Abstract
Purpose:
The molecular complexity of acute myeloid leukemia (AML) presents a considerable challenge to implementation of clinical genetic testing for accurate risk stratification. Identification of better biomarkers therefore remains a high priority to enable improving established stratification and guiding risk-adapted therapy decisions.
Experimental Design:
We systematically integrated and analyzed the genome-wide CRISPR-Cas9 data from more than 1,000 in vitro and in vivo knockout screens to identify the AML-specific fitness genes. A prognostic fitness score was developed using the sparse regression analysis in a training cohort of 618 cases and validated in five publicly available independent cohorts (n = 1,570) and our RJAML cohort (n = 157) with matched RNA sequencing and targeted gene sequencing performed.
Results:
A total of 280 genes were identified as AML fitness genes and a 16-gene AML fitness (AFG16) score was further generated and displayed highly prognostic power in more than 2,300 patients with AML. The AFG16 score was able to distill downstream consequences of several genetic abnormalities and can substantially improve the European LeukemiaNet classification. The multi-omics data from the RJAML cohort further demonstrated its clinical applicability. Patients with high AFG16 scores had significantly poor response to induction chemotherapy. Ex vivo drug screening indicated that patients with high AFG16 scores were more sensitive to the cell-cycle inhibitors flavopiridol and SNS-032, and exhibited strongly activated cell-cycle signaling.
Conclusions:
Our findings demonstrated the utility of the AFG16 score as a powerful tool for better risk stratification and selecting patients most likely to benefit from chemotherapy and alternative experimental therapies.
Translational Relevance.
Distinguishing AML's fitness genes from the constitutive genetic dependencies shared across all tumors holds the key to unlocking cancer-specific therapeutic vulnerabilities. Here, we systematically identified a set of fitness genes using large-scale in vitro and in vivo CRISPR-Cas9 screens, which serve as AML-enriched dependency genes. We extracted the core molecular effectors of fitness and developed a prognostic AFG16 score. The AFG16 score was validated in six independent cohorts across different analysis platforms, proving clinical applicability. The associations of the AFG16 score and genetic abnormalities were also revealed and corroborated by our muti-omics data from the RJAML cohort. The AFG16 score was able to substantially improve the European LeukemiaNet classification. The predictive power of the AFG16 score to chemotherapy and targeted agents further demonstrated a great potential for its clinical applications. Its direct relation to AML fitness rendered them as potential targets that deserved further exploration to define their therapeutic implications.
Introduction
Acute myeloid leukemia (AML) is a group of aggressive hematologic malignancies with highly genetical and clinical heterogeneity, characterized by the accumulation of immature myeloid precursors in the bone marrow (BM) and other tissues (1, 2). Despite improvements in the treatment of AML, most patients would succumb to this disease. The global 5-year survival rate remains significantly lower at approximately 30% resulting from common chemo-refractory disease and high rate of relapse (3). Discovery of genes and biological processes that participate in the pathogenesis and contribute to the therapeutic resistance is realistically needed to improve clinical outcomes. Importantly, therapeutic advances may also arise from better risk stratification for patients with AML.
Currently, risk stratification for AML relies on genetic testing for mutations and cytogenetic alterations. Both the World Health Organization (WHO; ref. 4) and European LeukemiaNet (ELN; ref. 5) provided recommendations to stratify AML into three risk categories: favorable, intermediate, and adverse. However, genomic data and clinical screening are increasingly being inadequate for risk stratification (2), highlighting the need for improved risk stratification of patients to guide risk-adapted therapies. Accordingly, transcriptome-based models using either gene expression (6–8) or alternative mRNA splicing (9, 10) that represent key biological measurements with the potential to contribute to disease progression have been extensively proposed. Especially, the core transcriptional components of leukemic stem cells (LSC; refs. 11, 12) have garnered enormous interest, partly because these self-renewing LSCs are considered as critical players in AML initiation, resistance to chemotherapy as well as relapse (13, 14), and therefore hold great potential for therapy. However, more recent studies have revealed that persistence of LSCs could not fully account for chemoresistance and relapse in AML and leukemic-regenerating cells arise post chemotherapy presented distinct molecular features from therapy-naive LSCs (15, 16). Indeed, increased proliferation and inhibition of differentiation are hallmarks of AML (2, 17). Therefore, the fitness genes whose perturbations lead to proliferation defects of cells (18) lie in the center of transcriptomic programs with potential prognostic value.
The CRISPR-Cas9 genome editing system has been extensively utilized for high-throughput loss-of-function screens to identify essential genes required for the growth and proliferation of human cells in specific genetic context (18, 19). The power of the CRISPR screen has also been demonstrated by researches investigating synthetic lethality (19) and seeking for genes involved in cancer metastasis (20), drug resistance (21) as well as immune response (22). With the rapid accumulation of genome-wide screening data in a broad spectrum of human cancer cell lines, it has been possible to systematically identify cancer-specific fitness genes to fill the goal of precision medicine (23, 24). Distinguishing a cancer's fitness genes, and delineating them from the constitutive genetic dependencies shared across all tumors, holds the key to unlocking cancer-specific therapeutic vulnerabilities (18, 23).
In this study, we comprehensively analyzed the genome-wide CRISPR-Cas9 data from over one thousand in vitro and in vivo screens and systematically identified the AML-specific fitness genes, inactivation of which impairs survival and proliferation of AML cells. We further developed and validated a novel 16-gene fitness score that was highly prognostic in over 2,300 patients with AML and could complement the current ELN2017 scheme. We also investigated the mutational landscape and biological features associated with this fitness score, demonstrating the power of the score to predict therapeutic responses to both chemotherapy and targeted agents.
Materials and Methods
Patient cohorts
We collected BM specimens from a total of 157 patients with AML of the RJAML cohort diagnosed at Ruijin Hospital affiliated to Shanghai Jiao Tong University School of Medicine between June 2019 and September 2020. All samples were collected at diagnosis and patients received the similar intensive induction regimens according to national guidelines. Patients whose BM specimens were of poor quality were excluded. The collection of the specimens was approved by the Institutional Review Boards from Ruijin Hospital, and the written informed consent for specimen collection and research was obtained following the Declaration of Helsinki.
The HOVON cohort (n = 618; ref. 25) was retrieved from Array Express (Dataset ID: E-MTAB-3444) and served as the training set. The gene expression profiles and survival information of the GSE37642 (n = 553; ref. 26), GSE106291 (n = 250; ref. 27), GSE12417 (n = 240; ref. 28), and GSE71014 (n = 104; ref. 29) cohorts were obtained from Gene Expression Omnibus (GEO). The raw RNA-sequencing (RNA-seq) data along with clinical information and processed mutational variants of the TCGA-LAML (n = 179; ref. 1) and BeatAML (n = 244; ref. 30) cohorts were obtained from the GDC data portal. Only treatment-naïve samples from adult patients with AML were retained in the BeatAML cohort and the patients without survival information in these cohorts were excluded.
RNA-seq and targeted gene sequencing
The genomic DNA/mRNA extraction, library preparation, and analysis procedures for RNA-seq, targeted gene sequencing on 100 commonly mutated genes (Supplementary Table S1), and microarray data are listed in the Supplementary Materials and Methods.
Generation of AML-specific dependency library
AML-enriched dependencies were first identified by analyzing the large-scale of in vitro CRISPR-Cas9 screen dataset consisting of 26 AML cell lines and 1028 non-AML cancer cell lines from the Dependency Map (DepMap) portal. Genes with lower gene effect scores (indicating greater dependency) in AML cell lines (FDR < 0.2) compared with other cancer cell lines were determined as candidate AML dependencies. These candidates were further intersected to the AML-enriched dependencies in the in vitro CRISPR-Cas9 screen dataset including 14 AML cell lines (19), which were identified as the averaged CRISPR score < −1 across these cell lines. Notably, sgRNA raw counts for only 12 of these 14 human AML cell lines were available and used for visualization (Supplementary Table S2). Finally, the gene dependencies from the above two-way overlap were also filtered to include candidate dependencies (fold change of initial/final sgRNA abundance > 2.5) in the genome-wide in vivo CRISPR screen (31). AML-specific gene dependencies were independently validated by the DEMETER2 RNAi screen datasets with an FDR < 0.2 using the same two-class comparison (23 AML and 689 non-AML cell lines) (32). The in vitro and in vivo depletion scores from a focused CRISPR screen in U937, MV4-11 cell lines and a PDX model were obtained from the publication (33) for validation, which performed in vitro and in vivo CRISPR screens in parallel. Full details are provided as Supplementary Materials and Methods.
Data availability statement
The data generated in this study are publicly available in GEO at GSE201492. All other data generated in this study are available within the article and its Supplementary Data files.
Results
In vitro and in vivo CRISPR screens identify a 16-gene AML fitness (AFG16) score associated with the patient survival
To identify the specific dependency genes of AML growth, we extensively analyzed the in vitro CRISPR-Cas9 screen dataset and determined the genes that AML cells are specifically required for survival by comparing data from 26 AML cell lines with 1,028 cell lines of other cancer types (Fig. 1A). Approximately 2,000 genes were identified as AML-enriched dependencies including critical signal transducers, transcriptional factors, and cofactors that have been revealed to participate in the pathogenesis of AML (Supplementary Fig. S1A). These genes were intersected with additional AML genetic screen datasets, including in vitro CRISPR screens in 14 AML cell lines (19) and an in vivo screen in a BCR-ABL/NUP98-HOXA9–driven model of myeloid leukemia (Fig. 1A; ref. 31). We defined these 280 genes in the three-way overlap as AML fitness genes (AFG; Supplementary Fig. S1B; Supplementary Table S3). In this study, we define an AFG as any gene whose knockout decreases AML cell growth and proliferation in vitro and in vivo as previously reported (18). These fitness genes are also deemed as essential genes that are required for the survival of AML cells, consistent with prior work (19). To independently validate these 280 AFGs, we also analyzed the differential essentiality profile between 23 AML cells and 689 non-AML cell lines based on the RNA interference (RNAi) in vitro screens. This analysis identified 3,070 AFGs, which were significantly overlapped with these 280 AFGs identified using in vitro and in vivo CRISPR screen datasets (Supplementary Fig. S1C and S1D).
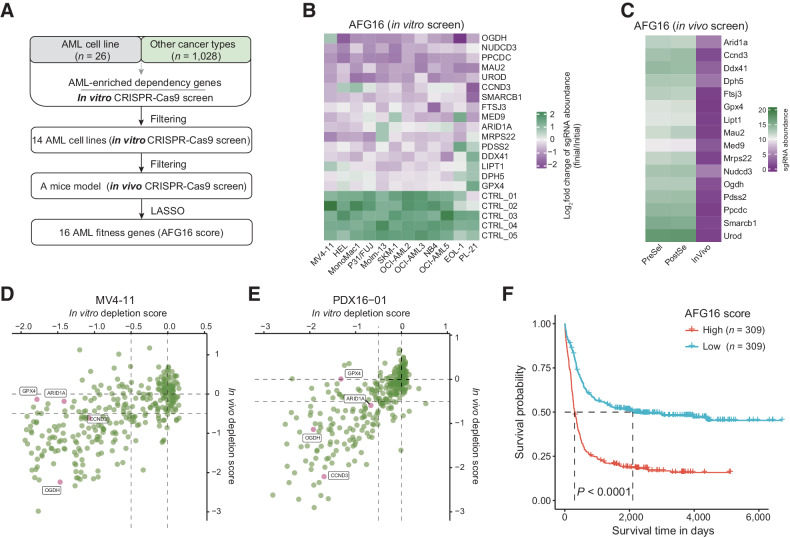
Figure 1.
Analysis of genome-wide CRISPR-Cas9 knockout screens identify an optimal 16-gene prognostic signature. A, Flowchart for the identification of AML-specific fitness genes and development of the 16-gene AML fitness (AFG16) signature. B, Heatmap showing the fold change of normalized read counts of sgRNAs targeting AFG16 signature genes across 12 AML cells transfected with the CRISPR-Cas9/sgRNA library before and after population doublings. Of note, sgRNA raw counts for only 12 of 14 human AML cell lines were available and used for visualization. C, Heatmap showing impact on the AFG16 signature genes PreSel, PostSel and in vivo. PreSel, PostSel, and InVivo represented preselection, postselection, and in vivo samples, respectively, as previously defined in the in vivo screen strategy (31). D and E, Scatter plots showing the in vitro and in vivo depletion scores of four out of the 16 signature genes GPX4, ARID1A, CCND3, and OGDH at a gene level in two AML models. Data points representing the median value of all targeting sgRNAs on each gene. F, Kaplan–Meier estimates of overall survival according to the AFG16 score in the HOVON cohort.
To extract the core molecular effectors of fitness that relate to patient outcomes across a wide spectrum of AML cytogenetic subtypes, we interrogated the HOVON dataset (25), in which 230 of the 280 AFGs were captured. We applied statistical regression algorithms based on the univariate CPH regression and the LASSO to relate AFGs to patient survival in this discovery cohort. This yielded an optimal signature comprising 16 genes (AFG16 score), which were significantly depleted in vitro (Fig. 1B) and in vivo (Fig. 1C) screens. Four of the 16 genes were also targeted by a focused CRISPR screen (33), which further confirmed the strong depletion of these fitness genes, especially in the in vitro and in vivo screening performed in the MV4-11 cell line and a PDX model (Fig. 1D and E) and only showed depletion in the in vitro screening from the U937 cell line (Supplementary Fig. S1E).
We further elaborated an AFG16 score for each patient described as the weighted sum of expression of the 16 genes to dichotomize patients with AML (Supplementary Table S4). Kaplan–Meier analysis demonstrated that high AFG16 risk score was significantly associated with inferior overall survival (OS; Fig. 1F). In addition, patients with high AFG16 scores were older and had higher incidences of unfavorable cytogenetics and the FLT3 internal tandem duplication (FLT3-ITD) mutation, as well as lower complete remission (CR) rates to standard induction chemotherapy, reflecting a potential link between AML fitness-associated gene expression programs and clinical outcomes (Supplementary Table S5).
The AFG16 score demonstrates robustness across technology platforms and patient subtypes
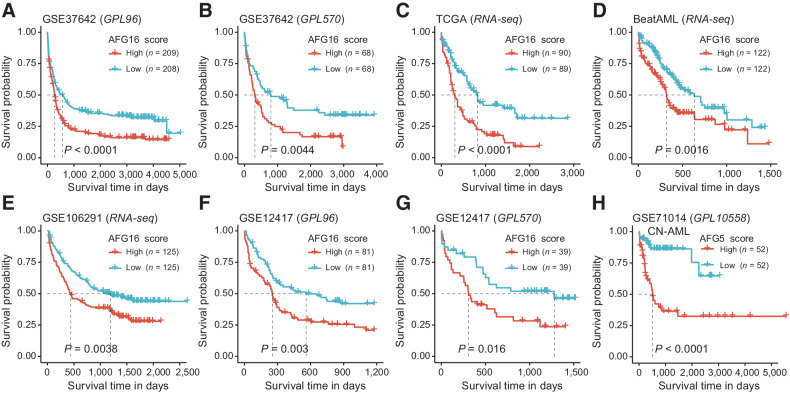
We next comprehensively evaluated the association of the AFG16 score with patient survival in five independent AML cohorts from three microarray platforms and the Illumina RNA-seq platform, consisting of 1,570 patients. In the GSE37642 cohort (n = 553), patients with higher AFG16 scores had significantly shorter OS than patients with lower scores (Fig. 2A and B). When applied to RNA-seq datasets for the TCGA (n = 179), BeatAML (n = 244), and GSE106291 (n = 250) cohorts, the AFG16 signature remained highly associated with clinical outcome, demonstrating superior robustness across technology platforms (Fig. 2C–E). These survival differences were also observed for the subsets of cytogenetically normal AML (CN-AML; Supplementary Fig. S2A–S2C) as well as in the independent GSE12417 cohort (n = 240) of patients with CN-AML (Fig. 2F and G). In multivariate survival analysis using CPH models, the AFG16 score retained independent prognostic value in training and validation cohorts independent of known markers of outcome including patient age, cytogenetic risk group, type of AML (de novo vs. secondary), presenting white blood cell (WBC) count, and the presence of NPM1 and FLT3-ITD mutations (Supplementary Table S6).
Figure 2.
The AFG16 score is robustly associated with OS in multiple independent AML cohorts across distinct analysis platforms and in patient subsets with diverse genetic lesions. Kaplan–Meier estimates of OS according to the AFG16 score in the GSE37642 cohort quantified on GPL96 (A) and GPL570 (B) microarray platforms as well as in the RNA-seq–based cohorts TCGA (C), BeatAML (D), and GSE106291 (E). Kaplan–Meier estimates of OS according to the AFG16 score in the cytogenetically normal AML cohorts quantified on GPL96 (F) and GPL570 (G) platforms. H, Kaplan–Meier estimates of OS according to the AFG5 score in the GSE71014 cohort quantified on GPL10558 microarray platform. Red and blue lines represent OS of patients with prognostic scores above and below the median cutoff, respectively.
The AFG16 signature was initially trained using the HOVON dataset, which included only a small subset (n = 89/618, 14.4%) of patients with CN-AML; therefore, survival differences within this subset might not have been optimally captured by the LASSO algorithm applied to the full cohort. We therefore retrained the 16 AFGs against OS for only the patients with CN-AML in the HOVON cohort, and derived a reweighted, optimized sub-signature in which only five of the 16 fitness genes contributed to the gene sum-value signature (AFG5). A high AFG5 score could identify patients with poor prognosis in the independent GSE71014 (n = 104) cohort of CN-AML cases (Fig. 2H). These results demonstrated the feasibility of optimizing the AFG16 score for selected patient subsets.
AFG16 score outperforms other gene expression-based signatures
We next compared the AFG16 score with the powerful prognostic signatures derived from gene expression analysis of stem cell subsets defined phenotypically (LSC17; ref. 11) or by multiple pairwise comparisons between AML subgroups and healthy controls (CODEG22; ref. 7), or generated by capturing intratumor heterogeneity of AML (GENE4; ref. 8). Noteworthy, the UROD gene in our AFG16 signature was also included in the CODEG22, which has recently been reported to be upregulated in AML and predicts poor prognosis (34). The HOVON, TCGA, and BeatAML cohorts with broad range of clinical annotations were used for comparisons. LSC17, CODEG22, and GENE4 scores were prognostic in some of these three cohorts when adjusting for common clinical covariates (Supplementary Tables S7–S9), whereas the AFG16 score displayed significantly prognostic relevance across all these three cohorts, indicating a broader applicability (Supplementary Table S6). The AFG16 score also showed a slightly higher Harrell C-index across these three cohorts, despite a lower value than the CODEG22 score in the TCGA cohort (Supplementary Fig. S3). Furthermore, when the AFG16 score and other three signatures were introduced in one multivariate model, only the AFG16 score remained prognostic value in all three cohorts (Supplementary Table S10). Overall, these results demonstrated that the AFG16 score was independent of other published gene expression-based signatures and could be a robust prognostic predictor of AML survival.
The AFG16 score is able to distill downstream consequences of specific genetic abnormalities and can complement the ELN2017 classification scheme
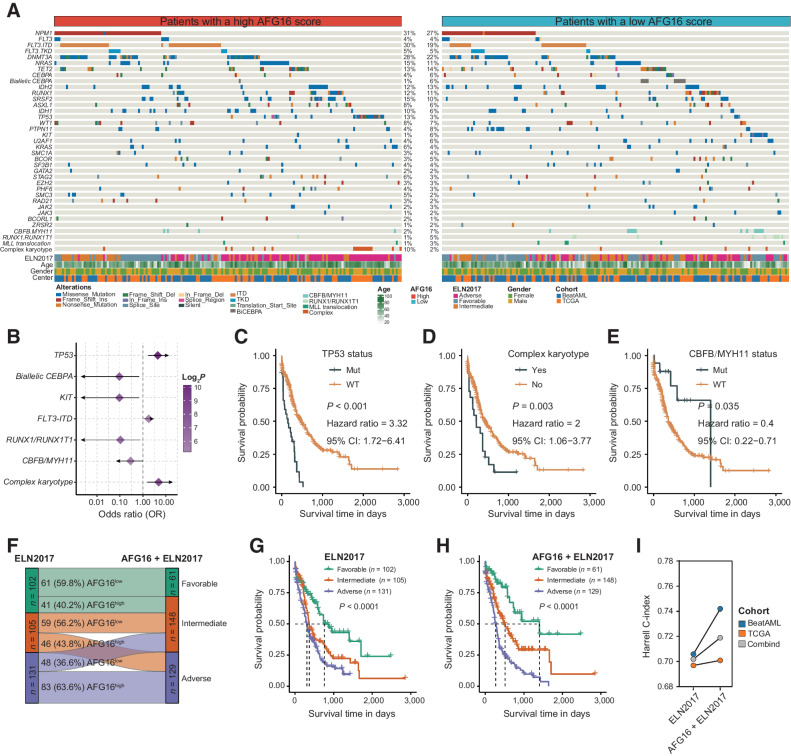
Recurrent mutations and cytogenetics in AML carry independent prognostic information, which are critical prognostic markers intrinsic to the ELN scheme (5). To obtain more comprehensive insights into the mutational landscape associated with the AFG16 score, we analyzed recurrently mutated somatic driver genes in the TCGA and BeatAML cohorts (Fig. 3A). Notably, a high AFG16 score was significantly associated with poor OS in this combined dataset (Supplementary Fig. S4A). Moreover, seven molecular markers were occurred at significantly different frequencies between patients with AFG16high and AFG16low scores (Fig. 3B). TP53, FLT3-ITD mutations, and complex karyotype were more frequent in the AFG16high group of patients. In contrast, patients with a AFG16low score more frequently harbored mutations in biallelic CEBPA and KIT genes and core-binding factor (CBF) fusions, RUNX1/RUNX1T1 and CBFB/MYH11, which were consistent with the findings from the HOVON cohort (Supplementary Table S5). TP53 mutation and complex karyotype were significantly associated with poor prognosis while the CBFB/MYH11 fusion was suggestive of relatively better clinical outcome as single variate (Fig. 3C–E). However, in a multivariate CPH regression model that included all seven of these genetic abnormalities and well-known clinical parameters as well as the AFG16 score, only TP53 retained prognostic significance, whereas the AFG16 score remained a significant independent prognostic factor (Supplementary Table S11).
Figure 3.
Differences in the incidences of genomic abnormalities between AFG16high and AFG16low patients. A, Heatmap showing somatic mutations and clinical information between AFG16high and AFG16low patient groups. B, Forest plot showing the mutations and cytogenetic abnormalities that differ in frequencies between AFG16high and AFG16low groups. Kaplan–Meier estimates of OS according to the status of TP53 mutation (C), complex karyotype (D), and CBFB/MYH11 fusion (E). F, Reassignment of patient risks from the three ELN2017 schema (favorable, intermediate, and adverse risk) to the ELN plus AFG16 categories. ELN2017 favorable/AFG16high and ELN2017 adverse/AFG16low patients were reclassified to the intermediate risk, and ELN2017 intermediate/AFG16high patients were reassigned to the adverse risk. Kaplan–Meier estimates of OS according to the risk categories of patients with AML in the ELN2017 (G) and AFG16 plus ELN2017 (H) schema. I, Harrell C-index of risk classification by ELN2017 and AFG16 plus ELN2017 in the TCGA, BeatAML, and combined dataset.
We next evaluated the prognostic value of the AFG16 score in the context of the state-of-the-art ELN2017 classification. A multivariable CPH regression analysis revealed that the AFG16 score was independent of the patient age, gender, WBC count, and ELN2017 classification (Supplementary Table S12). We also found significant differences in the ELN2017 risk-group distribution between patients with AFG16high and AFG16low scores. Nearly half of patients with a AFG16high score were determined as having adverse risk, whereas 24% and 27% were classified as having favorable and intermediate risk, respectively. In the patients with AFG16low scores, 36%, 35%, and 29% were classified in the favorable-, intermediate-, and adverse-risk groups, respectively (Supplementary Fig. S4B). The AFG16 score was also able to strongly discriminate patients with longer and shorter OS in the three ELN2017 subgroups (Supplementary Fig. S4C–S4E). By incorporating the AFG16 signature into the ELN2017 scheme, three new risk groups were generated as follows: patients with ELN2017-favorable/AFG16low were reclassified as the favorable-risk group, whereas ELN2017-favorable/AFG16high, ELN2017-intermediate/AFG16low and ELN2017-adverse/AFG16low patients were reclassified as the intermediate-risk group, and ELN2017-intermediate/AFG16high patients were reassigned to the adverse-risk group (Fig. 3F). The refined risk scheme contributed to an improved risk classification of patients with AML (Fig. 3G and H), evidenced by a higher Harrell C-index in the combined cohort or in the two cohorts, respectively (Fig. 3I). These results demonstrated that the AFG16 score could provide additional prognostic power to the ELN2017 classification scheme.
The AFG16 score is a clinically applicable gene expression–based signature
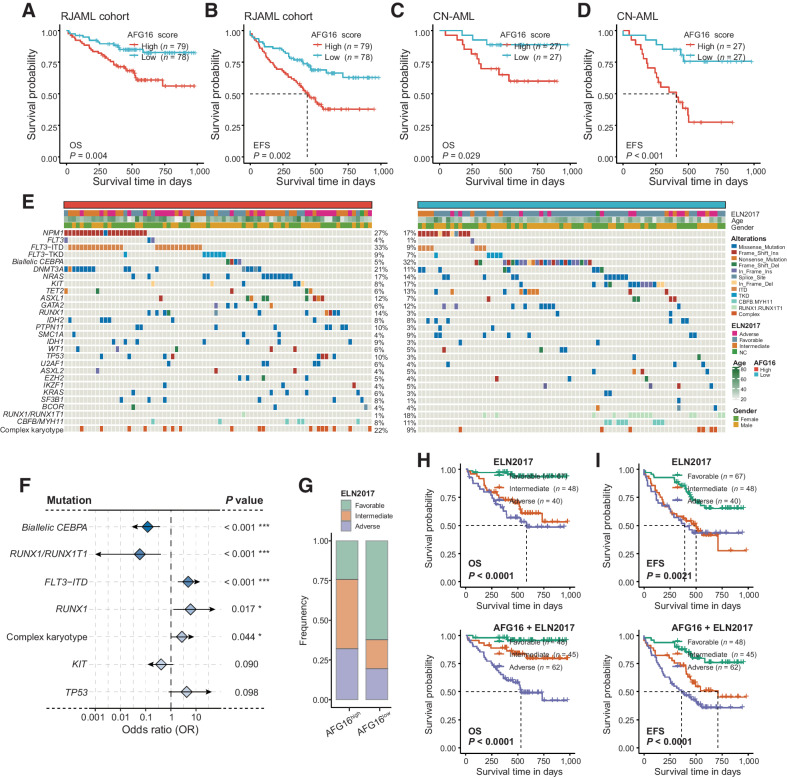
With the rapid evolution of high-throughput next-generation sequencing (NGS) technologies, RNA-seq, and targeted gene sequencing hold expanded promise for improving diagnosis, prognosis, and treatment of human cancer (3, 35, 36). We sought to validate the AFG16 score retrospectively by using RNA-seq and high-depth targeted gene sequencing on a de novo AML cohort from the Ruijin Hospital, including a total of 157 patients (Supplementary Table S13). Consistent with our findings based on public transcriptomics datasets, patients with AFG16high scores had significantly inferior OS than patients with AFG16low scores (Fig. 4A). Similarly, a high AFG16 score was strongly associated with shorter EFS (Fig. 4B). In a multivariate OS model or EFS model including established clinical factors, the AFG16 score retained independent prognostic value (Supplementary Table S14). The association between a high AFG16 score and shorter OS and EFS was also significant in the subset of patients with CN-AML (Fig. 4C and D). We also compared the mutational landscape of AFG16high and AFG16low patients in the RJAML cohort (Fig. 4E). In addition to FLT3-ITD mutation and complex karyotype, a high AFG16 score was also associated with higher incidence of RUNX1 mutation. Biallelic CEBPA mutation and RUNX1/RUNX1T1 fusion were more frequent in the AFG16low group (Fig. 4F). Despite no statistical significance, the TP53 and KIT mutations exhibited expected mutational trends in frequency between these two patient groups. A high AFG16 score was associated with unfavorable ELN2017 risk (Fig. 4G), and predicted relatively poorer survival in the ELN2017 subgroups, respectively. Despite no statistical significance in some of the subgroups considering OS or EFS, possibly due to the small sample size, a clear separation of survival curves between the AFG16high and AFG16low patients was observed (Supplementary Fig. S5A and S5B). More importantly, addition of the AFG16 signature into the ELN2017 scheme significantly improved the risk classification of patients with AML (Fig. 4H and I; Supplementary Fig. S5C and S5D). Overall, these results demonstrated the strong prognostic significance and broad applicability of the AFG16 score on the clinically serviceable NGS platform.
Figure 4.
Validation of the prognostic value of the AFG16 score and its associated mutational landscape in the RJAML cohort. Kaplan–Meier estimates of OS (A) and EFS (B) according to the AFG16 score in the full RJAML cohort and in the subset of patients with cytogenetically normal AML (C and D). E, Heatmap showing somatic mutations and clinical information between AFG16high and AFG16low patient groups in the RJAML cohort. F, Forest plot showing the mutations and cytogenetic abnormalities that differs in frequencies between AFG16high and AFG16low groups. G, Bar plot showing the distribution of ELN2017 risks in AFG16high and AFG16low patients with AML. Kaplan–Meier estimates of OS (H) and EFS (I) according to the risk categories of patients with AML in the ELN2017 and AFG16 plus ELN2017 schema.
AFG16 score improves the prediction of chemotherapy resistance in AML
We next evaluated the ability of the AFG16 score to predict therapeutic resistance, which were defined as failure to achieve morphological CR after initial induction chemotherapy as previously reported (11). In the HOVON and RJAML cohorts, the AFG16 score and cytogenetic risk had comparable predictive ability for chemotherapy resistance in single-variable models [Supplementary Fig. S6A and S6B; HOVON cohort: area under the receiver operating characteristic curve (AUROC) = 0.695 vs. 0.657, P = 0.194; RJAML cohort: AUROC = 0.692 vs. 0.635, P = 0.137]. In multivariate logistic regression model that included common clinical parameters (age, NPM1 and FLT3-ITD mutations or WBC count), further adding the AFG16 score into the model had better predictive power for chemotherapy resistance than that including the cytogenetic risk in the HOVON cohort (Supplementary Fig. S6C, AUROC = 0.737 vs. 0.690, P = 0.011), whereas the finding did not reach statistical significance in the RJAML cohort (Supplementary Fig. S6D, AUROC = 0.774 vs. 0.729, P = 0.166). Multivariate logistic regression models that included common clinical parameters and cytogenetic risk, inclusion of the AFG16 score significantly improved the predictive power (Fig. 5A and B; HOVON cohort: AUROC = 0.738 vs. 0.690, P = 0.007; RJAML cohort: AUROC = 0.771 vs. 0.701, P = 0.038). The AFG16 score ranked the most significant covariate in multivariate logistic regression models as measured by the Wald χ2 statistic in both cohorts (Fig. 5C and D). Together, these results indicated that the AFG16 score can assist the prediction of chemotherapy resistance in patients with AML.
Figure 5.
The AFG16 score predicts chemotherapy response. ROC curves for prediction of initial chemotherapy resistance utilizing multivariate logistic regression models that include patient age, cytogenetic risk as well as NPM1 and FLT3-ITD mutations or WBC count as covariates, with or without AFG16 score in the HOVON cohort (A) and the RJAML cohort (B). P values are generated by DeLong's test. Wald χ2 statistic of each covariate for prediction of chemotherapy resistance in the multivariate regression models that include the AFG16 score in the HOVON cohort (C) and RJAML cohort (D). The size of dot represents −log2P value.
The AFG16 score captures specific transcriptomic properties and confers promising therapeutic strategies to AML
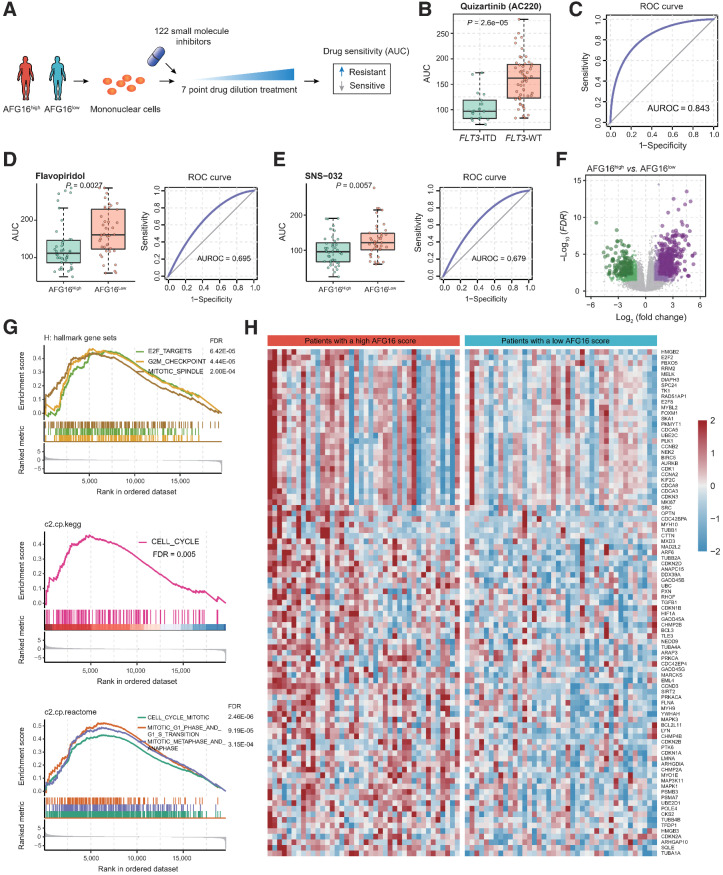
Given the significant associations between a high AFG16 score and lower CR rate to induction chemotherapy as well as inferior patient outcome, we further investigated whether the patients with AFG16high score could benefit from alternative therapeutic strategies in addition to chemotherapy. We then analyzed the ex vivo drug sensitivity profile of patients with AML to a panel of small-molecule inhibitors in the BeatAML cohort (Fig. 6A). To test the reliability of this analysis, we used the agent quizartinib as a positive control, which is a clinically effective drug known to markedly improve the prognosis of relapsed/refractory AML patients with FLT3-ITD mutation (37). Consistent with previous study, we found that mononuclear cells from FLT3-ITD–mutated AML patients were more sensitive to the quizartinib-mediated killing than those from patients without FLT3-ITD mutation, evidenced by relatively lower AUC values (Fig. 6B). The receiver operating characteristic (ROC) curve also suggested that patients with FLT3-ITD mutation will have lower drug sensitivity values than patients without FLT3 mutations (Fig. 6C). We next compared the drug sensitivity profile of the upper quartile of patients with the highest AFG16 scores to those of the bottom quartile of patients with the lowest AFG16 scores. This identified two drugs, flavopiridol and SNS-032, both of which significantly affected cell viability of the AFG16high patient group, showing with lower AUC values than that of the AFG16low patient group (Fig. 6D and E; Supplementary Table S15).
Figure 6.
Ex vivo drug sensitivity landscape of AFG16high and AFG16low AML. A, Schematic diagram for analyzing ex vivo drug sensitivity data in AML. The freshly isolated mononuclear cells from patients were exposed to 122 small-molecule inhibitors with 7-point drug dilution treatment, respectively. The drug sensitivity of these patient-derived cells was further determined. B, Boxplot showing treatment response between samples of FLT3-ITD and FLT3 wild-type (WT) AML patients against quizartinib (AC220). C, The ROC curve indicates the significance of the quizartinib effect on cells isolated from FLT3-ITD and FLT3-WT patient samples. D and E, Boxplot and ROC curve showing treatment response between samples of AFG16high and AFG16low patients against flavopiridol (D) and SNS-032 (E). F, Volcano plot of differentially expressed genes between the AFG16high and AFG16low patients. Significant genes were determined using the threshold of |log2(fold change) | ≥ 1 and FDR < 0.05. G, GSEA showing upregulation of cell-cycle signaling in AFG16high patients. H, Heatmap showing the expression level of the cell-cycle signaling genes with core enrichment from the GSEA.
Interestingly, flavopiridol and SNS-032 are both novel potent cell-cycle inhibitors of CDK family members, and showed encouraging clinical activities in AML (38, 39). We reasoned that a high AFG16 score probably reflected biological properties of AML blasts that confer higher sensitivity to these two targeted therapeutic agents. To find evidence in support of this, we carried out differential gene expression analysis between the AFG16high and AFG16low patient groups. This identified 1,194 genes as upregulated and 642 genes as downregulated in the AFG16high group (Fig. 6F; Supplementary Table S16). Gene set enrichment analysis (GSEA) using the canonical gene sets from the Molecular Signatures Database (MSigDB) further revealed significantly elevated cell-cycle signaling (Fig. 6G) and associated genes (Fig. 6H), which explained the stronger inhibitory effect of flavopiridol and SNS-032 agents for the AFG16high patients. Collectively, these findings suggested that the AFG16 score could be employed to facilitate more rational use of potent drug candidates like flavopiridol and SNS-032, not only used for patients with relapsed/refractory AML but also in newly diagnosed AML with a high probability to poorly respond to conventional induction chemotherapy.
Discussion
The state-of-the-art risk stratification system, such as the ELN2017 scheme, provides a broadly accepted and reliable classification of patients with AML (5), which rely heavily on cytogenetic screening and mutational profiling on specific genes (NPM1, FLT3, CEBPA, TP53, RUNX1, and ASXL1). However, the molecular complexity of AML presents a considerable barrier to clinical implementation of such approaches (1, 3). Clinically serviceable transcriptomics technologies with the reproducibility and analytic validity hold the great potential to uncover prognostic transcriptome information for AML (6–11, 35). Such an approach will allow rapid risk assessment for all patients indiscriminately, including those not bearing mutations intrinsic to the ELN classification and with a normal karyotype, and therefore it could provide additional prognostic value and complement the ELN guidelines.
Because the implementation of CRISPR-Cas9–based genome editing system in cancer research, numerous candidate AML targets have been revealed largely by in vitro CRISPR screens (19, 40). Recent studies further developed an in vivo screening strategy to not only identify leukemia dependencies that are essential for cancer progression but also define the microenvironment-responsive signals required for leukemogenesis (31, 33). Here, we carried out an integrated analysis of large-scale in vitro and genome-wide in vivo CRISPR screens to comprehensively identify the fitness genes for AML. Given the critical importance of fitness genes for functional phenotypes of AML cells and the direct link between leukemia cell survival and disease progression, we speculated that these AML-specific fitness genes may also harbor key prognostic information. We thus devised a powerful prognostic signature composed of 16 fitness genes (AFG16) and subsequently validated the prognostic impact in six independent validation sets across the spectrum of AML genotypes and distinct analysis platforms employed in data generation. Therefore, our study also indicated that a common prognostic signature can be derived, regardless of the underlying genetic lesions found in AML. The dysregulated expression of these fitness genes contained in the signature likely perturb survival signals and lead to distinct clinical outcomes among various subgroups of AML.
The AFG16 score mainly captured the cell fitness of leukemic blasts that were intrinsically related to molecular and clinical features of AML. A greater understanding of the established risk factors associated with fitness could contribute to better clinical assessment and improved outcomes. At a basic level, unfavorable-risk AML cells are commonly more malignant or more likely to evade chemotherapeutics with enhanced fitness. Consistent with this, the high AFG16 score was associated with adverse-risk factors including older age (41), presence of TP53 and FLT3-ITD mutations (1, 3) as well as complex karyotype (42). These adverse-risk factors may confer a fitness advantage to leukemia cells, accompanied by enhanced stress adaptation (43) and competitive strength over other cells (44), to fully engage in oncogenic transformation and therapy resistance. Conversely, patients with a AFG16low score more frequently harbored biallelic CEBPA and KIT mutations as well as CBF fusions. As previously reported, patients with biallelic CEBPA mutation (45) and CBF-AML (46) had relatively favorable clinical outcome compared with patients with other genetic lesions. The elevated frequency of KIT mutations in AFG16low patients can be explained, at least in part, by the higher incidence of CBF-AML, since KIT mutations are frequently associated with CBF-AML (47). The strong prognostic impact of the AFG16 score was independent of these prognostic molecular abnormalities, which suggested that perturbations introduced by these driver mutations coalesced on changes in fitness properties, and that the AFG16 score had the ability to distill these downstream consequences.
More importantly, we demonstrated that the AFG16 score captured additional prognostic information and could substantially complement the ELN2017 risk classification system. The new integrated risk classification schema after incorporating the AFG16 score provided a better stratification and prognosis prediction, which would facilitate the reassign of patient risk at diagnosis and adopt a more appropriate consolidation of therapy regimen. A high AFG16 score also probably reflected part of the biological properties of LSCs that drive chemoresistance in AML (14), evidenced by the significant depletion of these 16 genes in this signature using the in vivo CRISPR screen, which was performed in a physiologically relevant context using LSCs (31). We indeed found that the AFG16 score was a strong prognosticator that can significantly assist the prediction of chemotherapy resistance in patients with AML. The AFG16 RNA-seq assay will allow rapid risk assessment for newly diagnosed patients, enabling recommendation of more pioneering investigational treatments to be directed to AFG16high patients with a high predicted likelihood to have resistant disease, while avoiding unnecessary added toxicity for AFG16low patients.
Several powerful prognostic signatures including LSC17 (11), CODEG22 (7), and GENE4 (8) scores have recently been proposed to predict outcome in patients with AML. The relatively better performance of the AFG16 score compared to these signatures indicated that molecular markers conferring fitness defects in cells contain potential valuable prognostic information that underpin the prediction of patient outcome and should not be underestimated. A few possible reasons could explain the better performance of AFG16 signature. Most importantly, the genome-wide CRISPR screens, especially the in vivo screens offer a strategy for identifying critical dependencies of AML cells within the physiologically relevant microenvironment, allowing for the capture of core transcriptional programs associated with survival. Furthermore, the fitness genes were screened out across a broad spectrum of AML cell lines and AFG16 score was trained using a large scale of patient cohort consisting of multiple cytogenetic/genotype subgroups, reflecting the common transcriptional dependencies among various subsets of AML. In line with previous reports (7, 11), the AFG16 genes were implicated in a multi-layer and coordinated biological program important for AML cells, which render not only their prognostic relevance but also promising therapeutic targets for patients with AML. For example, the shared gene with the CODEG22 signature, UROD, has been reported to mediate the oncogenicity of MYCN by increasing activity of the heme/porphyrin pathway, endowing leukemia cells with survival advantages (34). The AFG16 genes were also involved in fundamental leukemia biology including alternative splicing, ferroptosis, chromatin remodeling, myeloid differentiation, and transcription regulation. Perturbations mediated by these biological processes coalesced on changes in fitness properties, contributing to the functional phenotypes of AML cells, thus informing the prognosis for patients. We also demonstrated the prognostic impact and clinical applicability of our signature through a retrospective cohort using clinically serviceable RNA-seq and targeted gene sequencing. With the emergence of NGS technologies in clinical routine (35, 36), the expression level of signature genes can be easily evaluated, the AFG16 score thus can be rapidly calculated for individual patient for risk assessment in clinical practice.
Furthermore, our study also serves as a proof-of-concept for the utility of the AFG16 score to identify associated biological processes activated or inactivated, potentially enabling determination of therapeutic susceptibility for patients with AML to specific targeted agents. We carried out an ex vivo drug sensitivity analysis based on a small panel of small-molecule inhibitors, and suggested that the AFG16high patients exhibited higher sensitivity to cell-cycle inhibitors, flavopiridol (48) and SNS-032 (38). Further pathway enrichment analysis confirmed the significant activation of cell-cycle signaling in high-score patients. Despite an ex vivo study, our results indicated the potential use of flavopiridol and SNS-032 to effectively treat AFG16high patients. In fact, flavopiridol has shown encouraging clinical activity in both newly diagnosed and relapsed/refractory patients with AML as single agent (49) or combinational therapies (50). These findings suggested that the AFG16 score may be utilized to facilitate the more rational use of flavopiridol in patients most likely to benefit, such as AFG16high patients, while sparing low-score patients who derive little benefit any unnecessary potential toxicities.
In summary, using large-scale in vitro and in vivo CRISPR screens, we identified a set of fitness genes for AML, which serve as critical candidates of leukemia that deserve further explorations to define their therapeutic implications. Importantly, we derived a 16-gene fitness score for rapid risk assessment for newly diagnosed patients with AML. Incorporation of the AFG16 score into the ELN2017 classification schema could significantly improve AML risk stratification, thus guiding risk-adapted therapy decisions. The predictive power of the AFG16 score to therapeutic responses further demonstrated a great potential for its clinical applications.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81870110), the National Key Research and Development Program of China (2019YFA0905900), Shanghai Science Technology and Innovation Action Plan-Key Program on Medical Innovation Research (21Y31920400), and Youth Development Program of Ruijin Hospital (KY00305).
The computational analysis was run on the π 2.0 cluster supported by the Center for High Performance Computing at Shanghai Jiao Tong University. We thank the researchers of the TCGA-LAML (phs000178) and BeatAML (phs001657) projects for making their multi-omics data and clinical data available.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
P. Jin: Visualization, methodology, writing–original draft. Q. Jin: Visualization, methodology, writing–original draft. X. Wang: Writing–original draft. M. Zhao: Methodology. F. Dong: Writing–review and editing. G. Jiang: Collect patient samples. Z. Li: Collect patient samples. J. Shen: Collect clinical samples. W. Zhang: Writing–review and editing. S. Wu: Writing–review and editing. R. Li: Collect patient samples. Y. Zhang: Supervision, writing–review and editing, followed up the patients and acquired the clinical data. X. Li: Supervision, writing–original draft, writing–review and editing. J. Li: Supervision, funding acquisition, writing–review and editing.
References
- 1. The Cancer Genome Atlas Research Network, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med 2015;373:1136–52. [DOI] [PubMed] [Google Scholar]
- 3. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 5. Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017;129:424–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Docking TR, Parker JDK, Jadersten M, Duns G, Chang L, Jiang J, et al. A clinical transcriptome approach to patient stratification and therapy selection in acute myeloid leukemia. Nat Commun 2021;12:2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nehme A, Dakik H, Picou F, Cheok M, Preudhomme C, Dombret H, et al. Horizontal meta-analysis identifies common deregulated genes across AML subgroups providing a robust prognostic signature. Blood Adv 2020;4:5322–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen Z, Song J, Wang W, Bai J, Zhang Y, Shi J, et al. A novel 4-mRNA signature predicts the overall survival in acute myeloid leukemia. Am J Hematol 2021;96:1385–95. [DOI] [PubMed] [Google Scholar]
- 9. Anande G, Deshpande NP, Mareschal S, Batcha AMN, Hampton HR, Herold T, et al. RNA splicing alterations induce a cellular stress response associated with poor prognosis in acute myeloid leukemia. Clin Cancer Res 2020;26:3597–607. [DOI] [PubMed] [Google Scholar]
- 10. Jin P, Tan Y, Zhang W, Li J, Wang K. Prognostic alternative mRNA splicing signatures and associated splicing factors in acute myeloid leukemia. Neoplasia 2020;22:447–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016;540:433–7. [DOI] [PubMed] [Google Scholar]
- 12. Bill M, Nicolet D, Kohlschmidt J, Walker CJ, Mrózek K, Eisfeld AK, et al. Mutations associated with a 17-gene leukemia stem cell score and the score's prognostic relevance in the context of the European LeukemiaNet classification of acute myeloid leukemia. Haematologica 2020;105:721–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dick JE. Stem cell concepts renew cancer research. Blood 2008;112:4793–807. [DOI] [PubMed] [Google Scholar]
- 14. Vetrie D, Helgason GV, Copland M. The leukaemia stem cell: similarities, differences and clinical prospects in CML and AML. Nat Rev Cancer 2020;20:158–73. [DOI] [PubMed] [Google Scholar]
- 15. Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov 2017;7:716–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boyd AL, Aslostovar L, Reid J, Ye W, Tanasijevic B, Porras DP, et al. Identification of chemotherapy-induced leukemic-regenerating cells reveals a transient vulnerability of human AML recurrence. Cancer Cell 2018;34:483–98. [DOI] [PubMed] [Google Scholar]
- 17. Thomas D, Majeti R. Biology and relevance of human acute myeloid leukemia stem cells. Blood 2017;129:1577–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 2015;163:1515–26. [DOI] [PubMed] [Google Scholar]
- 19. Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic ras. Cell 2017;168:890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang J, Li Y, Liu H, Zhang J, Wang J, Xia J, et al. Genome-wide CRISPR/Cas9 library screen identifies PCMT1 as a critical driver of ovarian cancer metastasis. J Exp Clin Cancer Res 2022;41:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bester AC, Lee JD, Chavez A, Lee YR, Nachmani D, Vora S, et al. An Integrated genome-wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell 2018;173:649–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Crowther MD, Dolton G, Legut M, Caillaud ME, Lloyd A, Attaf M, et al. Genome-wide CRISPR-Cas9 screening reveals ubiquitous T cell cancer targeting via the monomorphic MHC class I-related protein MR1. Nat Immunol 2020;21:178–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Behan FM, Iorio F, Picco G, Gonçalves E, Beaver CM, Migliardi G, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 2019;568:511–6. [DOI] [PubMed] [Google Scholar]
- 24. Dempster JM, Pacini C, Pantel S, Behan FM, Green T, Krill-Burger J, et al. Agreement between two large pan-cancer CRISPR-Cas9 gene dependency data sets. Nat Commun 2019;10:5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 2004;350:1617–28. [DOI] [PubMed] [Google Scholar]
- 26. Li Z, Herold T, He C, Valk PJ, Chen P, Jurinovic V, et al. Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J Clin Oncol 2013;31:1172–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Herold T, Jurinovic V, Batcha AMN, Bamopoulos SA, Rothenberg-Thurley M, Ksienzyk B, et al. A 29-gene and cytogenetic score for the prediction of resistance to induction treatment in acute myeloid leukemia. Haematologica 2018;103:456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood 2008;112:4193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang YH, Lin CC, Hsu CL, Hung SY, Yao CY, Lee SH, et al. Distinct clinical and biological characteristics of acute myeloid leukemia with higher expression of long noncoding RNA KIAA0125. Ann Hematol 2021;100:487–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bajaj J, Hamilton M, Shima Y, Chambers K, Spinler K, Van Nostrand EL, et al. An in vivo genome-wide CRISPR screen identifies the RNA-binding protein Staufen2 as a key regulator of myeloid leukemia. Nat Cancer 2020;1:410–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McFarland JM, Ho ZV, Kugener G, Dempster JM, Montgomery PG, Bryan JG, et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun 2018;9:4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin S, Larrue C, Scheidegger NK, Seong BKA, Dharia NV, Kuljanin M, et al. An in vivo CRISPR screening platform for prioritizing therapeutic targets in AML. Cancer Discov 2022;12:432–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fukuda Y, Wang Y, Lian S, Lynch J, Nagai S, Fanshawe B, et al. Upregulated heme biosynthesis, an exploitable vulnerability in MYCN-driven leukemogenesis. JCI Insight 2017;2:e92409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet 2016;17:257–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buzdin A, Sorokin M, Garazha A, Glusker A, Aleshin A, Poddubskaya E, et al. RNA sequencing for research and diagnostics in clinical oncology. Semin Cancer Biol 2020;60:311–23. [DOI] [PubMed] [Google Scholar]
- 37. Cortes JE, Khaled S, Martinelli G, Perl AE, Ganguly S, Russell N, et al. Quizartinib versus salvage chemotherapy in relapsed or refractory FLT3-ITD acute myeloid leukaemia (QuANTUM-R): a multicentre, randomised, controlled, open-label, phase 3 trial. Lancet Oncol 2019;20:984–97. [DOI] [PubMed] [Google Scholar]
- 38. Walsby E, Lazenby M, Pepper C, Burnett AK. The cyclin-dependent kinase inhibitor SNS-032 has single agent activity in AML cells and is highly synergistic with cytarabine. Leukemia 2011;25:411–9. [DOI] [PubMed] [Google Scholar]
- 39. Boffo S, Damato A, Alfano L, Giordano A. CDK9 inhibitors in acute myeloid leukemia. J Exp Clin Cancer Res 2018;37:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang E, Lu SX, Pastore A, Chen X, Imig J, Chun-Wei Lee S, et al. Targeting an RNA-binding protein network in acute myeloid leukemia. Cancer Cell 2019;35:369–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Juliusson G, Antunovic P, Derolf A, Lehmann S, Möllgård L, Stockelberg D, et al. Age and acute myeloid leukemia: real world data on decision to treat and outcomes from the Swedish acute leukemia registry. Blood 2009;113:4179–87. [DOI] [PubMed] [Google Scholar]
- 42. Stölzel F, Mohr B, Kramer M, Oelschlägel U, Bochtler T, Berdel WE, et al. Karyotype complexity and prognosis in acute myeloid leukemia. Blood Cancer J 2016;6:e386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Biancon G, Joshi P, Zimmer JT, Hunck T, Gao Y, Lessard MD, et al. Precision analysis of mutant U2AF1 activity reveals deployment of stress granules in myeloid malignancies. Mol Cell 2022;82:1107–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Van Egeren D, Escabi J, Nguyen M, Liu S, Reilly CR, Patel S, et al. Reconstructing the lineage histories and differentiation trajectories of individual cancer cells in myeloproliferative neoplasms. Cell Stem Cell 2021;28:514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dufour A, Schneider F, Metzeler KH, Hoster E, Schneider S, Zellmeier E, et al. Acute myeloid leukemia with biallelic CEBPA gene mutations and normal karyotype represents a distinct genetic entity associated with a favorable clinical outcome. J Clin Oncol 2010;28:570–7. [DOI] [PubMed] [Google Scholar]
- 46. Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom medical research council trials. Blood 2010;116:354–65. [DOI] [PubMed] [Google Scholar]
- 47. Paschka P, Marcucci G, Ruppert AS, Mrózek K, Chen H, Kittles RA, et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a cancer and leukemia group B study. J Clin Oncol 2006;24:3904–11. [DOI] [PubMed] [Google Scholar]
- 48. Chao SH, Fujinaga K, Marion JE, Taube R, Sausville EA, Senderowicz AM, et al. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J Biol Chem 2000;275:28345–8. [DOI] [PubMed] [Google Scholar]
- 49. Zeidner JF, Karp JE. Clinical activity of alvocidib (flavopiridol) in acute myeloid leukemia. Leuk Res 2015;39:1312–8. [DOI] [PubMed] [Google Scholar]
- 50. Zeidner JF, Lee DJ, Frattini M, Fine GD, Costas J, Kolibaba K, et al. Phase I study of alvocidib followed by 7+3 (Cytarabine + Daunorubicin) in newly diagnosed acute myeloid leukemia. Clin Cancer Res 2021;27:60–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are publicly available in GEO at GSE201492. All other data generated in this study are available within the article and its Supplementary Data files.