Abstract
Plant immunity is the result of multiple distinct cellular processes cooperating with each other to generate immune responses. Autophagy is a conserved cellular recycling process and has well established roles in nutrient starvation responses and cellular homeostasis. Recently, the role of autophagy in immunity has become increasingly evident. However, our knowledge about plant autophagy remains limited and how this fundamental cellular process is involved in plant immunity is still somewhat perplexing. Here, we summarize the current understanding of the positive and negative roles of autophagy in plant immunity and how different microbes exploit this process to their own advantage. The dualistic role of autophagy in plant immunity emphasizes that much remains to be explored in this area.
Keywords: Autophagy, ETI, PTI, Pathogen effectors, HR-PCD, Salicylic acid, Reactive oxygen species, Immunity
Graphical Abstract
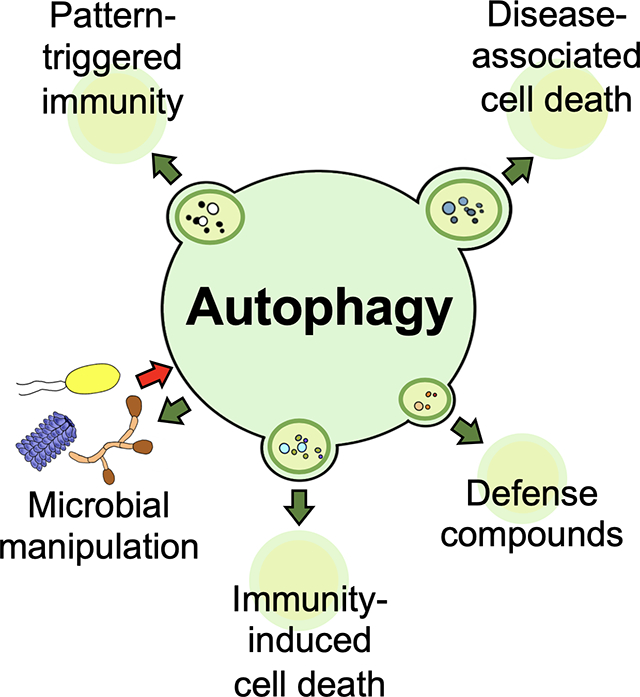
Significant insight into how a system for cellular degradation and recycling such as autophagy is involved in plant immunity has recently emerged. Many host pathways can positively or negatively modulate autophagy and the associated immune responses. Additionally, multiple plant microbes are able to manipulate autophagy. This review discusses recent advances in our understanding of the interplay between autophagy and plant defense.
Introduction
Plants possess a sophisticated immune network to combat pathogen infections [1–4]. Plants pattern triggered immunity (PTI) involves recognition of conserved microbial components, such as fungal chitin or bacterial flagellin, by cell-surface localized pattern recognition receptors (PRRs). This results in a cascade of defense responses including the production of reactive oxygen species (ROS), phytohormones, callose deposition, and transcriptional reprogramming of defense-related genes [3]. Consequently, some pathogens have evolved strategies to overcome PTI by secreting effector proteins into host cells [5]. Such effector proteins modulate various cellular and molecular activities to suppress PTI thereby promoting pathogenesis [5]. Not to be outdone, plants can deploy nucleotide-binding leucine-rich repeat (NLR) class of intracellular immune receptors that detect the presence or activity of effectors. This effector-triggered immunity (ETI) results in the activation of immune signaling that culminates in localized programmed cell death, known as the hypersensitive response (HR-PCD). The initiation and control of HR-PCD can be mediated by various signaling molecules, including salicylic acid (SA) and ROS. This localized programmed cell death restricts pathogens from spreading to adjacent cells [1,6]. In general, plants have two classes of NLRs: Toll-interleukin-1 receptor homology (TIR) domain containing NLRs (TNLs) and coiled-coil (CC) domain containing NLRs (CNLs). Despite having distinct triggers and characteristics, the overlap between PTI and ETI is becoming increasingly evident, and the cooperation between these two immunity programs is crucial in the perpetual fight against plant diseases [7–10].
Autophagy is a conserved eukaryotic process in which cytoplasmic materials and damaged organelles are recycled or degraded inside a lytic cellular compartment in order to maintain homeostasis [11,12]. While multiple types of autophagy exist, macroautophagy has been the most extensively explored and is commonly referred to simply as autophagy in the literature (as well as hereinafter). The primary distinguishing characteristic of autophagy is the formation of autophagosomes, which are specialized double-membrane vesicles capable of delivering cytoplasmic components into either the plant vacuole or animal lysosomes for degradation [11,13]. Selective autophagy occurs when only specific types of organelles or molecules are targeted [14,15]. In plants, more than 40 autophagy-related (ATG) genes have been identified, which have distinct yet collaborative roles in mediating autophagy [14,16]. Disruptions of ATGs can not only impair autophagy but can also impact other cellular and developmental processes [12,17]. Although the history and mechanisms of autophagy are not the focus of this review (more information can be found in [11–14,16,18]), the process of autophagy includes initiation, nucleation, elongation, completion, and ultimately fusion of autophagosomes with the vacuole or lysosome for the delivery and subsequent breakdown or recycling of cargoes.
Autophagy has been extensively researched in animal systems, yet plant autophagy has only begun to be explored. Several studies have demonstrated that plant autophagy is indispensable for proper function of plant immunity (Figure 1 and Table 1). Moreover, pathogens possess different strategies to target autophagy in order to compromise the host immunity (Figure 2). In this review, we explore various dimensions of the relationships between autophagy and plant immunity, including the roles of autophagy in plant defense and strategies that pathogens have evolved to manipulate autophagy, with a focus on bacterial and oomycete pathogens. A more complete review regarding plant viruses and their ways to manipulate autophagy can be found in this issue [19].
Figure 1. Functions of autophagy in plant immunity.
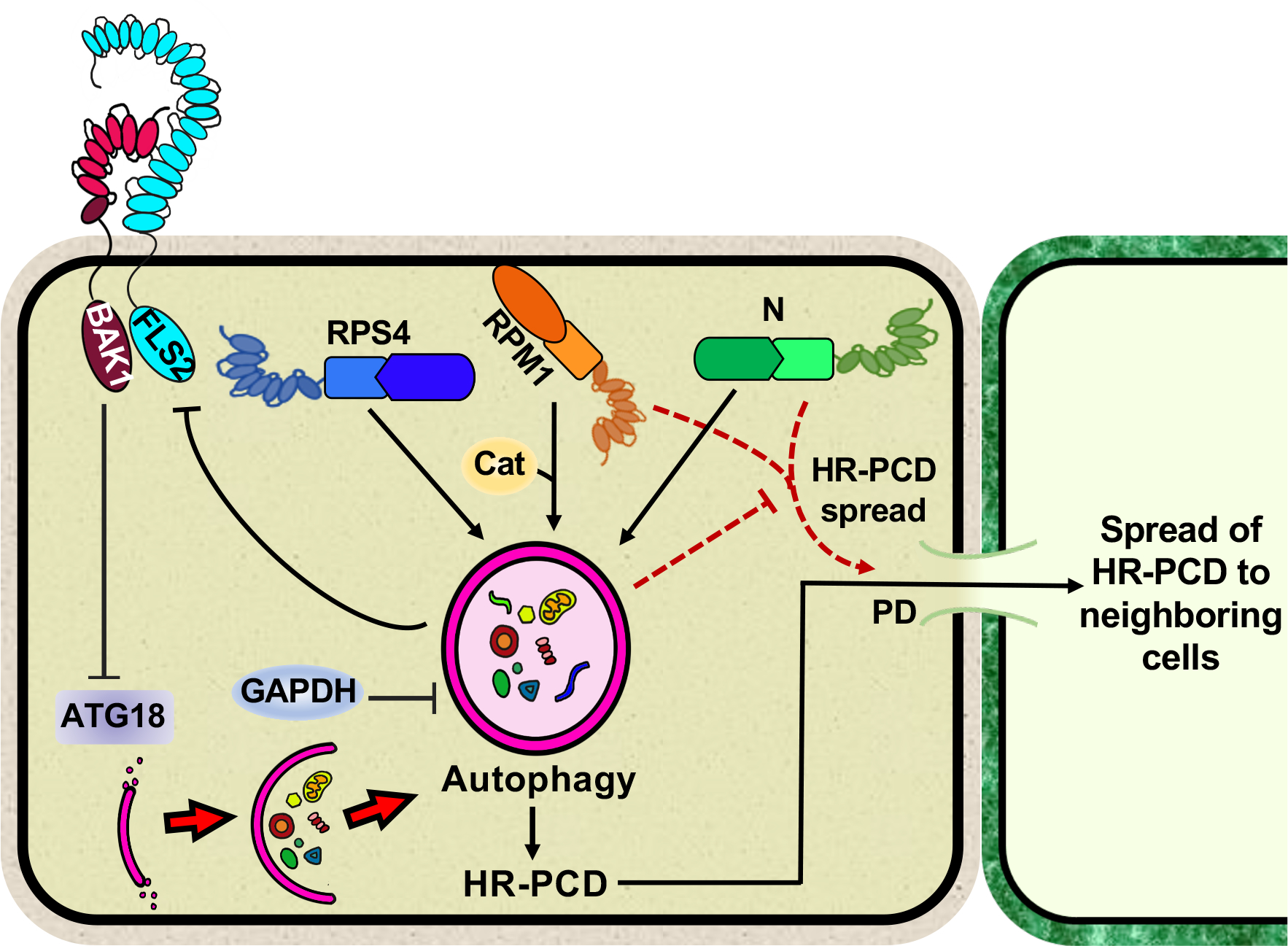
Autophagy negatively regulates FLS2 PRR levels, whereas BAK1 co-receptor inhibits the function of ATG18, which is required for the formation of the phagophore. Autophagy is required for the initiation of HR-PCD triggered by the RPM1 CNL and the RPS4 TNL. Additionally, autophagy is required for restricting HR-PCD to the infection site triggered by RPM1- or N-mediated immunity (represented by red dashed lines). Catalase (Cat) functions upstream of autophagy in RMP1-mediated HR-PCD. Gyceraldehyde-3-phosphate dehydrogenases (GAPDH) is a negative regulator of autophagy. PD, plasmodesmata.
Table 1.
Roles of autophagy (ATG) genes in plant defense and susceptibility.
| Function in autophagy | Gene | Knockdown or knockout phenotype | References |
|---|---|---|---|
|
| |||
| Nucleation of autophagosomes | ATG6 | - Growth defects - Unrestricted HR-PCD during N- and RPM1- mediated resistance - Susceptibility to Pst DC3000 |
[26,27] |
| VPS34 | - Unrestricted HR-PCD during N-mediated resistance | [26] | |
|
| |||
| Delivery of lipids for expansion of autophagosomal membrane | ATG2 | - Autoimmune phenotype - Increased SA signaling and ROS accumulation - Resistance to powdery mildew in Arabidopsis and P. syringae pv. glycinea in soybeans |
[28,37,46] |
| ATG9 | - Suppression of HR-PCD during RPS4- and RPP1- mediated resistance | [33–35] | |
| ATG18 | - Resistance to Pst DC3000 - Susceptibility to B. cinerea and A. brassicicola |
[23,36] | |
|
| |||
| A part of ATG8 conjugation system mediating lipidation of ATG8 and promoting expansion of autophagosomal membrane | ATG3 | - Unrestricted HR-PCD during N-mediated resistance | [26] |
| ATG5 | - Early senescence - Increased SA and ROS accumulation - Suppression of HR-PCD in young plants and unrestricted HR-PCD in older plants during RPM1- mediated resistance - Susceptibility to B. cinerea and A. brassicola |
[23,28,33–37] | |
| ATG7 | - Unrestricted HR-PCD during N-mediated resistance - Suppression of HR-PCD in young plants and unrestricted HR-PCD in older plants during RPM1- mediated resistance - Resistance to powdery mildew - Susceptibility to B. cinerea and A. brassicola |
[23,28,33–37] | |
| ATG10 | - Increased SA accumulation - Resistance to Pst DC3000 and powdery mildew - Susceptibility to A. brassicola |
[36,37] | |
Figure 2. Manipulations of autophagy by phytopathogens.
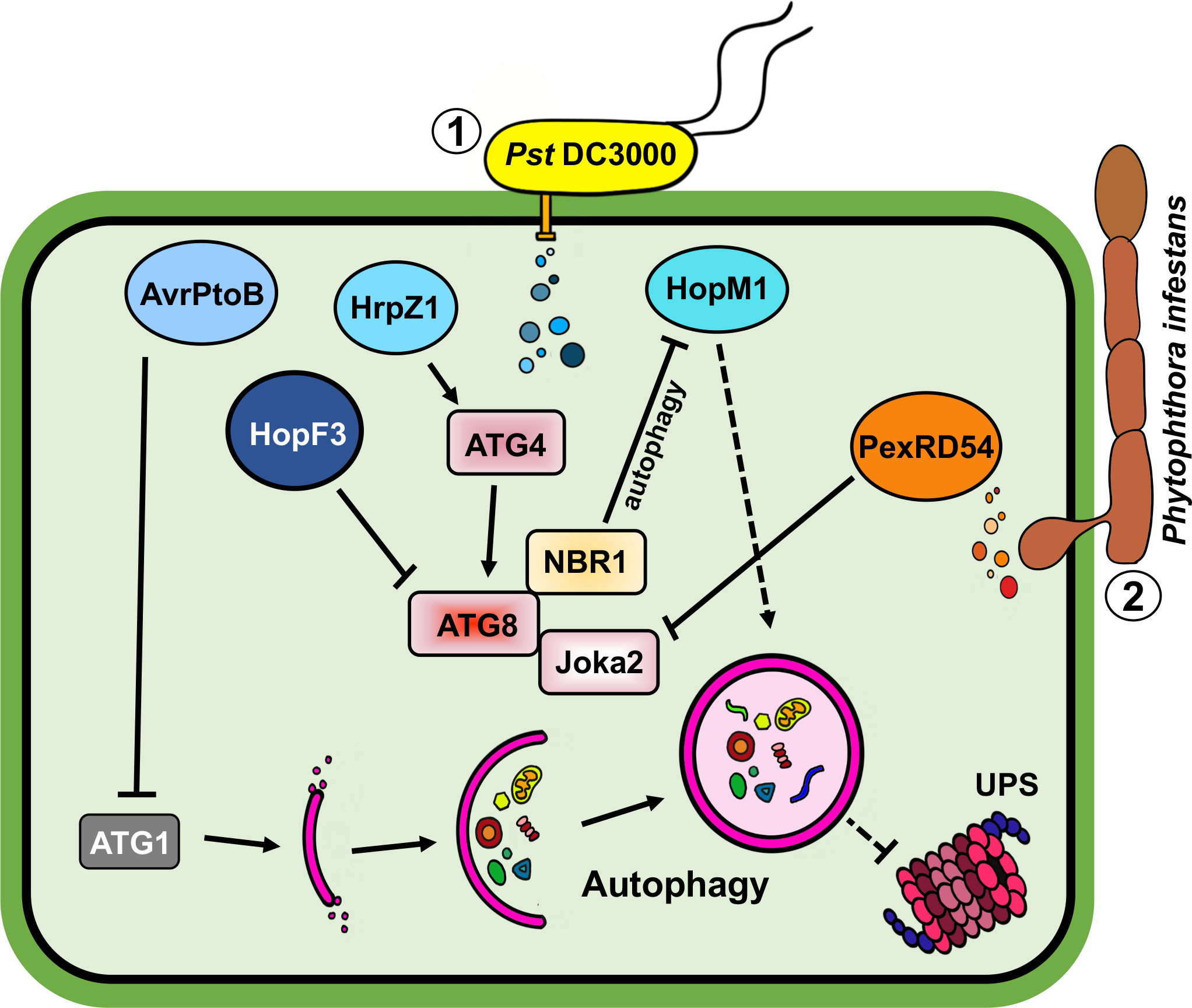
Phytopathogens employ different effectors to promote pathogenicity in host plants. Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) (1) utilizes a set of effectors to enhance virulence. HopF3 and AvrPtoB inhibit autophagy through their interactions with ATG8 and ATG1, respectively, whereas HrpZ1 activates autophagy by promoting ATG4 activity. HopM1 enhances autophagy to mediate degradation of ubiquitin-proteasome system (UPS) in plants, thus compromising host defense (dotted lines). However, the effects of HopM1 are antagonized by the NBR1-mediated selective autophagy. Phytophthora infestans (2) also secretes effectors that disrupt host immunity. PexRD54, for example, specifically and competitively binds to an ATG8 ortholog and prevents ATG8 from interacting with the autophagy cargo receptor Joka2 (NBR1 homolog), which initiates the formation of defense-related autophagosomes during P. infestans infection.
Roles of autophagy in PRR-mediated defense
In Arabidopsis, autophagy regulates levels of FLAGELLIN-SENSING 2 (FLS2), a PRR receptor kinase that recognizes bacterial flagellin and activates PTI [20] through orosomucoid (ORM) proteins [21] (Figure 1). ORMs can act as autophagy receptors, allowing FLS2 to be targeted for autophagic degradation. Both ORM RNAi and CRISPR knockout orm1 and orm2 plants exhibited over-accumulation of FLS2 and hyperactive PTI after infection with Pseudomonas syringae pv. tomato strain DC3000 (Pst DC3000). In contrast, overexpression of ORMs resulted in reduced FLS2 accumulation and enhanced susceptibility to Pst DC3000. Furthermore, overexpression of ORMs in atg7-2 and atg10-1 mutants had no effect on FLS2 accumulation and resulted in resistance to Pst DC3000 compared to that of wild-type plants [21]. Overall, this study shows the negative role of autophagy on FLS2-mediated PTI by mediating autophagic degradation of FLS2. Since ORMs had no effect on other PRR-mediated signaling pathways tested, it will be interesting to learn if autophagy also plays a role in maintaining levels of PRRs levels through other modes of targeting and selective degradation.
BAK1 is a receptor-like kinase (RLK) co-receptor for multiple PRRs, which is crucial for activation of immune signaling [3]. Recently, BAK1 has been shown to negatively regulate ATG18a activities upon Botrytis cinerea infection [22] (Figure 1). ATG18a is essential for host defense against B. cinerea likely through its roles in activating autophagy-mediated degradation and expression of the defense-related transcription factor WRKY33 [23]. BAK1 phosphorylated and suppressed ATG18a activity during resistance against B. cinerea. Loss-of-function in BAK1 revealed low levels of phosphorylated ATG18a and strong induction of autophagy, resulting in enhanced resistance to B. cinerea [22]. Together, this study has discovered a novel connection between PRR-mediated defense and autophagy, in which the immune system modulates autophagy in order to keep the pathogen-induced defense responses in check.
Dual role of autophagy in immunity-induced cell death
Autophagy can play a dual role in the plant immune system, contributing to both pro-cell survival and pro-cell death activities (Figure 1). Evidence suggests that this depends on multiple factors, including types of pathogens, plant age, and the defense mechanisms invoked. Generally, recognition of pathogen-encoded effectors (also known as avirulent [Avr] protein) by a host NLR, triggers the ETI response, leading to lesion of cell death at the infection site and containment of pathogens [1,6]. The restriction of HR-PCD to the infection site is necessary to prevent spread of HR-PCD to neighboring cells and distal tissues.
The tobacco N protein is a TNL that confers resistance to Tobacco mosaic virus (TMV) [24]. In Nicotiana benthamiana plants expressing the N TNL, TMV infection induces HR-PCD and limits TMV to the infection site [25]. However, silencing of plant ortholog of ATG6/Beclin1 that is required for nucleation of autophagosomes [11,12] in N-containing plants resulted in spreading of HR-PCD into surrounding healthy tissue and systemic leaves [26]. Similar results could be seen after silencing other key genes involved with autophagy, such as ATG3, ATG7, and VPS34 [26]. These findings indicate that in autophagy-deficient cells, the pro-death signals that cause HR-PCD are no longer restricted, and therefore support a pro-survival role for immunity-induced autophagy. A similar spreading cell death phenomenon was observed in four-week old Arabidopsis ATG6 RNAi plants when infected with the hemibiotroph Pst DC3000 harboring AvrRpm1 effector (Pst-AvrRpm1) [27]. Additionally, Arabidopsis atg5-1 knockout plants exhibited unrestricted HR-PCD in response to Pst-AvrRpm1 infection [28]. These findings overall suggest that immunity-induced autophagy plays an important pro-survival role by eliminating the pro-death signals associated with HR-PCD (Figure 1 and Table 1).
In mammalian systems, anti-apoptotic B-cell lymphoma 2 (Bcl-2) family members and pro-apoptotic BAX (BCL2-associated X) and BAK (BCL2 antagonist/killer) proteins regulate autophagy and cell death [17]. Plants lack Bcl-2, BAX and BAK homologs but contain the evolutionarily conserved cell death suppressor, Bax inhibitor 1 (BI-1) like protein [29]. Plant BI-1 interacts with ATG6 and this interaction is required for induction of autophagy during N TNL-mediated resistance to TMV [30]. Silencing of BI-1 resulted in increased accumulation of TMV-GFP and enhanced cell death, indicating that BI-1 is required for induction of autophagy to negatively regulate cell death. Contrary to the cell death suppressing role of BI-1, overexpression of BI-1 induced cell death in plants and BI-1 induced cell death requires autophagy. These findings provide evidence for both the death promoting and inhibiting role of plant BI-1. Although how BI-1 shifts between these functions remains elusive, it is likely that autophagy, which is also modulated by BI-1, plays a pivotal role in this process.
Cytoplasmic gyceraldehyde-3-phosphate dehydrogenases (GAPDH) has also been proven to modulate autophagy in plants [31]. In N. benthamiana, GAPDH acts as a suppressor of autophagy and its function could be carried out by its interaction with ATG3 (Figure 1). Additionally, silencing of GAPDH led to enhanced HR-PCD during N-TMV interaction and also increased resistance to the virulent Pst DC3000 and P. syringae pv. tabaci [31]. Similarly, Arabidopsis GAPDH knockout plants accumulated increased levels of ROS and exhibited constitutive autophagy. The enhanced HR-PCD in response to Pst-AvrRpt2 and basal resistance against Pst DC3000 infection were also observed in the mutant plants [32]. Together, GAPDH can function as a negative regulator of immunity-mediated cell death and basal resistance, which could link to its inhibitory role on plant autophagy.
Plant autophagy can also operate in a pro-cell death manner during some plant-pathogen interactions (Figure 1). The Arabidopsis RPS4 and RPP1 TNLs recognize Pst DC3000 harboring the AvrRps4 effector (Pst-AvrRps4), and the AvrAtr1 effector of the oomycete Hyaloperonospora arabidopsidis, respectively. The Arabidopsis RPM1 and RPS2 CNLs recognize Pst-AvrRpm1 and Pst-AvrRpt2, respectively. Successful recognition of these effectors induces HR-PCD; however, HR-PCD becomes inhibited in atg7-1 and atg9-1 mutants after infection with Pst-AvrRps4 and H. arabidopsidis race Noco2, as measured by an electrolyte leakage assay [33]. This pro-death function of autophagy may possess a level of specificity, since very little reduction of electrolyte leakage was observed in atg7-1 and atg9-1 mutants after infection with Pst-AvrRpm1 or in atg5-1 and atg7-2 mutants after infection with Pst-AvrRpt2 [33]. However, in a single-cell death assay, HR-PCD induced during Pst-AvrRpm1 was suppressed in two-week old atg5-1 and atg18a mutant plants [34]. Furthermore, catalase, an antioxidant enzyme, seems to function upstream of autophagy in Pst-AvrRpm1-induced cell death [35]. Although cell death was compromised in the cases described above, there was no effect on the bacterial growth in atg2, atg5-1, atg7-1 or atg18a mutants compared to wild-type plants [33–35]. Together, these findings provide support for the role of autophagy in cell death triggered by certain NLRs when young plants are challenged with pathogens.
Dual role of autophagy during disease-associated cell death
The disease-associated cell death generally refers to necrotic cell death that is induced by necrotrophic pathogens such as B. cinerea as a result of host susceptibility. In addition to its pro-survival role in immunity-induced cell death, autophagy can play a role in the regulation of disease-associated cell death (Table 1). During infection with virulent Pst DC3000, Arabidopsis ATG6 RNAi lines displayed unrestricted spread of disease-induced cell death [27]. Arabidopsis atg5-1, atg10-1 and atg18a-1 and ATG18a RNAi (atg18a-2) plants exhibited spread of disease-associated cell death and enhanced susceptibility upon infection with necrotrophic fungi Alternaria brassicicola [36]. Similarly, Arabidopsis atg5-1, atg7-2, atg7-3, and atg18a-1 and atg18a-2 lines showed increased disease-associated cell death and susceptibility to the necrotrophic fungi B. cinerea [23]. Together, these studies support a positive role for autophagy in necrotrophic pathogen defense.
In contrast, atg2-2 plants displayed enhanced disease resistance to the powdery mildew Golovinomyces cichoracearum, an obligate biotrophic fungi [37]. Consistent with the enhanced resistance phenotype, atg2-2 plants had increased expression of defense-related genes, including PR1, PR2, and PR5, and increased levels of SA and ROS. Additionally, other atg mutants such as atg5-1, atg7-1, and atg10-1 also acquired enhanced G. cichoracearum resistance, similar to that of atg2-2. These findings suggest that autophagy additionally plays a negative role in resistance against this obligate biotroph.
Bcl-2-associated athanogene (BAG) family members have been known to play an important role in cell death regulation [38]. In Arabidopsis, BAG6 plays a role in disease-associated cell death in response to B. cineria infection [38,39]. Wild-type Col-0 Arabidopsis plants generally induce symptomatic cell death at the site of B. cineria inoculation. However, in Arabidopsis bag6 mutants, cell death rapidly spreads beyond the inoculation site and promotes enhanced susceptibility to B. cineria [39]. Interestingly, the ability of BAG6 to confer immunity to B. cineria requires cleavage of BAG6 by aspartyl protease APCB1 (Aspartyl Protease Cleaving BAG) [39]. Overexpression of a cleavage-resistant mutant of BAG6 in bag6 mutant failed to rescue resistance against B. cinerea. Both infection of plants by B. cinerea and expression of cleaved BAG6 can induce autophagy that is crucial for immune activation and autophagic cell death to limit B. cineria to the infection site [39]. These studies highlight functions of BAG6 as a positive regulator of plant immunity through its ability to modulate host autophagy and subsequently pathogen-induced cell death.
The role of autophagy in SA and ROS modulation
SA and ROS are pro-defense compounds in plant that are tightly controlled [6]. In plants, SA is crucial defense signaling hormone, and pathogen perception can trigger SA biosynthesis and accumulation. While ROS is induced upon pathogen recognition and is critical for defense signaling, uncontrolled ROS accumulation could have detrimental effects, including cellular damage. Both SA and ROS have been linked to the formation and regulation of HR-PCD. Autophagy has been shown to negatively regulate SA and ROS accumulation [28]. During infection with Pst DC3000, Arabidopsis atg5-1 plants accumulated three-fold higher SA compared to wild-type plants. Consistent with this, expression of the SA-responsive genes PR1 and PR2 were elevated in atg2-1 and atg5-1 plants. Furthermore, these atg mutants accumulated higher levels of hydrogen peroxide (H2O2). The spreading cell death phenotype observed in atg5-1 in response to Pst-AvrRpm1 was suppressed when SA-related pathways were inactivated in the atg5-1 plants. These findings indicate that over-accumulated SA and ROS may play a role in pathogen-induced cell death spread in the absence of autophagy [28].
Similar results were found when analyzing immunity in atg2-2 plants in response to powdery mildew G. cichoracearum [37]. Besides the enhanced resistance to the fungal pathogen, both atg2-1 and atg2-2 displayed autoimmune phenotypes, designated by stunted growth, early senescence and spontaneous cell death. Further analysis revealed that the key defense-related factors such as PR1, PR2, PR5 and ROS were upregulated in the atg2-2 mutants [37], which was consistent with the previous report of atg2-1 [28]. Although hyperaccumulation of SA in both atg2-1 and atg2-2 was not reported in both studies, the upregulation of SA responsive genes might imply high levels of SA in the mutant plants. Additionally, inactivation of SA signaling in the atg2-2 background suppressed the autoimmune phenotypes as well as the powdery mildew resistance based on the analysis of fungal growth [37]. Since SA and ROS are essential for regulation of both senescence and immunity in plants [40], the increase in SA, ROS and PR genes likely explain the autoimmune phenotype observed on the atg2 mutants and also enhanced resistance to pathogen infection. However, many other atg mutants are normal and they induce cell death similar to wild-type plants but the cell death spreads. This suggests a role for autophagy in eliminating the SA and ROS signals after host-induced HR-PCD, and the SA-ROS amplification signaling that mediates HR-PCD could be a target of active downregulation by autophagy.
Despite coordination of SA and ROS accumulation in Arabidopsis atg mutants during host-microbe interactions, Lenz et al., [36] further explored this relationship across multiple types of pathogens. Arabidopsis atg5-1, atg10-1, atg18a-1 and atg18a-2 were found to be more resistant to Pst DC3000 infection compared to wild-type plants. Phytohormone quantification in some of these atg mutants revealed that atg5-1 and atg10-1 accumulated two-fold higher amounts of SA during the infection of Pst DC3000 than the wild-type plants, while wild-type levels of PTI responses were still found in these mutants. This report was consistent with a previous study [28], supporting the negative regulatory roles of autophagy on SA-dependent defense response to biotrophic pathogens. However, these mutants responded differently to the necrotrophic pathogen A. brassicicola. During infection with A. brassicicola, the atg mutants used previously for Pst DC3000 disease assays exhibited significantly enlarged necrotic lesions without an increase in senescence molecular markers. Surprisingly, ROS accumulation, which is often associated with cell death phenotype, was not altered in the atg mutants in comparison to the wild-type plants [36]. The results show that the misregulation of host autophagy leads to the increased vulnerability of these mutants to the necrotrophic pathogen without significant alterations in ROS production. It should be noted that an interplay exists between SA and the phytohormone jasmonic acid (JA), where JA often mediates resistance to necrotrophic pathogens and antagonizes SA-mediated resistance to biotrophic pathogens. However, there were no significant changes in levels of JA and PDF1.2b expression, a JA-responsive gene, between the wild-type and atg mutants upon A. brassicicola infection [36]. Overall, this study emphasizes the dynamic roles of autophagy in modulating two primary defense compounds in plants, SA and ROS, and the regulatory functions of autophagy depend on specific lifestyles of pathogens and their interactions with host plants.
Microbial manipulation of autophagy
Multiple effectors from diverse pathogens appear to target the autophagy pathway and molecular machinery to promote pathogenesis, suggesting a fundamental role for autophagy in the determination of infection outcomes and the pathogen-plant arms race. Here, effectors from bacterial and oomycete pathogens are discussed (Figure 2).
The success of plant bacterial infection relies on the pathogens ability to subvert host immunity. While multiple bacterial effectors and their targeted biological pathways in plants have been extensively studied, effectors capable of manipulating plant autophagy are only recently being identified. To promote virulence, type 3 effectors from Pst DC3000 inhibit the ubiquitin-proteasome system (UPS), which is a major protein degradation pathway in eukaryotes [41] (Figure 2). However, Pst effectors failed to inhibit UPS in atg5-1 knockout mutant suggesting the importance of the pathogen-induced autophagy in interfering with plant UPS. Moreover, HopM1 was identified as an effector that activates host autophagy [42]. However, NBR1, an autophagic cargo receptor, antagonized the HopM1-induced water-soaked lesions and bacterial growth [42]. Collectively, Pst DC3000 facilitates host autophagy through the HopM1 effector to mediate degradation of UPS, which promotes bacterial growth and infection. Meanwhile, plants combat the effects of HopM1 by NBR1-mediated selective autophagy utilizing target proteins that have yet to be identified.
The HrpZ1 effector of Pst DC3000 also activates plant autophagy and supports disease development [43] (Figure 2). HrpZ1 interacts with multiple Arabidopsis ATG8 isoforms both in vitro and in vivo, insinuating that HrpZ1 might function to manipulate the host autophagy pathway. Further functional analysis revealed that HrpZ1 enhanced autophagy through increasing the activity of ATG4b protease to processes ATG8 at the conserved C-terminal glycine residue, which is a vital step in autophagosome biogenesis [43].
In addition to inducing autophagy, some bacterial effectors have been shown to inhibit autophagy as a strategy to promote bacterial virulence. HopF3 is a Pseudomonas effector that interacts directly and selectively with a subset of Arabidopsis ATG8s. Unlike HrpZ1, HopF3 attenuates autophagy (Figure 2). Expression of HopF3 in Arabidopsis atg5-1 mutant diminished the enhanced Pst DC3000 virulence observed in wild-type plants, further suggesting that host autophagy is required for HopF3-mediated virulence [43]. The AvrPtoB effector of Pst DC3000 has also been shown to suppress autophagy similarly to HopF3. However, instead of targeting ATG8s, AvrPtoB interacts with ATG1 kinase, a key initiator of the autophagy process (Figure 2). Strong interaction was detected between AvrPtoB and the microtubule interaction and transportation (MIT) domain of ATG1, and this interaction depended on the previously uncharacterized N-terminal domain of AvrPtoB. Biochemical assay showed that AvrPtoB inhibited phosphorylation of ATG1 which limits autophagy but promotes bacterial virulence [43].
Like bacteria, oomycete pathogens also employ effector proteins to impede host immunity (Figure 2). The oomycete effector PexRD54 from Phytophthora infestans contains two predicted ATG8 interacting motifs (AIM) [44]. One of these AIMs and the host small GTPase Rab8a, a key player in the vesicle trafficking pathway, were necessary for the interaction between PexRD54 and ATG8CL. This interaction allows the effector to be loaded into autophagosomes, eventually perturbing the interaction between ATG8CL and the autophagy cargo receptor Joka2 in tobacco, which is an NBR1 homolog. This ultimately confers defense against P. infestans infection [44,45].
Considering the evolutionary arms race between plants and pathogens, it is not surprising that pathogens can manipulate or hijack the autophagy pathway to promote pathogenesis. This microbial manipulation is often executed using microbial effectors. While some effector proteins induce autophagy, others function to suppress it. Additionally, some effectors compete to interact with host autophagy components without altering autophagic flux. Despite differences in modes of function, enhanced pathogen virulence remains a shared goal. Nevertheless, only a few effectors have been shown to specifically interfere with autophagy. Several effectors from pathogens across different kingdoms were found to interact with ATG proteins [43]. However, how they function to modulate autophagy remains unclear. Understanding their functions and how plants counteract these effectors would provide a more complete picture of the interplays between autophagy and plant immunity.
Conclusions and perspectives
Autophagy is a vital recycling pathway responding to stresses, especially nutrient deprivation. Impairment of autophagy causes abnormality in eukaryotic organisms both developmentally and physiologically. There is growing evidence supporting a connection between autophagy and immunity against pathogens in plants. Indeed, many studies have shown that defects in different ATG genes affect how plants interact with pathogens both via the PTI and the ETI branches of host defense. In terms of PTI and basal resistance, plant lines with either loss-of-function mutations or gene silencing of different ATG genes revealed both enhanced and dampened host resistance to different types of virulent pathogens. The alterations of host defense were found to be linked to changes in hallmarks of basal resistance and homeostasis of immune receptors. In relation to ETI, autophagy is required for proper regulation of HR-PCD. Pro-death and pro-survival roles of autophagy in ETI-mediated PCD have been found upon pathogen infection. It should however be noted that different types of pathogens and plant atg mutant genotypes certainly contribute to the discrepancies observed in both PTI and ETI studies, which may suggest potential roles for individual ATGs in other biological pathways intertwined with autophagy. A plethora of host factors involved in the route from pathogen perception to induction of autophagy and HR-PCD have been established. However, understanding their exact roles and mechanisms of action within this pathway, as well as whether or not other unknown players exist, will require further research that piece together the puzzle. Despite some seemingly contradictions, pathogens have found ways to promote their virulence by manipulating plant autophagy. Overall, the dualistic role of autophagy in plant immunity emphasizes how intricate this relationship is and how much remains to be explored in this field.
Acknowledgments
The immunity and autophagy work in SPDK laboratory is supported by US National Institute of Health grant R01GM132582 and was supported by NSF-IOS-1354434 and NSF-MCB-1549580.
Abbreviations:
- AIM
ATG8 interacting motifs
- APCB1
Aspartyl Protease Cleaving BAG
- ATG
autophagy-related genes or proteins
- BAG
Bcl-2-associated athanogene
- BAK
BCL2 antagonist/killer
- BAX
BCL2-associated X
- Bcl-2
B-cell lymphoma 2
- BI-1
Bax inhibitor 1
- CC
coiled-coil
- CNLs
coiled-coil (CC) domain containing NLRs
- ETI
effector-triggered immunity
- FLS2
FLAGELLIN-SENSING 2
- GAPDH
gyceraldehyde-3-phosphate dehydrogenases
- H2O2
hydrogen peroxide
- HR-PCD
hypersensitive response-programmed cell death
- MIT
microtubule interaction and transportation
- NLR
nucleotide-binding leucine-rich repeat
- ORM
orosomucoid
- PRRs
pattern recognition receptors
- Pst DC3000
Pseudomonas syringae pv. tomato strain DC3000
- PTI
pattern triggered immunity
- RLK
receptor-like kinase
- ROS
reactive oxygen species
- SA
salicylic acid
- TIR
Toll-interleukin-1 receptor homology
- TMV
Tobacco mosaic virus
- TNLs
Toll-interleukin-1 receptor homology (TIR) domain containing NLRs
- UPS
ubiquitin-proteasome system
References
- [1].Jones JD and Dangl JL (2006). The plant immune system. Nature 444, 323–9. [DOI] [PubMed] [Google Scholar]
- [2].Dangl JL, Horvath DM and Staskawicz BJ (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].DeFalco TA and Zipfel C (2021). Molecular mechanisms of early plant pattern-triggered immune signaling. Mol Cell 81, 4346. [DOI] [PubMed] [Google Scholar]
- [4].Ngou BPM, Ding P and Jones JD (2022). Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell online: 10.1093/plcell/koac041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Toruno TY, Stergiopoulos I and Coaker G (2016). Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol 54, 419–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang X and Dong X (2022). Life-or-death decisions in plant immunity. Curr Opin Immunol 75, 102169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yuan M et al. (2021). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ngou BPM, Ahn HK, Ding P and Jones JDG (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. [DOI] [PubMed] [Google Scholar]
- [9].Pruitt RN et al. (2021). The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature 598, 495–499. [DOI] [PubMed] [Google Scholar]
- [10].Tian H et al. (2021). Activation of TIR signalling boosts pattern-triggered immunity. Nature 598, 500–503. [DOI] [PubMed] [Google Scholar]
- [11].Yin Z, Pascual C and Klionsky DJ (2016). Autophagy: machinery and regulation. Microb Cell 3, 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Morishita H and Mizushima N (2019). Diverse cellular roles of autophagy. Annu Rev Cell Dev Biol 35, 453–475. [DOI] [PubMed] [Google Scholar]
- [13].Hu Y and Reggiori F (2022). Molecular regulation of autophagosome formation. Biochem Soc Trans 50, 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Marshall RS and Vierstra RD (2018). Autophagy: The master of bulk and selective recycling. Annu Rev Plant Biol 69, 173–208. [DOI] [PubMed] [Google Scholar]
- [15].Gubas A and Dikic I (2022). A guide to the regulation of selective autophagy receptors. FEBS J 289, 75–89. [DOI] [PubMed] [Google Scholar]
- [16].Tang J and Bassham DC (2018). Autophagy in crop plants: what’s new beyond Arabidopsis? Open Biol 8, 180162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Levine B and Kroemer G (2019). Biological functions of autophagy genes: A disease perspective. Cell 176, 11–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Su T, Li X, Yang M, Shao Q, Zhao Y, Ma C and Wang P (2020). Autophagy: An intracellular degradation pathway regulating plant survival and stress response. Front Plant Sci 11, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yang M and Liu Y (2022). Autophagy in plant viral infection. FEBS Lett 10.1002/1873-3468.14349 [DOI] [PubMed] [Google Scholar]
- [20].Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G and Boller T (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–7. [DOI] [PubMed] [Google Scholar]
- [21].Yang F, Kimberlin AN, Elowsky CG, Liu Y, Gonzalez-Solis A, Cahoon EB and Alfano JR (2019). A plant immune receptor degraded by selective autophagy. Mol Plant 12, 113–123. [DOI] [PubMed] [Google Scholar]
- [22].Zhang B, Shao L, Wang J, Zhang Y, Guo X, Peng Y, Cao Y and Lai Z (2021). Phosphorylation of ATG18a by BAK1 suppresses autophagy and attenuates plant resistance against necrotrophic pathogens. Autophagy 17, 2093–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lai Z, Wang F, Zheng Z, Fan B and Chen Z (2011). A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J 66, 953–68. [DOI] [PubMed] [Google Scholar]
- [24].Whitham S, Dineshkumar SP, Choi D, Hehl R, Corr C and Baker B (1994). The product of the Tobacco mosaic-virus resistance gene-N - Similarity to Toll and the interleukin-1 receptor. Cell 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- [25].Liu Y, Schiff M, Marathe R and Dinesh-Kumar SP (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30, 415–29. [DOI] [PubMed] [Google Scholar]
- [26].Liu Y, Schiff M, Czymmek K, Talloczy Z, Levine B and Dinesh-Kumar SP (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–77. [DOI] [PubMed] [Google Scholar]
- [27].Patel S and Dinesh-Kumar SP (2008). Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4, 20–7. [DOI] [PubMed] [Google Scholar]
- [28].Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y and Shirasu K (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Henke N, Lisak DA, Schneider L, Habicht J, Pergande M and Methner A (2011). The ancient cell death suppressor BAX inhibitor-1. Cell Calcium 50, 251–60. [DOI] [PubMed] [Google Scholar]
- [30].Xu G, Wang S, Han S, Xie K, Wang Y, Li J and Liu Y (2017). Plant Bax Inhibitor-1 interacts with ATG6 to regulate autophagy and programmed cell death. Autophagy 13, 1161–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Han S, Wang Y, Zheng X, Jia Q, Zhao J, Bai F, Hong Y and Liu Y (2015). Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 27, 1316–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Henry E, Fung N, Liu J, Drakakaki G and Coaker G (2015). Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet 11, e1005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hofius D et al. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137, 773–83. [DOI] [PubMed] [Google Scholar]
- [34].Coll NS, Smidler A, Puigvert M, Popa C, Valls M and Dangl JL (2014). The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ 21, 1399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hackenberg T et al. (2013). Catalase and NO CATALASE ACTIVITY1 promote autophagy-dependent cell death in Arabidopsis. Plant Cell 25, 4616–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lenz HD et al. (2011). Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J 66, 818–30. [DOI] [PubMed] [Google Scholar]
- [37].Wang Y, Nishimura MT, Zhao T and Tang D (2011). ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J 68, 74–87. [DOI] [PubMed] [Google Scholar]
- [38].Thanthrige N, Jain S, Bhowmik SD, Ferguson BJ, Kabbage M, Mundree S and Williams B (2020). Centrality of BAGs in plant PCD, stress responses, and host defense. Trends Plant Sci 25, 1131–1140. [DOI] [PubMed] [Google Scholar]
- [39].Li Y, Kabbage M, Liu W and Dickman MB (2016). Aspartyl protease-mediated cleavage of BAG6 is necessary for autophagy and fungal resistance in plants. Plant Cell 28, 233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhang Y, Wang HL, Li Z and Guo H (2020). Genetic network between leaf senescence and plant immunity: Crucial regulatory nodes and new insights. Plants (Basel) 9, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ustun S, Sheikh A, Gimenez-Ibanez S, Jones A, Ntoukakis V and Bornke F (2016). The proteasome acts as a hub for plant immunity and is targeted by Pseudomonas type III effectors. Plant Physiol 172, 1941–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ustun S, Hafren A, Liu Q, Marshall RS, Minina EA, Bozhkov PV, Vierstra RD and Hofius D (2018). Bacteria exploit autophagy for proteasome degradation and enhanced virulence in plants. Plant Cell 30, 668–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lal NK, Thanasuwat B, Huang PJ, Cavanaugh KA, Carter A, Michelmore RW and Dinesh-Kumar SP (2020). Phytopathogen effectors use multiple mechanisms to manipulate plant autophagy. Cell Host Microbe 28, 558–571 e6. [DOI] [PubMed] [Google Scholar]
- [44].Dagdas YF et al. (2016). An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife 5, e10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pandey P et al. (2021). An oomycete effector subverts host vesicle trafficking to channel starvation-induced autophagy to the pathogen interface. eLife 10, e65285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hashimi SM, Wu NN, Ran J and Liu JZ (2021). Silencing Autophagy-Related Gene 2 (ATG2) results in acelerated senescence and enhanced immunity in soybean. Int J Mol Sci 22 [DOI] [PMC free article] [PubMed] [Google Scholar]


