Abstract
Purpose:
Liquid biopsy offers an attractive platform for noninvasive tumor diagnosis, prognostication, and prediction of glioblastoma clinical outcomes. Prior studies report that 30% to 50% of GBM lesions characterized by EGFR amplification also harbor the EGFRvIII mutation.
Experimental Design:
A novel digital droplet PCR (ddPCR) assay for high GC content amplicons was developed and optimized for sensitive detection of EGFRvIII in tumor tissue and circulating extracellular vesicle RNA (EV RNA) isolated from the plasma of patients with glioma.
Results:
Our optimized qPCR assay detected EGFRvIII mRNA in 81% [95% confidence interval (CI), 68%–94%] of EGFR-amplified glioma tumor tissue, indicating a higher than previously reported prevalence of EGFRvIII in glioma. Using the optimized ddPCR assay in discovery and blinded validation cohorts, we detected EGFRvIII mutation in 73% (95% CI, 64%–82%) of patients with a specificity of 98% (95% CI, 87%–100%), compared with qPCR tumor tissue analysis. In addition, upon longitudinal monitoring in 4 patients, we report detection of EGFRvIII in the plasma of patients with different clinical outcomes, rising with tumor progression, and decreasing in response to treatment.
Conclusions:
This study demonstrates the feasibility of detecting EGFRvIII mutation in plasma using a highly sensitive and specific ddPCR assay. We also show a higher than previously reported EGFRvIII prevalence in glioma tumor tissue. Several features of the assay are favorable for clinical implementation for detection and monitoring of EGFRvIII-positive tumors.
Translational Relevance.
In this study, we investigate the clinical feasibility of liquid biopsy assessment of the EGFRvIII mutation as a diagnostic and predictive blood-based biomarker in patients with glioma. EGFRvIII detection in patient plasma complements a multimodality approach (radiology, clinical features) in achieving reliable diagnosis and disease surveillance of EGFRvIII+ gliomas. Tissue biopsy, the current gold standard for glioma diagnosis, is invasive and samples tumor focally and therefore poses challenges in monitoring the dynamic genomic alterations in tumors over the course of treatment and progression. Our extracellular vesicle RNA-based ddPCR method can detect a canonical tumor mutation in EGFRvIII+ gliomas, and suggests the potential for serial, longitudinal assessment of tumor mutational burden even in the context of intratumoral heterogeneity. The cost effectiveness and safety of our proposed method provides an alternative strategy to repetitive, invasive tissue-based brain biopsies.
Introduction
Glioblastoma (GBM) is the most common and aggressive primary brain tumor with a global incidence of 0.5 to 5 per 100,000 persons (1, 2, 3) and a median survival time of 15 months from the time of diagnosis (4, 5). Despite increasing advances in therapeutics, prognosis remains poor with high rates of treatment failure and relapse. The current gold standard diagnostic tool, tissue biopsy, in addition to being invasive only provides a single time point evaluation of the tumor genomic profile, thereby limiting its utility in exploring the tumor heterogeneity spatially and over time. Therefore, there remains the need for development and clinical implementation of alternative sensitive, noninvasive modalities to facilitate serial sampling and real-time monitoring of evolving genetic and epigenetic alterations. In this regard, liquid biopsy, defined as sampling and analysis of nonsolid biological fluids (CSF, plasma), offers new opportunities for GBM molecular characterization and detection of prognostic biomarkers (6). Tumors including gliomas have been shown to shed biological material into the circulation which in turn reflects the transcriptomic and proteomic profile of cell of origin. Examples of analytes include extracellular vesicles (EV), circulating tumor cells (CTC), cell-free DNA (cfDNA), proteins, and metabolites. Tumor-derived EVs are particularly promising for biomarker discovery because they encapsulate and retain tumor-specific mRNAs in a stable form in a range of biofluids. In addition, the enclosed cargo can vary in response to diverse stimuli and tumor stage (7).
Previous work has established detection of several key molecular markers in biofluids from patients with glioma, including EGFRvIII mRNA, IDH1 RNA/DNA, TERT promoter, and PTEN deletion (8, 9, 10, 11). Epidermal growth factor receptor (EGFR) amplification represents one of the hallmark alterations of GBM and is present in about 57% of primary GBMs and 8% of GBMs arising from recurrent low-grade gliomas (12, 13). Approximately, 30% to 50% of EGFR-amplified GBMs harbor an in-frame deletion of exons 2–7, forming a truncated receptor with loss of extracellular ligand binding domain (14). This deleted EGFR variant III (EGFRvIII) is capable of constitutive signaling in a ligand-independent manner, thereby promoting tumorigenesis and overall tumor growth. In contrast to EGFR wild type (EGFRwt), EGFRvIII is only expressed in tumor cells, thereby making it a viable therapeutic and diagnostic target in precision oncology (15).
Clinical testing and validation of EGFRvIII from biofluids has been hampered by several challenges, namely limited understanding of structural biology of EGFRvIII, use of suboptimal methods of tumor-specific EV isolation, low sensitivity of employed PCR technologies, and overall low sensitivity and/or specificity of reported methods.
Interestingly, we observed that the EGFRvIII mRNA transcript contains several four guanine (4G) repeat sequences, especially close to the junction site (exon1:exon8), which is also the target region of interest. These 4G sequences have a well-documented role in the formation of G-quadruplexes as well as other 3D secondary structures (16, 17), which are known to play an inhibitory role in reverse transcription and PCR amplification (18, 19). Addition of PCR additives may destabilize secondary structures, as previously reported for the amplification of the highly GC-rich TERT promoter region (20).
To the best of our knowledge, RNA secondary structures in EGFRvIII mRNA have not been previously discussed in the literature. Here, we sought to establish a sensitive plasma-based droplet digital PCR (ddPCR) assay for the detection of the EGFRvIII mutation in EV-derived RNA. To achieve this, we devised novel protocols with different optimization variables, including minimizing the adverse effects of secondary structures at the reverse transcription and PCR steps, by exploring the use of different PCR additives. Furthermore, to improve amplification efficiency and specificity we tested thermocycling conditions, temperature gradients, and different purification protocols. Using the overall optimized protocol, we determined the prevalence of EGFRvIII mRNA in tumor tissue from n = 37 tumor tissue samples. Finally, we tested the novel plasma assay in n = 54 plasma samples that included tissue confirmed EGFRvIII (n = 30), EGFRwt (n = 10) as well as age-matched healthy controls (n = 14).
Materials and Methods
Tumor tissue processing
Tumor tissue was microdissected and suspended in RNAlater (Ambion) or flash-frozen and stored at −80°C.
Patient plasma processing
Whole blood was collected using K2 EDTA tubes with an inert gel barrier (BD Vacutainer Blood Collection Tubes), from preoperatively placed arterial lines or venipuncture. Within 2 hours of collection, samples were centrifuged at 1,100 × g for 10 minutes at 20°C to separate the plasma from the hematocrit and filtered using 0.8 μmol/L filters. Aliquots (1 mL) were stored at −80°C for later downstream analysis. Except for the longitudinal samples, all baseline samples were collected prior to surgical resection.
Synthetic RNA
Nucleotide sequence of the EGFRvIII mRNA transcript around the region of interest (120 bp) was purchased as Ultramer RNA oligonucleotide (Integrated DNA Technologies). Amount of Oligo was 68.49 nmol and purified using standard desalting. The quality of the synthetic RNA was not optimal due to the high GC content. The ultramer RNA oligo was then resuspended in nuclease-free water to make up to 100 μmol/L. Initial quantification using NanoDrop Spectrophotometer (Thermo Fisher Scientific) demonstrated RNA concentration of 4,430 ng/μL. Aliquots were prepared and stored at −80.0°C. Accurate qualitative and quantitative analysis of the RNA was performed using Agilent RNA 6000 Pico Kit run on Agilent Technologies 2100 Bioanalyzer. For this purpose, the sample was diluted 2,000-fold (based on Nanodrop measurement). Electropherograms obtained from Bioanalyzer revealed two peaks and we manually selected for the concentration (area under the curve) of the peak of interest based on the RNA size (120 bp). The resulting nanograms were then converted to EGFRvIII copy number using the standard copy number formula for single-strand RNA as shown below:
 |
Where: X = amount of amplicon (ng)
N = length of ssRNA amplicon
660 g/mole = average mass of 1 bp ssRNA
Study population
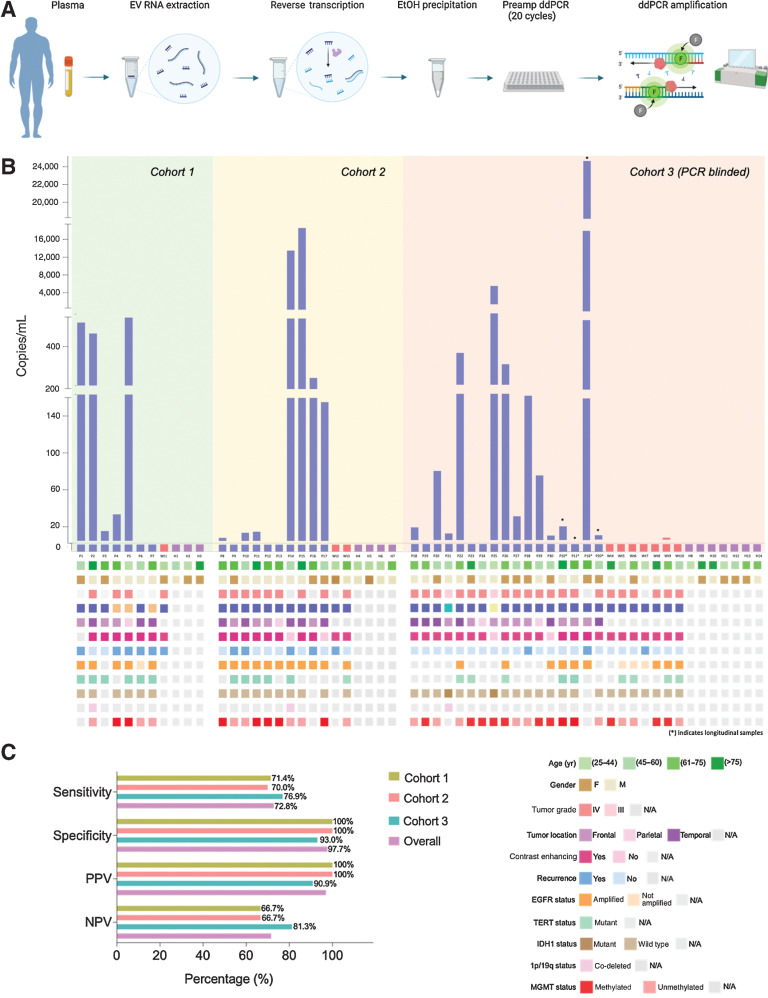
The study population (n = 54, n = 30 EGFRvIII positive, n = 10 EGFR wild type, n = 14 age-matched healthy controls) included 54 patients aged 18 years or older who underwent surgery at the Massachusetts General Hospital (MGH) for biopsy or resection of a primary brain lesion. Clinical EGFRvIII status was established with either immunohistochemistry (IHC) or by the MGH SNaPShot Panel. Additional inclusion criteria for the study population included histopathologic confirmation of disease. Healthy control patients with a history of oncologic, neurologic, or ongoing infectious conditions were excluded from the study. All samples were collected under IRB-approved protocol no. 2017P001581 with informed patient consent. Patient demographics are depicted in oncoprint format in Fig. 4B and in table format in Table 1.
Figure 4.
Detection of EGFRvIII mutation in plasma samples of patient cohort (n = 3). A, Schematic depicting experimental workflow, including isolation of plasma, extraction platform, reverse transcription, purification, and subsequent ddPCR amplification. B, Copies/mL of EGFRvIII mutation in tested plasma samples are graphed according to sample number. Oncoprint depicting the demographic characteristics and genomic landscape of each sample are plotted underneath. C, Contingency tables were constructed from data obtained, and sensitivity and specificity were calculated and graphed above. Overall sensitivity and specificity across all three cohorts are also reported.
Table 1.
Patient demographics.
| Cohort 1 | % | Cohort 2 | % | Cohort 3 | % | |
|---|---|---|---|---|---|---|
| Total | 11 | 16 | 27 | |||
| Age median (y) | 53 | 62 | 64 | |||
| Sex | ||||||
| Male | 8 | 72.7 | 11 | 68.8 | 16 | 59.3 |
| Female | 3 | 27.3 | 5 | 31.3 | 11 | 40.1 |
| Race, n (%) | ||||||
| White | 8 | 72.7 | 12 | 75 | 19 | 70.4 |
| Other | ||||||
| Unavailable | 3 | 27.3 | 4 | 25 | 8 | 29.6 |
| Ethnicity, n (%) | ||||||
| Hispanic | ||||||
| Non-Hispanic | 8 | 72.7 | 12 | 75 | 19 | 70.4 |
| Unavailable | 3 | 27.3 | 4 | 25 | 8 | 29.6 |
| WHO Grade, n (%) | 8 | 12 | 20 | |||
| III | 1 | 8.3 | 2 | 10 | ||
| IV | 7 | 87.5 | 11 | 91.7 | 17 | 85 |
| N/A | 1 | 12.5 | 1 | 5 | ||
| Disease status | ||||||
| Newly diagnosed | 1 | 6 | 16 | |||
| Recurrent | 6 | 3 | 4 | |||
| N/A | 1 | 3 | ||||
| Medications, n (%; from diseased total) | ||||||
| Anticonvulsants (levetiracetam, clonazepam, lacosamide, carbamazepine, lamotrigine) | 5 | 62.5 | 9 | 75 | 10 | 50 |
| Steroids (dexamethasone) | 7 | 87.5 | 5 | 41.7 | 4 | 20 |
| Blood thinner (aspirin, coumadin) | 3 | 25 | 5 | 25 | ||
| Prior radiotherapy | ||||||
| Prior chemotherapy | ||||||
| Clinical trial participant | 3 | 37.5 | 1 | 8.3 | 1 | 5 |
RNA isolation from tumor tissue
RNA was isolated using the RNeasy Kit (Qiagen) per the manufacturer's recommendations and eluted in 20 μL of nuclease-free water (Qiagen). Frozen tissue was thawed and lysed in 1 to 2 mL of ice-cold Trizol Reagent (Thermo Fisher Scientific). Lysate was homogenized by passing through a 20-gauge RNAse-free needle 10 times. Total RNA was then extracted as per the manufacturer's protocol and eluted in 20 μL of nuclease-free water (Invitrogen). All RNA samples were assessed for purity with the NanoDrop One spectrophotometer (Thermo Fisher Scientific). Agilent RNA 6000 Pico Kit was used with Agilent Technologies 2100 Bioanalyzer to determine the concentration and RIN (RNA Integrity Number) value of the samples. Isolated RNA was stored at −80°C prior to downstream analysis.
EV RNA extraction from plasma samples
In order to isolate the analyte of interest, 2 mL plasma was obtained from each clinical sample and thawed on ice. EV RNA was isolated using the ExoRNeasy Maxi Kit (Qiagen), per the manufacturer's recommendations. EV RNA was eluted in 17 μL of nuclease-free water (Qiagen) and assessed for concentration and purity with the NanoDrop One spectrophotometer (Thermo Fisher Scientific).
Optimized reverse transcription
Messenger RNA (mRNA) isolated from tumor tissue and EV RNA isolated from plasma samples, was reverse transcribed into cDNA using the SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific). The protocol was optimized to maximize cDNA yield. In the first part, 1.0 μL 50 μmol/L Oligo d(T)20, 1.0 μL 50 μmol/L random hexamers, 1.5 μL 10 mmol/L dNTP mix, and 1.0 μL 7-deaza-dGTP (New England BioLabs) were combined with template RNA in a 0.2 mL PCR reaction tube. Nuclease-free water was then added to reach a final reaction volume of 13 μL. The RNA–primer mix was then incubated at 65°C for 5 minutes, immediately followed by incubation on ice for 1 minute. In the second part, reverse transcription (RT) reaction mix was prepared by combining 4.0 μL 5× SSIV Buffer, 1.0 μL 100 mmol/L DTT, 1.0 μL ribonuclease inhibitor, and 1.0 μL SuperScript IV Reverse Transcriptase (200 U/μL) in a reaction tube to a final volume of 7 μL. The contents were briefly vortexed and centrifuged. The RT reaction mix was then added to annealed RNA from the first part of the protocol. Combined annealed RNA-RT reaction mix was then incubated at 23°C for 10 minutes, 53°C for 10 minutes, 80°C for 10 minutes. The resulting 20 μL cDNA was then purified using ethanol precipitation.
Ethanol precipitation
Ethanol precipitation was used to concentrate the cDNA from reverse transcription and remove potential downstream inhibitors. The choice of appropriate salt was made based on which preparation would give maximum yield without inhibition of polymerase activity and removal of dNTPs. Consequently, 3 mol/L, pH 5.2 sodium acetate (Invitrogen) was used as the salt. Reverse transcription product, cDNA (20 μL), was combined with 0.1 volume of 3 mol/L sodium acetate (i.e., 2.0 μL) and 1.0 μL of 20 mg/mL glycogen (Thermo Fisher Scientific). Furthermore, 3 volumes (i.e., 60 μL) of ice-cold, 100% ethanol was added and mixed well. The ethanolic solution was stored at −20°C for 1 hour, as determined by optimization experiments. Following this, cDNA was recovered by centrifugation at 16,000 × g for 30 minutes at 4°C. The supernatant was carefully aspirated without disturbing the pellet. Subsequently, the pellet was washed with 0.5 mL of ice-cold, freshly prepared 70% ethanol. This was followed by centrifugation at maximum speed for 10 minutes at 4°C. The supernatant was removed and the tube was left open at room temperature to ensure that last traces of fluid have evaporated. The pellet was then dissolved and resuspended in 6 μL of nuclease-free water (Invitrogen).
Primers and probes
Through the course of optimization and preliminary testing, at least 10 different primer pairs specific to EGFRvIII deletion detection were designed and tested. The primer sets generated amplicons of varying lengths, ranging from 34 to 428 bp. The key criteria in selecting the optimal primer set included; target amplicon size <100 bp, minimum formation of primer dimers, and absence of nonspecific products. Tools used to design and assess different parameters include Primer-BLAST, Serial Cloner, and Primer3Plus. Different primer sets were also compared on the basis of GC content, Tm (°C) mismatch, self-dimerization, and hairpin formation. The final primer set selected for further optimization produced a target amplicon of 96 bp. Sequence for primers and probe are as follows: forward mRNA primer (5′-GGCTCTGGAGGAAAAGAAAGGTAATT-3′), reverse mRNA primer (5′-CCGTCTTCCTCCATCTCATAGC-3′), EGFRvIII-mutant probe (5′-FAM-TGACAGATCACGGCTC-MGBNFQ -3′). All oligonucleotide preparations were synthesized by Thermo Fisher Scientific. Primers were purified by desalting and resuspended in DEPC-treated water, at a final concentration of 100 μmol/L. Probe was HPLC purified and resuspended in 1× TE 100 pmol/μL.
Optimized droplet digital PCR assay in tumor tissue
Droplet digital PCR (ddPCR) was performed using purified cDNA obtained from reverse transcription of RNA isolated from tumor tissue. The ddPCR reaction was performed in the Applied Biosystems 96-well Thermal Cycler (Thermo Fisher Scientific) in a final volume of 20 μL and was prepared using 10 μL of ddPCR SuperMix for Probes (no dUTP; Bio-Rad), 1,500 nmol/L forward and reverse primers and 350 nmol/L probe, and 2 μL of 5 mol/L betaine solution (Sigma-Aldrich). The prepared reaction mixture was transferred to the wells of DG8 cartridge. To generate the droplets, 70 μL of Droplet generation oil for probes (Bio-Rad) was added and the plate was loaded into the QX200 Droplet generator (Bio-Rad). The droplet emulsions were then carefully transferred to a semi-skirted, PCR-clean 96-well plate (Eppendorf) using a multichannel pipette. Finally, the plate was sealed using PX1 PCR plate sealer (Bio-Rad). Based on the annealing temperature gradient optimization experiment, the final thermal cycling conditions were as follows: initial enzyme activation at 95°C (51% ramp) for 10 minutes, then 40 cycles of denaturation at 94°C (51% ramp) for 30 seconds, and annealing/extension at 55.5°C for 1 minute, followed by enzyme inactivation at 98°C for 10 minutes and final hold at 4°C until analysis. Droplets were analyzed using QX 200 droplet reader (Bio-Rad) and data were acquired and analyzed with QuantaSoft analysis software (Bio-Rad).
Optimized quantitative (qPCR) assay in tumor tissue
Quantitative PCR (qPCR) was performed using purified cDNA obtained from reverse transcription of RNA isolated from tumor tissue. The qPCR reaction was performed in the Applied Biosystems StepOnePlus Real-Time PCR (Thermo Fisher Scientific) in a final volume of 20 μL and was prepared using 10 μL of TaqMan Universal Master Mix II, no UNG (Thermo Fisher Scientific), 1,500 nmol/L forward and reverse primers and 350 nmol/L probe, and 2 μL of 5 mol/L betaine solution (Sigma-Aldrich). The reaction mixture was prepared in the wells of 96-well, nonskirted PCR plate (Thermo Fisher Scientific). Finally, the plate was sealed using the Adhesive PCR plate sealer (Thermo Fisher Scientific). On the basis of the annealing temperature gradient optimization experiment, the final thermal cycling conditions were as follows: initial enzyme activation at 95°C (100% ramp) for 10 minutes, then 40 cycles of denaturation at 94°C (100% ramp) for 30 seconds, annealing at 55.5°C for 30 seconds, and extension at 60.0°C for 1 minute. Quantitative analysis was performed using Comparative Ct (ΔΔCt) method with StepOne Software (Applied Biosystems, Thermo Fisher Scientific). Data were acquired and interpreted using amplification plots. To calculate amplification efficiency, Ct values were plotted on a logarithmic scale along with corresponding concentrations of RNA input. Next, a linear regression curve through the data points was generated and slope of the trend line calculated. Finally, efficiency was calculated using the equation: E = −1+10(−1/slope).
Optimized ddPCR assay in plasma-derived EV RNA and preamplification step
ddPCR was performed using purified cDNA obtained from reverse transcription of EV RNA isolated from plasma samples. For plasma samples, detection of low levels of mutant EV RNA was enhanced via PCR preamplification. The preamplification ddPCR was performed in the Applied Biosystems 96-well Thermal Cycler (Thermo Fisher Scientific) in a final volume of 20 μL and was prepared using 6 μL of purified cDNA, 10 μL of ddPCR SuperMix for Probes (no dUTP; Bio-Rad), 1,500 nmol/L forward and reverse primers, and 1.0 μL of 10 mmol/L dNTP (Thermo Fisher Scientific). The thermocycling conditions were as follows: initial enzyme activation at 95°C (51% ramp) for 5 minutes, then 20 cycles of denaturation at 94°C (51% ramp) for 30 seconds, and annealing/extension at 55.5°C for 1 minute, followed by enzyme inactivation at 98°C for 10 minutes. The final ddPCR was performed using the preamplified product in a final reaction volume of 20 μL. The reaction mixture was prepared using 2 μL of preamplified product, 10 μL of ddPCR SuperMix for Probes (no dUTP; Bio-Rad), 1,500 nmol/L forward and reverse primers and 350 nmol/L probe, and 2 μL of 5 mol/L betaine solution (Sigma-Aldrich). The final thermal cycling conditions were as follows: initial enzyme activation at 95°C (51% ramp) for 10 minutes, then 40 cycles of denaturation at 94°C (51% ramp) for 30 seconds, and annealing/extension at 55.5°C for 1 minute, followed by enzyme inactivation at 98°C for 10 minutes and final hold at 4°C until analysis. Droplets were analyzed using QX 200 droplet reader (Bio-Rad) and data were acquired and analyzed with QuantaSoft analysis software (Bio-Rad).
ddPCR dMIQE 2020 guideline compliance
This section of the Materials and Methods describes our compliance with the updated 2020 dMIQE Guidelines for the technical development of a plasma-based ddPCR assay for the EGFRvIII mutation detection. Specimen type numbers, sampling procedure, aliquotation, conditions, and duration are provided in the following sections of the Materials and Methods: Study Population, Tumor Tissue Processing, and Plasma Processing. Details about the specific extraction techniques, nucleic acid assessment/storage, and reverse transcription are provided in sections: RNA Isolation from tumor tissue, and Exosomal RNA Isolation from plasma samples. Information about the ddPCR oligonucleotides and its target sequences are provided in Fig. 1A and in the section titled Primers and Probe. Details about the assay analytic validation are provided in Fig. 2. Finally, information regarding data analysis is provided both in Results and Fig. 4.
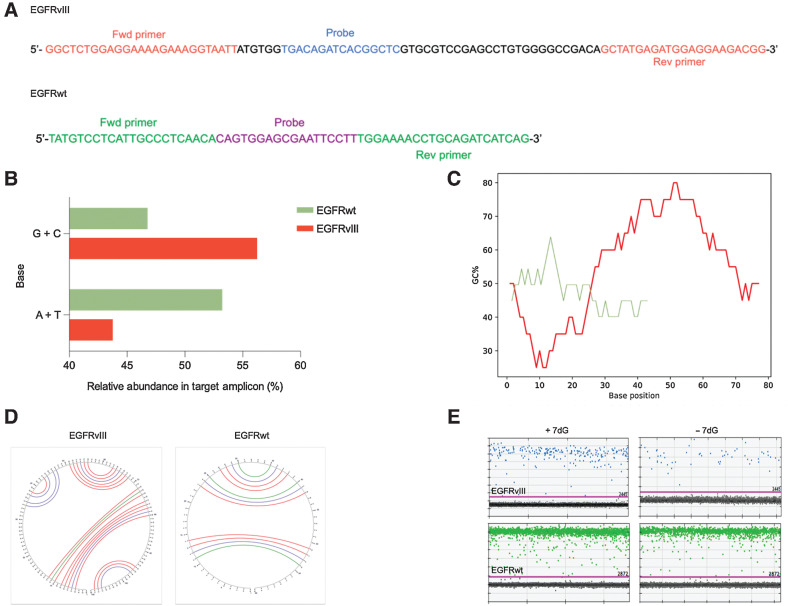
Figure 1.
RNA folding and secondary structures occurring in the EGFR transcript variant III. A, Diagram depicting nucleotide sequence of the target amplicon in the deleted variant of the EGFR transcript (EGFRvIII; top) and wild-type EGFR (bottom). Forward and reverse primers as well as probe sequence specific to each amplicon are illustrated. B, A bar graph comparing the relative abundance (percentage) of different nitrogenous bases (G +C, A +T) in the target amplicons (green = EGFRvIII, red = EGFRwt). C, Graphical representation of GC content distribution across the length of the target amplicon in both sequences (EGFRvIII, red = 96 bp, EGFRwt, green = 62 bp). Calculation of GC content (percentage) is performed using the formula, (G + C)/(A + T + G + C) *100 with the window size set to 20 bases. D, Output of secondary structure analysis using Mfold. The individual base pair interactions in EGFRvIII and EGFRwt target amplicon are illustrated using circle graphs. Each color denoted arc represents a unique base pair interaction (blue = A-U, A-T, red = G-C, green = G-U, G-T). E, Effect of 7dG on the secondary structures and the copy number of EGFRvIII (top) and EGFRwt (bottom) in tumor tissue as shown in dd PCR 1D plots.
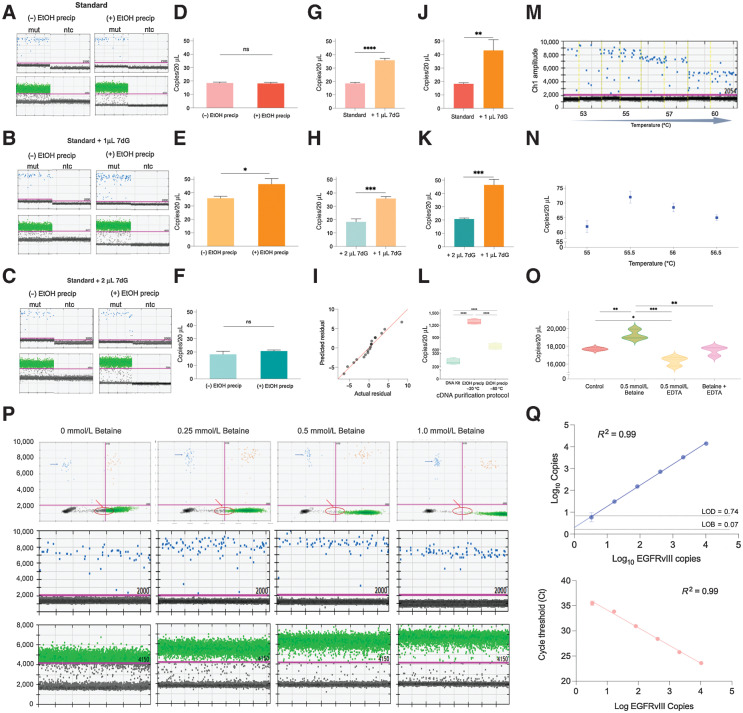
Figure 2.
EGFR transcript variant vIII droplet digital PCR (ddPCR) optimization. A–C, ddPCR 1D plots with mutant channel amplitude (blue, top) and GAPDH channel amplitude (green, bottom), run in parallel with (right) and without (left) ethanol precipitation of EGFRvIII cDNA from three different reverse transcription protocols: (a) standard, (b) standard + 1 μL 7-deaza-GTP, (c) standard + 2 μL. D–F, Two-tailed t test results depicting the difference in copies/20 μL from ddPCR using EGFRvIII cDNA treated with and without ethanol precipitation from different reverse transcription protocols; (d) standard, (e) standard + 1 μL 7-deaza-GTP, (f) standard + 2 μL. G, Two-tailed t test results comparing the statistically significant difference in copies/20 μL in EGFRvIII cDNA from standard and (standard + 1 μL 7-deaza-dGTP) RT protocols, both untreated with ethanol precipitation. H, Two-tailed t test results comparing the statistically significant difference in copies/20 μL in EGFRvIII cDNA from (standard + 2 μL 7-deaza-GTP) and (standard + 1 μL 7-deaza-dGTP) RT protocols, both untreated with ethanol precipitation. I, QQ plot obtained from one-way ANOVA results depicting statistically significant difference in average copies/20 μL in EGFRvIII cDNA across different conditions overall. J, Two-tailed t test results comparing the statistically significant difference in copies/20 μL in EGFRvIII cDNA from standard and (standard + 1 μL 7-deaza-dGTP) RT protocols, treated with ethanol precipitation. K, Two-tailed t test results comparing the statistically significant difference in copies/20 μL in EGFRvIII cDNA from (standard + 2 μL 7-deaza-GTP) and (standard + 1 μL 7-deaza-dGTP) RT protocols, treated with ethanol precipitation. L, Two-tailed t test results depicting statistically significant difference in copies/20 μL in EGFRvIII cDNA (standard + 1 μL 7-deaza-dGTP, RT protocol) purified using different cleanup protocols. M, ddPCR 1D plots demonstrating change in separation of mutant events from the baseline at different annealing/extension temperatures (low vs. high). N, Quantitative difference in copies/20 μL of mutant events across different annealing/extension temperatures. O, Violin plots demonstrating statistically significant difference in copies/20 μL with the addition of 0.5 mmol/L betaine, 0.5 mmol/L EDTA, 0.5 mmol/L (betaine + EDTA) versus no addition of ddPCR additive (control). P, ddPCR 2D plots (top row) and 1D plots depicting cluster density, tightness, and separation of mutant events (blue) and GAPDH events (green) at different concentrations of betaine versus no betaine addition to ddPCR. Q, Varying numbers of EGFRvIII synthetic RNA were spiked into the reverse transcription reaction. The resulting cDNA was then amplified using the optimized ddPCR (top) and qPCR (bottom) cycling conditions. For ddPCR, copies per 20 μL are plotted against EGFRvIII copies spike-in (top). Limit of detection (LOD, dashed line) is plotted, defined as 3 standard deviations over average copies per 20 μL obtained when only a small concentration of EGFRvIII was run along with blanks. Limit of blank (LOB, dashed line) is plotted, defined as the apparent highest copy number expected to be found when replicates of a blank sample containing no EGFRvIII (or small concentration of EGFRvIII) are tested. For qPCR cycle threshold (Ct) is plotted against EGFRvIII copies spike-in (bottom).
Quantification of EGFRvIII mutation in plasma
The number of EGFRvIII copies per mL of plasma was calculated from QuantaSoft data as follows: copies/mL plasma = C EV/TV/P where C = copies per 20 μL, EV = exNA elution volume (μL), TV = exNA input into ddPCR reaction (μL) and p = plasma volume (mL). Only samples with >10,000 droplets/well were included in the analysis. A positive sample is considered one with at least 1 event on at least two wells. Four wells are analyzed in parallel for each sample.
Statistical analysis
Statistical analysis was performed using the unpaired two-tailed Student t test in GraphPad Prism 8 software and P < 0.05 was considered to be statistically significant. The results are presented as mean ± SD.
Institutional Review Board statement
Our studies were conducted in accordance with principles for human experimentation as defined in the U.S. Common Rule and were approved by the Partners Institutional Review Board (IRB)–approved protocol number 2017P001581. All healthy control subjects were screened for pertinent oncologic and neurologic medical histories. Individuals with a history of cancer, neurologic disorders, and infectious diseases were excluded from the study.
Informed consent statement
All samples were collected with written informed consent after the patient was advised of the potential risks and benefits, as well as the investigational nature of the study.
Illustrations
Figures 3A and 4A were generated using www.BioRender.com.
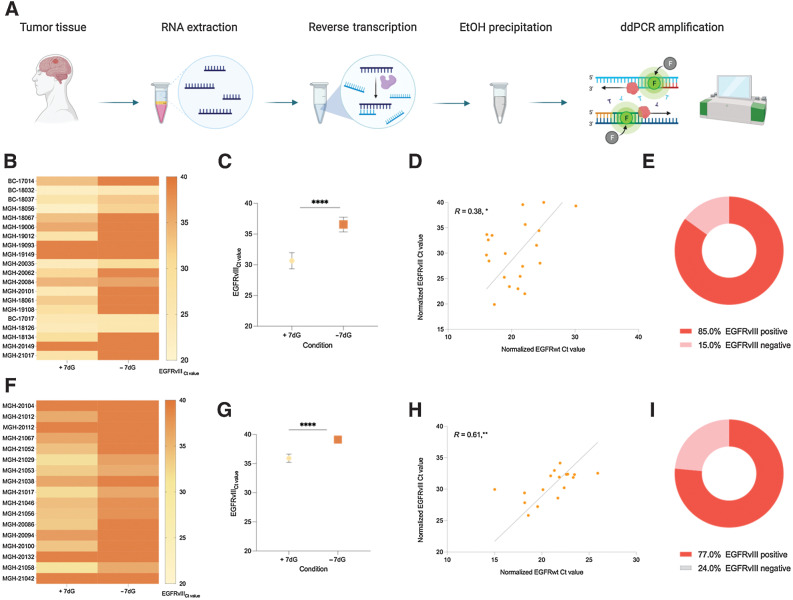
Figure 3.
Higher prevalence of EGFRvIII in tumor tissue using 7-deaza-dGTP (7dG). A, Experimental workflow for detecting EGFRvIII mutation in tumor tissue. B, Heatmap of EGFRvIII Ct values obtained by qPCR in tumor tissue RNA from cohort 2, in parallel, with and without 7-deaza-dGTP. C, Two-tailed t test depicting the statistically significant difference in the Ct value obtained, with and without 7-deaza-dGTP. D, Pearson correlation of normalized EGFRwt (x-axis) and normalized EGFRvIII (y-axis; reference gene, GAPDH). E,EGFRvIII prevalence in cohort 2. F, Heatmap of EGFRvIII Ct values obtained by qPCR in tumor tissue RNA from cohort 3, in parallel, with and without 7-deaza-dGTP. G, Two-tailed t test depicting the statistically significant difference in the Ct value obtained, with and without 7-deaza-dGTP. H, Pearson correlation of normalized EGFRwt (x-axis) and normalized EGFRvIII (y-axis; reference gene, GAPDH). I,EGFRvIII prevalence in cohort 3. ****, P ≤ 0.00001; **, P ≤ 0.0047; *, P ≤ 0.0493.
Data availability statement
The data presented in this study are available on request from the corresponding author. The raw ddPCR runs are not publicly available because raw PCR runs are not commonly shared, but they are available upon request.
Results
EGFRvIII mRNA is rich in GC regions and G repeats leading to secondary structures
Epidermal growth factor receptor variant III (EGFRvIII) is a result of an 801-bp deletion in the extracellular domain of the EGFR gene (exons 2–7). Custom primers and TaqMan probes were designed for targeted detection of a 96-bp amplicon in EGFRvIII and a 62-bp amplicon in EGFRwt mRNA sequence and used in all experiments (Fig. 1A). The composition of nitrogenous bases in the EGFRvIII amplicon was compared with the EGFRwt amplicon. To account for the difference in amplicon length, relative abundance of different bases was calculated as a percentage. Interestingly, the GC content of EGFRvIII amplicon was 1.2-fold higher compared with the counterpart EGFRwt (56% and 46% respectively, Fig. 1B). Further comparison of the GC content (%) distribution across the length of two target amplicons was performed by calculating average GC content of smaller segments (20-bp) in each RNA sequence. This revealed segments within the EGFRvIII amplicon with GC content as high as 80%. Furthermore, the majority sequence of EGFRvIII amplicon had a GC content in the range of 60% to 80%. Conversely, the RNA sequence of EGFRwt amplicon using the same analytic parameters had a lower GC content (40%–50%) with only one peak seen at 60% (Fig. 1C).
High GC content is one of the factors contributing to secondary structure formation. To further understand the structure, we used Mfold (21, 22), an RNA folding prediction tool to visualize potential secondary structures in the target amplicons. The software is based on minimum free energy (MFE) algorithms that consider the complex interactions between RNA secondary structures and free energy. We determined four distinct Minimum Free Energy (MFE) secondary structures in EGFRvIII amplicon with ΔG (Gibbs free energy) ranging from −32.9 kcal/mol to −37.7 kcal/mol, while the EGFRwt amplicon had only three foldings with a significantly lower ΔG between −4.3 kcal/mol and −7.2 kcal/mol, suggesting a higher thermostability of 3D secondary structures for EGFRvIII RNA (Supplementary Fig. S1A–S1C). Furthermore, the circle graphs for both EGFRvIII (Fig. 1D, left) and EGFRwt (Fig. 1D, right) indicate individual base pair interactions instrumental in stabilizing the predicted secondary structures. This is represented by an arc with each color indicating a distinct base pair (Watson Crick and non-Watson Crick) interaction; blue (A–U, A–T), red (G–C), and green (G–U, G–T). This phenomenon also contributes to the formation of predicted pseudoknots in EGFRvIII amplicon (Supplementary Fig. S1b). Overall, the predicted three-dimensional secondary structures are thermodynamically more stable in EGFRvIII(∆G: EGFRvIII −30.7 kcal/mol, EGFRwt −7.2 kcal/mol) compared with EGFRwt. To investigate this further, the RNA sequence unique to EGFRvIII (exon 1:exon 8, 467-bp) was also analyzed via Mfold using the same metrics. This analysis further revealed the presence of close to 90% of GC or G repeat region (Supplementary Fig. S2A and S2B) and at least 20 distinct secondary structures with a high ΔG (approximately, −200 kcal/mol), indicating high thermal stability of the predicted foldings (Supplementary Fig. S2B–S2D).
Based on these results, we examined the potential of using 7-deaza-2′-deoxyguanosine 5′-triphosphate (7dG) to overcome the negative impact on downstream analysis of this higher level organization. Reverse transcription and ddPCR amplification of EGFRvIII and EGFRwt target amplicons was performed with and without the addition of 7dG. We observed that addition of the additive resulted in a notable improvement in the amplification of EGFRvIII cDNA with higher number of EGFRvIII copies and no significant effect on the amplification of EGFRwt (Fig. 1E).
ddPCR assay design and optimization
One of the main barriers to successful clinical implementation of EV-based liquid biopsy for brain tumors is the significantly low concentration of mutant EV RNA isolated from circulating EVs in patient plasma combined with the high signal–noise ratio typical of a complex biofluid such as plasma. The goal was to maximize the sensitivity of the assay in patient plasma derived EV RNA and minimize nonspecific signal using an optimal combination of different measures. We first optimized the reverse transcription reaction by testing a series of concentrations of Oligo d(T)20 and random hexamers (Supplementary Fig. S3) with the final concentration set at 50 μmol/L each of both oligo d(T)20 and random hexamers.
This was followed by testing the effect of addition of increasing amounts (1 or 2 μL) of 7dG at the reverse transcription step. Total RNA extracted from EGFRvIII tumor tissue, was diluted to 1 ng and used as a starting input for three different reverse transcription conditions (two replicates per condition) in a reaction volume of 20 μL; standard (mock 7dG), standard + 1 μL 7dG, standard + 2 μL 7dG. The resulting cDNA from one replicate of each condition was subject to ethanol precipitation and resuspended in 20 μL of nuclease-free water prior to ddPCR amplification. The second replicate was run in parallel without ethanol precipitation. Across all reverse transcription conditions when cDNA was not further purified by ethanol precipitation, we observed an elevation of baseline in the positive well, located above the baseline of no template control (NTC; Fig. 2A–C, left). On the other hand, post ethanol precipitation, this inhibition was overcome leading to a significant improvement in the baseline correction and alignment to the reference NTC well (Fig. 2A–C, right). Again, this effect was observed across all three reverse transcription conditions. A similar trend was also seen in GAPDH, which was used as a reference gene in channel 2 (Fig. 2A–C). This, therefore, highlighted the role of potential ddPCR inhibitors, which can be removed using ethanol precipitation.
In addition, comparison of the ddPCR results across the three reverse transcription conditions continued to demonstrate the favorable effect of adding 7dG. Reverse transcription preparation with 1 μL 7dG resulted in the highest number of EGFRvIII copies with a statistically significant higher detection post-ethanol precipitation of 7dG-treated cDNA (Fig. 2D–F). Furthermore, we report a statistically significant increase in the number of mutant copies with the addition of 1 μl, as opposed to 2 μL, of 7dG at the reverse transcription step (Fig. 2G, H, J, and K). The significantly higher yield of EGFRvIII copies post-ethanol precipitation (Fig. 2E) pointed to the potential carryover and inhibitory effect of excess 7dG at the reverse transcription and PCR level. All ddPCR conditions were run in triplicates and the results of ANOVA across all conditions were statistically significant, at R2 value 0.98 (Fig. 2I). We also compared several clean up methods including ethanol precipitation and commercial kits and determined the highest recovery of EGFRvIII mRNA using ethanol precipitation at −20°C (Fig. 2L).
To achieve maximum cluster separation on ddPCR, a temperature gradient was conducted (53°C–60°C) to determine the optimal annealing temperature (Fig. 2M). Better cluster separation and tightly formed droplets were seen at lower as compared to higher temperature. The fluorescence of the mutant signal was observed to decrease significantly in amplitude after 57°C. Furthermore, at temperature <55°C, the clusters were more diffuse. To investigate this further, a second round of temperature gradient was conducted at 0.5°C increment within the optimal range (55°C–56.5°C) determined from the preliminary findings (Fig. 2N). Based on results of the second temperature gradient, 55.5°C was selected as the optimal annealing temperature, because it resulted in best cluster separation and maximum number of tightly formed mutant droplets (Fig. 2N).
Finally, we also explored the effect of employing different ddPCR additives: betaine (Fig. 3P) and ethylenediaminetetraacetic acid (EDTA; Supplementary Fig. S6F) at serial dilutions (0.25, 0.5, and 1.0 mmol/L). The analysis demonstrated a significant improvement in mutant cluster separation at 0.5 mmol/L concentration each of EDTA and betaine when compared with control (Supplementary Fig. S6F and Fig. 2P, respectively). To determine the quantitative effect of the additives on measured EGFRvIII copies, 0.5 mmol/L each of betaine and EDTA were run individually and in combination (Fig. 2O). At the chosen concentration of 0.5 mmol/L betaine alone, the measured EGFRvIII concentration was consistently higher than all other conditions (Fig. 2O). Moreover, we continued to see a statistically significant decrease in mutant signal with the addition of 0.5 mmol/L EDTA (both, individually and when used in combination with betaine). Furthermore, as previously observed, use of 0.5 mmol/L betaine concentration also improved the mutant cluster separation from baseline and cluster tightness as evidenced by increase in fluorescence amplitude on 1D and 2D ddPCR plots (Fig. 2P).
Optimized assay conditions were then evaluated using EGFRvIII synthetic RNA copies spiked into the reverse transcription reaction (see Materials and Methods). The resulting cDNA from each RNA concentration was amplified using ddPCR (top, Fig. 2Q) and quantitative PCR (qPCR; bottom, Fig. 2Q). Reverse transcription conversion is highly efficient and linear. For ddPCR, we report analytical parameters including limit of detection (LOD, 5.53 copies of EGFRvIII) and limit of blank (LOB, 1.18 copies of EGFRvIII; top, Fig. 2Q). For qPCR, we report the linearity of cycle threshold (Ct) across a range of EGFRvIII copies spike-in. Overall efficiency of qPCR is 92.2% (calculation described in Materials and Methods). EGFRvIII synthetic RNA was also reverse transcribed into cDNA with and without 7dG to determine the difference in detection potential by downstream ddPCR amplification. We show a significant difference in amplification efficiency when comparing 7dG with no 7dG (Supplementary Fig. S6C). The experiments with serial dilution of tumor tissue RNA reverse transcribed into cDNA followed by PCR amplification were also run in parallel (Supplementary Fig. S6A, S6B, and S6D). The final optimized assay was then ready to be applied to plasma derived EV RNA from patients with glioma.
Increased prevalence of EGFRvIII mRNA detected in glioma tumor tissue upon addition of 7dG
Having established the value of 7dG addition, we sought to determine the assay performance in RNA extracted from tumor tissue using quantitative PCR (qPCR). Two separate cohorts with tissue availability were analyzed to test the rigor and reproducibility of the assay. We first determined optimal qPCR conditions for a range of variables, including primer (Supplementary Fig. S4A and S4B) and probe concentrations (Supplementary Fig. S4C), choice of master mix, annealing temperature gradient (Supplementary Fig. S4D), and two-step or three-step cycling conditions (Supplementary Fig. S4E–S4H). The final protocol with the lowest Ct value was used for tumor tissue testing.
Two independent cohorts were compiled for testing. Cohort 2 included 20 patients with histopathologically confirmed GBM and EGFR-amplified status. Cohort 3 included 17 patients with histopathologically confirmed GBM and EGFR amplification confirmed by qPCR (Supplementary Table S2). No tumor tissue was available from cohort 1 and the EGFRvIII status was histopathologically confirmed (Supplementary Table S1). Total RNA input for reverse transcription of all samples was normalized to 1 ng, based on quantification data from Agilent Technologies 2100 Bioanalyzer. Extracted RNA from each sample was then reverse transcribed to cDNA using two different reverse transcription conditions: with and without 7dG (See Materials and Methods). The resulting cDNA from the two reverse transcription conditions was then amplified in two replicates per condition using the optimized qPCR protocol (Fig. 3A). Results were tabulated by comparing Ct values across both conditions (i.e., with and without 7dG; Fig. 3B, C, F, and G; Supplementary Table S3).
Across all 37 tumor tissue RNA samples (cohorts 2 and 3), 7dG-treated cDNA was consistently associated with significantly lower Ct values and hence higher EGFRvIII concentration (Fig. 3B and F respectively; Supplementary Table S3; average Ct; cohort 2 = 30.65 vs. 36.55, cohort 3 = 35.92 vs. 39.12). This difference was statistically significant (Fig. 3C and G; P < 0.0001). The data confirmed and validated our preliminary findings that the addition of 7dG to the reverse transcription significantly improved EGFRvIII detection. Using the assay without 7dG resulted in a statistically significantly lower EGFRvIII detection with only 7 of 20 (35%) patients testing positive in cohort 2 and 6 of 17 (35%) in cohort 3 (Fig. 3E and I). To confirm EGFR amplification status, we normalized EGFRwt using GAPDH and performed Pearson correlation test against EGFRvIII (also normalized to GAPDH) and confirmed a statistically significant positive correlation between EGFRvIII and EGFRwt in cohorts 2 and cohort 3 (Fig. 3D and H, respectively).
In summary, use of 7dG was associated with a significant improvement in tumor tissue assay sensitivity. This statistically significant finding was rigorous and reproducible in two independent tumor tissue cohorts.
Detection of EGFRvIII mutation in plasma-derived EV RNA from three independent cohorts
Finally, the optimized ddPCR workflow (Fig. 4A) was applied to the detection of the EGFRvIII mutation in EV RNA isolated from patient plasma (Supplementary Table S1). Baseline plasma collected prior to surgical resection was used for testing. For this purpose, three independent cohorts were analyzed (Table 1). Overall, our study population (n = 54) comprised of n = 30 histopathologically confirmed GBM (cohort 1, n = 7, cohort 2, n = 10, cohort 3, n = 13) patients, n = 10 EGFRwt controls (cohort 1 n = 1, cohort 2, n = 2, cohort 3, n = 7), and n = 14 age matched healthy controls (cohort 1, n = 3, cohort 2, n = 4, cohort 3, n = 7). In addition, four GBM patients with longitudinal plasma available were included in cohort 3 to evaluate the clinical utility of our assay in disease monitoring and surveillance. Plasma samples were analyzed and gated using predefined criteria (see Materials and Methods).
In cohort 1, we report positivity in 5 of 7 EGFRvIII-mutant samples and none of the 4 controls (EGFRwt, healthy controls), with a sensitivity of 71.4% and specificity of 100% (Fig. 4B and C). We then verified the assay performance in a second independent cohort (cohort 2) of 16 plasma samples (EGFRvIII mutant n = 10, EGFRwt n = 2, healthy controls n = 4; Fig. 4B and C). Here, we report positivity in 7 of 10 EGFRvIII-mutant samples and none of the 6 controls (EGFRwt, healthy controls), with a sensitivity of 70.0% and specificity of 100% (Fig. 4C).
Finally, the assay performance was validated in a third independent cohort, blinded to the operator (total n = 27, EGFRvIII mutant n = 13, EGFRwt n = 7, healthy controls n = 7; Fig. 4B). Here we report a positivity in 10 of 13 EGFRvIII-mutant samples and only 1 of the 14 controls (EGFRwt sample, false positive), with a sensitivity of 76.9% and specificity of 93% (Fig. 4C). Importantly, the sample reported as false positive had on average 1 mutant copy in the four replicates (only two out of four replicates had positive events) and the patient was confirmed to have an EGFR amplified glioblastoma. The blinded cohort using a large number of disease and non-disease controls, verifies and validates the clinically relevant sensitivity and specificity of our assay for the detection of EGFRvIII mutation in patient plasma derived EV RNA.
In summary, combining all three cohorts (n = 54), we present a ddPCR assay with overall sensitivity of 72.77% (95% CI, 63.71%–81.83%), specificity of 97.67% (CI, 87.63%–100%), positive predictive value (PPV) of 97.90%, and negative predictive value (NPV) of 70.04%. Overall, the plasma detection of EGFRvIII did not significantly correlate with different clinical parameters (Supplementary Fig. S5A and S5B). However, the copies/mL did show a tendency to correlation (P value not statistically significant) with tumor volume, age, and progression-free survival with higher copies likely to be detected in higher tumor volumes, older patients, and more aggressive disease.
Utility of EGFRvIII ddPCR assay in longitudinal monitoring using patient plasma
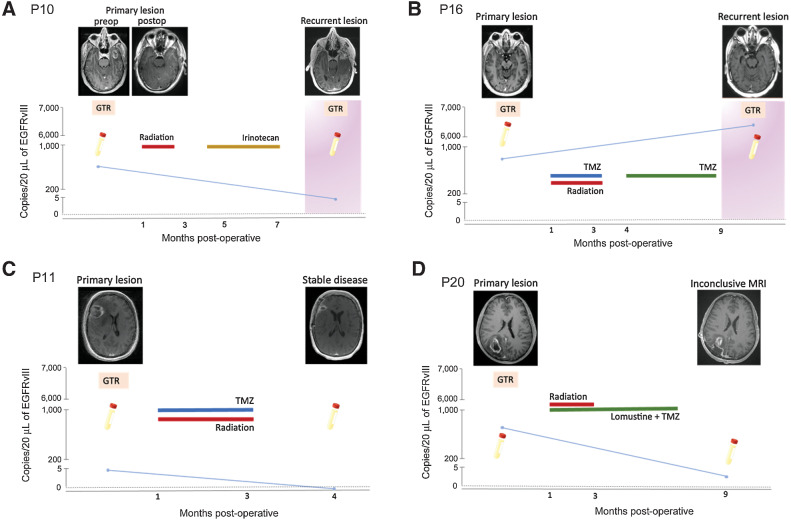
To assess the performance of EGFRvIII assay in a longitudinal setting, we isolated and analyzed EV RNA from serial plasma samples of four patients with glioma, tissue confirmed to harbor the EGFRvIII mutation (Fig. 5; Supplementary Fig. S5B, Supplementary Table S1). EGFRvIII-mutant copies correlated with the clinical and radiologic findings. Each of the patients had detectable EGFRvIII copies prior to the initial surgical resection. On follow-up, P11 had stable disease following chemoradiation with no evidence of progression on radiology. Neurologic examination in clinical settings was also unremarkable. This was supported by the ddPCR results with no detectable EGFRvIII-mutant copies in plasma samples collected at the same time (Fig. 5C). Patients P10 and P16 developed tumor recurrence with significant progression of contrast enhancement on surveillance MRI. Furthermore, there was clinical deterioration warranting a second surgery. In both patients, plasma collected prior to second surgery demonstrated EGFRvIII-mutant copies coincident with the radiologic and clinical findings (Fig. 5A and B, respectively). Finally, patient P20 is on tumor surveillance posttreatment with dual alkylating chemotherapy and radiation. The surveillance MRI demonstrates an irregular increase in contrast enhancement extending beyond the margins of the resection cavity. In other words, tumor progression cannot be ruled out with the possibility of viable tumor along the margins of the initial reaction cavity. As per clinical documentation the patient also reports occasional nonspecific neurologic signs. Plasma analysis of EV RNA corresponding to this time point via our ddPCR assay demonstrates detectable EGFRvIII-mutant copies (Fig. 5D). Given the radiologic findings, this patient will be followed to monitor clinical symptoms and progression versus stability of the MRI findings.
Figure 5.
Detection of EGFRvIII mutation using optimized plasma-based assay in patients with different clinical outcomes. A–D,EGFRvIII mutation (copies/20 μL) in serial plasma samples obtained from four glioma patients are plotted against time (months postoperatively). Patient cases represent different clinical outcomes: recurrent disease (A and B), stable disease (C), and inconclusive MRI (D), that is, inability to clearly delineate pseudo-progression from true tumor progression. T1-weighted, contrast-enhanced MRI images are provided for different clinical timepoints. Surgical procedures are indicated using an orange square (GTR = gross total resection). Disease progression (tumor recurrence) is indicated using a pink background. Treatment courses for each patient are outlined.
Discussion
This study demonstrates for the first time, the detection of EGFRvIII mutation in tumor tissue and glioma patients’ plasma samples using a novel optimized ddPCR assay. We report an overall sensitivity of 72.8% and a specificity of 97.7% for detecting EGFRvIII in plasma compared with tumor tissue analysis. We also report a higher prevalence of EGFRvIII in EGFR amplified tumor tissues (∼80%) as compared with what previously reported in the literature. Using this platform, we report the potential clinical utility of this technique in patients with glioma with several applications: diagnosis, monitoring tumor progression, and assessing response to therapy.
Development of a robust and sensitive ddPCR assay for detecting EGFRvIII mutation posed substantial technical and structural challenges. Secondary structures in the EGFRvIII RNA sequence have not been previously described in literature. There are two main types of algorithms commonly used for RNA secondary structure prediction: deterministic dynamic programming algorithm, and minimum free energy algorithm. The former is based mainly on the measurement of frequency of base pair interactions with the assumption that the formed base pairs are discontinuous, thereby having low accuracy (23). Based on the Nussinov algorithm, Zuker proposed a minimum free energy algorithm, a more robust prediction tool based on the principle of interaction of secondary structures with free energy and neighboring base pairs (21, 22). Hence, this was also used for predicting secondary structures in EGFRvIII RNA. Structural analysis of the predicted 3D foldings using crystallography and other sophisticated modalities is beyond the scope of this study. However, it is worth mentioning that thermodynamically stable secondary structures were also found to be highly prevalent close to the junction site (fusion of exon 1 and exon 8), a nucleotide sequence unique to EGFRvIII mutation. This presented a challenge in primer design and selection due to primer dimers, and Tm mismatch. This could potentially lead to non-specific products thereby limiting the specificity of the assay. In addition, secondary structures have a well-documented role in reducing the efficiency of polymerase activity and overall efficacy of PCR amplification. Hence, we sought to overcome these challenges through reverse transcription enhancement via 7dG and development of an amplification protocol that effectively utilized betaine as a potent reaction buffer. 7dG, a modified nucleotide, is capable of selectively blocking Hoogsten bond formation, without interfering with normal Watson–Crick base pair interactions (24). Role of betaine in improving amplification of GC rich regions in TERT promoter mutation has also been reported previously where it was used in conjunction with EDTA (25). However, while we did see an improvement in separation with EDTA, there was a simultaneous lower mutant fluorescent signal. Hence, we finally used betaine alone at optimized concentration of 0.5 mmol/L. Based on our findings of the prevalence of secondary structures in EGFRvIII, future studies are necessary to further elucidate EGFRvIII structural biology. Additional optimization measures included amplicon length and ddPCR optimization. Short amplicon length, optimized for fragmented plasma nucleic acid (26), combined with betaine, known to act as an iso-stabilizing agent, significantly improved the binding of oligonucleotides to the target region (27, 28).
Using the optimized assay, we were also able to detect EGFRvIII mutation with a higher sensitivity in tumor tissue. In this study, overall tumor tissue EGFRvIII prevalence measured in two cohorts was 81%. The estimated EGFRvIII tissue expression in existing literature is about 24% to 67% of GBMs (29). Our study population included patients with confirmed EGFR amplified status. As per literature, approximately 46% of GBM lesions are EGFR amplified, with EGFRvIII present in about 50% of such cases (30, 31, 32). However, our results showed a significantly higher prevalence of EGFRvIII in EGFR amplified tumor tissue. This not only highlights the sensitivity of our assay but also raises an important question on the true prevalence of EGFRvIII in GBM patients and our findings suggest that it is higher than previously thought. Because EGFRvIII expression is tumor cell specific, this makes it a potentially useful therapeutic target (33, 34, 35), which has led to a growing interest in developing anti-EGFR/EGFRvIII therapeutics including antibodies (cetuximab, panitumumab, and nimotuzumab), vaccines, chimeric antigen receptor (CAR) T cells, and RNA-based treatments (13). Hence, more accurate clinical classification of patients in clinical settings and their subsequent enrollment in relevant targeted clinical trials can potentially lead to better clinical outcomes and lower rate of treatment resistance, offering new opportunities in precision oncology.
We note that our sensitivity of 72.77% and a specificity of 97.67% in the plasma of 30 patients with EGFRvIII confirmed tumors, which approximates findings previously achieved by our group using CSF as a biofluid for EGFRvIII detection with clinically relevant sensitivity (10). However, lumbar puncture (LP), an invasive procedure is required to obtain CSF. In this regard, plasma is preferable with the ability to do serial sampling. This proposed method, once fully validated, has multiple clinical applications: (i) the potential of circumventing tissue biopsy in patients who are not favorable candidates for surgery or have a tumor in an inaccessible location; (ii) monitoring disease burden and treatment response; (iii) patient stratification and selection of potential candidates for clinical trials and/or targeted therapeutics; (iv) companion surveillance tool to aid in clinical decision making and differentiate pseudo-progression and/or treatment induced effects from true disease. While a focus of future research, we have observed examples in a small number of patients with longitudinal samples, where changes in EGFRvIII detection at different time points appeared concordant with different clinical outcomes: stable disease, tumor progression, and treatment effectiveness, including a patient where increased plasma EGFRvIII was detected at a follow-up time point observed where there is no clear radiologic delineation between radiation necrosis versus tumor progression. Another patient with effective treatment response and stable MRI findings showed no detectable EGFRvIII in follow-up plasma analysis. To further explore the question of longitudinal assay performance, larger scale future studies, should assess whether EGFRvIII-mutant allele frequency in plasma correlates with disease progression and treatment outcomes. Exploration of the correlation of EGFRvIII positivity in plasma with tumor progression may have important implications as previous studies targeting EGFRvIII via rindopepimut vaccine have reported the ability of tumor to escape through selection and preferential proliferation of cells that are EGFRvIII negative. It is also worth mentioning that one false positive sample detected in our blinded cohort has a recurrent contrast-enhancing lesion with confirmed EGFR amplification. Our tissue analysis did not reveal EGFRvIII mutation. However, in plasma, we detected 2 mutant events in one well and 1 mutant event in the second well (2/4 wells), which highlights the sensitivity of our assay, and potential tumor heterogeneity and the variable EGFRvIII expression in tumoral versus peritumoral areas in tissue (36). We hypothesize that in some cases liquid biopsy may provide a more sensitive sampling of the entire tumor compared with a spatially limited tissue biopsy.
In short, we present here a promising EGFRvIII assay for diagnosis and longitudinal monitoring and prognostication in glioma. The current assay presented in this work is a proof of concept and not ready for clinical diagnostics in current form. Future experiments aimed at testing the performance of our assay in larger patient cohorts as well as parallel comparisons with other gold standard assays in clinically defined patient populations will be instrumental in exploring this further and understanding the molecular expression pattern in glioma patient population. This work, along with previous work on development of a plasma-based assay for TERT promoter mutation detection, also serves to advocate and expand the potential of liquid biopsy. A combination of these multiple key oncogenic mutation assays in plasma as a multi-analyte modality may offer the potential to improve clinical decision making.
Supplementary Material
Acknowledgments
This work is supported by grants U01 CA230697 (B.S. Carter, L. Balaj), P01 CA069246 (B.S. Carter), and R01 CA239078, CA237500 (B.S. Carter, L. Balaj), Rappaport Foundation (L. Balaj). The funding sources had no role in the writing of the manuscript or decision to submit the manuscript for publication. The authors have not been paid to write this article by any entity. The corresponding author has full access and assumes final responsibility for the decision to submit for publication.
We would like to thank all MGH Neurosurgery clinicians and staff that assisted with the collection of samples. We are also deeply appreciative to the patients and their families for participating in the study.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
L. Balaj reports grants from the National Cancer Institute and the Rappaport Foundation during the conduct of the study. B.S. Carter reports personal fees from Koh Young outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
S.M. Batool: Data curation, formal analysis, validation, investigation, visualization, writing–original draft, writing–review and editing. K. Muralidharan: Conceptualization, formal analysis, investigation, writing–review and editing. T. Hsia: Formal analysis, methodology, writing–review and editing. S. Falotico: Investigation, writing–review and editing. A.S. Gamblin: Formal analysis, methodology, writing–review and editing. Y.B. Rosenfeld: Data curation, formal analysis, methodology. S.K. Khanna: Validation, investigation, writing–review and editing. L. Balaj: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. B.S. Carter: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Agnihotri S, Burrell KE, Wolf A, Jalali S, Hawkins C, Rutka JT, et al. Glioblastoma, a brief review of history, molecular genetics, animal models and novel therapeutic strategies. Arch Immunol Ther Exp 2013;61:25–41. [DOI] [PubMed] [Google Scholar]
- 2. Messali A, Villacorta R, Hay JW. A review of the economic burden of glioblastoma and the cost effectiveness of pharmacologic treatments. Pharmacoeconomics 2014;32:1201–12. [DOI] [PubMed] [Google Scholar]
- 3. Taylor OG, Brzozowski JS, Skelding KA. Glioblastoma multiforme: an overview of emerging therapeutic targets. Front Oncol 2019;9:963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol 2015;17Suppl 4;iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baid U, Rane SU, Talbar S, Gupta S, Thakur MH, Moiyadi A, et al. Overall survival prediction in glioblastoma with radiomic features using machine learning. Front Comput Neurosci 2020;14:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saenz-Antoñanzas A, Auzmendi-Iriarte J, Carrasco-Garcia E, Moreno-Cugnon L, Ruiz I, et al. Liquid biopsy in glioblastoma: opportunities, applications and challenges. Cancers 2019;11:950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Santiago-Dieppa DR, Steinberg J, Gonda D, Cheung VJ, Carter BS, Chen CC. Extracellular vesicles as a platform for ‘liquid biopsy’ in glioblastoma patients. Expert Rev Mol Diagn 2014;14:819–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garcia-Romero N, Carrion-Navarro J, Esteban-Rubio S, Lazaro-Ibanez E, Peris-Celda M, Alonso MM, et al. DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget 2017;8:1416–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Skog J, Wurdinger T, Van Rijn S, Meijer DH, Gainche L, Curry WT, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008;10:1470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Figueroa JM, Skog J, Akers J, Li H, Komotar R, Jensen R, et al. Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro Oncol 2017;19:1494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Manda SV, Kataria Y, Tatireddy BR, Ramakrishnan B, Ratnam BG, Lath R, et al. Exosomes as a biomarker platform for detecting epidermal growth factor receptor-positive high-grade gliomas. J Neurosurg 2018;128:1091–101. [DOI] [PubMed] [Google Scholar]
- 12. Oprita A, Baloi S-C, Staicu G-A, Alexandru O, Tache DE, Danoiu S, et al. Updated insights on EGFR signaling pathways in glioma. Int J Mol Sci 2021;22:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. An Z, Aksoy O, Zheng T, Fan Q-W, Weiss WA. Epidermal growth factor receptor and EGFRvIII in glioblastoma: signaling pathways and targeted therapies. Oncogene 2018;37:1561–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gan HK, Kaye AH, Luwor RB. The EGFRvIII variant in glioblastoma multiforme. J Clin Neurosci 2009;16:748–54. [DOI] [PubMed] [Google Scholar]
- 15. Kalman B, Szep E, Garzuly F, Post DE. Epidermal growth factor receptor as a therapeutic target in glioblastoma. Neuromolecular Med 2013;15:420–34. [DOI] [PubMed] [Google Scholar]
- 16. Varshney D, Spiegel J, Zyner K, Tannahill D, Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol 2020;21:459–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brierley I, Pennell S, Gilbert RJC. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Microbiol 2007;5:598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fan H, Wang J, Komiyama M, Liang X. Effects of secondary structures of DNA templates on the quantification of qPCR. J Biomol Struct Dyn 2019;37:2867–74. [DOI] [PubMed] [Google Scholar]
- 19. Lyubetsky VA, Rubanov LI, Seliverstov AV, Pirogov SA. Model of gene expression regulation in bacteria via formation of RNA secondary structures. Mol Biol 2006;40:440–53. [PubMed] [Google Scholar]
- 20. Muralidharan K, Yekula A, Small JL, Rosh ZS, Kang KM, Wang L, et al. Promoter mutation analysis for blood-based diagnosis and monitoring of gliomas. Clin Cancer Res 2021;27:169–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res 1981;9:133–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003;31:3406–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nussinov R, Pieczenik G, Griggs JR, Kleitman DJ. Algorithms for loop matchings. SIAM J Appl Math 1978;35:68–82. [Google Scholar]
- 24. Frey UH, Bachmann HS, Peters J¼, Siffert W. PCR-amplification of GC-rich regions: ‘slowdown PCR’. Nat Protoc 2008;3:1312–7. [DOI] [PubMed] [Google Scholar]
- 25. Corless BC, Chang GA, Cooper S, Syeda MM, Shao Y, Osman I, et al. Development of novel mutation-specific droplet digital PCR assays detecting TERT promoter mutations in tumor and plasma samples. J Mol Diagn 2019;21:274–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun K, Jiang P, Cheng SH, Cheng THT, Wong J, Wong VWS, et al. Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin. Genome Res 2019;29:418–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Musso M, Bocciardi R, Parodi S, Ravazzolo R, Ceccherini I. Betaine, dimethyl sulfoxide, and 7-deaza-dGTP, a powerful mixture for amplification of GC-rich DNA sequences. J Mol Diagn 2006;8:544–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rees WA, Yager TD, Korte J, Von Hippel PH. Betaine can eliminate the base pair composition dependence of DNA melting. Biochemistry 1993;32:137–44. [DOI] [PubMed] [Google Scholar]
- 29. Heimberger AB, Hlatky R, Suki D, Yang D, Weinberg J, Gilbert M, et al. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res 2005;11:1462–6. [DOI] [PubMed] [Google Scholar]
- 30. Felsberg JR, Hentschel B, Kaulich K, Gramatzki D, Zacher A, Malzkorn B, et al. Epidermal growth factor receptor variant III (EGFRvIII) positivity in -amplified glioblastomas: prognostic role and comparison between primary and recurrent tumors. Clin Cancer Res 2017;23:6846–55. [DOI] [PubMed] [Google Scholar]
- 31. Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A 1990;87:8602–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ekstrand AJ, Sugawa N, James CD, Collins VP. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc Natl Acad Sci 1992;89:4309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rutkowska A, Stoczyå„Ska-Fidelus E, Janik K, WÅ‚Odarczyk A, Rieske P. EGFRvIII: An oncogene with ambiguous role. J Oncol 2019;2019:1092587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci U S A 1992;89:2965–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Del Vecchio CA, Giacomini CP, Vogel H, Jensen KC, Florio T, Merlo A, et al. EGFRvIII gene rearrangement is an early event in glioblastoma tumorigenesis and expression defines a hierarchy modulated by epigenetic mechanisms. Oncogene 2013;32:2670–81. [DOI] [PubMed] [Google Scholar]
- 36. Nozawa T, Okada M, Natsumeda M, Eda T, Abe H, Tsukamoto Y, et al. EGFRvIII is expressed in cellular areas of tumor in a subset of glioblastoma. Neurol Med Chir 2019;59:89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The raw ddPCR runs are not publicly available because raw PCR runs are not commonly shared, but they are available upon request.