Summary
The central step in the initiation of eukaryotic DNA replication is the loading of the minichromosome maintenance 2–7 (MCM2-7) complex, the core of the replicative DNA helicase, onto chromatin at replication origin. Here, we reported the cryo-EM structure of endogenous human single hexameric MCM2-7 complex with a resolution at 4.4 Å, typically an open-ring hexamer with a gap between Mcm2 and Mcm5. Strikingly, further analysis revealed that human MCM2-7 can self-associate to form a loose double hexamer which potentially implies a novel mechanism underlying the MCM2-7 loading in eukaryote. The high-resolution cryo-EM structure of human MCM2-7 is critical for understanding the molecular mechanisms governing human DNA replication, especially the MCM2-7 chromatin loading and pre-replicative complex assembly.
Subject areas: Molecular biology, Cell biology, Structural biology
Graphical abstract
Highlights
-
•
A Twin-Strep-Tag II tag was fused to Mcm4 by using CRISPR-Cas9 technique
-
•
The endogenous human MCM2-7 complex was successfully purified
-
•
The high-resolution cryo-EM structure of human hexameric MCM2-7 complex
-
•
The human single MCM2-7 hexamer can self-associate to form a double hexamer
Molecular biology; Cell biology; Structural biology
Introduction
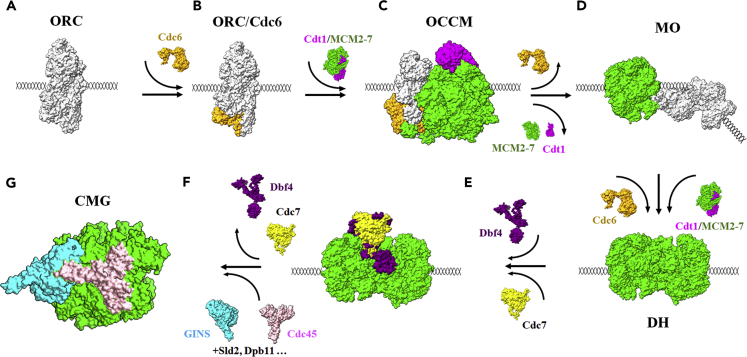
DNA replication is the heart of cell proliferation and has been the subject of intense investigations over many years. In eukaryotes, DNA replication starts with ATP-dependent association of the origin recognition complex (ORC) consisting of Orc1-6 subunits and binding with DNA at a particular position in chromosomes called “origin” (Bell and Stillman, 1992). In G1 phase, ORC binding to replication origin serves as an origin marker and recruits the initiation factor Cdc6 and Cdt1 to origin for the chromatin loading of the minichromosome maintenance 2–7 (MCM2-7) heterohexamer onto double-stranded DNA to form the ORC/Cdc6/Cdt1/MCM2-7 (OCCM) complex (Feng et al., 2021; Fernandez-Cid et al., 2013; Li et al., 2018; Sun et al., 2013; Yuan et al., 2017) (Figures 1A, 1B, and 1C). Subsequently, an ordered release of Cdc6 and Cdt1 results in the formation of an MCM2-7/ORC (MO) intermediate (Miller et al., 2019; Ticau et al., 2015) (Figure 1D). Following the recruitment of a second Cdc6, the formation of the ORC/Cdc6/MCM2-7 (OCM) intermediate rapidly recruits a second MCM2-7 hexamer in a Cdt1-dependent manner yielding a salt-stable, head-to-head MCM2-7 double hexamer (DH) that encircles dsDNA (Evrin et al., 2009, 2013; Fernandez-Cid et al., 2013; Miller et al., 2019; Remus et al., 2009; Ticau et al., 2015) (Figure 1E). The double hexamer loading to form the pre-replication complex (pre-RC) is also known as DNA licensing. The resultant double hexamer lacks helicase activity until the cell enters S phase. At the G1-S transition of a yeast cell, Dbf4-dependent kinase and S-phase-specific cyclin-dependent kinase coordinating with a large number of activation factors including Sld3, Cdc45, Sld2, Dpb11, polymerase ε, and Mcm10 transform the inactive double hexamer into two active Cdc45/MCM2-7/GINS (CMG) helicases (Cheng et al., 2022; Georgescu et al., 2017; Heller et al., 2011; Labib, 2010; Saleh et al., 2022; Sheu and Stillman, 2006; Yeeles et al., 2017) (Figures 1F and 1G). The CMG complex is thought to serve in S phase as the replicative helicase component of the replisome for daughter-strand synthesis.
Figure 1.
Eukaryotic initiation of DNA replication
The known complexes in yeast that have been solved by Cryo-EM are shown in surface view.
(A) ORC is first bound to DNA (PDB: 5ZR1).
(B) Cdc6 is loaded onto ORC to form the landing platform for Cdt1/MCM2-7 (PDB: 7MCA 5381).
(C) The association of Cdt1/MCM2-7 (PDB: 5XF8) with ORC/Cdc6 results in the OCCM (PDB: 5V8F) formation.
(D) With the release of Cdt1 and Cdc6 from the OCCM, MO is an essential intermediate for the recruitment of a second Cdt1/MCM2-7 heptamer (PDB: 6RQC).
(E) In cooperation of the Cdc6, a MCM2-7 DH is formed which is activated in S phase (PDB: 6F0L).
(F) In S phase, relying on Dbf4-Cdc7 kinase (PDB: 7V3V) and coordinating with other activation factors such as Cdc45, Sld2, Dpb11 etc. to form the active Cdc45/MCM2-7/GINS (CMG) complex (PDB: 5U8S).
The central complex of DNA replication is MCM2-7 which includes six homologous proteins, Mcm2, 3, 4, 5, 6, and 7, to form a heterohexamer (Bochman and Schwacha, 2009; Forsburg, 2004). Recently, with the technical evolution of cryo-electron microscopy (cryo-EM), a few of critical components of pre-RC containing MCM2-7, such as OCCM, MO, and DH involved in DNA replication of Saccharomyces cerevisiae, became available at sub-nanometer resolution (Riera et al., 2017; Zhai et al., 2017b). All of these structures provide a series of snapshots showing a glimpse of the conformational changes of MCM2-7 during DNA replication licensing in yeast. Although structural studies of eukaryotic-isolated MCM2-7 complexes including Encephalitozoon cuniculi MCM2-7 (Lyubimov et al., 2012), S. cerevisiae MCM2-7 (Li et al., 2015; Remus et al., 2009; Samel et al., 2014), and Homo sapiens MCM2-7 (Boskovic et al., 2016; Hesketh et al., 2015) have been reported, until now, the high-resolution structure of eukaryotic-isolated MCM2-7 complex remains elusive. In this study, we purified endogenous human MCM2-7 complex and determined the cryo-EM structure of human single hexameric MCM2-7 complex with a resolution at 4.4 Å, which shows a similar architecture with yeast MCM2-7 stabilized by Cdt1, typically an open-ring hexamer with a gap between Mcm2 and Mcm5. Strikingly, further biochemistry and EM analysis revealed that human MCM2-7 can self-associate to form a loose double hexamer when compared with the loaded yeast MCM2-7 DH. The high-resolution cryo-EM structure of human MCM2-7 is critical for the understanding of the molecular mechanisms governing human DNA replication and may ultimately contribute to facilitate the structure-based drug design for cancer treatments.
Results
Purification of the endogenous human MCM2-7 complex
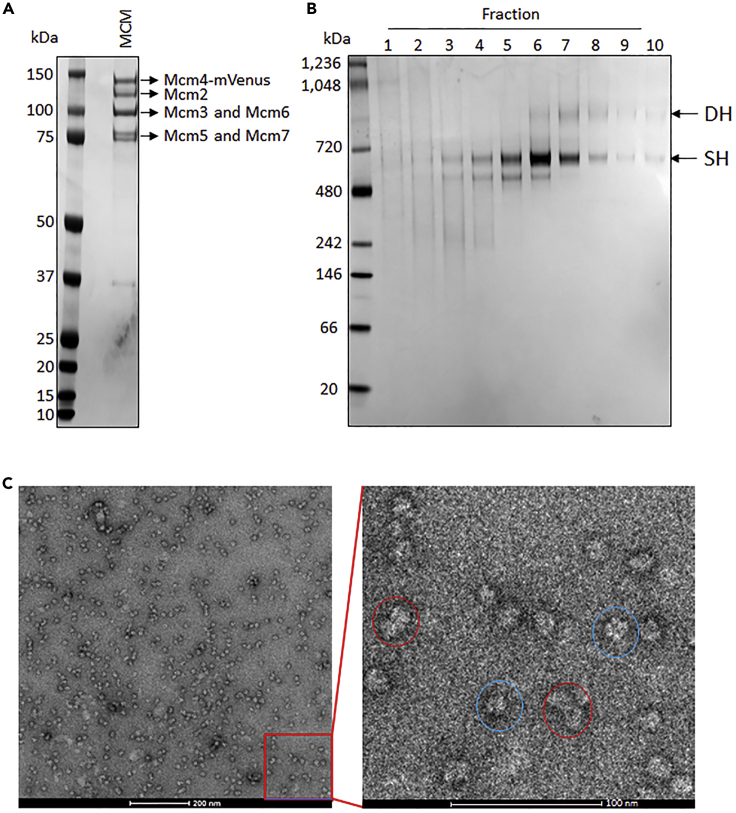
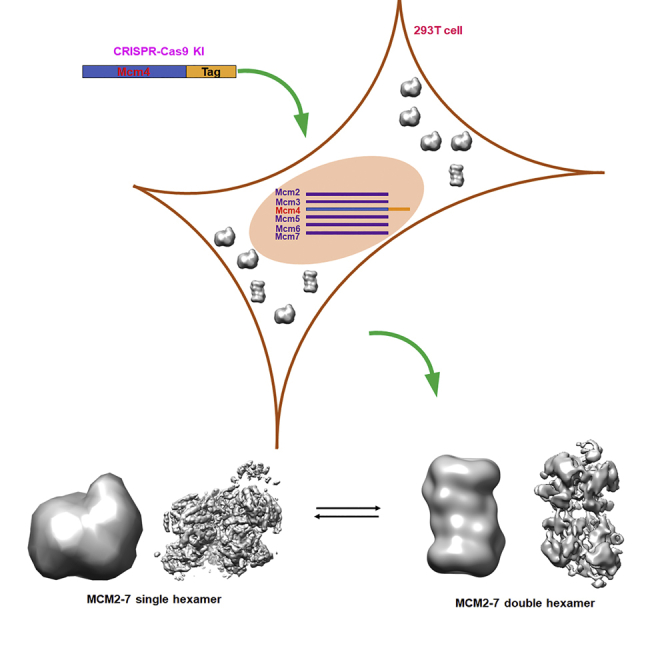
To obtain the endogenous human MCM2-7 complex, a Twin-Strep-Tag II tag was fused to Mcm4 by using CRISPR-Cas9 technique as described in STAR Methods. The strep-tagged protein was purified by affinity column and analyzed by SDS-PAGE (Figure 2A), which showed four groups of protein bands with similar intensity. The further mass spectrometer analysis showed that these bands correspond to Mcm4 together with Mcm2, 3, 5, 6, and 7 indicating we have successfully obtained the endogenous human MCM2-7 complex (Figures 2A and S1).
Figure 2.
Purification of human MCM2-7 complex
(A) Analysis of human MCM2-7 complex by SDS-PAGE.
(B) MCM2-7 was grafixed and analyzed by native gel. DH, MCM2-7 double hexamer. SH, MCM2-7 single hexamer.
(C) Negative staining of MCM2-7 complex. The Fraction 6 in (B) was collected for negative staining test and the DH and SH are shown in red and blue circle, respectively.
Endogenous human MCM2-7 complex shows the co-existence of single and double hexamers
Subsequently, the gradient fixation was performed using the purified MCM2-7 complex by affinity column. Interestingly, two separated bands were observed on native PAGE gel indicating two types of conformation are adopted by human MCM2-7 complexes (Figure 2B). Interestingly, the subsequent negative staining examination reveals that the band with slow migration rate corresponds to MCM2-7 double hexamer (DH) and the other band corresponds to MCM2-7 single hexamer (SH) (Figures 2C and S1). Notably, the band position corresponding to MCM2-7 DH migrated between 720 and 1,048 kDa on native PAGE, which is smaller than the theoretic molecular weight ∼1,200 kDa, potentially caused by its particular shape (non-globular shape) and/or dynamic property such as conformational equilibrium.
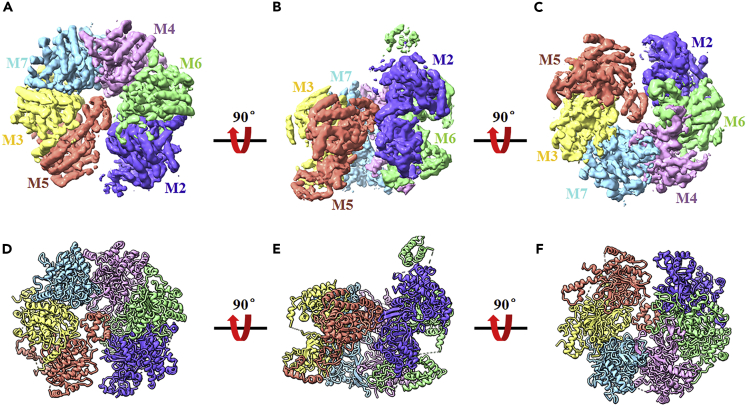
Overall cryo-EM structures of the endogenous human MCM2-7 complex
The image acquisition was carried out on a Titan Krios microscope equipped with a K3 camera operating at 300 keV (Figure S2A and Table S1). The subsequent 2D and 3D classifications and refinements rendered a 4.4 Å map of MCM2-7 from a set of 1,135,993 particles (Figures S2B–S2D and S3). The excellent quality and high resolution of the map allowed us to assign and build models for each component of MCM2-7 (Figure 3).
Figure 3.
Overall structure of the human MCM2-7 single hexamer
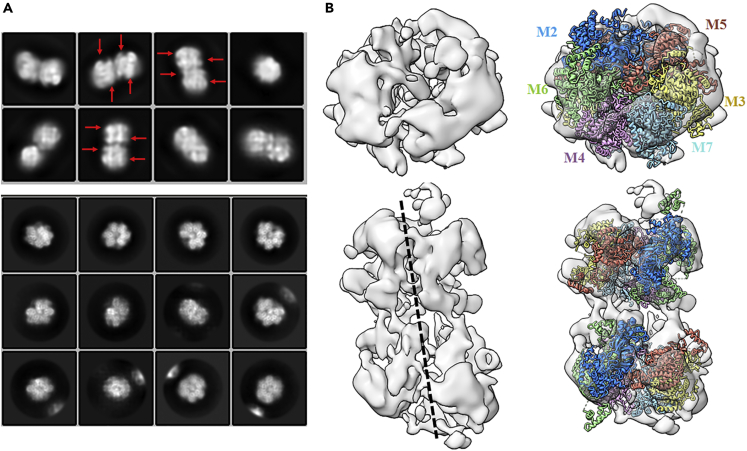
(A–C) The cryo-EM map of the MCM2-7 single hexamer shown in surface representation, with subunits color-coded as indicated by the labels. Shown are the top, side, and bottom views (A–C).
(D–F) The atomic model of human MCM2-7 single hexamer shown in cartoon mode. Mcm2 blue, Mcm3 yellow, Mcm4 purple, Mcm5 red, Mcm6 green, and Mcm7 cyan.
The six-subunit assembly of the human MCM2-7 shows a left-handed spiral configuration, proceeding Mcm5→Mcm3→Mcm7→Mcm4→Mcm6→Mcm2, with a gate between Mcm2 and Mcm5 (Figure 3). This hexameric open-ring structure shares similar structure with the cryo-EM structure of yeast MCM2-7 in complex with Cdt1 (Cdt1/MCM2-7 complex), with RMSD value ∼2.22 Å (Figure S4) (Zhai et al., 2017a). When compared to the active state of human MCM2-7 in the Cdc45/MCM2-7/GINS (CMG) complex and the inactive state of MCM2-7 in yeast double hexamer, the corresponding RMSD values of structural alignments are 7.29 and 6.01 Å, indicating the conformational changes of human MCM2-7 in the process of loading and activation (Figure S4). Additionally, the distance of the gap between Mcm2 and Mcm5 is ∼10–15 Å, which is similar to the gap observed in yeast Cdt1/MCM2-7 complex.
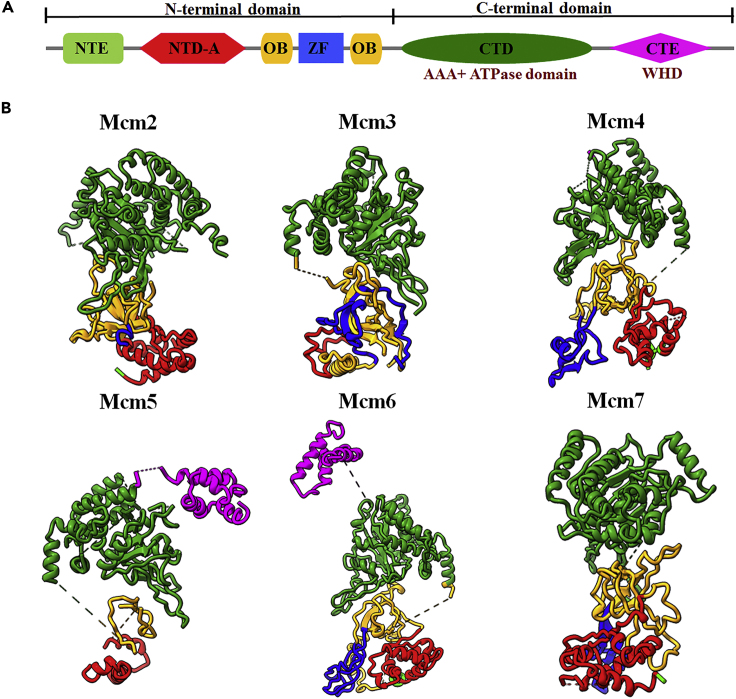
Structure of the endogenous human hexameric MCM2-7 complex
The six homologous subunits making up MCM2-7 are very well conserved from yeast to human (Zhai et al., 2017b). The MCM2-7 proteins are composed of two domains: (1) the N-terminal domain which comprises N-terminal extension (NTE), N-terminal domain A (NTD-A), oligonucleotide-binding fold (OB), and zinc finger (ZF) contributing to assembly and DNA binding and (2) the C-terminal domain containing the highly conserved AAA + ATPase domain (CTD) and C-terminal extension (CTE) which is composed of wing helix domain (WHD) (Figure 4A). The length of NTE varies among the six subunits. The NTEs of Mcm2 and Mcm4 include 191 and 163 amino acids, most of which are not visible in our structure indicating these regions are flexible (Figure 4B). Although the length of NTE in other four subunits is less than 25 amino acids, several residues of these regions can be observed except Mcm5. The NTD-A, OB, and ZF of each subunit are well resolved except Mcm5 (Figures 4B and S5A). The N-terminal domain of Mcm5 is not well resolved possibly due to its high flexibility as one of the gate-forming subunits. As shown in Figure 4, the CTD of all six subunits are well resolved based on the excellent quality of the map (Figures 4B and S5B). A domain-swapped helix in each CTD tethers each MCM to its neighboring subunit (Figure S5C). In addition to CTD, each MCM subunit contains a CTE composed of a WHD. However, only the WHDs of Mcm2, Mcm5, and Mcm6 were well resolved in electron density map and could be modeled (Figure 3B). The WHD of Mcm5 could be identified and deposited in central channel of MCM2-7 and the WHD of Mcm6 sits on the rim of CTD (Figure S5D) which is involved in the binding to Cdt1 from our previous NMR structure studies (Liu et al., 2012; Wei et al., 2010).
Figure 4.
The conformation of each MCM2-7 subunit
(A) Schematic illustration of domain organization and subunit-specific features of MCM2-7 subunits. NTE, N-terminal extension; NTD-A, N-terminal domain A; OB, oligonucleotide-binding fold; ZF, zinc finger; CTD, C-terminal domain; CTE, C-terminal extension which contains the WHD, wing helix domain.
(B) Individual MCM subunits.
The double hexamer of endogenous human MCM2-7 complex
Surprisingly, in addition to the expected fully assembled human MCM2-7 single hexamer, we found an additional complex which was a MCM2-7 double hexamer (DH) with a ∼30% of population. As shown in Figure 5A, particles containing four layers, corresponding to DH, have been observed in the 2D class averages of cryo-EM data. The raw image data of DH particles allowed a 3D EM reconstruction at 16 Å resolution (Figures S2 and S3). The 3D map reveals that the MCM2-7 DH has an approximately cylindrical structure with 260 Å long and 150 Å wide (Figure S6). The two hexamers slightly tilt from the cylindrical axis similar to the observation in EM structure of yeast DH (Figure 5B). Interestingly, the DH displays C1 symmetry in our model, while the yeast DH shows C2 symmetry. However, the quality and resolution of the map did not permit us to build a model at atomic resolution of the DH of MCM2-7. Specifically, the EM images revealed that the DHs did not further interact to form higher-order oligomers indicating the existence SH-DH equilibrium of MCM2-7 in solution. Interestingly, the sparse electron density of inter-hexamer interface indicated that the head-to-head stacking is weaker than that in the yeast DH loaded onto DNA, as the length of human DH is ∼45 Å longer than that of yeast DH (Figure S6). This result further demonstrated that DH of human MC2-7 observed in this study is potentially a transient state of the SH which may be a relative stable form in vivo.
Figure 5.
The double hexamer exists with single hexamer
(A) 2D class averages of cryo-EM data. The red arrows indicate the four layers in DH.
(B) The left panel is cryo-EM map of human double hexamer, the right panel is the map fit with human single hexamer, produced by Chimera, with subunits color-coded as indicated by the labels.
Discussion
Here, we reported the cryo-EM structure of endogenous human MCM2-7 single hexamer at an atomic resolution. This structure indicates that the endogenous human MCM2-7 single hexamer is almost identical to that of yeast MCM2-7 in complex with Cdt1 (Zhai et al., 2017a), typically an open-ring hexamer with a gap between Mcm2 and Mcm5 (all atom RMSD ∼ 2.22 Å based on structural alignment, Figure S4).
Until recently, four complexes, the Cdt1/MCM2-7 (CM) heptamer (7.1 Å) (Zhai et al., 2017a), the ORC/Cdc6/Cdt1/MCM2-7 (OCCM) intermediate (3.9 Å) (Yuan et al., 2017), the MCM2-7 double hexamer (MCM-DH) (3.8 Å) (Li et al., 2015), and the MCM2-7/ORC (MO) intermediate (4.4 Å) (Miller et al., 2019), representing prominent stages of DNA replication in yeast, have been structurally characterized by cryo-EM at high resolution. The conformations of MCM2-7 in these complexes are different, revealing the distinct functional states (Riera et al., 2017; Zhai et al., 2017b). Comparing with MCM2-7 in these yeast complexes, no changes are observed in structural core/fold of each MCM subunit in our human MCM2-7 complex (Figure S7). However, the NTDs of human MCM2-7 SH in our study are not well resolved due to the high flexibility in comparison with the NTDs in the yeast Cdt1/MCM2-7 (CM) heptamer (Figures S7 and S8), indicating the NTDs of Mcm2, Mcm4, and Mcm6 are stabilized through the interaction with Cdt1 in the yeast Cdt1/MCM2-7 (CM) heptamer (Zhai et al., 2017a). Especially, the NTD of human Mcm5 is poorly resolved and almost non-discernible potentially due to the lack of other components for stabilizing the conformation (Figure S8). Although there are no direct interactions between Mcm5 and Cdt1 in the yeast Cdt1/MCM2-7 (CM) heptamer, the network of interactions induced by Cdt1 potentially plays a major role in stabilization of the NTD of Mcm5 (Figure S8).
The previous biochemical experiments have shown the important role of the CTE of MCM subunits in pre-RC formation (Fernandez-Cid et al., 2013; Frigola et al., 2013). In this structure, the CTEs of human Mcm5 and Mcm6 are well resolved. The CTE of Mcm5 occludes in the central channel in which other subunits fix the position of CTE making it visible. In the previous study, the CTE of Mcm6 shows to interact with Cdt1 (Liu et al., 2012; Wei et al., 2010) and displays an auto-inhibitory function preventing MCM2-7 interaction with ORC-Cdc6 in the absence of Cdt1 (Fernandez-Cid et al., 2013). Although no interaction between CTE of Mcm6 and Cdt1 is observed in the yeast Cdt1-MCM2-7 (CM) (Zhai et al., 2017a), the CTE of Mcm6 is observable in our human MCM2-7 indicating potentially an unknown functional state. Furthermore, in order to visualize the differences of the human MCM2-7 single hexamer in apo form and CMG, a structural alignment was performed separately for the CTDs and NTDs of each MCM subunit (Figures S9 and S10 and Videos S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, and S12). This analysis revealed that the NTDs and CTDs of MCM2-7 hexamer in the CMG twisted in the same direction in comparison with the ones in the single hexamer reported here.
Until now, only two human MCM2-7 structures at an atomic level are reported, human MCM2-7 single hexamer (termed apo form in this work) and Cdc45/MCM2-7/GINS complex (CMG) (Rzechorzek et al., 2020). Because of the highly structural similarity between the human and yeast MCM2-7 single hexamer, potentially the conformational changes and functional states of MCM2-7 in different stages of DNA replication are similar and conserved from yeast to human.
The previous studies by EM method showed that MCM2-7 appears to exist as a mixture of open and closed rings (Costa et al., 2011; Frigola et al., 2017) and only closed rings were detected after the treatment of ATPγS (Frigola et al., 2017). Although it was reported that the interaction of yeast MCM2-7 with ATPγS promotes closure of Mcm2-Mcm5 gate and subsequently Cdt1 stabilizes the Mcm2-Mcm5 gate, only the open ring form of human MCM2-7 is observed after treatment of ATPγS in the absence of Cdt1 (Figure 3). Recently, it was reported that the MCM-binding protein (MCMBP) binds to Mcm3 through its N-terminal domain, which results in promoting the assembly of MCM2-7 complex in human cells (Saito et al., 2022). However, we did not observe both MCMBP and Cdt1 during the preparation of MCM2-7 complex, indicating that the MCM2-7 hexamer is very stable once the six subunits assembled together.
Strikingly, our EM analysis reveals an MCM-DH structure with 16 Å resolution (Figure 4) indicating the human MCM2-7 hexamer may self-associate to form a double hexamer in solution. Interestingly, it is reminiscent of the Archaeal MCM proteins which form a tight double hexamer without loading in comparison with the yeast MCM proteins (Kelman et al., 2020). By docking the cryo-EM structure of the human MCM2-7 hexamer solved here into the DH density, we found the self-associated DH is not compact in comparison with the yeast DH loaded onto DNA (Abid Ali et al., 2017; Li et al., 2015) indicating a weaker interaction at the inter-hexamer interface (Figure 4). Currently, there are three different models that have been proposed for description of MCM2-7 loading onto the replication origin based on the biochemical and structural studies (Kyei Barffour and Acheampong, 2021). In each model, the MCM2-7 complex is loaded onto DNA through the cooperation with ORC, Cdc6, and Cdt1 in a single hexameric form. The self-assembly of human MCM2-7 observed here could then be recruited by a single ORC complex, revealing a potential different model for the initiation of DNA replication.
Limitations of the study
In this study, we determined the high-resolution cryo-EM structure of human MCM2-7 SH and revealed that the endogenous human MCM2-7 can self-associate to form a well-defined hexameric structure and a loose double hexamer (DH). However, the resolution of human MCM2-7 DH reported here was not sufficient to provide detailed packing interactions of the two human MCM2-7 complexes, which is important for understanding the mechanism of the self-assembly of human MCM2-7 complex.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Guang Zhu (gzhu@ust.hk).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
293T cells were maintained in DMEM media with 1% penicillin-streptomycin and 10% fetal bovine serum (Life Technologies) and cultured in a humidified incubator at 37°C with 5% CO2. HEK293T Mcm4-2xStrepII cells were maintained in Union293 media (Union-biotech) and shaken at 120 rpm in a humidified orbital shaker at 37°C with 8% CO2.
Method details
Generation of stable cell line expressing Mcm4-2xStrepII
The Twin-Strep-Tag II (2xStrepII, WSHPQFEKGGGSGGGSGGSAWSHPQFEK) was knocked into the C-terminus of Mcm4 by using CRISPR-Cas9 technique. Briefly, sgRNA was designed to target 55-bp-region immediate after the stop codon of Mcm4. In the template donor plasmid, the C-terminus of Mcm4 was in-framed fused with mVenus and 2xStrepII. The sgRNA-targeted sequence (55 bp) was deleted from the template donor plasmid. The sgRNA together with the template donor plasmid were co-transfected into 293T cells by using Lipofectamine 3000 (ThermoFisher). One day after transfection, 293T cells were diluted into 96-well plate at the density of 0.5 cell/well. Two weeks later, the mVenus-positive colonies were selected. The insertion of mVenus and 2xStrepII sequences was verified by PCR and sequencing. Finally, one Mcm4-2xStrepII stable cell line was chosen for MCM2-7 complex purification.
Purification of Mcm4-2xStrepII
The Mcm4-2xStrepII stable cell line was adapted to suspension culture. One-liter stable cell culture was centrifuged, collected, lysed and sonicated with 50 mL lysis buffer (Tris 10 mM pH7.4, 150 mM NaCl, 5 mM MgCl2, 0.25% Triton X-100, and protease and phosphatase inhibitors). The resultant cell lysate was treated with Benzonase and incubated with Strep-Tactin Superflow resin. After washing with lysis buffer, the protein was eluted with 30 mM biotin. The final purified proteins range from 200 to 300 μg per liter culture. ATPγS was added in the buffer during the preparation for negative staining and cryo-EM.
Negative staining
MCM2-7 sample was diluted to 0.01 mg/mL with PBS. Aliquot of 7 μL was applied to glow-discharged carbon-coated copper grids. After incubation for 1 min, excess sample solution was removed by filter paper. The grid was stained with 7 μL 2% uranyl acetate for 30 s for 3 times. The stained grid was examined using an FEI Tecnai T20 electron microscope operated at 120 kV.
Gradient fixation
The concentrated sample then applied to the top of a 20–40% glycerol gradient in buffer (20 mM HEPES, pH 7.5, 150 mM NaCl), which contains 0.3% glutaraldehyde in the denser solution. The gradients were centrifuged in a TLS-55 rotor (Beckman Optima TLX ultracentrifuge) at 250,000g for 5 h. The fractions were collected and the fixation reactions were stopped by 50 mM Tris at the terminal concentration.
Grids preparation and data collection
To prepare cryo-EM grids, the glycerol was removed by buffer exchange through centrifugation with 20 mM HEPES at pH 7.5 and 150 mM NaCl. Aliquots of 3 μL sample at concentration of 0.3 mg/mL were applied to glow discharged holey carbon grids (Quantifoil, Cu 200 mesh, R2/2). The grids were blotted for 2.5 s in 100% humidity at 8°C and were then immediately plunged into liquid ethane by using a Vitrobot Mark IV (Thermo Fisher). The grids were examined on a Titan Krios microscope G3i (Thermo Fisher) operated at 300 kV equipped with Gatan BioQuantum energy filter and K3 direct electron detector (Gatan). Micrographs were collected in electron counting mode, at a nominal magnification of 1,050,00x, which yielded a pixel size of 1.06 Å at objective scale. Defocus values were set in a range from −1.5 to −2.5 μm.
Image processing
The original image stacks for drift and beam-induced motion at the micrograph level were corrected by using MotionCorr2 (Li et al., 2013). Contrast-transfer-function parameters were estimated with CTFFIND4 (Rohou and Grigorieff, 2015). Particles were automatically selected using the Gautomatch program (http://www.mrc-lmb.cam.ac.uk/kzhang/). Particles extraction, 2D and 3D classification and refinement were done with RELION 3.0.8 (Zivanov et al., 2018). A total of 1,261,069 raw particles (with a binning factor of four) from 12,196 micrographs were subjected to reference-free 2D classification. After the 2D-classification-based particle screening, 1,135,993 particles were subjected to 3D classification. It shown that 30% was double hexamer, and 70% was single hexamer. For single hexamer, the reference for 3D classification was converted from the MCM2-7 model (PDB 5XF8) and filtered to 60 Å. After 3D classification, four out of nine models were selected to produce the multi-reference models to perform 3D classification and the models were not filtered. Subsequently, the most reasonable model was selected to perform 3D classification further. Finally, 186,918 particles were assigned to the hexamer complex. These particles were then subjected to reference-based refinement. The maps were further processed with the post-processing options of RELION with a negative B-factor of 104 Å2, and the modulation transfer function of the detector was also corrected. The final resolutions of the hexamer complex were 4.4 Å (gold-standard FSC 0.143 criteria). The map for model building was further processed by deepEMhancer (Sanchez-Garcia et al., 2021). For double hexamer, the reference for 3D classification was converted from the map (EMD_6338) and filtered to 60 Å. Finally, 68,909 particles were subjected to reference-based refinement. Subsequent post-processing, the final resolution of double hexamer was 16 Å. Local resolution variations were estimated using ResMap (Kucukelbir et al., 2014).
Model building
The MCM2-7 in Saccharomyces cerevisiae Cdt1/MCM2–7 structure (PDB 5XF8) was used for homologous modeling through the SWISS-MODEL server (Bertoni et al., 2017). The generated models were initially fitted into the human MCM2-7 density map using UCSF Chimera (Pettersen et al., 2004), respectively. After that, the fitted model was rebuilt using RosettaCM (Song Y, 2013) and the best output model was selected according to the energy and fitness of the model to the EM density map. To build the density map completely, Chimera UCSF was used to select and save the C terminal domain of Mcm2 (aa857 - 904) and Mcm6 (aa719 – 770) from the human CMG model (PDB 6XTX), then these domains were merged into the corresponding monomers by COOT (Emsley P, 2010). The merged model was rebuilt by RosettaCM. Finally, the generated model was selected and refined by the PHENIX cryoEM Real-space Refinement tool (Adams et al., 2010), with rotamer restraints, Ramachandran plot restraints enabled. The superposition of models and RMSD values were calculated by COOT and figures were prepared using Chimera UCSF (Pettersen et al., 2004) and ChimeraX (Goddard et al., 2018). Statistics for data collection, processing and refinement are reported in Table S1.
Quantification and statistical analysis
This study does not include quantification or statistical analysis.
Acknowledgments
We thank the cryo-EM platform of Tsinghua University, Cryo-EM Center of Southern University of Science and Technology, and the biological Cryo-EM Center at the Hong Kong University of Science and Technology (HKUST) for data collection. This work was supported by the National Natural Science Foundation of China (32071188 to C.L.; 31925023, 21827810 to Y.X.), Hong Kong Special Administrative Region (GRF16103719, GRF16101120, GRF16101121, SZSTI19SC02, SMSEGL20SC01-H, AoE/M-403-16, AoE/M-401/20, VPRDO19RD03-6 to G.Z.), the Ministry of Science and Technology of the People's Republic of China (2021YFA1300204 to Y.X.), the Beijing Frontier Research Center for Biological Structure and the Beijing Advanced Innovation Center for Structure Biology to Y.X., Guangdong Basic and Applied Basic Research Foundation (2020A1515010034 to C.L., 2021A1515220104 to N.X.).
Author contributions
N.X. and H.T. expressed and purified the MCM2-7 complex and prepared the cryo-EM sample. C.M.S. contributed to purify the MCM2-7 protein. Q.L. H.Y and P.W. collected cryo-EM data. Q.L. determined the cryo-EM structure. Q.L. and C.L. performed structural analysis and interpreted data with H.T. and N.X. N.X., H.T., Q.L., and C.L. wrote the manuscript. Y.X. and G.Z. supervised and oversaw the project.
Declaration of interests
The authors declare no competing interests.
Published: September 16, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104976.
Contributor Information
Ye Xiang, Email: yxiang@mail.tsinghua.edu.cn.
Guang Zhu, Email: gzhu@ust.hk.
Supplemental information
Data and code availability
-
•
Data: The cryo-EM map and structure coordinates of the single MCM2-7 hexamer produced in this study have been deposited at EMDB (https://www.emdataresource.org/) and PDB (https://www.rcsb.org/) with the accession codes of EMD-32326 and 7W68, respectively.
-
•
Code: This paper does not report original code.
-
•
Additional information: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abid Ali F., Douglas M.E., Locke J., Pye V.E., Nans A., Diffley J.F.X., Costa A. Cryo-EM structure of a licensed DNA replication origin. Nat. Commun. 2017;8:2241. doi: 10.1038/s41467-017-02389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/s0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Bertoni M., Kiefer F., Biasini M., Bordoli L., Schwede T. Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-09654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman M.L., Schwacha A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 2009;73:652–683. doi: 10.1128/MMBR.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovic J., Bragado-Nilsson E., Saligram Prabhakar B., Yefimenko I., Martínez-Gago J., Muñoz S., Méndez J., Montoya G. Molecular architecture of the recombinant human MCM2-7 helicase in complex with nucleotides and DNA. Cell Cycle. 2016;15:2431–2440. doi: 10.1080/15384101.2016.1191712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Li N., Huo Y., Dang S., Tye B.K., Gao N., Zhai Y. Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase. Nat. Commun. 2022;13:1396. doi: 10.1038/s41467-022-29070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A., Ilves I., Tamberg N., Petojevic T., Nogales E., Botchan M.R., Berger J.M. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/s0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C., Clarke P., Zech J., Lurz R., Sun J., Uhle S., Li H., Stillman B., Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci. USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C., Fernández-Cid A., Zech J., Herrera M.C., Riera A., Clarke P., Brill S., Lurz R., Speck C. In the absence of ATPase activity, pre-RC formation is blocked prior to MCM2-7 hexamer dimerization. Nucleic Acids Res. 2013;41:3162–3172. doi: 10.1093/nar/gkt043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Noguchi Y., Barbon M., Stillman B., Speck C., Li H. The structure of ORC-Cdc6 on an origin DNA reveals the mechanism of ORC activation by the replication initiator Cdc6. Nat. Commun. 2021;12:3883. doi: 10.1038/s41467-021-24199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Cid A., Riera A., Tognetti S., Herrera M.C., Samel S., Evrin C., Winkler C., Gardenal E., Uhle S., Speck C. An ORC/Cdc6/MCM2-7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol. Cell. 2013;50:577–588. doi: 10.1016/j.molcel.2013.03.026. [DOI] [PubMed] [Google Scholar]
- Forsburg S.L. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 2004;68:109–131. doi: 10.1128/mmbr.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J., He J., Kinkelin K., Pye V.E., Renault L., Douglas M.E., Remus D., Cherepanov P., Costa A., Diffley J.F.X. Cdt1 stabilizes an open MCM ring for helicase loading. Nat. Commun. 2017;8 doi: 10.1038/ncomms15720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigola J., Remus D., Mehanna A., Diffley J.F.X. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–343. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu R., Yuan Z., Bai L., de Luna Almeida Santos R., Sun J., Zhang D., Yurieva O., Li H., O'Donnell M.E. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc. Natl. Acad. Sci. USA. 2017;114:E697–E706. doi: 10.1073/pnas.1620500114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller R.C., Kang S., Lam W.M., Chen S., Chan C.S., Bell S.P. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh E.L., Parker-Manuel R.P., Chaban Y., Satti R., Coverley D., Orlova E.V., Chong J.P.J. DNA induces conformational changes in a recombinant human minichromosome maintenance complex. J. Biol. Chem. 2015;290:7973–7979. doi: 10.1074/jbc.M114.622738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman L.M., O'Dell W.B., Kelman Z. Unwinding 20 Years of the archaeal minichromosome maintenance helicase. J. Bacteriol. 2020;202 doi: 10.1128/JB.00729-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyei Barffour I., Acheampong D.O. Prospect of reprogramming replication licensing for cancer drug development. Biomed. Pharmacother. 2021;136 doi: 10.1016/j.biopha.2020.111190. [DOI] [PubMed] [Google Scholar]
- Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Lam W.H., Zhai Y., Cheng J., Cheng E., Zhao Y., Gao N., Tye B.K. Structure of the origin recognition complex bound to DNA replication origin. Nature. 2018;559:217–222. doi: 10.1038/s41586-018-0293-x. [DOI] [PubMed] [Google Scholar]
- Li N., Zhai Y., Zhang Y., Li W., Yang M., Lei J., Tye B.K., Gao N. Structure of the eukaryotic MCM complex at 3.8 angstrom. Nature. 2015;524:186–191. doi: 10.1038/nature14685. [DOI] [PubMed] [Google Scholar]
- Li X., Mooney P., Zheng S., Booth C.R., Braunfeld M.B., Gubbens S., Agard D.A., Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wu R., Zhou B., Wang J., Wei Z., Tye B.K., Liang C., Zhu G. Structural insights into the Cdt1-mediated MCM2-7 chromatin loading. Nucleic Acids Res. 2012;40:3208–3217. doi: 10.1093/nar/gkr1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimov A.Y., Costa A., Bleichert F., Botchan M.R., Berger J.M. ATP-dependent conformational dynamics underlie the functional asymmetry of the replicative helicase from a minimalist eukaryote. Proc. Natl. Acad. Sci. USA. 2012;109:11999–12004. doi: 10.1073/pnas.1209406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T.C.R., Locke J., Greiwe J.F., Diffley J.F.X., Costa A. Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM. Nature. 2019;575:704–710. doi: 10.1038/s41586-019-1768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Remus D., Beuron F., Tolun G., Griffith J.D., Morris E.P., Diffley J.F.X. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera A., Barbon M., Noguchi Y., Reuter L.M., Schneider S., Speck C. From structure to mechanism-understanding initiation of DNA replication. Genes Dev. 2017;31:1073–1088. doi: 10.1101/gad.298232.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A., Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzechorzek N.J., Hardwick S.W., Jatikusumo V.A., Chirgadze D.Y., Pellegrini L. CryoEM structures of human CMG-ATP gamma S-DNA and CMG-AND-1 complexes. Nucleic Acids Res. 2020;48:6980–6995. doi: 10.1093/nar/gkaa429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito Y., Santosa V., Ishiguro K.I., Kanemaki M.T. MCMBP promotes the assembly of the MCM2-7 hetero-hexamer to ensure robust DNA replication in human cells. Elife. 2022;11 doi: 10.7554/eLife.77393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A., Noguchi Y., Aramayo R., Ivanova M.E., Stevens K.M., Montoya A., Sunidhi S., Carranza N.L., Skwark M.J., Speck C. The structural basis of Cdc7-Dbf4 kinase dependent targeting and phosphorylation of the MCM2-7 double hexamer. Nat. Commun. 2022;13:2915. doi: 10.1038/s41467-022-30576-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samel S.A., Fernández-Cid A., Sun J., Riera A., Tognetti S., Herrera M.C., Li H., Speck C. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Genes Dev. 2014;28:1653–1666. doi: 10.1101/gad.242404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garcia R., Gomez-Blanco J., Cuervo A., Carazo J.M., Sorzano C.O.S., Vargas J. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. Commun. Biol. 2021;4:874. doi: 10.1038/s42003-021-02399-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu Y.J., Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol. Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., DiMaio F., Wang R.Y.R., Kim D., Miles C., Brunette T., Thompson J., Baker D. High-resolution comparative modeling with RosettaCM. Structure. 2013;21:1735–1742. doi: 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Evrin C., Samel S.A., Fernández-Cid A., Riera A., Kawakami H., Stillman B., Speck C., Li H. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat. Struct. Mol. Biol. 2013;20:944–951. doi: 10.1038/nsmb.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticau S., Friedman L.J., Ivica N.A., Gelles J., Bell S.P. Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell. 2015;161:513–525. doi: 10.1016/j.cell.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Liu C., Wu X., Xu N., Zhou B., Liang C., Zhu G. Characterization and structure determination of the Cdt1 binding domain of human minichromosome maintenance (Mcm) 6. J. Biol. Chem. 2010;285:12469–12473. doi: 10.1074/jbc.C109.094599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles J.T.P., Janska A., Early A., Diffley J.F.X. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol. Cell. 2017;65:105–116. doi: 10.1016/j.molcel.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z., Riera A., Bai L., Sun J., Nandi S., Spanos C., Chen Z.A., Barbon M., Rappsilber J., Stillman B., et al. Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat. Struct. Mol. Biol. 2017;24:316–324. doi: 10.1038/nsmb.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y., Cheng E., Wu H., Li N., Yung P.Y.K., Gao N., Tye B.K. Open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer. Nat. Struct. Mol. Biol. 2017;24:300–308. doi: 10.1038/nsmb.3374. [DOI] [PubMed] [Google Scholar]
- Zhai Y., Li N., Jiang H., Huang X., Gao N., Tye B.K. Unique roles of the non-identical MCM subunits in DNA replication licensing. Mol. Cell. 2017;67:168–179. doi: 10.1016/j.molcel.2017.06.016. [DOI] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/elife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data: The cryo-EM map and structure coordinates of the single MCM2-7 hexamer produced in this study have been deposited at EMDB (https://www.emdataresource.org/) and PDB (https://www.rcsb.org/) with the accession codes of EMD-32326 and 7W68, respectively.
-
•
Code: This paper does not report original code.
-
•
Additional information: Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.