Abstract
Background
In vitro, animal model and clinical evidence suggests that tuberculosis is not a monomorphic disease, and that host response to tuberculosis is protean with multiple distinct molecular pathways and pathologies (endotypes). We applied unbiased clustering to identify separate tuberculosis endotypes with classifiable gene expression patterns and clinical outcomes.
Methods
A cohort comprised of microarray gene expression data from microbiologically confirmed tuberculosis patients was used to identify putative endotypes. One microarray cohort with longitudinal clinical outcomes was reserved for validation, as were two RNA-sequencing (seq) cohorts. Finally, a separate cohort of tuberculosis patients with functional immune responses was evaluated to clarify stimulated from unstimulated immune responses.
Results
A discovery cohort, including 435 tuberculosis patients and 533 asymptomatic controls, identified two tuberculosis endotypes. Endotype A is characterised by increased expression of genes related to inflammation and immunity and decreased metabolism and proliferation; in contrast, endotype B has increased activity of metabolism and proliferation pathways. An independent RNA-seq validation cohort, including 118 tuberculosis patients and 179 controls, validated the discovery results. Gene expression signatures for treatment failure were elevated in endotype A in the discovery cohort, and a separate validation cohort confirmed that endotype A patients had slower time to culture conversion, and a reduced cure rate. These observations suggest that endotypes reflect functional immunity, supported by the observation that tuberculosis patients with a hyperinflammatory endotype have less responsive cytokine production upon stimulation.
Conclusion
These findings provide evidence that metabolic and immune profiling could inform optimisation of endotype-specific host-directed therapies for tuberculosis.
Short abstract
The host immune response to tuberculosis (TB) is not uniform. Unbiased bioinformatics identify distinct host immune responses (endotypes) associated with different clinical outcomes and different predicted beneficial host-directed therapy. https://bit.ly/3JbMhQL
Introduction
Molecular host-directed therapies could improve the efficacy and shorten the duration of treatment regimens, or ameliorate tuberculosis (TB)-induced lung pathology. However, the molecular pathology in TB is not homogenous. The fields of asthma, COPD and most cancers have identified biological endotypes: distinct molecular and cellular pathologies leading to similar disease phenotypes. Consequently, treatment for these diseases depends on the specific molecular pathways that are disturbed, with endotype-specific therapies improving clinical outcomes [1]. For example, asthma endotypes can generally be divided into neutrophilic versus eosinophilic disease, with the former being more responsive to macrolide therapy and the latter being corticosteroid-responsive [1]. Leprosy is also treated based on immune endotypes, with the cell-mediated and paucibacillary form requiring less antibiotics for a shorter duration, while the anergic and multibacillary endotype requires more antibiotics for a longer duration. No comparable categorisation system is available to guide TB host-directed therapy.
Studies have identified incongruous immune responses that can lead to TB [2–6]. To further test the premise that there is not a single stereotypical immune response to TB, we sought to provide evidence for the diversity of host responses during TB. Mycobacterial immunity requires a balanced, well-regulated response from multiple cell types. For example, immune control of Mycobacterium tuberculosis requires tightly regulated interferon (IFN)-γ and tumour necrosis factor (TNF)-α, with decreased IFN-γ or TNF-α resulting in decreased M. tuberculosis intracellular killing capacity [4], but with exuberant IFN-γ or TNF-α inducing macrophage and tissue necrosis with extracellular M. tuberculosis survival [2, 3]. To better characterise TB endotypes, we implemented an unbiased clustering of publicly available gene expression data, then validated the results using two external cohorts, eventually identifying a hyperinflammatory TB endotype associated with worse clinical outcomes.
Methods
Study inclusion
A systematic review and meta-analysis were implemented according to Preferred Reporting Items for Systematic reviews and Meta-Analyses guidelines (described in detail in the supplementary methods). Publicly available data were identified using PubMed and the National Center for Biotechnology Information Gene Expression Omnibus repository. Studies without microbiological confirmation, without description of the methods of microbiological confirmation or without evaluation of whole blood were excluded. 10 transcriptomic (eight using microarray and two using RNA-sequencing (seq)) studies using whole blood were identified that included participants with microbiologically confirmed pulmonary TB (supplementary methods). Studies or datasets that included only cases without controls or evaluated <10 000 genes were excluded. Studies using microarray were used for a discovery cohort and RNA-seq datasets for a validation cohort (table 1). One microarray cohort (GSE147689, GSE147691) was reserved for validation because it contained longitudinal clinical outcomes (tables 1 and 2). Normalisation, processing, clustering and development of a gene expression based classifier are described in full in the supplementary methods. Clinical outcomes for this cohort [14] were defined using the TBnet criteria: cure is defined as culture-negative at 6 months, with no positive cultures thereafter and no disease relapse within a year after treatment completion. Treatment failure was based on a positive culture 6 months after treatment initiation or disease relapse within 1 year after treatment completion.
TABLE 1.
Publicly available tuberculosis (TB) transcriptomic studies
| GEO ID | TB | Healthy controls | Countries | Age range (years) | HIV? | Endotype A | Endotype B | |
| Discovery data (microarray) | ||||||||
| Anderson, 2014 [7] | GSE39939 | 35 | 14 | Kenya | <15 | Yes | 23 | 12 |
| Berry, 2010 [8] | GSE19435 | 7 | 12 | United Kingdom | 21–51 | No | 6 | 1 |
| GSE19439 | 13 | 29 | United Kingdom | 19–72 | No | 9 | 4 | |
| GSE19442# | 20 | 31 | South Africa | 18–48 | No | 14 | 6 | |
| GSE19444¶ | 21 | 33 | United Kingdom | 18–78 | No | 11 | 10 | |
| Blankley, 2016 [9] | GSE83456 | 45 | 61 | United Kingdom | 20–80 | No | 27 | 18 |
| Bloom, 2012 [10] | GSE40553 | 29 | 38 | South Africa | >17 | No | 22 | 7 |
| Bloom, 2013 [11] | GSE42825 | 8 | 23 | United Kingdom | >18 | No | 8 | 0 |
| GSE42826 | 11 | 52 | United Kingdom, France | >18 | No | 11 | 0 | |
| GSE42830 | 16 | 38 | United Kingdom | >18 | No | 13 | 3 | |
| Kaforou, 2013 [12] | GSE37250 | 195 | 167 | South Africa, Malawi | 19–53 | Yes | 108 | 87 |
| Walter, 2016 [13] | GSE73408 | 35 | 35 | United States | 20–86 | No | 17 | 18 |
| Borstel validation data (microarray) | ||||||||
| Heyckendorf, 2021 [14] | GSE147689-91 | 121 | 14 | Germany, Romania | 18–85 | No | 64 | 57 |
| RNA-seq validation data | ||||||||
| Leong, 2018 [15] | GSE101705 | 28 | 16 | India | 16–65 | No | 7 | 21 |
| Singhania, 2018 [16] | GSE107991¶ | 21 | 33 | United Kingdom | 18–78 | No | 8 | 13 |
| GSE107992# | 16 | 31 | South Africa | 18–48 | No | 10 | 6 | |
| GSE107994 | 53 | 99 | United Kingdom | 16–84 | No | 40 | 13 |
Data are presented as n. Seven studies, including 533 healthy controls and 435 TB patients, had microarray transcriptomic profiles from whole blood and were used as discovery cohort. Two RNA-sequencing (seq) studies including 118 TB patients and 179 controls were used as a validation dataset. An eighth microarray cohort from Germany and Romania (“Borstel cohort”) was reserved as an additional validation cohort because it contained clinical outcomes. GEO: Gene Expression Omnibus. #: GSE107992 reanalysed samples from GSE19442; ¶: GSE107991 reanalysed samples from GSE19444.
TABLE 2.
Epidemiological characteristics of the German and Romanian (Borstel) validation cohort
| All | Endotype A | Endotype B | p-value # | |
| Participants | 64 | 57 | ||
| Age¶ (years) | 37.95 | 39.78 | 37.95 | 0.6948 |
| Male | 62.2 (56/90) | 62.5 (30/48) | 61.9 (26/42) | 0.4768 |
| BMI¶ (kg·m−2) | 20.75 | 19.49 | 21.28 | 0.0009 |
| Current or previous smoker | 52.0 (51/98) | 46.2 (24/52) | 58.7 (27/46) | 0.1074 |
| Cavitary disease | 71.7 (66/92) | 76.5 (39/51) | 65.9 (27/41) | 0.1305 |
| MDR-TB | 62.0 (75/121) | 54.7 (35/64) | 70.2 (40/57) | 0.0399 |
| Baseline TTP (days) | 17 | 14 | 21 | 0.0169 |
| TCC¶ (days) | 54.0 | 64.5 | 33.5 | 0.0005 |
| Cure | 81.0 (51/63) | 74.4 (29/39) | 91.7 (22/24) | 0.0447 |
| Mortality | 6.3 (4/63) | 10.3 (4/39) | 0 (0/24) | 0.0525 |
Data are presented as n or % (n/N), unless otherwise stated. Not all data were available for all participants. Mann–Whitney test was performed for continuous variables and one-sided Chi-squared test evaluated differences in populations. BMI: body mass index; MDR-TB: multidrug-resistant tuberculosis; TTP: time to positivity; TCC: time to culture conversion. #: Mann–Whitney U-test or one-sided Chi-squared (A worse than B) were used to determine significance, as appropriate; ¶: median.
Immunology validation cohort
Multiplex ELISA (LegendPlex) was implemented with and without overnight mitogen stimulation in a cohort of pulmonary TB patients (n=40) or their asymptomatic household contacts (n=39) from Eswatini. The study protocol was reviewed by institutional review boards and all participation was voluntary and in concordance with the Declaration of Helsinki. TB patients were defined by both the presence of symptoms and microbiological confirmation by MGIT liquid culture and/or Gene Xpert. 22 (55%) of the TB cases and nine (23%) of the household contacts were HIV co-infected.
Statistics and clustering
Chi-squared test with a one-sided tail assessed incidence of clinical variables between endotypes. Rank-sum of the cytokines/chemokines from the ELISA validation assay was used to stratify TB patients. Differences between subgroups were analysed using Mann–Whitney rank sum test. ANOVA or Kruskal–Wallis was implemented with corrections for multiple comparison testing. Full description of normalisation, pre-processing and clustering are included in the supplementary methods. In brief, after ComBat was used to normalise the microarray data, Seurat clustering was applied across a range of resolutions (supplementary figure S1). Enriched pathways were evaluated were evaluated using gene set enrichment analysis (GSEA). To quantify pathway activity scores, gene expression was scaled via z-score transformation with mean of zero and standard deviation of 1 for each gene, then summed z-scores were determined for each pathway.
Results
Discovery and validation cohort selection of TB patient with whole-blood transcriptomic profiles
The discovery cohort included seven studies that profiled whole blood and included both cases and controls by microarray gene expression analysis. The studies used in the discovery cohort included 435 individuals with microbiologically confirmed TB and 533 healthy controls [7–13]. These seven studies were used for unbiased clustering to identify TB endotypes, based on 12 468 commonly evaluated genes (figure 1; table 1). Two additional studies with both cases and controls used RNA-seq transcriptome profiling and included 118 TB patients and 179 controls [15, 16]. These studies were reserved for creation of an RNA-seq validation cohort (table 1). A cohort from Germany and Romania including 121 TB patients and 14 healthy controls using microarrays was reserved as an additional external validation dataset because it included clinical outcome data (tables 1 and 2) [14]. A fourth cohort of TB patients and healthy controls with multiplex ELISA at baseline and phytohaemagglutinin-stimulated whole blood (figure 1; supplementary table S3).
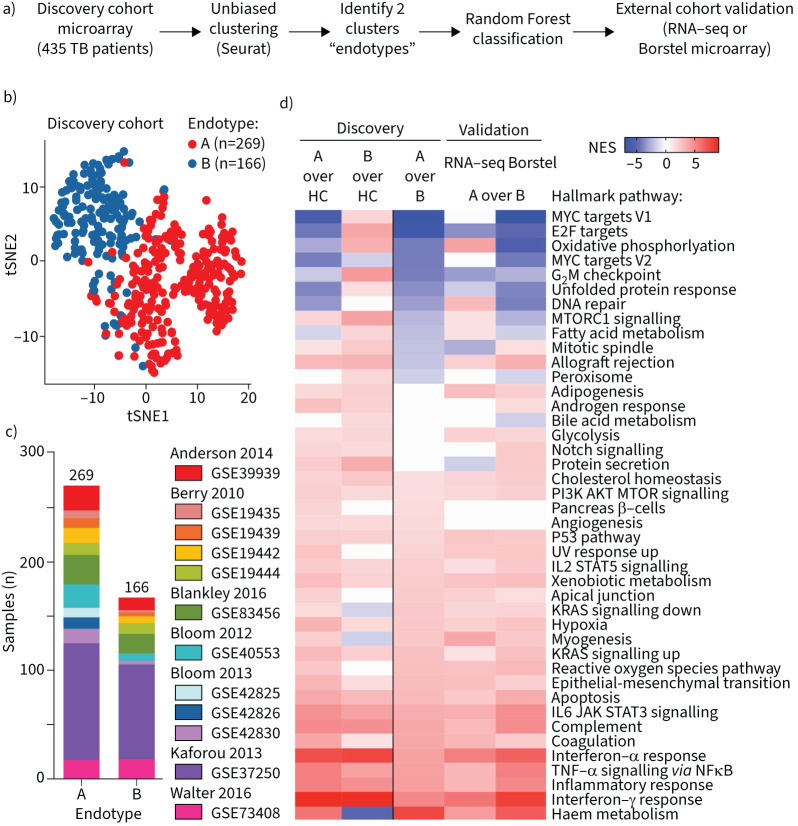
FIGURE 1.
Overview of study. Using unbiased clustering of tuberculosis (TB) patients from seven publicly available microarray studies, a Random Forest (RF) gene classifier was derived to predict TB endotype A versus B. This was validated on two publicly available studies using RNA-sequencing (seq) and on one microarray patient cohort that included longitudinal clinical outcomes data. Immunological validation was evaluated by multiplex ELISA using a separate cohort from Eswatini. Finally, similarity to endotype gene signatures was used to assess and rank previously evaluated host-directed therapy candidates. TCC: time to culture conversion; LINCS: Library of Integrated Network-based Cellular Signatures.
Identification and clinical data characterisation of TB endotypes
To identify potential TB endotypes, unbiased clustering of the microarray transcriptome of 435 TB patients was performed (figure 2a and b). Clustering was evaluated at various resolutions (supplementary figure S1a) with final analysis being performed at resolution 0.4 due to limited discriminatory capacity at higher resolutions. At resolution 0.4, two distinct endotypes were identified. Endotype A consisted of 269 (54.8%) TB patients, and endotype B included 166 (45.1%) TB patients. Patients from each country and each study were well distributed in both endotype clusters (figure 2c; table 1). In the discovery cohort, only two datasets included individuals co-infected with HIV (GSE37250 and GSE39939; table 1). The 108 TB patients living with HIV clustered into endotype A and endotype B, with 57 and 51 in each, respectively. Only one study included children aged <15 years (GSE39939), with 23 children clustering in endotype A and 12 clustering in endotype B (table 1).
FIGURE 2.
Unbiased clustering identifies unique tuberculosis (TB) endotypes. a) Unbiased clustering was implemented on discovery cohort of seven studies (table 1), identifying two major endotypes; next, a Random Forest gene classifier was developed and applied to two external validation datasets. b) Network-based unbiased clustering using the Louvain method identifies two major endotypes of TB. c) Distribution of individual studies into endotype A or B. d) TB endotypes were compared to healthy controls and against each other, then pathway enrichment via gene set enrichment analysis was carried out against the Hallmark pathway compendium. Discovery and validation cohorts are described in table 1. seq: sequencing; tSNE: t-distributed stochastic neighbour embedding; HC: healthy controls; NES: normalised enrichment score; mTORC: mammalian target of rapamycin complex; PI3K: phosphoinositide 3-kinase; UV: ultraviolet; IL: interleukin; STAT: signal transducer and activator of transcription; JAK: Janus kinase; TNF: tumour necrosis factor.
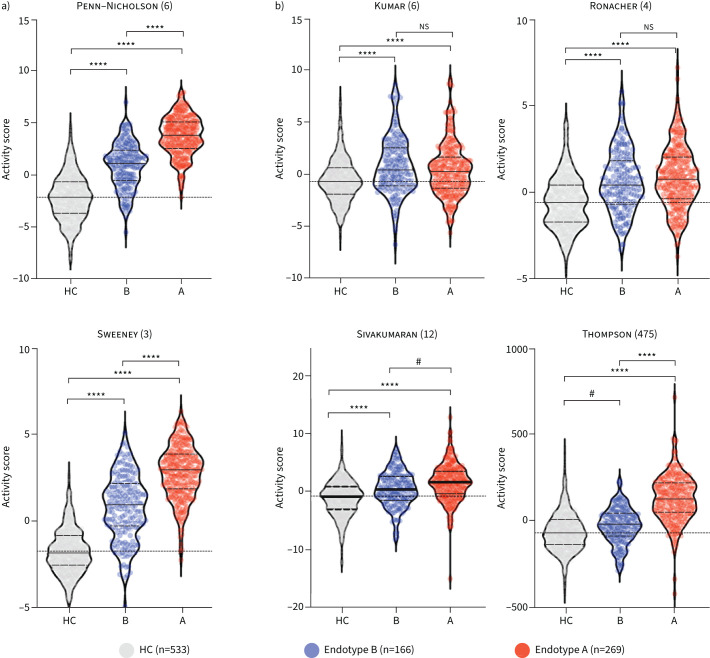
Using the Random Forest machine-learning algorithm, a classifier was derived to categorise endotype A versus endotype B based on the discovery cohort. Genes were ranked by their individual classification score, then accuracy of a Random Forest gene classifier was evaluated across a range of the top informative genes using the average (out-of-bag) classification error, determined by repeated subsampling (supplementary methods; supplementary figure S2a). A 40-gene classifier had both a low misclassification rate and comprised a low gene count. We applied the endotype classifier to a validation cohort comprised of 118 TB patients and 179 controls from two previously published RNA-seq studies (table 1). In the RNA-seq validation cohort, 65 TB samples classified as endotype A (55%) and 53 as endotype B (45%; table 1). We computed pathway enrichment using GSEA between each of the predicted endotypes and the control samples, and for the comparison between the endotypes. Random Forest classification recapitulated the significant enrichments for immune-related pathways (figure 2d, supplementary figure S2b). For one study, microarray profiles (GSE19442 and GSE19444) were used in the discovery dataset, whereas RNA-seq profiles (GSE107991) for the same samples were also used in the RNA-seq validation cohort. Our classifier achieved good concordance between the microarray discovery cohort endotypes and the predicted RNA-seq endotypes (supplementary figure S3). Similar to the discovery cohort, compared to both healthy controls and endotype B, endotype A enriched for inflammation, IFN-γ signalling, TNF-α and haem metabolism. Compared to endotype A, endotype B enriched for pathways related to cellular proliferation including E2F, G2M and mitosis (figure 2d). Compared to endotype A, gene targets of the transcription factors E2F, ELK1 and NRF1 were increased in endotype B (supplementary table S2; supplementary figure S5). The endotypes were evaluated against six previously published scores that identified risk of treatment failure, with two of the scores also identifying risk for TB disease severity. (Gene signature activity score computation via z-score normalisation is described in the supplementary methods.) Compared to both healthy controls and endotype B, endotype A exhibited higher scores for risk of treatment failure and more severe disease compared to healthy controls (figure 3).
FIGURE 3.
Evaluation of tuberculosis (TB) risk signatures over TB endotypes. Previous studies identified gene signatures (number of signatures given in parentheses) associated with a) disease severity and treatment failure [17, 18] or b) risk of treatment failure [19–22]. These signatures were evaluated in healthy controls (HC), endotype A and endotype B. Activity scores (summed z-scores across all signature genes) were computed for each risk signature across HC, endotype A and endotype B; ANOVA. ns: nonsignificant. ****: p<0.0001, #: p<0.0002. Data are presented as median (interquartile range), with dotted line at median of HC.
Differential clinical outcome between TB endotypes
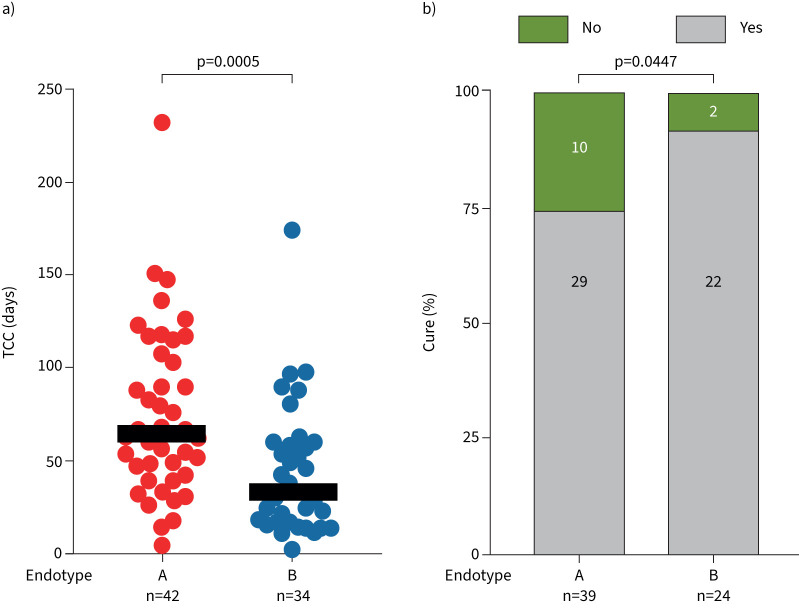
The classifier was applied to a cohort of TB patients assayed using microarray from a prospective, multicentre trial in Germany and Romania. This cohort contained information on baseline bacillary burden, time to culture conversion, and clinical outcomes defined by the TBnet criteria (table 2). Out of 121 TB patients, 64 were classified as endotype A (53%), while 57 were classified as endotype B (47%). Similar to the RNA-seq validation cohort, endotype A and B demonstrated distinct enriched pathway profiles (figure 2d, supplementary figure S2b). Based on the increased predicted treatment failure and disease severity signatures (figure 3), we hypothesised that endotype A patients would display worse clinical outcome compared to endotype B patients. Whereas endotype A had a lower rate of multidrug resistance (54.7% versus 70.2%), endotype A had slightly increased rates of cavitary disease (endotype A 76.5% versus endotype B 65.9%, p=0.1305), higher initial bacterial load (14 versus 21 days; p=0.0169), slower times to culture conversion (64.6 days versus 33.5 days, p=0.0005; figure 4a and table 2) and decreased rates of cure outcomes (74.4% versus 91.7%, p=0.0447; figure 4b and table 2). All deaths occurred in endotype A (11.7% versus 0%, p=0.0525). With antibiotics, the predominance of individuals classified as endotype A at baseline were reclassified as endotype B after weeks of therapy (supplementary figure S4).
FIGURE 4.
Endotype evaluation of tuberculosis (TB) clinical outcomes. Using the clinical annotations of the Borstel TB cohort, outcome differences between endotypes and association of pathway scores with outcomes were evaluated. a) Time to culture conversion (TCC) in TB patients identified as endotype A or B (p=0.0005 by Mann–Whitney U-test). b) Rates of cure in TB patients identified as endotype A or B (p=0.0447 by one-sided Chi-squared test).
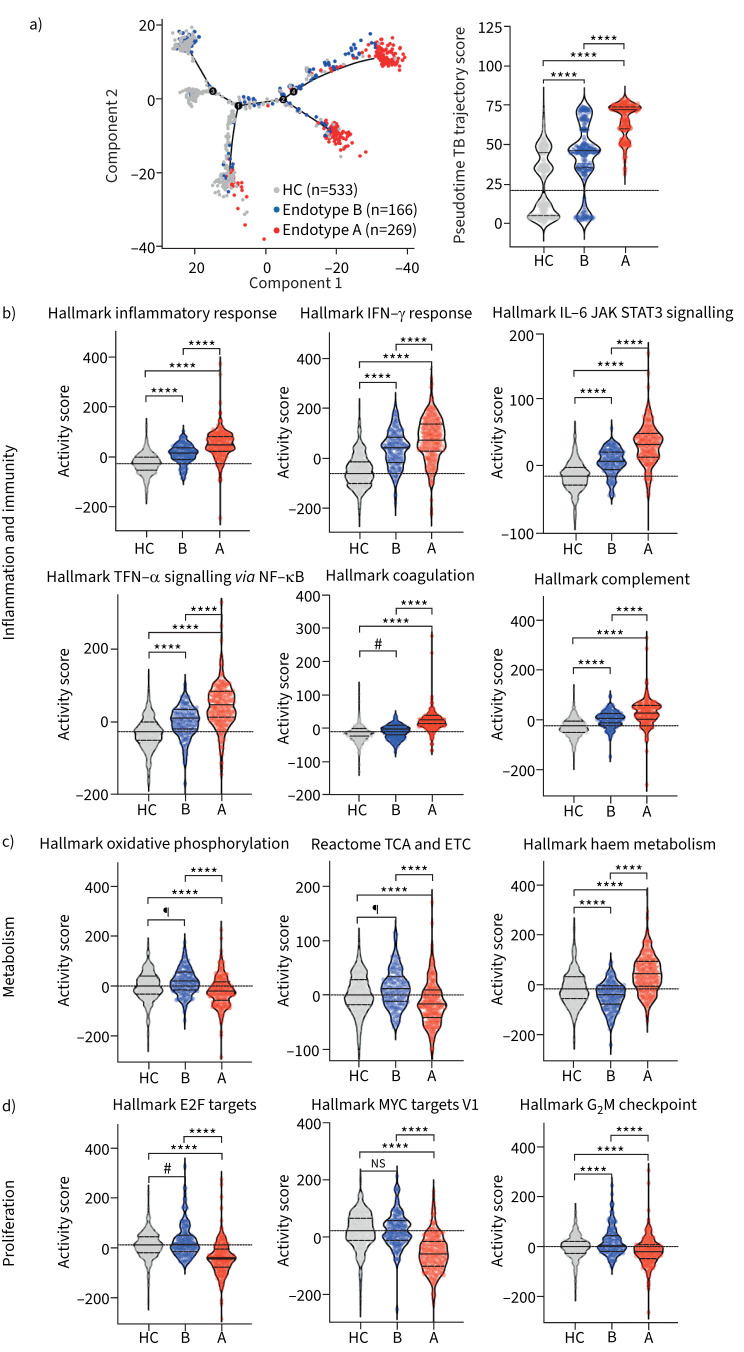
Characterising transcriptome trajectories across endotypes
To understand the relationships between controls and endotypes A and B, we employed cell trajectory (also termed pseudotime trajectory) based on transcriptomic profiles. The trajectory score increased from healthy controls to endotype B to endotype A (figure 5a); this result led to a search for specific molecular properties that follow the predicted disease trajectory. We computed pathway activity scores for each patient in the discovery cohort. The results demonstrate that, in general, pathways related to inflammation and immunity increase in a monotonic manner from healthy controls to endotype B to endotype A (figure 5b). Upon acute infection, increased glycolysis, the tricarboxylic acid (TCA) cycle and one-carbon metabolism provide metabolites requisite to fuel cellular proliferation; however, if infection is chronic, cells become metabolically exhausted and proliferation decreases [23–26]. Compared to healthy controls, endotype B has increased expression of pathways related to metabolism and proliferation (oxidative phosphorylation, electron transport chain (ETC) and G2M; figure 5c and d and supplementary table S2). In contrast, endotype A patients exhibit decreased activity scores for pathways related to cellular proliferation (G2M, MYC and E2F; p<0.001) and metabolism (oxidative phosphorylation, the TCA cycle and the ETC; p<0.001; figure 5c and d).
FIGURE 5.
Tuberculosis (TB) endotypes display distinct immune and metabolic gene expression activity scores. a) Pseudotime TB trajectory score in discovery cohort. Pathway activity scores were evaluated between healthy controls (HC), endotype B and endotype A. b) Inflammation and immunity pathways; c) metabolic pathways; and d) proliferation pathways. Specific gene changes are presented in supplementary table S2. ANOVA was used. Data are presented as median (interquartile range), with dotted line at median of HC. IFN: interferon; JAK: Janus kinase; STAT: signal transducer and activator of transcription; TNF: tumour necrosis factor; TCA: tricarboxylic acid; ETC: electron transport chain; ns: nonsignificant. ****: p<0.0001, #: p<0.0002, ¶: p<0.0021.
Hyperinflammatory, hyporesponsive TB endotype
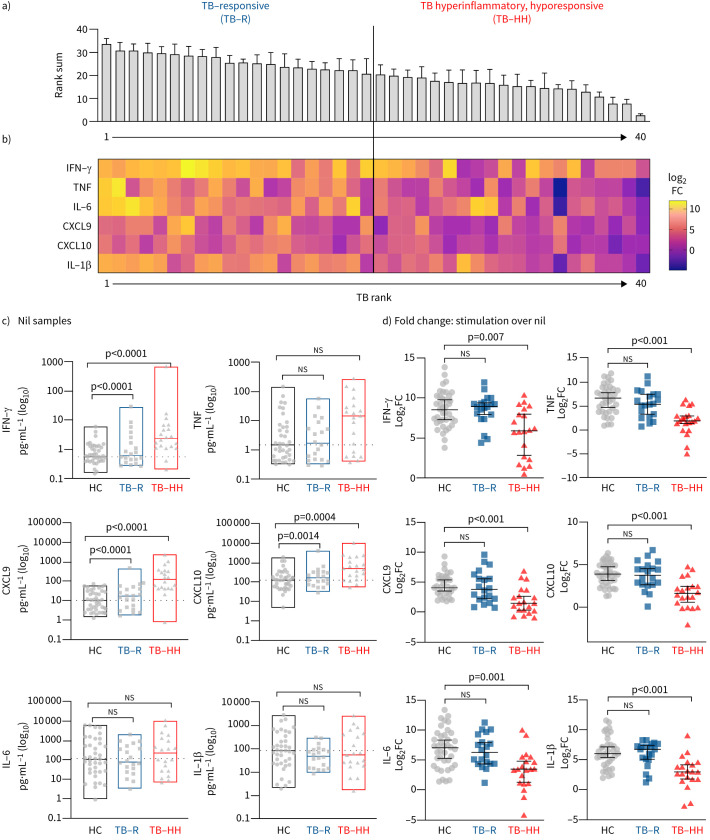
At the gene transcription level, TB endotype A is characterised by increased inflammation and increased interferon, TNF-α and interleukin (IL)-6 signalling in nonstimulated blood (figure 2d). To evaluate the functional response upon stimulation, an independent cohort from Eswatini with mitogen-stimulated whole-blood samples was analysed by ELISA (supplementary table S3). Rank-sum analysis was implemented to stratify the patients into immune-responsive versus less-responsive groups based on their response to stimulation with mitogen (phytohaemagglutinin) (figure 6a and b). The two TB patient subgroups were compared to healthy controls. The hyporesponsive group demonstrated a baseline hyperinflammatory condition, similar to endotype A, but decreased capacity to upregulate IFN-γ, TNF-α, IL-1β, IL-6, CXCL9 and CXCL10 upon stimulation with mitogen (figure 6c and d) (p<0.007). The immune-responsive group was similar to the healthy controls with regard to the capacity to respond to stimulation. Among the 40 TB patients, there were four deaths; three in the hyperinflammatory, hyporesponsive group, and one in the immune-responsive group (Chi-squared p=0.27).
FIGURE 6.
Identification of hyperinflammatory, hyporesponsive cytokine production in tuberculosis (TB) patient endotypes. Whole blood from TB patients (n=40) and healthy controls (n=39) was stimulated overnight with or without mitogen (phytohaemagglutinin), followed by measurement of cytokines and chemokines. a) Samples were ranked for upregulation of six cytokines to determine an overall rank sum (1 lowest, 40 highest). Using the rank-sum value, TB patients were then split in half into “hyporesponsive” and “responsive” groups. b) Heatmap of cytokine expression as log2 fold change (FC) relative to controls. c) Absolute protein expression of the nonstimulated plasma. d) Cytokine protein expression (log2 FC) is graphed for each subgroup. Significance determined by Kruskal–Wallis with Dunn's multiple comparison test. IFN: interferon; TNF: tumour necrosis factor; IL: interleukin.
Comparison of chemical compound signatures to TB endotype signatures
The Library of Integrated Network-based Cellular Signatures enables the identification and prioritisation of putative drugs to treat a pathological condition by countering its transcriptome signature. Comparing gene signatures for endotypes A and B to healthy controls, we obtained ranked lists of >5000 chemical compounds for each endotype and performed a comparative analysis. Previously identified candidates for host-directed therapy, including vitamin D, glucocorticoids, nonsteroidal anti-inflammatory drugs (NSAIDs) and retinoids, demonstrated connectivity scores that suggest a putative benefit for one endotype and either an inconsequential or contradictory response for the other endotype (supplementary figure S6). For example, histone deacetylase inhibitors such as vorinostat and phenylbutyrate demonstrated transcriptomic signatures similar to endotype A, but dissimilar to endotype B.
Discussion
In the pre-antibiotic era, a fifth of humans with active TB survived >10 years [27], but present knowledge is inadequate to describe the underlying mechanisms of a sufficient immune response to overcome TB and contain M. tuberculosis infection. Furthermore, to date, a single adjuvant host-directed therapy has not been identified, probably because the immune response to TB is protean and polymorphic. In this study, we identified clinically relevant TB endotypes by using unbiased clustering of unstimulated blood transcriptomes. Compared to controls, both endotypes displayed elevated gene expression related to pathways for inflammation and immunity, with higher levels among endotype A. Compared to controls, endotype B enriched for oxidative phosphorylation, the TCA cycle and pathways related to cellular proliferation, while endotype A demonstrated decreased pathways related to proliferation. Haem metabolism was upregulated in endotype A and downregulated in endotype B compared to controls, as described previously [6]. We derived a concise Random Forest classifier for TB endotypes, then used it to predicted endotypes in a validation cohort with richly annotated clinical outcomes; endotype A demonstrated slower times to bacterial clearance, and reduced incidence of disease cure.
Patients with TB are currently treated based on studies examining large heterogeneous groups. However, it is reasonable to hypothesise that subgroups exist within these large populations and that stratified and precision medicine strategies may improve outcomes [28]. These data provide support for individually stratified treatment approaches. Considering the animal model, in vitro and human evidence, additional subtypes and endotypes will become identifiable when more robust epidemiology, strain characterisations and functional immune analyses are integrated with transcriptomic results. It is notable that both endotypes exhibit elevated unstimulated gene expression levels of IFN-γ and TNF-α; however, in functional studies the TB patients with elevated basal IFN-γ and TNF-α were less likely to upregulate IFN-γ and TNF-α upon stimulation. For endotypes to help guide host-directed therapy, future pair-wise transcriptomic and immune function studies are needed to confirm that endotype A displays characteristics of immune exhaustion (hyperinflammatory, but hyporesponsive), and that circulating endotypes correlate with the tissue-specific immune function.
The trajectory analyses suggest that pathways related to immunity and inflammation monotonically progress from healthy controls to first endotype B and subsequently to endotype A. In contrast, cellular proliferation and oxidative phosphorylation and the TCA cycle increase in endotype B, but decrease in endotype A. Similarly, pathways related to proliferation decrease from controls to endotype A. This is a pattern similar to murine models of TB and other chronic infections [23, 29, 30], and therefore suggests that a stage-specific intervention can prevent the progression to terminal immune exhaustion in TB.
This study is limited in its capacity to determine appropriateness of the host immune response due to limited metadata and suboptimal means to quantify bacillary burden. Immunity to M. tuberculosis is tightly regulated to avoid pathological inflammation [2, 26, 31, 32]. Animal models have demonstrated that both IFN-γ and TNF-α require delicate homeostatic regulation with deficient responses allowing disease progression and exuberant responses resulting in immune-mediated pathology [2, 3, 26, 31, 33]. The validation cohort included used the best available measurement of bacillary burden (liquid culture time to positivity) and suggests that endotype A has a hyperinflammatory response with delayed culture conversion. Prospective studies need to combine gene expression analysis with functional immunology and quantitative measures of bacillary burden to clarify the appropriateness of host immunity in respect to bacillary burden. We anticipate that once endotypes are analysed using robust multi-omic platforms, effective and pragmatic classifiers could use a minimal complement of informative features. Capitalising on this minimised complement, cost-effective diagnostics could be developed and deployed at point-of-care in TB high burden settings.
The link between metabolism, particularly glycolysis, and immune function has been appreciated for >90 years [24–26]. Initially upon immune activation, there is an increase in both glycolysis and oxidative phosphorylation; however, with sustained activation, immune cells become metabolically exhausted, leading to transcriptional and epigenetic changes that drive immune exhaustion [23, 29, 30, 34–36]. Therefore, it is interesting that endotypes displayed incongruent regulation of genes and pathways related to metabolism, proliferation and immune response. Many host-directed therapy candidates target these pathways. For example, metformin mediates the AKT–mammalian target of rapamycin (mTOR) pathway, blunting cellular glycolysis and the TCA cycle, leading to inhibition of chromatin conformational changes that drive antigen-induced immune function [34, 37]. Everolimus, another inhibitor of mTOR, decreased TB-induced lung damage [32]. In silico evidence suggests the most pronounced benefit of everolimus for endotype A.
Previously identified candidates for host-directed therapies include IFN-γ, granulocyte–macrophage colony-stimulating factor, TNF-α, TNF-α inhibitors, NSAIDs, vitamin D, glucocorticoids, histone deacetylase inhibitors, mTOR modulators, retinoids and statins [32, 38]. The in silico analysis demonstrated that previously identified host-directed therapies would perform better if applied in an endotype-specific manner. If functional studies validate one endotype to have decreased immune responsiveness, then vitamin D or exogenous recombinant IFN-γ may be an appropriate host-directed therapy option. In contrast, if future validation studies demonstrate one endotype to have pathological, exuberant immunity, then NSAID, TNF-α inhibitor or glucocorticoid treatment would be appropriate. Animal and in vitro models that recapitulate the clinically relevant endotypes are also needed to better evaluate endotype-specific host-directed therapies.
All included studies evaluated unstimulated host gene expression. TB is a chronic infection resulting in immune suppression. While many genes downstream of IFN-γ are elevated in TB patients at baseline [7, 9, 39, 40], they have decreased antigen-induced immune upregulation [41–44]. The multiplex ELISA data highlight the limitations of inferring immune function based on nonstimulated gene expression measurements; in fact, the group with elevated baseline cytokines (hyperinflammatory) was hyporesponsive upon stimulation. Additional gene expression subclusters were visible at higher resolution; however, biological relevance was not readily obvious in these subclusters. We speculate that the integration of transcriptomics with functional immune analysis, more robust epidemiology and strain characterisation will identify more than two endotypes.
Progression to TB is related to interactions among host, pathogen and environmental factors. Progression to a specific endotype of TB is likely similarly related to as-yet unappreciated interactions. Unlike the Cancer Genome Atlas, very limited epidemiology is available in existing public data repositories. Epidemiological predispositions probably drive the divergent endotypes, including malnutrition, HIV, helminths, tobacco use and/or indoor biomass fuel exposure. For example, despite successful deworming, previous schistosomiasis infection ablates mycobacterial immunity, leaving long-lasting immune suppression. We speculate that individuals with pre-existing immune suppression progress rapidly to endotype A, in contrast to previously healthy individuals.
In conclusion, this unbiased clustering provides additional evidence that there are multiple molecular host pathways modulated during TB [2–6, 28]. This analysis of transcriptome and protein data from TB patients provides additional evidence for biologically distinctive TB endotypes that differentially affect clinical outcomes. Specifically, host gene expression in TB patient clusters into at least two endotypes with differential immune and metabolic transcriptomic signatures. These observations suggest that different endotypes display responses that are likely to have clinical and pathological relevance and provides the basis for studies to evaluate endotype-specific host-directed therapies.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary methods. ERJ-02263-2021.Supplement (122.6KB, pdf)
Supplementary table S1. a) Differential expressed genes in the discovery cohort at resolution 0.4. b) genes comprising the random forest classifier. ERJ-02263-2021.Table_S1 (55KB, xlsx)
Supplementary table S2. Enriched gene sets by gene set enrichment analysis (GSEA). a) Enriched hallmark pathways. b) Enriched pathway analysis from KEGG, reactome, and gene ontology biological process. c) Enriched transcription factor targets. ERJ-02263-2021.Table_S2 (926KB, xlsx)
Supplementary table S3. Demographic information from cohort 4 multiplex ELISA (BioLegend LegendPlex). ERJ-02263-2021.Table_S3 (19.1KB, xlsx)
Supplementary figure S1. a) Cluster tree of the discovery cohort based on different resolutions of the Louvain method. Evaluation occurred at resolution 0.4, which identified 2 clusters, labeled endotype A and B. Up to four sub-clusters are visible at resolution 1.0. b) tSNE plot of the discovery cohort including healthy controls, endotype A, and endotype B. ERJ-02263-2021.Figure_S1 (737.1KB, jpg)
Supplementary figure S2. a) Classification error rate for random forest classifiers developed across a range of the top informative genes. Classification error rate is the average prediction error determined by repeated subsampling with replacement of the endotype A and B samples. b) Heatmap of GSEA normalized enrichment scores (NES) for Hallmark pathways using the 40, 50 and 500 gene TB endotype classifiers over discovery and validation cohorts. ERJ-02263-2021.Figure_S2 (747.2KB, jpg)
Supplementary figure S3. Thirty-four samples were included both in the Berry 2010 microarray and Singhania 2018 RNA-seq studies. tSNE plot demonstrating the low discordant (green in a and red in b) sample classification. ERJ-02263-2021.Figure_S3 (878.1KB, jpg)
Supplementary figure S4. Longitudinal analysis of endotype classification after time on antimicrobial therapy. ERJ-02263-2021.Figure_S4 (296.2KB, jpg)
Supplementary figure S5. Enriched transcription factors targets of between endotype A and endotype B. Bars represent the normalised enrichment score for endotype A versus endotype B. Comparison using Gene set enrichment analysis with an FDR <0.05. Red bars indicate higher expression in endotype A and blue bars indicate higher expression in endotype B. Full data can be found in supplementary table. ERJ-02263-2021.Figure_S5 (671.1KB, jpg)
Supplementary figure S6. Heatmap of connectivity scores for select chemical compounds within the TB endotypes A and B based on the Library of Integrated Network-based Cellular Signatures (LINCS). Positive connectivity scores represent compounds inducing gene expression profiles similar to the endotype, while negative connectivity scores represent compounds inducing gene expression profiles antithetical to the endotype. ERJ-02263-2021.Figure_S6 (512.1KB, jpg)
Shareable PDF
Footnotes
Members of the DZIF-TB cohort study group: Jan Heyckendorf (Division of Clinical Infectious Diseases, Research Center Borstel, Borstel, Germany; German Center for Infection Research (DZIF), Germany; International Health/Infectious Diseases, University of Lübeck, Lübeck, Germany), Sebastian Marwitz (Pathology of the Universal Medical Center Schleswig-Holstein (UKSH) and the Research Center Borstel, Campus Borstel, Airway Research Center North (ARCN); German Center for Lung Research (DZL), Germany), Maja Reimann (Division of Clinical Infectious Diseases, Research Center Borstel, Borstel, Germany; German Center for Infection Research (DZIF), Germany; International Health/Infectious Diseases, University of Lübeck, Lübeck, Germany), Korkut Avsar (Asklepios Fachkliniken München-Gauting, Munich, Germany), Andrew DiNardo (The Global Tuberculosis Program, Texas Children's Hospital, Immigrant and Global Health, Department of Pediatrics, Baylor College of Medicine, Houston, USA), Gunar Günther (Department of Medicine, University of Namibia School of Medicine, Windhoek, Namibia; Inselspital Bern, Department of Pulmonology, Bern, Switzerland), Michael Hoelscher (Division of Infectious Diseases and Tropical Medicine, University Hospital, LMU Munich, Munich, Germany; German Center for Infection Research (DZIF), partner site Munich, Germany), Elmira Ibraim (Institutul de Pneumoftiziologie “Marius Nasta”, MDR-TB Research Department, Bucharest, Romania), Barbara Kalsdorf (Division of Clinical Infectious Diseases, Research Center Borstel, Borstel, Germany; German Center for Infection Research (DZIF), Germany; International Health/Infectious Diseases, University of Lübeck, Lübeck, Germany), Stefan H.E. Kaufmann (Max Planck Institute for Infection Biology, Berlin, Germany; Max Planck Institute for Biophysical Chemistry, Göttingen, Germany; Hagler Institute for Advanced Study, Texas A&M University, College Station, USA), Irina Kontsevaya (Division of Clinical Infectious Diseases, Research Center Borstel, Borstel, Germany; German Center for Infection Research (DZIF), Germany; International Health/Infectious Diseases, University of Lübeck, Lübeck, Germany), Frank van Leth (Department of Global Health, Amsterdam University Medical Centres, Location AMC, Amsterdam, The Netherlands; Amsterdam Institute for Global Health and Development, Amsterdam, The Netherlands), Anna Maria Mandalakas (The Global Tuberculosis Program, Texas Children's Hospital, Immigrant and Global Health, Department of Pediatrics, Baylor College of Medicine, Houston, USA), Florian P. Maurer (National and WHO Supranational Reference Center for Mycobacteria, Research Center Borstel, Borstel, Germany; Institute of Medical Microbiology, Virology and Hygiene, University Medical Center Hamburg-Eppendorf, Hamburg, Germany), Marius Müller (Sankt Katharinen-Krankenhaus, Frankfurt, Germany), Dörte Nitschkowski (Pathology of the Universal Medical Center Schleswig-Holstein (UKSH) and the Research Center Borstel, Campus Borstel, Airway Research Center North (ARCN); German Center for Lung Research (DZL), Germany), Ioana D. Olaru (London School of Hygiene and Tropical Medicine, London, UK; Biomedical Research and Training Institute, Harare, Zimbabwe), Cristina Popa (Institutul de Pneumoftiziologie “Marius Nasta”, MDR-TB Research Department, Bucharest, Romania), Andrea Rachow (Division of Infectious Diseases and Tropical Medicine, University Hospital, LMU Munich, Munich, Germany; German Center for Infection Research (DZIF), partner site Munich, Germany), Thierry Rolling (Division of Infectious Diseases, I. Department of Internal Medicine, German Center for Infection Research (DZIF); University Medical Centre Hamburg-Eppendorf, Hamburg, Germany, Department of Clinical Immunology of Infectious Diseases, Bernhard-Nocht-Institute for Tropical Medicine, Hamburg, Germany), Jan Rybniker (Department I of Internal Medicine, Division of Infectious Diseases, University of Cologne, Cologne, Germany; German Center for Infection Research (DZIF), Partner Site Bonn-Cologne, Cologne, Germany; Center for Molecular Medicine Cologne, University of Cologne, Cologne, Germany), Helmut J.F. Salzer (Department of Pulmonology, Kepler University Hospital, Linz, Austria), Patricia Sanchez-Carballo (Division of Clinical Infectious Diseases, Research Center Borstel, Borstel, Germany; German Center for Infection Research (DZIF), Germany; International Health/Infectious Diseases, University of Lübeck, Lübeck, Germany), Maren Schuhmann (Universitäts Thoraxklinik-Heidelberg, Heidelberg, Germany), Dagmar Schaub (Division of Clinical Infectious Diseases, Research Center Borstel, Borstel, Germany; German Center for Infection Research (DZIF), Germany; International Health/Infectious Diseases, University of Lübeck, Lübeck, Germany), Victor Spinu (Institutul de Pneumoftiziologie “Marius Nasta”, MDR-TB Research Department, Bucharest, Romania), Isabelle Suárez (German Center for Infection Research (DZIF), Partner Site Bonn-Cologne, Cologne, Germany), Elena Terhalle (Division of Clinical Infectious Diseases, Research Center Borstel, Borstel, Germany; German Center for Infection Research (DZIF), Germany; International Health/Infectious Diseases, University of Lübeck, Lübeck, Germany), Markus Unnewehr (Department of Respiratory Medicine and Infectious Diseases, St. Barbara-Klinik, Hamm, Germany; University of Witten-Herdecke, Witten, Germany), January Weiner 3rd (Berlin Institute of HealthCUBI (Core Unit Bioinformatics), Berlin, Germany), Torsten Goldmann (Pathology of the Universal Medical Center Schleswig-Holstein (UKSH) and the Research Center Borstel, Campus Borstel, Airway Research Center North (ARCN); German Center for Lung Research (DZL), Germany), Christoph Lange (Division of Clinical Infectious Diseases, Research Center Borstel, Borstel, Germany; German Center for Infection Research (DZIF), Germany; International Health/Infectious Diseases, University of Lübeck, Lübeck, Germany; The Global Tuberculosis Program, Texas Children's Hospital, Immigrant and Global Health, Department of Pediatrics, Baylor College of Medicine, Houston, USA)..
Conflict of interest: The authors have declared that no conflict of interest exists.
Support statement: A.R. DiNardo is supported by NIAID K23 AI141681-02. S.L. Grimm, T. Gandhi and C. Coarfa are supported by the Cancer Prevention Institute of Texas (CPRIT) RP170005, RP200504 and RP210227, NIH/NIAID 1U19AI144297, NIH/NCI P30 shared resource grant CA125123, and NIEHS grants P30 ES030285 and P42 ES027725. J. Heyckendorf and C. Lange are supported by the German Center for Infection Research (DZIF). J.D. Cirillo is funded in part from funds provided by the Texas A&M University System and National Institutes of Health grant AI104960. M.G. Netea is supported by an ERC Advanced Grant (#833247) and a Spinoza grant of the Netherlands Organization for Scientific Research. R. van Crevel is supported by National Institute of Health (R01AI145781). A.M. Mandalakas is supported by NIH R01AI137527, U01GH002278 and DoD W81XWH1910026. Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Svenningsen S, Nair P. Asthma endotypes and an overview of targeted therapy for asthma. Front Med 2017; 4: 158. doi: 10.3389/fmed.2017.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tobin DM, Roca FJ, Oh SF, et al. Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell 2012; 148: 434–446. doi: 10.1016/j.cell.2011.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sakai S, Kauffman KD, Sallin MA, et al. CD4 T cell-derived IFN-γ plays a minimal role in control of pulmonary Mycobacterium tuberculosis infection and must be actively repressed by PD-1 to prevent lethal disease. PLoS Pathog 2016; 12: e1005667. doi: 10.1371/journal.ppat.1005667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bustamante J, Boisson-Dupuis S, Abel L, et al. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin Immunol 2014; 26: 454–470. doi: 10.1016/j.smim.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ní Cheallaigh C, Sheedy FJ, Harris J, et al. A common variant in the adaptor Mal regulates interferon gamma signaling. Immunity 2016; 44: 368–379. doi: 10.1016/j.immuni.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrade BB, Pavan Kumar N, Amaral EP, et al. Heme oxygenase-1 regulation of matrix metalloproteinase-1 expression underlies distinct disease profiles in tuberculosis. J Immunol 2015; 195: 2763–2773. doi: 10.4049/jimmunol.1500942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heyckendorf J, Marwitz S, Reimann M, et al. Prediction of anti-tuberculosis treatment duration based on a 22-gene transcriptomic model. Eur Respir J 2021; 58: 2003492. doi: 10.1183/13993003.03492-2020 [DOI] [PubMed] [Google Scholar]
- 8.Berry MP, Graham CM, McNab FW, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010; 466: 973–977. doi: 10.1038/nature09247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloom CI, Graham CM, Berry MP, et al. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS One 2012; 7: e46191. doi: 10.1371/journal.pone.0046191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bloom CI, Graham CM, Berry MP, et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One 2013; 8: e70630. doi: 10.1371/journal.pone.0070630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walter ND, Miller MA, Vasquez J, et al. Blood transcriptional biomarkers for active tuberculosis among patients in the United States: a case-control study with systematic cross-classifier evaluation. J Clin Microbiol 2016; 54: 274–282. doi: 10.1128/JCM.01990-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson ST, Kaforou M, Brent AJ, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med 2014; 370: 1712–1723. doi: 10.1056/NEJMoa1303657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaforou M, Wright VJ, Oni T, et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med 2013; 10: e1001538. doi: 10.1371/journal.pmed.1001538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blankley S, Graham CM, Turner J, et al. The transcriptional signature of active tuberculosis reflects symptom status in extra-pulmonary and pulmonary tuberculosis. PLoS One 2016; 11: e0162220. doi: 10.1371/journal.pone.0162220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singhania A, Verma R, Graham CM, et al. A modular transcriptional signature identifies phenotypic heterogeneity of human tuberculosis infection. Nat Commun 2018; 9: 2308. doi: 10.1038/s41467-018-04579-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leong S, Zhao Y, Joseph NM, et al. Existing blood transcriptional classifiers accurately discriminate active tuberculosis from latent infection in individuals from south India. Tuberculosis 2018; 109: 41–51. doi: 10.1016/j.tube.2018.01.002 [DOI] [PubMed] [Google Scholar]
- 17.Penn-Nicholson A, Mbandi SK, Thompson E, et al. RISK6, a 6-gene transcriptomic signature of TB disease risk, diagnosis and treatment response. Sci Rep 2020; 10: 8629. 10.1038/s41598-020-65043-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sweeney TE, Braviak L, Tato CM, et al. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir Med 2016; 4: 213–224. 10.1016/S2213-2600(16)00048-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar NP, Moideen K, Nancy A, et al. Plasma chemokines are baseline predictors of unfavorable treatment outcomes in pulmonary tuberculosis. Clin Infect Dis 2021; 73: e3419–e3427. 10.1093/cid/ciaa1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ronacher K, Chegou NN, Kleynhans L, et al. Distinct serum biosignatures are associated with different tuberculosis treatment outcomes. Tuberculosis. 2019; 118: 101859. 10.1016/j.tube.2019.101859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sivakumaran D, Jenum S, Vaz M, et al. Combining host-derived biomarkers with patient characteristics improves signature performance in predicting tuberculosis treatment outcomes. Commun Biol 2020; 3: 359. 10.1038/s42003-020-1087-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson EG, Du Y, Malherbe ST, et al. Host blood RNA signatures predict the outcome of tuberculosis treatment. Tuberculosis. 2017; 107: 48–58. 10.1016/j.tube.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Russell SL, Lamprecht DA, Mandizvo T, et al. Compromised metabolic reprogramming is an early indicator of CD8+ T cell dysfunction during chronic Mycobacterium tuberculosis infection. Cell Rep 2019; 29: 3564–3579. doi: 10.1016/j.celrep.2019.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ratzan KR, Musher DM, Keusch GT, et al. Correlation of increased metabolic activity, resistance to infection, enhanced phagocytosis, and inhibition of bacterial growth by macrophages from Listeria- and BCG-infected mice. Infect Immun 1972; 5: 499–504. doi: 10.1128/iai.5.4.499-504.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol 1927; 8: 519–530. doi: 10.1085/jgp.8.6.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair S, Huynh JP, Lampropoulou V, et al. Irg1 expression in myeloid cells prevents immunopathology during M. tuberculosis infection. J Exp Med 2018; 215: 1035–1045. doi: 10.1084/jem.20180118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tattersall WH. The survival of sputum-positive consumptives; a study of 1,192 cases in a county borough between 1914 and 1940. Tubercle 1947; 28: 107–114. doi: 10.1016/S0041-3879(47)80088-1 [DOI] [PubMed] [Google Scholar]
- 28.DiNardo AR, Nishiguchi T, Grimm SL, et al. Tuberculosis endotypes to guide stratified host-directed therapy. Med 2021; 2: 217–232. doi: 10.1016/j.medj.2020.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bengsch B, Johnson AL, Kurachi M, et al. Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8+ T cell exhaustion. Immunity 2016; 45: 358–373. doi: 10.1016/j.immuni.2016.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scharping NE, Menk AV, Moreci RS, et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 2016; 45: 374–388. doi: 10.1016/j.immuni.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravimohan S, Auld SC, Maenetje P, et al. Lung injury on antiretroviral therapy in adults with human immunodeficiency virus/tuberculosis. Clin Infect Dis 2020; 70: 1845–1854. doi: 10.1093/cid/ciz560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wallis RS, Ginindza S, Beattie T, et al. Adjunctive host-directed therapies for pulmonary tuberculosis: a prospective, open-label, phase 2, randomised controlled trial. Lancet Respir Med 2021; 9: 897–908. doi: 10.1016/S2213-2600(20)30448-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roca FJ, Ramakrishnan L. TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell 2013; 153: 521–534. doi: 10.1016/j.cell.2013.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arts RJW, Carvalho A, La Rocca C, et al. Immunometabolic pathways in BCG-induced trained immunity. Cell Rep 2016; 17: 2562–2571. doi: 10.1016/j.celrep.2016.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barili V, Fisicaro P, Montanini B, et al. Targeting p53 and histone methyltransferases restores exhausted CD8+ T cells in HCV infection. Nat Commun 2020; 11: 604. doi: 10.1038/s41467-019-14137-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abhimanyu, Ontiveros CO, Guerra-Resendez RS, et al. Reversing post-infectious epigenetic-mediated immune suppression. Front Immunol 2021; 12: 688132. doi: 10.3389/fimmu.2021.688132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Böhme J, Martinez N, Li S, et al. Metformin enhances anti-mycobacterial responses by educating CD8+ T-cell immunometabolic circuits. Nat Commun 2020; 11: 5225. doi: 10.1038/s41467-020-19095-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaufmann SHE, Dorhoi A, Hotchkiss RS, et al. Host-directed therapies for bacterial and viral infections. Nat Rev Drug Discov 2018; 17: 35–56. doi: 10.1038/nrd.2017.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zak DE, Penn-Nicholson A, Scriba TJ, et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 2016; 387: 2312–2322. doi: 10.1016/S0140-6736(15)01316-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupta RK, Turner CT, Venturini C, et al. Concise whole blood transcriptional signatures for incipient tuberculosis: a systematic review and patient-level pooled meta-analysis. Lancet Respir Med 2020; 8: 395–406. doi: 10.1016/S2213-2600(19)30282-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harari A, Rozot V, Enders FB, et al. Dominant TNF-α+ Mycobacterium tuberculosis-specific CD4+ T cell responses discriminate between latent infection and active disease. Nat Med 2011; 17: 372–376. doi: 10.1038/nm.2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahiratmadja E, Alisjahbana B, de Boer T, et al. Dynamic changes in pro- and anti-inflammatory cytokine profiles and gamma interferon receptor signaling integrity correlate with tuberculosis disease activity and response to curative treatment. Infect Immun 2007; 75: 820–829. doi: 10.1128/IAI.00602-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh A, Mohan A, Dey AB, et al. Inhibiting the programmed death 1 pathway rescues Mycobacterium tuberculosis-specific interferon γ-producing T cells from apoptosis in patients with pulmonary tuberculosis. J Infect Dis 2013; 208: 603–615. doi: 10.1093/infdis/jit206 [DOI] [PubMed] [Google Scholar]
- 44.DiNardo A, Rajapakshe K, Nishiguchi T, et al. DNA hypermethylation during tuberculosis dampens host immune responsiveness. J Clin Invest 2020; 130: 3113–3123. doi: 10.1172/JCI134622 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary methods. ERJ-02263-2021.Supplement (122.6KB, pdf)
Supplementary table S1. a) Differential expressed genes in the discovery cohort at resolution 0.4. b) genes comprising the random forest classifier. ERJ-02263-2021.Table_S1 (55KB, xlsx)
Supplementary table S2. Enriched gene sets by gene set enrichment analysis (GSEA). a) Enriched hallmark pathways. b) Enriched pathway analysis from KEGG, reactome, and gene ontology biological process. c) Enriched transcription factor targets. ERJ-02263-2021.Table_S2 (926KB, xlsx)
Supplementary table S3. Demographic information from cohort 4 multiplex ELISA (BioLegend LegendPlex). ERJ-02263-2021.Table_S3 (19.1KB, xlsx)
Supplementary figure S1. a) Cluster tree of the discovery cohort based on different resolutions of the Louvain method. Evaluation occurred at resolution 0.4, which identified 2 clusters, labeled endotype A and B. Up to four sub-clusters are visible at resolution 1.0. b) tSNE plot of the discovery cohort including healthy controls, endotype A, and endotype B. ERJ-02263-2021.Figure_S1 (737.1KB, jpg)
Supplementary figure S2. a) Classification error rate for random forest classifiers developed across a range of the top informative genes. Classification error rate is the average prediction error determined by repeated subsampling with replacement of the endotype A and B samples. b) Heatmap of GSEA normalized enrichment scores (NES) for Hallmark pathways using the 40, 50 and 500 gene TB endotype classifiers over discovery and validation cohorts. ERJ-02263-2021.Figure_S2 (747.2KB, jpg)
Supplementary figure S3. Thirty-four samples were included both in the Berry 2010 microarray and Singhania 2018 RNA-seq studies. tSNE plot demonstrating the low discordant (green in a and red in b) sample classification. ERJ-02263-2021.Figure_S3 (878.1KB, jpg)
Supplementary figure S4. Longitudinal analysis of endotype classification after time on antimicrobial therapy. ERJ-02263-2021.Figure_S4 (296.2KB, jpg)
Supplementary figure S5. Enriched transcription factors targets of between endotype A and endotype B. Bars represent the normalised enrichment score for endotype A versus endotype B. Comparison using Gene set enrichment analysis with an FDR <0.05. Red bars indicate higher expression in endotype A and blue bars indicate higher expression in endotype B. Full data can be found in supplementary table. ERJ-02263-2021.Figure_S5 (671.1KB, jpg)
Supplementary figure S6. Heatmap of connectivity scores for select chemical compounds within the TB endotypes A and B based on the Library of Integrated Network-based Cellular Signatures (LINCS). Positive connectivity scores represent compounds inducing gene expression profiles similar to the endotype, while negative connectivity scores represent compounds inducing gene expression profiles antithetical to the endotype. ERJ-02263-2021.Figure_S6 (512.1KB, jpg)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-02263-2021.Shareable (538.2KB, pdf)