Abstract
N6-methyladenosine (m6A) modification regulates RNA translation, splicing, transport, localization, and stability at the post-transcriptional level. The m6A modification has been reported to have a wide range of effects on the nervous system, including neurogenesis, cerebellar development, learning, cognition, and memory, as well as the occurrence and development of neurological disorders. In this review, we aim to summarize the findings on the role and regulatory mechanism of m6A modification in the nervous system, to reveal the molecular mechanisms of neurodevelopmental processes, and to promote targeted therapy for nervous system-related diseases.
Keywords: m6A methylation, nervous system, development, neurological disorders, mechanism
Introduction
Epigenetic modification is the regulation of gene expression through a series of complex modifications without changing the gene sequence, and mainly includes DNA modification, RNA modification, histone modification, and chromatin remodeling (Jones et al., 2016). Among these, RNA modification is the most common. Different types of RNA modifications have been identified, including N6-methyladenosine (m6A) (Dominissini et al., 2012), 5-methylcytidine (m5c) (Edelheit et al., 2013) and N1-methyladenosine modification (m1A) (Dominissini et al., 2016; Li and Liu, 2016). Among these, m6A is the most abundant and reversible chemical modification of eukaryotic mRNA (Liu and Pan, 2016). An increasing number of studies have shown that m6A plays an important role in RNA stability, mRNA translation, alternative splicing, and subcellular RNA localization (Zhao et al., 2017; Yang et al., 2018; Shi et al., 2019).
The m6A modification and dynamic changes are regulated by the cooperation of m6A methyltransferase (writer), m6A demethylase (eraser), and m6A methylation binding protein (reader). Multiple studies have shown that m6A modification is highly expressed in the brain tissue and plays a critical role in the functions of the nervous system. Therefore, in this review, we mainly focus on the function and regulatory mechanisms of m6A methylation in the nervous system.
m6A modification-related proteins
A variety of RNA methyltransferases and demethylases as well as various m6A-binding proteins are present in the cells. Different types of RNAs undergo dynamic changes in methylation and demethylation, which, in turn, participate in the regulation of various physiological processes. With the development of molecular biology detection technology, an increasing number of enzymes and regulatory proteins related to m6A modifications have been discovered. Moreover, their roles in the development, homeostasis, and pathological state of the nervous system have been revealed (Figures 1, 2; Table 1).
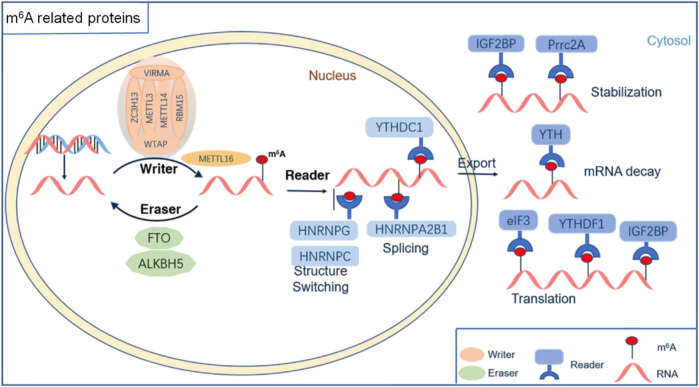
FIGURE 1.
The molecular mechanism of RNA m6A modification. m6A methylation was catalyzed by the writer complex, including METTL3, METTL14, METTL16, WTAP, KIAA1429, RBM15/15B, and ZC3H13. The m6A modification was erased by demethylases, including FTO and ALKBH5. The m6A-modified RNA reader proteins included YTHDF1/2/3, YTHDC1/2, IGF2BP1/2/3, HNRNPC/G, HNRNPA2B1, Prrc2A, and eIF3.
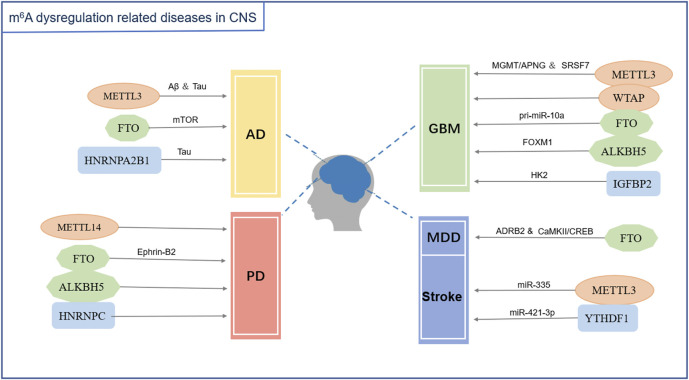
FIGURE 2.
m6A modification dysfunction related diseases in the central nervous system. Abbreviations: AD (Alzheimer’s disease), PD (Parkinson’s disease), GBM (Glioblastoma), MDD (Major depressive disorder), Aβ (amyloid-β), Tau (Tau protein), mTOR (Mammalian target of rapamycin signaling pathway), Ephrin-B2 (erythropoietin-producing hepatocyte receptor-interacting-B2), MGMT (O6-methylguanine-DNA methyltransferase), APNG (alkylpurine-DNA-N-glycosylase), SRSF7 (Serine/arginine-rich splicing factor 7), pri-miR-10a (primary microRNA-10a), FOXM1 (transcription factor FOXM1), HK2 (Hexokinase 2), ADRB2 (Adrenoceptor beta 2), CaMKII (Calcium-calmodulin-dependent protein kinase II), CREB (cAMP response element-binding protein), miR-335 (microRNA-335), miR-421-3P (microRNA-421-3p).
TABLE 1.
Functions of m6A modification-related proteins.
| Type | Genetic name | Function | References |
|---|---|---|---|
| m 6 A writer | METTL3 | Identify the conservative base sequence of m6A, catalytic methylation modification | Geula et al. (2015) |
| METTL14 | Assist METTL3 to catalytic methylation modifications | Wang et al. (2016) | |
| WTAP | Promote METTL3 and METTL14 heterogeneous dilate | Agarwala et al. (2012) | |
| VIRMA | Special modification of the 3′UTR region | Yue et al. (2018) | |
| RBM15 | Combined with the m6A complex and recruit it to a special RNA site | Patil et al. (2016) | |
| ZC3H13 | Connect to WTAP to mRNA binding factor Nito | Wen et al. (2018) | |
| METTL16 | Catalytic methylation modification | Pendleton et al. (2017) | |
| m 6 A eraser | FTO | Delete methylation modification | Jia et al. (2011) |
| ALKBH5 | Delete methylation modification | Zheng et al. (2013) | |
| m 6 A reader | YTHDC1 | Recruit splicing factors to splicing target mRNAs, promoting mRNA output degradation and specific transcription | (Xiao et al., 2016; Roundtree et al., 2017; Shima et al., 2017) |
| YTHDC2 | Adjust the stability and translation of mRNA | Hsu et al. (2017) | |
| YTHDF1 | Recruit translation start factor to promote the translation of mRNA | Wang et al. (2015) | |
| YTHDF2 | Combined with m6A modifiers to recruit CCR4-NOT deadenylase complex acceleration destination mRNA degradation | Du et al. (2016) | |
| YTHDF3 | Promote mRNA degradation or medium translation | (Li et al., 2017a; Shi et al., 2017) | |
| HNRNPA2B1 | Promote primary microRNA processing | Wu et al. (2018) | |
| HNRNPC/G | Mediate mRNA splicing | (Liu et al., 2015; Wang et al., 2015) | |
| IGF2BP1/2/3 | Improve the stability of mRNA | Huang et al. (2018) | |
| eIF3 | Combined with m6A to start translation | Meyer et al. (2015) | |
| FMRP | Adjust the stability, translation and nuclear output of mRNA | Santoro et al. (2012) | |
| Prrc2A | Improve the stability of mRNA | Wu et al. (2019) |
m6A methyltransferase
m6A methylation in mRNA is generally completed by two methyltransferases: METTL3/METTL14 and Wilms’ tumor 1-associating protein (WTAP) (Liu et al., 2014; Ping et al., 2014). METTL14 can assist METTL3 with catalytic methylation modifications (Wang et al., 2016) and WTAP promotes the formation of the WTAP/METTL3/METTL14 complex (Agarwala et al., 2012). Other constituent proteins of the complex include virus-like m6A methyltransferase-related protein (KIAA1429/VIRMA) (Yue et al., 2018), zinc finger CCCH domain-containing protein 13 (ZC3H13) (Wen et al., 2018), RNA-binding motif protein 15 and its paralog RBM15B (RBM15/15B) (Patil et al., 2016). Another methyltransferase is METTL16 (Pendleton et al., 2017).
METTL3 is the first discovered m6A methyltransferase component and acts as a core component of the methyltransferase complex. METTL3 is an S-adenosylmethionine-dependent methyltransferase that specifically recognizes the conserved m6A motif RRACH (R = A or G; H = A, C, or U) (Narayan and Rottman, 1988; Harper et al., 1990). METTL14 is a homologous protein of METTL3, which can form a dimer with METTL3 in a ratio of 1:1 (Liu et al., 2014). Knockout of METTL3 depletes m6A modifications in mRNA, which have been shown to cause early embryonic lethality in mice (Geula et al., 2015). Similar to METTL3, deletion of METTL14 leads to an equivalent reduction in m6A modifications in mRNA, and exhibits resistance to differentiation as seen in METTL3 knockout ESCs (Geula et al., 2015), suggesting a critical role of m6A modification in vivo (Liu et al., 2014). The depletion of WTAP significantly reduces the abundance of m6A in cellular mRNA, and its absence strongly reduces the binding ability of METTL3 to other components of the methyltransferase complex, suggesting that WTAP might target RNA by recruiting m6A methyltransferase complex (Ping et al., 2014).
An increasing number of proteins that interact with the core methyltransferase complex have been identified. VIRMA, a new subunit of the m6A methyltransferase complex, was found to regulate m6A methylation in the region near the 3′UTR and stop codon and is thus associated with alternative polyadenylation (Yue et al., 2018). RNA binding motif protein 15 (RBM15) and its paralog RBM15B can bind to the m6A methyltransferase complex and recruit it to specific sites in RNA. Knockdown of RBM15 and RBM15B impairs gene silencing mediated by X-inactive specific transcript (XIST, a long non-coding RNA that mediates the silencing of genes on the X chromosome) (Patil et al., 2016). In addition, zinc finger CCCH domain-containing protein 13 (ZC3H13) has been reported to be essential for methyltransferase complex localization. Knockdown of ZC3H13 in mouse embryonic stem cells was shown to significantly decrease global m6A mRNA levels (Wen et al., 2018). METTL16 is another indispensable methyltransferase, which regulates U6 small nuclear RNA (snRNA) methyltransferase and adenosyl transferase 2A levels (Pendleton et al., 2017; Doxtader et al., 2018). Unlike the METTL3 preferred m6A modification motif RRACH, METTL16 uses a structured RNA carrying a specific nonameric sequence of UACAGAGAA (Mendel et al., 2018).
m6A demethylase enzyme
m6A modification is a dynamic, reversible process. Demethylases, including fat mass and obesity-associated protein (FTO) and alkylation repair homologue 5 (ALKBH5), both belong to the non-heme Fe (II)- and α-KG-dependent dioxygenase ALKB dioxygenase family, and are responsible for eliminating m6A methylation modification in RNA (Jia et al., 2011; Zheng et al., 2013).
FTO is the first discovered m6A demethylase. It was originally recognized as a demethylase that mediates demethylation of 3-methylthymine in single-stranded DNA (Gerken et al., 2007) and 3-methyluracil in single-stranded RNA (Jia et al., 2008). Overexpression of FTO significantly decreases m6A content, whereas FTO knockdown increases m6A mRNA levels (Jia et al., 2011).
ALKBH5 affects mRNA export and RNA metabolism, as well as the assembly of mRNA processing factors in nuclear speckles. ALKBH5 deficiency increases m6A mRNA levels and impairs fertility in mice (Zheng et al., 2013). In addition to FTO and ALKBH5, additional m6A demethylases remain to be discovered.
m6A binding proteins
The functioning of m6A methylation requires the recognition and binding of specific RNA-binding proteins. At present, the known m6A-binding proteins in mammals include YTH N6-Methyladenosine RNA Binding Protein 1 (YTHDF1), YTHDF2, YTHDF3, YTH domain containing 1 (YTHDC1), and YTHDC2 in the cytoplasm, heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNPA2B1), Heterogeneous Nuclear Ribonucleoprotein C (HNRNPC), and Heterogeneous Nuclear Ribonucleoprotein G (HNRNPG) in the nucleus.
YTHDF1 is a cytoplasmic m6A-binding protein that does not have a critical effect on the stability of mRNA, but mainly promotes the translation of target mRNA by recruiting translation initiation factors (Wang et al., 2015). YTHDF2 is an m6A-binding protein that mediates mRNA translation, thereby affecting RNA half-life, accelerating RNA degradation, and mRNA stability (Wang et al., 2014). Upon binding to the m6A modification site, YTHDF2 accelerates target mRNA degradation by recruiting the CCR4-NOT deadenylase complex (Du et al., 2016). YTHDF3 either interacts with YTHDF2 to promote mRNA degradation or interacts with YTHDF1 to promote translation (Li et al., 2017a; Shi et al., 2017). Moreover, YTHDC2 is a cytoplasmic m6A-binding protein that regulates mRNA stability and translation and regulates spermatogenesis (Hsu et al., 2017; Wojtas et al., 2017). YTHDC1 is a nuclear m6A-binding protein that promotes exon inclusion in targeted mRNAs by recruiting the pre-mRNA splicing factor Ser/Arg-rich splicing factor 3 (SRSF3) (Xiao et al., 2016). YTHDC1 knockdown results in the accumulation of transcripts in the nucleus and subsequent depletion in the cytoplasm, suggesting a critical role in the metabolism of mRNAs (Roundtree et al., 2017). Interestingly, it has been reported that cytoplasmic YTHDC1 is involved in the mRNA maturation process, involving MAT2A (Shima et al., 2017).
Moreover, HNRNPA2B1, an m6A reader in the nucleus, has been shown to recognize and bind m6A-modified RNA, and regulate the maturation, transport, and metabolism of mRNA and the maturation of gene regulation of long noncoding RNAs (Alarcón et al., 2015; Wu et al., 2018), suggesting its role as a critical reader in m6A marking and its subsequent effects. In addition, insulin-like growth factor binding proteins (IGF2BP1/2/3), another set of nuclear m6A readers, have been shown to enhance the stability and translation of mRNA (Huang et al., 2018).
HNRNPC and HNRNPG are the nuclear RNA-binding proteins responsible for pre-mRNA processing. The binding ability of HNRNPC and HNRNPG is regulated in an m6A-switch dependent manner, illustrating a new mechanism of RNA-modification-coded cellular biology (Liu et al., 2015; Liu et al., 2017). In addition to the above, there are other m6A-binding proteins that can recognize and bind m6A modified sites, such as the translation initiation factor eukaryotic initiation factor 3 (eIF3), which selectively binds to m6A-modified RNAs. Upon binding with eIF3, 5′UTR m6A residues stimulate translation initiation (Meyer et al., 2015). Furthermore, proline rich coiled-coil 2A (Prrc2A), a novel m6A specific binding protein, has been shown to bind with m6A modified mRNA and regulate myelination of oligodendrocytes (Wu et al., 2019).
The RNA-binding protein, fragility X mental retardation protein (FMRP), is encoded by FMR1. FMRP is critical in metabotropic glutamate receptor-dependent long-term depression and synaptic plasticity. The absence of FMRP causes excessive and persistent protein synthesis in postsynaptic dendrites and dysregulated synaptic functions (Santoro et al., 2012).
The role of m6A in the development of the nervous system
The process of nervous system development is highly coordinated by different cell types, and has the potential for self-renewal and the ability to produce many types of differentiated cells. The gene expression of neural cells is regulated by many factors, including various epigenetic modifications, such as m6A modification. As reported, a high abundance of m6A modifications exist in the nervous system (Livneh et al., 2020). In the brain, the level of m6A increases from embryonic stage to adulthood (Jiang et al., 2021), and nearly half of the mRNAs stably expressing cerebral cortex genes in adult mice have m6A methylation modification (Chang et al., 2017), suggesting that m6A methylation modification plays an important role in the nervous system.
m6A methylation in neurogenesis
Neurogenesis is the process of producing neurons from different types of neural progenitor cells (NPCs, including neural stem cells), most of which occurs during embryonic development (Florio and Huttner, 2014). Neurogenesis is regulated by multiple factors, among which m6A methylation is particularly important for the regulation of neurogenesis, and an imbalance in m6A methylation levels can cause neurogenesis abnormalities.
The m6A methyltransferase, METTL3, affects neurogenesis and neuronal development, and its deletion significantly reduces m6A levels, affects the proliferation and cell cycle process of adult neural stem cells (aNSCs), and inhibits the morphological maturation of newborn neurons. Meanwhile, overexpression of the histone methyltransferase Ezh2 was shown to rescue neurogenesis and neuronal development defects caused by METTL3 deletion (Chen et al., 2019a). METTL14, one of the core molecules of the m6A methyltransferase complex, mediates m6A modification to participate in neurogenesis by modulating cell cycle progression of cortical neural progenitors. Loss of METTL14 results in the nuclear accumulation of mRNA associated with neural differentiation and delayed differentiation of mouse neural progenitor cells (Yoon et al., 2017; Edens et al., 2019). FTO is widely expressed and highly enriched in neuronal and adult neural stem cells. It is closely related to the proliferation and differentiation of aNSCs, and its deficiency can reduce proliferation and neuronal differentiation of aNSCs (Li et al., 2017b). Another study has reported that FTO dynamically influences neurogenesis by modulating the Pdgfra/Socs5-Stat3 pathway (Cao et al., 2020).
Conditional depletion of YTHDF2 leads to embryonic lethality, suggesting that YTHDF2 plays a critical role in early embryonic development. YTHDF2-mediated m6A epitranscriptomics regulates cortical neurogenesis during embryonic neural development by regulating RNA degradation of m6A-tagged genes associated with neural development and differentiation (Li et al., 2018a). Moreover, YTHDF2 can destabilize m6A-modified neural-specific RNA, thus inhibiting the differentiation of induced pluripotent stem cells (Heck et al., 2020). Furthermore, decline in Imp (IGF2BP) levels, a highly conserved RNA-binding protein, has been shown to limit the stability of myc mRNA and restrict neuroblast growth and division in Drosophila (Samuels et al., 2020). Another study reported that deletion of Fmr1 is associated with abnormalities in cortical development and dendritic spine formation, which leads to abnormal learning and behavior in fragile X syndrome (Castrén et al., 2005; Saffary and Xie, 2011; La Fata et al., 2014). Loss of Fmr1 results in nuclear retention of m6A-modified FMRP target mRNAs that regulate neural differentiation, ultimately resulting in defective neuronal function (Edens et al., 2019).
In addition to being essential for neuronal development, m6A also plays an important role in the development of glial cells in mammals. Oligodendrocytes are glial cells in the central nervous system that are responsible for myelination of axons (Nave and Werner, 2014; Simons and Nave, 2015). METTL14-mediated m6A methylation is critical for regulation of oligodendrocyte development and myelination of the central nervous system. Conditional inactivation of METTL14 leads to a decrease in the number of oligodendrocytes and CNS hypomyelination. Moreover, dynamic changes in m6A transcription affect the development of oligodendrocyte lineage (Xu et al., 2020a). In our study, we found that Prrc2a regulates the myelination of oligodendrocytes. Conditional knockout of Prrc2a in mice leads to pathological features, such as inadequate myelination, shortened lifespan, and impaired motor and cognitive function. Mechanistically, Prrc2a increases the stability of oligo2 mRNA in an m6A-dependent manner, thereby regulating the proliferation and fate of oligodendrocytes (Wu et al., 2019).
m6A methylation in the formation and function of neuronal system
m6A methylation plays an important role in axonal and synaptic growth, and dysregulation of m6A modification leads to abnormal synaptic function. For example, loss of METTL14 or YTHDF1 attenuates injury-induced protein translation in adult dorsal root ganglion neurons (DRGs) and decreases functional axon regeneration in the peripheral nervous system (Weng et al., 2018). FTO is highly expressed in neuronal axons, suggesting that it may play an important role in neuronal axon development and function (Li et al., 2017b). FTO knockdown in axons increases m6A levels, decreases local translation and expression of growth-associated protein 43 mRNA, and markedly inhibits axonal growth (Yu et al., 2018).
The YTHDF family of m6A methylation-binding proteins also limit the growth and development of axons by regulating the translation of related transcripts (Worpenberg et al., 2021). YTHDF1 is closely associated with axonal growth, development, and function. It physiologically regulates the translation of Robo3.1, which controls the guidance of pre-crossing axons in the embryonic spinal cord (Zhuang et al., 2019). In addition, YTHDF1/2 is closely related to axonal growth in granulosa cells. Knockdown of YTHDF1/2 promotes axonal growth in granule cells by regulating the local translation of Wnt5a signaling components (Yu et al., 2021). Knockdown of YTHDF1 in hippocampal neurons results in synaptic dysfunction, including immature dendritic spine morphology and inhibition of excitatory synaptic transmission, accompanied by a reduction in postsynaptic density 95 (PSD-95) and decreased surface expression of GluA1 of AMPA receptors (Merkurjev et al., 2018).
m6A in learning cognitive function
METTL3-mediated m6A methylation is involved in the formation of long-term memory in the hippocampus. Depletion of METTL3 in the mouse hippocampus reduces memory consolidation. Moreover, adequate training or restoration of METTL3 could rescue learning memory defect (Zhang et al., 2018). In addition, m6A methylation modification in glutamatergic neurons has been reported to be important for context fear memory generalization (Chang et al., 2022). Deletion of METTL3 in hippocampal glutamatergic neurons results in more freezing behavior, suggesting a lower discrimination index.
Another indispensable component of the m6A methyltransferase complex is METTL14, which has been shown to influence learning in mice. It was found that METTL14 was essential for striatal function and learning-related transcriptional regulation, and conditional neuron-specific striatal deletion of METTL14 reduced m6A methylation levels and impaired striatum-mediated behavior and learning abilities (Koranda et al., 2018). Additionally, FTO plays an important role in the formation of memory in the cerebral cortex and hippocampus. FTO deficiency could reduce the proliferation of aNSCs and neuronal differentiation, resulting in impaired neurogenesis as well as learning and memory defects (Li et al., 2017b). The level of m6A modification in the prefrontal cortex of mice increases with behavioral experience, and targeted knockdown of FTO in the prefrontal cortex significantly enhances the consolidation of cued fear memory (Widagdo et al., 2016). In addition, m6A-binding proteins play a key role in learning and memory processes. YTHDF1 facilitates learning and memory, and deletion of YTHDF1 impairs synaptic transmission and long-term potentiation. YTHDF1 facilitates the translation of m6A-methylated neuronal mRNAs in response to neuronal stimulation, which is necessary for learning and memory. (Shi et al., 2018). Moreover, the YTHDF family mediated m6A modification was also shown to affect learning and memory formation in Drosophila (Kan et al., 2021).
m6A modification in cerebellar development
m6A modification is also indispensable in the development of the cerebellum. The m6A-related genes METTL3, METTL14, WTAP, FTO, and ALKBH5 in mice showed a decreasing trend with age during cerebellar development, with the highest level in P7, which decreased and stabilized after P14. Dynamic regulation of METTL3-mediated m6A methylation modification is associated with cerebellar development (Ma et al., 2018; Wang et al., 2018).
Conditional deletion of METTL3 using Nestin-Cre in the nervous system causes severe developmental defects in the brain. Mechanistically, deletion of METTL3 significantly increases the apoptosis of newborn cerebellar granulocytes (CGCs) and impairs the maturation of Purkinje cells (Wang et al., 2018). Consistently, knockdown of METTL3 by lentivirus was shown to result in a severe alteration in Purkinje cell numbers, laminal structure, and stunted dendrites and caused defects in mouse cerebellar development. In addition, ALKBH5 deletion affects RNA nuclear output, impairs RNA metabolism, and affects cerebellar development (Ma et al., 2018). Furthermore, WTAP has been found to be closely related with cerebellar development in mice, and WTAP deletion leads to Purkinje cell degeneration, cerebellar ataxia, and cerebellar atrophy (Yang et al., 2022). The m6A-binding proteins also play an important role in the development of the cerebellum, and specific knockout of YTHDF1 or YTHDF2 in granule cells promotes the growth of parallel fibers and the formation of cerebellar synapses, thereby improving motor coordination in mice (Yu et al., 2021). Together, these studies suggest that m6A plays a critical role in neurogenesis, cerebellar development, and cognitive function, which are indispensable for proper physiological functioning of the nervous system (Table 1).
m6A methylation in neurological disorders
m6A methylation is widely present in mammalian transcripts that affect the basic metabolism of RNA and participate in the dynamic regulation of various biological processes. In recent years, multiple studies have shown that m6A methylation is closely related to nervous system diseases. The level of m6A methylation changes in the pathological state. Simultaneously, the dysregulation of m6A methylation modification also causes neurodevelopmental abnormalities and neurological diseases (Figure 2; Table 2).
TABLE 2.
Functions of m6A modification-related proteins in neurological diseases.
| Disease type | m6A regulator | Function | References |
|---|---|---|---|
| Alzheimer’s disease | METTL3 | Involve in the pathogenesis of AD | Zhao et al. (2021) |
| FTO | regulate the mTOR signaling pathway and affected the development of AD | Li et al. (2018b) | |
| HNRNPA2B1 | Involved in the regulation of AD | Kolisnyk et al. (2016) | |
| Parkinson’s Disease | METTL14 | Affect the production of key enzymes required for dopamine synthesis | Teng et al. (2021) |
| FTO | Maintain dopaminergic neuron function | (Hess et al., 2013; Qi et al., 2022) | |
| HNRNPC | regulate dopaminergic proliferation | Quan et al. (2021) | |
| ALKBH5 | Relate to the risk of PD | Qiu et al. (2020) | |
| Major depressive disorder | FTO | Target ADRB2 induces depression-like behaviors | Liu et al. (2021a) |
| FTO | Target the CaMKII/CREB signaling pathway modulates hippocampal synaptic plasticity | Shen et al. (2021) | |
| FTO | Affect resistance to tricyclic antidepressants | Wu et al. (2021) | |
| FTO | regulate the function of astrocytes | Huang et al. (2020b) | |
| Stroke | METTL3 | Involve in the formation of early stress granule cells | Si et al. (2020) |
| YTHDF1 | Involve in inflammatory response in stroke model mice | Zheng et al. (2020) | |
| YTHDC1 | Influence the degradation of PTEN mRNA | Zhang et al. (2020) | |
| Glioblastoma | METTL3 | Influence self-renewal capacity of GBM cells | Li et al. (2019) |
| METTL3 | Influence the development process of GBM | Cui et al. (2017) | |
| METTL3 | Influence tumor growth and development | Visvanathan et al. (2018) | |
| METTL3 | Maintain specific gene expression in GSCs and regulate oncogenic-related signaling pathways | Visvanathan et al. (2019) | |
| METTL3 | Regulates DNA repair factors and affects cellular drug resistance | Shi et al. (2021a) | |
| WTAP | Involve in carcinogenesis | Jin et al. (2012) | |
| FTO | Affect GBM cell proliferation, migration and invasion | Zhang et al. (2022) | |
| ALKBH5 | Influence the GSCs self-renewal and tumorigenesis | Zhang et al. (2017) | |
| ALKBH5 | Influence the radioresistance and invasive ability of GSCs | Kowalski-Chauvel et al. (2020) | |
| YTHDF2 | Influence the development process of GBM | Dixit et al. (2021) | |
| YTHDF2 | Influence the GBM cell proliferation, invasion, and tumorigenesis | Fang et al. (2021) | |
| YTHDF2 | Involve in UBXN1 mRNA degradation | Chai et al. (2021) | |
| IGFBP1/2/3 | Affects the formation of GBM microenvironment, regulates carcinogenic factors, affects GBM aerobic glycolysis, and affects GBM prognosis | (Liu et al., 2021b; Sun et al., 2021; Xiong et al., 2022) |
m6A methylation in Alzheimer’s disease (AD)
AD is a neurodegenerative disease characterized by progressive cognitive dysfunction and impaired learning and memory (Lane et al., 2018). As mentioned above, m6A plays a critical role in learning functions, such as cognitive and memory formation and consolidation (Weng et al., 2018; Zhang et al., 2018). It has been reported that m6A methylation also occurs during the occurrence and development of AD. The m6A modification levels were shown to be significantly reduced in pyramidal neurons in AD patients, whereas the m6A level in glial cells was significantly increased (Zhao et al., 2021). The abundance of m6A modification was shown to be significantly lower in 5×FAD mouse brain than in control mice (Shafik et al., 2021). However, it was reported that the levels of m6A methylation in the cortex and hippocampus of APP/PS1 transgenic mice were higher than those in the control group (Han et al., 2020). Differences in mouse models and brain tissue might contribute to these contradictory results. In addition, m6A methyltransferases METTL3, METTL14, and WTAP; demethylase FTO; and m6A binding protein YTHDF1 were found to be significantly reduced in the frontal cortex tissues of patients with AD. Notably, the expression of METTL3 was significantly downregulated in the brains of patients with mild cognitive impairment (Zhao et al., 2021). In addition, METTL3 accumulates in amyloid-β (Aβ) plaques and intracellular tau nerve fiber tangles, and its accumulation levels are positively correlated with tau protein levels (Huang et al., 2020a). Abnormal expression and distribution of m6A modification-related proteins in the hippocampus of patients with AD suggests that m6A modification is associated with the pathogenesis of AD. In an AD mouse model, METTL3 knockdown in the hippocampus resulted in cognitive dysfunction, synaptic loss, neuronal death, increased oxidative stress, and abnormal cell cycle events. Overexpression of METTL3 in neurons salvaged Aβ-induced synaptic damage and cognitive impairment (Zhao et al., 2021). METTL3 levels were elevated in the cortical and hippocampal brain regions of AD mice, whereas FTO levels were decreased in the hippocampal brain region (Han et al., 2020). However, one study reported that FTO levels were upregulated in AD mouse models, and overexpression of FTO activated the mTOR signaling pathway to increase phosphorylated tau protein levels, which promoted the development of AD (Li et al., 2018b). Moreover, the m6A-binding protein HNRNPA2B1 was shown to participate in the pathological process of AD (Kolisnyk et al., 2016). Although the role of m6A methylation in AD is not yet exhaustive, the study of m6A modification provides a new perspective target for the treatment of AD.
m6A methylation in Parkinson’s disease (PD)
PD is a common age-related neurodegenerative disease, but its pathogenesis has not been fully elucidated. The loss of dopaminergic neurons in the substantia nigra of the midbrain leads to a decrease in the content of dopamine in the striatum, resulting in a decrease in the activity of the substantia nigra–striatum dopamine transmitter system and relative hyperactivity of cholinergic neurons in the striatum (Schapira and Jenner, 2011). m6A methylation modifications are highly abundant in the brain and play a vital role in dopaminergic neuronal pathways in the midbrain. Specific deletion of FTO in dopaminergic neurons causes abnormal dopaminergic neuron function (Hess et al., 2013). FTO was shown to play a protective role in manganese-mediated PD models by antagonizing the downregulation of the axon-guided molecule ephrin-B2 (Qi et al., 2022). Specific knockout of methyltransferase METTL14 in the substantia nigra region was demonstrated to cause a decrease in the key enzyme of dopamine synthesis, tyrosine hydroxylase, which in turn caused motor dysfunction in mice (Teng et al., 2021). These studies suggest that an imbalance in the dynamic regulation of m6A is one of the causes of pathological changes in PD. Moreover, reduced levels of m6A modification in neurons were shown to increase the expression of N-methyl-d-aspartate receptor l, thereby increasing the inflow of calcium and oxidative stress, ultimately leading to apoptosis of dopaminergic neurons (Chen et al., 2019b). In addition, the m6A methylation recognition protein HNRNPC is significantly downregulated in PD, and it may be involved in the process of regulating PD by inhibiting the proliferation of dopaminergic neurons (Quan et al., 2021). In addition to animal experiments, m6A-associated mononucleotide polymorphisms in the rs1378602, rs4924839, and rs8071834 genes encoding the demethylase ALKBH5 in PD patients were found to be closely related to the risk of PD (Qiu et al., 2020). Although numerous studies have shown an important relationship between m6A modification and PD, the characteristics of m6A modification and the mechanisms that regulate the development of PD require further exploration.
m6A methylation in depression
Major depressive disorder (MDD) is a common, chronic, and recurrent psychiatric disorder. Epigenetic RNA modifications are also involved in the pathogenesis of MDD. The RNA demethylase ALKBH5 was found to be closely related to MDD, suggesting that m6A methylation plays an important role in MDD (Du et al., 2015; Barbon and Magri, 2020). FTO expression has been shown to be downregulated in the hippocampus of MDD patients and mouse models of depression, and FTO in mouse hippocampal tissue was shown to induce depressive-like behavior in mice by targeting ADRB2 (Liu et al., 2021a). Similarly, FTO was demonstrated to target the CaMKII/CREB signaling pathway to regulate hippocampal synaptic plasticity, and activation of FTO in the hippocampus alleviated chronic restraint stress-induced depression-like behaviors in mice (Shen et al., 2021). In addition, the absence of FTO in the ventral covered area (VTA) of the midbrain was shown to increase the susceptibility of mice to stress and resistance to tricyclic antidepressants. Consistently, overexpression of FTO in the VTA significantly alleviated depression-like behavior in mice (Wu et al., 2021). Moreover, FTO deficiency has been shown to reduce anxiety- and depression-like behaviors (Sun et al., 2019). These contradictory results may be due to differences in the role of FTO in different brain regions. In addition, the cyclic RNA STAG1 (circSTAG1) was shown to alleviate astrocyte dysfunction by capturing ALKBH5, which in turn played an antidepressant role (Huang et al., 2020b). Together, these studies demonstrate that m6A methylation is involved in the regulation of MDD and provide new ideas for the treatment of MDD.
m6A methylation in stroke
Several studies have shown that m6A methylation is closely related to ischemic stroke (Xu et al., 2020b; Yi et al., 2021). m6A methylation has been shown to be involved in post-stroke pathophysiological processes, such as inflammation, apoptosis, and transcriptional regulation, but the specific regulatory mechanisms need to be explored further (Chokkalla et al., 2019). The m6A methyltransferase METTL3 was shown to be associated with the formation of stress granules (SGs) in early acute ischemic stroke and reducing the apoptosis of injured neurons and cells by increasing the maturation of miR-335 through m6A methylation (Si et al., 2020). The m6A methylation-binding protein YTHDF1 was demonstrated to play an important role in cerebral ischemia-reperfusion injury. After cerebral ischemia reperfusion injury, the expression of miR-421-3p was significantly reduced, which inhibited the translation of p65 by targeting YTHDF1, thereby inhibiting the inflammatory response (Zheng et al., 2020). Another study found that knockdown of YTHDC1 aggravated ischemic brain injury, whereas overexpression of YTHDC1 increased phosphorylation of Akt by promoting the degradation of PTEN mRNA, thereby saving neurons after ischemia and protecting rats from cerebral ischemic damage (Zhang et al., 2020).
m6A methylation in glioblastoma
Studies have shown that m6A modifications are widely involved in the occurrence, progression, and immune regulation of various cancers (Deng et al., 2018; Geng et al., 2020; Jiang et al., 2021; Uddin et al., 2021). Glioblastoma (GMB) is the most common primary malignant glioma in the central nervous system (Omuro and DeAngelis, 2013; Lapointe et al., 2018). m6A not only affects the clinical prognosis of GBM (Du et al., 2020) but is also closely related to GBM mesenchymal conversion and angiogenesis (Tao et al., 2022). In addition, m6A regulatory proteins are also associated with immune infiltration in GBM (Pan et al., 2021).
Several biological information analyses have shown that m6A regulatory proteins affect the prognosis and survival rate of GBM patients (Cai et al., 2021; Wang et al., 2021). METTL3-mediated m6A methylation promotes proliferation and self-renewal of GBM cells (Li et al., 2019). METTL3 knockout promotes the growth and self-renewal of human glioblastoma stem cells (GSCs) and promotes GBM development (Cui et al., 2017). In contrast, it was reported that METTL3 knockout in mouse GBM models inhibited tumor growth and development, and METTL3 knockdown in GSCs led to increased sensitivity of cells to radiation exposure (Visvanathan et al., 2018). In addition, METTL3 is indispensable in maintaining GSC-specific gene expression and positive regulation of carcinogenic-related signaling pathways (Visvanathan et al., 2019). These contradictory findings might be attributed to different GBM cell lines, differences in culture conditions, and genetic heterogeneity. In addition to influencing the occurrence and development of GBM, METTL3 is also related to resistance to chemotherapy drugs. METTL3 and m6A methylation levels have been shown to affect the sensitivity of temozolomide-resistant GBM cells. METTL3 regulates m6A methylation levels of DNA repair factors MGMT and APNG, thereby affecting glioma resistance to temozolomide (Shi et al., 2021a). Furthermore, SRSF7 plays an oncogenic role in various cancers. SRSF7 in GBM patients specifically targets and promotes the methylation of m6A sites in genes involved in cell proliferation and migration by recruiting a methyltransferase complex (Cun et al., 2021). Additionally, the horizontal expression of WTAP, a component of the m6A methyltransferase complex, is also strongly associated with poor prognosis in GBM (Xi et al., 2016). WTAP acts as a nucleoprotein associated with proliferation and apoptosis regulation and plays a carcinogenic role in gliomas (Jin et al., 2012). In addition to m6A methyltransferase, m6A demethylase and m6A recognition proteins are involved in regulating and influencing the occurrence and development of GBM. Reportedly, inhibition of FTO prevented neurosphere formation in patient-derived GSCs without inhibiting the growth of healthy neural stem cell-derived neurospheres (Huff et al., 2021). However, it was found that a decreased FTO level is closely related with poor prognosis. FTO regulates the maturation of miR-10a in an m6A-dependent manner and recruits the microRNA microprocessor complex protein DGCR8 through the recognition protein HNRNPA2B1, thereby promoting the proliferation, migration, and invasion of GBM cells (Zhang et al., 2022). ALKBH5 is highly expressed in GSCs. ALKBH5 in GBM stem-like cells enhances self-renewal and tumorigenesis by regulating FOXM1 (Zhang et al., 2017). In addition, ALKBH5 promotes radiation resistance and invasion of GSCs (Kowalski-Chauvel et al., 2020). In addition, the m6A methylation-binding protein YTHDF2 was found to affect tumor growth in mice through the YTHDF2-MYC-IGFBP3 pathway, providing a new target for the treatment of GBM (Dixit et al., 2021). YTHDF2 was shown to be associated with poor prognosis in glioma patients, and is required for GBM cell proliferation, invasion, and tumorigenesis (Fang et al., 2021). In addition, highly expressed YTHDF2 accelerates the degradation of UBXN1 mRNA by recognizing m6A modifications, thereby promoting the activation of NF-κB. UBXN1 overexpression attenuates YTHDF2 to promote malignant progression of glioma (Chai et al., 2021), suggesting that YTHDF2 might be involved in the occurrence and malignant progression of GBM. In addition, the m6A methylation binding proteins, including IGFBP1/2/3 are closely related to GBM. IGFBP1 was significantly positively correlated with most immune cells and matrix-related pathways, indicating that m6A methylation modification plays a role in the formation of the GBM microenvironment. IGF2BP2 interacts with the CASC9/IGF2BP2 complex and accelerates aerobic glycolysis in GBM by regulating the mRNA of HK2. It was reported that high expression of IGF2BP3 in patients with glioma significantly reduced survival time, which was also associated with poor prognosis, suggesting that IGF2BP3 might be used as a potential prognostic biomarker for gliomas (Liu et al., 2021b; Sun et al., 2021; Xiong et al., 2022). The important role of modified m6A-related proteins in GBM provides new insights into the prognosis and treatment of GBM.
m6A methylation in other neuropathies
In addition to participating in the regulation of the above diseases, m6A is also closely related with other neurological diseases, such as amyotrophic lateral sclerosis, epilepsy, intellectual disability, schizophrenia, and traumatic brain injury (Batista, 2017; Jung and Goldman, 2018; Livneh et al., 2020). Mutations related to amyotrophic lateral sclerosis in the prion-like domain of hnRNPA could promote protein fibrosis in cells, suggesting that hnRNPA might be involved in the pathogenesis of atrophic lateral sclerosis (Kim et al., 2013). FTO is involved in regulating microRNAs and plays an important role in the physiology and pathology of the central nervous system (Rowles et al., 2012). METTL3-mediated m6A modification is involved in the dysregulation of NRIP1 expression in Down syndrome (Shi et al., 2021b). Deletion of the m6A methylated binding protein FMRP leads to fragile X syndrome (Santoro et al., 2012). In a traumatic brain injury rat model, it was shown that FTO plays an important role in maintaining neural function but has no effect on spatial learning and memory ability (Yu et al., 2020). In addition, m6A modification is involved in regulating drug toxicity-related nervous system damage. In a study in young mice, sevoflurane was shown to impair m6A-mediated mRNA translation and cause motor defects. YTHDF1 salvaged sevoflurane-induced suppression of protein synthesis and motor disorders in the young mice (Zhang et al., 2021). FTO has also been found to alleviate dopaminergic neurotransmission defects caused by exposure to arsenite and improve the progression of arsenic-related neurological disorders (Bai et al., 2018).
Summary and outlook
In summary, as a very important epigenetic modification, m6A methylation regulates RNA translation, splicing, transport, localization, and stability at the post-transcriptional level. m6A methylation has a wide range of effects on nervous system development and function, including neurogenesis, cerebellar development, learning, cognition, and memory. It also plays an important role in the regulation of the occurrence and development of nervous system diseases. These studies provide new ideas and directions for revealing the molecular mechanisms of neurodevelopmental processes and promoting targeted therapies for neurological diseases.
Although research on m6A methylation has rapidly developed in recent years, there are still many unknowns. For example, the mechanism and function of some known m6A-related proteins in the nervous system have not been fully clarified, and there may be some undiscovered m6A methyltransferases, demethylases, and methylation recognition enzymes.
Author contributions
LJ conceived the review topic, reviewed the literature, wrote the manuscript, and prepared the figure; XL and SW reviewed the manuscript; ZY and JC performed a comprehensive review of the literature.
Funding
This work was supported by the grants from the National Nature Science Foundation of China (82071218 and 81930029).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Agarwala S. D., Blitzblau H. G., Hochwagen A., Fink G. R. (2012). RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 8 (6), e1002732. 10.1371/journal.pgen.1002732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón C. R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S. F. (2015). HNRNPA2B1 is a mediator of m(6)a-dependent nuclear RNA processing events. Cell. 162 (6), 1299–1308. 10.1016/j.cell.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L., Tang Q., Zou Z., Meng P., Tu B., Xia Y., et al. (2018). m6A demethylase FTO regulates dopaminergic neurotransmission deficits caused by arsenite. Toxicol. Sci. 165 (2), 431–446. 10.1093/toxsci/kfy172 [DOI] [PubMed] [Google Scholar]
- Barbon A., Magri C. (2020). RNA editing and modifications in mood disorders. Genes. (Basel) 11 (8), E872. 10.3390/genes11080872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista P. J. (2017). The RNA modification N(6)-methyladenosine and its implications in human disease. Genomics Proteomics Bioinforma. 15 (3), 154–163. 10.1016/j.gpb.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Zhang J., Liu Z., Su J., Xu J., Li Z., et al. (2021). Identification of an N6-methyladenosine (m6A)-related signature associated with clinical prognosis, immune response, and chemotherapy in primary glioblastomas. Ann. Transl. Med. 9 (15), 1241. 10.21037/atm-21-3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Zhuang Y., Chen J., Xu W., Shou Y., Huang X., et al. (2020). Dynamic effects of Fto in regulating the proliferation and differentiation of adult neural stem cells of mice. Hum. Mol. Genet. 29 (5), 727–735. 10.1093/hmg/ddz274 [DOI] [PubMed] [Google Scholar]
- Castrén M., Tervonen T., Karkkainen V., Heinonen S., Castren E., Larsson K., et al. (2005). Altered differentiation of neural stem cells in fragile X syndrome. Proc. Natl. Acad. Sci. U. S. A. 102 (49), 17834–17839. 10.1073/pnas.0508995102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai R. C., Chang Y. Z., Chang X., Pang B., An S. Y., Zhang K. N., et al. (2021). YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m(6)A modification to activate NF-κB and promote the malignant progression of glioma. J. Hematol. Oncol. 14 (1), 109. 10.1186/s13045-021-01124-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., Lv H., Zhang W., Ma C., He X., Zhao S., et al. (2017). Region-specific RNA m(6)A methylation represents a new layer of control in the gene regulatory network in the mouse brain. Open Biol. 7 (9), 170166. 10.1098/rsob.170166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y., Zhang J., Zhang J., Zhu W., Zheng Q., Qian Z., et al. (2022). N(6)-methyladenosine RNA modification of glutamatergic neurons is associated with contextual fear discrimination. Physiol. Behav. 248, 113741. 10.1016/j.physbeh.2022.113741 [DOI] [PubMed] [Google Scholar]
- Chen J., Zhang Y. C., Huang C., Shen H., Sun B., Cheng X., et al. (2019). m(6 A regulates neurogenesis and neuronal development by modulating histone methyltransferase Ezh2. Genomics Proteomics Bioinforma. 17 (2), 154–168. 10.1016/j.gpb.2018.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Yu C., Guo M., Zheng X., Ali S., Huang H., et al. (2019). Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem. Neurosci. 10 (5), 2355–2363. 10.1021/acschemneuro.8b00657 [DOI] [PubMed] [Google Scholar]
- Chokkalla A. K., Mehta S. L., Kim T., Chelluboina B., Kim J., Vemuganti R. (2019). Transient focal ischemia significantly alters the m(6)A epitranscriptomic tagging of RNAs in the brain. Stroke 50 (10), 2912–2921. 10.1161/STROKEAHA.119.026433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., et al. (2017). m(6 A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell. Rep. 18 (11), 2622–2634. 10.1016/j.celrep.2017.02.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cun Y., An S., Zheng H., Lan J., Chen W., Luo W., et al. (2021). Specific regulation of m(6)A by SRSF7 promotes the progression of glioblastoma. Genomics Proteomics Bioinforma. 10.1016/j.gpb.2021.11.001 [DOI] [PubMed] [Google Scholar]
- Deng X., Su R., Feng X., Wei M., Chen J. (2018). Role of N(6)-methyladenosine modification in cancer. Curr. Opin. Genet. Dev. 48, 1–7. 10.1016/j.gde.2017.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit D., Prager B. C., Gimple R. C., Poh H. X., Wang Y., Wu Q., et al. (2021). The RNA m6A reader YTHDF2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discov. 11 (2), 480–499. 10.1158/2159-8290.CD-20-0331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485 (7397), 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Dominissini D., Nachtergaele S., Moshitch-Moshkovitz S., Peer E., Kol N., Ben-Haim M. S., et al. (2016). The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 530 (7591), 441–446. 10.1038/nature16998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxtader K. A., Wang P., Scarborough A. M., Seo D., Conrad N. K., Nam Y. (2018). Structural basis for regulation of METTL16, an S-adenosylmethionine homeostasis factor. Mol. Cell. 71 (6), 1001–1011.e4. 10.1016/j.molcel.2018.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., et al. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7, 12626. 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Hou K., Mi S., Ji H., Ma S., Ba Y., et al. (2020). Malignant evaluation and clinical prognostic values of m6A RNA methylation regulators in glioblastoma. Front. Oncol. 10, 208. 10.3389/fonc.2020.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T., Rao S., Wu L., Ye N., Liu Z., Hu H., et al. (2015). An association study of the m6A genes with major depressive disorder in Chinese Han population. J. Affect. Disord. 183, 279–286. 10.1016/j.jad.2015.05.025 [DOI] [PubMed] [Google Scholar]
- Edelheit S., Schwartz S., Mumbach M. R., Wurtzel O., Sorek R. (2013). Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 9 (6), e1003602. 10.1371/journal.pgen.1003602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens B. M., Vissers C., Su J., Arumugam S., Xu Z., Shi H., et al. (2019). FMRP modulates neural differentiation through m(6)a-dependent mRNA nuclear export. Cell. Rep. 28 (4), 845–854.e5. e5. 10.1016/j.celrep.2019.06.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R., Chen X., Zhang S., Shi H., Ye Y., Shi H., et al. (2021). EGFR/SRC/ERK-stabilized YTHDF2 promotes cholesterol dysregulation and invasive growth of glioblastoma. Nat. Commun. 12 (1), 177. 10.1038/s41467-020-20379-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florio M., Huttner W. B. (2014). Neural progenitors, neurogenesis and the evolution of the neocortex. Development 141 (11), 2182–2194. 10.1242/dev.090571 [DOI] [PubMed] [Google Scholar]
- Geng Y., Guan R., Hong W., Huang B., Liu P., Guo X., et al. (2020). Identification of m6A-related genes and m6A RNA methylation regulators in pancreatic cancer and their association with survival. Ann. Transl. Med. 8 (6), 387. 10.21037/atm.2020.03.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T., Girard C. A., Tung Y. C. L., Webby C. J., Saudek V., Hewitson K. S., et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318 (5855), 1469–1472. 10.1126/science.1151710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A. A., Kol N., Salmon-Divon M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347 (6225), 1002–1006. 10.1126/science.1261417 [DOI] [PubMed] [Google Scholar]
- Han M., Liu Z., Xu Y., Liu X., Wang D., Li F., et al. (2020). Abnormality of m6A mRNA methylation is involved in alzheimer's disease. Front. Neurosci. 14, 98. 10.3389/fnins.2020.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. E., Miceli S. M., Roberts R. J., Manley J. L. (1990). Sequence specificity of the human mRNA N6-adenosine methylase in vitro . Nucleic Acids Res. 18 (19), 5735–5741. 10.1093/nar/18.19.5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck A. M., Russo J., Wilusz J., Nishimura E. O., Wilusz C. J. (2020). YTHDF2 destabilizes m(6)A-modified neural-specific RNAs to restrain differentiation in induced pluripotent stem cells. Rna 26 (6), 739–755. 10.1261/rna.073502.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess M. E., Hess S., Meyer K. D., Verhagen L. A. W., Koch L., Bronneke H. S., et al. (2013). The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 16 (8), 1042–1048. 10.1038/nn.3449 [DOI] [PubMed] [Google Scholar]
- Hsu P. J., Zhu Y., Ma H., Guo Y., Shi X., Liu Y., et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell. Res. 27 (9), 1115–1127. 10.1038/cr.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Camats-Perna J., Medeiros R., Anggono V., Widagdo J. (2020). Altered expression of the m6A methyltransferase METTL3 in alzheimer's disease. eNeuro 7 (5), ENEURO.0125–20.2020. 10.1523/ENEURO.0125-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell. Biol. 20 (3), 285–295. 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R., Zhang Y., Bai Y., Han B., Ju M., Chen B., et al. (2020). N(6)-Methyladenosine modification of fatty acid amide hydrolase messenger RNA in circular RNA STAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol. Psychiatry 88 (5), 392–404. 10.1016/j.biopsych.2020.02.018 [DOI] [PubMed] [Google Scholar]
- Huff S., Tiwari S. K., Gonzalez G. M., Wang Y., Rana T. M. (2021). m(6 A-RNA demethylase FTO inhibitors impair self-renewal in glioblastoma stem cells. ACS Chem. Biol. 16 (2), 324–333. 10.1021/acschembio.0c00841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7 (12), 885–887. 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Yang C. G., Yang S., Jian X., Yi C., Zhou Z., et al. (2008). Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 582 (23-24), 3313–3319. 10.1016/j.febslet.2008.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Liu B., Nie Z., Duan L., Xiong Q., Jin Z., et al. (2021). The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 6 (1), 74. 10.1038/s41392-020-00450-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D. I., Lee S. W., Han M. E., Kim H. J., Seo S. A., Hur G. Y., et al. (2012). Expression and roles of Wilms' tumor 1-associating protein in glioblastoma. Cancer Sci. 103 (12), 2102–2109. 10.1111/cas.12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A., Issa J. P., Baylin S. (2016). Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 17 (10), 630–641. 10.1038/nrg.2016.93 [DOI] [PubMed] [Google Scholar]
- Jung Y., Goldman D. (2018). Role of RNA modifications in brain and behavior. Genes. Brain Behav. 17 (3), e12444. 10.1111/gbb.12444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan L., Ott S., Joseph B., Park E. S., Dai W., Kleiner R. E., et al. (2021). Publisher correction: A neural m6A/ythdf pathway is required for learning and memory in Drosophila. Nat. Commun. 12 (1), 1743. 10.1038/s41467-021-22219-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Kim N. C., Wang Y. D., Scarborough E. A., Moore J., Diaz Z., et al. (2013). Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495 (7442), 467–473. 10.1038/nature11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolisnyk B., Al-Onaizi M. A., Xu J., Parfitt G. M., Ostapchenko V. G., Hanin G., et al. (2016). Cholinergic regulation of hnRNPA2/B1 translation by M1 muscarinic receptors. J. Neurosci. 36 (23), 6287–6296. 10.1523/JNEUROSCI.4614-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koranda J. L., Dore L., Shi H., Patel M. J., Vaasjo L. O., Rao M. N., et al. (2018). Mettl14 is essential for epitranscriptomic regulation of striatal function and learning. Neuron 99 (2), 283–292.e5. e5. 10.1016/j.neuron.2018.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski-Chauvel A., Lacore M. G., Arnauduc F., Delmas C., Toulas C., Cohen-Jonathan-Moyal E., et al. (2020). The m6A RNA demethylase ALKBH5 promotes radioresistance and invasion capability of glioma stem cells. Cancers (Basel) 13 (1), E40. 10.3390/cancers13010040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fata G., Gartner A., Dominguez-Iturza N., Dresselaers T., Dawitz J., Poorthuis R. B., et al. (2014). FMRP regulates multipolar to bipolar transition affecting neuronal migration and cortical circuitry. Nat. Neurosci. 17 (12), 1693–1700. 10.1038/nn.3870 [DOI] [PubMed] [Google Scholar]
- Lane C. A., Hardy J., Schott J. M. (2018). Alzheimer's disease. Eur. J. Neurol. 25 (1), 59–70. 10.1111/ene.13439 [DOI] [PubMed] [Google Scholar]
- Lapointe S., Perry A., Butowski N. A. (2018). Primary brain tumours in adults. Lancet 392 (10145), 432–446. 10.1016/S0140-6736(18)30990-5 [DOI] [PubMed] [Google Scholar]
- Li A., Chen Y. S., Ping X. L., Yang X., Xiao W., Yang Y., et al. (2017). Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell. Res. 27 (3), 444–447. 10.1038/cr.2017.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Yi Y., Miao Y., Long W., Long T., Chen S., et al. (2019). N(6)-Methyladenosine modulates nonsense-mediated mRNA decay in human glioblastoma. Cancer Res. 79 (22), 5785–5798. 10.1158/0008-5472.CAN-18-2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ren Y., Mao K., Hua F., Yang Y., Wei N., et al. (2018). FTO is involved in Alzheimer's disease by targeting TSC1-mTOR-Tau signaling. Biochem. Biophys. Res. Commun. 498 (1), 234–239. 10.1016/j.bbrc.2018.02.201 [DOI] [PubMed] [Google Scholar]
- Li L., Zang L., Zhang F., Chen J., Shen H., Shu L., et al. (2017). Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 26 (13), 2398–2411. 10.1093/hmg/ddx128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Zhao X., Wang W., Shi H., Pan Q., Lu Z., et al. (2018). Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 19 (1), 69. 10.1186/s13059-018-1436-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu S. (2016). Suppression of HBXIP reduces cell proliferation, migration and invasion in vitro, and tumorigenesis in vivo in human urothelial carcinoma of the bladder. Cancer biother. Radiopharm. 12 (5), 311–316. 10.1089/cbr.2016.2038 [DOI] [PubMed] [Google Scholar]
- Liu H., Qin S., Liu C., Jiang L., Li C., Yang J., et al. (2021). m(6 A reader IGF2BP2-stabilized CASC9 accelerates glioblastoma aerobic glycolysis by enhancing HK2 mRNA stability. Cell. Death Discov. 7 (1), 292. 10.1038/s41420-021-00674-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10 (2), 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518 (7540), 560–564. 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Pan T. (2016). N6-methyladenosine–encoded epitranscriptomics. Nat. Struct. Mol. Biol. 23 (2), 98–102. 10.1038/nsmb.3162 [DOI] [PubMed] [Google Scholar]
- Liu N., Zhou K. I., Parisien M., Dai Q., Diatchenko L., Pan T. (2017). N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45 (10), 6051–6063. 10.1093/nar/gkx141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Xiu J., Zhu C., Meng K., Li C., Han R., et al. (2021). Fat mass and obesity-associated protein regulates RNA methylation associated with depression-like behavior in mice. Nat. Commun. 12 (1), 6937. 10.1038/s41467-021-27044-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livneh I., Moshitch-Moshkovitz S., Amariglio N., Rechavi G., Dominissini D. (2020). The m(6)A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 21 (1), 36–51. 10.1038/s41583-019-0244-z [DOI] [PubMed] [Google Scholar]
- Ma C., Chang M., Lv H., Zhang Z. W., Zhang W., He X., et al. (2018). RNA m(6)A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 19 (1), 68. 10.1186/s13059-018-1435-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel M., Chen K. M., Homolka D., Gos P., Pandey R. R., McCarthy A. A., et al. (2018). Methylation of structured RNA by the m(6)A writer METTL16 is essential for mouse embryonic development. Mol. Cell. 71 (6), 986–1000.e11. e11. 10.1016/j.molcel.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkurjev D., Hong W. T., Iida K., Oomoto I., Goldie B. J., Yamaguti H., et al. (2018). Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat. Neurosci. 21 (7), 1004–1014. 10.1038/s41593-018-0173-6 [DOI] [PubMed] [Google Scholar]
- Meyer K. D., Patil D. P., Zhou J., Zinoviev A., Skabkin M. A., Elemento O., et al. (2015). 5' UTR m(6)A promotes cap-independent translation. Cell. 163 (4), 999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan P., Rottman F. M. (1988). An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science 242 (4882), 1159–1162. 10.1126/science.3187541 [DOI] [PubMed] [Google Scholar]
- Nave K. A., Werner H. B. (2014). Myelination of the nervous system: Mechanisms and functions. Annu. Rev. Cell. Dev. Biol. 30, 503–533. 10.1146/annurev-cellbio-100913-013101 [DOI] [PubMed] [Google Scholar]
- Omuro A., DeAngelis L. M. (2013). Glioblastoma and other malignant gliomas: A clinical review. Jama 310 (17), 1842–1850. 10.1001/jama.2013.280319 [DOI] [PubMed] [Google Scholar]
- Pan Y., Xiao K., Li Y., Li Y., Liu Q. (2021). RNA N6-methyladenosine regulator-mediated methylation modifications pattern and immune infiltration features in glioblastoma. Front. Oncol. 11, 632934. 10.3389/fonc.2021.632934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil D. P., Chen C. K., Pickering B. F., Chow A., Jackson C., Guttman M., et al. (2016). m(6 A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537 (7620), 369–373. 10.1038/nature19342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton K. E., Chen B., Liu K., Hunter O. V., Xie Y., Tu B. P., et al. (2017). The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 169 (5), 824–835.e14. e14. 10.1016/j.cell.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping X. L., Sun B. F., Wang L., Xiao W., Yang X., Wang W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell. Res. 24 (2), 177–189. 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Wang S., Li J., Wen Y., Cui R., Zhang K., et al. (2022). Protective role of mRNA demethylase FTO on axon guidance molecules of nigro-striatal projection system in manganese-induced parkinsonism. J. Hazard. Mat. 426, 128099. 10.1016/j.jhazmat.2021.128099 [DOI] [PubMed] [Google Scholar]
- Qiu X., He H., Huang Y., Wang J., Xiao Y. (2020). Genome-wide identification of m(6)A-associated single-nucleotide polymorphisms in Parkinson's disease. Neurosci. Lett. 737, 135315. 10.1016/j.neulet.2020.135315 [DOI] [PubMed] [Google Scholar]
- Quan W., Li J., Liu L., Zhang Q., Qin Y., Pei X., et al. (2021). Influence of N6-methyladenosine modification gene HNRNPC on cell phenotype in Parkinson's disease. Park. Dis. 2021, 9919129. 10.1155/2021/9919129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree I. A., Luo G. Z., Zhang Z., Wang X., Zhou T., Cui Y., et al. (2017). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6, e31311. 10.7554/eLife.31311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowles J., Wong M., Powers R., Olsen M. (2012). FTO, RNA epigenetics and epilepsy. Epigenetics 7 (10), 1094–1097. 10.4161/epi.21977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffary R., Xie Z. (2011). FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J. Neurosci. 31 (4), 1427–1439. 10.1523/JNEUROSCI.4854-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels T. J., Jarvelin A. I., Ish-Horowicz D., Davis I. (2020). Imp/IGF2BP levels modulate individual neural stem cell growth and division through myc mRNA stability. Elife 9, e51529. 10.7554/eLife.51529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M. R., Bray S. M., Warren S. T. (2012). Molecular mechanisms of fragile X syndrome: A twenty-year perspective. Annu. Rev. Pathol. 7, 219–245. 10.1146/annurev-pathol-011811-132457 [DOI] [PubMed] [Google Scholar]
- Schapira A. H., Jenner P. (2011). Etiology and pathogenesis of Parkinson's disease. Mov. Disord. 26 (6), 1049–1055. 10.1002/mds.23732 [DOI] [PubMed] [Google Scholar]
- Shafik A. M., Zhang F., Guo Z., Dai Q., Pajdzik K., Li Y., et al. (2021). N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer's disease. Genome Biol. 22 (1), 17. 10.1186/s13059-020-02249-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Yang L., Wei W. (2021). Role of Fto on CaMKII/CREB signaling pathway of hippocampus in depressive-like behaviors induced by chronic restraint stress mice. Behav. Brain Res. 406, 113227. 10.1016/j.bbr.2021.113227 [DOI] [PubMed] [Google Scholar]
- Shi H., Wang X., Lu Z., Zhao B. S., Ma H., Hsu P. J., et al. (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell. Res. 27 (3), 315–328. 10.1038/cr.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wei J., He C. (2019). Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell. 74 (4), 640–650. 10.1016/j.molcel.2019.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Zhang X., Weng Y. L., Lu Z., Liu Y., Lu Z., et al. (2018). m(6 A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 563 (7730), 249–253. 10.1038/s41586-018-0666-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Chen G., Dong X., Li H., Li S., Cheng S., et al. (2021). METTL3 promotes the resistance of glioma to temozolomide via increasing MGMT and ANPG in a m(6)A dependent manner. Front. Oncol. 11, 702983. 10.3389/fonc.2021.702983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Yang F., Dai R., Sun Y., Chu Y., Liao S., et al. (2021). METTL3-Mediated N6-methyladenosine modification is involved in the dysregulation of NRIP1 expression in Down syndrome. Front. Cell. Dev. Biol. 9, 621374. 10.3389/fcell.2021.621374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H., Matsumoto M., Ishigami Y., Ebina M., Muto A., Sato Y., et al. (2017). S-adenosylmethionine synthesis is regulated by selective N(6)-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell. Rep. 21 (12), 3354–3363. 10.1016/j.celrep.2017.11.092 [DOI] [PubMed] [Google Scholar]
- Si W., Li Y., Ye S., Li Z., Liu Y., Kuang W., et al. (2020). Methyltransferase 3 mediated miRNA m6A methylation promotes stress granule formation in the early stage of acute ischemic stroke. Front. Mol. Neurosci. 13, 103. 10.3389/fnmol.2020.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M., Nave K. A. (2015). Oligodendrocytes: Myelination and axonal support. Cold Spring Harb. Perspect. Biol. 8 (1), a020479. 10.1101/cshperspect.a020479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Zheng X., Sun Y., Yu J., Sheng M., Yan S., et al. (2021). Identification of IGF2BP3 as an adverse prognostic biomarker of gliomas. Front. Genet. 12, 743738. 10.3389/fgene.2021.743738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Ma L., Zhang H., Cao Y., Wang C., Hou N., et al. (2019). Fto deficiency reduces anxiety- and depression-like behaviors in mice via alterations in gut microbiota. Theranostics 9 (3), 721–733. 10.7150/thno.31562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao M., Li X., He L., Rong X., Wang H., Pan J., et al. (2022). Decreased RNA m(6)A methylation enhances the process of the epithelial mesenchymal transition and vasculogenic mimicry in glioblastoma. Am. J. Cancer Res. 12 (2), 893–906. [PMC free article] [PubMed] [Google Scholar]
- Teng Y., Liu Z., Chen X., Liu Y., Geng F., Le W., et al. (2021). Conditional deficiency of m6A methyltransferase Mettl14 in substantia nigra alters dopaminergic neuron function. J. Cell. Mol. Med. 25 (17), 8567–8572. 10.1111/jcmm.16740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M. B., Wang Z., Yang C. (2021). The m(6)A RNA methylation regulates oncogenic signaling pathways driving cell malignant transformation and carcinogenesis. Mol. Cancer 20 (1), 61. 10.1186/s12943-021-01356-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan A., Patil V., Abdulla S., Hoheisel J. D., Somasundaram K. (2019). N⁶-Methyladenosine landscape of glioma stem-like cells: METTL3 is essential for the expression of actively transcribed genes and sustenance of the oncogenic signaling. Genes. (Basel) 10 (2), E141. 10.3390/genes10020141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan A., Patil V., Arora A., Hegde A. S., Arivazhagan A., Santosh V., et al. (2018). Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene 37 (4), 522–533. 10.1038/onc.2017.351 [DOI] [PubMed] [Google Scholar]
- Wang C. X., Cui G. S., Liu X., Xu K., Wang M., Zhang X. X., et al. (2018). METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 16 (6), e2004880. 10.1371/journal.pbio.2004880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Doxtader K. A., Nam Y. (2016). Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 63 (2), 306–317. 10.1016/j.molcel.2016.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Li J., Lin F., Guo J., Zhao J. (2021). Identification of N(6)-methyladenosine-related lncRNAs for patients with primary glioblastoma. Neurosurg. Rev. 44 (1), 463–470. 10.1007/s10143-020-01238-x [DOI] [PubMed] [Google Scholar]
- Wang X., Lu Z., Gomez A., Hon G. C., Yue Y., Han D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505 (7481), 117–120. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao B. S., Roundtree I. A., Lu Z., Han D., Ma H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 161 (6), 1388–1399. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J., Lv R., Ma H., Shen H., He C., Wang J., et al. (2018). Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 69 (6), 1028–1038.e6. 10.1016/j.molcel.2018.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y. L., Wang X., An R., Cassin J., Vissers C., Liu Y., et al. (2018). Epitranscriptomic m(6)A regulation of axon regeneration in the adult mammalian nervous system. Neuron 97 (2), 313–325.e6. 10.1016/j.neuron.2017.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widagdo J., Zhao Q. Y., Kempen M. J., Tan M. C., Ratnu V. S., Wei W., et al. (2016). Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 36 (25), 6771–6777. 10.1523/JNEUROSCI.4053-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtas M. N., Pandey R. R., Mendel M., Homolka D., Sachidanandam R., Pillai R. S. (2017). Regulation of m(6)A transcripts by the 3'→5' RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell. 68 (2), 374–387.e12. e12. 10.1016/j.molcel.2017.09.021 [DOI] [PubMed] [Google Scholar]
- Worpenberg L., Paolantoni C., Longhi S., Mulorz M. M., Lence T., Wessels H. H., et al. (2021). Ythdf is a N6-methyladenosine reader that modulates Fmr1 target mRNA selection and restricts axonal growth in Drosophila. Embo J. 40 (4), e104975. 10.15252/embj.2020104975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Su S., Patil D. P., Liu H., Gan J., Jaffrey S. R., et al. (2018). Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 9 (1), 420. 10.1038/s41467-017-02770-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P. F., Han Q. Q., Chen F. F., Shen T. T., Li Y. H., Cao Y., et al. (2021). Erasing m(6)A-dependent transcription signature of stress-sensitive genes triggers antidepressant actions. Neurobiol. Stress 15, 100390. 10.1016/j.ynstr.2021.100390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Li A., Sun B., Sun J. G., Zhang J., Zhang T., et al. (2019). A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell. Res. 29 (1), 23–41. 10.1038/s41422-018-0113-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z., Xue Y., Zheng J., Liu X., Ma J., Liu Y. (2016). WTAP expression predicts poor prognosis in malignant glioma patients. J. Mol. Neurosci. 60 (2), 131–136. 10.1007/s12031-016-0788-6 [DOI] [PubMed] [Google Scholar]
- Xiao W., Adhikari S., Dahal U., Chen Y. S., Hao Y. J., Sun B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 61 (4), 507–519. 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- Xiong W., Li C., Wan B., Zheng Z., Zhang Y., Wang S., et al. (2022). N6-Methyladenosine regulator-mediated immue patterns and tumor microenvironment infiltration characterization in glioblastoma. Front. Immunol. 13, 819080. 10.3389/fimmu.2022.819080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Dzhashiashvili Y., Shah A., Kunjamma R. B., Weng Y. L., Elbaz B., et al. (2020). m(6 A mRNA methylation is essential for oligodendrocyte maturation and CNS myelination. Neuron 105 (2), 293–309.e5. e5. 10.1016/j.neuron.2019.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Mo Y., Li D., Yu Q., Wang L., Lin F., et al. (2020). N(6)-methyladenosine demethylases Alkbh5/Fto regulate cerebral ischemia-reperfusion injury. Ther. Adv. Chronic Dis. 11, 2040622320916024. 10.1177/2040622320916024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Hsu P. J., Chen Y. S. (2018). Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell. Res. 28 (6), 616–624. 10.1038/s41422-018-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Huang G., Jiang X., Li X., Sun K., Shi Y., et al. (2022). Loss of Wtap results in cerebellar ataxia and degeneration of Purkinje cells. J. Genet. Genomics. 10.1016/j.jgg.2022.03.001 [DOI] [PubMed] [Google Scholar]
- Yi D., Wang Q., Zhao Y., Song Y., You H., Wang J., et al. (2021). Alteration of N (6) -methyladenosine mRNA methylation in a rat model of cerebral ischemia-reperfusion injury. Front. Neurosci. 15, 605654. 10.3389/fnins.2021.605654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K. J., Ringeling F. R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., et al. (2017). Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell. 171 (4), 877–889.e17. 10.1016/j.cell.2017.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Chen M., Huang H., Zhu J., Song H., Zhu J., et al. (2018). Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 46 (3), 1412–1423. 10.1093/nar/gkx1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., She Y., Yang L., Zhuang M., Han P., Liu J., et al. (2021). The m(6) A readers YTHDF1 and YTHDF2 synergistically control cerebellar parallel fiber growth by regulating local translation of the key Wnt5a signaling components in axons. Adv. Sci. 8 (22), e2101329. 10.1002/advs.202101329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zhang Y., Ma H., Zeng R., Liu R., Wang P., et al. (2020). Epitranscriptomic profiling of N6-methyladenosine-related RNA methylation in rat cerebral cortex following traumatic brain injury. Mol. Brain 13 (1), 11. 10.1186/s13041-020-0554-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell. Discov. 4, 10. 10.1038/s41421-018-0019-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Cheng Y., Xue Z., Li J., Wu N., Yan J., et al. (2021). Sevoflurane impairs m6A-mediated mRNA translation and leads to fine motor and cognitive deficits. Cell. Biol. Toxicol. 38, 347–369. 10.1007/s10565-021-09601-4 [DOI] [PubMed] [Google Scholar]
- Zhang S., Zhao B. S., Zhou A., Lin K., Zheng S., Lu Z., et al. (2017). m(6 A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 31 (4), 591–606.e6. 10.1016/j.ccell.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Zhao S., Qi Y., Li B., Wang H., Pan Z., et al. (2022). SPI1-induced downregulation of FTO promotes GBM progression by regulating pri-miR-10a processing in an m6A-dependent manner. Mol. Ther. Nucleic Acids 27, 699–717. 10.1016/j.omtn.2021.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wang M., Xie D., Huang Z., Zhang L., Yang Y., et al. (2018). METTL3-mediated N(6)-methyladenosine mRNA modification enhances long-term memory consolidation. Cell. Res. 28 (11), 1050–1061. 10.1038/s41422-018-0092-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Wang Q., Zhao X., Shao L., Liu G., Zheng X., et al. (2020). YTHDC1 mitigates ischemic stroke by promoting Akt phosphorylation through destabilizing PTEN mRNA. Cell. Death Dis. 11 (11), 977. 10.1038/s41419-020-03186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B. S., Roundtree I. A., He C. (2017). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell. Biol. 18 (1), 31–42. 10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Xu Y., Gao S., Qin L., Austria Q., Siedlak S. L., et al. (2021). METTL3-dependent RNA m(6)A dysregulation contributes to neurodegeneration in Alzheimer's disease through aberrant cell cycle events. Mol. Neurodegener. 16 (1), 70. 10.1186/s13024-021-00484-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C. M., Li C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 49 (1), 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Tang X., Lu M., Sun S., Xie S., Cai J., et al. (2020). microRNA-421-3p prevents inflammatory response in cerebral ischemia/reperfusion injury through targeting m6A Reader YTHDF1 to inhibit p65 mRNA translation. Int. Immunopharmacol. 88, 106937. 10.1016/j.intimp.2020.106937 [DOI] [PubMed] [Google Scholar]
- Zhuang M., Li X., Zhu J., Zhang J., Niu F., Liang F., et al. (2019). The m6A reader YTHDF1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res. 47 (9), 4765–4777. 10.1093/nar/gkz157 [DOI] [PMC free article] [PubMed] [Google Scholar]