Summary
Background
Primary Ovarian Insufficiency (POI), a public health problem, affects 1-3.7% of women under 40 yielding infertility and a shorter lifespan. Most causes are unknown. Recently, genetic causes were identified, mostly in single families. We studied an unprecedented large cohort of POI to unravel its molecular pathophysiology.
Methods
375 patients with 70 families were studied using targeted (88 genes) or whole exome sequencing with pathogenic/likely-pathogenic variant selection. Mitomycin-induced chromosome breakages were studied in patients’ lymphocytes if necessary.
Findings
A high-yield of 29.3% supports a clinical genetic diagnosis of POI. In addition, we found strong evidence of pathogenicity for nine genes not previously related to a Mendelian phenotype or POI: ELAVL2, NLRP11, CENPE, SPATA33, CCDC150, CCDC185, including DNA repair genes: C17orf53(HROB), HELQ, SWI5 yielding high chromosomal fragility. We confirmed the causal role of BRCA2, FANCM, BNC1, ERCC6, MSH4, BMPR1A, BMPR1B, BMPR2, ESR2, CAV1, SPIDR, RCBTB1 and ATG7 previously reported in isolated patients/families. In 8.5% of cases, POI is the only symptom of a multi-organ genetic disease. New pathways were identified: NF-kB, post-translational regulation, and mitophagy (mitochondrial autophagy), providing future therapeutic targets. Three new genes have been shown to affect the age of natural menopause supporting a genetic link.
Interpretation
We have developed high-performance genetic diagnostic of POI, dissecting the molecular pathogenesis of POI and enabling personalized medicine to i) prevent/cure comorbidities for tumour/cancer susceptibility genes that could affect life-expectancy (37.4% of cases), or for genetically-revealed syndromic POI (8.5% of cases), ii) predict residual ovarian reserve (60.5% of cases). Genetic diagnosis could help to identify patients who may benefit from the promising in vitro activation-IVA technique in the near future, greatly improving its success in treating infertility.
Funding
Université Paris Saclay, Agence Nationale de Biomédecine.
Keywords: Primary ovarian insufficiency, Personalized medicine, NF-KB, Post - translational regulation, Meiosis/DNA repair genes, Mitophagy
Research in context.
Evidence before this study
Infertility is a major public health problem, affecting 10% of couples worldwide. Primary Ovarian Insufficiency (POI) is one of the most common syndromes implicated in female infertility, affecting ∼1-3.7% of women under 40 years. POI most often leads to permanent sterility and severe complications: osteoporosis, cardiovascular disease and even neurologic degenerative diseases. Remarkably, a decrease in longevity was also observed which could be due to specific causes. But most causes are currently unknown. Despite the technological leap in genetics, the clinical impact on the diagnosis and management of POI remains unknown, as well as the molecular mechanisms disturbed in POI.
Added value of this study
To get insight into this disorder, we studied the feasibility of a first line genetic diagnosis in a large cohort of 375 patients including 70 families and looked for new responsible genes and altered signalling pathways. We have sequenced either the whole coding part of the genome or 88 known causing genes. A high-yield diagnosis of 29.3 % was obtained supporting the use of genetics routinely to diagnose all unexplained POI. Interestingly, we identified 9 genes not previously related to POI or Mendelian disease and confirm 13 others previously reported in isolated patients or families. We showed how the molecular dissection of the pathways involved leads to personalized management of patients.
The main family is the DNA repair/meiosis/mitosis gene family (37.4% of cases), but it is also a tumour/cancer susceptibility gene family. Lifelong monitoring is therefore necessary to prevent or treat the appearance of these tumours. The second major family involved is that of follicular growth genes (35.4%). Strikingly, in 8.5% of cases, POI is the only single visible expression of a complex multi-organ genetic disease. A full patient assessment is required.
Other novel pathways identified could yield novel therapeutic targets: i.e. immunity, gene regulation, mitochondrial autophagy.
Three genes had been implicated in the large variance in the age of natural menopause, confirming a genetic link and a continuum between the two conditions, the difference may be related to the severity of the genetic variants involved, major in POI.
Implications of all the available evidence
We describe here a high-performance genetic diagnosis of POI, critical to understand the pathogenesis of POI and leading to personalized medicine. These results have major implications for preventing/curing comorbidities related to tumour/cancer susceptibility genes that could affect lifespan or multi-organ disease revealed by genetics. Apart from the infertility of patients, it is therefore their entire state of health that must be assessed and treated as soon as the patient consults for POI. This should be done according to the underlying cause which must imperatively be identified beforehand.
Genetics is also critical to establish a fertility prognosis for the promising technique of in vitro follicular activation in the future by predicting a residual ovarian reserve (60.5% of cases). The selection of patients who could benefit from this technique, the genetic cause highlighting existing follicles in the ovaries blocked in their growth, could clearly improve its success in the treatment of infertility of POI. The pathways identified could also provide future therapeutic targets.
Alt-text: Unlabelled box
Introduction
Infertility affects 15% of couples worldwide and is a public health problem. Primary Ovarian Insufficiency (POI) affects ∼1-3.7% of the women before 40 years1,2 with cessation of ovarian functions.1 About 60–70% of cases remain idiopathic.1 Women with POI are usually definitively infertile but spontaneous pregnancies have been described in 3.5% of patients with secondary amenorrhea or spaniomenorhea.3 Moreover, various epidemiological studies have shown a decreased lifespan or an increased risk of all-cause mortality in infertile women.4,5 It is thus a priority to identify the causes involved. Next generation sequencing (NGS) especially whole exome sequencing (WES) has recently identified several genetic causes of POI, although in single or rare families, mostly consanguineous.1 Due to this high genetic heterogeneity, the clinical impact of the etiological diagnosis of POI remains unknown. In clinical practice, only karyotype and FMR1 studies are routinely performed with respective diagnosis yields of 7–10% and 3–5%.1
In this study, we describe the genetic landscape of POI in a large cohort of 375 patients, using WES or targeted NGS comprising all human POI genes known to date, and with a classification of variants using the American College of Medical Genetics and Genomics (ACMG) criteria.6 Our study revealed that a powerful etiological diagnosis is possible in a high proportion of patients, namely: 29.3%. We show here that personalized genetic counselling and management of patients and family is possible based on the identification of the genetic cause. Our study also highlights genes not previously related to Mendelian phenotype, pathways and mechanisms in POI, potentially allowing identification of new therapeutic targets.
Methods
Ethics statement
The study was approved by all the institutions involved and by the Agence de Biomedecine (reference number PFS12-002). Written informed consent was received from participants prior to inclusion in the study.
Patients
Between 2017 and 2022, 375 patients were referred to our university hospital laboratory by different hospitals in Europe, Turkey, Africa, and Asia. We performed NGS analysis in patients who lacked the FMR1 premutation and had a normal karyotype. POI was diagnosed in the presence of primary amenorreha (PA), secondary amenorrhea (SA) or spaniomenorrhea (SP) for more than 4 months associated with follicle-stimulating hormone (FSH) plasma level ≥ 25 IU/L before the age of 40 years. Patients with known etiology of POI, such as chemotherapy, radiotherapy, or extensive ovarian surgery, were excluded from the study. The following clinical data were requested before the NGS analysis: menstrual cycle (PA, SA, SP), pubertal development, ethnicity, spontaneous pregnancies or pregnancies after in vitro fertilization (IVF), and familial history. Information on autoimmune disease or the presence of extraovarian symptoms—such as intellectual disability, hyperandrogenism, neurological and ophthalmic disorders, deafness, tumour/cancer, in the patient and/or family, family history of male or female infertility, was also requested. We also requested ultrasonography to assess ovarian and follicular sizes, the number of follicles, as well as a complete hormonal assay in particular the serum level of follicle stimulating hormone (FSH), luteinizing hormone (LH), estradiol, anti-Mullerian Hormone (AMH), thyroid stimulating hormone (TSH), and autoantibodies (anti-TPO, anti-TG, and anti-21-hydroxylase antibodies).
Molecular genetics studies
-
a)
A custom-made targeted NGS including all known genes (88) involved in POI to date (supplementary material and methods and Table S1)7,8 was performed. WES was performed by IntegraGen SA (Evry, France) in case of consanguineous or familial cases (See supplementary material and methods).7,9 We only considered variants classified as pathogenic or likely pathogenic according to ACMG guidelines.6 Genes of unknown significance not previously related to Mendelian phenotype and identified in this study have been prioritized using in silico prediction softwares, ovarian expression, segregation studies, animal models, and interactions with other known genes causing POI.
-
b)Copy Number Variation (CNV) were studied using two methods:
-
-For patients studied with WES, we used the Bioconductor DNACopy package (DNAcopy 1.32.0) and compared control exome data to reference samples pool (DOI: 10.18129/B9.bioc.DNAcopy). It implements the circular binary segmentation (CBS) algorithm to segment DNA copy number data. All changes were annotated with the catalog of the Database of Genomic Variants (DGV) to provide a comprehensive summary of structural variation in the Human genome.
-
-For patients studied with targeted NGS, we used an in-house coverage-based pipeline to detect CNV.10 This method is based on the Read Depth (RD) or the Read Count (RC), which reflects the abundance of the genome segment. RD is defined as the number of reads covering a given position in the sequence and RC is defined as the number of reads in a given region. Using these concepts and hypothesizing that the RD or RC is proportional to the initial abundance of the genetic material used for sequencing, we can detect potential CNV.
-
-
Chromosomal analysis
Hypersensitivity to the chromosome-breaking effect of crosslinking agents is a reliable marker of homologous recombination (HR) efficiency.11 For POI genes involved in DNA repair and not expressed exclusively in germ cells, chromosomal breakage studies were performed in cultures of peripheral lymphocytes obtained from the patient, the mother when available, and a healthy woman as a control. The experiments were performed at the Gustave Roussy Institute (Villejuif, France), following a standard in-house protocol.7, 8, 9,12 Fresh peripheral blood lymphocytes were cultured under standard conditions for karyotyping. DNA damage was induced by treating the cells with mitomycin C (MMC; Sigma) for 48 h to examine cellular hypersensitivity to DNA crosslinking agents. Three treatment conditions were used: without MMC to analyze spontaneous DNA damage and with 150 or 300 nM MMC as recommended.11 Chromosome breaks were scored by an experienced cytogeneticist using at least 20 metaphases.
Role of the funding source
The study funders had no role in the study design of this study, data collection, data analysis, data interpretation, or report writing.
Results
Cohort characteristics
The characteristics of the patients recruited in this study are summarized in Figure 1, Figure S1, and Table S2. The cohort of POI comprises 375 patients (344 index patients and 31 affected mothers or sisters): 26.7% were familial and 73.3% were sporadic, 24.8% of patients had PA, 65.3% SA, and 9.9% SP. Targeted NGS was performed in 345 patients and WES in 30 patients. Twelve patients (12/375; 3.2%) had syndromic POI with additional extra-ovarian features (Figure S1, Table S3).
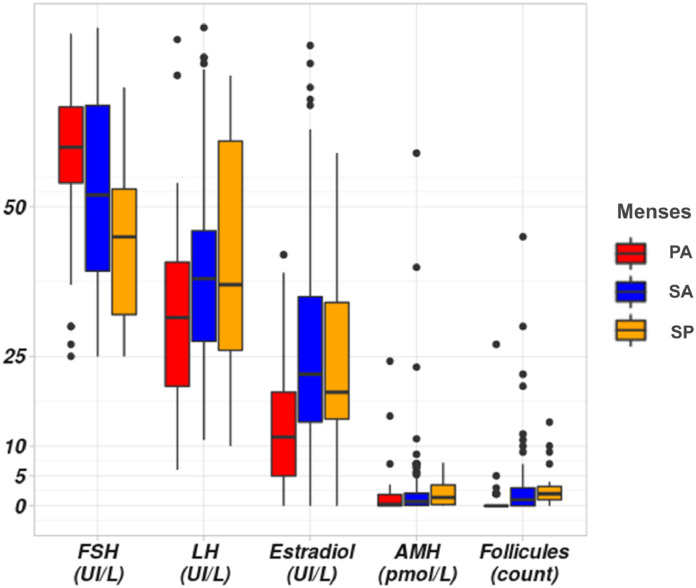
Figure 1.
Hormonal and ultrasonographical datas in our cohort of patients with POI. Hormonal assays (FSH, LH, Estradiol and AMH) in patients according to menses. PA: Primary Amenorrhea, SA: Secondary Amenorrhea, SP: Spaniomenorrhea. The Y scale unit is variable according to each hormonal assay represented in the X-axis. Normal range: AMH (7-20.7 pmol/l); Respectively follicular, ovulatory, luteal phases and menopause: FSH (IU/L): (2.9-12), (6.3-24), (1.5-7), (17-95); LH: (IU/L) (1.5-8), (9.6-80), (0.2-6.5), (8-33); E2 (ng/l): (19.5-144.2),(63.9-356.7),(55.8-214.2),(≤ 32.2).
Molecular findings
In our whole cohort, we identified 216 variants in 215 patients (out of 375) (Figure 2, Tables S4-S7). In fourteen patients, we found strong evidence of pathogenicity for nine genes not previously related to Mendelian phenotype or POI (Table 1). Using the ACMG criteria for genes known to be implicated in POI, pathogenic (PV) or likely pathogenic (LPV) were identified in 110 patients. The diagnostic performance of our NGS study with the ACMG criteria6 including only PV/LPV was 29.3% (110/375) for the whole cohort and 26.3% (61/232) for European patients (n = 232, 61.9% of the cohort) (Table S4). For isolated POI it was 28.4% (103/363 patients), and 58.3% for syndromic POI (7/12) (Table S3).
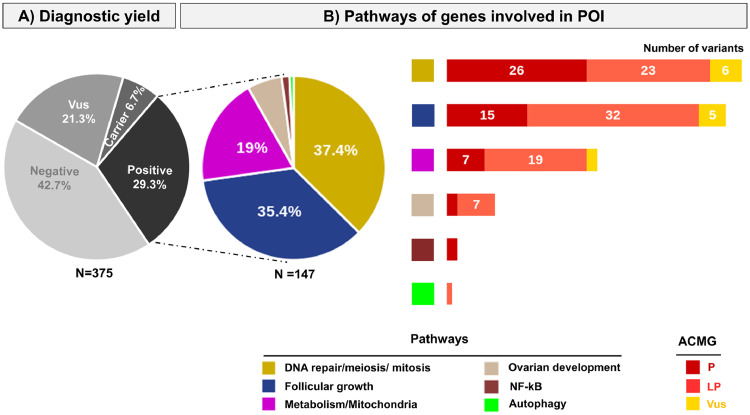
Figure 2.
Genetic studies of the cohort of patients with POI. Patients were studied by a custom-made targeted NGS comprising 88 genes or by whole exome sequencing (see methods). A) Diagnostic yield using ACMG criteria: Variants are classified according to the American College of Medical Genetics (ACMG) guidelines. N= 375: the whole cohort comprises 375 patients with POI. Vus: Variant of unknown significance. Carrier: patients harbouring a heterozygous pathogenic variant in a known autosomal recessive POI gene. Positive: the diagnostic yield corresponds to patients carrying pathogenic (P) or likely pathogenic (LP) variants and is 29.3%. B) Pathways of genes involved in POI. The different pathways are indicated with different colors. The pie chart represents the proportion of patients with P or LP variants in a specific pathway: DNA repair meiosis and mitosis (37.4%), Follicular growth (35.4%), Mitochondria and Metabolism (19%), Ovarian development (6.1%), NF-kB (1.4%), Autophagy (0.7%). The histograms show the number and type of variants detected in each pathway.
Table 1.
Variants in genes not previously related to Mendelian phenotype or POI.
| ID | Ethnicity | Age | Menses | Gene | variant | Status | Pathway | Expression | Animal or in vivo models |
|---|---|---|---|---|---|---|---|---|---|
| 258 | Turkish | 15 | SA | C18orf53 (HROB) | NM_024032.5: c.502delG:p.Glu168ArgfsTer35 | Hom | DNA repair & Meiosis | Ubiquitous | C18orf53-/- mice are infertile18 |
| 311 | Turkish | 16 | PA | ||||||
| 310 | North-African | 24 | SA | HELQ | NM_133636.5 : c.3095delA: p.Tyr1032SerfsTer4 | Hom | DNA repair & Meiosis | Ubiquitous | Helq-/- mice are infertile19 |
| 373 | North-African | 20 | SA | ||||||
| 317 | North-African | 16 | PA | SWI5 | NM_001318092.1 : c.261-1G>C:p.? | Hom | DNA repair & Meiosis | Ubiquitous | Impaired meiosis in in yeast20 |
| 302 | Asian | 29 | SA | CENPE | NM_001813.2 : c.2023C>T:Q675Ter | Het | Cell Cycle | Ubiquitous | Embryonic lethality of Cenpe-/- mice23 |
| 303 | Asian | 14 | SA | ||||||
| 304 | Asian | 34 | SA | ||||||
| 311 | North-African | 16 | PA | SPATA33 | NM_153025.2: c.34dupT:p.Cys12LeufsTer2 | Hom | Autophagy | Gonadal | Spata33 knockout suppresses mitophagy in germ cells24 |
| 270 | North-African | 20 | SP | NLRP11 | NM_145007.4: c.1867A>T : p.Arg623Ter/ c.2206G>A:p.E736K | Comp Het | Immunity-Inflammation | Ubiquitous | Reduced fertility in Nrlp11/-mice16 |
| 306 | North-African | 15 | SA | CCDC150 | NM_001080539.2 :c.291_292delTG :p.Cys97Ter | Hom | Unkown | Gonadal | No available animal model |
| 314 | Turkish | 17 | SA | CCDC185 | NM_152610.3: c.1174C>T :p.Gln392Ter | Hom | Unkown | Gonadal | No available animal model |
| 318 | European | 20 | SA | ELAVL2 | NM_001351472.2:c.448C>T / :p. Arg150Cys / c.313C>T :Leu105Phe | Comp Het | Post-transcriptional regulation | Gonadal & neurons | Female Elavl2-/- mice are infertile devoid of follicles17 |
| 319 | 24 | SA |
PA: Primary amenorrhea; SA: secondary amenorrhea; Hom: homozygous; Het : heterozygous: Comp Het : compound heterozygous.
The diagnostic yield of targeted NGS is 28.7% (99/345) in the whole cohort, and 25.8% (57/221) in the European population. The diagnostic yield of WES is 36.7 % (11/30) in the whole cohort and 36.4% (4/11) in the European population.
Most genes (n = 24) were identified in single patients. Only nine genes were found in two different patients/families and seventeen “recurrent” genes were found in at least three patients/families (Figure S2) confirming the high genetic heterogeneity in POI.
In 25 other patients (25/375: 6.7%), only heterozygous PV/LPV were found in known autosomal recessive POI genes (Figure 2). This may either indicate a new mode of inheritance for these variants (as for BMP15 or FOXL2 known to have both autosomal dominant and recessive inheritance1), or most likely correspond to a carrier status (Table S7).
The genes displaying PV/LPV are involved in different pathways (Figure 2). Remarkably, 37.4 % of genes are involved in meiosis/DNA repair or mitosis making this family the major family involved in POI, 35.4% are involved in follicular growth, 19% in metabolism and mitochondrial functions, Ovarian development (6.1%), NF-kB pathway (1.4%), Autophagy (0.7%). (Figure 2).
We detected a CNV in a patient with POI and primary amenorrhea (ID 6). It was a 3.2kb-heterozygous deletion encompassing exons 10-17 of STAG3 (Figure S3). The patient carried a heterozygous pathogenic variant of STAG3: NM_012447.4:c.3046delT:p.Ser1016ProfsTer16. Parental samples were not available to perform segregation studies. We thus considered this patient as a presumed compound heterozygous.
For all other patients, especially those identified as carriers in our study, carrying a pathogenic or likely pathogenic heterozygous variant in known autosomal recessive POI genes (n=19,) no CNV was detected.
I. Identification of genes and pathways not previously related to Mendelian phenotype or POI
Twelve candidate genes were identified in our cohort. Nine candidate genes are not previously related to a Mendelian phenotype or POI. For XPNPEP2, there is to date only a balanced translocation (X; 12) presumed to interrupt this gene that has been described in a POI patient and her affected mother.13 The variants are mentioned in Table 1 and the family pedigrees are shown in Figure 3. We previously reported two genes, FANCM and BRCA2, involved in the RAD51 pathway causing non-syndromic POI.9,12
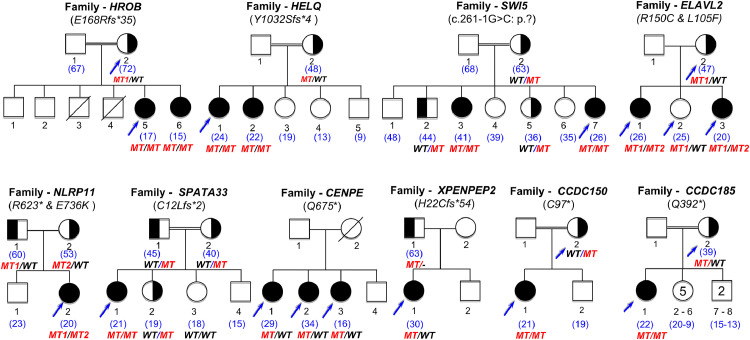
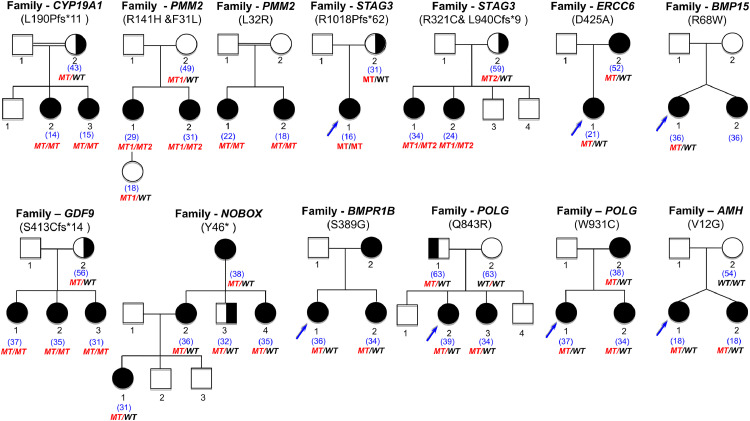
Figure 3.
Pedigrees of the families with genes not previously involved in Mendelian phenotype or POI. On top of each family, the name of the gene is indicated, with the variant found. Double lines indicate consanguineous unions. The age of the patient at diagnosis is indicated in blue between brackets. Blue arrows indicate the patients and relatives studied with NGS (see text and Methods chapter). All family members available were studied by Sanger Sequencing. The segregation of the variants is shown under each individual sequenced. MT: mutated. MT1: first mutation. MT2: second mutation. WT: wild type. Family CCDC185: the number inside each symbol indicates the number of siblings in the family, either sisters (5) or brothers (2).
a) NF-kB signalling and regulation of inflammatory responses
NLRP11: In a Tunisian patient with POI and SP at the age of 20 years, we identified two compound heterozygous variants of NLRP11 encoding the Nod-like-receptor protein (NLRP)11: a nonsense substitution in exon 6 (NM_145007.4: c.1867A>T), introducing a stop codon (p.Arg623Ter) and a missense substitution in exon 8 (NM_145007.4: c.2206G>A: p.Glu736Lys). The NLRP protein family regulates the immune response to infection through NF-kB and type I interferon signalling pathways.14 NLRP11 is highly expressed in the testes and ovary as are many other NLRP proteins.14 Remarkably, NLRP5 has been involved in POI in human and mice.15 Moreover, a very recent study based on mouse models highlights the important role of inactivated NF–KB in POI.16
b) RNA binding protein and posttranscriptional/translational regulations
ELAVL2: In a Polish family, two sisters developed POI with SA at 20 and 24 years. WES revealed compound heterozygous variants in the RNA binding domain of ELAVL2 (NM_001351472.2): c.448C>T, Arg150Cys and c.313C>T, Leu105Phe, the latter transmitted by the mother. ELAVL2 encodes a RNA binding protein, mainly expressed in ovaries, testes and neurons. It is involved in post-transcriptional and post-translational regulations.17 Mice deficient in Elavl2 are sterile and ovaries are devoid of follicle.17
c) Meiosis/ DNA repair genes/ mitosis
C17orf53 (HROB): In two sisters from a consanguineous Turkish family, both presenting PA and streak gonads, we identified a homozygous truncated variant of HROB (NM_024032.5: c.502delG; p.Glu168ArgfsTer35), found also homozygous in her affected sister and heterozygous in both parents. HROB (C17orf53) encodes a newly identified protein involved in homologous recombination (HR).18 It corresponds to a factor containing an oligonucleotide/oligosaccharide (OB) fold that recruits MCM8 and MCM9—two proteins that, when defective, cause POI—to the site of DNA damage. Mice deficient in Hrob are infertile with small uteri and ovaries completely devoid of follicle.18 Chromosome breakage studies revealed high chromosomal instability with spontaneous breaks, enhanced in the presence of mitomycin (MMC), and numerous radial figures, similar to what is observed in patients with Fanconi Anemia (FA) strongly supporting a defect in HR (Figure 4, Table S8).
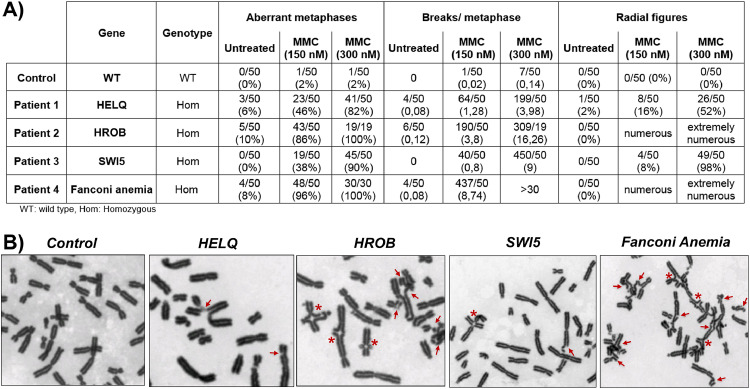
Figure 4.
Chromosomal instability in patients with molecular defects of DNA repair genes. A). Chromosomal breaks analysis of the patient's lymphocytes, a control (a WT fertile woman), and a patient with Fanconi anemia, in the presence of increasing concentration of mitomycin (MMC). B) Selected figures of metaphases from the patients with molecular defects of SWI5, HELQ, HROB compared to Fanconi anemia and control cells in the presence of 300 nM of MMC. Chromosomal breaks are shown with red arrows and radial figures with asterisks.
In the absence of MMC, while no spontaneous breaks are observed in cells of the patient with the SWI5 homozygous splice variant, respectively 6% and 10 % of cells of the patients with homozygous truncated variants of HELQ and HROB presented increased breaks, similarly to cells of the patient with Fanconi anemia (8%). In the presence of 150nM MMC, 86% of cells with the HROB pathogenic variant presented breaks with 3.8 breaks per metaphase very similarly to cells of the patient with Fanconi anemia (96%) and radial figures were observed in numerous cells of both types. Increased breaks were also observed in 46% of cells with the HELQ pathogenic variant (1.28 breaks per metaphase) and 38% of cells with the homozygous SWI5 variant (0.8 breaks per metaphase). Radial figures were observed in 16 % and 8 % of cells respectively. At 300 nM MMC, HROB-cells mimic Fanconi anemia cells, all presenting breaks (16.26 breaks per metaphase) with numerous radial figures, while 90% of SWI5-cells presented breaks with a mean of 9 breaks per metaphase and 82% of HELQ-cells with a mean of 3.98 breaks per metaphase. HROB-cells mimic Fanconi anemia cells with spontaneous chromosomal instability and marked hypersensitivity to MMC while HELQ-cells and SWI5-cells have a milder phenotype, but clearly with increased breaks when compared to control cells, and with radial figures, a pathognomonic feature of Fanconi anemia.
HELQ: A patient born to consanguineous Moroccan parents presented SA at the age of 20 years. WES identified a homozygous truncated variant in exon 17 of HELQ (NM_133636.5: c.3095delA; p.Tyr1032SerfsTer4), heterozygous in her mother, who still had regular menses at the age of 50 years and homozygous in her affected sister. HELQ encodes a DNA helicase that plays a pivotal role in DNA replication, recombination and inter-strand crosslink repair.19 HELQ deficient mice exhibit subfertility and tumour predisposition.19 The orthologues of this protein in Drosophila melanogaster and Caenorhabditis elegans are required for meiotic double-strand break repair.19 Concordantly, chromosomal studies in the patient revealed spontaneous chromosomal breaks markedly enhanced with MMC (Figure 4, Table S8).
SWI5: The proposita (25 years) and her sister (41 years), born to consanguineous Moroccan parents, had POI with PA and streak ovaries devoid of follicles. The mother had menopause at the age of 52 years. WES in the proposita identified a homozygous splice variant in the canonical GT acceptor splice site of intron 3 of SWI5 (NM_001318092.1: c.261-1G>C), also homozygous in the affected sister and heterozygous in unaffected siblings and parents. This variant is absent in all available databases. In silico predictions with all available softwares (Human Splice Finder, dbscSNV, Alamut and SpliceAI) of the splice variant predicts a high impact on splicing. SWI5 encodes a protein part of an auxiliary complex, Swi5-Sfr1 that regulates HR through the RAD51 pathway, synergistically with the HOP2-MND1 complex.20 Interestingly, PSMC3IP (encoding HOP2) and MND1 cause POI in humans.1,21 Furthermore, an orthologue of SWI5-SFR1 in yeast (Schizosaccharomyces pombe)— Sae3-Mei5—is required for meiotic HR.20 Concordantly, the patient's cells exhibit high MMC-induced chromosomal breaks (Figure 4, Table S8). All available studies on Swi5-Sfr1 have only been performed in vitro or in yeast, and so far, it is not known whether SWI5 plays a role in mammalian HR and meiosis.20,22 The family we report here provides a unique opportunity to observe the in vivo effect of a high impact bi-allelic molecular defect of SWI5, confirming its functional importance in humans. The role of SWI5 in meiosis is consistent with the phenotype of both sisters in our family, with early POI and ovarian atrophy. SWI5 could also act in a complex with SFR1 in humans, since the interaction between the two molecules is predicted to be abolished in our patients. Our results thus reinforce the crucial role of the Swi5-Sfr1 complex in meiosis and provide a strong argument for extending its role to the human species.
CENPE: This study includes a family with mixed origin (Asian and European) in which three sisters presented POI with SA at 16, 29 and 34 years. WES identified a heterozygous, stop-gain variant of CENPE (NM_001813.2: c.2023C>T; Q675Ter). CENPE encodes the centromeric protein E, a kinesin-like motor protein essential for correct chromosome segregation in both meiosis and mitosis and in genome stability. It is highly expressed in germ cells. In mice this protein has an essential role in meiosis and in maturing oocytes.23
d) Mitochondrial autophagy (Mitophagy)
SPATA33: In a patient with POI and PA born to consanguineous Algerian parents, WES studies identified a homozygous insertion of a single nucleotide in the exon 1 of SPATA33 (NM_153025.2: c.34dup; p.Cys12LeufsTer2), heterozygous in her parents and one unaffected sister. The other unaffected sister inherited both wild-type alleles. SPATA33 is a protein exclusively expressed in mitochondria germ cells.24 Recent in vitro studies performed on mouse testes revealed that this protein is localized in mitochondria and has been described as a novel autophagy mediator for mitophagy. This suggests a potential role of mitophagy in POI (mitochondrial autophagy).24
e) Gene involved in peptide processing
XPNPEP2: In a French patient presented POI and SA at the age of 30 years, we identified a paternal heterozygous frameshift deletion in exon 2 of XPNPEP2 (NM_003399: c.63_94del; p.His22CysfsTer54). XPNPEP2 encodes APP2, an aminopeptidase, ubiquitously expressed that removes N-terminal amino-acids from peptides with a penultimate prolyl residue (as Arg-Pro-Pro)25 notably collagen that regulate follicle growth.25 In the rat ovary, it was suggested that XPNPEP2 regulates collagens formation.25 To date, there is only a single report of a balanced translocation (X;12) that interrupts XPNPEP2 in a patient and her mother, who both presented with SA and POI.13 This point PV of this gene that we identified here validates its causal role in POI.
f) Genes with unknown function
Interestingly two genes belonging to the coiled-coil family with unknown function were identified in two unrelated patients.
CCDC150: In a consanguineous Moroccan patient with primo-secondary amenorrhea at the age of 15 years, WES identified a homozygous two-base-pairs deletion in exon 2 of CCDC150 (containing 28 exons): NM_001080539.2: c.291_292delTG: p.Cys97Ter. This gene is highly expressed in the testes, according to the GTex database. Of note, only adult ovaries are included in GTex. Other genes exclusively expressed in testes according to GTex were in fact also expressed in foetal ovaries such as STAG3.1 Very little is known about this protein. However, a strong interaction is predicted between CCDC150 and RAD50, a major protein involved in homologous recombination, using String software (https://string-db.org/).
CCDC185 (C1orf65): A Turkish patient born to consanguineous parents had POI and early SA at the age of 17 years. WES identified a homozygous stop codon in the unique exon of CCDC185 (NM_152610.3: c.1174C>T: p.Gln392Ter), heterozygous in the mother. This gene is exclusively expressed in the testes according to the GTex database.
II. Involvement of genes responsible for syndromic POI in patients with apparently isolated forms
Unexpectedly, in 32 (8.5%) patients initially presenting as isolated POI, we identified PV/LPV in genes known to cause syndromic POI (Table S7, S9). One patient with POI and early SA had the G188Q homozygous PV of GALT, the most frequent mutation causing galactosemia.26 It is a severe, life-threatening metabolic disease often diagnosed at birth and fatal without specific regimen. Enzymatic assays confirmed the diagnosis. Reverse phenotyping revealed only a cataract during infancy treated by surgery. Our patient survived to adulthood without any specific medical care unlike all previously reported adult patients with an early childhood diagnosis who had a continuous adapted regimen. Eight patients inherited genetic defects in genes causing pulmonary arterial hypertension (PAH): CAV1(n=1), BMPR2 (n=2) or BMPR1B (n=5).27 BMPR2 and BMPR1B encode a transmembrane receptor, member of the bone morphogenetic protein (BMP) receptor family and CAV1 encodes caveolin that has a crucial role in signal transduction and vesicular trafficking.27, 28, 29 All these proteins are expressed both in lungs and ovaries especially in granulosa cells and display functional roles in both tissues.1,27,28 Deficient bmp15-/- mice are subfertile with decreased ovulation and fertilization rates while gdf9-/- mice are sterile.28 Cav1-/- null mice show reduced fertility.29 The absence of PAH in particular in patients with variants of BMRP2 may be explained by several mechanisms: i) the type of the variant; indeed, 50 to 75 % of the BMPR2 variants detected in patients presenting PAH were truncated variants while in POI only missense variants are found30,31; ii) the localization of the variant: the majority of the BMPR2 variants detected in patients with PAH are located in the kinase domain of the protein (encoded by exon 6-10)30 while three out of the four BMPR2 variants detected in POI patients are localized in the cytoplasmic tail of BMPR2 (the L656S and R597Q variants described here and the previously reported S987F variant).32 It was reported that patients carrying a mutation affecting the cytoplasmic tail of BMPR2 were characterized by an older age at diagnosis compared with other BMPR2 mutation carriers33 and iii) the low penetrance of the mutation observed for PAH genes in particular for BMPR2.27,31 The penetrance of BMPR2 mutation is estimated to be between 20 and 43%.27,31 A 20-year-old patient had a LPV in BMPR1A. This gene predisposes to hamartomatous polyposis, absent in our patient. Seven patients had genetic defects in POLG encoding the mitochondrial DNA polymerase, without associated neurological or ocular symptoms. We report four supplementary variants of TP63 (one splice variant and three missense variants), a gene causing both isolated and syndromic POI with dominant inheritance.34 TP63 variants described in POI are usually located in the C-terminal region.34 However, missense and truncated variants in the different domains of TP63 causing POI have been described.35 Our data confirm that pathogenic or likely pathogenic variants in both the 5’ end (c.192+2T>G) of TP63 or in the N-terminal (p.Ile225Thr, p.Lys214Glu) or the C-terminal (p.Thr566Met) regions of TP63 cause POI. One patient had a heterozygous LPV of TWNK, a mitochondrial helicase implicated in progressive external ophtalmoplegia with both dominant and recessive mode of inheritance.36 In all these cases, a comprehensive evaluation of patients and their families was recommended with appropriate follow-up in a multidisciplinary team.
III. Confirmation of the causal role of genes in POI
In 26 patients, we identified PV/LPV in thirteen POI genes previously described in single patients/families (Figure 5, Table 2). This confirmed their role in the pathogenesis of POI worldwide. We showed recently that a patient harbouring a homozygous SPIDR truncating variant exhibited a strong MMC-induced chromosomal fragility. This supported a role of SPIDR in the RAD51 pathway and HR.8
Figure 5.
Pedigrees of the family confirming the causal role of genes in POI. Double lines indicate consanguineous union. Blue arrows indicate the patients and relatives studied with NGS, The other family members available are studied by Sanger sequencing. The segregation of the variant is shown under each sequenced individual. MT: Mutated, MT1: first mutation, MT2, second mutation, WT: wild type. The age of patients at diagnosis is indicated in blue between brackets.
Table 2.
Variants confirming the causal role worldwide of genes reported in single patients/families.
| ID | Ethnicity | Menses (age) | Gene | Variant | Status | ACMG | Pathway |
|---|---|---|---|---|---|---|---|
| 321 | North-African | PA | BRCA2 | NM_000059.4:c.8350C>T :p.Arg2784Trp | Hom | P | DNA repair & Meiosis |
| 167 | European | SA (18) | FANCM | NM_020937.4:c.5101C>T :p.Gln1701Ter / c.1528G>A : p.Gly510Ser | Pres Comp Het | P/LP | DNA repair & Meiosis |
| ATM | NM_000051.4:c.6596_6597delCT :p.Ser2199Ter | Het | P | DNA repair & Meiosis | |||
| 192 | Finnish | PA | FANCM | NM_020937.4:c.5101C>T :p.Gln1701Ter | Hom | P | DNA repair & Meiosis |
| 305 | Finnich | PA | FANCM | NM_020937.4:c.5101C>T :p.Gln1701Ter | Hom | P | DNA repair & Meiosis |
| 306 | Finnich | PA | FANCM | NM_020937.4:c.5101C>T :p.Gln1701Ter | Hom | P | DNA repair & Meiosis |
| 326 | European | SP (20) | FANCM | NM_020937.4:c.5101C>T :p.Gln1701Ter / c.575A>T : p.Gln192Leu | Comp Het | P/LP | DNA repair & Meiosis |
| 323 | European | SP (17) | MSH4 | NM_002440.4:c.2198C>A : p.Ser733Ter | Hom | P | DNA repair & Meiosis |
| 59 | Asian | PA | SPIDR | NM_001080394.4: c.814C>T: p.Arg272Ter | Hom | LP | DNA repair & Meiosis |
| 55 | North-African | SA (39) | BNC1 | NM_001717.4: c.2319C>A:p.Asn773Lys | Het | LP | DNA repair & Meiosis |
| 88 | European | SP (18) | ERCC6 | NM_000124.4: c.1996C>T : p.Arg666Cys | Het | LP | DNA repair & Meiosis |
| 109 | European | SP (27) | ERCC6 | NM_000124.4: c.1996C>T : p.Arg666Cys | Het | LP | DNA repair & Meiosis |
| 79 | European | SA (19) | ERCC6 | NM_000124.4: c.1274A>C :p.Asp425Ala | Het | LP | DNA repair & Meiosis |
| 105 | European | SA (30) | ERCC6 | NM_000124.4: c.1274A>C :p.Asp425Ala | Het | LP | DNA repair & Meiosis |
| 211 | Middle East | SA (37) | ERCC6 | NM_000124.4: c.1274A>C :p.Asp425Ala | Het | LP | DNA repair & Meiosis |
| 140 | European | PA | BMPR1A | NM_004329.3: c.1327C>T : p.Arg443Cys | Het | LP | Follicular growth |
| 155 | European | SA (22) | BMPR1B | NM_001256793.2: c.761G>A : p.Arg254His | Het | LP | Follicular growth |
| 156 | European | SA (22) | BMPR1B | NM_001256793.2: c.761G>A : p.Arg254His | Het | LP | Follicular growth |
| 267 | North-African | SA (33) | BMPR1B | NM_001256793.2: c.1165A>G : p.Ser389Gly | Het | LP | Follicular growth |
| 268 | North-African | SA (35) | BMPR1B | NM_001256793: c.1165A>G:p.Ser389Gly | Het | LP | Follicular growth |
| PCCB | NM_000532: c.646A>G:p.Met216Val | Het | LP | Metabolism | |||
| 38 | European | SA (16) | BMPR1B | NM_001256793.2 : c.836A>T:p.Gln279Leu | Het | LP | Follicular growth |
| 163 | European | SA (21) | BMPR2 | NM_001204.7: c.1967T>C : p.Leu656Ser | Het | LP | Follicular growth |
| 275 | European | PA | BMPR2 | NM_001204.7:c.1790G>A : p.Arg597Gln | Het | LP | Follicular growth |
| AMH | NM_000479.5:c.35T>G :p.Val12Gly | Het | LP | Follicular growth | |||
| 325 | European | SP (24) | ESR2 | NM_001437.2:c.335C>A :p.Ser112Ter | Het | LP | Follicular growth |
| 112 | Turkish | SA (14) | CAV1 | NM_001172895.1: c.1A>G: p.Met1Val | Het | P | Follicular growth |
| 272 | European | PA | RCBTB1 | NM_018191.4: c.1271T>G :p.Phe424Cys / c.962C>T : p.Pro321Leu | Pres Comp Het | LP/LP | Follicular growth |
| 168 | North-African | SA (31) | ATG7 | NM_006395.3: c.1478A>G : p.Lys493Arg | Het | LP | Autophagy |
PA: Primary amenorrhea; SA: secondary amenorrhea; Hom: homozygous; Het: heterozygous; Comp Het: compound heterozygous; Pres Comp Het: Presumed compound heterozygous (parents not available); ACMG: Classification of variant according to American college of Medical genetic; P: Pathogenic, LP, Likely pathogenic. Patient 105 is the mother of patient 79; Patient 267 and 268 are sisters.
IV. Syndromic POI
- Known syndromic Genes identified
In our cohort, 12 patients (12/375 =3.2%) had syndromic POI (Table S3). We identified the genes involved in seven patients (7/12; 58.3%). In all these cases, genetic diagnosis led to a complete screening for other signs potentially associated with each syndrome, and to familial segregation studies.
-
-
Possible syndromic POI not previously described
Interestingly, our series included four patients with new syndromic phenotypes indicating previously unreported syndromes (Table S3). They include: 1) keratoconus, 2) horseshoe kidney and epilepsy, 3) Marfanoid habitus, scoliosis and hyperlaxity and 4) single kidney, Hashimoto's thyroiditis, ptosis, and scoliosis. Genes causing these syndromic POI phenotypes remain to be discovered.
V. Patients with variants in two different genes
In a small proportion of patients (8/375; 2.1%), we identified P/LPV in two different genes (Table S10). In these patients, however, one of the mutated genes alone was sufficient to cause POI. Therefore, we did not find evidence of di/multigenic inheritance of POI in our cohort. Moreover, each exome is known to contain at least hundreds of rare variants, and it is not surprising to find heterozygous pathogenic or likely pathogenic variants in more than one POI gene. A recent study based on 6,447 exomes of healthy, genetically unrelated Europeans of two distinct ancestries estimates that each individual is a carrier of at least two pathogenic variants in currently known autosomal recessive genes.37
Discussion
Pathways and candidate genes involved in POI not previously described in humans
Our study shows the powerful approach to identify genetic etiology and pathways not previously implicated in POI. This allows better understanding of the pathophysiology of POI and opens the way to the development of novel therapeutic targets.
The immune pathway has a major role in the ovary and may explain 4–31% of POI cases.1 However, no specific gene has been found in isolated POI. AIRE causes the APECED syndrome (Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy).1 We involve in this study NLRP11 a gene of the immune NF-kB pathway in isolated POI not previously implicated. Environmental factor especially phthalates, bisphenol A, pesticides have a negative impact on ovarian function, yielding increased follicular depletion.1 Interestingly, the NF-κB pathway is involved in the production of TNF-α and IL-6 in response to bisphenol A and may also be a therapeutic target.38
We also describe novel mechanisms in POI revealed by the genetic defect of ELAVL2, involving post-transcriptional and translational regulations in the ovaries, known to play a major physiological role.39 Elavl2-/- mice are infertile. Interestingly, these two candidate genes (ELAVL2 and NLRP11) have been recently associated with the age of natural menopause (ANM)40 reinforcing the genetic link between POI and the physiological variance of the ANM (see further).
We found strong evidence of a causal role in POI for four DNA repair/meiosis/mitosis genes based on mice/yeast models, and/or chromosomal instability. A high chromosomal fragility was found in our patients with defects in three new DNA repair genes: HROB, HELQ, SWI5, the highest found for the homozygous truncation of HROB, similar to FA cells (Figure 3, Table S8). However, there was no sign of FA in our patients (anaemia, cancer microcephaly, small size or café-au-lait spots). This chromosomal fragility may lead to an increased risk of malignancies and therefore a long-term follow-up by a multidisciplinary team is indicated.
In our study, we only performed MMC-induced chromosome breakage studies in patients with possible genetic alteration of double-strand DNA repair genes like HELQ, SWI5 and HROB (C17orf53). Indeed, MMC-induced chromosomal breakage study only detects defects in homologous DNA repair and does not allow evaluating defects in other DNA repair pathways.41 Thus, we suggest a combined two-step approach in POI patients using sequentially an NGS study followed by a chromosomal breakage study in patients with variants in a gene known or supposed to be involved in homologous recombination. This can improve variant classification.
Interestingly, we identified other candidate genes, CCDC155 and CCDC185, exclusively expressed in gonads but with unknown function. Mice models are needed to unravel unknown pathophysiological mechanisms involved in POI. SPATA33 has been recently suggested to be involved in autophagy. If confirmed in rodent models, it would support the implication of this crucial pathway in human follicular physiology and provides a future perspective on curing autophagy-related reproductive infertility diseases as for other linked diseases.42
Performance of genetic diagnosis and impact on patients’ management
Despite the large-scale use of NGS analysis in the era of genomic medicine, the study of genes involved in POI is often limited to the screening of the FMR1 premutation (3-5% of cases).1 We show here in our large cohort of 375 patients that NGS is an effective diagnostic tool for patients with POI. Using ACMG criteria, the rate of detection of pathogenic variants was 29.3% in the whole cohort and 26.3% in the European population (232 patients) (Including only P/ LP variants in known POI-genes. Variants in genes not previously related to POI or a Mendelian phenotype are considered of unknown significance-VUS as recommended6). This high yield supports a thorough genetic study of all unexplained POI patients in clinical practice. Very few studies were previously reported including either a small cohort (20-60 patients), a very limited number of POI genes (n=18),43, 44, 45 or reporting only heterozygous variants, the pathogenicity of which could not be formally demonstrated. Indeed no familial segregation studies were performed46 and the rodent models deficient in these genes have a recessive mode of transmission.46 Moreover, in these studies, authors also considered VUS or likely benign variants that cannot be used for a genetic diagnosis.43, 44, 45, 46, 47 Only variants classified as pathogenic or likely pathogenic can guide management of patients. Accordingly, we could not confirm the high involvement described up to 6.2% for some POI genes notably NOBOX or BMP15.43, 44, 45 Of note, the R117W-NOBOX variant is classified benign according to ACMG criteria and has a high Minor Allele Frequency-MAF up to 8.17 % in the African population in GnomAD exome and genome databases. Efforts to improve adherence to these guidelines will be important to prevent erroneous misclassification of non-pathogenic variants in POI genetic testing and inappropriate diagnosis.
To date, both homozygous and heterozygous variants of NOBOX have been reported in patients with POI, some of which have been validated with in vitro functional studies.1,45,48, 49, 50 We report an additional truncating variant of NOBOX (Y46X) in two unrelated patients (ID 143 and 331) (Figure 5). Unlike previous reports, we were able to perform a large familial transmission study in three generations in one family (family of the proposita with ID 331). The patient's mother, grandmother and maternal aunt, all with POI and secondary amenorrhea, harboured the Y46X variant of NOBOX. This strongly supports an autosomal dominant transmission of this truncated variant. Altogether these data argue that inheritance pattern of NOBOX is either autosomal dominant1 or autosomal recessive49, 50 with a genotype-phenotype correlation. Biallelic truncated variants of NOBOX cause POI and primary amenorrhea with a recessive mode of inheritance while heterozygous variants of NOBOX yield POI and secondary amenorrhea with an autosomal dominant transmission or haploinsufficiency.
The identification of the genetic cause has a major impact on genetic counselling and personalized patient care, with:
-
i)
Psychological consequences for patients and their family: as it reveals the causal mechanism for their infertility. Furthermore, offering a presymptomatic genetic testing in siblings may enable fertility preservation, if appropriate.
-
ii)
Important information on the fertility prognosis by predicting a possible residual OR. There is no direct method to evaluate this OR. AMH assays are negative when only small follicles are present in the ovaries.1
We show that in 60.5% of our patients the genetic defect allows prediction of a persistent OR at an early stage: this includes patients with defects in genes involved in follicular growth (35.4%), metabolism and mitochondrial functions (19%), ovarian developement (6.1%), autophagy (1 patient). Indeed, previous studies have reported positive AMH values and even pregnancies in patients with defects of these gene families, and the OR is also conserved in the corresponding rodent models.1,51 It should be noted that, concordantly, previous histological study revealed the presence of an OR in 57% of patients with POI.52
For instance, we identified a mild form of galactosemia with a homozygous PV of GALT in a patient initially presenting with isolated POI. Such patients have 48% of chances of being spontaneously pregnant in two years26 (90% in the general population). The patient stopped the oocyte donation program she had started. Other patients with complete FSHR or BMP15 inactivation are known to have a preserved OR, although a rapid decrease can occur.1,52,53 Similarly, patients with POI and mild mutations of some meiosis/DNA repair genes: FANCM, ERCC6, BNC1 may have spontaneous pregnancies, as found in our cohort.1,9 This may be due to a hypomorphic mutation or alternative compensatory pathways or to the fact that heterozygous mutations are found with possible escape of oocytes to DNA damage. The genetic diagnosis of POI is thus a medical priority as soon as all other causes have been eliminated. Fertility preservation in a multidisciplinary team should be discussed without delay in these patients and their affected siblings when appropriate to avoid the rapid follicular atresia that may occur in such patients.1,53 The prediction of a residual OR could allow these patients to benefit from new in vitro activation- IVA techniques in the near future.54 Variable results were obtained in small cohorts of unselected POI with a yield of 3.7 up to 21% of healthy newborns.54 A genetic pre-screening of patients could markedly improve the success of IVA by excluding patients whose ovarian reserve is predicted to be absent or severely impaired.
Similarily, in male infertility55 it has been proposed that a genetic diagnosis could predict positive testicular sperm extraction (TESE) or, on the contrary, may avoid unnecessary surgical interventions in case of major defects of DNA repair and meiosis genes.55
By contrast, in 29.4% of our patients, often with PA, major defects in meiosis/DNA repair genes are found. A major alteration of the OR is predicted with a very poor fertility prognosis, as in previously reported patients.1 This is also confirmed by the corresponding animal models.51 A rapid referral of these patients to egg donation or adoption could avoid repeated IVF failures and unnecessary delay in their parental desire.
-
iii)Prevention or treatment of possible comorbidities. In our cohort, 45.9% of patients may have, or are at risk to develop, associated comorbidities, requiring a comprehensive assessment by a multidisciplinary team:
-
α)Other organic defects are found in genetically revealed syndromic POI (8.5% of cases): in 32 patients initially referred as isolated POI, we identified pathogenic variants of genes causing syndromic POI. Similarly, Alhathal et al found that in 3.1% (9/275 patients presenting male infertility with azoospermia or severe oligozoospermia), infertility can be the sole or major phenotypic expression of genes that are known to cause multisystemic manifestations in humans.56 In the same way, Kraucz et al found that 3/29 patients presenting infertility with Sertoli cell-only syndrome had biallelic variants of FANCA with only subtle haematological abnormalities corresponding in fact to occult FA.57 The existence of potential defects of other organs, undiagnosed, requires complete evaluation of the patients and most often a long-term follow-up of patients and their families.
-
β)Tumour/cancer in POI linked to meiosis/DNA repair genes (37.4% of cases):37.4% of patients had alteration of meiosis/DNA repair genes or mitosis. Although this gene family is the main contributor to POI, unexpectedly only six genes so far are known to cause POI and tumours/cancers in humans: BRCA2, FANCM, MCM8/9, PSMC3IP,STAG3.1,7,9,12 The majority of the corresponding deficient mice, however, developed tumours.51 Prospective studies in larger cohorts of patients will allow a more precise evaluation of the risk of tumours/cancers in these patients. Genetic counselling should be offered to patients and their families with long-term follow-up when adequate.The identification of this latter subpopulation of patients with POI due to PV/LPV variants in cancer susceptibility genes is therefore a medical and scientific priority.
-
α)
Overlap between genes implicated in azoospermia and POI
There is a remarkable overlap between genes involved in azoospermia and POI.56 It is interesting to note that genes belonging to the meiosis/DNA repair family including genes of the synaptonemal complex is the leading family shared by POI and male infertility with azoospermia. This is the case for novel variants in genes identified in our study: STAG3,58, 59, 60 SPIDR,8,56,61 PSMC3IP,62,63 HFM1,56,64 FANCM.9,65 We can therefore expect the discovery of other genes causing both male and female infertility in the near future, belonging especially to the meiosis and DNA repair gene family.
Link between genes involved in POI and genes involved in the physiological reproductive lifespan
Epidemiological studies in various populations have evidenced a link between the age of natural menopause and lifespan or the odd ratio of all-cause mortality, including cancer.1,4,5 WES and genome-wide association studies (GWAS) have highlighted a genetic link between POI and the ANM, through the DNA repair/meiosis gene family, the latter also being involved in aging.4,5 Recently, a large GWAS study involving more than 200 000 women worldwide identified 290 loci involved in the ANM with an effect ranging from 3.5 weeks to 74 weeks (∼1.2 years).40 Very interestingly, three genes involved in POI in our study: HELQ, ELAVL2 and NLRP11 were also found to be associated with the ANM. This markedly supports the role of these candidate genes and pathways in the age of physiological and pathological menopause/POI, especially ELAVL2 and NLRP11.
Conclusion
We describe here a highly successful genetic screening in a large cohort of patients with POI, allowing a genetic diagnosis in 29.3% of the patients. We showed that in 8.5% of cases, POI is the only phenotypic expression of a complex genetic disorder affecting other organs. This led to an adapted genetic counselling and a personalized management of patients and families either for prevention/treatment of potential comorbidities (other organ defects, or tumours/cancers when DNA repair genes are involved) or for early fertility preservation when an OR can be predicted in the light of the future development of innovative treatments of IVA. Strong evidence involved 9 new genes in POI and, three previously undescribed pathways that may provide new therapeutic targets for the infertility of POI in the future. Identifying the causes of POI will greatly expand our knowledge of the mechanisms controlling fertility in women.
Limitations
This work involves the largest cohort of patients with POI studied by next generation sequencing to date. However, due to the relatively high prevalence of this condition (1 to 3.7% of women before the age of 40),1,2 a larger cohort could be studied in the future to better define the monogenic part of POI, ∼30 % as shown in this study. This will: i) confirm the proportion of POI that can benefit from genetic diagnosis, ii) definitively implicate the nine genes identified here as candidate genes in POI. Indeed, these genes had never been associated with POI or a Mendelian disease, iii) to study the impact of a genetic diagnosis on the management of patients. Our study could encourage physicians to systematically prescribe NGS studies to all patients without an identified etiology, and not be limited solely to karyotyping and screening for the FMR1 premutation, the only genetic studies currently performed in most centres across the world. The Guidelines of International Societies in the field of Reproduction and Fertility may be updated in the light of these results. It should be noted that in our study, the segregation of the identified variants could only be achieved in 40% of cases because the parents were not available for the other patients. The absence of available trios has hampered the assessment of de novo mutations that might explain a subset of POI as has been observed in infertile men66 or other conditions67 and may improve the diagnostic performance of NGS studies in POI.
Contributors
Conceptualization: MM, CO. Formal analysis: AH, MM, NA. Funding acquisition: MM.
Data Collection and Patient's Recruitments: AR, CO, SDC, IB, LBR, NF, CB, RR, KM, SL, MMa, ICD, AC, MP, BDH, SP, CD, CNB, HE, MK, CRL, MF, ELB, PL, EG,AZ, FA, FP, EBB, AB, LG, MBe, NG, SO, BGD, BI, LL, SH, AMGj, ABP, HF, MP, HL, SE, PBG, SW, SH, KA, SCJ. Resources: JP, CC. Clinical Investigation: AH, AMG, KA. Data Curation: AH, MBr, SC, MLRS. Methodology: AH, MM. Project administration: AH, MM, CO. Software: AH. Supervision: MM. Validation: MM. Visualization: AH. Writing-original draft: AH, MM. Writing-review & editing: AH, MM and KA. Verification of the underlying data: AH, MM. All authors read and approved the final version of the manuscript.
Data sharing statement
Non-identifying data supporting the claims made in this paper are available upon request after its publication. Requests should be addressed to the corresponding authors of this paper: Prof Micheline Misrahi (micheline.misrahi@aphp.fr).
Declaration of interests
The authors declare no conflict of interest.
Acknowledgements
This study was supported by Université Paris Sud-Paris Saclay, Hôpitaux Universitaires Paris Saclay (AH, MM), the Institut National de la Santé et de la Recherche Médicale-INSERM (AH, MM), and by the Agence Nationale de Biomédecine (AH, MM). We thank the molecular & genetics Platform facility of GHU APHP Paris-Saclay for access to instruments and technical facilities.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104246.
Appendix. Supplementary materials
Table S6: List of variants detected in the cohort.
Table S7: Molecular findings in the cohort.
References
- 1.Huhtaniemi I, Hovatta O, La Marca A, et al. Advances in the molecular pathophysiology, genetics, and treatment of primary ovarian insufficiency. Trends Endocrinol Metab. 2018;29:400–419. doi: 10.1016/j.tem.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 2.Golezar S, Ramezani Tehrani F, Khazaei S, Ebadi A, Keshavarz Z. The global prevalence of primary ovarian insufficiency and early menopause: a meta-analysis. Climacteric. 2019;22:403–411. doi: 10.1080/13697137.2019.1574738. [DOI] [PubMed] [Google Scholar]
- 3.Bachelot A, Nicolas C, Bidet M, et al. Long-term outcome of ovarian function in women with intermittent premature ovarian insufficiency. Clin Endocrinol. 2017;86:223–228. doi: 10.1111/cen.13105. [DOI] [PubMed] [Google Scholar]
- 4.Stentz NC, Koelper N, Barnhart KT, Sammel MD, Senapati S. Infertility and mortality. Am J Obstet Gynecol. 2020;222:251.e1–251.e10. doi: 10.1016/j.ajog.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y-X, Farland LV, Wang S, et al. Association of infertility with premature mortality among US women: Prospective cohort study. Lancet Reg Health Am. 2021;7:100122. doi: 10.1016/j.lana.2021.100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heddar A, Beckers D, Fouquet B, Roland D, Misrahi M. A novel phenotype combining primary ovarian insufficiency growth retardation and pilomatricomas with MCM8 mutation. J Clin Endocrinol Metab. 2020;105:1973–1982. doi: 10.1210/clinem/dgaa155. [DOI] [PubMed] [Google Scholar]
- 8.Heddar A, Guichoux N, Auger N, Misrahi M. A SPIDR homozygous nonsense pathogenic variant in isolated primary ovarian insufficiency with chromosomal instability. Clin Genet. 2021;101:242–246. doi: 10.1111/cge.14080. [DOI] [PubMed] [Google Scholar]
- 9.Fouquet B, Pawlikowska P, Caburet S, et al. A homozygous FANCM mutation underlies a familial case of non-syndromic primary ovarian insufficiency. Elife. 2017;6:e30490. doi: 10.7554/eLife.30490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gordeeva V, Sharova E, Arapidi G. Progress in methods for copy number variation profiling. Int J Mol Sci. 2022;23:2143. doi: 10.3390/ijms23042143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oostra AB, Nieuwint AWM, Joenje H, de Winter JP. Diagnosis of fanconi anemia: chromosomal breakage analysis. Anemia. 2012;2012 doi: 10.1155/2012/238731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caburet S, Heddar A, Dardillac E, et al. Homozygous hypomorphic BRCA2 variant in primary ovarian insufficiency without cancer or Fanconi anaemia trait. J Med Genet. 2020;58:125–134. doi: 10.1136/jmedgenet-2019-106672. [DOI] [PubMed] [Google Scholar]
- 13.Prueitt RL, Ross JL, Zinn AR. Physical mapping of nine Xq translocation breakpoints and identification of XPNPEP2 as a premature ovarian failure candidate gene. Cytogenet Cell Genet. 2000;89:44–50. doi: 10.1159/000015560. [DOI] [PubMed] [Google Scholar]
- 14.Ellwanger K, Becker E, Kienes I, et al. The NLR family pyrin domain-containing 11 protein contributes to the regulation of inflammatory signaling. J Biol Chem. 2018;293:2701–2710. doi: 10.1074/jbc.RA117.000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stolk L, Perry JRB, Chasman DI, et al. Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat Genet. 2012;44:260–268. doi: 10.1038/ng.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo X, Xu J, Zhao R, et al. The role of inactivated NF-κB in premature ovarian failure. Am J Pathol. 2022;192:468–483. doi: 10.1016/j.ajpath.2021.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Kato Y, Iwamori T, Ninomiya Y, et al. ELAVL2-directed RNA regulatory network drives the formation of quiescent primordial follicles. EMBO Rep. 2019;20:e48251. doi: 10.15252/embr.201948251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hustedt N, Saito Y, Zimmermann M, et al. Control of homologous recombination by the HROB-MCM8-MCM9 pathway. Genes Dev. 2019;33:1397–1415. doi: 10.1101/gad.329508.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han X, Zhao L, Li X. HELQ in cancer and reproduction. Neoplasma. 2016;63:825–835. doi: 10.4149/neo_2016_601. [DOI] [PubMed] [Google Scholar]
- 20.Tsubouchi H, Argunhan B, Ito K, Takahashi M, Iwasaki H. Two auxiliary factors promote Dmc1-driven DNA strand exchange via stepwise mechanisms. Proc Natl Acad Sci USA. 2020;117:12062–12070. doi: 10.1073/pnas.1917419117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jolly A, Bayram Y, Turan S, et al. Exome sequencing of a primary ovarian insufficiency cohort reveals common molecular etiologies for a spectrum of disease. J Clin Endocrinol Metab. 2019;104:3049–3067. doi: 10.1210/jc.2019-00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Argunhan B, Murayama Y, Iwasaki H. The differentiated and conserved roles of Swi5-Sfr1 in homologous recombination. FEBS Lett. 2017;591:2035–2047. doi: 10.1002/1873-3468.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duesbery NS, Choi T, Brown KD, et al. CENP-E is an essential kinetochore motor in maturing oocytes and is masked during mos-dependent, cell cycle arrest at metaphase II. Proc Natl Acad Sci USA. 1997;94:9165–9170. doi: 10.1073/pnas.94.17.9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Xu X, Hu M, Wang X, Cheng H, Zhou R. SPATA33 is an autophagy mediator for cargo selectivity in germline mitophagy. Cell Death Differ. 2021;28:1076–1090. doi: 10.1038/s41418-020-00638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banu SK, Stanley JA, Sivakumar KK, Arosh JA, Barhoumi R, Burghardt RC. Identifying a novel role for X-prolyl aminopeptidase (Xpnpep) 2 in CrVI-induced adverse effects on germ cell nest breakdown and follicle development in rats. Biol Reprod. 2015;92:67. doi: 10.1095/biolreprod.114.125708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Erven B, Berry GT, Cassiman D, et al. Fertility in adult women with classic galactosemia and primary ovarian insufficiency. Fertil Steril. 2017;108:168–174. doi: 10.1016/j.fertnstert.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Morrell NW, Aldred MA, Chung WK, et al. Genetics and genomics of pulmonary arterial hypertension. Eur Respir J. 2019;53 doi: 10.1183/13993003.01899-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Persani L, Rossetti R, Di Pasquale E, Cacciatore C, Fabre S. The fundamental role of bone morphogenetic protein 15 in ovarian function and its involvement in female fertility disorders. Hum Reprod Update. 2014;20:869–883. doi: 10.1093/humupd/dmu036. [DOI] [PubMed] [Google Scholar]
- 29.Huang K, Dang Y, Zhang P, et al. CAV1 regulates primordial follicle formation via the Notch2 signalling pathway and is associated with premature ovarian insufficiency in humans. Hum Reprod. 2018;33:2087–2095. doi: 10.1093/humrep/dey299. [DOI] [PubMed] [Google Scholar]
- 30.Girerd B, Montani D, Jaïs X, et al. Genetic counselling in a national referral centre for pulmonary hypertension. Eur Respir J. 2016;47:541–552. doi: 10.1183/13993003.00717-2015. [DOI] [PubMed] [Google Scholar]
- 31.Hamid R, Cogan JD, Hedges LK, et al. Penetrance of pulmonary arterial hypertension is modulated by the expression of normal BMPR2 allele. Hum Mutat. 2009;30:649–654. doi: 10.1002/humu.20922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patiño LC, Silgado D, Laissue P. A potential functional association between mutant BMPR2 and primary ovarian insufficiency. Syst Biol Reprod Med. 2017;63:145–149. doi: 10.1080/19396368.2017.1291767. [DOI] [PubMed] [Google Scholar]
- 33.Girerd B, Coulet F, Jaïs X, et al. Characteristics of pulmonary arterial hypertension in affected carriers of a mutation located in the cytoplasmic tail of bone morphogenetic protein receptor type 2. Chest. 2015;147:1385–1394. doi: 10.1378/chest.14-0880. [DOI] [PubMed] [Google Scholar]
- 34.Luan Y, Xu P, Yu S-Y, Kim S-Y. The role of mutant p63 in female fertility. Int J Mol Sci. 2021;22:8968. doi: 10.3390/ijms22168968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tucker EJ, Gutfreund N, Belaud-Rotureau M-A, et al. Dominant TP63 missense variants lead to constitutive activation and premature ovarian insufficiency. Hum Mutat. 2022 doi: 10.1002/humu.24432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodríguez-López C, García-Cárdaba LM, Blázquez A, et al. Clinical, pathological and genetic spectrum in 89 cases of mitochondrial progressive external ophthalmoplegia. J Med Genet. 2020;57:643–646. doi: 10.1136/jmedgenet-2019-106649. [DOI] [PubMed] [Google Scholar]
- 37.Fridman H, Yntema HG, Mägi R, et al. The landscape of autosomal-recessive pathogenic variants in European populations reveals phenotype-specific effects. Am J Hum Genet. 2021;108:608–619. doi: 10.1016/j.ajhg.2021.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu J, Jiang L, Liu Y, et al. MAPK and NF-κB pathways are involved in bisphenol A-induced TNF-α and IL-6 production in BV2 microglial cells. Inflammation. 2015;38:637–648. doi: 10.1007/s10753-014-9971-5. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Yan Z, Qin Q, et al. Transcriptome landscape of human folliculogenesis reveals oocyte and granulosa cell interactions. Mol Cell. 2018;72:1021–1034.e4. doi: 10.1016/j.molcel.2018.10.029. [DOI] [PubMed] [Google Scholar]
- 40.Ruth KS, Day FR, Hussain J, et al. Genetic insights into biological mechanisms governing human ovarian ageing. Nature. 2021;596:393–397. doi: 10.1038/s41586-021-03779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katari S, Aarabi M, Kintigh A, et al. Chromosomal instability in women with primary ovarian insufficiency. Hum Reprod. 2018;33:531–538. doi: 10.1093/humrep/dey012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klionsky DJ, Petroni G, Amaravadi RK, et al. Autophagy in major human diseases. EMBO J. 2021;40 doi: 10.15252/embj.2021108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eskenazi S, Bachelot A, Hugon-Rodin J, et al. Next generation sequencing should be proposed to every woman with “idiopathic” primary ovarian insufficiency. J Endocr Soc. 2021;5:bvab032. doi: 10.1210/jendso/bvab032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bouilly J, Beau I, Barraud S, et al. Identification of multiple gene mutations accounts for a new genetic architecture of primary ovarian insufficiency. J Clin Endocrinol Metab. 2016;101:4541–4550. doi: 10.1210/jc.2016-2152. [DOI] [PubMed] [Google Scholar]
- 45.Bouilly J, Bachelot A, Broutin I, Touraine P, Binart N. Novel NOBOX loss-of-function mutations account for 6.2% of cases in a large primary ovarian insufficiency cohort. Hum Mutat. 2011;32:1108–1113. doi: 10.1002/humu.21543. [DOI] [PubMed] [Google Scholar]
- 46.Gorsi B, Hernandez E, Moore MB, et al. Causal and candidate gene variants in a large cohort of women with primary ovarian insufficiency. J Clin Endocrinol Metab. 2021;107:685–714. doi: 10.1210/clinem/dgab775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heddar A, Misrahi M. Genetics of primary ovarian insufficiency: a careful step-by-step approach based on solid foundations to bring new knowledge. Fertil Steril. 2022;118:421–424. doi: 10.1016/j.fertnstert.2022.05.024. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Wang B, Zhang W, et al. A homozygous NOBOX truncating variant causes defective transcriptional activation and leads to primary ovarian insufficiency. Hum Reprod. 2017;32:248–255. doi: 10.1093/humrep/dew271. [DOI] [PubMed] [Google Scholar]
- 49.Sassi A, Désir J, Duerinckx S, et al. Compound heterozygous null mutations of NOBOX in sisters with delayed puberty and primary amenorrhea. Mol Genet Genomic Med. 2021;9:e1776. doi: 10.1002/mgg3.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.França MM, Funari MFA, Lerario AM, et al. A novel homozygous 1-bp deletion in the NOBOX gene in two Brazilian sisters with primary ovarian failure. Endocrine. 2017;58:442–447. doi: 10.1007/s12020-017-1459-2. [DOI] [PubMed] [Google Scholar]
- 51.Jiao S-Y, Yang Y-H, Chen S-R. Molecular genetics of infertility: loss-of-function mutations in humans and corresponding knockout/mutated mice. Hum Reprod Update. 2021;27:154–189. doi: 10.1093/humupd/dmaa034. [DOI] [PubMed] [Google Scholar]
- 52.Massin N, Méduri G, Bachelot A, Misrahi M, Kuttenn F, Touraine P. Evaluation of different markers of the ovarian reserve in patients presenting with premature ovarian failure. Mol Cell Endocrinol. 2008;282:95–100. doi: 10.1016/j.mce.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 53.Mayer A, Fouquet B, Pugeat M, Misrahi M. BMP15 “knockout-like” effect in familial premature ovarian insufficiency with persistent ovarian reserve. Clin Genet. 2017;92:208–212. doi: 10.1111/cge.12970. [DOI] [PubMed] [Google Scholar]
- 54.Vo KCT, Kawamura K. In vitro activation early follicles: from the basic science to the clinical perspectives. Int J Mol Sci. 2021;22:3785. doi: 10.3390/ijms22073785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krausz C, Riera-Escamilla A, Moreno-Mendoza D, et al. Genetic dissection of spermatogenic arrest through exome analysis: clinical implications for the management of azoospermic men. Genet Med. 2020;22:1956–1966. doi: 10.1038/s41436-020-0907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alhathal N, Maddirevula S, Coskun S, et al. A genomics approach to male infertility. Genet Med. 2020;22:1967–1975. doi: 10.1038/s41436-020-0916-0. [DOI] [PubMed] [Google Scholar]
- 57.Krausz C, Riera-Escamilla A, Chianese C, et al. From exome analysis in idiopathic azoospermia to the identification of a high-risk subgroup for occult Fanconi anemia. Genet Med. 2019;21:189–194. doi: 10.1038/s41436-018-0037-1. [DOI] [PubMed] [Google Scholar]
- 58.Caburet S, Arboleda VA, Llano E, et al. Mutant cohesin in premature ovarian failure. N Engl J Med. 2014;370:943–949. doi: 10.1056/NEJMoa1309635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heddar A, Dessen P, Flatters D, Misrahi M. Novel STAG3 mutations in a Caucasian family with primary ovarian insufficiency. Mol Genet Genomics. 2019;294:1527–1534. doi: 10.1007/s00438-019-01594-4. [DOI] [PubMed] [Google Scholar]
- 60.Riera-Escamilla A, Enguita-Marruedo A, Moreno-Mendoza D, et al. Sequencing of a “mouse azoospermia” gene panel in azoospermic men: identification of RNF212 and STAG3 mutations as novel genetic causes of meiotic arrest. Hum Reprod. 2019;34:978–988. doi: 10.1093/humrep/dez042. [DOI] [PubMed] [Google Scholar]
- 61.Smirin-Yosef P, Zuckerman-Levin N, Tzur S, et al. A biallelic mutation in the homologous recombination repair gene SPIDR is associated with human gonadal dysgenesis. J Clin Endocrinol Metab. 2017;102:681–688. doi: 10.1210/jc.2016-2714. [DOI] [PubMed] [Google Scholar]
- 62.Zangen D, Kaufman Y, Zeligson S, et al. XX ovarian dysgenesis is caused by a PSMC3IP/HOP2 mutation that abolishes coactivation of estrogen-driven transcription. Am J Hum Genet. 2011;89:572–579. doi: 10.1016/j.ajhg.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Al-Agha AE, Ahmed IA, Nuebel E, et al. Primary ovarian insufficiency and azoospermia in carriers of a homozygous PSMC3IP stop gain mutation. J Clin Endocrinol Metab. 2018;103:555–563. doi: 10.1210/jc.2017-01966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang J, Zhang W, Jiang H, Wu B-L, Primary Ovarian Insufficiency Collaboration Mutations in HFM1 in recessive primary ovarian insufficiency. N Engl J Med. 2014;370:972–974. doi: 10.1056/NEJMc1310150. [DOI] [PubMed] [Google Scholar]
- 65.Kasak L, Punab M, Nagirnaja L, et al. Bi-allelic recessive loss-of-function variants in FANCM Cause non-obstructive azoospermia. Am J Hum Genet. 2018;103:200–212. doi: 10.1016/j.ajhg.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oud MS, Smits RM, Smith HE, et al. A de novo paradigm for male infertility. Nat Commun. 2022;13:154. doi: 10.1038/s41467-021-27132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vissers LELM, de Ligt J, Gilissen C, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S6: List of variants detected in the cohort.
Table S7: Molecular findings in the cohort.