Abstract
Oxytocin regulates social behavior via direct modulation of neurons, regulation of neural network activity, and interaction with other neurotransmitter systems. The behavioral effects of oxytocin signaling are determined by the species-specific distribution of brain oxytocin receptors. The socially monogamous prairie vole has been a useful model organism for elucidating the role of oxytocin in social behaviors, including pair bonding, response to social loss, and consoling. However, there has been no comprehensive mapping of oxytocin receptor-expressing cells throughout the prairie vole brain. Here, we employed a highly sensitive in situ hybridization, RNAscope, to construct an exhaustive, brain-wide map of oxytocin receptor mRNA-expressing cells. We found that oxytocin receptor mRNA expression was widespread and diffuse throughout the brain, with specific areas displaying particularly robust expression. Comparing receptor binding with mRNA revealed that regions of the hippocampus and substantia nigra contained oxytocin receptor protein but lacked mRNA, indicating that oxytocin receptors can be transported to distal neuronal processes, consistent with presynaptic oxytocin receptor functions. In the nucleus accumbens, a region involved in oxytocin-dependent social bonding, oxytocin receptor mRNA expression was detected in both the D1 and D2 dopamine receptor-expressing subtypes of cells. Furthermore, natural genetic polymorphisms robustly influenced oxytocin receptor expression in both D1 and D2 receptor cell types in the nucleus accumbens. Collectively, our findings further elucidate the extent to which oxytocin signaling is capable of influencing brain-wide neural activity, responses to social stimuli, and social behavior.
Keywords: dopamine receptor, autoradiography, nucleus accumbens, phenotypic plasticity, Oxtr mRNA, in situ hybridization, Oxtr SNPs
Graphical Abstract
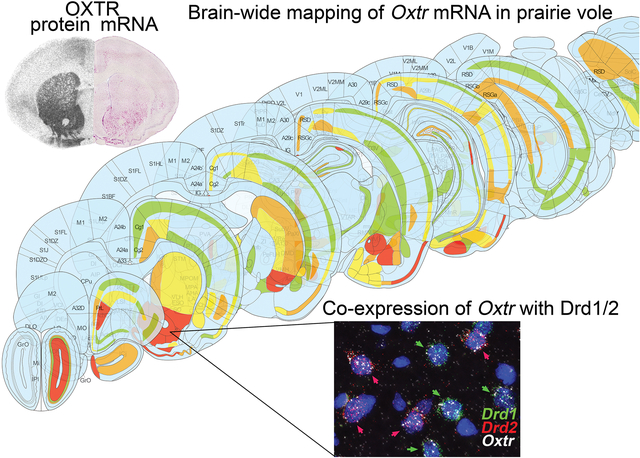
Using RNAscope in situ hybridization, we comprehensively mapped oxytocin receptor mRNA throughout the prairie vole brain. Comparison of mRNA and protein localization revealed localized mismatches suggesting both pre- and post-synaptic function for OXTRs. Co-expression of Oxtr mRNA with D1/D2 receptors in the nucleus accumbens was also examined.
Introduction:
Oxytocin is a neuromodulatory nonapeptide that regulates social behavior and reproductive physiology in a wide variety of taxa through its signaling via oxytocin receptors (OXTR) in the brain (Froemke & Young, 2021; Grinevich & Ludwig, 2021; Jurek & Neumann, 2018). Largely conserved through 700 million years of evolution (Donaldson & Young, 2008), oxytocin and its analogs control egg-laying in annelids (Oumi et al., 1996), social preference in fish (Landin et al., 2020), and flocking in birds (Goodson, Wilson, & Schrock, 2012). In mammals, oxytocin plays a critical role not only in reproductive and parenting behaviors (Numan, 2020; Rilling & Young, 2014) but also in complex social dynamics like consolation of conspecifics (Burkett et al., 2016), intergroup conflicts (Samuni et al., 2017), and interspecies bonds (Nagasawa et al., 2015). Oxytocin may also modulate social behaviors believed to be uniquely human, such as love (Walum & Young, 2018; L. Young & Alexander, 2012) and altruism. Perhaps most importantly, the oxytocin system is a promising therapeutic target for neuropsychiatric conditions with social dysfunction, including autism spectrum disorders, schizophrenia, and post-traumatic stress disorder (Andari, Hurlemann, & Young, 2018; Andari et al., 2021; DeMayo, Young, Hickie, Song, & Guastella, 2019; Charles L. Ford & Larry J. Young, 2021; C. L. Ford & L. J. Young, 2021).
Despite its evolutionary conservation, oxytocin mediates remarkably diverse behaviors across species via species-specific patterns of OXTR distribution throughout the brain (Johnson & Young, 2017) (Larry J. Young & Zhang, 2021). For instance, OXTR signaling in nucleus accumbens (NAc) and prelimbic cortex (PLC) is critical for pair bonding in monogamous prairie voles, and monogamous and polygamous vole species differ in OXTR expression in these regions (L. J. Young, Huot, Nilsen, Wang, & Insel, 1996; L. J. Young, Lim, Gingrich, & Insel, 2001; L. J. Young & Wang, 2004). Subsequent studies in rodents have supported the idea that interspecies variation in social behavior results from variation in OXTR distribution (Campbell, Ophir, & Phelps, 2009; Francis, Champagne, & Meaney, 2000; A. R. Freeman, Aulino, Caldwell, & Ophir, 2020; S. M. Freeman & Young, 2016; Rogers Flattery et al., 2021; Smeltzer, Curtis, Aragona, & Wang, 2006). A recent quantitative analysis of data from 13 rodent species compared in seven papers demonstrates that OXTR density in NAc and lateral septum predicts social group size, behavior towards conspecifics, and reproductive strategies (D. E. Olazábal & Sandberg, 2020). However, a recent study comparing OXTR distributions in monogamous and polygamous lemur species indicates that, in some primates, OXTR binding does not predict mating strategies, suggesting that other mechanisms also contribute to social behavioral diversity (Grebe et al., 2021). Nevertheless, differences among primates, including humans, in the brain regions innervated by oxytocinergic fibers or containing OXTR may contribute to certain behavioral differences across species (Rogers Flattery et al., 2021; Rogers Flattery et al., 2018).
Differences in OXTR distribution mediate intraspecies variations in behavior as well (Keebaugh, Barrett, Laprairie, Jenkins, & Young, 2015; Alexander G. Ophir, Gessel, Zheng, & Phelps, 2012; Ross et al., 2009). In prairie voles, single nucleotide polymorphisms (SNPs) in the OXTR gene, Oxtr, predict individual differences in OXTR density in the NAc, but not other areas, which, in turn, predicts social attachment behaviors and resilience to early-life neglect (Ahern, Olsen, Tudino, & Beery, 2021; Barrett, Arambula, & Young, 2015; King, Walum, Inoue, Eyrich, & Young, 2016). Additionally, decreased OXTR density in the lateral septum of male prairie voles is associated with higher rates of social investigation of females (A. G. Ophir, Zheng, Eans, & Phelps, 2009). In rat dams, higher densities of OXTR in the central nucleus of the amygdala and medial preoptic area (MPOA) are associated with increased maternal behaviors (Champagne, Diorio, Sharma, & Meaney, 2001; Francis et al., 2000). Many changes in an individual’s social behavior are also be mediated by changes in Oxtr expression. Rats undergo an estrogen-dependent increase in Oxtr mRNA expression in the ventromedial hypothalamus (VMH) during pregnancy and in the MPOA and the bed nucleus stria terminalis (BNST) at parturition (Meddle, Bishop, Gkoumassi, van Leeuwen, & Douglas, 2007; L. J. Young, Muns, Wang, & Insel, 1997). OXTR density in VMH is highly sensitive to sex steroids, with gonadal steroids having opposite effects on OXTR density in VMH in mice and rats (Insel, Young, Witt, & Crews, 1993). Paternal behavior is also regulated, at least in part, by changes in Oxtr expression. Male mandarin voles’ behavior toward pups changes from infanticidal to parental after they sire offspring, a change that requires an upregulation of OXTRs in the MPOA (Yuan et al., 2019).
OXTRs localized to specific regions have specific behavioral functions. In rodents, OXTRs are heavily expressed throughout olfactory pathways, including the anterior olfactory area (also known as anterior olfactory nucleus)(S. M. Freeman & Young, 2016; Freund-Mercier et al., 1987). Oxytocin signaling in the anterior olfactory area facilitates social recognition by enhancing the signal-to-noise ratio of olfactory information processing via top-down projections to inhibitory granule cells in the main olfactory bulb (Oettl et al., 2016). In primates, OXTRs are more densely expressed in areas responsible for visual information processing and attention, including the nucleus basalis of Meynert and superior colliculus (S. M. Freeman, Inoue, Smith, Goodman, & Young, 2014; S. M. Freeman & Young, 2016; Grebe et al., 2021). In the auditory cortex of maternal rats, OXTRs enhance the salience of pup calls and facilitate pup retrieval by regulating cortical inhibition (Marlin, Mitre, D’Amour J, Chao, & Froemke, 2015). OXTRs in insular cortex mediate the recognition of emotion in rats (M. M. Rogers-Carter et al., 2018), in the anterior cingulate cortex they mediate partner-directed consolation behavior in voles (Burkett et al., 2016), and in the PLC and NAc they regulate pair bonding in voles (Walum & Young, 2018).
Beyond its brain region-specific effects, oxytocin is a modulator of modulators; it coordinates activity across a network of social brain regions (Johnson et al., 2016) (Johnson & Young, 2017) and it influences the activity of other neurotransmitters, including serotonin and dopamine (Froemke & Young, 2021). OXTRs synthesized in dorsal raphe nucleus, a primary source of serotonergic projections, and in the ventral tegmental area, a primary source of dopaminergic projections, are both critical for social reward (Dölen, Darvishzadeh, Huang, & Malenka, 2013; Hung et al., 2017). In particular, the interplay of OXTRs, D1 dopamine receptors (D1R), and D2 dopamine receptors (D2R) on medium spiny neurons (MSNs) in the striatum mediates many complex social behaviors, including social reward learning, pair bonding, and selective aggression. Specifically, D1R activity appears to inhibit pair bonding, D2R activity appears to facilitate pair bonding, and an increase in the ratio of D1R:D2R expression appears to underlie selective aggression in pair-bonded male voles (Aragona et al., 2006).
Although OXTRs have been mapped in mice using immunohistochemistry (Mitre et al., 2016), the translational relevance of social neuroscience research in mice and rats is limited by these models’ social behavioral repertoire, particularly in relation to pair bonding, partner loss, biparental care, and empathy-based consoling behavior. In contrast, the prairie vole has emerged as the premier model organism for studying the neurochemistry and neural circuit mechanisms of these social behaviors (McGraw & Young, 2010). The prairie vole has already proven useful for elucidating the role of oxytocin in social behavior and cognition, and its utility as a model organism will continue increasing now that transgenic and gene-editing tools once only available in mice have become available in voles (Boender & Young, 2020; Kengo Horie, Inoue, Nishimori, & Young, 2020; K. Horie et al., 2019).
Despite the value of the prairie vole model for understanding the effects of oxytocin on behavior, there has been no comprehensive mapping of Oxtr mRNA expression throughout the vole brain. Previous OXTR localization studies in voles have relied on autoradiographic detection, which does not provide cellular resolution. Here, we used RNAscope in situ hybridization to label Oxtr mRNA throughout the prairie vole brain. We compared the subcellular localization of Oxtr mRNA to the localization of OXTR protein using autoradiography, which revealed localized mismatches suggesting a presynaptic function for OXTRs. We also examined the colocalization of Oxtr mRNA with mRNA for D1R (Drd1) and D2R (Drd2) on cells in NAc. Additionally, we leveraged the naturally-occurring genetic variation in our outbred prairie vole colony to examine how SNPs known to affect social behavior affect Oxtr mRNA expression in Drd1-expressing compared to Drd2-expressing cells in NAc.
Materials and Methods:
Subjects
A total of 16 male and 16 female adult (150 – 220 days old) prairie voles (Microtus ochrogaster) were selected from our outbred laboratory colony at Emory that was originally derived from field-caught voles in Champaign, Illinois. Eight males and eight females were homozygous for the high Oxtr expression allele (High-Oxtr; C/C at NT213739), and eight males and eight females were homozygous for the low Oxtr expression alleles (Low-Oxtr; T/T at NT213739) as described in King et al. (2016). Voles were group-housed at 22 °C under a 14:10 hr light/dark cycle in ventilated 26 × 18 × 19 cm Plexiglass cages containing Bedo’cobbs Laboratory Animal Bedding (The Andersons; Maumee, Ohio) with ad libitum access to water and food (Lab Rabbit Diet HF #5326, LabDiet). All experiments were done in accordance with the Institutional Animal Care and Use Committee at Emory University.
Brain sectioning
Voles were euthanized by isoflurane overdose followed by decapitation, and the brains were removed and immediately frozen on powdered dry ice. Brains were stored at −80 °C until they were sectioned coronally into 14 μm sections using a cryostat, mounted onto Superfrost Plus slides (Fisherbrand; Pittsburgh, Pennsylvania), and stored at −80 °C.
Chromogenic RNAscope in situ hybridization for mapping Oxtr mRNA
To map Oxtr mRNA for Fig. 1–7 and Supplementary Table 1, an RNAscope 2.5 High Definition Red Assay (Advanced Cell Diagnostics; Newark, California) was used to perform chromogenic in situ hybridization of Oxtr mRNA according to the manufacturer’s instructions on every ten sections from four brains (one High- and one Low-Oxtr male and one High- and one Low-Oxtr female). For regions included in sections used for quantitative analysis of Oxtr genotype effects (OB, NAc, CPu, Tu, VP and SNR) at least 6 brains per genotype per sex were used. Briefly, slide-mounted sections were thawed and fixed in 10% neutral buffered formalin for 15 min prior to 30 min of treatment with RNAscope Protease IV. Sections were then incubated in hybridization probe specific for prairie vole Oxtr (Cat. No. 500721-C3) for two hours at 40 °C. Following six amplification steps, hybridized probes were reacted with chromogenic RNAscope RED substrate for 10 minutes at room temperature and then counterstained with a 50% Hematoxylin staining solution for two minutes at room temperature. Slides were then dried for 15 minutes at 60 °C, dipped in xylene, and coverslipped with EcoMount mounting medium (Biocare Medical; Pacheco, California). Two negative controls, one performed without a probe and one performed with a bacterial DapB probe (Cat. No. 310043), appeared similar and showed no specific labeling. All sections shown in figures were taken from High-Oxtr voles, except where indicated otherwise.
Figure 1. Oxtr mRNA expression throughout prairie vole brain.
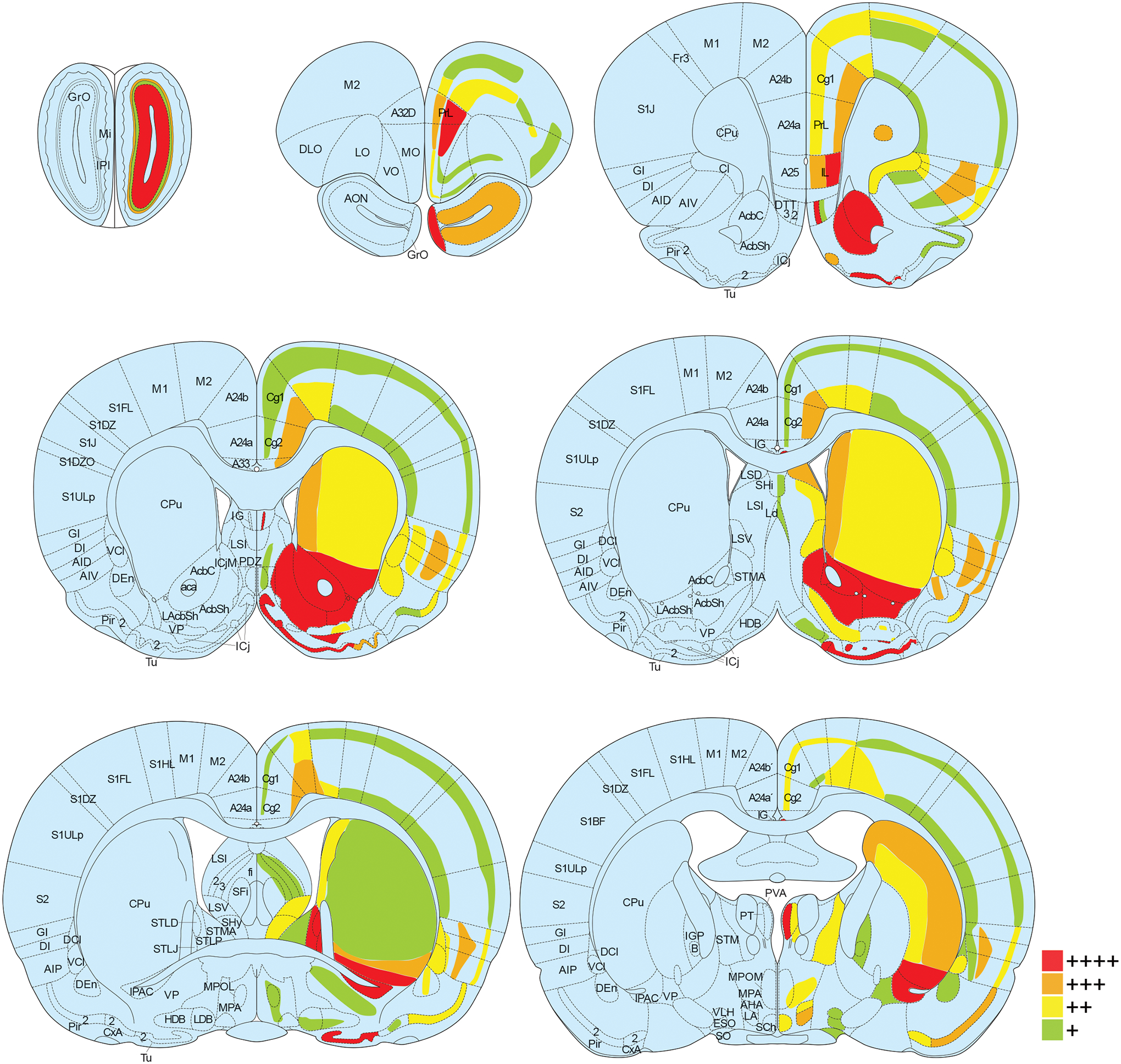
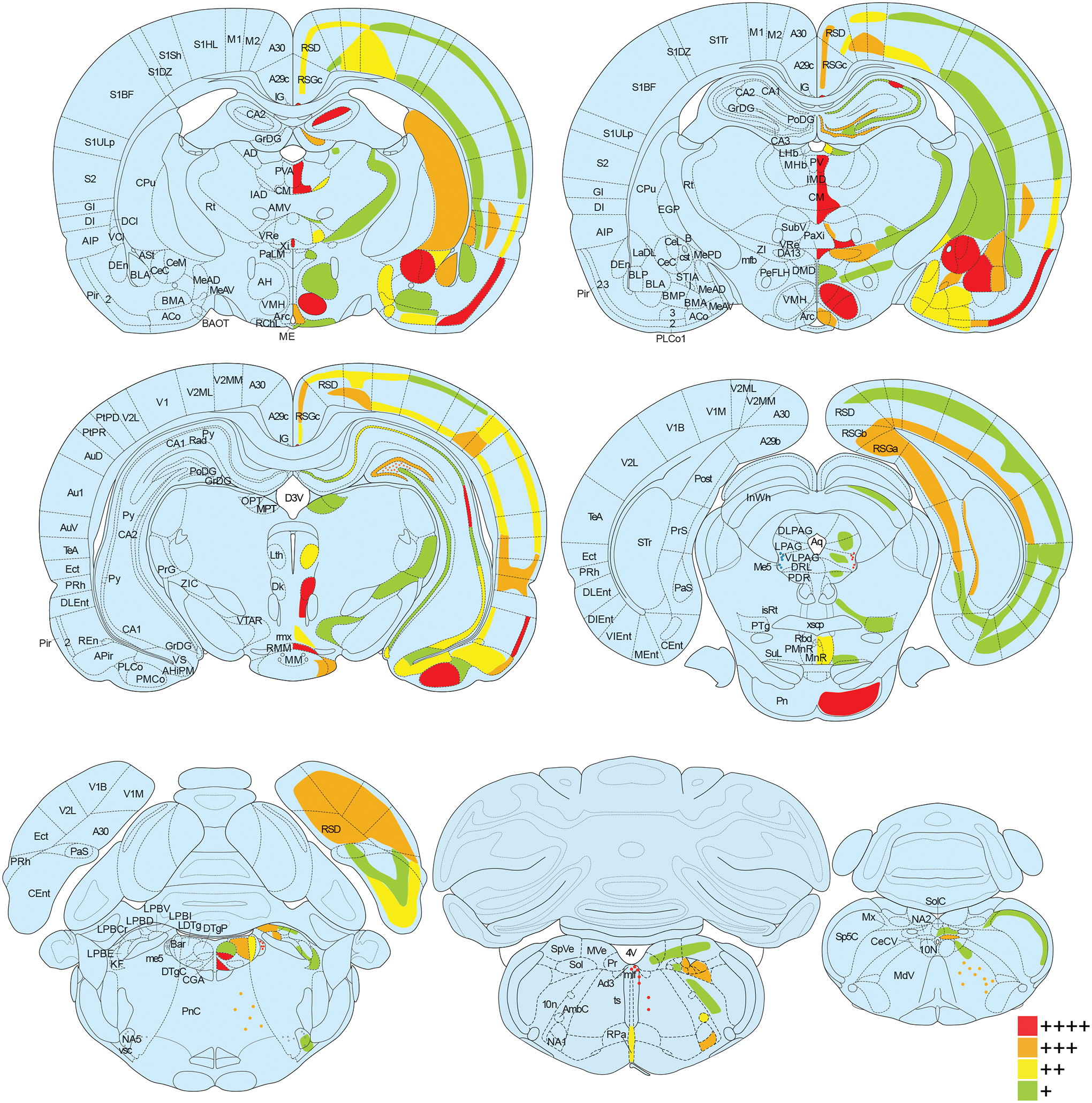
Oxtr mRNA was labeled with in situ hybridization and the percent of cells positive for Oxtr was calculated for each brain region. Brain regions are color coded according to Oxtr expression density (green for 1–25% of cells expressing Oxtr, yellow for 26–50%, orange for 51–75%, and red for 76–100%) in coronal sections from a rat brain atlas at 14 points along the rostral-caudal axis (Paxinos, 2014). For some regions (i.e. reticular formation, molecular layer of the hippocampus) with sparse cells containing exceptionally strong expression, red dots were used. Abbreviations on the left half of the images correspond to new nomenclature used in the seventh edition of “The Rat Brain Atlas in Stereotaxic Coordinates” (Paxinos, 2014), while the abbreviations on the right half of the images correspond to traditional nomenclature used in previous editions. See Supplementary Table 1 for definitions of abbreviations.
Figure 7. Genotype-dependent Oxtr mRNA and OXTR protein mismatch in substantia nigra pars reticulata.
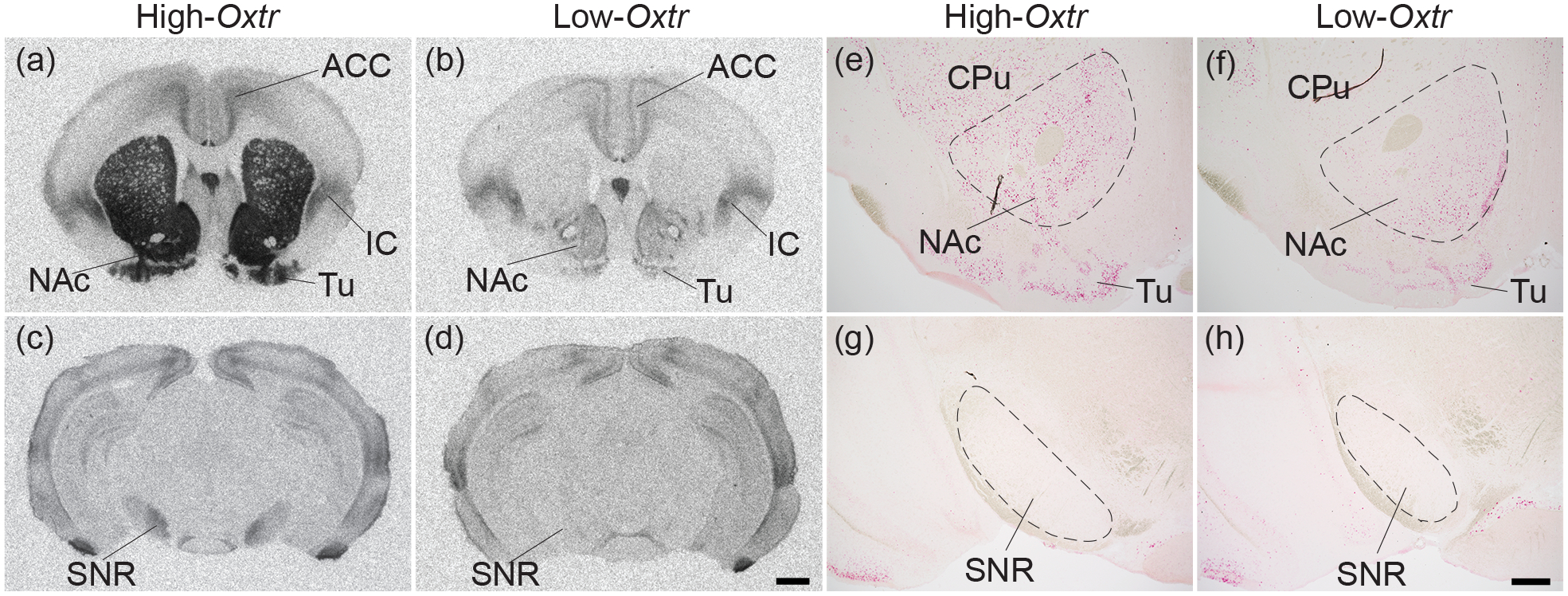
OXTR protein (black; a-d) was labeled with autoradiography and, in adjacent sections, Oxtr mRNA (red; e-h) was labeled with in situ hybridization against a Hematoxylin counterstain (purple) in High-Oxtr voles (a,c,e,g) and Low-Oxtr voles (b,d,f,h). High-Oxtr and Low-Oxtr genotypes predicted protein binding in the nucleus accumbens (NAc) and substantia nigra pars reticulata (SNR), and mRNA expression in NAc. However, no Oxtr mRNA was detected in the SNR of either genotype. CPu, caudate putamen; ACC, anterior cingulate cortex; Tu, olfactory tubercle; IS, insular cortex. Scale bars for (a-d) =1mm shown in (d), for (e-h) =400μm shown in (h).
Oxytocin receptor autoradiography for mRNA-protein comparison
To compare the location of Oxtr mRNA and OXTR protein, adjacent sections from the brains used for chromogenic in situ hybridization described above underwent OXTR autoradiography as previously described (Ross et al., 2009). Briefly, sections were thawed and then fixed for two min with 0.1% paraformaldehyde in PBS at room temperature. They were then incubated in 50 pM 125I-OVTA (2200 Ci/mmol; PerkinElmer; Boston, MA), a selective, radiolabeled OXTR ligand, for one hour. Unbound 125I-OVTA was then removed with Tris-MgCl2 buffer and sections were allowed to dry. Sections were exposed to BioMax MR film (Kodak; Rochester, New York) for five days and a QCAM CCD digital camera (Qimaging; Surrey, Canada) was used to capture 1200 dpi images in 8-bit gray scale. Adobe Photoshop CS6 (San Jose, California) was used to adjust the contrast and brightness of the images. Acetylcholine esterase staining was performed after OXTR binding as described previously to enable accurate identification of brain regions (Lim, Hammock, & Young, 2004). All sections shown in figures were taken from High-Oxtr males, except where noted otherwise.
Fluorescent RNAscope for colocalizing Oxtr mRNA within D1R- and D2R- expressing cells
An RNAscope Fluorescent Multiplex Reagent Kit (Advanced Cell Diagnostics; Newark, California) was used to perform fluorescent in situ hybridization triple labeling of Oxtr, Drd1, and Drd2 mRNA according to the manufacturer’s instructions on sections from 24 brains (Six High- and six Low-Oxtr males, and six High- and six Low-Oxtr females). Briefly, slide-mounted sections were thawed and fixed in 10% neutral buffered formalin for 15 min prior to 30 min of treatment with RNAscope Protease IV. Sections were then incubated in hybridization probes specific for prairie vole Oxtr (Cat. No. 500721-C3), Drd1 (Cat. No. 422581), and Drd2 (Cat. No. 534471-C2) mRNA for two hours at 40 °C. Sequences of prairie vole Drd1 and Drd2 and several other behaviorally relevant genes are available from BAC clones described previously (McGraw, Davis, Thomas, Young, & Thomas, 2012). Following three amplification steps, hybridized probes were labelled with fluorophores Alexa 488 (Drd1), Atto 550 (Drd2), and Atto 647 (Oxtr). Sections were coverslipped with ProLong Gold Antifade Mountant with DAPI (ThermoFisher Scientific; Waltham, Massachusetts) for nuclei labeling. Two negative controls, one performed without a probe and one performed with a bacterial dapB probe (Cat. No. 310043), appeared similar and showed no specific labeling. Autoradiography was performed on a small subset of sections from each brain to confirm that genotype predicted NAc OXTR density as expected. All sections shown in figures were taken from male voles.
Microscopy
A Keyence BZ-X710 microscope was used to take all in situ hybridization images. For chromogenic in situ hybridization, whole slides were imaged at 10x and stitched together using Keyence BZ X Analyzer Software. Fluorescent in situ hybridization images were taken with a 40x lens in four channels (blue, green, red, and infrared) using the Z-stack function. These images were processed using the Keyence BZ X Analyzer Software to merge the four channels and convert the Z-stack into a single two-dimensional image using the Full Focus function.
Image analysis and statistics
For the semi-quantitative brain-wide assessment of chromogenic RNAscope in situ hybridization, brain regions were defined according to the seventh edition of “The Rat Brain Atlas in Stereotaxic Coordinates” (Paxinos, 2014). For each brain region, 80 cells, detected by Hematoxylin staining, were randomly selected and assessed for positive labeling of Oxtr mRNA. The number of cells expressing Oxtr were counted and binned into four groups: 1–20 cells (green in Fig. 1, + in Supplementary Table 1), 21–40 cells (yellow in Fig. 1, ++ in Supplementary Table 1), 41–60 cells (orange in Fig. 1, +++ in Supplementary Table 1), and 61 or more cells (red in Fig. 1, ++++ in Supplementary Table 1). Brain regions in which Oxtr was unevenly distributed were subdivided and Oxtr was quantified separately for each subregion. Only the male High-Oxtr brain was used to generate Supplementary Table 1, though this expression pattern was consistent across the other three brains except for the lower expression in genotype-dependent regions such as NAc and CPu of Low-Oxtr voles.
ImageJ with Fiji (version 2.1.0/1.53c; Schindelin et al., 2012) was used to quantify fluorescent RNAscope in situ hybridization of Oxtr mRNA. Sections were selected from the anterior portion of the NAc immediately posterior to the formation of the genu of the corpus collosum. The area selected for analysis was approximately midway between the anterior commissure and the ventral tip of the lateral ventricle. In each of 24 voles, 20 Drd1-expressing and 20 Drd2-expressing cells (D1R and D2R cells, respectively) were randomly selected for analysis in each hemisphere, yielding a total of 40 D1R and 40 D2R cells from each animal. Each of the selected cells was outlined and the mean gray value for the infrared (Oxtr) channel measured to quantify the level of Oxtr mRNA. These values were averaged for D1R cells and D2R cells to yield one value per cell type in each animal. The statistical software Prism (version 9.2.0; GraphPad; San Diego, California) was used to conduct a three-way mixed model ANOVA with cell type (D1R vs. D2R) as a repeated-measures, within-subjects factor and with sex (male vs. female) and genotype (High- vs. Low-Oxtr) as between-subjects factors. A post hoc Šidák’s multiple comparisons test was used to make pairwise comparisons.
Results:
Brain-wide mapping of Oxtr mRNA
To investigate expression of Oxtr mRNA throughout the prairie vole brain, we performed chromogenic RNAscope in situ hybridization on sections extending from the olfactory bulb to the brainstem. Labeling reflected specific binding of the Oxtr mRNA probe as a negative control using an mRNA probe for the bacterial gene dapB produced no signal. In total, we identified Oxtr mRNA in over 250 distinct brain regions, which were assessed in a semi-quantitative manner and catalogued in Supplementary Table 1. Semi-quantitative schematic illustrations of Oxtr expression at 14 points along the rostral-caudal axis are provided in Figure 1, where colors reflect the percentage of cells expressing Oxtr. These illustrations were made by overlaying representative images of mRNA labeling onto schematics from a rat brain atlas (Paxinos, 2014). Diffuse expression of Oxtr can be seen in numerous regions throughout the brain, with robust expression concentrated in specific regions.
Regions involved in social salience and memory
Oxtr mRNA expression was particularly pronounced in areas involved in social recognition, reward, valence, and memory, processes thought to be important for pair bonding (Figure 2) (Walum & Young, 2018). Olfaction is a highly social sensory modality for rodents, and both the olfactory bulb and anterior olfactory area expressed Oxtr (Figure 2a,b). Oxtr mRNA was detected throughout the amygdala, which processes olfactory information and modulates valence, and expression was particularly dense in the central and basolateral regions (Figure 2e) and the posteromedial cortical amygdala (Figure 1). Both the core and shell of the NAc, a critical reward center, contained high levels of Oxtr mRNA (Figure 2d), as did all regions of the PFC including prelimbic and infralimbic cortex, although strong expression was more prominent in deeper cortical layers, particularly 5 and 6, in the prelimbic cortex (Figure 1,2c). Certain regions of the hippocampal formation, including CA2, ventral CA1, and the dentate gyrus, contained high levels of Oxtr mRNA, though it was primarily restricted to the stratum pyramidale of CA fields and the granule cell and polymorph layers of the dentate gyrus (Figure 6c,d, 4d, 2f).
Figure 2. Oxtr expression in regions involved in social salience and memory.
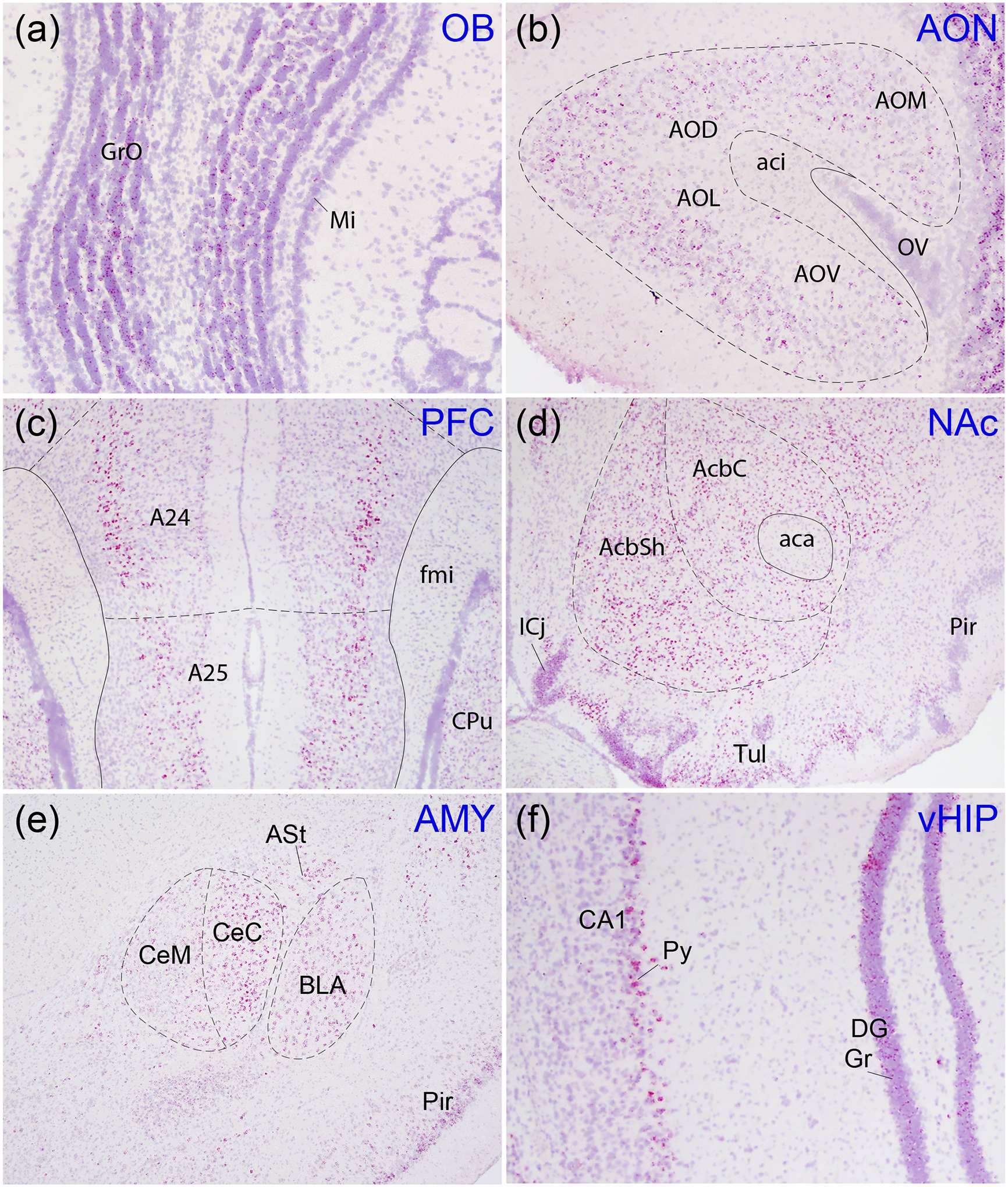
Oxtr mRNA (red) was labeled with in situ hybridization against a Hematoxylin counterstain (purple). The olfactory bulb (a), anterior olfactory area (AO; AOV, ventral; AOL, lateral; AOD, dorsal; AOM, medial; b), prefrontal cortex (c), nucleus accumbens (AcbSh, shell; AcbC, core; d), amygdala (e), and ventral hippocampus (f) all showed strong labeling. GrO, granule cell layer of olfactory bulb; Mi, mitral cell layer of olfactory bulb; aci, anterior commissure, intrabulbar; OV, olfactory ventricle; A24, cingulate cortex previously known as prelimbic cortex; A25, cingulate cortex previously known as infralimbic cortex; fmi, forceps minor of the corpus callosum; CPu, caudate putamen; ICj, island of Calleja; Tul, olfactory tubercle; aca, anterior commissure, anterior part; CeC, central amygdaloid nucleus, capsular; CeM, central amygdaloid nucleus, medial; BLA, basolateral amygdaloid nucleus, anterior; ASt, amygdalostriatal transition area; Pir, piriform cortex; Py, stratum pyramidale; DG, dentate gyrus; Gr, granule cell layer of the dentate gyrus.
Figure 6. Oxtr mRNA and OXTR protein mismatch in hippocampus.
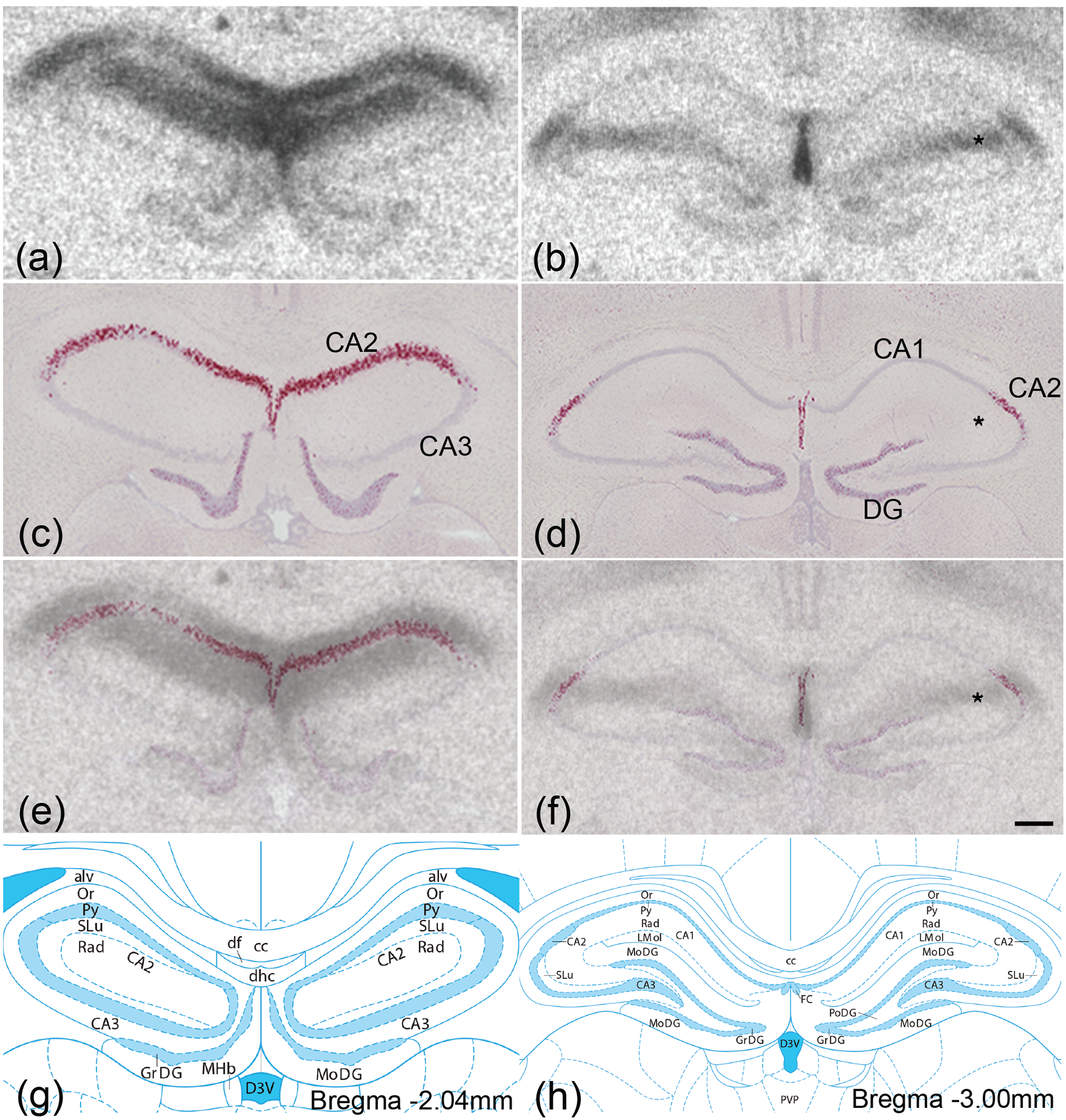
OXTR protein (black; a,b) was labeled with autoradiography and, in adjacent sections, Oxtr mRNA (red; c,d) was labeled with in situ hybridization against a Hematoxylin counterstain (purple). Images from adjacent autoradiography and in situ hybridization sections were merged (e,f) to compare protein and mRNA localization. Oxtr mRNA was restricted to stratum pyramidale, the principal cell layer containing the somas of pyramidal cells, while robust OXTR protein binding was present superficial and basal to stratum pyramidale, most likely in the stratum radiatum, stratum lacunosum-moleculare, and stratum oriens. Illustrations from the Rat brain Atlas indicate that Oxtr mRNA was enriched in the CA2 in the Ammon’s horn (g,h). DG, dentate gyrus; * denotes OXTR protein in the absence of Oxtr mRNA. Scale bar =300μm shown in (f).
Figure 4. Oxtr expression in neuromodulatory areas.
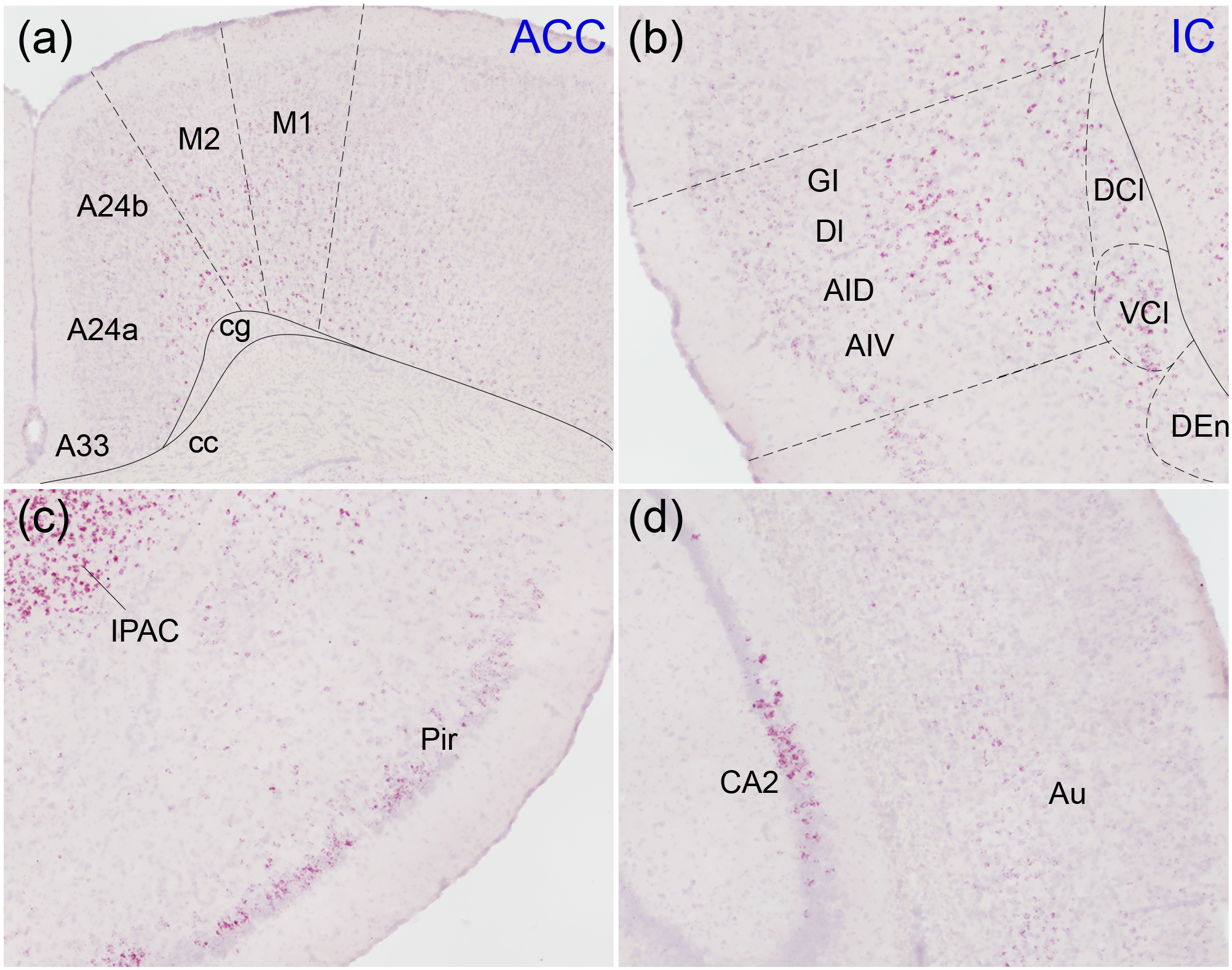
Oxtr mRNA (red) was labeled with in situ hybridization against a Hematoxylin counterstain (purple). The anterior cingulate cortex (A24a, A24b, A33; a), insular cortex (b), piriform cortex (Pir; c), and secondary auditory cortex (Au; d) all contained Oxtr primarily restricted to cortical layers 2, 5, and 6. M1, primary motor cortex; M2, secondary motor cortex; cg, cingulum; cc, corpus callosum; GI, granular insular cortex; DI, dysgranular insular cortex; AID, agranular insular cortex, dorsal; AIV, agranular insular cortex, ventral; DCl, dorsal claustrum; VCl, ventral claustrum; DEn, dorsal endopiriform nucleus; IPAC, interstitial nucleus of the posterior limb of the anterior commissure.
Hypothalamic, thalamic, and septal regions
Oxtr mRNA-producing cells were identified in certain hypothalamic areas and related structures involved in social and reproductive behavior including the VMH (Figure 3f), lateral septum (Figure 3a), medial preoptic area (Figure 3c), paraventricular thalamus (Figure 3d), paraventricular nucleus of the hypothalamus (Figure 3e), and BNST, with more intense signal in the lateral division (Figure 3b). The BNST, VMH, and paraventricular thalamic nucleus displayed exceptionally strong expression, while the medial preoptic area and lateral septum contained more moderate levels of Oxtr mRNA. The paraventricular nucleus of the hypothalamus and supraoptic nucleus, two primary site of oxytocin synthesis, contained only sparse labeling (Supplemental table 1).
Figure 3. Oxtr expression in hypothalamic regions.
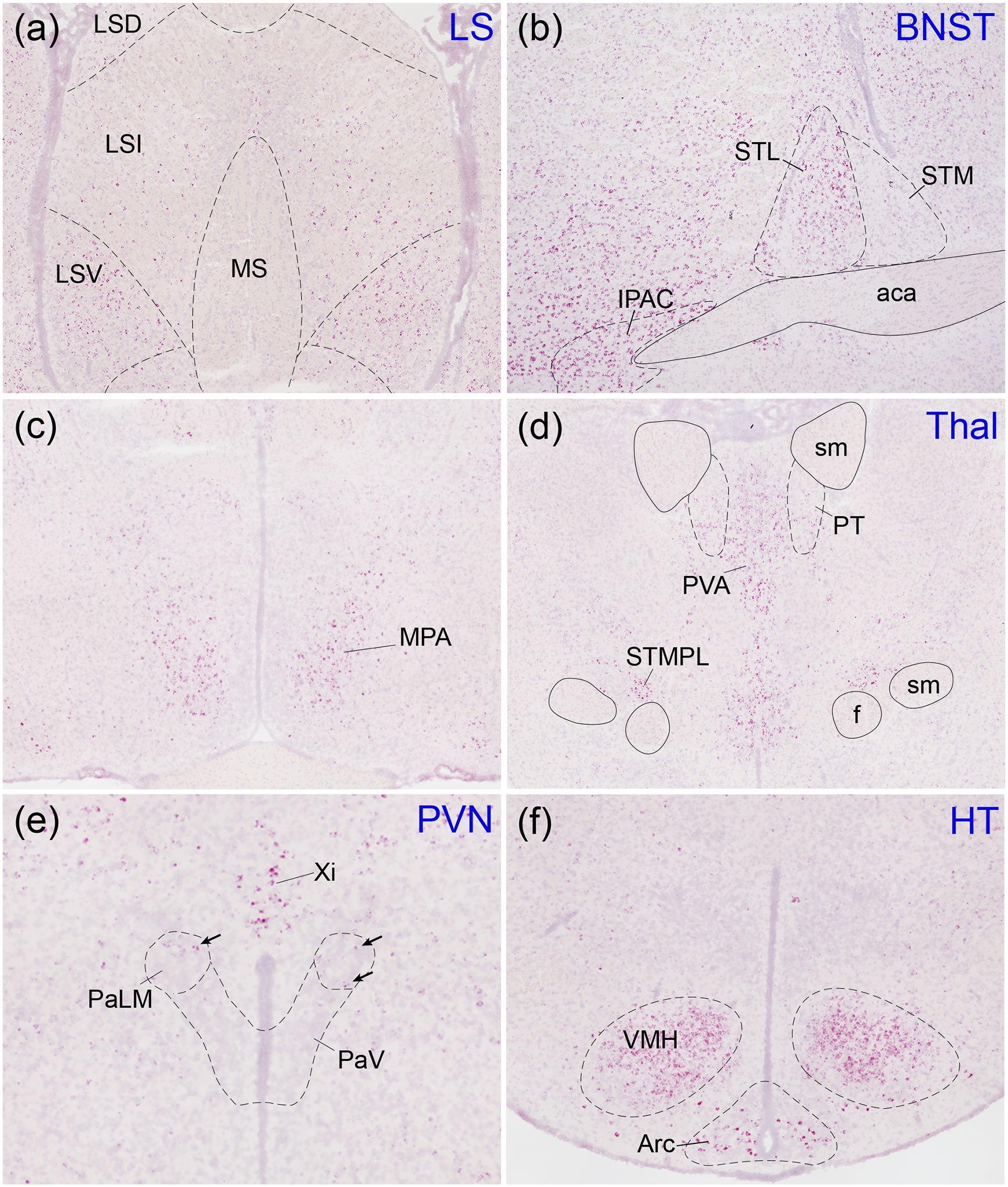
Oxtr mRNA (red) was labeled with in situ hybridization against a Hematoxylin counterstain (purple). The lateral septum (a) contained moderate expression concentrated mostly in the ventral (LSV) portion. Oxtr was robustly expressed in the lateral division of the bed nucleus of the stria terminalis (STL) and the interstitial nucleus of the posterior limb of the anterior commissure (IPAC; b) as well as the ventromedial nucleus of the hypothalamus (VMH; f). The medial preoptic area (MPA; c), thalamus (d), and arcuate nucleus of the hypothalamus (Arc; f) displayed moderate expression while the paraventricular nucleus of the hypothalamus (PaLM, lateral magnocellular; PaV, ventral part; e; arrows indicate Oxtr-positive cells) displayed sparse expression. LSI, lateral septal nucleus, intermediate; LSD, lateral septal nucleus, dorsal; MS, medial septal nucleus; STM, bed nucleus of the stria terminalis, medial division; aca, anterior commissure, anterior part; PT, paratenial thalamic nucleus; PVA, paraventricular thalamic nucleus, anterior; sm, stria medullaris; f, fornix; STMPL, bed nucleus of the stria terminalis, medial, posterolateral; Xi, xiphoid thalamic nucleus.
Cortical regions
Oxtr mRNA expression in cerebral cortex was relatively diffuse, with certain regions containing moderate-to-high levels of expression (Figure 1). These signals were primarily located in cortical layers 2, 5, and 6 (Figure 4). In piriform cortex, expression was mostly restricted to posterior piriform cortex, with the anterior portion displaying only sparse labeling (Figure 1, 4c). In contrast, Oxtr mRNA coursed through both the anterior and posterior insular cortex (Figure 4b). Both the anterior cingulate cortex (Figure 4a) and auditory cortex (Figure 4d) showed relatively modest levels of Oxtr mRNA labeling overall, though expression was mostly restricted to, and relatively dense in, layers 5 and 6. The interstitial nucleus of the posterior limb of the anterior commissure, a part of the extended amygdala receiving dense innervation from tyrosine hydroxylase-containing fibers, can be seen in these images and contained very high levels of Oxtr mRNA.
Neuromodulatory areas
Certain areas in the hindbrain displayed robust Oxtr expression, including the hypothalamic retromamillary nucleus (Figure 5a), posterior hypothalamic nucleus (Figure 5b), and the alpha part of the central gray (Figure 5d). The ventral tegmental area (Figure 5a), periaqueductal gray area (Figure 5c), dorsal raphe nucleus (Figure 5c), the laterodorsal tegmental nucleus (Figure 5d), and the nucleus basalis of Meynert (not shown) contained relatively sparse Oxtr expression. Barrington’s nucleus and the locus coeruleus displayed strong Oxtr expression, though no signal was observed in the caudal portion of the locus coeruleus (Figure 5d). The vagus nerve nucleus (Figure 1) also displayed strong expression.
Figure 5. Oxtr expression in other regions of interest.
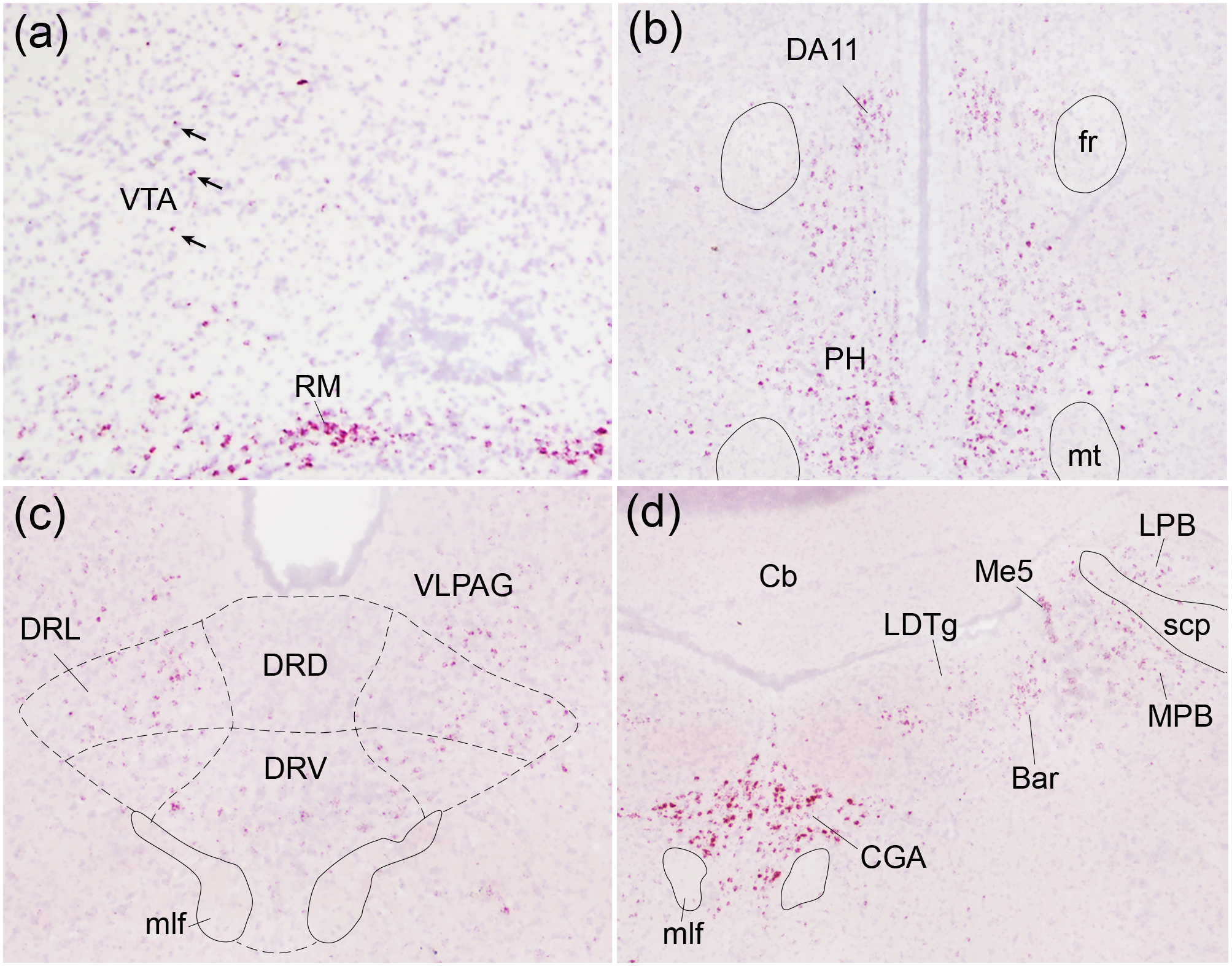
Oxtr mRNA (red) was labeled with in situ hybridization against a Hematoxylin counterstain (purple). The ventral tegmental area (VTA; a) and dorsal raphe nucleus (DR; c) displayed spare Oxtr labeling while the retromammillary nucleus (RM; a), posterior hypothalamic nucleus (PH; b), and central gray, alpha part (CGA; d) displayed robust labeling. Arrows indicate Oxtr-positive cells within VTA; DA11, DA11 dopamine cells; fr, fasciculus retroflexus; mt, mammillothalamic tract; DRD, dorsal raphe nucleus, dorsal part; DRL, dorsal raphe nucleus, lateral part; DRV, dorsal raphe nucleus, ventral part; VLPAG, ventrolateral periaqueductal gray; mlf, medial longitudinal fasciculus; CG, central gray, LDTg, laterodorsal tegmental nucleus; LPB, lateral parabrachial nucleus; MPB, medial parabrachial nucleus; Me5, mesencephalic trigeminal nucleus; Bar, Barrington’s nucleus; Cb, cerebellum; scp, superior cerebellar peduncle.
Mismatch of Oxtr mRNA and OXTR protein
Next, we performed autoradiography on adjacent sections to compare the locations of OXTR protein and Oxtr mRNA. Protein and mRNA expression were closely aligned in most brain regions, but not all. Both OXTR protein (Figure 6a,b) and Oxtr mRNA (Figure 6c,d) were strongly expressed in stratum pyramidale of hippocampal CA2, but even stronger protein labeling in the absence of any mRNA signal was evident immediately superficial and deep to the stratum pyramidale (Figure 6e,f). The resolution of receptor autoradiography is too low to identify with certainty which layers contained this protein labeling, but the two layers immediately superficial to the stratum pyramidale, the stratum radiatum and stratum lacunosum-moleculare, as well as the stratum oriens immediately deep to the stratum pyramidale, are plausible options. This disparity between the locations of OXTR protein and Oxtr mRNA suggests that OXTRs may be synthesized in the cell body in one location and transported to projections in another, enabling synaptic transmission.
We also examined differences in mRNA and protein expression between voles with genetically determined differences in Oxtr expression (Figure 7). High-Oxtr voles expressed far more OXTR protein (Figure 7a) and Oxtr mRNA (Figure 7e) in NAc than Low-Oxtr voles (Figure 7b,f), as expected. Interestingly, a similar genotype effect on OXTR binding was observed in the substantia nigra pars reticulata (SNR; Figure 7c,d), which is heavily innervated by the striatum. However, no Oxtr mRNA-positive cells were detected in the SNR in voles of either genotype (Figure 7g,h), suggesting that OXTR binding in the SNR is on projections from another brain area, most likely the striatum where Oxtr expression is genotype-dependent (King et al., 2016)(Figure 7).
Colocalization of Oxtr with Drd1 and Drd2 mRNA in NAc
Finally, we investigated the colocalization of Oxtr with Drd1 and Drd2 mRNA in the NAc of male and female High- and Low-Oxtr voles (Figure 8a–d and Figure 8e–h, respectively). As in other rodents, Drd1 (Figure 8a,e) and Drd2 (Figure 8b,f) were largely expressed in separate cells. Oxtr mRNA labeling (Figure 8c,g) was detected in approximately 80% of cells. To determine how Oxtr expression varied between cell types (D1R vs. D2R), genotypes (High- vs. Low-Oxtr), and sexes, we measured the intensity of Oxtr labeling in a subset of D1R and D2R cells and analyzed the data using a three-way mixed model ANOVA with cell type as a repeated-measures, within-subjects factor and with sex and genotype as between-subjects factors. There was a main effect of genotype (F (1, 20) = 74.20, p < 0.0001) accounting for 70.81% of variation in Oxtr expression (Figure 9), which aligns closely with previous studies (King et al., 2016)(King et al., 2016). There was no main effect of sex, but there was a main effect of cell type (F (1, 20) = 20.90, p = 0.0002); although slightly more Oxtr signal was detected in D2R cells than in D1R cells, cell type accounted for just 2.18% of variation. Weak genotype-by-sex (F (1, 20) = 4.41, p = 0.0485) and genotype-by-cell type (F (1, 30) = 4.80, p = 0.0405) interactions were also detected, accounting for 4.21% and 0.50% of variation, respectively, in which genotype had slightly larger effects on females than on males and on D2R cells than on D1R cells. A post hoc Šidák’s multiple comparisons test revealed significant differences in Oxtr expression between High- and Low-Oxtr genotypes in each sex-by-cell-type comparison (p < 0.01). There was also a significant difference between D1R and D2R cells in High-Oxtr males (p = 0.0053) with D2R cells expressing slightly more Oxtr, but there was no significant difference between D1R and D2R cells in Low-Oxtr males or in females of either genotype.
Figure 8. Oxtr mRNA colocalized with both Drd1 and Drd2 mRNA in NAc.
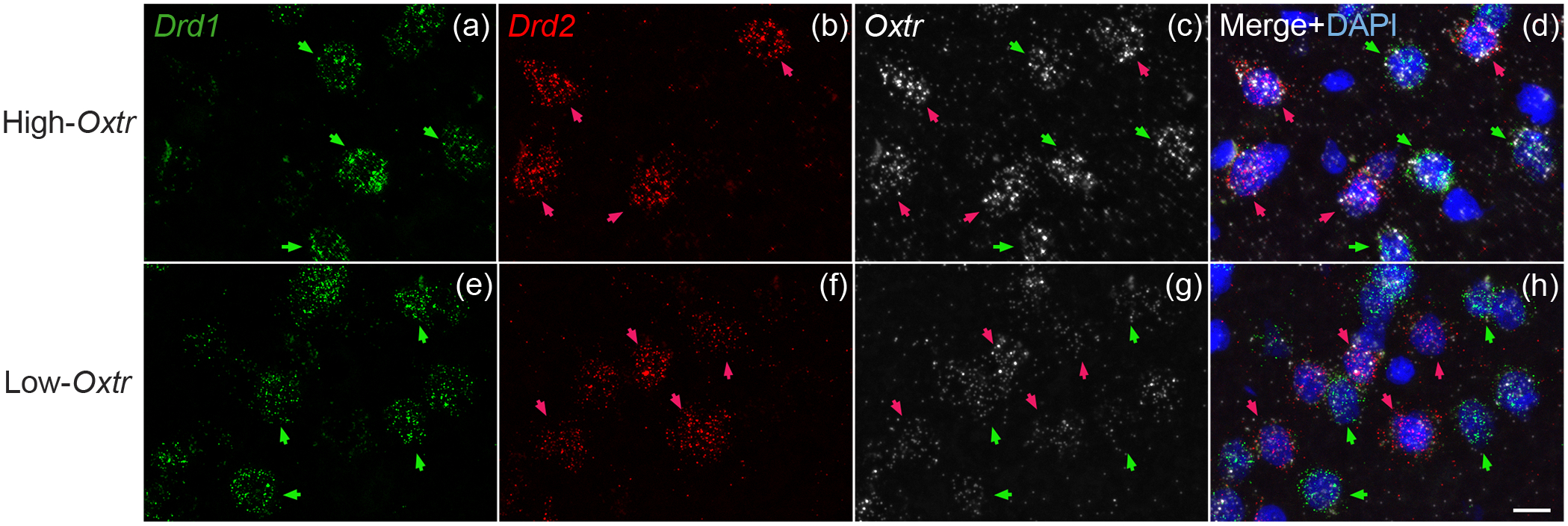
Drd1 (green; a,e), Drd2 (red; b,f), and Oxtr (white; c,g) mRNA were labeled with in situ hybridization in both High- and Low-Oxtr genotypes (a-d, e-h, respectively). Oxtr was expressed in both Drd1- and Drd2-expressing cells (d,h) in both genotypes. Scale bar =10μm shown in (h).
Figure 9. Genotype predicts Oxtr mRNA expression in both D1R and D2R cells in NAc.
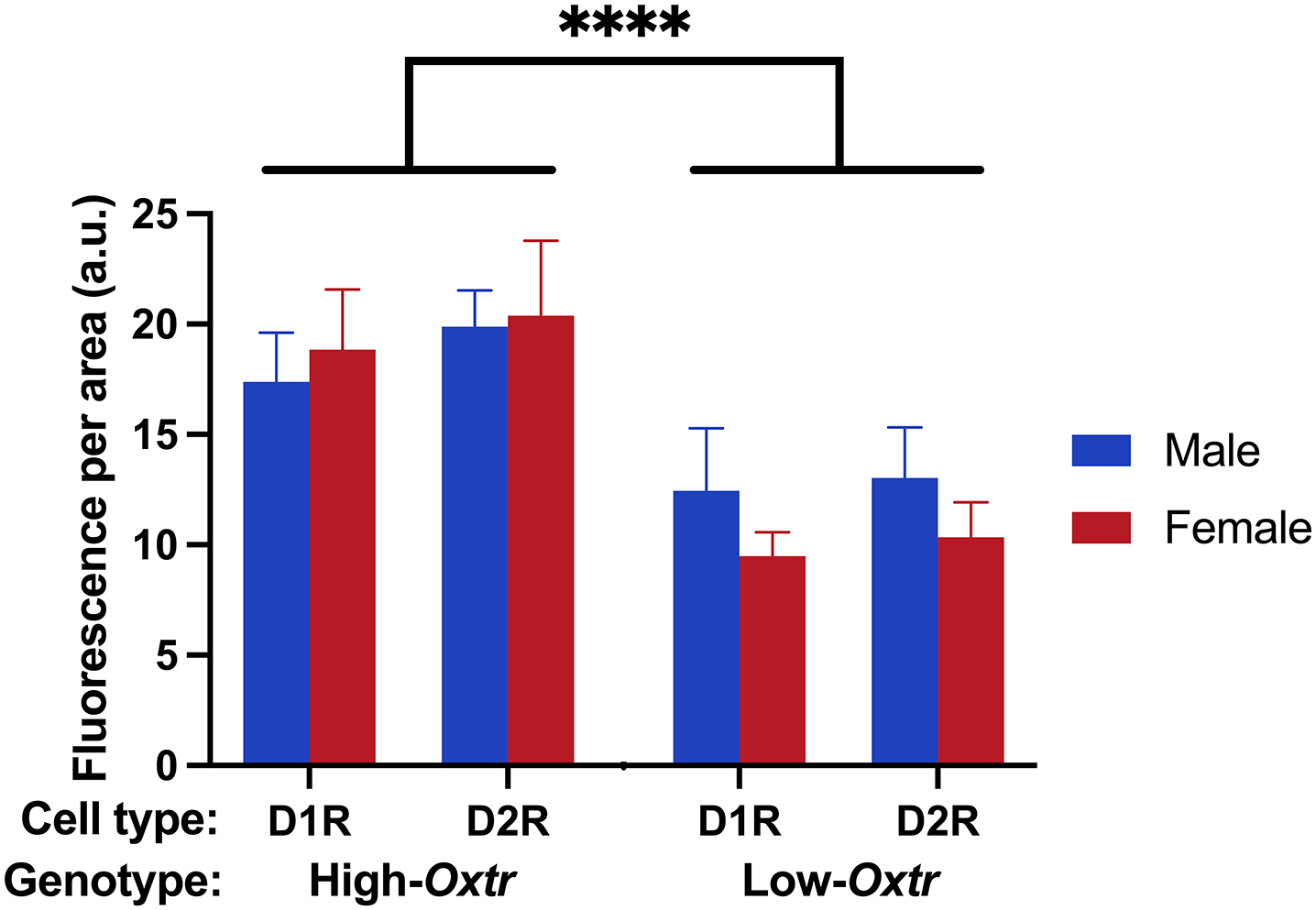
The intensity of Oxtr labeling was quantified in D1R and D2R cells of both genotypes (High-Oxtr and Low-Oxtr) and sexes. A three-way mixed model ANOVA revealed a significant main effect of genotype accounting for 70.81% of variation in Oxtr expression. Post hoc tests revealed significant differences in Oxtr expression between High- and Low-Oxtr genotypes in each sex-by-cell-type comparison. ****, p < 0.0001.
Discussion:
Here we provide the first detailed description of the distribution of Oxtr-producing cells in the prairie vole brain, and the first information on the colocalization of Oxtr mRNA with Drd1 and Drd2. Our comprehensive analysis of Oxtr mRNA revealed widespread expression of Oxtr across the brain, with enrichment in particular brain regions, indicating that OXTR signaling may exert more profound modulation of brain function than expected from earlier receptor binding studies. Oxytocin is released in response to a myriad of social stimuli including social touch (Tang et al., 2020), visual observation of maternal behavior (Carcea et al., 2021), infant vocalizations (Valtcheva et al., 2021), and even interspecies eye contact between humans and dogs (Nagasawa et al., 2015). Once released, oxytocin is capable of modulating the activity of neural networks at three levels: i) by directly depolarizing projection neurons, ii) by affecting the activity of interneurons that regulate local circuit activity, and iii) by modulating the activity of other neuromodulator systems like dopamine, acetylcholine, and serotonin (Froemke & Young, 2021; Putnam, Young, & Gothard, 2018). Our findings further expand the potential influence of oxytocin signaling as the widespread Oxtr expression we observed, coupled with the diverse social triggers of oxytocin release and the many neural processes oxytocin has been shown to affect, suggests that social encounters can profoundly impact brain function through oxytocin-dependent modulation of vast neural networks.
Our observations are consistent with previous studies labeling Oxtr mRNA in prairie voles, although these earlier efforts analyzed fewer anatomical regions and utilized in situ hybridization techniques that were far less sensitive and had lower resolution than those we employed in the present study (Lim, Murphy, & Young, 2004; L. J. Young et al., 1996). Although species differences exist, our findings are generally consistent with protein and mRNA analyses of Oxtr expression patterns in other species. Mitre et al. mapped OXTR protein in mouse brain using immunohistochemistry and showed labeling in many of the same regions we found Oxtr mRNA in prairie voles, including hippocampus, amygdala, BNST, certain hypothalamic nuclei, and certain cortical regions (Mitre et al., 2016). There were also some differences with our findings. Intermediate to high level of OXTR immunolabeling was detected in the mouse PVN, SON and SCN, whereas only small population of cells expressed Oxtr mRNA in these nuclei of the prairie vole. Although Mitre et al.’s labeling in mice is convincing, antibodies targeting OXTRs are notorious for producing non-specific labeling (Yoshida et al., 2009). In humans, Oxtr mRNA expression appears similarly widespread and diffuse with isolated regions of high expression including the striatum, thalamus, hypothalamus, hippocampus, amygdala, and olfactory bulbs (Bethlehem et al., 2017; Quintana et al., 2019). Notably, the high level of OTXR mRNA expression in human CPu and NAc is consistent with prairie vole, but not mouse or rat brain. There is also notable overlap of OXTR mRNA with dopamine receptor mRNA expression (Quintana et al., 2019).
It should be noted that species- and brain region-specific developmental changes in OXTR density have been reported in rodents, which our study cannot address since only adult animals were used. For instance, in the NAc, CPu, BLA and PVA, OXTR binding increases from juvenile to the adulthood in prairie voles, whereas OXTR binding is significantly decreased in several of the same regions as rats mature into adults (Prounis, Thomas, & Ophir, 2018; Smith et al., 2016). Future studies should examine developmental changes in Oxtr mRNA in prairie vole brain, particularly since developmental oxytocin signaling is thought to influence later life social behaviors in adults (Barrett et al., 2015).
Oxytocin receptor mRNA expression in select areas influencing behavior
Although widespread, it is evident that OXTRs are concentrated in certain areas that subserve specific functions. We observed Oxtr mRNA in many subregions of the hippocampus, but expression was particularly strong in dorsal CA2 and ventral CA1, regions studied extensively for their role in social memory. Multi-sensory social information is believed to be processed in dorsal CA2, then transmitted to ventral CA1 for social memory storage (Watarai, Tao, Wang, & Okuyama, 2021). Dorsal CA2 and its projections to ventral CA1 are critical for social memory encoding, consolidation, and recall (Meira et al., 2018; Oliva, Fernández-Ruiz, Leroy, & Siegelbaum, 2020), and OXTRs in CA2 are necessary for discrimination of social stimuli and formation of social recognition memory (Lin et al., 2018; Raam, McAvoy, Besnard, Veenema, & Sahay, 2017). Indeed, dorsal CA2 provides strong excitatory input to ventral CA1 (Meira et al., 2018) and in mice, OXTR binding depolarizes CA2 pyramidal cells, which regulates the short-term plasticity of CA2-to-CA1 synaptic transmission (Tirko et al., 2018). Ventral CA1 is believed to house social engrams, or memory traces for specific individuals (Okuyama, 2018; Okuyama, Kitamura, Roy, Itohara, & Tonegawa, 2016), and oxytocin signaling in CA1 fast-spiking interneurons enhances signal-to-noise ratio in CA1 pyramidal cells (Owen et al., 2013). OXTR binding in hippocampal subfields, particularly dorsal CA2 and ventral CA1, therefore appears to modulate hippocampal network activity and facilitate the flow of social information through the hippocampus, thus enabling social discrimination and memory.
Oxytocin signaling in the medial amygdala is also essential for social recognition in mice (Ferguson, Aldag, Insel, & Young, 2001), and in the central amygdala, oxytocin signaling modulates emotion discrimination (Ferretti et al., 2019). In prairie voles, we observed extensive labeling in the amygdala, with particularly strong expression in the basolateral and central nuclei. Interestingly, while prairie voles have high densities of OXTR binding in the basolateral amygdala, mice do not (Larry J. Young & Zhang, 2021). Strong Oxtr labeling was detected in the anterior olfactory area and granule cell layer of the olfactory bulb as well. OXTR signaling in the anterior olfactory area enhances social recognition in mice by increasing excitatory output to inhibitory granule cell interneurons in the main olfactory bulb (Oettl et al., 2016). This increased inhibition enhances the signal-to-noise ratio of social olfactory information conveyed via mitral and tufted cell projection neurons from the olfactory bulb to olfactory cortex (Oettl et al., 2016). OXTR binding in the prairie vole anterior olfactory area is highly sensitive to estrogen, suggesting that oxytocin-dependent salience of olfactory social cues is enhanced during mating, which facilitates pair bonding (Witt, Carter, & Lnsel, 1991). Very strong Oxtr expression was also observed in the posteromedial cortical amygdaloid nucleus, which is the only cortical target of the direct projection from the accessory olfactory bulb. The accessory olfactory bulb relays vomeronasal information. Thus, oxytocin signaling might be involved in the information processing of the accessory olfactory system and the influence of this system on sociosexual behavior.
We observed very strong Oxtr mRNA expression in the NAc, where oxytocin signaling is necessary for the formation of monogamous pair bonds (L. J. Young et al., 2001), and decreases in oxytocin signaling appear to mediate some of the effects of partner loss, such as passive stress coping (Bosch et al., 2016; Pohl, Young, & Bosch, 2019). There is emerging evidence that the neurophysiological effects of OXTR signaling in the NAc dynamically change with social experience. OXTR signaling in the NAc of virgins decreases the amplitude of spontaneous excitatory postsynaptic currents (EPSCs), while it increases the amplitude of electrically evoked EPSCs in an endocannabinoid-dependent manner in the NAc of pair-bonded voles (Borie et al., 2022). The BNST also showed very strong labeling. Oxytocin signaling in the BNST and NAc mediates social vigilance and the regulation of social approach behaviors (Duque-Wilckens et al., 2020; Williams et al., 2020). We only detected a sparse labeling of Oxtr mRNA in the VTA and PAG. Although there are some reports of OT behavioral effects on these regions (Hung et al., 2017; Song, Borland, Larkin, O’Malley, & Albers, 2016), to our knowledge, there is no report of robust expression of Oxtr in these regions. Consistent with our sparse Oxtr labeling in the adult prairie vole VTA, Oxtr expression in the rat VTA was decreased during development and only weak expression was detected in the adult (Yoshimura, Kimura, Watanabe, & Kiyama, 1996). Oxtr mRNA in the mouse PAG also showed decline from juvenile to adulthood (Daniel E. Olazábal & Alsina-Llanes, 2016). We speculate that either few Oxtr expressing neurons in the VTA are physiologically sufficient to exert behavioral effects in rodents, or mice have greater numbers of Oxtr neurons than prairie voles and OXTR signaling plays a lesser role in prairie vole behavior compared to mice. Notably, the vagus nerve nucleus showed strong labeling as well, as peripheral oxytocin signaling in the vagus nerve can influence feeding and drug self-administration behaviors (Everett, Turner, Costa, Baracz, & Cornish, 2021; Iwasaki et al., 2015), and therefore could potentially mediate some of the behavioral effects of intranasally administered oxytocin in humans.
We detected Oxtr mRNA expression in numerous hypothalamic nuclei. Oxytocin is synthesized and released primarily by the magnocellular and parvocellular neurons of the paraventricular and supraoptic nuclei as well as accessory nuclei (Althammer & Grinevich, 2017; Grinevich & Ludwig, 2021). Via their projections into the posterior pituitary, magnocellular neurons are the primary source of oxytocin released as a hormone into peripheral circulation. The anatomical origins and physiological regulation of oxytocin release in the central nervous system are more complicated and not fully understood, but certainly rely on parvocellular projections, magnocellular axon collaterals, and extra-synaptic oxytocin release (Althammer & Grinevich, 2017; Grinevich & Ludwig, 2021). Intra-hypothalamic oxytocin signaling appears particularly important. For example, somatodendritic release of oxytocin in the paraventricular and supraoptic nuclei is believed to have local autocrine and paracrine effects with important behavioral consequences (Mike Ludwig, Apps, Menzies, Patel, & Rice, 2016; M. Ludwig & Leng, 2006; Modi et al., 2015). Additionally, a recent study found that a small population of parvocellular neurons in the paraventricular nucleus is selectively activated by social touch, and that these cells project to and regulate the activity of magnocellular neurons (Tang et al., 2020). High levels of OXTR immunolabeling was reported in the mouse paraventricular and supraoptic nuclei, whereas Oxtr mRNA expression in these nuclei in prairie voles was notably less robust than in many other hypothalamic nuclei. Despite the small proportion of neurons expressing Oxtr, they may be sufficient to trigger functional effects through collateral activation of interconnected magnocellular neurons. The VMH, which is critical for satiety as well as defensive behavior (Wang et al., 2019) and aggressive behavior in both males (Karigo et al., 2021) and females (Hashikawa et al., 2017), contained particularly high levels of Oxtr mRNA. Notably, expression in the prairie vole VMH was least dense in the ventrolateral portion. In contrast, rats have robust Oxtr expression in the ventrolateral portion of the VMH, and this expression influences sexual behavior and is regulated by estrogen and progesterone (Bale, Dorsa, & Johnston, 1995; Schumacher, Coirini, Pfaff, & McEwen, 1990). The medial preoptic area, which is critical for sexual behavior and both maternal and paternal parenting behaviors, also contained robust levels of Oxtr mRNA (Kohl et al., 2018; Numan, 2020; Pedersen, Caldwell, Walker, Ayers, & Mason, 1994; Yuan et al., 2019).
Many cortical areas displayed sparse labeling of Oxtr mRNA, but some, including the anterior cingulate and insular cortices, showed strong labeling in restricted layers, primarily layers two, five, and six. Oxytocin signaling in the anterior cingulate cortex is critical for consoling and helping behaviors in prairie voles (Burkett et al., 2016; Kitano, Yamagishi, Horie, Nishimori, & Sato, 2020), mandarin voles (L. F. Li et al., 2020), and rats (Yamagishi, Lee, & Sato, 2020). In the insular cortex, OXTR activity is necessary for rats to demonstrate appropriate social affective preference; that is, to approach stressed juveniles and avoid stressed adults (M. M. Rogers-Carter et al., 2018). The anterior cingulate and insular cortices share reciprocal connectivity, suggesting a mechanism by which OXTRs in these regions could coordinate activity between two distinct cortical areas and thus regulate emotion detection and behavioral responses. Notably, in humans, the dorsal anterior cingulate cortex and anterior insular cortex constitute the key nodes of a salience network, which is critical for the detection of behaviorally relevant stimuli and commonly disrupted in neuropsychiatric conditions (Seeley et al., 2007; Uddin, 2015). Moreover, insular cortex has been implicated as a central node linking social affective cues to social decision-making network (SDMN) which consists of brain regions such as MeA, BNST, MPOA, LS, HIP, NAc and BLA (Morgan M. Rogers-Carter & Christianson, 2019). Oxtr was consistently expressed in the insular cortex and all the nodes of the SDMN.
Mismatches in the localization of Oxtr mRNA and OXTR protein
We compared Oxtr mRNA expression to OXTR protein distribution by applying in situ hybridization and receptor autoradiography to adjacent sections. To our knowledge, this is the first brain-wide comparison of OXTR protein (i.e., binding) and Oxtr mRNA distribution with cellular resolution in any species. Throughout most of the brain, protein and mRNA expression overlapped. There were, however, two notable instances in which we observed OXTR binding (i.e. protein) in the absence of mRNA, which provide compelling evidence that Oxtr mRNA can be located in cell somas in one region while the cells’ OXTR proteins reside in projections elsewhere in the brain. This has important implications for the application of targeted OXTR knockdown techniques, such as viral vector-mediated CRISPR, as genome editing in one area could eliminate OXTR signaling on distal projections (Boender & Young, 2020).
The first instance of misaligned protein and mRNA localization was in the hippocampus. The CA2 region displayed strong expression of both Oxtr mRNA and OXTR protein. The mRNA was restricted to the stratum pyramidale while protein labeling was evident both superficial and basal to the stratum pyramidale. It is important to note that OXTR binding patterns have been shown to vary widely between rodent species (Raggenbass, Tribollet, Dubois-Dauphin, & Dreifuss, 1989; Larry J. Young & Zhang, 2021). Nevertheless, in mice, CA2 receives afferent projections from the paraventricular nucleus of the hypothalamus, and CA2 pyramidal cells express robust levels of OXTRs, which maintain the long-term potentiation of synapses with entorhinal cortex projections (Cui, Gerfen, & Young, 2013; Lin et al., 2018; Yoshida et al., 2009). It is therefore plausible that the OXTR protein labeling we observed superficial and basal to the stratum pyramidale is located on the dendritic arbors of pyramidal neurons. However, other neurons could also be responsible for this protein signal; numerous types of interneurons throughout the hippocampus express OXTRs, some interneurons with somas in stratum pyramidale send projections into strata radiatum and lacunosum-moleculare, and the retromamillary nucleus, which had very high expression, has reciprocal connections with CA2 (Botcher, Falck, Thomson, & Mercer, 2014; W. S. Young & Song, 2020). Although the origin of the OXTR protein signal we observed cannot be determined definitively from our images, it is clear that the proteins reside and act in receptive fields a considerable distance from the somas in which the mRNA resides. Interestingly, hippocampal OXTR binding has been reported to vary with sex and reproductive physiology, and speculated to be involved in sex differences in spatial memory and fidelity in voles (Alexander G. Ophir, 2017; Zheng, Larsson, Phelps, & Ophir, 2013), therefore future studies using Cre-dependent retrograde viral tracers in Oxtr-Cre prairie voles may provide behaviorally relevant insight into neural circuitry contributing to mating tactics by identifying the sources of OXTR protein in the hippocampus (Kengo Horie et al., 2020).
The second instance of misaligned protein and mRNA localization was in the SNR, but this misalignment was contingent on the animal’s Oxtr genotype. We previously identified a set of SNPs in linkage disequilibrium in the prairie vole Oxtr gene that predicts both the animal’s ability to form a pair bond and the density of OXTR protein expressed in the NAc, CPu, OB, and anterior olfactory area (King et al., 2016). Levels of Oxtr mRNA in the NAc also varied in accordance with the protein (King et al., 2016). Here, we again observed that OXTR protein and Oxtr mRNA levels in the NAc varied with genotype. We also observed that High-Oxtr voles had considerable OXTR binding in the SNR, while Low-Oxtr voles showed no detectable binding there. More interestingly, no Oxtr mRNA signals was detected either in the SNR of Low-Oxtr or High-Oxtr voles. As NAc MSNs send significant projections to the SNR (Matamales et al., 2009), it is likely that the OXTR protein labeling we observed in the SNR is from OXTRs located on the presynaptic terminals of NAc MSNs. This observation supports the idea that OXTRs can serve presynaptic functions far from the anatomical location of the cells expressing the Oxtr gene (Dölen et al., 2013; Mairesse et al., 2015; Mitre et al., 2016; Putnam et al., 2018). It also expands our understanding of how SNPs in the Oxtr gene are capable of influencing oxytocin signaling throughout the brain. A similar genotype-dependent pattern of OXTR binding was also observed in the ventral pallidum, which had a few sparse Oxtr mRNA-positive cells. Like the SNR, the ventral pallidum receives strong direct projections from the NAc, and we suspect that the genotype-dependent OXTR binding we observed in the ventral pallidum of High-Oxtr voles is from OXTR proteins on the terminals of axons from NAc. However, as the ventral pallidum receives projections from both D1R and D2R MSNs while the SNR receives projections from only D1R MSNs (Pardo-Garcia et al., 2019), our observation of genotype-dependent OXTR expression in these brain regions raises the question of whether the High- and Low-Oxtr alleles influence Oxtr expression in both D1R and D2R MSNs or only D1R MSNs. Given that D1R in NAc inhibits, while D2R facilitates pair bond formation (see below), and both MSN types express Oxtr, it would be important to determine whether presynaptic OXTR signaling in the VP and SNR modulates pair bonding and parental care as well, particularly in relation to proposed circuit mechanisms of pair bonding involving the release of VP from NAc inhibition to promote maternal care and pair bonding (Numan & Young, 2016).
Comparing localization of Oxtr mRNA with Drd1 and Drd2 mRNA in NAc
The primary projection neurons of the NAc, GABAergic MSNs, constitute approximately 95% of all neurons in the NAc and can be subdivided into those that express Drd1 and those that express Drd2. Both D1R and D2R MSNs receive afferent input from a wide range of brain regions (Z. Li et al., 2018). Although oversimplified, the canonical notion of D1R and D2R MSNs is that they project to the SNR either directly via the monosynaptic “direct pathway” or indirectly via the multi-synaptic “indirect pathway,” respectively (Calabresi, Picconi, Tozzi, Ghiglieri, & Di Filippo, 2014). In the direct pathway, D1R MSNs project to and inhibit the SNR, which sends inhibitory projections to the thalamus, resulting in disinhibition of thalamocortical projections. More recent experimental evidence has shown that D1R MSNs can project to the ventral tegmental area and ventral pallidum as well (Yang et al., 2018). In the indirect pathway, D2R MSNs project indirectly to the SNR through a multi-synaptic pathway including the globus pallidus externa and subthalamic nucleus. This pathway excites the inhibitory SNR neurons that project to the thalamus, resulting in inhibition of thalamocortical projections.
Activation of D1R versus D2R MSNs are associated with antagonistic behavioral effects including reward (D1R) and punishment (D2R) (Soares-Cunha, Coimbra, Sousa, & Rodrigues, 2016) and pair bond formation (D2R) and maintenance (D1R) (Aragona et al., 2006). In prairie voles, D2R activation facilitates pair bond formation, and either blocking D2Rs or activating D1Rs inhibits pair bond formation (Aragona et al., 2006). However, two weeks after pair bonds are formed, male voles display an upregulation of D1R in the NAc (Aragona et al., 2006). This upregulation of D1R promotes selective aggression toward unfamiliar conspecifics, which serves to help maintain the pair bond (Aragona et al., 2006). Most importantly, pair bond formation requires activation of both D2R and OXTR in the NAc (Liu & Wang, 2003).
Given the influence of Oxtr gene SNPs on pair bond formation and Oxtr expression in the NAc, and the critical interaction of oxytocin and D1R versus D2R signaling in this region, we quantified and compared Oxtr mRNA expression in Drd1- and Drd2-expressing cells in the NAc of males and females homozygous for either the High-Oxtr or Low-Oxtr alleles. We found that 71% of variation in Oxtr expression in NAc was determined by genotype, which is remarkably close to our previous estimate of 74% (King et al., 2016). Although D2R cells expressed slightly more Oxtr than D1R cells, the vast majority of variation in Oxtr expression is determined by SNPs in the Oxtr gene. Of critical importance, these SNPs affect Oxtr expression more or less equally in both D1R and D2R cells.
Conclusion and future directions
Our comprehensive, brain-wide analysis of Oxtr mRNA expression revealed diffuse, widespread distribution throughout the brain as well as robust expression restricted to specific brain regions. These regions of concentrated expression suggest a mechanism by which oxytocin signaling can modulate the activity of neural networks throughout the brain in response to social stimuli. Furthermore, our comparison of Oxtr mRNA and OXTR protein localization in adjacent slices suggests that OXTRs may act in receptive fields after being transported a considerable distance from somas containing Oxtr mRNA, consistent with both pre- and post-synaptic signaling mechanisms. Lastly, we determined that SNPs in the Oxtr gene, which are predictive of an animal’s ability to pair bond, influence Oxtr expression in both D1R and D2R cells in the NAc. Comparative studies are needed to determine how these expression patterns differ from those of other organisms, especially other rodents and organisms with both similar and divergent behavioral repertoires. However, these data should inform future work in voles, particularly experiments attempting region-specific manipulations of Oxtr expression. Indeed, tracing experiments could help identify which of these oxytocin-sensitive brain regions communicate with each other, and site-specific knockdown of Oxtr expression could determine how oxytocin signaling in these regions and networks influences behavior. Additionally, we have identified many brain regions that express Oxtr mRNA in which the function of oxytocin signaling has not been studied. Exploring oxytocin signaling in these under-studied regions could yield new insights into how social stimuli and oxytocin signaling affect the brain and behavior.
Supplementary Material
Supplementary Table 1. Regions expressing Oxtr mRNA in prairie vole brain. Oxtr mRNA was labeled with in situ hybridization and the percent of cells positive for Oxtr was calculated for each brain region expressing Oxtr (+, 1–25% of cells expressing Oxtr; ++, 26–50%; +++, 51–75%; ++++, 76–100%). Brain regions were defined according to the seventh edition of “The Rat Brain Atlas in Stereotaxic Coordinates” (Paxinos, 2014).
Key points:
Oxytocin receptor mRNA is diffusely expressed throughout the brain, with strong expression concentrated in certain areas involved in social behavior.
Oxytocin receptor mRNA expression and protein localization are misaligned in some areas, indicating that the receptor protein may be transported to distal processes.
In the nucleus accumbens, oxytocin receptors are expressed on cells expressing both D1 and D2 dopamine receptor subtypes, and the majority of variation in oxytocin receptor expression between animals is attributable to polymorphisms in the oxytocin receptor gene.
Acknowledgments:
This work was supported by NIH grants R01MH112788 and P50MH100023 to LJY, and P51OD11132 to Emory National Primate Research Center
Footnotes
Conflict of interest: The authors declare no conflicts of interest.
Data availability statement:
Data supporting the brain wide quantification of Oxtr mRNA are available in the supplementary material of this article. The raw data used to generate Figure 9 of this study and any other data are available from the corresponding author upon reasonable request.
References
- Ahern TH, Olsen S, Tudino R, & Beery AK (2021). Natural variation in the oxytocin receptor gene and rearing interact to influence reproductive and nonreproductive social behavior and receptor binding. Psychoneuroendocrinology, 128, 105209. doi: 10.1016/j.psyneuen.2021.105209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althammer F, & Grinevich V (2017). Diversity of oxytocin neurons: beyond magno- and parvocellular cell types? J Neuroendocrinol. doi: 10.1111/jne.12549 [DOI] [PubMed] [Google Scholar]
- Andari E, Hurlemann R, & Young LJ (2018). A Precision Medicine Approach to Oxytocin Trials. Curr Top Behav Neurosci, 35, 559–590. doi: 10.1007/7854_2017_29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andari E, Massa NM, Fargotstein MD, Taylor NB, Halverson DM, Owens AV, … Duncan EJ (2021). Effects of Oxytocin on Emotion Recognition in Schizophrenia: A Randomized Double-Blind Pilot Study. J Clin Psychopharmacol, 41(2), 103–113. doi: 10.1097/jcp.0000000000001367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona BJ, Liu Y, Yu YJ, Curtis JT, Detwiler JM, Insel TR, & Wang Z (2006). Nucleus accumbens dopamine differentially mediates the formation and maintenance of monogamous pair bonds. Nat Neurosci, 9(1), 133–139. doi: 10.1038/nn1613 [DOI] [PubMed] [Google Scholar]
- Bale TL, Dorsa DM, & Johnston CA (1995). Oxytocin receptor mRNA expression in the ventromedial hypothalamus during the estrous cycle. The Journal of neuroscience : the official journal of the Society for Neuroscience, 15(7 Pt 1), 5058–5064. doi: 10.1523/JNEUROSCI.15-07-05058.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett CE, Arambula SE, & Young LJ (2015). The oxytocin system promotes resilience to the effects of neonatal isolation on adult social attachment in female prairie voles. Translational psychiatry, 5(7), e606–e606. doi: 10.1038/tp.2015.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethlehem RAI, Lombardo MV, Lai MC, Auyeung B, Crockford SK, Deakin J, … Baron-Cohen S (2017). Intranasal oxytocin enhances intrinsic corticostriatal functional connectivity in women. Translational psychiatry, 7(4), e1099–e1099. doi: 10.1038/tp.2017.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boender AJ, & Young LJ (2020). Oxytocin, vasopressin and social behavior in the age of genome editing: A comparative perspective. Horm Behav, 124, 104780. doi: 10.1016/j.yhbeh.2020.104780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borie AM, Agezo S, Lunsford P, Boender AJ, Guo JD, Zhu H, … Liu RC (2022). Social experience alters oxytocinergic modulation in the nucleus accumbens of female prairie voles. Curr Biol, 32(5), 1026–1037.e1024. doi: 10.1016/j.cub.2022.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch OJ, Dabrowska J, Modi ME, Johnson ZV, Keebaugh AC, Barrett CE, … Young LJ (2016). Oxytocin in the nucleus accumbens shell reverses CRFR2-evoked passive stress-coping after partner loss in monogamous male prairie voles. Psychoneuroendocrinology, 64, 66–78. doi: 10.1016/j.psyneuen.2015.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botcher NA, Falck JE, Thomson AM, & Mercer A (2014). Distribution of interneurons in the CA2 region of the rat hippocampus. Front Neuroanat, 8, 104. doi: 10.3389/fnana.2014.00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkett JP, Andari E, Johnson ZV, Curry DC, de Waal FB, & Young LJ (2016). Oxytocin-dependent consolation behavior in rodents. Science, 351(6271), 375–378. doi: 10.1126/science.aac4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabresi P, Picconi B, Tozzi A, Ghiglieri V, & Di Filippo M (2014). Direct and indirect pathways of basal ganglia: a critical reappraisal. Nat Neurosci, 17(8), 1022–1030. doi: 10.1038/nn.3743 [DOI] [PubMed] [Google Scholar]
- Campbell P, Ophir AG, & Phelps SM (2009). Central vasopressin and oxytocin receptor distributions in two species of singing mice. J Comp Neurol, 516(4), 321–333. doi: 10.1002/cne.22116 [DOI] [PubMed] [Google Scholar]
- Carcea I, Caraballo NL, Marlin BJ, Ooyama R, Riceberg JS, Mendoza Navarro JM, … Froemke RC (2021). Oxytocin neurons enable social transmission of maternal behaviour. Nature, 596(7873), 553–557. doi: 10.1038/s41586-021-03814-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne F, Diorio J, Sharma S, & Meaney MJ (2001). Naturally occurring variations in maternal behavior in the rat are associated with differences in estrogen-inducible central oxytocin receptors. Proc Natl Acad Sci U S A, 98(22), 12736–12741. doi: 10.1073/pnas.221224598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z, Gerfen CR, & Young WS 3rd. (2013). Hypothalamic and other connections with dorsal CA2 area of the mouse hippocampus. J Comp Neurol, 521(8), 1844–1866. doi: 10.1002/cne.23263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMayo MM, Young LJ, Hickie IB, Song YJC, & Guastella AJ (2019). Circuits for social learning: A unified model and application to Autism Spectrum Disorder. Neurosci Biobehav Rev, 107, 388–398. doi: 10.1016/j.neubiorev.2019.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölen G, Darvishzadeh A, Huang KW, & Malenka RC (2013). Social reward requires coordinated activity of nucleus accumbens oxytocin and serotonin. Nature, 501(7466), 179–184. doi: 10.1038/nature12518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson ZR, & Young LJ (2008). Oxytocin, vasopressin, and the neurogenetics of sociality. Science, 322(5903), 900–904. doi: 10.1126/science.1158668 [DOI] [PubMed] [Google Scholar]
- Duque-Wilckens N, Torres LY, Yokoyama S, Minie VA, Tran AM, Petkova SP, … Trainor BC (2020). Extrahypothalamic oxytocin neurons drive stress-induced social vigilance and avoidance. Proc Natl Acad Sci U S A, 117(42), 26406–26413. doi: 10.1073/pnas.2011890117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett NA, Turner AJ, Costa PA, Baracz SJ, & Cornish JL (2021). The vagus nerve mediates the suppressing effects of peripherally administered oxytocin on methamphetamine self-administration and seeking in rats. Neuropsychopharmacology, 46(2), 297–304. doi: 10.1038/s41386-020-0719-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson JN, Aldag JM, Insel TR, & Young LJ (2001). Oxytocin in the medial amygdala is essential for social recognition in the mouse. J Neurosci, 21(20), 8278–8285. doi: 10.1523/jneurosci.21-20-08278.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti V, Maltese F, Contarini G, Nigro M, Bonavia A, Huang H, … Papaleo F (2019). Oxytocin Signaling in the Central Amygdala Modulates Emotion Discrimination in Mice. Curr Biol, 29(12), 1938–1953.e1936. doi: 10.1016/j.cub.2019.04.070 [DOI] [PubMed] [Google Scholar]
- Ford CL, & Young LJ (2021). Refining oxytocin therapy for autism: context is key. Nature Reviews Neurology, 18(2), 67–68. doi: 10.1038/s41582-021-00602-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford CL, & Young LJ (2021). Translational opportunities for circuit-based social neuroscience: advancing 21st century psychiatry. Curr Opin Neurobiol, 68, 1–8. doi: 10.1016/j.conb.2020.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis DD, Champagne FC, & Meaney MJ (2000). Variations in maternal behaviour are associated with differences in oxytocin receptor levels in the rat. J Neuroendocrinol, 12(12), 1145–1148. doi: 10.1046/j.1365-2826.2000.00599.x [DOI] [PubMed] [Google Scholar]
- Freeman AR, Aulino EA, Caldwell HK, & Ophir AG (2020). Comparison of the distribution of oxytocin and vasopressin 1a receptors in rodents reveals conserved and derived patterns of nonapeptide evolution. J Neuroendocrinol, 32(4), e12828. doi: 10.1111/jne.12828 [DOI] [PubMed] [Google Scholar]
- Freeman SM, Inoue K, Smith AL, Goodman MM, & Young LJ (2014). The neuroanatomical distribution of oxytocin receptor binding and mRNA in the male rhesus macaque (Macaca mulatta). Psychoneuroendocrinology, 45, 128–141. doi: 10.1016/j.psyneuen.2014.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman SM, & Young LJ (2016). Comparative Perspectives on Oxytocin and Vasopressin Receptor Research in Rodents and Primates: Translational Implications. J Neuroendocrinol, 28(4). doi: 10.1111/jne.12382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund-Mercier MJ, Stoeckel ME, Palacios JM, Pazos A, Reichhart JM, Porte A, & Richard P (1987). Pharmacological characteristics and anatomical distribution of [3H]oxytocin-binding sites in the Wistar rat brain studied by autoradiography. Neuroscience, 20(2), 599–614. doi: 10.1016/0306-4522(87)90113-8 [DOI] [PubMed] [Google Scholar]
- Froemke RC, & Young LJ (2021). Oxytocin, Neural Plasticity, and Social Behavior. Annu Rev Neurosci, 44, 359–381. doi: 10.1146/annurev-neuro-102320-102847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson JL, Wilson LC, & Schrock SE (2012). To flock or fight: neurochemical signatures of divergent life histories in sparrows. Proc Natl Acad Sci U S A, 109 Suppl 1(Suppl 1), 10685–10692. doi: 10.1073/pnas.1203394109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe NM, Sharma A, Freeman SM, Palumbo MC, Patisaul HB, Bales KL, & Drea CM (2021). Neural correlates of mating system diversity: oxytocin and vasopressin receptor distributions in monogamous and non-monogamous Eulemur. Sci Rep, 11(1), 3746. doi: 10.1038/s41598-021-83342-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinevich V, & Ludwig M (2021). The multiple faces of the oxytocin and vasopressin systems in the brain. Journal of Neuroendocrinology, 33(11). doi: 10.1111/jne.13004 [DOI] [PubMed] [Google Scholar]
- Hashikawa K, Hashikawa Y, Tremblay R, Zhang J, Feng JE, Sabol A, … Lin D (2017). Esr1(+) cells in the ventromedial hypothalamus control female aggression. Nat Neurosci, 20(11), 1580–1590. doi: 10.1038/nn.4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie K, Inoue K, Nishimori K, & Young LJ (2020). Investigation of Oxtr-expressing Neurons Projecting to Nucleus Accumbens using Oxtr-ires-Cre Knock-in prairie Voles (Microtus ochrogaster). Neuroscience, 448, 312–324. doi: 10.1016/j.neuroscience.2020.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie K, Inoue K, Suzuki S, Adachi S, Yada S, Hirayama T, … Nishimori K (2019). Oxytocin receptor knockout prairie voles generated by CRISPR/Cas9 editing show reduced preference for social novelty and exaggerated repetitive behaviors. Horm Behav, 111, 60–69. doi: 10.1016/j.yhbeh.2018.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LW, Neuner S, Polepalli JS, Beier KT, Wright M, Walsh JJ, … Malenka RC (2017). Gating of social reward by oxytocin in the ventral tegmental area. Science, 357(6358), 1406–1411. doi: 10.1126/science.aan4994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insel TR, Young L, Witt DM, & Crews D (1993). Gonadal steroids have paradoxical effects on brain oxytocin receptors. J Neuroendocrinol, 5(6), 619–628. doi: 10.1111/j.1365-2826.1993.tb00531.x [DOI] [PubMed] [Google Scholar]
- Iwasaki Y, Maejima Y, Suyama S, Yoshida M, Arai T, Katsurada K, … Yada T (2015). Peripheral oxytocin activates vagal afferent neurons to suppress feeding in normal and leptin-resistant mice: a route for ameliorating hyperphagia and obesity. Am J Physiol Regul Integr Comp Physiol, 308(5), R360–369. doi: 10.1152/ajpregu.00344.2014 [DOI] [PubMed] [Google Scholar]
- Johnson ZV, Walum H, Jamal YA, Xiao Y, Keebaugh AC, Inoue K, & Young LJ (2016). Central oxytocin receptors mediate mating-induced partner preferences and enhance correlated activation across forebrain nuclei in male prairie voles. Horm Behav, 79, 8–17. doi: 10.1016/j.yhbeh.2015.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ZV, & Young LJ (2017). Oxytocin and vasopressin neural networks: Implications for social behavioral diversity and translational neuroscience. Neurosci Biobehav Rev, 76(Pt A), 87–98. doi: 10.1016/j.neubiorev.2017.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurek B, & Neumann ID (2018). The Oxytocin Receptor: From Intracellular Signaling to Behavior. Physiol Rev, 98(3), 1805–1908. doi: 10.1152/physrev.00031.2017 [DOI] [PubMed] [Google Scholar]
- Karigo T, Kennedy A, Yang B, Liu M, Tai D, Wahle IA, & Anderson DJ (2021). Distinct hypothalamic control of same- and opposite-sex mounting behaviour in mice. Nature, 589(7841), 258–263. doi: 10.1038/s41586-020-2995-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keebaugh AC, Barrett CE, Laprairie JL, Jenkins JJ, & Young LJ (2015). RNAi knockdown of oxytocin receptor in the nucleus accumbens inhibits social attachment and parental care in monogamous female prairie voles. Soc Neurosci, 10(5), 561–570. doi: 10.1080/17470919.2015.1040893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King LB, Walum H, Inoue K, Eyrich NW, & Young LJ (2016). Variation in the Oxytocin Receptor Gene Predicts Brain Region-Specific Expression and Social Attachment. Biol Psychiatry, 80(2), 160–169. doi: 10.1016/j.biopsych.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano K, Yamagishi A, Horie K, Nishimori K, & Sato N (2020). Helping Behavior in Prairie Voles: A Model of Empathy and the Importance of Oxytocin. bioRxiv, 2020.2010.2020.347872. doi: 10.1101/2020.10.20.347872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl J, Babayan BM, Rubinstein ND, Autry AE, Marin-Rodriguez B, Kapoor V, … Dulac C (2018). Functional circuit architecture underlying parental behaviour. Nature, 556(7701), 326–331. doi: 10.1038/s41586-018-0027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landin J, Hovey D, Xu B, Lagman D, Zettergren A, Larhammar D, … Westberg L (2020). Oxytocin Receptors Regulate Social Preference in Zebrafish. Sci Rep, 10(1), 5435. doi: 10.1038/s41598-020-61073-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LF, Yuan W, He ZX, Ma H, Xun YF, Meng LR, … Tai FD (2020). Reduced Consolation Behaviors in Physically Stressed Mandarin Voles: Involvement of Oxytocin, Dopamine D2, and Serotonin 1A Receptors Within the Anterior Cingulate Cortex. Int J Neuropsychopharmacol, 23(8), 511–523. doi: 10.1093/ijnp/pyz060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen Z, Fan G, Li A, Yuan J, & Xu T (2018). Cell-Type-Specific Afferent Innervation of the Nucleus Accumbens Core and Shell. Front Neuroanat, 12, 84. doi: 10.3389/fnana.2018.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim MM, Hammock EA, & Young LJ (2004). A method for acetylcholinesterase staining of brain sections previously processed for receptor autoradiography. Biotech Histochem, 79(1), 11–16. doi: 10.1080/10520290410001671344 [DOI] [PubMed] [Google Scholar]
- Lim MM, Murphy AZ, & Young LJ (2004). Ventral striatopallidal oxytocin and vasopressin V1a receptors in the monogamous prairie vole (Microtus ochrogaster). J Comp Neurol, 468(4), 555–570. doi: 10.1002/cne.10973 [DOI] [PubMed] [Google Scholar]
- Lin YT, Hsieh TY, Tsai TC, Chen CC, Huang CC, & Hsu KS (2018). Conditional Deletion of Hippocampal CA2/CA3a Oxytocin Receptors Impairs the Persistence of Long-Term Social Recognition Memory in Mice. J Neurosci, 38(5), 1218–1231. doi: 10.1523/jneurosci.1896-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, & Wang ZX (2003). Nucleus accumbens oxytocin and dopamine interact to regulate pair bond formation in female prairie voles. Neuroscience, 121(3), 537–544. doi: 10.1016/s0306-4522(03)00555-4 [DOI] [PubMed] [Google Scholar]
- Ludwig M, Apps D, Menzies J, Patel JC, & Rice ME (2016). Dendritic Release of Neurotransmitters. 235–252. doi: 10.1002/cphy.c160007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M, & Leng G (2006). Dendritic peptide release and peptide-dependent behaviours. Nat Rev Neurosci, 7(2), 126–136. doi: 10.1038/nrn1845 [DOI] [PubMed] [Google Scholar]
- Mairesse J, Gatta E, Reynaert ML, Marrocco J, Morley-Fletcher S, Soichot M, … Maccari S (2015). Activation of presynaptic oxytocin receptors enhances glutamate release in the ventral hippocampus of prenatally restraint stressed rats. Psychoneuroendocrinology, 62, 36–46. doi: 10.1016/j.psyneuen.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Marlin BJ, Mitre M, D’Amour JA, Chao MV, & Froemke RC (2015). Oxytocin enables maternal behaviour by balancing cortical inhibition. Nature, 520(7548), 499–504. doi: 10.1038/nature14402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamales M, Bertran-Gonzalez J, Salomon L, Degos B, Deniau JM, Valjent E, … Girault JA (2009). Striatal medium-sized spiny neurons: identification by nuclear staining and study of neuronal subpopulations in BAC transgenic mice. PLoS One, 4(3), e4770. doi: 10.1371/journal.pone.0004770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw LA, Davis JK, Thomas PJ, Young LJ, & Thomas JW (2012). BAC-based sequencing of behaviorally-relevant genes in the prairie vole. PLoS One, 7(1), e29345. doi: 10.1371/journal.pone.0029345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw LA, & Young LJ (2010). The prairie vole: an emerging model organism for understanding the social brain. Trends Neurosci, 33(2), 103–109. doi: 10.1016/j.tins.2009.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meddle SL, Bishop VR, Gkoumassi E, van Leeuwen FW, & Douglas AJ (2007). Dynamic changes in oxytocin receptor expression and activation at parturition in the rat brain. Endocrinology, 148(10), 5095–5104. doi: 10.1210/en.2007-0615 [DOI] [PubMed] [Google Scholar]
- Meira T, Leroy F, Buss EW, Oliva A, Park J, & Siegelbaum SA (2018). A hippocampal circuit linking dorsal CA2 to ventral CA1 critical for social memory dynamics. Nat Commun, 9(1), 4163. doi: 10.1038/s41467-018-06501-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitre M, Marlin BJ, Schiavo JK, Morina E, Norden SE, Hackett TA, … Froemke RC (2016). A Distributed Network for Social Cognition Enriched for Oxytocin Receptors. J Neurosci, 36(8), 2517–2535. doi: 10.1523/jneurosci.2409-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi ME, Inoue K, Barrett CE, Kittelberger KA, Smith DG, Landgraf R, & Young LJ (2015). Melanocortin Receptor Agonists Facilitate Oxytocin-Dependent Partner Preference Formation in the Prairie Vole. Neuropsychopharmacology, 40(8), 1856–1865. doi: 10.1038/npp.2015.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa M, Mitsui S, En S, Ohtani N, Ohta M, Sakuma Y, … Kikusui T (2015). Social evolution. Oxytocin-gaze positive loop and the coevolution of human-dog bonds. Science, 348(6232), 333–336. doi: 10.1126/science.1261022 [DOI] [PubMed] [Google Scholar]
- Numan M (2020). The Parental brain : mechanisms, development, and evolution.
- Numan M, & Young LJ (2016). Neural mechanisms of mother–infant bonding and pair bonding: Similarities, differences, and broader implications. Hormones and Behavior, 77, 98–112. doi: 10.1016/j.yhbeh.2015.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettl LL, Ravi N, Schneider M, Scheller MF, Schneider P, Mitre M, … Kelsch W (2016). Oxytocin Enhances Social Recognition by Modulating Cortical Control of Early Olfactory Processing. Neuron, 90(3), 609–621. doi: 10.1016/j.neuron.2016.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama T (2018). Social memory engram in the hippocampus. Neurosci Res, 129, 17–23. doi: 10.1016/j.neures.2017.05.007 [DOI] [PubMed] [Google Scholar]
- Okuyama T, Kitamura T, Roy DS, Itohara S, & Tonegawa S (2016). Ventral CA1 neurons store social memory. Science, 353(6307), 1536–1541. doi: 10.1126/science.aaf7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olazábal DE, & Alsina-Llanes M (2016). Are age and sex differences in brain oxytocin receptors related to maternal and infanticidal behavior in naïve mice? Hormones and Behavior, 77, 132–140. doi: 10.1016/j.yhbeh.2015.04.006 [DOI] [PubMed] [Google Scholar]
- Olazábal DE, & Sandberg NY (2020). Variation in the density of oxytocin receptors in the brain as mechanism of adaptation to specific social and reproductive strategies. Gen Comp Endocrinol, 286, 113337. doi: 10.1016/j.ygcen.2019.113337 [DOI] [PubMed] [Google Scholar]
- Oliva A, Fernández-Ruiz A, Leroy F, & Siegelbaum SA (2020). Hippocampal CA2 sharp-wave ripples reactivate and promote social memory. Nature, 587(7833), 264–269. doi: 10.1038/s41586-020-2758-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ophir AG (2017). Navigating Monogamy: Nonapeptide Sensitivity in a Memory Neural Circuit May Shape Social Behavior and Mating Decisions. Frontiers in Neuroscience, 11. doi: 10.3389/fnins.2017.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ophir AG, Gessel A, Zheng D-J, & Phelps SM (2012). Oxytocin receptor density is associated with male mating tactics and social monogamy. Hormones and Behavior, 61(3), 445–453. doi: 10.1016/j.yhbeh.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ophir AG, Zheng DJ, Eans S, & Phelps SM (2009). Social investigation in a memory task relates to natural variation in septal expression of oxytocin receptor and vasopressin receptor 1a in prairie voles (Microtus ochrogaster). Behav Neurosci, 123(5), 979–991. doi: 10.1037/a0016663 [DOI] [PubMed] [Google Scholar]
- Oumi T, Ukena K, Matsushima O, Ikeda T, Fujita T, Minakata H, & Nomoto K (1996). Annetocin, an annelid oxytocin-related peptide, induces egg-laying behavior in the earthworm, Eisenia foetida. J Exp Zool, 276(2), 151–156. doi: [DOI] [PubMed] [Google Scholar]
- Owen SF, Tuncdemir SN, Bader PL, Tirko NN, Fishell G, & Tsien RW (2013). Oxytocin enhances hippocampal spike transmission by modulating fast-spiking interneurons. Nature, 500(7463), 458–462. doi: 10.1038/nature12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Garcia TR, Garcia-Keller C, Penaloza T, Richie CT, Pickel J, Hope BT, … Heinsbroek JA (2019). Ventral Pallidum Is the Primary Target for Accumbens D1 Projections Driving Cocaine Seeking. J Neurosci, 39(11), 2041–2051. doi: 10.1523/jneurosci.2822-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos GWC (2014). Paxinos and Watson’s the rat brain in stereotaxic coordinates.
- Pedersen CA, Caldwell JD, Walker C, Ayers G, & Mason GA (1994). Oxytocin activates the postpartum onset of rat maternal behavior in the ventral tegmental and medial preoptic areas. Behav Neurosci, 108(6), 1163–1171. doi: 10.1037//0735-7044.108.6.1163 [DOI] [PubMed] [Google Scholar]
- Pohl TT, Young LJ, & Bosch OJ (2019). Lost connections: Oxytocin and the neural, physiological, and behavioral consequences of disrupted relationships. Int J Psychophysiol, 136, 54–63. doi: 10.1016/j.ijpsycho.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prounis GS, Thomas K, & Ophir AG (2018). Developmental trajectories and influences of environmental complexity on oxytocin receptor and vasopressin 1A receptor expression in male and female prairie voles. Journal of Comparative Neurology, 526(11), 1820–1842. doi: 10.1002/cne.24450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam PT, Young LJ, & Gothard KM (2018). Bridging the gap between rodents and humans: The role of non-human primates in oxytocin research. American journal of primatology, 80(10), e22756. doi: 10.1002/ajp.22756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana DS, Rokicki J, van der Meer D, Alnæs D, Kaufmann T, Córdova-Palomera A, … Westlye LT (2019). Oxytocin pathway gene networks in the human brain. Nat Commun, 10(1), 668. doi: 10.1038/s41467-019-08503-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raam T, McAvoy KM, Besnard A, Veenema AH, & Sahay A (2017). Hippocampal oxytocin receptors are necessary for discrimination of social stimuli. Nat Commun, 8(1), 2001. doi: 10.1038/s41467-017-02173-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raggenbass M, Tribollet E, Dubois-Dauphin M, & Dreifuss JJ (1989). Correlation between oxytocin neuronal sensitivity and oxytocin receptor binding: an electrophysiological and autoradiographical study comparing rat and guinea pig hippocampus. Proc Natl Acad Sci U S A, 86(2), 750–754. doi: 10.1073/pnas.86.2.750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rilling JK, & Young LJ (2014). The biology of mammalian parenting and its effect on offspring social development. Science, 345(6198), 771–776. doi: 10.1126/science.1252723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers Flattery CN, Coppeto DJ, Inoue K, Rilling JK, Preuss TM, & Young LJ (2021). Distribution of brain oxytocin and vasopressin V1a receptors in chimpanzees (Pan troglodytes): comparison with humans and other primate species. Brain Structure and Function. doi: 10.1007/s00429-021-02369-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers Flattery CN, Ross AP, Sahu SP, Siegel ER, Dooyema JM, Cree MA, … Preuss TM (2018). Oxytocin- and arginine vasopressin-containing fibers in the cortex of humans, chimpanzees, and rhesus macaques. American journal of primatology, 80(10), e22875–e22875. doi: 10.1002/ajp.22875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers-Carter MM, & Christianson JP (2019). An insular view of the social decision-making network. Neuroscience & Biobehavioral Reviews, 103, 119–132. doi: 10.1016/j.neubiorev.2019.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers-Carter MM, Varela JA, Gribbons KB, Pierce AF, McGoey MT, Ritchey M, & Christianson JP (2018). Insular cortex mediates approach and avoidance responses to social affective stimuli. Nat Neurosci, 21(3), 404–414. doi: 10.1038/s41593-018-0071-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross HE, Freeman SM, Spiegel LL, Ren X, Terwilliger EF, & Young LJ (2009). Variation in oxytocin receptor density in the nucleus accumbens has differential effects on affiliative behaviors in monogamous and polygamous voles. J Neurosci, 29(5), 1312–1318. doi: 10.1523/jneurosci.5039-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuni L, Preis A, Mundry R, Deschner T, Crockford C, & Wittig RM (2017). Oxytocin reactivity during intergroup conflict in wild chimpanzees. Proc Natl Acad Sci U S A, 114(2), 268–273. doi: 10.1073/pnas.1616812114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher M, Coirini H, Pfaff DW, & McEwen BS (1990). Behavioral effects of progesterone associated with rapid modulation of oxytocin receptors. Science, 250(4981), 691–694. doi: 10.1126/science.2173139 [DOI] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, … Greicius MD (2007). Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci, 27(9), 2349–2356. doi: 10.1523/jneurosci.5587-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeltzer MD, Curtis JT, Aragona BJ, & Wang Z (2006). Dopamine, oxytocin, and vasopressin receptor binding in the medial prefrontal cortex of monogamous and promiscuous voles. Neurosci Lett, 394(2), 146–151. doi: 10.1016/j.neulet.2005.10.019 [DOI] [PubMed] [Google Scholar]
- Smith CJW, Poehlmann ML, Li S, Ratnaseelan AM, Bredewold R, & Veenema AH (2016). Age and sex differences in oxytocin and vasopressin V1a receptor binding densities in the rat brain: focus on the social decision-making network. Brain Structure and Function, 222(2), 981–1006. doi: 10.1007/s00429-016-1260-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares-Cunha C, Coimbra B, Sousa N, & Rodrigues AJ (2016). Reappraising striatal D1- and D2-neurons in reward and aversion. Neurosci Biobehav Rev, 68, 370–386. doi: 10.1016/j.neubiorev.2016.05.021 [DOI] [PubMed] [Google Scholar]
- Song Z, Borland JM, Larkin TE, O’Malley M, & Albers HE (2016). Activation of oxytocin receptors, but not arginine-vasopressin V1a receptors, in the ventral tegmental area of male Syrian hamsters is essential for the reward-like properties of social interactions. Psychoneuroendocrinology, 74, 164–172. doi: 10.1016/j.psyneuen.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Benusiglio D, Lefevre A, Hilfiger L, Althammer F, Bludau A, … Grinevich V (2020). Social touch promotes interfemale communication via activation of parvocellular oxytocin neurons. Nat Neurosci, 23(9), 1125–1137. doi: 10.1038/s41593-020-0674-y [DOI] [PubMed] [Google Scholar]
- Tirko NN, Eyring KW, Carcea I, Mitre M, Chao MV, Froemke RC, & Tsien RW (2018). Oxytocin Transforms Firing Mode of CA2 Hippocampal Neurons. Neuron, 100(3), 593–608.e593. doi: 10.1016/j.neuron.2018.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ (2015). Salience processing and insular cortical function and dysfunction. Nat Rev Neurosci, 16(1), 55–61. doi: 10.1038/nrn3857 [DOI] [PubMed] [Google Scholar]
- Valtcheva S, Issa HA, Martin KA, Jung K, Kwon H-B, & Froemke RC (2021). Neural circuitry for maternal oxytocin release induced by infant cries. bioRxiv, 2021.2003.2025.436883. doi: 10.1101/2021.03.25.436883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walum H, & Young LJ (2018). The neural mechanisms and circuitry of the pair bond. Nat Rev Neurosci, 19(11), 643–654. doi: 10.1038/s41583-018-0072-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Talwar V, Osakada T, Kuang A, Guo Z, Yamaguchi T, & Lin D (2019). Hypothalamic Control of Conspecific Self-Defense. Cell Rep, 26(7), 1747–1758.e1745. doi: 10.1016/j.celrep.2019.01.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watarai A, Tao K, Wang MY, & Okuyama T (2021). Distinct functions of ventral CA1 and dorsal CA2 in social memory. Curr Opin Neurobiol, 68, 29–35. doi: 10.1016/j.conb.2020.12.008 [DOI] [PubMed] [Google Scholar]
- Williams AV, Duque-Wilckens N, Ramos-Maciel S, Campi KL, Bhela SK, Xu CK, … Trainor BC (2020). Social approach and social vigilance are differentially regulated by oxytocin receptors in the nucleus accumbens. Neuropsychopharmacology, 45(9), 1423–1430. doi: 10.1038/s41386-020-0657-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt DM, Carter CS, & Lnsel TR (1991). Oxytocin receptor binding in female prairie voles: endogenous and exogenous oestradiol stimulation. J Neuroendocrinol, 3(2), 155–161. doi: 10.1111/j.1365-2826.1991.tb00258.x [DOI] [PubMed] [Google Scholar]
- Yamagishi A, Lee J, & Sato N (2020). Oxytocin in the anterior cingulate cortex is involved in helping behaviour. Behav Brain Res, 393, 112790. doi: 10.1016/j.bbr.2020.112790 [DOI] [PubMed] [Google Scholar]
- Yang H, de Jong JW, Tak Y, Peck J, Bateup HS, & Lammel S (2018). Nucleus Accumbens Subnuclei Regulate Motivated Behavior via Direct Inhibition and Disinhibition of VTA Dopamine Subpopulations. Neuron, 97(2), 434–449.e434. doi: 10.1016/j.neuron.2017.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Takayanagi Y, Inoue K, Kimura T, Young LJ, Onaka T, & Nishimori K (2009). Evidence that oxytocin exerts anxiolytic effects via oxytocin receptor expressed in serotonergic neurons in mice. J Neurosci, 29(7), 2259–2271. doi: 10.1523/jneurosci.5593-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura R, Kimura T, Watanabe D, & Kiyama H (1996). Differential expression of oxytocin receptor mRNA in the developing rat brain. Neurosci Res, 24(3), 291–304. doi: 10.1016/0168-0102(95)01003-3 [DOI] [PubMed] [Google Scholar]
- Young L, & Alexander B (2012). The Chemistry Between us: Love, Sex and the Science of Attraction: Penguin Press. [Google Scholar]
- Young LJ, Huot B, Nilsen R, Wang Z, & Insel TR (1996). Species differences in central oxytocin receptor gene expression: comparative analysis of promoter sequences. J Neuroendocrinol, 8(10), 777–783. doi: 10.1046/j.1365-2826.1996.05188.x [DOI] [PubMed] [Google Scholar]
- Young LJ, Lim MM, Gingrich B, & Insel TR (2001). Cellular mechanisms of social attachment. Horm Behav, 40(2), 133–138. doi: 10.1006/hbeh.2001.1691 [DOI] [PubMed] [Google Scholar]
- Young LJ, Muns S, Wang Z, & Insel TR (1997). Changes in oxytocin receptor mRNA in rat brain during pregnancy and the effects of estrogen and interleukin-6. J Neuroendocrinol, 9(11), 859–865. doi: 10.1046/j.1365-2826.1997.00654.x [DOI] [PubMed] [Google Scholar]
- Young LJ, & Wang Z (2004). The neurobiology of pair bonding. Nat Neurosci, 7(10), 1048–1054. doi: 10.1038/nn1327 [DOI] [PubMed] [Google Scholar]
- Young LJ, & Zhang QI (2021). On the Origins of Diversity in Social Behavior. 動物心理学研究, 71(1), 45–61. doi: 10.2502/janip.71.1.4 [DOI] [Google Scholar]
- Young WS, & Song J (2020). Characterization of Oxytocin Receptor Expression Within Various Neuronal Populations of the Mouse Dorsal Hippocampus. Front Mol Neurosci, 13, 40. doi: 10.3389/fnmol.2020.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, He Z, Hou W, Wang L, Li L, Zhang J, … Tai F (2019). Role of oxytocin in the medial preoptic area (MPOA) in the modulation of paternal behavior in mandarin voles. Horm Behav, 110, 46–55. doi: 10.1016/j.yhbeh.2019.02.014 [DOI] [PubMed] [Google Scholar]
- Zheng D-J, Larsson B, Phelps SM, & Ophir AG (2013). Female alternative mating tactics, reproductive success and nonapeptide receptor expression in the social decision-making network. Behavioural Brain Research, 246, 139–147. doi: 10.1016/j.bbr.2013.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Regions expressing Oxtr mRNA in prairie vole brain. Oxtr mRNA was labeled with in situ hybridization and the percent of cells positive for Oxtr was calculated for each brain region expressing Oxtr (+, 1–25% of cells expressing Oxtr; ++, 26–50%; +++, 51–75%; ++++, 76–100%). Brain regions were defined according to the seventh edition of “The Rat Brain Atlas in Stereotaxic Coordinates” (Paxinos, 2014).
Data Availability Statement
Data supporting the brain wide quantification of Oxtr mRNA are available in the supplementary material of this article. The raw data used to generate Figure 9 of this study and any other data are available from the corresponding author upon reasonable request.


