Abstract
The effects of the rulAB operon of Pseudomonas syringae on mutagenic DNA repair and the transcriptional regulation of rulAB following irradiation with UV-B wavelengths were determined. For a rulB::Km insertional mutant constructed in P. syringae pv. syringae B86-17, sensitivity to UV-B irradiation increased and UV mutability decreased by 12- to 14-fold. rulAB-induced UV mutability was also tracked in phyllosphere populations of B86-17 for up to 5 days following plant inoculation. UV mutability to rifampin resistance (Rifr) was detected at all sampling points at levels which were significantly greater than in nonirradiated controls. In P. aeruginosa PAO1, the cloned rulAB determinant on pJJK17 conferred a 30-fold increase in survival and a 200-fold increase in mutability following a UV-B dose of 1,900 J m−2. In comparative studies using defined genetic constructs, we determined that rulAB restored mutability to the Escherichia coli umuDC deletion mutant RW120 at a level between those of its homologs mucAB and umuDC. Analyses using a rulAB::inaZ transcriptional fusion in Pseudomonas fluorescens Pf5 showed that rulAB was rapidly induced after UV-B irradiation, with expression levels peaking at 4 h. At the highest UV-B dose administered, transcriptional activity of the rulAB promoter was elevated as much as 261-fold compared to that of a nonirradiated control. The importance of rulAB for survival of P. syringae in its phyllosphere habitat, coupled with its wide distribution among a broad range of P. syringae genotypes, suggests that this determinant would be appropriate for continued investigations into the ecological ramifications of mutagenic DNA repair.
The involvement of bacterial plasmids in increasing the survival of their hosts following irradiation with UV wavelengths was first reported in 1965 by Howarth (13), who was working with the ColIb-P9 plasmid in Salmonella enterica serovar Typhimurium LT2. Howarth also noted that the frequency of mutants in irradiated cultures of serovar Typhimurium LT2(ColIb-P9) was increased (14). These two initial observations have been followed by the discovery that a large number of bacterial plasmids from many incompatibility groups confer phenotypes of increased UV survival and mutability (43, 52). Genes conferring the UV mutability phenotype are also chromosomally located in some cases, an important example being the umuDC operon of Escherichia coli.
The umuDC operon is one component of the SOS regulon of E. coli, a set of approximately 20 unlinked genes which are coordinately regulated in response to DNA damage (42). The umuDC operon is regulated by the recA and lexA gene products, with the UmuDC and RecA proteins alone required for UV mutability (42). LexA functions as a repressor through binding to a conserved DNA sequence (SOS box) located within the promoter region of SOS regulon genes (3, 25). The irradiation of E. coli cells with UV wavelengths results in the occurrence of DNA lesions of which the cyclobutane pyrimidine dimer and the pyrimidine(6-4)pyrimidinone photoproduct are the most typical (8); these lesions can result in a blockage of DNA polymerase activity, leading to a stalling of replication. Cellular perception of DNA damage is thought to occur through the binding of RecA to single-stranded DNA immediately downstream of a DNA lesion; during this process, RecA is converted to an activated form (RecA*) (42). RecA* then mediates a self-cleavage reaction of LexA, resulting in the removal of LexA and allowing the expression of the SOS response genes (24).
Following the expression of umuDC, a posttranslational self-modification of UmuD (removal of the first 24 amino acids of UmuD) mediated by RecA* is required to generate the active UmuD′ protein (5, 40). UmuD′ forms a homodimer (UmuD′2) which complexes with UmuC, resulting in the mutagenically active UmuD′2C complex (4). The function of the UmuD′2C complex in mutagenic DNA repair (MDR) occurs as a result of translesion DNA synthesis (42, 50). The model for replicative lesion bypass includes DNA pol III holoenzyme complex, the UmuD′2C complex, and RecA* (48). However, recent evidence has shown that the UmuD′2C complex itself may be sufficient in translesion synthesis with the possibility of either the UmuD′2C complex or UmuC itself functioning as a DNA polymerase (36, 49).
To date, five distinct plasmid-carried umuDC homologs have been characterized at the sequence level. These include impCAB, mucAB, rulAB, rumABR391, and samAB (19, 26, 31, 33, 46). Each of these sequences contains a consensus LexA-binding site within the respective promoter regions and a conserved internal cleavage site within the umuD homolog. With the exception of rulAB, the other known MDR systems were isolated from enterobacteria and have not been well characterized in terms of their contribution to the ecological fitness of their hosts. The rulAB operon was originally cloned from pPSR1, an indigenous plasmid from Pseudomonas syringae pv. syringae A2, and was initially characterized for its role in UV radiation (UVR) tolerance (46). Although not absolutely required for survival, the UVR tolerance phenotype conferred by rulAB was subsequently shown to increase P. syringae populations by 10- to 30-fold in its leaf surface (phyllosphere) habitat (47). rulAB is the most distantly related to umuDC of the other plasmid-carried umuDC homologs, as rulA and rulB share only 30.9% and 41.5% amino acid similarity to umuD and umuC, respectively (42). However, rulAB does share the important features of this group, including its function in UVR tolerance and its lack of expression and activity in a P. syringae recA background (46). Recent evidence has also shown that rulAB is widely distributed among strains within and among pathovars of P. syringae (39, 47), suggesting the importance of this determinant to a wide range of genotypes.
Our laboratory is interested in further elucidating the role of the rulAB system in the population biology of P. syringae. To that end, we are fully characterizing the functional roles of rulAB (UVR tolerance and MDR), along with analyzing the regulation of this determinant so that we can ultimately address the biological significance of the rulAB system to P. syringae in its natural environment. Our studies reported here utilize UV-B (290 to 320 nm) wavelengths, which is in contrast to most analyses of DNA repair and UVR-induced mutagenesis in microorganisms, where higher-energy UV-C (254 nm) wavelengths are used. In nature, UV-C wavelengths are screened by the stratospheric ozone layer and do not reach the earth's surface; thus, the use of UV-B wavelengths is relevant from an ecological standpoint. The most important types of UV-B-induced DNA lesions appear to be cyclobutane pyrimidine dimers and pyrimidine(6-4)pyrimidinone photoproducts (30). Ecological studies have shown that solar UV-B radiation has profound deleterious effects on microorganisms dwelling in locations with high sunlight exposure, including surface aquatic habitats and the phyllosphere (11, 15, 45).
In this study, we report an analysis of the MDR capacity of rulAB and an analysis of mutability in a P. syringae strain containing a stable insertional mutation within rulAB. We also examined the MDR potential of rulAB-containing P. syringae in planta and show that UVR-induced mutability occurs in leaf surface populations at rates similar to those observed in vitro. We further demonstrate that rulAB complements an E. coli umuDC mutant and compare the MDR activity of rulAB with that of mucAB and umuDC in E. coli and P. aeruginosa by using defined genetic constructs. Finally, we analyzed the regulation of rulAB in response to increasing doses of UV-B (290 to 320 nm) radiation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains, plasmids, and specific oligonucleotides utilized which were relevant to this study are listed in Table 1. All bacterial strains were grown in Luria-Bertani (LB) medium (Difco) or King's medium B (KB) (17); E. coli strains and Pseudomonas aeruginosa PAO1 were grown at 37°C, and Pseudomonas fluorescens Pf5 and P. syringae pv. syringae B86-17 were grown at 28°C. Plasmid transfer from E. coli to Pseudomonas strains was accomplished by triparental mating using the helper plasmid pRK2013. The exconjugants were selected on MG medium (16) or Pseudomonas isolation agar (Difco) supplemented with appropriate antibiotics. Antibiotics were added to the media in the following concentrations: ampicillin, 75 μg ml−1; carbenicillin, 150 μg ml−1; cycloheximide, 100 μg ml−1; gentamicin (Gm), 20 μg ml−1; kanamycin, 50 μg ml−1; rifampin, 100 μg ml−1; and spectinomycin, 40 μg ml−1.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotide primers utilized in this study and their relevant characteristics
| Strain, plasmid, or oligonucleotide primer | Relevant characteristicsa or nucleotide sequenceb | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli | ||
| DH10B | Plasmid-free strain used for cloning | 9 |
| RW120 | lexA+ recA+ Δ(umuDC)595::cat | 12 |
| P. aeruginosa | ||
| PAO1 | UVs, no detectable plasmids | A. M. Chakrabarty |
| P. fluorescens | ||
| Pf5 | UVs, ina-negative | 27 |
| P. syringae pv. syringae | ||
| B86-17 | UVr, contains rulAB on pB8617A | 21 |
| GWS242 | As B86-17 but also rulB::Km | This study |
| Plasmids | ||
| pBluescript SK(+) | Apr, cloning vector | Stratagene |
| pBSL86 | Source of Kmr cassette | 1 |
| pCR2.1 | Apr Kmr, direct cloning vector for PCR products | Invitrogen |
| pET-5a | Apr, source of Shine-Dalgarno sequence | Promega |
| pGem7zf− | Apr, cloning vector | Promega |
| pJB321 | Cbr, broad-host-range cloning vector | 2 |
| pJQ200SK | GmrsacB, suicide gene replacement vector | 34 |
| pPROBE KI′ | Kmr, broad-host-range, inaZ reporter vector | S. E. Lindow |
| pRK2013 | Helper plasmid for triparental matings | 7 |
| pRW144 | Spr, 2.4-kb mucAB as BamHI in pGB2 | 12 |
| pRW154 | Spr, 2.8-kb umuDC as EcoRI in pGB2 | 12 |
| pB8617A | rulAB+, native plasmid from P. syringae pv. syringae B86-17 | This study |
| pGWS140 | 0.7-kb HindIII-PstI from pSM1 in pBluescript SK(+) | 47 |
| pJJK1 | 6.4-kb rulAB as BamHI from pB8617A in pBluescript SK(+) | This study |
| pJJK5 | 1.7-kb mucAB as NdeI-BamHI from pRW144 in pET-5a | This study |
| pJJK12 | 3.8-kb rulAB as partial EcoRI from pJJK1 in pGem7zf− | This study |
| pJJK15 | 1.2-kb Kmr cassette as HincII from pBSL86 in pJJK12 at blunt-ended BssHII | This study |
| pJJK16 | 4.4-kb rulA, rulB::Km as XbaI-BamHI in pJQ200SK | This study |
| pJJK17 | 3.8-kb rulAB as XbaI-BamHI from pJJK12 in pJB321 | This study |
| pJJK20 | 0.75-kb umuDC promoter as SphI-XbaI in pET-5a | This study |
| pJJK21 | 1.7-kb rulAB as NdeI-BamHI from pJJK12 in pJJK20 | This study |
| pJJK22 | 0.75-kb umuDC promoter as SphI-XbaI in pJJK5 | This study |
| pJJK23 | 1.7-kb umuDC as NdeI-EcoRI from pRW154 in pET-5a | This study |
| pJJK24 | 0.75-kb umuDC promoter as SphI-XbaI in pJJK23 | This study |
| pJJK25 | 2.45-kb umuDC promoter + rulAB as SalI-BamHI in pJB321 | This study |
| pJJK26 | 2.45-kb umuDC promoter + mucAB as SphI-BamHI in pJB321 | This study |
| pJJK27 | 2.45-kb umuDC promoter + umuDC as SphI-EcoRI in pJB321 | This study |
| pJJK40 | 1.1-kb rulAB promoter region in pCR2.1 | This study |
| pJJK41 | 0.9-kb rulAB promoter as HindIII-EcoRI from pJJK40 in pPROBE KI′ | This study |
| Oligonucleotide primers | ||
| mucAB Nde 5′ | 5′-GGAATTCCATATGAAGGTCGATATTTTTG-3′ | This study |
| mucAB Bam 3′ | 5′-GATCGGATCCTTATTTGATGGTGGCTATTGG-3′ | This study |
| rulAB Nde 5′ | 5′-GGAATTCCATATGAACGTCAAAATACTCGGGCGG-3′ | This study |
| 3′ rulB TAA BamHI | 5′-GATCGGATCCTTACTTTACAACCCACAGCTG-3′ | This study |
| rul PX | 5′-GGATTGGGAAAACCGGCAGGG-3′ | This study |
| umuDC Nde 5′ | 5′-GATCCATATGTTGTTTATCAAGCCTGCGG-3′ | This study |
| umuDC Eco 3′ | 5′-GATCGAATTCTTATTTGACCCTCAGTAAATCAG-3′ | This study |
| umu Pro 5′ SphI | 5′-GATCGCATGCGAGCAATTGCGTCGC-3′ | This study |
| umu Pro 3′ | 5′-GTACTCTAGACTGCCTGAAGTTATACTG-3′ | This study |
Abbreviations: Ap, ampicillin; Cb, carbenicillin; Gm, gentamicin; Km, kanamycin; Sp, spectinomycin.
Restriction sites incorporated in primers are underlined. GGATCC, BamHI; GAATTC, EcoRI; CATATG, NdeI; GAGCTC, SacI; GCATGC, SphI; TCTAGA, XbaI.
Molecular biology techniques.
Plasmid isolation from P. syringae was accomplished using the technique of Crosa and Falkow (6). Restriction digestions, isolation of DNA fragments from agarose gels, PCR amplifications, ligations, and Southern transfer to nylon membranes were performed using standard techniques (38). DNA fragments used as probes were labeled with digoxigenin-11-dUTP (Genius kit; Boehringer Mannheim, Indianapolis, Ind.) following the instructions of the manufacturer. Hybridizations at 65°C followed by high-stringency washes were performed as described previously (44).
Construction of P. syringae pv. syringae GWS242 (B86-17 rulB::Km).
The rulAB gene disruption on the native plasmid pB8617A of strain B86-17 was created via gene replacement using a homologous recombination strategy. The intact rulAB sequence on pB8617A was initially cloned into pBluescript II SK(+) as a 6.4-kb BamHI fragment (pJJK1) and subcloned as a partial 3.8-kb EcoRI fragment into pGem 7zf−, creating pJJK12. This plasmid was then digested with BssHII, resulting in the removal of a 0.6-kb segment from within rulB. Following BssHII digestion, the ends were made blunt by incubation with the Klenow fragment of DNA polymerase (Life Technologies), and the 1.2-kb kanamycin resistance (Kmr) gene cassette (released from pBSL86 as a HincII fragment) was ligated in. The entire DNA region was excised as a 4.4-kb XbaI-BamHI fragment and ligated into the suicide gene replacement vector pJQ200SK, creating pJJK16. pJJK16 contained approximately 1.0 kb of flanking DNA sequences on each side of the rulAB (rulB::Km) sequence. Following transfer of pJJK16 into P. syringae pv. syringae B86-17, those P. syringae cells in which a plasmid integration event had occurred were selected on MG supplemented with Gm. Several isolated Gmr colonies were then cultured in LB broth containing Km. After 2 days of incubation, 0.1-ml aliquots were plated onto LB containing Km and 5% sucrose to counterselect against the sacB gene encoded on pJQ200SK. The sucrose-resistant (Sucr) colonies recovered were subsequently tested for sensitivity to Gm as a phenotypic assay for loss of the vector sequences. Since there are no BamHI sites within rulAB or the Kmr cassette, the increased size of pB8617A rulB::Km was visualized by comparing the sizes of the BamHI fragment from pB8617A and pB8617A rulB::Km, which hybridized to an internal rulAB gene probe consisting of the 0.7-kb HindIII-PstI fragment from pGWS140. Further confirmatory hybridizations were done using the Kmr cassette from pBSL86 as a probe both to the BamHI plasmid digests and to BamHI digests of total genomic DNA from P. syringae pv. syringae B86-17 and GWS242 to ensure that there was only one insertion of the Kmr cassette within the GWS242 genome.
UV-B irradiation and MDR assays.
Bacterial strains were grown to late log phase in LB medium containing the appropriate antibiotics; 1 ml of the cultures was pelleted, washed with an equal volume of sterile saline (0.85% NaCl) solution, and resuspended in an equal volume of saline. The cell suspension was then diluted with an additional 9 ml of saline and placed in a sterile glass petri dish. Cell suspensions were irradiated with UV-B (peak, 302 nm) wavelengths using an XX-15M model UV-B lamp (UVP Products, San Gabriel, Calif.) filtered through cellulose diacetate (Kodacel; Eastman Kodak, Rochester, N.Y.) to eliminate any UV-C wavelengths (<290 nm) given off. The UV-B lamp was turned on 15 min prior to use to allow for stabilization of the UVR output. The energy output of the lamp was monitored with a UV-X radiometer (UVP Products) and determined to be 2.8 J m−2 s−1. Cell suspensions were mixed continuously while receiving the UV-B dose to eliminate survival as a result of shading. After irradiation, surviving cells were enumerated by dilution plating conducted under conditions that minimized photoreactivation. For the MDR assays, 0.1 ml of untreated cells and cells from all UV-B treatments were diluted in 0.9 ml of sterile saline and added to 1 ml of 2× LB broth. Following overnight incubation under dark conditions, appropriate dilutions of cell suspensions were plated on LB alone and LB containing rifampin. The frequency of mutation to rifampin resistance (Rifr) was calculated as the number of Rifr mutants per 108 cells.
Analysis of MDR in phyllosphere populations of P. syringae pv. syringae B86-17.
P. syringae pv. syringae B86-17 was grown on LB agar medium for 18 h before inoculation. The cells were washed and then resuspended in 0.1 M potassium phosphate buffer (pH 7.0), and cell suspensions were adjusted turbidimetrically to approximately 5 × 109 cells ml−1. Bush bean plants (Phaseolus vulgaris cv. “Blue Lake 214”) were grown under controlled conditions in a growth chamber (24°C, 80% relative humidity, 240-μmol m−2 s−1 light intensity, and 12-h photoperiod) with care taken during watering to prevent nonspecific leaf surface colonization. Experiments consisted of spraying inoculum of strain B86-17 onto the primary bean leaves using an air brush (Badger model 350; Badger Co., Franklin Park, Ill.) until the leaf surfaces were uniformly coated with the cell suspension. Immediately and at 24, 72, and 120 h after inoculation, four of the leaves were excised and individually irradiated with a 500-J m−2 dose of UV-B by using the XX-15M lamp as described above. An additional four leaves were also removed, and these leaves were not irradiated as a control. The leaves were placed onto sterile, moist paper towels in a covered sterile Bio-Assay dish (Nunc Products) and incubated under dark conditions for 12 h at 25°C. After incubation, each leaf was sonicated for 7 min in 20 ml of sterile washing buffer (0.1 M potassium phosphate [pH 7.0] and 0.1% peptone). The combined washing buffer from four leaves per treatment was reduced to 1 ml by centrifugation, and appropriate serial dilutions of the leaf washings were plated onto KB agar medium amended with cycloheximide alone (KBc) and KBc containing rifampin. A total of six independent experiments were conducted. The data were normalized based on total leaf surface populations and reported as the number of Rifr cells per 108 total cells. A one-way analysis of variance based on UV-B irradiation was done to compare the mean number of Rifr cells per 108 total cells at each sampling time; means were differentiated using a t test.
Phenotypic comparison of rulAB with mucAB and umuDC.
The individual coding sequences of the mucAB, rulAB, and umuDC MDR operons were amplified via PCR using the following oligonucleotide primers and source DNA: (i) mucAB Nde 5′, mucAB Bam 3′, and pRW144 for mucAB; (ii) rulAB Nde 5′, 3′ rulB TAA BamHI, and pJJK12 for rulAB; and (iii) umuDC Nde 5′, umuDC Eco 3′, and pRW154 for umuDC. The amplified product from pRW144 was confirmed as having the correct size on an agarose gel, digested with the appropriate restriction enzymes, and ligated into pET-5a, creating pJJK5. A 0.75-kb region upstream of umuDC on the E. coli chromosome was next amplified from pRW154 using the primers umu Pro SphI 5′ and umu Pro 3′, digested, and ligated upstream of the mucAB coding sequence in pJJK5, creating pJJK22. The other genetic constructs were made by first ligating the umuDC promoter into pET-5a, creating pJJK20, and then ligating the respective rulAB or umuDC amplified coding sequence into pJJK20. In each case, the translational start site of the MDR coding sequences was the ATG sequence within the NdeI sequence of pET-5a, allowing optimal spacing from a Shine-Dalgarno sequence present on the pET-5a vector. The final constructs, including the umuDC promoter sequences, Shine-Dalgarno sequence from pET-5a, and the respective MDR coding sequences were ligated into pJB321, creating pJJK25, pJJK26, and pJJK27. Each of these plasmids was transferred into E. coli RW120 and P. aeruginosa PAO1 and utilized in MDR assays as described above.
Determination of rulAB promoter activity using the inaZ reporter gene.
The intergenic sequences between rulAB and repA on pB8617A were fused to the promoterless ice nucleation gene (inaZ) in pPROBE KI′ in order to quantify rulAB expression following UV-B irradiation. The primers T7 (Promega) and rul PX were used to amplify a 1.1-kb fragment from pJJK12; the PCR product was directly ligated into pCR2.1, creating pJJK40. A 0.9-kb HindIII-EcoRI fragment was subsequently excised from pJJK40 and ligated into pPROBE KI′, resulting in a transcriptional fusion of the rulAB promoter region with inaZ in the final construct, pJJK41. Ice nucleation activity (INA) was chosen as a reporter because of the sensitivity with which INA can be detected and the large range of INAs which can be quantified (28). INA is also easily quantified in situ, and our future experiments would be geared toward rulAB expression analysis in the phyllosphere. Unfortunately, most strains of P. syringae produce the ice nucleation protein naturally; however, we felt that the sensitivity of the reporter would enable us to detect differences in expression resulting after small differences in the UV-B dose and that it justified the use of inaZ even though a different Pseudomonas species would be used in the assay.
The rulAB::inaZ expression analyses were performed in P. fluorescens Pf5, an ice nucleation-negative strain, following the confirmation of rulAB activity (increased UV-B survival and MDR) in strain Pf5. Cells of P. fluorescens Pf5(pJJK41) were grown overnight in KB broth containing Km, pelleted by centrifugation, resuspended in 0.01 M potassium phosphate buffer (pH 7.0), and exposed to various UV-B doses using the methods described above. Twenty-five ml of UV-B-irradiated or nonirradiated cells was then cultured in 25 ml of 2× KB broth under dark conditions. Aliquots were withdrawn immediately upon culture and at designated intervals up to 18 h following irradiation and then resuspended in a small volume of 0.01 M potassium phosphate buffer for assessment of INA. The number of ice nuclei per cell was estimated by the droplet-freezing assay as described by Lindow (23), with the number of ice nuclei calculated as the fraction of droplets that froze within 5 min. Dilution plating on KB was used to count the number of viable cells so that the data could be normalized on the basis of numbers of ice nuclei per cell.
RESULTS
Construction and analysis of a rulB insertional mutant of P. syringae pv. syringae B86-17.
The rulAB determinant was initially isolated from the indigenous plasmid pPSR1 from P. syringae pv. syringae A2, a pathogen of ornamental pear trees (46). In this and subsequent studies, it was of interest to work with the rulAB determinant of a P. syringae strain which was pathogenic on bean, a plant host which is more easily utilized in growth chamber studies. A bean-pathogenic strain, P. syringae pv. syringae B86-17, was chosen for use; this strain has a phenotype of UV tolerance and encodes rulAB on its large indigenous plasmid (47). For our experimental analyses, the rulAB determinant from pB8617A, the indigenous plasmid harbored by P. syringae pv. syringae B86-17, was subcloned as a 3.8-kb XbaI-BamHI fragment in the broad-host-range low-copy-number vector pJB321, creating the plasmid pJJK17.
The function of rulAB in elevating UV survival and MDR was evaluated by using P. syringae pv. syringae GWS242 (B86-17 rulB::Km), a strain that was constructed by gene replacement using a sucrose-mediated counterselection system. Confirmation of the insertion of a kanamycin cassette within rulB was done using Southern hybridization analysis (data not shown). The UV-B sensitivity and mutability of P. syringae pv. syringae GWS242 were then compared with those of the wild-type B86-17 strain. The rulB::Km mutant GWS242 showed an increased UV-B sensitivity of approximately threefold at the highest dose level utilized (Fig. 1A). UV-B survival was restored to wild-type levels by the rulAB-containing plasmid pJJK17 (Fig. 1B). The increased sensitivity of the rulB::Km mutant GWS242 was of a lower magnitude than those observed previously by using two other P. syringae pv. syringae strains containing rulAB insertional mutations created by Campbell integration (47). This observation could be due to the insertional mutation occurring only within rulB of pB8617A, leaving the rulA sequence intact, or because P. syringae pv. syringae B86-17 contains additional UV-B-protective mechanisms which are not present in the strains utilized in the previous study.
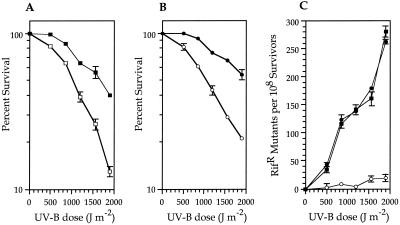
FIG. 1.
(A) Survival of P. syringae pv. syringae B86-17 (■) and GWS242 (□); (B) survival of GWS242(pJB321) (○) and GWS242(pJJK17) (●) after UV-B irradiation. Each data point represents the mean (±the standard error of the mean) from three replicate UV sensitivity experiments. (C) Effect of the rulB::Km mutation in GWS242 on UV-inducible mutagenesis. Rifs strains were irradiated with different doses of UV-B, samples were removed to initiate cultures that were incubated in LB for 18 h, and the number of Rifr colonies was determined. The number of spontaneous mutations conferring Rifr in the absence of UV-B irradiation has been subtracted. Each data point represents the mean (± the standard error of the mean) from three replicate experiments. Symbols: ■, P. syringae pv. syringae B86-17; ●, GWS242(pJJK17); ○, GWS242(pJB321).
An examination of UV-B mutability showed that the mutation frequency of spontaneous Rifr in P. syringae pv. syringae B86-17 is elevated following UV-B irradiation (Fig. 1C). This increase in mutation frequency is not observed in cultures of strain B86-17 which are not exposed to UV-B irradiation, suggesting that the phenotype is inducible (Fig. 1C). A reduction in mutability of 12- to 14-fold at UV-B doses of ≥850 J m−2 was observed in the rulB::Km strain GWS242; mutability at wild-type rates was restored when GWS242 was complemented with rulAB on the plasmid pJJK17 (Fig. 1C). It should be noted that the mutability of strain GWS242 (Fig. 1C) was also increased to a small degree at each UV-B dose compared to that of nonirradiated GWS242 (≤1 Rifr mutant per 108 survivors recovered at each UV-B dose). In our experiments, we typically observed between 10 and 30 Rifr mutants per 108 survivors at the higher (1,500 and 1,900 J m−2) UV-B doses (Fig. 1C). This observation raises the possibility that an additional source of UV-B mutability is present within the B86-17 genome.
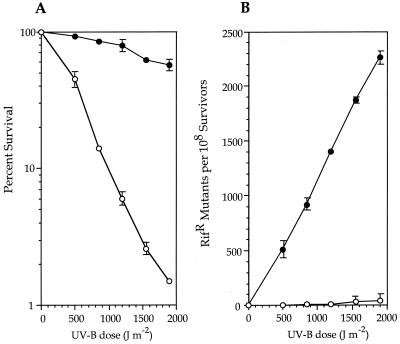
Analyses of additional, natural P. syringae strains which did not contain rulAB also indicated a phenotype of UV-B mutability following irradiation with 1,500-J m−2 UV-B (J. J. Kim and G. W. Sundin, unpublished data), albeit at a significantly lower rate than that of rulAB-containing strains. Thus, to fully evaluate the contribution of rulAB to UV-B survival and mutability, we utilized P. aeruginosa PAO1, a naturally UV-sensitive strain which, according to previous studies, does not contain a functional MDR system within its chromosome (29, 41). We transferred either pJJK17 or the vector pJB321 into strain PAO1 by triparental mating and subsequently determined that rulAB conferred an increase in UV-B survival to this strain of as much as 30-fold at the highest UV-B dose (Fig. 2A). UV-B mutability was also significant (up to a 200-fold increase in Rifr mutants following irradiation at 1,900 J m−2) and clearly inducible by UV-B irradiation (Fig. 2B).
FIG. 2.
(A) Survival of P. aeruginosa PAO1(pJB321) (○) and PAO1(pJJK17) (●) after UV-B irradiation. Each data point represents the mean (±the standard error of the mean) from three replicate UV sensitivity experiments. (B) Analysis of rulAB-mediated MDR in PAO1. Rifs strains were irradiated with different doses of UV-B, samples were removed to initiate cultures that were incubated in LB for 18 h, and the number of Rifr colonies was determined. The number of spontaneous mutations conferring Rifr in the absence of UV-B irradiation has been subtracted. Each data point represents the mean (±the standard error of the mean) from three replicate experiments. Symbols: ○, PAO1(pJB321); ●, PAO1(pJJK17).
In planta UV-B mutability of P. syringae pv. syringae B86-17.
The role of the rulAB determinant in enabling P. syringae strains to maintain and increase population size in their natural leaf surface (phyllosphere) habitat has been previously established (47). However, we have not previously assessed UV-B mutability in planta and, to our knowledge, the occurrence of MDR has not been previously shown using any bacteria in their natural habitat. Our studies were done by inoculating populations of P. syringae pv. syringae B86-17 onto bean leaves, excising the leaves at various sampling times after inoculation, irradiating the leaves with UV-B at 500 J m−2, and subsequently incubating the excised leaves for 12 h in a sterile, high-humidity environment. Following the incubation, bacteria were removed from the leaves by sonication and enumerated through plating on KBc alone and KBc containing rifampin. A total of six experiments were done, with the frequency of mutation to Rifr shown to be significantly greater from irradiated leaves than from nonirradiated control leaves at all sampling points (Table 2). Over time, the ratio of Rifr cells recovered from irradiated leaves to those recovered from nonirradiated leaves decreased slightly (4.8- to 2.9-fold) (Table 2). The levels of increase in mutability (at 500 J m−2) observed immediately and at 24 h following inoculation were similar to those observed in the previous experiments performed in vitro (Fig. 1C).
TABLE 2.
Analysis of rulAB-mediated MDR in P. syringae pv. syringae B86-17 at each sampling point following inoculation to the phyllosphere of bean
| Category of leaf | No. of Rifr mutants per 108 cells recovered at each time point (in h) after inoculation:a
|
|||
|---|---|---|---|---|
| 0 h | 24 h | 72 h | 120 h | |
| Nonirradiated control | 3.8 | 3.3 | 3.6 | 4.5 |
| Irradiated at 500 J m−2 | 18.3*** | 16.3** | 13.0* | 13.0* |
Values reported are the means from six independent experiments. Significant differences between means at each sampling time are indicated (∗, P = 0.05; ∗∗, P = 0.01; ∗∗∗, P = 0.001).
Phenotypic comparison of rulAB with mucAB and umuDC.
Since rulAB was found to be distantly related to umuDC at the amino acid level (42), it was of interest to determine if rulAB was functionally similar to other well-characterized MDR operons and especially to determine if rulAB could restore mutability functions to an E. coli umuDC mutant. In order to facilitate phenotypic-analysis studies of rulAB, it was desirable to choose hosts in which there was minimal interfering background activity in terms of mutability. We chose two hosts for these experiments, E. coli RW120, in which the chromosomal source of MDR, the umuDC determinant, was deleted, and P. aeruginosa PAO1, a strain which does not encode an MDR determinant. Comparisons of rulAB with its homologs mucAB and umuDC were done using the defined genetic constructs pJJK25, pJJK26, and pJJK27. These constructs only differed by the inclusion of the individual MDR systems; the promoter, Shine-Dalgarno sequences, and vector were identical among them.
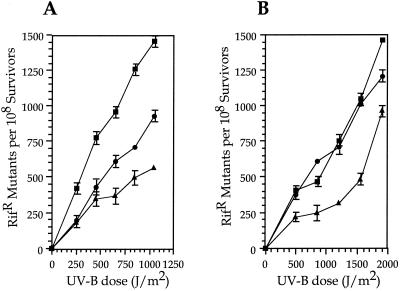
Restoration of UV-B mutability was first assessed in E. coli RW120 containing pJJK25, pJJK26, pJJK27, or the pJB321 vector. As expected, the use of the umuDC promoter sequences to control the expression of the MDR systems resulted in UV-B mutability being inducible. The mucAB determinant resulted in the largest frequency of mutation to Rifr (Fig. 3A), confirming previous observations of mutability with mucAB using UV-C radiation in strain RW120 (19). Mutability of strain RW120 containing rulAB on pJJK25 was at an intermediate level between those of mucAB and umuDC (Fig. 3A). The additional significance of these observations is that rulAB complemented the umuDC mutation in E. coli RW120. When P. aeruginosa PAO1 was used as the host, the positions of each MDR system remained the same, with mucAB resulting in the highest mutation frequency (Fig. 3B). However, little difference in mutability was observed between mucAB and rulAB at four of the five UV-B levels utilized, with a small increase in mucAB mutability observed only at the largest UV-B dose (Fig. 3B).
FIG. 3.
Ability of defined genetic constructs encoding rulAB, mucAB, and umuDC to restore mutagenesis functions to an E. coli umuDC strain and to induce mutagenesis in P. aeruginosa PAO1. (A) E. coli strain RW120 [lexA+ recA+ Δ(umuDC)595::cat]. The number of spontaneous mutations conferring Rifr in the absence of UV-B irradiation has been subtracted. Symbols: ●, rulAB (pJJK25); ■, mucAB (pJJK26); ▴, umuDC (pJJK27). (B) P. aeruginosa PAO1. As with E. coli RW120, the number of spontaneous mutations conferring Rifr in the absence of UV-B irradiation has been subtracted. Symbols: ●, rulAB (pJJK25); ■, mucAB (pJJK26); ▴, umuDC (pJJK27). In panels A and B, each data point represents the mean (±the standard error of the mean) from three replicate UV sensitivity experiments.
Regulation of the rulAB promoter.
We used the ice nucleation gene, inaZ, as a reporter in our regulation analyses because of the sensitivity of ice nucleation as a reporter (28) and the relative ease of measurement of this reporter when used for in planta studies, which will be reported elsewhere. We chose to use P. fluorescens Pf5 for our studies, since Pf5 does not possess INA, and also because Pf5 grows at 25°C, a temperature at which INA is readily expressed (28). Prior to the utilization of P. fluorescens Pf5 in our studies, we determined that the rulAB determinant was active in this strain, functioning in the increase of both UV-B survival and mutability (data not shown).
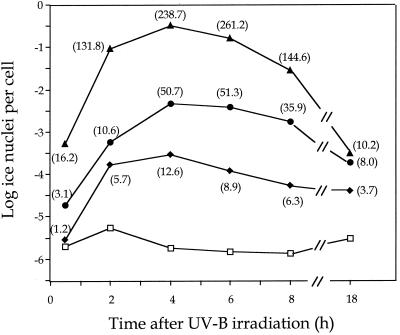
Since P. fluorescens Pf5 was more sensitive to UV-B irradiation than P. syringae pv. syringae B86-17, we irradiated with lower doses of UV-B in the regulation experiments. Following the irradiation of Pf5 containing the rulAB promoter-inaZ fusion construct pJJK41 with UV-B at 75, 150, and 300 J m−2, cultures were initiated using the irradiated cells and samples were removed at designated times for INA analysis. An increase in expression was observed within 30 min at the two higher UV-B doses and within 2 h at the 75-J m−2 dose (Fig. 4). The increase in rulAB promoter activity continued until 4 h after irradiation with the observed levels of INA ranging as high as 105 greater than those for the nonirradiated control (Fig. 4). There is an exponential relationship between INA and the abundance of ice nuclei, with INA increasing with the square of InaZ concentration until INA reaches 10−1 nuclei per cell (22). Between 10−1 and 100 ice nuclei per cell, INA increases with the third power (cube) of InaZ concentration (28). Thus, quantification of InaZ levels in our experiments reflects the transcriptional activity of the rulAB promoter and, when compared to InaZ levels detected in the control strain containing the pPROBE KI′ vector, indicates that rulAB transcription was increased up to 261-fold at 4 h after irradiation with the 300-J m−2 dose (Fig. 4). Because of the exponential relationship between INA and the abundance of ice nuclei, calculations of fold increases in promoter activity are sensitive to the INA measurements in the nonirradiated control treatment. Thus, the decrease in INA between 4 and 6 h after irradiation in the control treatment was large enough to result in an elevated increase in InaZ levels (Fig. 4, 150- and 300-J m−2 treatments) even though the INA observed at these treatments had actually decreased. Overall, a stair-step effect was consistently observed, with the lower UV-B doses resulting in lower promoter activity at all time points. INA levels expressed from the rulAB promoter were still elevated through 18 h after irradiation, although this observation could also be due to the stability of the InaZ protein (Fig. 4).
FIG. 4.
Analysis of the activity of a rulAB promoter fusion with a promoterless inaZ gene in P. fluorescens Pf5 following UV-B irradiation. P. fluorescens Pf5(pJJK41) was irradiated with different doses of UV-B, samples were removed to initiate cultures which were incubated in KB, and at the designated time points, cells were removed and analyzed for INA using the droplet-freezing assay. Each data point represents the mean from three replicate experiments. Values in parentheses indicate the fold increase in InaZ concentration at each data point relative to that of the nonirradiated control. Symbols for amounts of UV-B irradiation of Pf5(pJJK41): ⧫, 75 J m−2; ●, 150 J m−2; ▴, 300 J m−2; □, nonirradiated control.
DISCUSSION
The rulAB operon, which is widely distributed among diverse strains of P. syringae isolated from many plant hosts, plays an important functional role in protecting strains from the DNA-damaging effects of UV-B radiation. Exposure of P. syringae strains to UV-B wavelengths present in solar radiation would be predicted to occur with regularity in the phyllosphere habitat of this organism. Analysis of the UV-B mutability of the rulAB operon in E. coli RW120 indicated that rulAB is functionally similar to umuDC and other previously characterized MDR operons, such as mucAB and rumABR391. Indeed, rulAB shares many other features with the known MDR systems, including the presence of a binding site for the LexA repressor within its putative promoter region, possession of a conserved internal cleavage site within the rulA sequence, and lack of function in a recA background (46). Each of the known umu-like systems varies in its ability to promote mutagenesis in different cellular backgrounds. For example, the approximately threefold increase in our experiments of mutagenesis of mucAB in E. coli RW120 as compared to that of umuDC has been observed by others as well and is attributed to a more efficient processing of MucA to the truncated MucA′ form in an E. coli host (10). It is interesting to note that the difference in mutation frequency between rulAB and mucAB was reduced in the P. aeruginosa PAO1 background (Fig. 3B) due to a larger decrease in MucAB-mediated MDR in P. aeruginosa than in E. coli. This is probably due to a decreased affinity of the MucAB system with P. aeruginosa RecA or DNA polymerase.
In the Pseudomonas backgrounds, rulAB has a large effect on UV-B survival for P. syringae as noted previously (46, 47) and for P. aeruginosa in the current study. Although the rates of survival of P. aeruginosa PAO1(pJJK17) and P. syringae pv. syringae B86-17 (rulAB+) were similar for UV-B doses up to 1,900 J m−2, the frequency of mutation to Rifr was approximately sevenfold higher in P. aeruginosa. The increased mutation frequency in P. aeruginosa could be due to the relative contribution of other non-MDR systems and rulAB to the overall repair effort. At the 1,900-J m−2 UV-B dose, the survival rate of P. aeruginosa PAO1(pJB321) is approximately 20-fold lower than that of P. syringae pv. syringae GWS242(pJB321), suggesting that P. syringae is more efficient in non-MDR than P. aeruginosa.
Through analysis of UV-B mutability in the rulB::Km strain GWS242, we demonstrated a small (10- to 20-fold) increase in mutability to Rifr, an observation that we have also made when examining other natural P. syringae strains which do not contain rulAB (Kim and Sundin, unpublished). These observations are consistent with the occurrence of another source of UV mutability within the P. syringae genome. Hybridizations of BamHI-digested genomic DNA of the rulAB-negative strain P. syringae pv. syringae FF5 with a rulAB probe under low-stringency conditions resulted in the observation of a single hybridizing band (Kim and Sundin, unpublished). We are currently attempting to clone this chromosomal sequence and determine its role in UV survival and mutability. Other examples of MDR systems cooccurring on plasmids and chromosomes in a single strain have been examined in serovar Typhimurium (31) and more recently in Shigella flexneri (37). In serovar Typhimurium, the chromosomally encoded umuDCST determinant is active in UV-inducible mutagenesis, while the plasmid-encoded samAB determinant does not contribute to UV mutability (18). The situation is reversed in S. flexneri, as the plasmid-encoded impCAB determinant is required for UV mutability while the chromosomally encoded umuDC operon is not expressed following UV irradiation and therefore is apparently unable to promote UV-induced mutagenesis (37).
Thus, in each of these situations, only one of the MDR operons plays the important role in UV mutability, which leads to the obvious question—why are two operons present? Recently, Opperman et al. (32) showed that the E. coli umuDC gene products play an additional role in cell cycle control following DNA damage. These authors demonstrated individual roles for the UmuD and UmuD′ proteins and proposed a model in which the UmuD and UmuC proteins were involved in a delay in the recovery of DNA replication following DNA damage (32). RecA-mediated cleavage of UmuD and the formation of the UmuD′2C complex would then be required for MDR mediated by translesion synthesis; this would be followed by the resumption of DNA replication (32, 35). Cell cycle control and a delay in replication restart following DNA damage are presumably important because they allow for non-MDR processes such as excision repair to occur, ultimately lowering the cellular mutational load. Organisms which encode two MDR systems in which one is functionally dominant in terms of actual repair may utilize the other system in cell cycle control. Alternatively, the formation of chimeric MDR complexes may also be involved in the posttranslational regulation of these systems. We are interested in examining the chromosomal source of UV mutability in P. syringae in order to understand the contribution of this system and rulAB to the overall repair process and UV survival.
The regulation analyses performed in this study indicate that rulAB is expressed in a UV-inducible manner with increased promoter activity in response to increasing UV-B dosage. The rapid induction of expression, maintenance of transcriptional activity at high levels for 4 to 6 h following irradiation, and overall importance of the rulAB determinant to UV-B survival suggest that MDR could play a major role in P. syringae survival in the environment. As more studies show the significance of DNA repair processes to organismal survival in habitats with high solar radiation exposure (e.g., reference 15), it would be important to determine the occurrence and role of MDR in organisms in these habitats.
It was previously shown that rulAB-containing P. syringae strains maintained significantly larger phyllosphere populations following UV-B irradiation (47). Our current results from assays involving phyllosphere populations of P. syringae pv. syringae B86-17 indicate that MDR occurs and is detectable during the period when inoculated cells are establishing an infection on their host. Populations of P. syringae pv. syringae inoculated to bean leaf surfaces typically initiate disease, which is manifested by the occurrence of leaf spots, within 4 to 6 days. In our experiments, we observed leaf spot symptoms at the 5-day sampling time. Our data indicate that UV-B mutability occurs at levels similar to those recorded in vitro when UV-B irradiation is administered immediately after inoculation. Over time, as strain B86-17 established an infection on its bean host, the frequency of UV-B mutability to Rifr was slightly reduced (from 4.8- to 2.9-fold) but still significantly greater than in a nonirradiated control. As P. syringae strains become established in the phyllosphere, it is thought that a proportion of the cell population colonizes sites on leaves which are protected from external stresses (51). However, our data on UV-B mutability in the phyllosphere and previous data on rulAB-mediated UV-B survival in the phyllosphere (47) also clearly indicate the important contribution of the rulAB determinant to relative fitness during colonization and establishment of infection in planta.
The importance of rulAB to UV tolerance and the potential recurring necessity of this determinant for survival in the phyllosphere seem to distinguish rulAB from the other umu-like operons from an ecological standpoint. Each of the other umu-like operons characterized to date has been isolated from enteric organisms, whose exposure to UV radiation or chemical mutagens would probably be sporadic. One example of the consequence of a limited necessity for MDR in enteric organisms may be an attenuation of activity of the corresponding MDR determinant. For example, when located on its natural plasmid R46, the activity of mucAB is repressed by another gene located approximately 2 kb away (20). The role of some of the enteric-organism umu-like operons in UV protection is also unclear, as inactivation of either or both umuDCST and samAB in serovar Typhimurium has no effect on UV survival (18). Thus, the distinguishing features of the rulAB system in terms of activity when present on its native plasmid, function in UV tolerance, and potentially daily expression in response to solar UV damage make this system appropriate for continued analysis of the ecological and evolutionary ramifications of UV-induced mutagenesis.
ACKNOWLEDGMENTS
We thank Michael Hynes, Steve Lindow, Svein Valla, and Roger Woodgate for bacterial strains and plasmids and thank two anonymous reviewers for insightful comments on the manuscript.
This work was supported by the U.S. Department of Agriculture (NRICGP 9702832 and NRICGP 1999-02518) and the Texas Agricultural Experiment Station.
REFERENCES
- 1.Alexeyev M F. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques. 1995;18:52–54. [PubMed] [Google Scholar]
- 2.Blatny J M, Brautaset T, Winther-Larsen H C, Haugan K, Valla S. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl Environ Microbiol. 1997;63:370–379. doi: 10.1128/aem.63.2.370-379.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brent R, Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci USA. 1981;78:4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruck I, Woodgate R, McEntee K, Goodman M F. Purification of a soluble UmuD′C from Escherichia coli: cooperative binding of UmuD′C to single-stranded DNA. J Biol Chem. 1996;271:10767–10774. doi: 10.1074/jbc.271.18.10767. [DOI] [PubMed] [Google Scholar]
- 5.Burckhardt S E, Woodgate R, Scheuermann R H, Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci USA. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crosa J H, Falkow S. Plasmids. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. pp. 266–282. [Google Scholar]
- 7.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;79:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 9.Grant S G, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauser J, Levine A S, Ennis D G, Chumakov K, Woodgate R. The enhanced mutagenic potential of the MucAB proteins correlates with the highly efficient processing of the MucA protein. J Bacteriol. 1992;174:6844–6851. doi: 10.1128/jb.174.21.6844-6851.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herndl G J, Muller-Niklas G, Frick J. Major role of ultraviolet-B in controlling bacterioplankton growth in the surface layer of the ocean. Nature. 1993;361:717–719. [Google Scholar]
- 12.Ho C, Kulaeva O I, Levine A S, Woodgate R. A rapid method for cloning mutagenic DNA repair genes: isolation of umu-complementing genes from multidrug resistance plasmids R391, R446b, and R471a. J Bacteriol. 1993;175:5411–5419. doi: 10.1128/jb.175.17.5411-5419.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howarth S. Resistance to the bactericidal effects of ultraviolet radiation conferred on enterobacteria by the colicin factor ColI. J Gen Microbiol. 1965;40:43–55. doi: 10.1099/00221287-40-1-43. [DOI] [PubMed] [Google Scholar]
- 14.Howarth S. Increase in frequency of ultraviolet-induced mutation brought about by the colicine factor, Col-I in Salmonella typhimurium. Mutat Res. 1966;3:129–134. doi: 10.1016/0027-5107(66)90026-1. [DOI] [PubMed] [Google Scholar]
- 15.Jeffrey W H, Aas P, Lyons M M, Coffin R B, Pledger R J, Mitchell D L. Ambient solar radiation-induced photodamage in marine bacterioplankton. Photochem Photobiol. 1996;64:419–427. [Google Scholar]
- 16.Keane P J, Kerr A, New P B. Crown gall of stone fruit. II. Identification and nomenclature of Agrobacterium isolates. Aust J Biol Sci. 1970;23:585–595. [Google Scholar]
- 17.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 18.Koch W H, Cebula T A, Foster P L, Eisenstadt E. UV mutagenesis in Salmonella typhimurium is umuDC dependent despite the presence of samAB. J Bacteriol. 1992;174:2809–2815. doi: 10.1128/jb.174.9.2809-2815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulaeva O I, Wootton J C, Levine A S, Woodgate R. Characterization of the umu-complementing operon from R391. J Bacteriol. 1995;177:2737–2743. doi: 10.1128/jb.177.10.2737-2743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langer P J, Perry K L, Walker G C. Complementation of a pKM101 derivative that decreases resistance to UV killing but increases susceptibility to mutagenesis. Mutat Res. 1985;150:147–158. doi: 10.1016/0027-5107(85)90112-5. [DOI] [PubMed] [Google Scholar]
- 21.Legard D E, Aquadro C F, Hunter J E. DNA sequence variation and phylogenetic relationships among strains of Pseudomonas syringae pv. syringae inferred from restriction site maps and restriction fragment length polymorphism. Appl Environ Microbiol. 1993;59:4180–4188. doi: 10.1128/aem.59.12.4180-4188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindgren P B, Frederick R, Govindarajan A G, Panopolous N J, Staskawicz B J, Lindow S E. An ice nucleation reporter gene system: identification of inducible pathogenicity genes in Pseudomonas syringae pv. phaseolicola. EMBO J. 1989;8:1291–1301. doi: 10.1002/j.1460-2075.1989.tb03508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindow S E. Bacterial ice nucleation activity. In: Klement S, Rudolf K, Sands D C, editors. Methods in phytobacteriology. Budapest, Hungary: Akadémiai Kiadó; 1990. pp. 185–198. [Google Scholar]
- 24.Little J W. LexA cleavage and other self-processing reactions. J Bacteriol. 1993;175:4199–4203. doi: 10.1128/jb.175.16.4943-4950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Little J W, Mount D W, Yanisch-Perron C R. Purified lexA protein is a repressor of the recA and lexA genes. Proc Natl Acad Sci USA. 1981;78:4199–4203. doi: 10.1073/pnas.78.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lodwick D, Owen D, Strike P. DNA sequence analysis of the imp UV protection and mutation operon of the plasmid TP110: identification of a third gene. Nucleic Acids Res. 1990;18:5045–5050. doi: 10.1093/nar/18.17.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loper J E, Lindow S E. A biological sensor for iron available to bacteria in their habitats on plant surfaces. Appl Environ Microbiol. 1994;60:1934–1941. doi: 10.1128/aem.60.6.1934-1941.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loper J E, Lindow S E. Reporter gene systems useful in evaluating in situ gene expression by soil- and plant-associated bacteria. In: Hurst C J, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C.: ASM Press; 1997. pp. 482–492. [Google Scholar]
- 29.McBeth D L. Effect of degradative plasmid CAM-OCT on responses of Pseudomonas bacteria to UV light. J Bacteriol. 1989;171:975–982. doi: 10.1128/jb.171.2.975-982.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell D L, Nairn R S. The biology of the (6-4) photoproduct. Photochem Photobiol. 1989;49:805–819. doi: 10.1111/j.1751-1097.1989.tb05578.x. [DOI] [PubMed] [Google Scholar]
- 31.Nohmi T, Hakura A, Nakai Y, Watanabe M, Murayama S Y, Sofuni T. Salmonella typhimurium has two homologous but different umuDC operons: cloning of a new umuDC-like operon (samAB) present in a 60-megadalton cryptic plasmid of S. typhimurium. J Bacteriol. 1991;173:1051–1063. doi: 10.1128/jb.173.3.1051-1063.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Opperman T, Murli S, Smith B T, Walker G C. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc Natl Acad Sci USA. 1999;96:9218–9223. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry K L, Elledge S J, Mitchell B B, Marsh L, Walker G C. umuDC and mucAB operons whose products are required for UV light- and chemical-induced mutagenesis: UmuD, MucA, and LexA proteins share homology. Proc Natl Acad Sci USA. 1985;82:4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 35.Rangarajan S, Woodgate R, Goodman M F. A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc Natl Acad Sci USA. 1999;96:9224–9229. doi: 10.1073/pnas.96.16.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reuven N B, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD′, RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 37.Runyen-Janecky L J, Hong M, Payne S M. The virulence plasmid-encoded impCAB operon enhances survival and induced mutagenesis in Shigella flexneri after exposure to UV radiation. Infect Immun. 1999;67:1415–1423. doi: 10.1128/iai.67.3.1415-1423.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sesma A, Sundin G W, Murillo J. Closely related plasmid replicons coexisting in the phytopathogen Pseudomonas syringae show a mosaic organization of the replication region and altered incompatibility behavior. Appl Environ Microbiol. 1998;64:3948–3953. doi: 10.1128/aem.64.10.3948-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinagawa H, Iwasaki H, Kato T, Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci USA. 1988;85:1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simonson C S, Kokjohn T A, Miller R V. Inducible UV repair potential of Pseudomonas aeruginosa PAO. J Gen Microbiol. 1990;136:1241–1249. doi: 10.1099/00221287-136-7-1241. [DOI] [PubMed] [Google Scholar]
- 42.Smith B T, Walker G C. Mutagenesis and more: umuDC and the Escherichia coli SOS response. Genetics. 1998;148:1599–1610. doi: 10.1093/genetics/148.4.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strike P, Lodwick D. Plasmid genes affecting DNA repair and mutation. J Cell Sci Suppl. 1987;6:303–321. doi: 10.1242/jcs.1984.supplement_6.20. [DOI] [PubMed] [Google Scholar]
- 44.Sundin G W, Bender C L. Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae. Appl Environ Microbiol. 1993;59:1018–1024. doi: 10.1128/aem.59.4.1018-1024.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sundin G W, Jacobs J L. Ultraviolet radiation (UVR) sensitivity analysis and UVR survival strategies of a bacterial community from the phyllosphere of field-grown peanut (Arachis hypogeae L.) Microb Ecol. 1999;38:27–38. doi: 10.1007/s002489900152. [DOI] [PubMed] [Google Scholar]
- 46.Sundin G W, Kidambi S P, Ullrich M, Bender C L. Resistance to ultraviolet light in Pseudomonas syringae: sequence and functional analysis of the plasmid-encoded rulAB genes. Gene. 1996;177:77–81. doi: 10.1016/0378-1119(96)00273-9. [DOI] [PubMed] [Google Scholar]
- 47.Sundin G W, Murillo J. Functional analysis of the Pseudomonas syringae rulAB determinant in tolerance to ultraviolet B (290–320 nm) radiation and distribution of rulAB among P. syringae pathovars. Environ Microbiol. 1999;1:75–87. doi: 10.1046/j.1462-2920.1999.00008.x. [DOI] [PubMed] [Google Scholar]
- 48.Tang M, Bruck I, Eritja R, Turner J, Frank E G, Woodgate R, O'Donnell M, Goodman M F. Biochemical basis of SOS-induced mutagenesis in Escherichia coli: reconstitution of in vitro lesion bypass dependent on the UmuD′2C mutagenic complex and RecA protein. Proc Natl Acad Sci USA. 1998;95:9755–9760. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang M, Shen X, Frank E G, O'Donnell M, Woodgate R, Goodman M F. UmuD′2C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker G C. SOS-regulated proteins in translesion DNA synthesis and mutagenesis. Trends Biochem Sci. 1995;20:416–420. doi: 10.1016/s0968-0004(00)89091-x. [DOI] [PubMed] [Google Scholar]
- 51.Wilson M, Hirano S S, Lindow S E. Location and survival of leaf-associated bacteria in relation to pathogenicity and potential for growth within the leaf. Appl Environ Microbiol. 1999;65:1435–1443. doi: 10.1128/aem.65.4.1435-1443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodgate R, Sedgwick S G. Mutagenesis induced by bacterial UmuDC proteins and their plasmid homologues. Mol Microbiol. 1992;6:2213–2218. doi: 10.1111/j.1365-2958.1992.tb01397.x. [DOI] [PubMed] [Google Scholar]