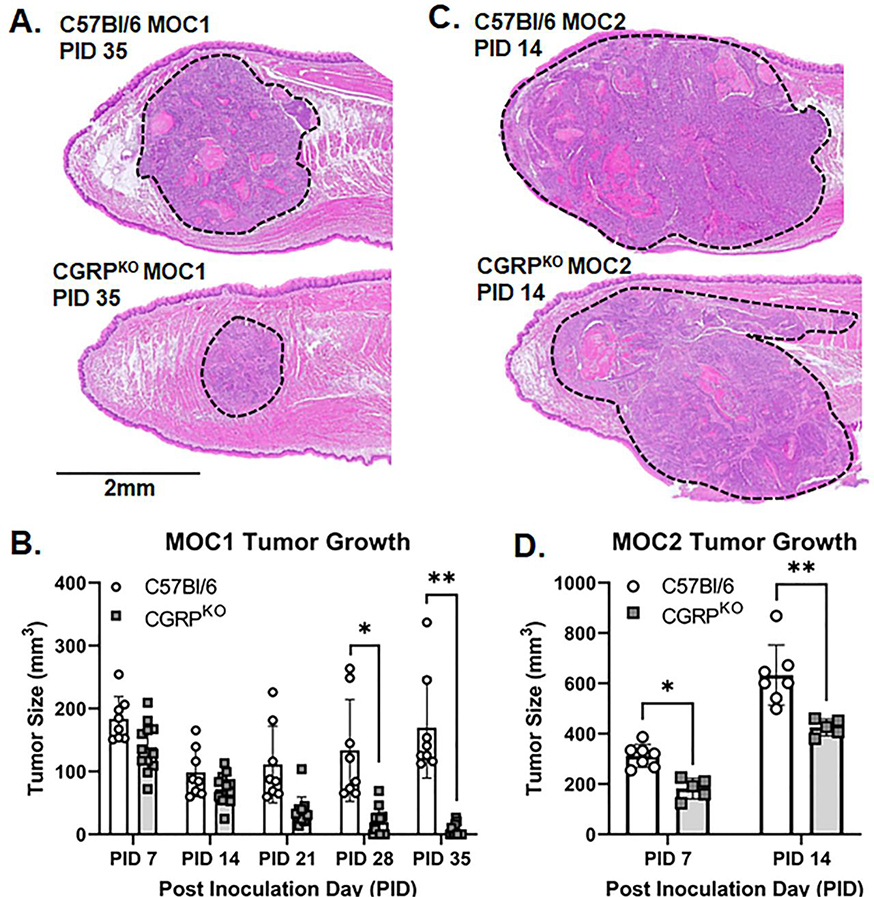
Next, we sought to test the hypothesis that αCGRP neurotransmission modulated oral cancer progression in vivo. Given the previous data that αCGRP protein was not significantly higher in tongue tissue from the HSC3 xenograft mouse model compared to sham, we hypothesize that the immune system might play a role; therefore, we proceeded with only the syngeneic orthotopic transplant mouse model using C57Bl/6 mice compared to Calca global knockout mice (CGRPKO). We chose to measure tumor growth in mice inoculated with either Matrigel alone (i.e. sham), MOC1 cells, or MOC2 cells. Given the aggressive nature of MOC2 cell growth compared to MOC1, tumor size was assessed at post inoculation day (PID) 7 and 14 for mice bearing MOC2 tumors, and weekly for 6 weeks for mice bearing MOC1 tumors. We found that both MOC1 and MOC2 tumor volume varied with the interaction of time and genotype (MOC1: F(4,72)=13.39, p<0.0001; MOC2: F(1, 10)=25.53, p=0.0005). MOC1 tumor volume in CGRPKO mice was significantly less at PID 28 (p=0.025) and 35 (p=0.003) compared to tumor volume in C57Bl/6 mice (Figure 3A,B). MOC2 tumor volume in CGRPKO mice was significantly less at PID 7 (p=0.014) and 14 (p=0.0002) compared to tumor volume in C57Bl/6 mice (Figure 3C,D). To evaluate whether the loss of CGRP affected the density of nerves in the tumor microenvironment, we quantified S100-IR nerve presence in sagittal sections from tumor-bearing tongue tissue from both wildtype and CGRPKO mice. We found no significant difference in total S100-IR area (t=1.422, p=0.208), bundle incidence (t=0.321, p=0.759), or percent of total S100-IR area relative to total tumor area measured between genotypes (t=0.988, p=0.361; Supplemental Figure 2).
Figure 3. Loss of CGRP signaling slows tumor growth.
A) Representative images of MOC1 tumor size in wildtype mice (top) and CGRPKO mice (global Calca knockout, bottom) at PID 35. Images were collected from hematoxylin and eosin stained 5μm sections at 4x magnification. Tumors are outlined in black dotted lines. B) Quantitative analysis of tumor size measured by caliper over time in MOC1-tumor bearing mice (n=11 wildtype, n=8 CGRPKO). Tumor size was calculated using ellipsoid volume formula. Two-way ANOVA *p<0.05, **p<0.01. C) Representative images of MOC2 tumor size in wildtype mice (top) and CGRPKO mice (bottom) at PID 14. Images were collected from hematoxylin and eosin stained 5μm sections at 4x magnification. Tumors are outlined in black dotted lines. D) Quantitative analysis of tumor size measured by caliper over time in MOC2-tumor bearing mice (n=7 wildtype, n=5 CGRPKO). Tumor size was calculated using ellipsoid volume formula. Two-way ANOVA *p<0.05, **p<0.01.
3.3. Human and mouse tumor tissue and oral cancer cell lines express CGRP receptor components
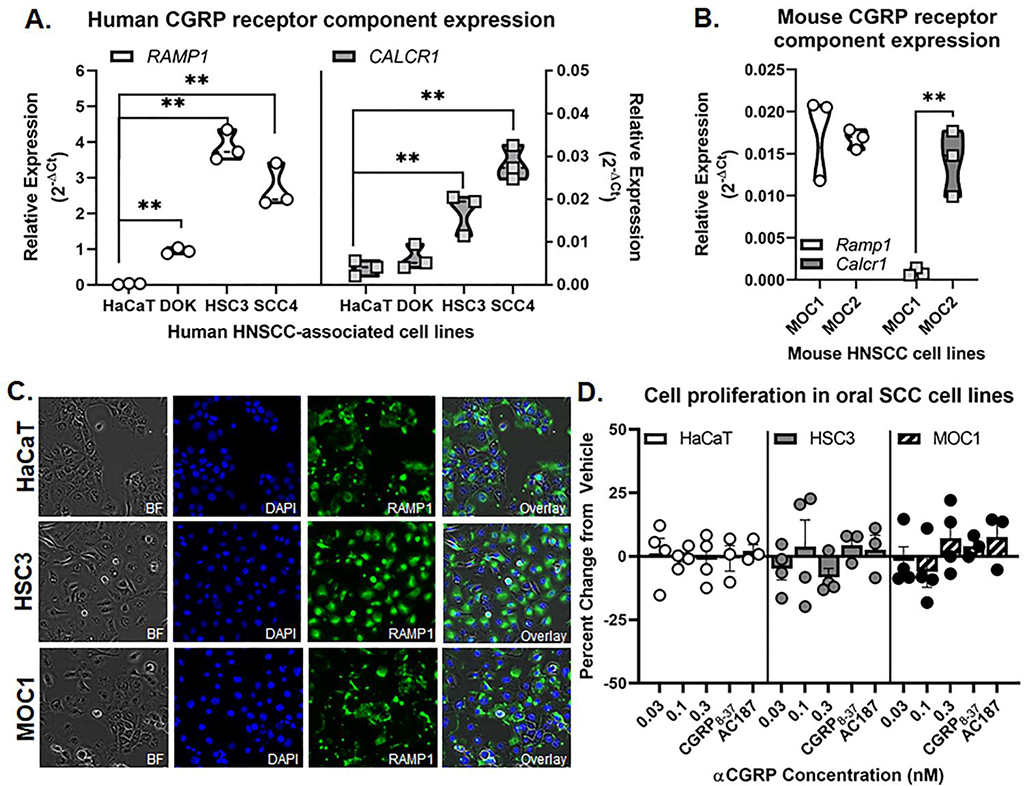
Previous studies have demonstrated that components for CGRP receptors, RAMP1 and CALCR1, are expressed in normal keratinocytes.[37] Furthermore, CGRP has been shown to exert migratory effects on cancer cell activity[38]; CGRP increased the invasiveness of prostate cancer and osteosarcoma cell lines.[39] We sought to determine if CGRP can influence oral cancer cell proliferation directly. First, we used qPCR to determine the relative expression of RAMP1 and CALCR1 in human oral cancer cell lines HSC3 and SCC-4. Immortalized non-tumorigenic skin keratinocyte cell line, HaCaT, and benign non-tumorigenic dysplastic oral keratinocyte cell line, DOK, were used as comparators. Three separate passages of each cell line were tested, and the expression relative to housekeeping gene GUSB was considered. RAMP1 and CALCR1 were highly expressed in oral cancer cell lines (HSC3, SCC-4) compared to HaCaT (Figure 4A). Consistent with human oral cancer lines, mouse derived cell lines, MOC1 and MOC2, also expressed both Ramp1 and Calcr1; however, MOC1 had significantly less expression of Calcr1 compared to MOC2 (Figure 4B). We confirmed RAMP1 protein expression in both in human-derived HaCaT and HSC3 cell lines as well as the mouse-derived MOC1 line using immunohistochemistry (Figure 4C). Lastly, to examine the effects of αCGRP on oral cancer cell proliferation in vitro, HaCaT, HSC3, and MOC1 cells were treated with different doses of αCGRP (0.03, 0.1, and 0.3nM) and proliferation was assayed at 48 hours by cell counting relative to vehicle treatment. There was no significant enhancement of cell proliferation in response to stimulation with any concentration of αCGRP or in response to CGRP inhibitors CGRP8–37 and AC187 for any cell line tested (F(4,39)=0.368, p=0.830; Figure 4D).
Figure 4. CGRP receptor expression in oral cancer cells.
RT-qPCR was used to quantify relative RAMP1 and CALCR1 gene expression in A) human oral cancer cell lines (HSC3, SCC-4) and dysplastic (DOK) cells compared to non-tumorigenic skin keratinocytes (HaCaT) as well as (B) mouse oral cancer cell lines (MOC1, MOC2). Data is presented as the relative gene expression (2−ΔCT) normalized to ACTB and represents three different cell passage determinations of gene expression. Two-way ANOVA **p<0.01. C) Representative images of RAMP1-IR (green) in HaCaT, HSC3, and MOC1 cell lines. DAPI (blue) was used to label cell nuclei. D) Cell proliferation assays were used to determine change in cell reproduction after αCGRP stimulation. The DAPI-labeled cell counts of non-stimulated vehicle-treated cells were set as 1, and the percent change in cell number of treated cells was determined relative to vehicle-treated cells (0.0005% water). CGRP8–37 and AC187 blocker treatment indicates co-treatment with 0.3nM αCGRP and 1nM antagonist. Data represent the mean (±SEM) of three different cell passage determinations of cell proliferation. One-way ANOVA within cell line, p>0.05.
3.4. Tumor infiltrating immune cells express more RAMP1 compared to healthy tissue
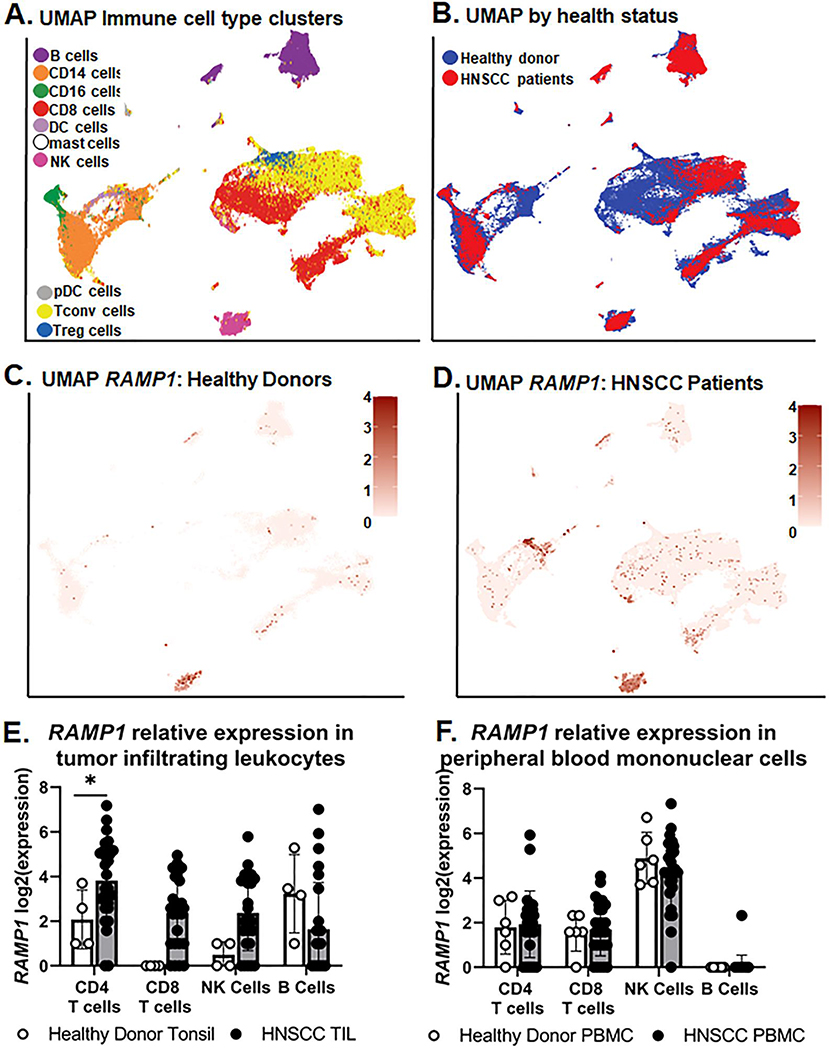
Data from oral cancer mouse models found tongue tumor growth significantly differed between genotypes and in vitro evidence suggests that αCGRP protein does not drive tumor cell proliferation directly. The immune system has been previously implicated as a direct recipient of CGRP action. We hypothesize CGRP signaling in the tumor microenvironment suppresses the adaptive immune response and is supportive of tumor growth. Under this hypothesis, a lack of CGRP signaling in CGRPKO mice thereby allowed for tumor clearance over time. Ramp1 receptor expression has been identified on lymphocyte populations in mouse models of inflammation[40]; however, RAMP1 gene expression in tumor infiltrating immune cells is currently unknown. Since CGRP receptor component RAMP1 is responsible for the specificity of the receptor complex to CGRP, we focused primarily on RAMP1 rather than CALCR1 in subsequent investigations. Using a recently publicly available single cell RNAseq dataset generated by Cillo and colleagues[29], we assessed the differential expression of CGRP-related genes in tumor infiltrating leukocytes (TIL) and peripheral blood mononuclear cells (PBMCs) isolated from 26 HNSCC patients (18 HPV- HNSCC, 8 HPV+ HNSCC) compared to healthy donors (i.e. tonsil tissue from 4 sleep apnea patients; PBMCs from 6 healthy donors). In total, 130,849 cells passed quality control measures and were assessed in the final dataset. Cell type annotations were included in the dataset; validation step of unsupervised clustering visualized 10 cell types based on canonical gene expression patterns (Figure 5A): CD19+ B cells, CD14+ monocytes, CD16+ myeloid derived suppressor cells, CD8+ cytotoxic T cells, dendritic cells, mast cells, cytotoxic NK cells, plasma dendritic cells, CD4+FOXP3– T conventional cells and CD4+FOXP3+ T regulatory cells. A UMAP was also generated to visualize distribution of samples by health status (Figure 5B). While minimal expression of CALCA and CALCB were found across all cell types, RAMP1 expression was well represented across many immune cell clusters isolated from both TIL and PBMCs (Figure 5C,D). Differentially expressed gene results from aggregated data demonstrated that RAMP1 was significantly increased in HNSCC TIL compared to healthy donor tonsil (Log(2)Fold Change = 2.033; Padj=0.0036) but not in PBMCs from HNSCC compared to healthy donor (Log(2)Fold Change = 0.175; Padj=0.944). Unfortunately, with only 4 healthy donor tonsil samples available, an abundance of zero-count transcripts resulted in the inability to conduct multiple testing across many of the immune cell subtypes; however, conventional T cells (CD3+CD4+FOXP3–) from HNSCC TIL had significantly higher RAMP1 expression compared to CD3+CD4+FOXP3– conventional T cells isolated from healthy donor tonsil (Log(2)Fold Change = 1.776; Padj=0.0263; Figure 5E). There was no significant difference in RAMP1 expression in PBMC subpopulations from HNSCC compared to healthy donors (Figure 5F).
Figure 5. Assessment of RAMP1 gene expression in human immune cells by scRNAseq.
Publicly available scRNAseq data was leveraged to assess differential gene expression changes in RAMP1. Distinct UMAPS were generated by A) immune cell subtype and B) tissue health status. The same unique cluster embedding was used to show RAMP1 gene expression represented as a heat map in samples from (C) healthy donor and (D) HNSCC patients. Tonsil/Tumor and peripheral blood mononuclear cell (PBMC) samples were combined for these plots. Relative expression is represented as Log2(expression). Quantitative analysis of RAMP1 Log2(expression) in lymphocyte subsets using the Wald Test between (E) HNSCC tumor-infiltrating cells (TIL) and subsets isolated from health donor tonsil as well as (F) PBMCs from HNSCC and healthy donor PBMCs. *p<0.05.
3.5. Loss of αCGRP signaling modulates immune cell infiltration into oral cancer
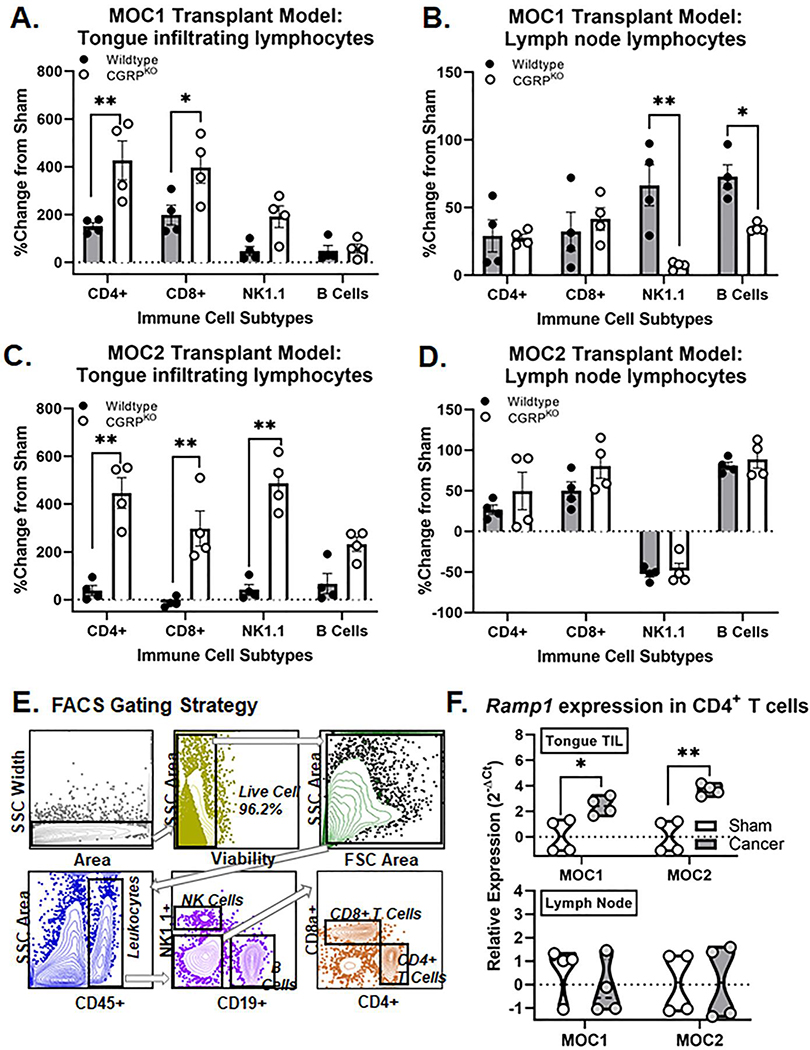
To determine if αCGRP signaling modulates the immune cell infiltration in oral cancer, we used flow cytometry to quantify MOC1 and MOC2 tumor infiltrating lymphocyte subsets compared sham tissue in wildtype C57Bl/6 mice and CGRPKO mice; submandibular and cervical (i.e. draining) lymph node was also assessed for comparison. We selected PID21 timepoint using the indolent MOC1 oral cancer model and PID 7 timepoint using the more aggressive MOC2 oral cancer model to assure a detectable tumor and prominent immune response would be present for both genotypes within each model. Based on the human scRNAseq analyses, four distinct lymphocyte subsets were assessed: CD4+CD8– T cells, CD4–CD8+ T cells, NK1.1+ NK cells, and CD19+ B cells. In sham mice, there was no significant difference in quantity of total CD45+ immune cells (t=6.173, p=0.102) or any immune subset relative to the total number of live cells between genotypes for either tongue (MOC1: F(3,24)=1.851, p=0.170; MOC2: F(3,24)=1.044, p=0.391) or lymph node (MOC1: F(3,24)=0.331, p=0.803; MOC2: F(3,24)=0.763, p=0.526) for either model. The percent change in tumor-infiltrating cells relative to sham was calculated for each subset isolated from tongue as well as draining lymph node. There was a significant interaction between genotype and MOC1 tumor-infiltrating immune cell subsets isolated from tongue (F(3,24)=3.085, p=0.046) (Figure 6A). There were significantly higher percentages of CD4+ T cells (p=0.001) and CD8+ T cells (p=0.018) in tumors from CGRPKO mice compared to tumors from wildtype mice. There was no significant difference in the percent of tumor infiltrating NK cells (p=0.127) or B cells (p=0.999) between genotypes. Additionally, there was a significant interaction between genotype and immune cell subsets isolated from draining lymph node (F(3,24)=5.590, p=0.005) (Figure 6B). There were significantly higher percentages of NK cells (p=0.001) and B cells (p=0.039) present in the lymph node tissue from wildtype mice compared to lymph node tissue from CGRPKO mice. Using the MOC2 oral cancer model, there was also a significant interaction between genotype and MOC2 tumor-infiltrating immune cell subsets isolated from tongue (F(3,24)=19.49, p<0.0001) (Figure 6C). There were 4-fold more CD4+ T cells (p=0.0003), 3-fold more CD8+ T cells (p=0.0054) and 5-fold more NK1.1+ NK cells (p<0.0001) in tumors from CGRPKO mice compared to sham, whereas these cell types showed almost no change from sham in wildtype tumors (Figure 6C). There was no significant difference in the percent of tumor infiltrating B cells between genotypes (p=0.993). Lastly, immune cell subsets isolated from draining lymph node did not differ between genotypes (F(3,24)=0.535, p=0.663) (Figure 6D). The percentages of total CD45+ immune cells quantified for each subpopulation from both sham and tumor-bearing mice are available in Table 2. These data suggest that the loss of CGRP signaling in the CGRPKO mice allowed for an increase in the anti-tumor associated immune response. The human scRNAseq data found that CD4+ T cells from human SCC tumors had higher RAMP1 expression compared to CD4+ T cells from healthy control tissue. To assess the Ramp1 gene expression of tumor infiltrating CD4+ T cells, we used FAC sorting to isolate the CD4+ lymphocyte subset in tongue tissue and associated draining lymph nodes from sham, MOC1-bearing and MOC2-bearing mice; purified CD4+ T cells were processed for RNA extraction and subsequent qPCR. Consistent with the human TIL, there was significantly higher relative Ramp1 expression in CD4+ T cells isolated from both MOC1 (t=3.170, p=0.019) and MOC2 (t=5.365, p=0.002) tumors compared to sham tongue tissue (Figure 6C,D). We found no significant difference in Ramp1 expression across treatment types in CD4+ T cells isolated from the draining lymph nodes for either cancer model (Figure 6D).
Figure 6. Increased immune cell infiltrate in CGRPKO mice.
Using flow cytometry, tumor infiltrating and lymph node lymphocyte subsets were quantified in dissociated tongue tissue from wildtype mice (black, n=4) and CGRPKO (white, n=4) from MOC1-tumor bearing mice at PID 21 (A,B) and MOC2-tumor bearing mice at PID 7 (C,D). Data are presented as a percent change from genotype-matched sham mice (n=4 per genotype). Two-way ANOVA, *p<0.05 **p<0.01. The percent of CD45+ immune cells quantified for both sham and tumor-bearing mice are available in Table 2. E) Representative gating strategy used to isolate tongue tumor infiltrating T cells by fluorescence-activated cell sorting (FACS). F) Quantification of Ramp1 expression in CD4+ T cells isolated from tongue or lymph node tissue from sham (white, n=4/time point), MOC1-tumor bearing mice (gray, n=4 PID 35), or MOC2-tumor bearing mice (gray, n=4, PID 14) relative to housekeeping gene Gapdh. Student’s t test *p<0.05, **p<0.01.
Table 2:
Immune cell subtypes from sham and tumor-bearing mice
| CD45+ cells | Wildtype Sham | Wildtype tumor-bear mice | Wildtype CGRPKO | CGRPKO tumor-bearing mice | ||||
|---|---|---|---|---|---|---|---|---|
| Tongue | Lymph node | Tongue | Lymph node | Tongue | Lymph node | Tongue | Lymph node | |
| MOC1 oral cancer model | ||||||||
| CD4+ | 0.93±0.03 | 16.5±2.28 | 2.34±0.27 | 20.44±5.04 | 0.71±0.07 | 18.52±2.39 | 3.72±1.15 | 23.75±3.95 |
| CD8+ | 0.74±0.16 | 12.83±1.75 | 2.38±0.95 | 16.97±3.66 | 0.69±0.39 | 12.26±1.66 | 3.44±0.92 | 17.37±2.04 |
| CD19+ | 1.38±0.18 | 32.44±3.08 | 1.73±0.43 | 53.99±9.78 | 0.72±0.02 | 48.00±5.17 | 1.07±0.33 | 48.70±4.09 |
| NK1.1+ | 0.27±0.12 | 0.82±0.06 | 0.55±0.15 | 0.37±0.04 | 0.23±0.01 | 0.72±0.04 | 0.55±0.17 | 0.25±0.05 |
| MOC2 oral cancer model | ||||||||
| CD4+ | 7.10±2.11 | 12.47±1.09 | 9.84±2.92 | 14.86±1.32 | 5.76±0.53 | 11.00±1.42 | 38.77±9.11 | 17.54±5.42 |
| CD8+ | 0.37±0.34 | 7.43±2.75 | 0.26±0.06 | 10.20±1.49 | 0.21±0.07 | 6.18±0.90 | 1.22±0.57 | 12.27±2.06 |
| CD19+ | 2.08±1.36 | 60.24±4.73 | 3.75±1.90 | 70.14±3.61 | 2.41±1.86 | 60.11±2.64 | 7.44±1.35 | 67.9±6.64 |
| NK1.1+ | 0.06±0.01 | 0.80±0.27 | 0.22±0.07 | 0.45±0.08 | 0.16±0.02 | 1.10±0.12 | 1.11±0.47 | 0.49±0.17 |
Data is presented as average percent of total CD45+ live cells counted ± standard deviation.
4.0. Discussion
We report that αCGRP neurotransmission modulates tumor growth and tumor-associated immune response in HNSCC. While pathologically denoted PNI has been implicated as an independent risk factor and is associated with a high recurrence rate in HNSCC[12], the impact of nerve presence in tumors generally (i.e. regardless of PNI diagnosis) and types of nerves involved (e.g., sensory, sympathetic, motor) are currently unknown. Many studies have suggested a potential diagnostic value for sensory neurotransmitter CGRP in various cancers.[38, 39, 41–43] Zhang and colleagues recently demonstrated that preoperative plasma CGRP levels, combined with preoperative pain status and T stage, could be used to predict lymph node metastasis in HNSCC patients.[38] We identified αCGRP-containing sensory nerves innervating human HNSCC and found an increase in CGRP receptor expression in tumor infiltrating lymphocytes isolated from human HNSCC tumor tissue. Using a syngeneic orthotopic transplant mouse model of oral cancer, we found both tumor-innervating CGRP-containing sensory nerves as well as an increase in αCGRP protein in tongue tissue compared to sham mice. We found no difference in αCGRP protein concentration in tongue tissue from the HSC3 xenograft mouse model compared to sham. There was also an increase in αCGRP gene and protein expression in trigeminal sensory neurons from tumor-bearing mice; we found no difference in Calca gene expression in TG from HSC3 xenograft mice compared to sham. Given the current hypothesis that inflammatory mediators released from tumor and immune cells directly activate sensory nerves in the tumor microenvironment to drive sensitization and subsequently cancer pain, we speculate that a fully intact immune system contributes to increased sensitization and subsequently more sensory neurotransmission in the tumor microenvironment. This is consistent with previous findings using a rat oral cancer model which demonstrated tumor-innervating CGRP nerve sprouting associated with upregulated CGRP expression in the maxillary and mandibular trigeminal branches.[22]
Previous literature had demonstrated that CGRP may be located in nonneuronal cells, such as endothelial cells and keratinocytes[44, 45] as well as several types of immune cells, including activated B cells, peripheral blood mononuclear cells, and macrophages.[46–48] We did not find meaningful CALCA or CALCB gene expression in immune cell subtypes isolated from tissue or blood from HNSCC patients or healthy controls, nor did we find detectable levels of αCGRP protein in MOC1 or MOC2 oral cancer cells. While we recognize that patient-reported pain can be multifaceted, our retrospective analyses found a positive correlation between the amount of αCGRP-containing sensory nerve presence and reported pain in HNSCC patients. These data suggest that sensory neurotransmission is likely present in tumor and a probable source for αCGRP. Furthermore, a recent study by Zhang and colleagues[38] linking αCGRP to metastasis in HNSCC found that αCGRP expression was higher in tumor tissues from HNSCC patients with PNI compared to adjacent normal tissues concluding that sensory innervation is the most likely contributor of αCGRP in this context. Still, additional experiments are needed to confirm that sensory nerves are the primary source of αCGRP in the tumor microenvironment.
While sensory neurotransmission has largely been considered only in the context of cancer symptoms (i.e. pain)[49, 50], CGRP signaling may also impact tumor progression through direct and indirect (e.g., immune) pathways. CGRP has been reported to promote tumor progression directly by affecting tumor cell function.[51] CGRP receptor components were identified at the gene and protein level in both human and mouse oral cancer cell lines. While we did not find any influence of CGRP stimulation on cell proliferation in either human or mouse oral cancer cell lines, others have demonstrated increase colony formation ability and wound healing ability of human oral cancer cell lines HN6 and Cal27.[38] Drissi et al found that human recombinant CGRP did not increase osteosarcoma cell proliferation but instead induced a rapid, transient, and dose-dependent increase in free cytosolic calcium levels not coupled to cAMP, suggesting downstream events driven by phospholipase C.[52] However, we found that tongue tumor-bearing CGRPKO mice had a significantly smaller tumor burden over time compared to wildtype mice using tumors generated by either aggressive (i.e. MOC2) and indolent (i.e. MOC1) mouse oral cancer cells. These findings are consistent with a previous study that found reduced tumor growth and tumor-associated angiogenesis in CGRPKO mice implanted with Lewis lung carcinoma (LLC) cells; furthermore, administration of CGRP receptor antagonist or denervation in tumor-bearing wildtype mice also markedly suppressed tumor growth.[51] CGRP receptor complex activation has also been shown to induce angiogenesis and lymphangiogenesis by upregulating vascular endothelial growth factor (VEGF) isoforms in both vascular endothelial cells and inflammatory cells.[53] Additionally, wound healing and wound-induced angiogenesis were also significantly suppressed in RAMP1 knockout mice.[54] While the exact mechanism of CGRP signaling on tumor growth is currently unknown, our data suggests that CGRP decreased tumor size at least in part through indirect mechanisms.
CGRP is a potent vasodilator and thought to regulate local inflammatory response to injury.[55] CGRP-containing nerve fibers have been shown to innervate the majority of immune-related tissues including bone, spleen, lymph nodes, and gut, and CGRP receptor components have been found on many hematopoietic cell types.[56] Using publicly available scRNAseq data, we demonstrate that RAMP1 gene expression was more prevalent in human tumor-infiltrating lymphocyte subsets, specifically CD3+CD4+FOXP3– T cells, compared to healthy donor tonsil tissue. There was no significant difference in RAMP1 expression in PBMCs from HNSCC compared to healthy donor suggesting the changes in gene expression are tumor specific. Overall conclusions from these data are restricted however, given the limitations in the analyses. HNSCC tumor immune landscape was markedly different from healthy donor tonsil immune landscape and an abundance of zero-count transcripts in tissue immune subtypes hindered multiple testing across most of the lymphocyte subtypes. Future directions will include comparisons of tumor immune landscape and RAMP1 expression in highly and weakly innervated tumors. However, supporting the claim that RAMP1 expression was increased in tumor infiltrating T cells, FAC sorted CD4+ T cells from MOC1- and MOC2-tumor bearing mice had significantly more Ramp1 gene expression compared to sham. While the functional implications of RAMP1 activation on T cells is unknown, available evidence suggests that CGRP has an inhibitory role on the immune response. CGRP limits damage during inflammation[40, 57], and RAMP1 signaling in immune cells, including macrophages and T cells, is important for suppression of inflammation.[58–60] Physiological concentrations of CGRP have been shown to inhibit proliferative responses of CD4+ and CD8+ T cells[61] while limiting B cell development and colony formation.[62] Additionally, antigen presentation is reduced on dendritic cells in response to CGRP stimulation, dampening the interactions with T cells.[63] In the present study, there was decreasing tumor size over time in MOC1-tumor bearing CGRPKO mice, suggesting an active anti-tumor immune response. Furthermore, the tumor bearing CGRPKO mice had a more anti-tumor immune presence in the tumor compared to wildtype mice in both the MOC1 and MOC2 models. We also found significant differences in NK and B cell populations in draining lymph node in the MOC1 model, suggesting different stages in the active tumor-associated immune response between genotypes at PID 21. However, due to the slow tumor progression that occurs in the MOC1 model (i.e. MOC1: 4–6 weeks growth, MOC2: 14 days growth), additional experiments are needed to fully understand the ongoing immune landscape over time. Together our data suggests that sensory nerves in the tumor microenvironment release CGRP which supports tumor-mediated immune suppression and immune evasion; furthermore, eliminating CGRP signaling allows for a more robust tumor-associated immune response and subsequently tumor clearance.
Neural influence of cancer progression is an emerging field.[64, 65] Clinically, nerve presence detected early in disease (stages I and II) is thought to have a serious impact on outcome in HNSCC[12]. Successful therapeutic strategies using humanized monoclonal antibodies directed against CGRP (e.g., Fremanezumab[66]) or non-peptide antagonists directed against the receptor (e.g., Rimegepant[67]) have recently been approved by the Food and Drug Administration (FDA) or are in clinical trials for the treatment of migraine; the impact on cancer is unknown. Additional experiments are warranted to investigate the link between sensory neurotransmission and HNSCC progression.
Supplementary Material
Acknowledgements
This work was supported by NIH NIDCR R00DE028019 and the Rita Allan Foundation. This project also used the UPMC Hillman Cancer Center and Tissue and Research Pathology/Pitt Biospecimen Core shared resource which is supported in part by award P30CA047904. We thank the Hillman Cancer Center Head and Neck Tissue Bank for suppling clinical specimens as well as Dr. Elizabeth Bilodeau DMD, MD, MSEd for her expertise in pathological assessment of the HNSCC tumor tissue. We thank the UPMC Hillman Cancer Center Cytometry Core for their expertise and technical support for FACsorting experiments.
Footnotes
Conflict of Interest
The authors have no conflicts of interest.
Bibliography
- 1.Cohen N, Fedewa S, Chen AY. Epidemiology and Demographics of the Head and Neck Cancer Population. Oral Maxillofac Surg Clin North Am 2018; 30: 381–395. [DOI] [PubMed] [Google Scholar]
- 2.Ernani V, Saba NF. Oral Cavity Cancer: Risk Factors, Pathology, and Management. Oncology 2015; 89: 187–195. [DOI] [PubMed] [Google Scholar]
- 3.Westra WH, Lewis JS Jr. Update from the 4th Edition of the World Health Organization Classification of Head and Neck Tumours: Oropharynx. Head Neck Pathol 2017; 11: 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- 5.Kamrani P, Sadiq NM. Anatomy, Head and Neck, Oral Cavity (Mouth). StatPearls. Treasure Island (FL)2022. [PubMed] [Google Scholar]
- 6.Liebig C, Ayala G, Wilks JA, Berger DH, Albo D. Perineural invasion in cancer: a review of the literature. Cancer 2009; 115: 3379–3391. [DOI] [PubMed] [Google Scholar]
- 7.Kolokythas A, Cox DP, Dekker N, Schmidt BL. Nerve growth factor and tyrosine kinase A receptor in oral squamous cell carcinoma: is there an association with perineural invasion? J Oral Maxillofac Surg 2010; 68: 1290–1295. [DOI] [PubMed] [Google Scholar]
- 8.Nair D, Mair M, Singhvi H, et al. Perineural invasion: Independent prognostic factor in oral cancer that warrants adjuvant treatment. Head Neck 2018; 40: 1780–1787. [DOI] [PubMed] [Google Scholar]
- 9.Schmitd LB, Scanlon CS, D’Silva NJ. Perineural Invasion in Head and Neck Cancer. J Dent Res 2018; 97: 742–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang X, Tian X, Wu K, et al. Prognostic impact of perineural invasion in early stage oral tongue squamous cell carcinoma: Results from a prospective randomized trial. Surg Oncol 2018; 27: 123–128. [DOI] [PubMed] [Google Scholar]
- 11.Rahima B, Shingaki S, Nagata M, Saito C. Prognostic significance of perineural invasion in oral and oropharyngeal carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2004; 97: 423–431. [DOI] [PubMed] [Google Scholar]
- 12.Laske RD, Scholz I, Ikenberg K, et al. Perineural Invasion in Squamous Cell Carcinoma of the Oral Cavity: Histology, Tumor Stage, and Outcome. Laryngoscope Investig Otolaryngol 2016; 1: 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kappos EA, Engels PE, Tremp M, et al. Denervation leads to volume regression in breast cancer. J Plast Reconstr Aesthet Surg 2018; 71: 833–839. [DOI] [PubMed] [Google Scholar]
- 14.Magnon C, Hall SJ, Lin J, et al. Autonomic nerve development contributes to prostate cancer progression. Science 2013; 341: 1236361. [DOI] [PubMed] [Google Scholar]
- 15.March B, Faulkner S, Jobling P, et al. Tumour innervation and neurosignalling in prostate cancer. Nat Rev Urol 2020; 17: 119–130. [DOI] [PubMed] [Google Scholar]
- 16.Qiao G, Chen M, Bucsek MJ, Repasky EA, Hylander BL. Adrenergic Signaling: A Targetable Checkpoint Limiting Development of the Antitumor Immune Response. Front Immunol 2018; 9: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saloman JL, Albers KM, Li D, et al. Ablation of sensory neurons in a genetic model of pancreatic ductal adenocarcinoma slows initiation and progression of cancer. Proc Natl Acad Sci U S A 2016; 113: 3078–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao CM, Hayakawa Y, Kodama Y, et al. Denervation suppresses gastric tumorigenesis. Sci Transl Med 2014; 6: 250ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheff NN, Ye Y, Bhattacharya A, et al. Tumor necrosis factor alpha secreted from oral squamous cell carcinoma contributes to cancer pain and associated inflammation. PAIN 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terenghi G, Polak JM, Rodrigo J, Mulderry PK, Bloom SR. Calcitonin gene-related peptide-immunoreactive nerves in the tongue, epiglottis and pharynx of the rat: occurrence, distribution and origin. Brain Res 1986; 365: 1–14. [DOI] [PubMed] [Google Scholar]
- 21.Russell FA, King R, Smillie SJ, Kodji X, Brain SD. Calcitonin gene-related peptide: physiology and pathophysiology. Physiol Rev 2014; 94: 1099–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagamine K, Ozaki N, Shinoda M, et al. Mechanical allodynia and thermal hyperalgesia induced by experimental squamous cell carcinoma of the lower gingiva in rats. J Pain 2006; 7: 659–670. [DOI] [PubMed] [Google Scholar]
- 23.Dottorini ME, Assi A, Sironi M, Sangalli G, Spreafico G, Colombo L. Multivariate analysis of patients with medullary thyroid carcinoma. Prognostic significance and impact on treatment of clinical and pathologic variables. Cancer 1996; 77: 1556–1565. [DOI] [PubMed] [Google Scholar]
- 24.McLatchie LM, Fraser NJ, Main MJ, et al. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature 1998; 393: 333–339. [DOI] [PubMed] [Google Scholar]
- 25.Honda M, Ito Y, Hattori K, et al. Inhibition of receptor activity-modifying protein 1 suppresses the development of endometriosis and the formation of blood and lymphatic vessels. J Cell Mol Med 2020; 24: 11984–11997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuru S, Ito Y, Matsuda H, et al. RAMP1 signaling in immune cells regulates inflammation-associated lymphangiogenesis. Lab Invest 2020; 100: 738–750. [DOI] [PubMed] [Google Scholar]
- 27.Umeda Y, Arisawa M. Inhibition of natural killer activity by calcitonin gene-related peptide. Immunopharmacol Immunotoxicol 1989; 11: 309–320. [DOI] [PubMed] [Google Scholar]
- 28.Scheff NN, Alemu RG, Klares R 3rd, et al. Granulocyte-Colony Stimulating Factor-Induced Neutrophil Recruitment Provides Opioid-Mediated Endogenous Anti-nociception in Female Mice With Oral Squamous Cell Carcinoma. Front Mol Neurosci 2019; 12: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cillo AR, Kurten CHL, Tabib T, et al. Immune Landscape of Viral- and Carcinogen-Driven Head and Neck Cancer. Immunity 2020; 52: 183–199 e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lun AT, Bach K, Marioni JC. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol 2016; 17: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy DJ, Campbell KR, Lun AT, Wills QF. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 2017; 33: 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal, complex systems 2006; 1695: 1–9. [Google Scholar]
- 33.Scheff NN, Bhattacharya A, Dowse E, et al. Neutrophil-Mediated Endogenous Analgesia Contributes to Sex Differences in Oral Cancer Pain. Front Integr Neurosci 2018; 12: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheff NN, Ye Y, Bhattacharya A, et al. Tumor necrosis factor alpha secreted from oral squamous cell carcinoma contributes to cancer pain and associated inflammation. Pain 2017; 158: 2396–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haberberger RV, Barry C, Dominguez N, Matusica D. Human Dorsal Root Ganglia. Front Cell Neurosci 2019; 13: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Messlinger K, Russo AF. Current understanding of trigeminal ganglion structure and function in headache. Cephalalgia 2019; 39: 1661–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu XJ, Li CY, Xu YH, Chen LM, Zhou CL. Calcitonin gene-related peptide increases proliferation of human HaCaT keratinocytes by activation of MAP kinases. Cell Biol Int 2009; 33: 1144–1148. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Chen M, Liu Z, Wang X, Ji T. The neuropeptide calcitonin gene-related peptide links perineural invasion with lymph node metastasis in oral squamous cell carcinoma. BMC Cancer 2021; 21: 1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagakawa O, Ogasawara M, Fujii H, et al. Effect of prostatic neuropeptides on invasion and migration of PC-3 prostate cancer cells. Cancer Lett 1998; 133: 27–33. [DOI] [PubMed] [Google Scholar]
- 40.Holzmann B Antiinflammatory activities of CGRP modulating innate immune responses in health and disease. Curr Protein Pept Sci 2013; 14: 268–274. [DOI] [PubMed] [Google Scholar]
- 41.Kelley MJ, Snider RH, Becker KL, Johnson BE. Small cell lung carcinoma cell lines express mRNA for calcitonin and alpha- and beta-calcitonin gene related peptides. Cancer Lett 1994; 81: 19–25. [DOI] [PubMed] [Google Scholar]
- 42.Nagakawa O, Ogasawara M, Murata J, Fuse H, Saiki I. Effect of prostatic neuropeptides on migration of prostate cancer cell lines. Int J Urol 2001; 8: 65–70. [DOI] [PubMed] [Google Scholar]
- 43.Takami H, Shikata J, Horie H, Horiuchi J, Sakurai H, Ito K. [Radioimmunoassay of plasma calcitonin gene-related peptide (CGRP) levels in patients with endocrine tumor]. Gan To Kagaku Ryoho 1989; 16: 2219–2225. [PubMed] [Google Scholar]
- 44.Cai WQ, Bodin P, Loesch A, Sexton A, Burnstock G. Endothelium of human umbilical blood vessels: ultrastructural immunolocalization of neuropeptides. J Vasc Res 1993; 30: 348–355. [DOI] [PubMed] [Google Scholar]
- 45.Hou Q, Barr T, Gee L, et al. Keratinocyte expression of calcitonin gene-related peptide beta: implications for neuropathic and inflammatory pain mechanisms. Pain 2011; 152: 2036–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bracci-Laudiero L, Aloe L, Buanne P, et al. NGF modulates CGRP synthesis in human B-lymphocytes: a possible anti-inflammatory action of NGF? J Neuroimmunol 2002; 123: 58–65. [DOI] [PubMed] [Google Scholar]
- 47.Linscheid P, Seboek D, Schaer DJ, Zulewski H, Keller U, Muller B. Expression and secretion of procalcitonin and calcitonin gene-related peptide by adherent monocytes and by macrophage-activated adipocytes. Crit Care Med 2004; 32: 1715–1721. [DOI] [PubMed] [Google Scholar]
- 48.Wang H, Xing L, Li W, Hou L, Guo J, Wang X. Production and secretion of calcitonin gene-related peptide from human lymphocytes. J Neuroimmunol 2002; 130: 155–162. [DOI] [PubMed] [Google Scholar]
- 49.Clouston PD, DeAngelis LM, Posner JB. The spectrum of neurological disease in patients with systemic cancer. Ann Neurol 1992; 31: 268–273. [DOI] [PubMed] [Google Scholar]
- 50.Giglio P, Gilbert MR. Neurologic complications of cancer and its treatment. Curr Oncol Rep 2010; 12: 50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toda M, Suzuki T, Hosono K, et al. Neuronal system-dependent facilitation of tumor angiogenesis and tumor growth by calcitonin gene-related peptide. Proc Natl Acad Sci U S A 2008; 105: 13550–13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Drissi H, Lieberherr M, Hott M, Marie PJ, Lasmoles F. Calcitonin gene-related peptide (CGRP) increases intracellular free Ca2+ concentrations but not cyclic AMP formation in CGRP receptor-positive osteosarcoma cells (OHS-4). Cytokine 1999; 11: 200–207. [DOI] [PubMed] [Google Scholar]
- 53.Majima M, Ito Y, Hosono K, Amano H. CGRP/CGRP Receptor Antibodies: Potential Adverse Effects Due to Blockade of Neovascularization? Trends Pharmacol Sci 2019; 40: 11–21. [DOI] [PubMed] [Google Scholar]
- 54.Kurashige C, Hosono K, Matsuda H, Tsujikawa K, Okamoto H, Majima M. Roles of receptor activity-modifying protein 1 in angiogenesis and lymphangiogenesis during skin wound healing in mice. FASEB J 2014; 28: 1237–1247. [DOI] [PubMed] [Google Scholar]
- 55.Engel MA, Khalil M, Siklosi N, et al. Opposite effects of substance P and calcitonin gene-related peptide in oxazolone colitis. Dig Liver Dis 2012; 44: 24–29. [DOI] [PubMed] [Google Scholar]
- 56.Assas BM, Pennock JI, Miyan JA. Calcitonin gene-related peptide is a key neurotransmitter in the neuro-immune axis. Front Neurosci 2014; 8: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim YJ, Granstein RD. Roles of calcitonin gene-related peptide in the skin, and other physiological and pathophysiological functions. Brain Behav Immun Health 2021; 18: 100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inoue T, Ito Y, Nishizawa N, et al. RAMP1 in Kupffer cells is a critical regulator in immune-mediated hepatitis. PLoS One 2018; 13: e0200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawashima-Takeda N, Ito Y, Nishizawa N, et al. RAMP1 suppresses mucosal injury from dextran sodium sulfate-induced colitis in mice. J Gastroenterol Hepatol 2017; 32: 809–818. [DOI] [PubMed] [Google Scholar]
- 60.Tsujikawa K, Yayama K, Hayashi T, et al. Hypertension and dysregulated proinflammatory cytokine production in receptor activity-modifying protein 1-deficient mice. Proc Natl Acad Sci U S A 2007; 104: 16702–16707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teresi S, Boudard F, Bastide M. Effect of calcitonin gene-related peptide and vasoactive intestinal peptide on murine CD4 and CD8 T cell proliferation. Immunol Lett 1996; 50: 105–113. [DOI] [PubMed] [Google Scholar]
- 62.Fernandez S, Knopf MA, McGillis JP. Calcitonin-gene related peptide (CGRP) inhibits interleukin-7-induced pre-B cell colony formation. J Leukoc Biol 2000; 67: 669–676. [DOI] [PubMed] [Google Scholar]
- 63.Carucci JA, Ignatius R, Wei Y, et al. Calcitonin gene-related peptide decreases expression of HLA-DR and CD86 by human dendritic cells and dampens dendritic cell-driven T cell-proliferative responses via the type I calcitonin gene-related peptide receptor. J Immunol 2000; 164: 3494–3499. [DOI] [PubMed] [Google Scholar]
- 64.Demir IE, Reyes CM, Alrawashdeh W, et al. Clinically Actionable Strategies for Studying Neural Influences in Cancer. Cancer Cell 2020; 38: 11–14. [DOI] [PubMed] [Google Scholar]
- 65.Demir IE, Reyes CM, Alrawashdeh W, et al. Future directions in preclinical and translational cancer neuroscience research. Nat Cancer 2021; 1: 1027–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferrari MD, Diener HC, Ning X, et al. Fremanezumab versus placebo for migraine prevention in patients with documented failure to up to four migraine preventive medication classes (FOCUS): a randomised, double-blind, placebo-controlled, phase 3b trial. Lancet 2019; 394: 1030–1040. [DOI] [PubMed] [Google Scholar]
- 67.Croop R, Goadsby PJ, Stock DA, et al. Efficacy, safety, and tolerability of rimegepant orally disintegrating tablet for the acute treatment of migraine: a randomised, phase 3, double-blind, placebo-controlled trial. Lancet 2019; 394: 737–745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.