Abstract
The genes for two different protocatechuate 3,4-dioxygenases (P34Os) were cloned from the 4-sulfocatechol-degrading bacterium Agrobacterium radiobacter strain S2 (DSMZ 5681). The pcaH1G1 genes encoded a P34O (P34O-I) which oxidized protocatechuate but not 4-sulfocatechol. These genes were part of a protocatechuate-degradative operon which strongly resembled the isofunctional operon from the protocatechuate-degrading strain Agrobacterium tumefaciens A348 described previously by D. Parke (FEMS Microbiol. Lett. 146:3–12, 1997). The second P34O (P34O-II), encoded by the pcaH2G2 genes, was functionally expressed and shown to convert protocatechuate and 4-sulfocatechol. A comparison of the deduced amino acid sequences of PcaH-I and PcaH-II, and of PcaG-I and PcaG-II, with each other and with the corresponding sequences from the P34Os, from other bacterial genera suggested that the genes for the P34O-II were obtained by strain S2 by lateral gene transfer. The genes encoding the P34O-II were found in a putative operon together with two genes which, according to sequence alignments, encoded transport proteins. Further downstream from this putative operon, two open reading frames which code for a putative regulator protein of the IclR family and a putative 3-carboxymuconate cycloisomerase were identified.
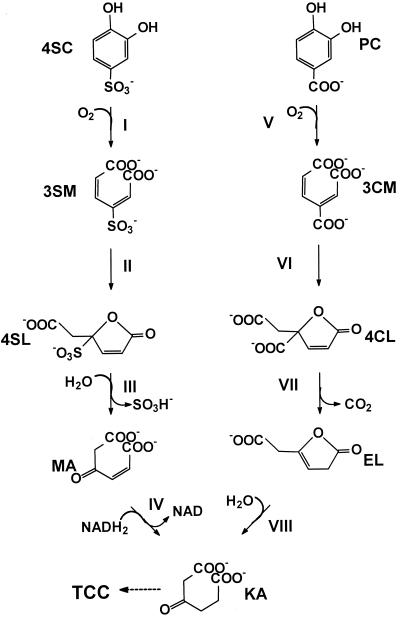
Aromatic compounds which carry sulfonic acid substituents are generally considered xenobiotic compounds which are rather recalcitrant against microbial degradation (1, 57). Nevertheless, several bacterial strains which utilize sulfonated benzenes or naphthalenes as sole sources of carbon and energy have been isolated (5, 7, 9, 31, 34, 35, 36, 55, 58, 59). Thus, it was shown that a coculture of Hydrogenophaga sp. strain S1 and Agrobacterium radiobacter strain S2 mineralized 4-aminobenzenesulfonate (sulfanilate) in a synthrophic association via 4-sulfocatechol (4SC), which was oxidized in both strains by an ortho-cleavage mechanism to 3-sulfomuconate (10, 17, 18) (Fig. 1). The 4SC-converting dioxygenase was purified to homogeneity from A. radiobacter strain S2, and it was shown that the purified enzyme also converted protocatechuate. The corresponding enzyme activity was therefore tentatively termed protocatechuate 3,4-dioxygenase type II (P34O-II) (18, 23). After growth with 4SC, strain S2 also induced a classical P34O which did not oxidize 4SC.
FIG. 1.
Proposed pathway for the degradation of 4SC and protocatechuate by A. radiobacter S2. Enzymes: I, P34O-II; II, 3-carboxymuconate cycloisomerase type II; III, “sulfolactone” hydrolase; IV, maleylacetate reductase; V, P34O-I; VI, 3-carboxymuconate cycloisomerase type I; VII, γ-carboxymuconolactone decarboxylase; VIII, β-ketoadipate enol-lactone hydrolase. Compounds: 3SM, 3-sulfomuconate; 4SL, “sulfolactone” (4-carboxymethyl-4-sulfobut-2-en-4-olide); MA, maleylacetate; KA, β-ketoadipate; PC, protocatechuate; 3CM, 3-carboxymuconate; 4CL, 4-carboxymuconolactone; EL, β-ketoadipate enol-lactone.
P34Os, (protocatechuate:oxygen 3,4-oxidoreductase; EC 1.13.11.3) are of central importance in the biodegradation of many aromatic compounds by bacteria (25). The enzymes catalyze the intradiol addition of molecular oxygen, cleaving the aromatic ring and forming 3-carboxy-cis,cis-muconate. P34Os from several bacterial sources have been intensively studied (for a review see reference 30). The cleavage of the sulfonated aromatic ring of 4SC by P34O-II is in contrast to almost all other degradative pathways described for the metabolism of sulfonated aromatics, which usually require the liberation of the sulfo group prior to ring fission (7, 32, 34, 37, 56, 59). Thus, the action of P34O-II leads to a new metabolic pathway for sulfonated benzenes. 4SC not only is a central metabolite in the degradation of sulfanilate but also is formed during the degradation of 1,3-benzenedisulfonate and presumably also during that of linear alkylbenzenesulfonates (8, 9). 4SC therefore appears to be a central intermediate for the microbial degradation of substituted sulfonated benzenes. Therefore, in the present study we attempted to compare the genes encoding both types of P34Os from A. radiobacter strain S2 in order to gain some insights into the evolution of a new pathway for the degradation of sulfonated compounds.
MATERIALS AND METHODS
Bacterial strains and media.
The isolation and characterization of A. radiobacter S2 (DSMZ 5681) has been reported before. The strain was routinely grown in SHPG medium as previously described (17, 18). Escherichia coli DH5α and E. coli BL21(DE3)/pLysS were used as host strains for recombinant DNA work. E. coli strains were routinely cultured in Luria-Bertani medium supplied with ampicillin (100 μg/ml) if appropriate.
Plasmids and DNA manipulation techniques.
The plasmid pBluescript II SK(+) (2) was used for most cloning experiments, and the plasmid vector pET11a (54) was used for high-level expression. Plasmid pARO162 (a PstI-HindIII deletion of pARO144) (43) was kindly provided by D. Parke (Yale University).
Genomic DNA was prepared after sodium dodecyl sulfate lysis and phenol extraction as described by Eulberg et al. (15). Plasmid DNA from E. coli DH5α was isolated with a Pharmacia GFX Micro Plasmid Prep kit. Digestion of DNA with restriction endonucleases (Gibco BRL and Boehringer), electrophoresis, and ligation with T4 DNA ligase (Gibco BRL) were performed by standard procedures (49). Transformation of E. coli was done by the method of Inoue et al. (26). For cloning of PCR products, a T-vector was prepared as described by Marchuk et al. (33).
PCR.
Oligonucleotides were custom synthesized according to the known or deduced amino-terminal amino acid sequences and conserved regions within the catalytic centers of various P34Os (Table 1). PCR mixtures (50 μl) for the amplification of genomic DNA contained 50 pmol of each primer, 0.1 to 0.2 μg of genomic template DNA, 0.1 mM each deoxynucleoside triphosphate, Taq DNA polymerase, and the corresponding reaction buffer (Stratagene).
TABLE 1.
Sequences of N termini, identified conserved regions within the P34Os, and deduced oligonucleotide primers
| Protein or peptide | Amino acid sequence | Deduced primer sequence |
|---|---|---|
| P34O type Ia | ||
| N terminus of PcaG-I | TPSQTVGP | 3′-TGG GGS AGM GTY TGG CAS CCG GG(S)-5′ |
| PcaH-I | TIKPGPYPW | 5′-ACS ATY AAR CCS GGY CCR TAY CCS TGG-3′ |
| P34O type IIb | ||
| N terminus of PcaG-II | TVGPFYAY | 3′-TGN CAN CCN GGN AAR ATR CGN TA(R)-5′ |
| N terminus of PcaH-II | EVPAEYP | 5′-GGN GTN CCN GCN GAR TAY CC(N)-3′ |
The oligonucleotides were deduced from the consensus sequences of different P34Os. The amino acid sequence for PcaH-I corresponded to amino acids 9 to 16, and that of PcaG-I corresponded to amino acids 141 to 149, of the respective subunits of the P34O from P. putida (21).
The oligonucleotides were deduced from the amino-terminal amino acid sequences of PcaG-II (amino acids 14 to 21) and PcaH-II (amino acids 9 to 15) from A. radiobacter strain S2 (23).
For the amplification of pcaH1 and pcaG1, the following PCR program was used: an initial denaturation (94°C for 3 min) was followed by 29 cycles consisting of annealing at 45°C (45 s), a polymerization step (72°C for 2 min), and denaturation (94°C for 30 s).
For the amplification of pcaH2 and pcaG2, PCR was performed with a touchdown thermocycle program which included an initial denaturation step (94°C for 3 min) before addition of the polymerase and then 29 cycles with decreasing annealing temperature (55 to 40°C for 45 s), polymerization (72°C for 1 min), and denaturation (94°C for 30 s). Finally, the product of the reaction was further amplified by the PCR program described above for the amplification of pcaH1 and pcaG1.
The PCR products were initially cloned into the T-tailed EcoRV site of pBluescript II SK(+) (33).
Partial inverse PCR.
For the determination of the complete sequence of pcaG1, a partial inverse PCR was performed (41). The template was prepared by digesting chromosomal DNA of strain S2 with HindIII and religating the fragments obtained with T4 DNA ligase. Thus, intramolecular ligation of these DNA fragments should result in circular DNA, which was then used as a template for the following PCR. Primers for this PCR were deduced from the partial sequence of pcaG1 present on the 11-kb PstI fragment previously obtained and were facing in both directions outwards from the known DNA sequence. This resulted in the amplification of an approximately 2-kb fragment, which was used to finally obtain an approximately 2.5-kb EcoRI fragment of chromosomal DNA after Southern hybridization. This fragment was cloned and finally sequenced.
Hybridization procedures.
A digoxigenin DNA labeling and detection kit was used according to the instructions of the supplier (Boehringer). The hybridization temperature was set to 63°C.
Nucleotide sequence analysis.
The DNA sequence was determined by dideoxy-chain termination with double-stranded DNAs of overlapping subclones in an automated DNA-sequencing system (ALF-Sequencer; Amersham-Pharmacia) with fluorescently labeled primers.
Sequence analysis, database searches, and comparisons were done with the PCGene software package (release 6.85) and the BLAST Search at the National Center for Biotechnology Information (BLASTX) (3). The alignments of the P34Os were obtained with the program CLUSTAL using the default parameters.
Expression of P34O-I and P34O-II in E. coli.
For expression in E. coli, pcaH1G1 and pcaH2G2 were inserted into pET11a (54) under the control of the phage T7 promoter. The DNA segments encompassing pcaH1G1 or pcaH2G2 were amplified by PCR with simultaneous introduction of an NdeI site upstream and a BamHI site downstream of pcaH1G1 or pcaH2G2. The oligonucleotide primers 5′-ACGC-CATATG-AGCAACCAGCCACCCGA-3′ and 5′-ATAA-GGATCC-CTCATGGCGTCGATATC-3′ were used for the amplification of pcaH1G1. The primers used for the amplification of pcaH2G2 were 5′-ATTT-CATATG-GCCTTGTTCCTCCCCGGG-3′ and 5′-ATAT-GGATCC-GTCAGAATTACCTTGCGC-3′. The amplified products were cleaved with NdeI and BamHI and ligated into pET11a. E. coli DH5α was transformed with the resulting plasmids. The plasmids were subsequently isolated and introduced into E. coli BL21(DE3)/pLysS by transformation.
Preparation of cell extracts.
Cell suspensions in 50 mM Tris-HCl buffer (pH 8.0) were disrupted by using a French press (Aminco; SLM Instruments Inc., Urbana, Ill.) at 1.1 × 108 Pa. Cells and cell debris were removed by centrifugation at 100,000 × g for 30 min at 4°C.
Protein estimation and enzyme assays.
The protein content of cell extracts was determined by the method of Bradford (6). Bovine serum albumin was used as a standard. One unit of enzyme activity is defined as the amount of enzyme that converts 1 μmol of substrate per min. P34O-I enzyme was measured by the procedure of Stanier and Ingraham (52) but using Tris-HCl buffer (50 mM, pH 8.0). For the assay of P34O-II activity, protocatechuate was replaced by 4SC as the substrate (18).
Nucleotide sequence accession number.
The nucleotide sequences of pcaQ, pcaH1G1, pcaH2G2, and the gene cluster connected to pcaH2G2 will appear in the GenBank nucleotide sequence data base under accession numbers AF230648, AF230649, AF230650, and AF282677, respectively.
RESULTS
Cloning of the genes for the P34O-I.
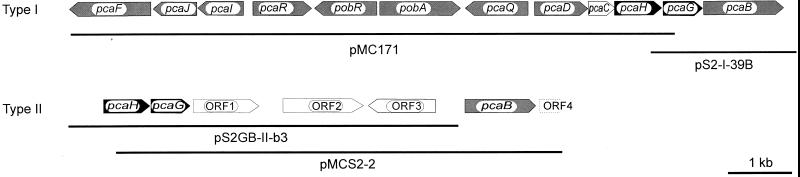
Previous biochemical studies suggested that A. radiobacter strain S2 synthesized a classical P34O (P34O-I), with the ability to oxidize protocatechuate but not 4SC, and also a second, modified type of P34O (P34O-II) which converted protocatechuate and 4SC. For both enzymes the amino-terminal amino acid sequences of the subunits (PcaH and PcaG) had been determined (23). The sequences determined for P34O-I served for the design of oligonucleotide primers, but no specific amplification was obtained. Therefore, the PCR experiments were repeated using oligonucleotide primers from amino-terminal amino acid sequences and conserved regions within the iron-binding sites of other known P34Os (Table 1). This resulted in the amplification of a fragment (409 bp) which showed significant sequence homology to other P34Os. The PCR fragment was labeled and used to clone an 11-kb PstI fragment from the genomic DNA of strain S2 in pBluescript II SK(+) after Southern hybridization, resulting in plasmid pMC171 (Fig. 2).
FIG. 2.
Structures of the two gene clusters (type I and type II genes) for the catabolism of protocatechuate and 4SC in A. radiobacter strain S2. The extents of the relevant subclones are shown below the maps.
The genes for a P34O (pcaHG) had previously been cloned from Agrobacterium tumefaciens A348 (43). A 405-bp EcoRI-EcoRV fragment was obtained from plasmid pARO162, encoding parts of pcaHG from A. tumefaciens A348. This fragment was also labeled and shown to hybridize with the 11-kb PstI fragment from strain S2 cloned in pMC171.
Determination of the nucleotide sequence of pcaH1G1 from strain S2.
DNA sequencing showed that approximately 200 bp of pcaG1 was missing at the 3′ end of the 11-kb PstI fragment cloned in pMC171. The missing part of pcaG1 was obtained after partial inverse PCR (41), resulting in plasmid pS2-I-39B. After sequencing of both inserts, the complete sequence of pcaH1G1 from A. radiobacter strain S2 was obtained (Fig. 2). A comparison of the deduced amino acid sequences of PcaH-I and PcaG-I from strain S2 with the sequences submitted recently by Parke to the National Center for Biotechnology Information data bank (accession number U32867) for the homologous proteins from A. tumefaciens A348 demonstrated 96.3 and 96.6% sequence identity, respectively.
The amino-terminal sequences of PcaH-I and PcaG-I from strain S2 which were deduced from the nucleotide sequences showed considerable differences from the sequences published previously by Hammer et al. (23) after amino acid sequencing. To test the identity of pcaH1G1 with the genes encoding the P34O-I, pcaH1G1 were functionally expressed in E. coli using a phage T7-promoter system. Cultures of E. coli BL21(DE3)/pLysS with the expression plasmid were grown at 37°C in 200 ml of Luria-Bertani medium plus 50 μg of ampicillin per ml and 10 mg of FeCl2 per ml to an optical density (510 nm) of about 1. The formation of the P34O was then induced by the addition of 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The cultures were further incubated at 30°C on a rotary shaker. Cells were harvested by centrifugation 90 min after the addition of IPTG, and the cells were resuspended in 1.8 ml of Tris-HCl buffer (pH 8, 50 mM) and disrupted with a French press. Enzyme activities were measured spectrophotometrically with protocatechuate and 4SC as described in Materials and Methods. The extract oxidized protocatechuate (specific activity, 0.09 U/mg of protein) but not 4SC (<0.001 U/mg of protein). It was therefore concluded that indeed the genes encoding the P34O-I had been cloned and sequenced. The recombinant E. coli strain expressed only low specific activities of P34O-I. This has been repeatedly observed for recombinant P34Os from various sources (14, 21, 46).
Analysis of the gene cluster with pcaH1G1.
A gene cluster including pcaHG had been previously described for A. tumefaciens, which differed in several aspects from the pathways for protocatechuate degradation in the well-studied gram-negative bacteria Acinetobacter calcoaceticus and Pseudomonas putida (45). Therefore, the organization of the pca genes in A. radiobacter strain S2 was determined by sequencing the 11-kb DNA fragment (partially only by single-strand sequencing), and it was found that the gene order within strain S2 appears to be identical to the one proposed by Parke (45) for A. tumefaciens. Furthermore, a comparison of the sequence of PcaQ determined by Parke (42, 44) with that of the corresponding gene product from strain S2 showed 91% sequence identity.
Cloning of the genes for P34O-II.
PCR using primers (Table 1) derived from the amino-terminal parts of PcaH-II and PcaG-II determined previously (23) resulted in the amplification of an approximately 700-bp DNA fragment. This fragment was used as a probe after digoxigenin labeling and gave a strong hybridization signal with an approximately 5-kb fragment of a SacII digest and a 4- to 5-kb fragment of a PstI digest of the total DNA from strain S2. Furthermore, a second, weaker signal with an approximately 11-kb fragment of the PstI digest was observed. This suggested that the probe also hybridized with the same 11-kb PstI fragment which was present in plasmid pMC171. The 5-kb SacII fragment was cloned in pBluescript II SK(+) (giving plasmid pMCS2-2 [Fig. 2]). DNA sequencing of the insert in pMCS2-2 showed that part of the 5′ end of pcaH2 was missing on that clone. Therefore the insert DNA was used to amplify a probe and to identify another plasmid from a gene library constructed with chromosomal DNA partially digested with PstI. The resulting plasmid (pS2GB-II-b3 [Fig. 2]) contained the complete pcaH2 gene. The pcaH2G2 genes were expressed using a T7 expression system as described above for the pcaH1G1 genes, and it was shown that the encoded dioxygenase was able to convert 4SC and protocatechuate (specific activities, 0.05 and 0.04 U/mg of protein, respectively). The sequences of pcaH2G2 demonstrated that both genes overlapped for 11 bp. This has not been previously observed for pcaHG genes from other sources. The deduced amino acid sequences of PcaH-I and PcaH-II shared only 40% sequence identity, and PcaG-I and PcaG-II shared only 38% sequence identity.
Analysis of the gene cluster with pcaH2G2.
The inserts in plasmids pMCS2-2 and pS2GB-II-b3 were completely sequenced. In this way the putative gene for a 3-carboxymuconate cycloisomerase (tentatively designated pcaB2) was identified. We are currently trying to express pcaB2 in order to demonstrate its function in the 4SC degradative pathway. Surprisingly, pcaH2G2 and pcaB2 were separated by approximately 4.3 kb which did not show any homology to known genes from the β-ketoadipate pathway. A BLASTX search with the sequences downstream of pcaG2 revealed two putative open reading frames (ORFs) (ORF1 and ORF2 in Fig. 2) with the highest degree of identity to two proteins (DctP and DctM) from Rhodobacter capsulatus. These proteins belong to a high-affinity transport system for C4-dicarboxylates (TRAP transporters) (20). Approximately 3,100 bp downstream of pcaG2 could be identified ORF3, which was transcribed in the opposite direction to pcaH2G2, ORF1, ORF2, and pcaB2 (Fig. 2). ORF3 coded for a putative protein which showed the highest degree of homology to transcriptional regulators of the IclR family. This group of regulators has already been described repeatedly as regulators in the protocatechuate metabolism of organisms such as A. calcoaceticus, A. tumefaciens, P. putida, and Rhodococcus opacus (13, 16, 22, 45, 48).
DISCUSSION
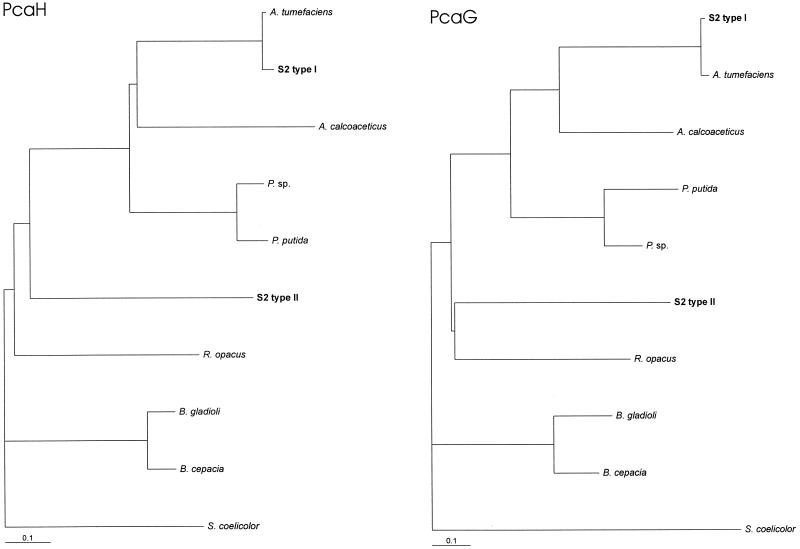
The results shown in the present study and the biochemical data published previously (18, 23) clearly demonstrate that A. radiobacter strain S2 synthesizes two different types of P34Os. The alignment of the amino acid sequences and the deduced dendrograms showed that both enzymes clearly clustered together with the previously determined sequences of the P34Os from organisms such as A. calcoaceticus, P. putida, Burkholderia cepacia, and R. opacus (Fig. 3 and 4) (16, 21, 24, 60) and were much more distantly related to other groups of intradiol dioxygenases such as catechol and chlorocatechol 1,2-dioxygenases. Thus, it appears that strain S2 synthesizes a distinct type of P34O (the type II enzyme) which is especially adapted to the degradation of the (xenobiotic) 4SC but which also shows significant activity with the natural substrate protocatechuate. The presence of two different P34Os in one bacterial strain has been previously shown for a Moraxella sp., which synthesized two different P34Os after growth on different substrates (53). From dendrograms comparing both subunits of the P34Os from strain S2 with the respective subunits of the P34Os from other organisms, it appears that the two isoenzymes from strain S2 are not more closely related to each other then to the P34Os from other organisms (Fig. 4). This suggests that the evolution of the P34O-II has not taken place in Agrobacterium but that the relevant genes have been acquired from another organism. This is also suggested by the different GC contents observed for the type I genes (58.6 and 60.5% for pcaH1 and pcaG1, respectively) and the type II genes (51.1 and 53.3% for pcaH2 and pcaG2, respectively). For members of the genus Agrobacterium, an average GC content of 57 to 63% has been found (28). Different origins of the two types of P34O were also suggested by the different organizations of the genes which accompany the two types of P34Os. While the protocatechuate operon which encodes the type I enzyme is obviously organized identically to the operon from A. tumefaciens A348 studied by Parke (43–45), the type II genes are found in a totally different type of genetic organization.
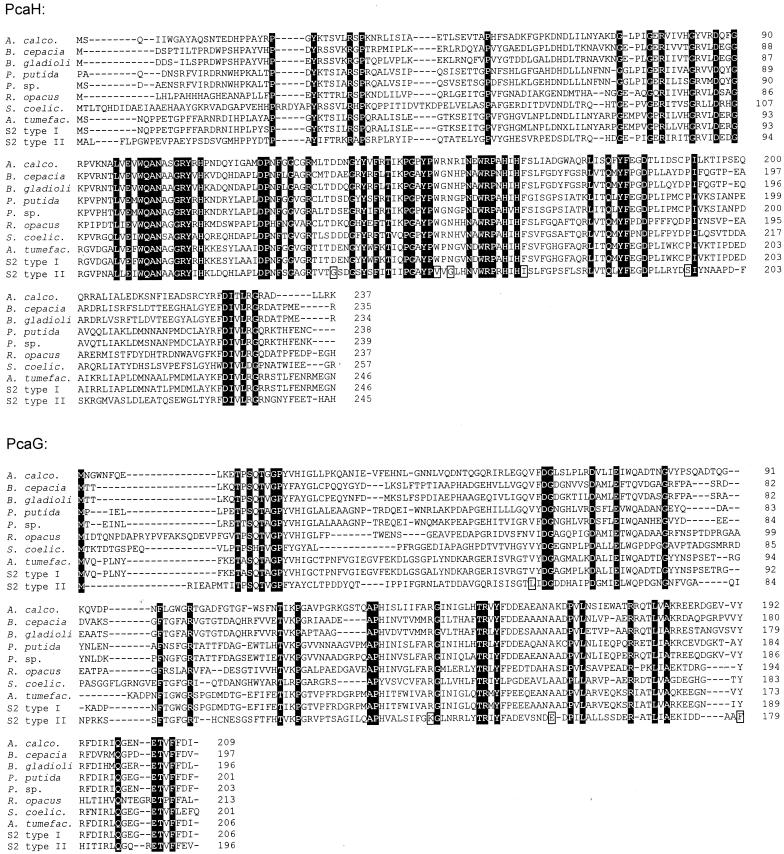
FIG. 3.
Sequence alignment of the subunits of P34Os. Positions that are identical in all sequences of PcaH and PcaG are highlighted by black boxes. Those identical in all P34O-Is but different in the sequences of P34O-II are marked by open boxes. The accession numbers (references) for the published sequences of the enzymes from A. calcoaceticus, A. tumefaciens, B. cepacia, P. putida, R. opacus, and Streptomyces coelicolor are L05770 (24), U32867, M30791 (60), L14836 (21), AF003947 (16), and AL079355 (47). (The function of the genes from S. coelicolor has not been proven experimentally.)
FIG. 4.
Dendrogram showing the relatedness of PcaH and PcaG from different bacterial sources. The dendrogram was produced using the programs PROTDIST and FITCH of the PHYLIP program package (19) and the program TREEVIEW (40). The accession numbers (references) for the published sequences of the P34Os from Burkholderia gladioli (“Pseudomonas marginata”) and Pseudomonas sp. are U33634 (46) and Y18527 (39). For the sources of the other sequences, see the legend to Fig. 3.
Although they are based on the analysis of just one example, a few first conclusions about the evolution of the new 4SC pathway may be drawn at the present state of the investigations. First, it appears that only small modifications are necessary for the adaptation of a classical P34O (and presumably also of a carboxymuconate cycloisomerase) to allow the transformation of the respective sulfonated structural analogues. This was surprising, as it has been suggested previously that sulfo substituents (like chloro or nitro groups) confer a xenobiotic character to synthetic compounds because the electron-withdrawing character of these substituents generates an electron deficiency and thus make these compounds less susceptible to oxidative catabolism (29). The clear homology of the P34O-I and P34O-II suggests that the two enzymes convert their substrates by basically identical catalytical mechanisms. This suggests that the oxidation of the sulfonated catechol is not limited by the general mechanism of enzymatic ring fission but that more probably simple steric problems limit the oxidation of 4SC. Whether this is also true for other sulfonated compounds or polysulfonated substrates remains to be investigated.
From the alignment of the P34O-II with the classical P34Os, only nine amino acids could be identified in both subunits which are conserved among all P34O-Is but which are different in the P34O-II (Fig. 3). For one of these residues (K134), which is a lysine in PcaG-II but an arginine in all other P34Os, it has been recently shown for the enzyme from A. calcoaceticus that a mutation of this residue (R133H) allowed the P34O to convert catechol, which is not oxidized by the wild-type enzyme (11). Furthermore, structural data from the crystal structure of the P34O from P. putida suggested that the exchange at position 154 of PcaH-II, which is a valine in PcaH-II but a tryptophan in all other P34Os, may also be relevant, because this residue is close to the carboxylic group of protocatechuate in the active center of the P34O from P. putida (38). We are currently trying to identify the amino acid exchanges which are responsible for the ability of the P34O-II to convert 4SC.
While the structural genes for pcaH2G2 and pcaB2 clearly resemble their classical counterparts from the degradative pathway of protocatechuate, the putative operon structure of the type II genes seems to be fundamentally different from the currently known (supra-)operonic structures of the pca genes in different organisms. This is not surprising, because the suggested pathway for the degradation of 4SC resembles the protocatechuate pathway only for the conversion of the aromatic compounds to the respective lactones, and the further steps are presumably different because of the differences in chemical reactivities of carboxylated and sulfonated muconolactones. Therefore, the “4-sulfolactone” is presumably hydrolytically desulfonated to maleylacetate (18). Thus, this pathway requires the combination of somehow-modified types of a P34O and a 3-carboxymuconate cycloisomerase with a hydrolase of currently unknown origin and a maleylacetate reductase, which is also involved in the degradation of dihydroxylated and chlorinated aromatics (4, 12, 27, 51).
The functional reasons for the linkage of pcaH2G2 with the putative transport proteins encoded by ORF1 and ORF2 is currently unknown. A similar linkage of pcaHG has not been observed previously in the classical protocatechuate degradation pathways. It was originally assumed by us that these genes could be involved in the uptake of the highly polar 4SC by A. radiobacter S2. Surprisingly, preliminary results with E. coli expressing the recombinant P34O-II in the absence of these genes did not indicate any limitations for the transport of 4SC compared to protocatechuate into the recombinant organisms. Nevertheless, it may be possible that the gene products from ORF1 and ORF2 are required for the uptake of 4SC or other intermediates of the 4SC pathway by A. radiobacter strain S2 (10).
The existence of a second type of P34O which is especially adapted to the degradation of the (xenobiotic) 4SC clearly resembles the situation observed for the degradation of chlorocatechols. It has been shown that some bacterial strains are able to synthesize a classical β-ketoadipate pathway for the degradation of catechol and a second set of evolutionarily related chlorocatechol 1,2-dioxygenases and chloromuconate cycloisomerases which are specifically adapted to the degradation of chlorinated catechols and muconates (50). Thus, it appears that the degradation of substituted benzenesulfonates is accomplished in nature by another novel variation of the β-ketoadipate pathway.
REFERENCES
- 1.Alexander M, Lustigman B K. Effect of chemical structure on microbial degradation of substituted benzenes. J Agric Food Chem. 1966;14:410–413. [Google Scholar]
- 2.Alting-Mees M A, Sorge J A, Short J M. pBluescript II: multifunctional cloning and mapping vectors. Methods Enzymol. 1992;216:483–495. doi: 10.1016/0076-6879(92)16044-k. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armengaud J, Timmis K N, Wittich R M. A functional 4-hydroxysalicylate/hydroxyquinol degradative pathway gene cluster is linked to the initial dibenzo-p-dioxin pathway genes in Sphingomonas sp. strain RW1. J Bacteriol. 1999;181:3452–3461. doi: 10.1128/jb.181.11.3452-3461.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balashov S V, Boronin A M. Sewage-sludge bacterial isolates decomposing sulfoaromatic compounds. Microbiology (Russia) 1996;65:549–552. [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Brilon C, Beckmann W, Knackmuss H-J. Catabolism of naphthalenesulfonic acids by Pseudomonas sp. A3 and Pseudomonas sp. C22. Appl Environ Microbiol. 1981;42:44–55. doi: 10.1128/aem.42.1.44-55.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contzen M, Wittich R-M, Knackmuss H-J, Stolz A. Degradation of benzene-1,3-disulfonate by a mixed bacterial culture. FEMS Microbiol Lett. 1996;136:45–50. doi: 10.1016/0378-1097(95)00483-1. [DOI] [PubMed] [Google Scholar]
- 9.Cook A M, Laue H, Junker F. Microbial desulfonation. FEMS Microbiol Rev. 1999;22:399–419. doi: 10.1111/j.1574-6976.1998.tb00378.x. [DOI] [PubMed] [Google Scholar]
- 10.Dangmann E, Stolz A, Kuhm A E, Hammer A, Feigel B, Noisommit-Rizzi N, Rizzi M, Reuß M, Knackmuss H-J. Degradation of 4-aminobenzenesulfonate by a two-species bacterial coculture. Physiological interactions between Hydrogenophaga palleronii S1 and Agrobacterium radiobacter S2. Biodegradation. 1996;7:223–229. doi: 10.1007/BF00058181. [DOI] [PubMed] [Google Scholar]
- 11.D'Argenio D A, Vetting M W, Ohlendorf D H, Ornston L N. Substitution, insertion, deletion, suppression, and altered substrate specificity in functional protocatechuate 3,4-dioxygenases. J Bacteriol. 1999;181:6478–6487. doi: 10.1128/jb.181.20.6478-6487.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daubaras D L, Saido K, Chakrabarty A M. Purification of hydroxyquinol 1,2-dioxygenase and maleylacetate reductase: the lower pathway of 2,4,5-trichlorophenoxyacetic acid metabolism by Burkholderia cepacia AC1100. Appl Environ Microbiol. 1996;62:4276–4279. doi: 10.1128/aem.62.11.4276-4279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DiMarco A A, Averhoff B, Ornston L N. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J Bacteriol. 1993;175:4499–4506. doi: 10.1128/jb.175.14.4499-4506.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doten R C, Ngai K-L, Mitchell D J, Ornston L N. Cloning and genetic organization of the pca genes from Acinetobacter calcoaceticus. J Bacteriol. 1987;169:3168–3174. doi: 10.1128/jb.169.7.3168-3174.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eulberg D, Golovleva L A, Schlömann M. Characterization of catechol catabolic genes from Rhodococcus opacus 1CP. J Bacteriol. 1997;179:370–381. doi: 10.1128/jb.179.2.370-381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eulberg D, Lakner S, Golovleva L A, Schlömann M. Characterization of a protocatechuate catabolic gene cluster from Rhodococcus opacus 1CP: evidence for a merged enzyme with 4-carboxymuconolactone-decarboxylating and 3-oxoadipate enol-lactone-hydrolyzing activity. J Bacteriol. 1998;180:1072–1081. doi: 10.1128/jb.180.5.1072-1081.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feigel B J, Knackmuss H-J. Bacterial catabolism of sulfanilic acid via catechol-4-sulfonic acid. FEMS Microbiol Lett. 1988;55:113–118. [Google Scholar]
- 18.Feigel B J, Knackmuss H-J. Syntrophic interactions during degradation of 4-aminobenzenesulfonic acid by a two species bacterial culture. Arch Microbiol. 1993;159:124–130. doi: 10.1007/BF00250271. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. PHYLIP (phylogeny interference package), version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 20.Forward J A, Behrendt M C, Wyborn N R, Cross R, Kelly D J. TRAP transporters: a new family of periplasmatic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J Bacteriol. 1997;179:5482–5493. doi: 10.1128/jb.179.17.5482-5493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frazee R W, Livingston D M, Laporte D C, Lipscomb J D. Cloning, sequencing, and expression of Pseudomonas putida protocatechuate 3,4-dioxygenase genes. J Bacteriol. 1993;175:6194–6202. doi: 10.1128/jb.175.19.6194-6202.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerischer U, Segura A, Ornston L N. PcaU, a transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J Bacteriol. 1998;180:1512–1524. doi: 10.1128/jb.180.6.1512-1524.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammer A, Stolz A, Knackmuss H-J. Purification and characterization of a novel type of protocatechuate 3,4-dioxygenase with the ability to oxidize 4-sulfocatechol. Arch Microbiol. 1996;166:92–100. doi: 10.1007/s002030050361. [DOI] [PubMed] [Google Scholar]
- 24.Hartnett C, Neidle E L, Ngai K-L, Ornston L N. DNA sequences of genes encoding Acinetobacter calcoaceticus protocatechuate 3,4-dioxygenase: evidence indicating shuffling of genes and of DNA sequences within genes during their evolutionary divergence. J Bacteriol. 1990;172:956–966. doi: 10.1128/jb.172.2.956-966.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harwood C S, Parales R E. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 26.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 27.Kaschabek S R, Reineke W. Degradation of chloroaromatics: purification and characterization of maleylacetate reductase from Pseudomonas sp. strain B13. J Bacteriol. 1993;175:6075–6081. doi: 10.1128/jb.175.19.6075-6081.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerr A. The genus Agrobacterium. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The procaryotes. 2nd ed. New York, N.Y: Springer; 1992. pp. 2214–2235. [Google Scholar]
- 29.Knackmuss H-J. Basic knowledge and perspectives of bioelemination of xenobiotic compounds. J Biotechnol. 1996;51:287–295. [Google Scholar]
- 30.Lipscomb J D, Orville A M. Mechanistic aspects of dihydroxybenzoate dioxygenases. Met Ions Biol Syst. 1992;28:243–298. [Google Scholar]
- 31.Locher H H, Thurnheer T, Leisinger T, Cook A M. 3-Nitrobenzenesulfonate, 3-aminobenzenesulfonate, and 4-aminobenzenesulfonate as sole carbon sources for bacteria. Appl Environ Microbiol. 1989;55:492–494. doi: 10.1128/aem.55.2.492-494.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Locher H H, Leisinger T, Cook A M. 4-Sulfobenzoate 3,4-dioxygenase. Biochem J. 1991;274:833–842. doi: 10.1042/bj2740833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchuk D, Drumm M, Saulino A, Collins F S. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nörtemann B, Baumgarten J, Rast H G, Knackmuss H-J. Bacterial communities degrading amino- and hydroxynaphthalene-2-sulfonates. Appl Environ Microbiol. 1986;52:1195–1202. doi: 10.1128/aem.52.5.1195-1202.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohe T, Watanabe Y. Degradation of 2-naphthylamine-1-sulfonic acid by Pseudomonas strain TA-1. Agric Biol Chem. 1986;50:1419–1426. [Google Scholar]
- 36.Ohe T, Watanabe Y. Microbial degradation of 1,6- and 2,6-naphthalenedisulfonic acid by Pseudomonas sp. S-1. Agric Biol Chem. 1988;52:2409–2414. [Google Scholar]
- 37.Ohe T, Ohmoto T, Kobayashi Y, Sato A, Watanabe Y. Metabolism of naphthalenesulfonic acids by Pseudomonas sp. TA-2. Agric Biol Chem. 1990;54:669–675. [Google Scholar]
- 38.Orville A M, Lipscomb J D, Ohlendorf D H. Crystal structure of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogeneous Fe3+ ligand displacement in response to substrate binding. Biochemistry. 1997;36:10052–10066. doi: 10.1021/bi970469f. [DOI] [PubMed] [Google Scholar]
- 39.Overhage J, Kresse A U, Priefert H, Sommer H, Krammer G, Rabenhorst J, Steinbüchel A. Molecular characterization of the genes pcaG and pcaH, encoding protocatechuate 3,4-dioxygenase, which are essential for vanillin catabolism in Pseudomonas sp. strain HR199. Appl Environ Microbiol. 1999;65:951–960. doi: 10.1128/aem.65.3.951-960.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comp Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 41.Pang K M, Knecht D A. Partial inverse PCR: a technique for cloning flanking sequences. BioTechniques. 1997;22:1046–1048. doi: 10.2144/97226bm07. [DOI] [PubMed] [Google Scholar]
- 42.Parke D. Positive regulation of phenolic catabolism in Agrobacterium tumefaciens by the pcaQ gene in response to β-carboxy-cis,cis-muconate. J Bacteriol. 1993;175:3529–3535. doi: 10.1128/jb.175.11.3529-3535.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parke D. Supraoperonic clustering of pca genes for catabolism of the phenolic compound protocatechuate in Agrobacterium tumefaciens. J Bacteriol. 1995;177:3808–3817. doi: 10.1128/jb.177.13.3808-3817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parke D. Characterization of PcaQ, a LysR-type transcriptional activator required for catabolism of phenolic compounds, from Agrobacterium tumefaciens. J Bacteriol. 1996;178:266–272. doi: 10.1128/jb.178.1.266-272.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parke D. Acquisition, reorganization, and merger of genes: novel management of the β-ketoadipate pathway in Agrobacterium tumefaciens. FEMS Microbiol Lett. 1997;146:3–12. [Google Scholar]
- 46.Petersen E I, Zuegg J, Ribbons D W, Schwab H. Molecular cloning and homology modeling of a protocatechuate 3,4-dioxygenase from Pseudomonas marginata. Microb Res. 1996;151:359–370. doi: 10.1016/s0944-5013(96)80004-8. [DOI] [PubMed] [Google Scholar]
- 47.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 48.Romero-Steiner S, Parales R E, Harwood C S, Houghton J E. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J Bacteriol. 1994;176:5771–5779. doi: 10.1128/jb.176.18.5771-5779.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Schlömann M. Evolution of chlorocatechol catabolic pathways. Biodegradation. 1994;5:301–321. doi: 10.1007/BF00696467. [DOI] [PubMed] [Google Scholar]
- 51.Seibert V, Stadler-Fritzsche K, Schlömann M. Purification and characterization of maleylacetate reductase from Alcaligenes eutrophus JMP134(pJP4) J Bacteriol. 1993;175:6745–6754. doi: 10.1128/jb.175.21.6745-6754.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanier R Y, Ingraham J I. Protocatechuic acid oxidase. J Biol Chem. 1954;210:799–808. [PubMed] [Google Scholar]
- 53.Sterjiades R, Pelmont J. Occurrence of two different forms of protocatechuate 3,4-dioxygenase in a Moraxella sp. Appl Environ Microbiol. 1989;55:340–347. doi: 10.1128/aem.55.2.340-347.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 55.Thurnheer T, Köhler T, Cook A M, Leisinger T. Orthanilic acid and analogues as carbon sources for bacteria: growth physiology and enzymic desulfonation. J Gen Microbiol. 1986;132:1215–1220. [Google Scholar]
- 56.Thurnheer T, Zürrer D, Höglinger O, Leisinger T, Cook A M. Initial steps in the degradation of benzene sulfonic acid, 4-toluene sulfonic acids, and orthanilic acid in Alcaligenes sp. strain O-1. Biodegradation. 1990;1:55–64. doi: 10.1007/BF00117051. [DOI] [PubMed] [Google Scholar]
- 57.Wellens H. Zur biologischen Abbaubarkeit mono- und disubstituierter Benzolderivate. Z Wasser-Abwasser Forsch. 1990;23:85–98. [Google Scholar]
- 58.Wittich R-M, Rast H G, Knackmuss H-J. Degradation of naphthalene-2,6- and naphthalene-1,6-disulfonic acid by a Moraxella sp. Appl Environ Microbiol. 1988;54:1842–1847. doi: 10.1128/aem.54.7.1842-1847.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zürrer D, Cook A M, Leisinger T. Microbial degradation of substituted naphthalenesulfonic acids and benzenesulfonic acids. Appl Environ Microbiol. 1987;53:1459–1463. doi: 10.1128/aem.53.7.1459-1463.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zylstra G J, Olsen R H, Ballou D P. Genetic organization and sequence of the Pseudomonas cepacia genes for the alpha and beta subunits of protocatechuate 3,4-dioxygenase. J Bacteriol. 1989;171:5915–5921. doi: 10.1128/jb.171.11.5915-5921.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]