Abstract
Background
The germline variant rs1047303 (HSD3B1[1245A/C]), restricting or enabling production of potent androgens and estrogens from adrenal precursors, affects outcomes of castration-resistant prostate cancer and is associated with estrogen receptor positivity in postmenopausal breast cancer. Like breast cancer, endometrial cancer is another malignancy with hormone-dependent and hormone-independent subtypes. We hypothesized that adrenal-restrictive HSD3B1 genotype would associate with hormone-independent cancer subtypes.
Methods
We employed a previously described classification of tumors in The Cancer Genome Atlas into genomic clusters. We determined HSD3B1 genotype frequencies by endometrial cancer genomic cluster and calculated the odds per adrenal-restrictive A allele for the largely hormone-independent copy-number (CN) high subtype vs other subtypes. An equivalent analysis was performed for the genomically similar, hormone-independent basal breast cancer subtype. Last, we performed survival analyses for UK Biobank participants with endometrial cancer by HSD3B1 genotype. All statistical tests were 2-sided.
Results
The adrenal-restrictive HSD3B1(1245A) allele was associated with the CN-high endometrial cancer subtype (odds ratio [OR] = 1.63, 95% confidence interval [CI] = 1.14 to 2.32; P = .007). Similarly, HSD3B1(1245A) was associated with the basal breast cancer subtype (OR = 1.54, 95% CI = 1.13 to 2.08; P = .006). In the UK Biobank, endometrial cancer patients homozygous for HSD3B1(1245A) had worse overall (hazard ratio [HR] = 1.39, 95% CI = 1.16 to 1.68; P < .001) and cancer-specific (HR = 1.39, 95% CI = 1.14 to 1.70; P = .001) survival, consistent with the A allele being enriched in the more aggressive CN-high subtype.
Conclusions
These findings suggest roles for adrenal-restrictive vs adrenal-permissive steroidogenesis, by way of rs1047303 genotype, in the development of and/or outcomes from at least 3 commonly hormone-associated types of cancer: prostate, breast, and endometrial.
A growing body of evidence links sex steroid synthesis from adrenal precursors, mediated by the steroid metabolism enzyme 3β-hydroxysteroid dehydrogenase-1 (3βHSD1), to the development of and/or outcomes from multiple types of hormone-associated cancer. 3βHSD1 catalyzes the first step in conversion from adrenally produced dehydroepiandrosterone (DHEA), which in its sulfated form is the most abundant steroid in circulation, into both potent androgens and estrogens (1,2). Notably, 3βHSD1 is 1 step upstream of 5α-reductase (to potent androgens) and aromatase (required for synthesis of estrone and estradiol) (3). The enzyme exists in 2 forms as determined by the distinct cellular metabolic phenotypes conferred by a functional germline variant (rs1047303) in the gene HSD3B1 that encodes 3βHSD1 (4). The adrenal-restrictive HSD3B1(1245A) limits conversion of DHEA to androgens and estrogens, whereas the adrenal-permissive HSD3B1(1245C) enhances conversion of DHEA to androgens and estrogens. This is a common germline variant, with adrenal-permissive C allele frequencies ranging from above 0.3 in most European populations to approximately 0.1 or below in most African and East Asian populations (5). Prostate cancer growth is driven by androgens, and the adrenal-permissive C allele has been consistently shown to confer worse outcomes in castration-resistant prostate cancer in at least 10 different cohorts (6-14). A more recent study showed an association between homozygous inheritance of the adrenal-permissive C allele, enabling greater estrogen production, and postmenopausal estrogen receptor (ER)–positive breast cancer in 3 cohorts (15). In the same study, in smaller sample sizes, there were also trends toward an association between the adrenal-restrictive A allele and ER-negative breast cancer in each of the cohorts.
Like breast cancer, which can be classified as ER positive or ER negative, endometrial cancer is another malignancy commonly divided into hormone-driven and less hormone-dependent subtypes. Historical classifications of endometrial cancer largely divide tumors based on the relation to obesity and a hyperestrogenic state, namely, type I and type II endometrial cancer (16). Type I endometrial cancer is largely composed of tumors with low histologic grade (grade I or II) and of endometrioid histology. Most often these are characterized by higher expression of both ER and progesterone receptor (PR) and improved clinical outcomes (17,18). Type II endometrial cancer is largely composed of tumors with high-grade histologies, such as grade III endometrioid, clear cell, and serous carcinomas. These tumors are characterized by lower ER and PR expression and worse clinical outcomes (19).
An integrated genomic, transcriptomic, and proteomic characterization of endometrial tumors in The Cancer Genome Atlas (TCGA) (20) research program’s TCGA–Uterine Corpus Endometrial Carcinoma (UCEC) project classified tumors into 4 genomic clusters (21). The copy-number (CN) high subtype was composed primarily of tumors with serous histology and a subset of high grade endometrioid histology tumors, roughly corresponding to the traditional type II classification. This cluster, labeled the “serous-like cluster,” was associated with a worse clinical outcome. A similar analysis was performed for breast cancer on TCGA–Breast Invasive Carcinoma (BRCA) project (22). The serous-like cluster in the endometrial cancer study was described as genomically similar to the basal breast subtype (21), largely composed of triple-negative (ie, negative for ER, PR, and HER2 expression) tumors (22).
Given the previously described associations between HSD3B1 genotype and prostate and breast cancer, along with the mechanistic links to tumor types with greater or lesser hormone dependence, we set out to determine whether HSD3B1 genotype would be associated with genomic clusters or subtypes of endometrial cancer and clinical outcomes.
Methods
TCGA
Germline genotype and clinical data for breast cancer and endometrial cancer patients were available from the National Cancer Institute’s TCGA, specifically projects TCGA-BRCA and TCGA-UCEC, respectively. Data were obtained from the National Cancer Institute’s Genomic Data Commons Legacy Archive. TCGA-UCEC data were available as a mutation annotation format file generated from Mutect2 variant calls. TCGA-BRCA data were available in mutation annotation format, generated from whole exome sequencing–based Mutect2 variant calling, as well as genotype array data (Affymetrix 6.0). To assess concordance between whole exome sequencing and array data for rs1047303 (ie, HSD3B1[1245A/C]), the array genotypes were harmonized with the TOPMed Freeze 5 panel using a modified version of the HRC-1000G-check-bim tool (https://www.well.ox.ac.uk/~wrayner/tools/#Checking), and chromosome 1 genotypes were imputed with the University of Michigan Imputation Server (phasing with Eagle version 2.4, imputation with Minimac4 version 1.2.4, using the TOPMed Freeze 5 panel) to generate germline genotype calls. Only samples with concordant genotypes were included in the analysis. Genomic subtype classifications for TCGA breast and endometrial tumors, as well as data on gene alterations, were obtained from the published analysis of Sanchez-Vega et al. (23). Genotypes and genomic subtypes were determined for all TCGA-BRCA and TCGA-UCEC patients with data of both categories available. Genetic ancestry information was obtained from the published analysis of Yuan et al. (24)
UK Biobank
Endometrial cancer survival analysis was performed on cases from the UK Biobank (25), a long-term study tracking medical information of more than 500 000 genotyped individuals. This study was approved by the National Health Services Research Ethics Committee (REC ref16/NW/0274) through the generic ethical approval for UK Biobank studies (approved UK Biobank application #44578). Genotyping was performed using the UK BiLEVE Axiom Array or UK Biobank Axiom Array by Applied Biosystems. Additional single-nucleotide polymorphisms (SNPs) were imputed with the IMPUTE4 program using the Haplotype Reference Consortium, UK10K, and 1000 Genomes phase III panels as reference panels. Rs1047303 (ie, HSD3B1[1245A/C]) was imputed with high accuracy (information score > 0.99). Endometrial cancer patients were identified using the UK Biobank’s cancer registry with additional diagnosed patients identified using inpatient hospital records (International Classification of Diseases version 10 [ICD10] code C54.1). Diagnosis dates were extracted from the cancer registry for patients in the cancer registry and from the earliest inpatient hospital record with ICD10 code C54.1 for patients not in the cancer registry. Treatment data were not available for most cancer patients in the UK Biobank and were therefore not included in analyses. The UK Biobank’s death register was used to determine which patients died, along with causes and dates of death. Deaths through March 23, 2021, were included. For the all-cause mortality analyses, deaths with any cause of death were included. For the cancer-specific mortality analyses, deaths with a cause of death of cancer (ICD10 code beginning with C) were included. This was done rather than endometrial cancer–specific mortality to account for deaths among endometrial cancer patients with listed causes such as ovarian cancer (C56), despite never having been diagnosed with ovarian cancer, or cancer with no site specified (C80).
Statistical Analysis
For TCGA analyses, odds ratios were calculated using logistic regression where outcome was defined by cancer-specific subtype (subtype = CN-high [for endometrial] or subtype = basal [for breast]) vs other cancer subtypes, separately for the BRCA and UCEC projects. An additive inheritance model was employed for the exposure of interest, the number of HSD3B1(1245A) alleles. Three regression models assessing the association between cancer-specific subtype and allelic count were used: a univariate regression including all individuals, a regression adjusted for genetic ancestry as determined by Yuan et al. (24), and a univariate regression limited to individuals of European ancestry. For the UK Biobank analyses, the Kaplan-Meier method and Cox proportional hazards models were used for analyses of all-cause and cancer-specific mortality, comparing cases of participants with AA genotype to the combined pool of cases of participants with AC and CC genotype (dominant inheritance model). Three models were used for the Cox proportional hazards analysis: a univariate regression; a regression adjusted for diagnosis age and body mass index (BMI); and a regression adjusted for diagnosis age, BMI, and the first 10 genetic principal components. Genetic principal components were obtained from the UK Biobank and computed by UK Biobank researchers using the fastPCA (26) algorithm. All analyses were performed in RStudio, and the survival analyses used the R package survminer. All statistical tests were 2-sided. Per guidance of the American Statistical Association (27), results of statistical tests were not dichotomized into significant vs nonsignificant.
Results
Three cohorts were used for the main analyses: endometrial cancer in TCGA (TCGA-UCEC project), female breast cancer in TCGA (TCGA-BRCA project), and participants who had endometrial cancer in the UK Biobank (25). Demographic information for the 3 cohorts is shown in Table 1. For the TCGA cohorts, genetic ancestry was used as determined by the analysis of Yuan et al. (24), because genetic ancestry should provide a more accurate reflection of the underlying genetics than self-described race and because self-described race was not available for approximately 6% (TCGA-UCEC) to 9% (TCGA-BRCA) of participants. Demographic information including self-reported race is shown in Supplementary Table 1 (available online). Analyses were performed using the genomic subtype classifications of Sanchez-Vega et al. (23); Table 1 and Supplementary Table 1 (available online) include columns with numbers restricted to participants whose samples had genomic subtype classified. Supplementary Tables 2 and 3 (available online) show demographic information broken down by genomic subtype for TCGA-UCEC and TCGA-BRCA cohorts, respectively.
Table 1.
Summary information of cohorts used in analysesa
| Characteristic | All genotyped patients | Patients with subtype classified |
|---|---|---|
| No. (%) | No. (%) | |
| TCGA-UCEC (endometrial cancer) | ||
| Genetic ancestry | ||
| African | 114 (20.9) | 107 (21.1) |
| East Asian | 30 (5.5) | 29 (5.7) |
| European | 388 (71.1) | 356 (70.4) |
| Native American | 12 (2.2) | 12 (2.4) |
| Other | 2 (0.4) | 2 (0.4) |
| Total No. of patients | 546 | 506 |
| Age, mean (range) | 63.9 (31-90) | 63.8 (31-90) |
| TCGA-BRCA, female patients | ||
| Genetic ancestry | ||
| African | 173 (16.5) | 153 (16.1) |
| East Asian | 55 (5.2) | 55 (5.8) |
| European | 789 (75.2) | 713 (75.1) |
| Native American | 24 (2.3) | 21 (2.2) |
| Other | 8 (0.8) | 7 (0.7) |
| Total No. of patients | 1049 | 949 |
| Age, mean (range) | 58.4 (26-90) | 58.2 (31-90) |
| UK Biobank participants with endometrial cancer diagnoses | ||
| Self-reported ethnicity | ||
| Asian or Asian British | 48 (1.8) | — |
| Black or Black British | 24 (0.9) | — |
| Chinese | 7 (0.3) | — |
| Do not know | 3 (0.1) | — |
| Mixed | 11 (0.4) | — |
| No answer or prefer not to answer | 11 (0.4) | — |
| Other ethnic group | 26 (1.0) | — |
| White | 2523 (95.1) | — |
| Total No. of patients | 2653 | — |
| Age, mean (range) | 61.5 (30-81) | — |
— = not applicable; BRCA = breast invasive carcinoma; TCGA = The Cancer Genome Atlas; UCEC = uterine corpus endometrial carcinoma.
The initial analysis was performed on endometrial cancer samples in TCGA-UCEC project. Table 2 shows the clinicopathological characteristics of the samples by HSD3B1(1245A/C) genotype. As expected, adrenal-permissive C allele frequencies were substantially higher in participants of European ancestry compared with other ancestries. Because this difference in allele frequencies by ancestry could confound results, analyses were performed on the entire cohort and specifically on the European ancestry cohort, which had the largest sample size. The adrenal-restrictive A allele frequency was somewhat higher in tumors of stage II or greater compared with stage I (P = .04 for all participants; P = .15 for European ancestry participants). The A allele frequency was also somewhat higher in tumors with serous histology compared with endometrioid histology (P = .04 for all participants; P = .10 for European ancestry participants). Both these results suggest the A allele could be associated with more aggressive and less hormone-dependent disease. Clinicopathological characteristics of samples from African ancestry participants are shown in Supplementary Table 4 (available online).
Table 2.
TCGA-UCEC clinicopathological characteristics by HSD3B1 genotype
| Variable | Genotype | All TCGA-UCEC |
European ancestry |
||
|---|---|---|---|---|---|
| No. (%) | P a | No. (%) | P a | ||
| Genetic ancestry | <.001b | n/a | |||
| African | AA | 97 (85.1) | — | ||
| AC | 14 (12.3) | — | |||
| CC | 3 (2.6) | — | |||
| East Asian | AA | 26 (86.7) | — | ||
| AC | 4 (13.3) | — | |||
| CC | 0 (0.0) | — | |||
| European | AA | 185 (47.7) | 185 (47.7) | ||
| AC | 176 (45.4) | 176 (45.4) | |||
| CC | 27 (7.0) | 27 (7.0) | |||
| Native American | AA | 11 (91.7) | — | ||
| AC | 1 (8.3) | — | |||
| CC | 0 (0.0) | — | |||
| Other | AA | 2 (100.0) | — | ||
| AC | 0 (0.0) | — | |||
| CC | 0 (0.0) | — | |||
| Stage | .04 | .15 | |||
| I | AA | 187 (54.8) | 113 (44.5) | ||
| AC | 135 (39.6) | 123 (48.4) | |||
| CC | 19 (5.6) | 18 (7.1) | |||
| ≥ II | AA | 134 (65.4) | 72 (53.7) | ||
| AC | 60 (29.3) | 53 (39.6) | |||
| CC | 11 (5.4) | 9 (6.7) | |||
| Histologic diagnosis | .04c | .10c | |||
| Endometrioid | AA | 232 (56.6) | 138 (45.8) | ||
| AC | 152 (37.1) | 140 (46.5) | |||
| CC | 26 (6.3) | 23 (7.6) | |||
| Mixed serous and endometrioid | AA | 14 (63.6) | 6 (46.2) | ||
| AC | 7 (31.8) | 6 (46.2) | |||
| CC | 1 (4.5) | 1 (7.7) | |||
| Serous | AA | 75 (65.8) | 41 (55.4) | ||
| AC | 36 (31.6) | 30 (40.5) | |||
| CC | 3 (2.6) | 3 (4.1) | |||
| Grade (endometrioid only) | .17 | .26 | |||
| 1 or 2 | AA | 120 (55.0) | 71 (44.4) | ||
| AC | 79 (36.2) | 73 (45.6) | |||
| CC | 19 (8.7) | 16 (10.0) | |||
| 3 | AA | 112 (58.3) | 67 (47.5) | ||
| AC | 73 (38.0) | 67 (47.5) | |||
| CC | 7 (3.6) | 7 (5.0) | |||
Two-sided P values from Wald tests on binomial logistic regression models are shown. — = not applicable; TCGA = The Cancer Genome Atlas; UCEC = uterine corpus endometrial carcinoma.
European vs other ancestries.
Serous vs endometrioid.
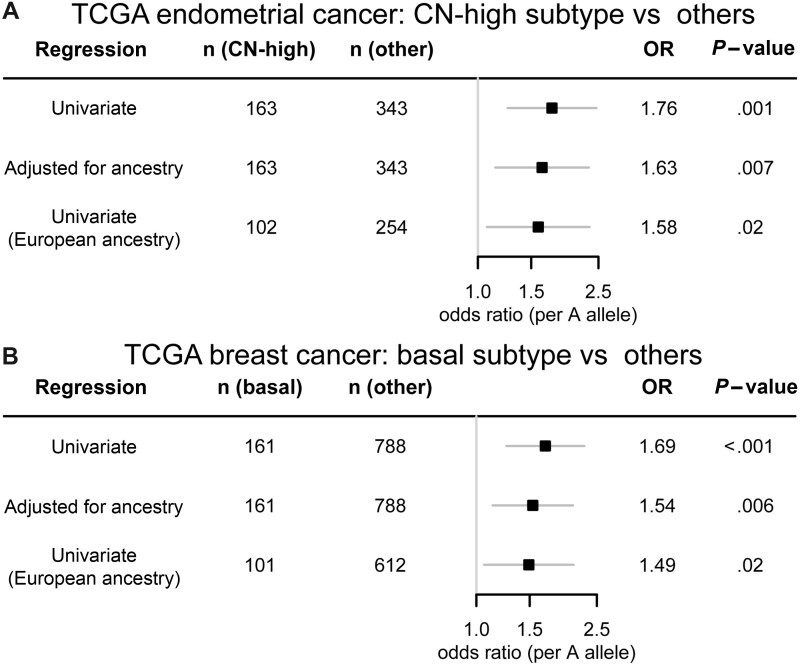
In the analysis published by Sanchez-Vega et al. (23), TCGA-UCEC samples were classified into 4 genomic subtypes: CN-high, CN-low, microsatellite instability hypermutated, and POLE ultramutated. We determined the HSD3B1 genotype frequencies for each subtype (Table 3), revealing higher rates of the A allele in the CN-high subtype, which is the subtype characterized by low ER and PR expression. Among samples from participants of all ancestries, the CN-high subtype had 66.9% AA genotype and only 1.2% CC genotype compared with 54.5% and 7.9%, respectively, for all other subtypes combined. Among samples from participants of European ancestry, the CN-high subtype had 53.9% AA genotype and only 2.0% CC genotype compared with 44.5% and 9.4%, respectively, for all other subtypes combined. Results from the logistic regression models revealed substantially elevated odds of CN-high subtype with the A allele in the univariate regression (odds ratio [OR] = 1.75, 95% confidence interval [CI] = 1.25 to 2.46; P = .001), adjusted for genetic ancestry (OR = 1.63, 95% CI = 1.14 to 2.32; P = .007), and limited to European ancestry (OR = 1.58, 95% CI = 1.07 to 2.34; P = .02) (Figure 1, A).
Table 3.
Breakdown of TCGA-UCEC samples by genomic subtype and HSD3B1 genotypea
| Genomic subtype | Genotype | All TCGA-UCEC | European ancestry |
|---|---|---|---|
| No. (%) | No. (%) | ||
| CN-high | AA | 109 (66.9) | 55 (53.9) |
| AC | 52 (31.9) | 45 (44.1) | |
| CC | 2 (1.2) | 2 (2.0) | |
| CN-low | AA | 82 (55.8) | 55 (48.2) |
| AC | 50 (34.0) | 46 (40.4) | |
| CC | 15 (10.2) | 13 (11.4) | |
| MSI | AA | 81 (55.1) | 49 (44.1) |
| AC | 57 (38.8) | 54 (48.6) | |
| CC | 9 (6.1) | 8 (7.2) | |
| POLE | AA | 24 (49.0) | 9 (31.0) |
| AC | 22 (44.9) | 17 (58.6) | |
| CC | 3 (6.1) | 3 (10.3) | |
| Non-CN-high subtypes combined | AA | 187 (54.5) | 113 (44.5) |
| AC | 129 (37.6) | 117 (46.1) | |
| CC | 27 (7.9) | 24 (9.4) | |
| All subtypes combined | AA | 296 (58.5) | 168 (47.2) |
| AC | 181 (35.8) | 162 (45.5) | |
| CC | 29 (5.7) | 26 (7.3) |
CN = copy number; MSI = microsatellite instability hypermutated; POLE = POLE ultramutated; TCGA = The Cancer Genome Atlas; UCEC = uterine corpus endometrial carcinoma.
Figure 1.
Adrenal-restrictive HSD3B1 genotype is associated with hormone-independent subtypes of endometrial and breast cancer. Odds ratios (per A allele) with 95% confidence intervals (error bars) for having (A) CN-high endometrial cancer vs other genomic subtypes of endometrial cancer and (B) basal breast cancer vs other genomic subtypes of breast cancer among participants in the TCGA-UCEC and TCGA-BRCA projects, respectively, whose tumors had genomic subtype classified. Results are shown from binomial logistic regressions: a univariate regression of (subtype = CN-high/basal) against number of A alleles, a regression adjusted for genetic ancestry classification, and a univariate regression restricted to participants classified as having European ancestry. Two-sided P values from Wald tests are shown. BRCA = breast invasive carcinoma; CN = copy number; OR = odds ratio; TCGA = The Cancer Genome Atlas; UCEC = uterine corpus endometrial carcinoma.
Like endometrial cancer, breast cancer is divided into hormone-dependent and hormone-independent subtypes, and the basal subtype of breast cancer is genomically similar to the CN-high subtype of endometrial cancer. The analysis of Sanchez-Vega et al. (23) classified breast cancer into 5 genomic subtypes: basal, HER2-enriched, luminal A, luminal B, and normal. For validation of the finding of an association between HSD3B1(1245A) and CN-high endometrial cancer, we tested for a similar association with basal breast cancer. The genotype frequencies of different breast cancer genomic subtypes are shown in Table 4. Among samples from participants of all ancestries, the basal subtype had 67.1% AA genotype and only 3.7% CC genotype compared with 53.2% and 9.5%, respectively, for all other subtypes combined. Among samples from participants of European ancestry, the basal subtype had 58.4% AA genotype and 5.9% CC genotype compared with 47.2% and 11.4%, respectively, for all other subtypes combined. The logistic regression models revealed substantially elevated odds of basal subtype with the A allele in the univariate regression (OR = 1.69, 95% CI = 1.26 to 2.27; P < .001), adjusted for genetic ancestry (OR = 1.54, 95% CI = 1.13 to 2.08; P = .006), and limited to European ancestry (OR = 1.49, 95% CI = 1.06 to 2.09; P = .02) (Figure 1, B). Thus, similar associations were found between adrenal-restrictive genotype and both CN-high endometrial cancer and the genomically similar basal breast cancer.
Table 4.
Breakdown of TCGA-BRCA samples by genomic subtype and HSD3B1 genotypea
| Genomic subtype | Genotype | All TCGA-BRCA | European ancestry |
|---|---|---|---|
| No. (%) | No. (%) | ||
| Basal | AA | 108 (67.1) | 59 (58.4) |
| AC | 47 (29.2) | 36 (35.6) | |
| CC | 6 (3.7) | 6 (5.9) | |
| Her2 | AA | 39 (51.3) | 19 (42.2) |
| AC | 30 (39.5) | 20 (44.4) | |
| CC | 7 (9.2) | 6 (13.3) | |
| LumA | AA | 257 (52.8) | 189 (47.7) |
| AC | 178 (36.6) | 158 (39.9) | |
| CC | 52 (10.7) | 49 (12.4) | |
| LumB | AA | 106 (56.1) | 73 (49.7) |
| AC | 71 (37.6) | 63 (42.9) | |
| CC | 12 (6.3) | 11 (7.5) | |
| Normal | AA | 17 (47.2) | 8 (33.3) |
| AC | 15 (41.7) | 12 (50.0) | |
| CC | 4 (11.1) | 4 (16.7) | |
| Non-basal subtypes combined | AA | 419 (53.2) | 289 (47.2) |
| AC | 294 (37.3) | 253 (41.3) | |
| CC | 75 (9.5) | 70 (11.4) | |
| All subtypes combined | AA | 527 (55.5) | 348 (48.8) |
| AC | 341 (35.9) | 289 (40.5) | |
| CC | 81 (8.5) | 76 (10.7) |
BRCA = breast invasive carcinoma; HER2 = HER2 enriched; LumA = luminal A; LumB = luminal B; TCGA = The Cancer Genome Atlas.
Because sex hormone concentrations change dramatically with age (eg, at menopause) and the study population included a wide age range, we additionally analyzed age-adjusted versions of each of the regression models. The associations between CN-high subtype and the A allele were marginally stronger in the age-adjusted endometrial cancer models than in the models without age adjustment (Supplementary Figure 1, A, available online, compared with Figure 1, A), and the associations between basal subtype and the A allele were similar in the age-adjusted breast cancer models to in the models without age adjustment (Supplementary Figure 1, B, available online, compared with Figure 1, B).
The CN-high subtype of endometrial cancer and the basal subtype of breast cancer are both characterized by high rates of alterations in the TP53 gene (21). Using data on presence of gene alterations from the analysis of Sanchez-Vega et al. (23), we tested whether presence of alterations in TP53 was associated with adrenal-restrictive genotype. In univariate regression models, we found modest associations between TP53 alteration and the A allele in endometrial (OR = 1.28, 95% CI = 0.94 to 1.73; P = .12) and breast cancer (OR = 1.27, 95% CI = 1.03. to 1.57; P = .02). However, these associations disappeared in regression models adjusted for genomic subtype (endometrial: OR = 0.79, 95% CI = 0.51 to 1.22; P = .28; breast: OR = 1.04, 95% CI = 0.80 to 1.37; P = .75), suggesting that adrenal-restrictive genotype is not associated with TP53 alteration independent of the association between genotype and genomic subtype.
CN-high endometrial cancer and basal breast cancer have also been described as genomically similar to serous ovarian cancer (21), so we assessed whether there was an association between adrenal-restrictive genotype and serous ovarian cancer in the TCGA-Ovarian (OV) project. Summary demographic information for the TCGA-OV cohort is shown in Supplementary Table 5 (available online). TCGA-UCEC and TCGA-BRCA projects contain samples from multiple tumor histologies and were classified into multiple genomic subtypes. By contrast, the TCGA-OV project contains only samples of serous histology, by far the most common histology of ovarian cancer (28), and the samples were not classified into genomic subtypes in the analysis of Sanchez-Vega et al. (23). Therefore, odds ratios for genomic subtype by HSD3B1 genotype could not be calculated for ovarian cancer. We determined the frequencies of each genotype for participants in the TCGA-OV project, by grade high (grade III) or low (grade I or II) as well as for all grades combined and did not observe notable associations with genotype (Supplementary Table 6, available online). The overall C allele frequency for TCGA-OV participants of European ancestry was 0.308, in line with typical C allele frequencies in European populations (5). Therefore, the associations found with CN-high endometrial cancer and basal breast cancer do not appear to be present for serous ovarian cancer.
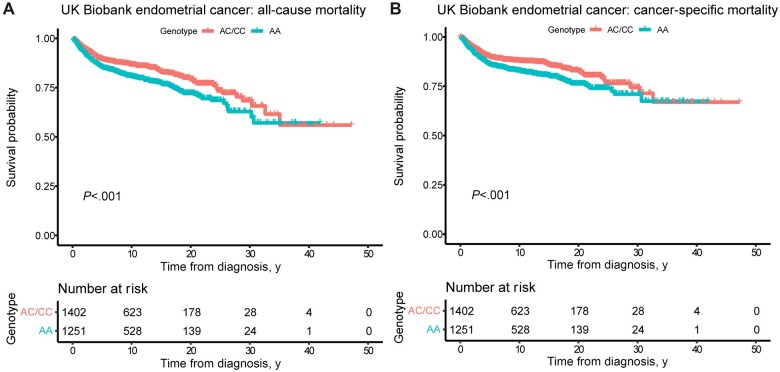
CN-high endometrial cancer has substantially worse outcomes than any of the other subtypes; therefore, based on the association between HSD3B1(1245A) and CN-high subtype, it could be predicted that the A allele would also be associated with worse outcomes. We performed survival analyses on a cohort with much more extensive survival data than that found in TCGA (26 739 patient-years of follow-up in the UK Biobank endometrial cancer cohort compared with 1690 patient-years in TCGA-UCEC): UK Biobank participants who were diagnosed with endometrial cancer. We analyzed all-cause mortality and cancer-specific mortality, and in both analyses, homozygous adrenal-restrictive genotype was associated with worse survival than heterozygous or homozygous adrenal-permissive genotypes (Figure 2; survival curves showing all 3 genotypes separately are shown in Supplementary Figure 2, available online). In Cox proportional hazards models for all-cause and cancer-specific mortality, AA genotype compared with other genotypes was associated with elevated hazard ratios in each of 3 regression models: an unadjusted model, a model adjusted for diagnosis age and BMI, and a model additionally adjusted for the first 10 genetic principal components (Table 5). In the fully adjusted model, which should largely account for potential confounding by ancestry, the hazard ratio for all-cause mortality was 1.39 (95% CI = 1.16 to 1.68; P < .001), and for cancer-specific mortality was also 1.39 (95% CI = 1.14 to 1.70; P = .001). Testing the proportional hazards assumptions did not identify violations of the assumptions for any of the Cox regression models (Supplementary Table 7, available online). Data on genomic subtypes were not available for the UK Biobank cohort, but the survival results are consistent with the A allele being associated with the more aggressive CN-high subtype.
Figure 2.
Homozygous adrenal-restrictive HSD3B1 genotype is associated with worse endometrial cancer outcomes. Kaplan-Meier curves of (A) all-cause mortality and (B) cancer-specific mortality for UK Biobank participants who were diagnosed with endometrial cancer. Endometrial cancer patients were identified using the UK Biobank’s cancer registry with additional diagnosed patients identified using inpatient hospital records (ICD10 code C54.1). Diagnosis dates were extracted from the cancer registry for patients in the cancer registry and from the earliest inpatient hospital record with ICD10 code C54.1 for patients not in the cancer registry. The UK Biobank’s death register was used to determine which patients died, along with causes and dates of death. Two-sided P values from log-rank tests are shown for the comparison of patients with AA genotype to patients of either AC or CC genotype pooled together. ICD10 = International Classification of Diseases, 10th revision.
Table 5.
Results from Cox proportional hazards models for survival of UK Biobank participants with endometrial cancer diagnoses
| Outcome | Regression model | HR (95% CI)a | P b |
|---|---|---|---|
| All-cause mortality | Univariate regression against genotype | 1.42 (1.18 to 1.71) | <.001 |
| Genotype + diagnosis age + BMI | 1.40 (1.16 to 1.69) | <.001 | |
| Genotype + diagnosis age + BMI + first 10 genetic principal components | 1.39 (1.16 to 1.68) | <.001 | |
| Cancer-specific mortality | Univariate regression against genotype | 1.40 (1.15 to 1.71) | <.001 |
| Genotype + diagnosis age + BMI | 1.39 (1.14 to 1.70) | .001 | |
| Genotype + diagnosis age + BMI + first 10 genetic principal components | 1.39 (1.14 to 1.70) | .001 |
AA genotype vs other genotypes. BMI = body mass index; CI = confidence interval; HR = hazard ratio.
Two-sided P values from Wald tests are shown.
Discussion
We have demonstrated, using data from TCGA, that there is an association between the adrenal-restrictive HSD3B1(1245A) and the CN-high subtype of endometrial cancer (Figure 1, A) and that there is a similar association between HSD3B1(1245A) and the basal subtype of breast cancer (Figure 1, B), which has been described as genomically similar to the CN-high subtype of endometrial cancer (21). Both these subtypes are considered to be less hormone dependent than other endometrial and breast cancer subtypes (21,22), consistent with the adrenal-restrictive form of HSD3B1 limiting the production of estrogens from adrenal precursors. We have additionally demonstrated, using data from the UK Biobank, that homozygous adrenal-restrictive HSD3B1 genotype is associated with worse overall survival (Figure 2, A) and cancer-specific survival (Figure 2, B) in endometrial cancer patients, consistent with the A allele being associated with the poor-prognosis CN-high subtype (21). Taken together, these results suggest that the limited estrogen production from adrenal precursors caused by the adrenal-restrictive HSD3B1 promotes development of hormone-independent endometrial and breast cancer subtypes (Figure 3).
Figure 3.
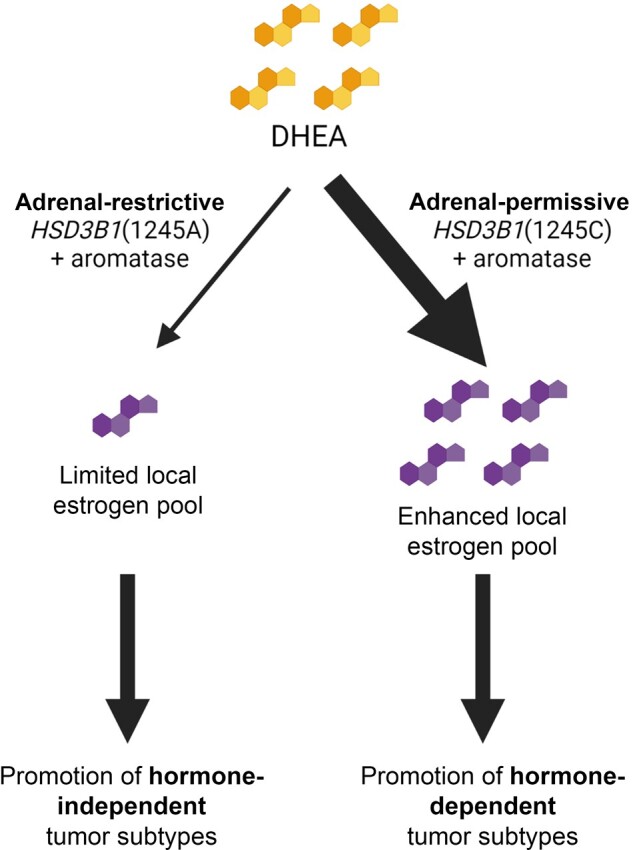
Proposed mechanism underlying associations between HSD3B1 genotype and subtypes of endometrial and breast cancer. DHEA taken up from the circulating pool is converted to downstream products in cells expressing HSD3B1, which in conjunction with aromatase leads to estrogen production. The adrenal-restrictive HSD3B1(1245A) limits local estrogen production, promoting largely hormone-independent tumor subtypes such as CN-high endometrial cancer and basal breast cancer. The adrenal-permissive HSD3B1(1245C) enhances local estrogen production, which could promote other, hormone-dependent tumor subtypes. Created using BioRender.com. DHEA = dehydroepiandrosterone.
Each of the main findings—associations between adrenal-restrictive genotype and CN-high endometrial cancer, between adrenal-restrictive genotype and basal breast cancer, and between adrenal-restrictive genotype and worse endometrial cancer outcomes—was identified in 1 cohort: TCGA-UCEC project, TCGA-BRCA project, and UK Biobank, respectively. It is important to validate all the findings in additional cohorts. At the same time, each finding is not only a novel finding but also complementary to the other findings. For example, the association between adrenal-restrictive genotype and CN-high endometrial cancer, which has the worst outcomes, suggests there could be an association between genotype and overall endometrial cancer outcomes, which indeed was found. Because each of the findings complements the others, the evidence taken together is stronger than if any of the findings was presented alone. Nonetheless, the findings coming from single cohorts is a limitation of the study, and seeking validation in additional cohorts is essential.
We focused our study on rs1047303 (ie, HSD3B1[1245A/C]) because of a clear consequential metabolic phenotype and mechanism that results from this SNP-encoded missense and subsequently described associations that fit the physiology of this SNP in prostate and breast cancer. This is not the only SNP in HSD3B1, but to our knowledge, it is the only one that has been clearly and functionally characterized (4). Other studies have examined other SNPs in HSD3B1; for example, an association between rs6203 and essential hypertension was reported (29), as was an association between rs6428829 and acne vulgaris (30).
Although serous ovarian cancer is also considered genomically similar to CN-high endometrial cancer and basal breast cancer (21), we did not observe a similar association between adrenal-restrictive genotype and serous ovarian cancer. Unlike in endometrial and breast cancer, in which CN-high and basal tumors are a smaller fraction of cases compared with other, more hormone-dependent subtypes, serous ovarian cancer comprises a large majority of ovarian cancer cases (28). Serous ovarian cancer is generally characterized by less hormone receptor expression than endometrioid ovarian cancer but by more than mucinous or clear cell (31), suggesting that whereas CN-high endometrial and basal breast cancer have less hormone receptor expression than most endometrial and breast cancers, serous ovarian cancer may have roughly average hormone receptor expression among ovarian cancers. Other ovarian cancer subtypes were not included in TCGA-OV project. Reproductive and hormonal factors have been reported to have heterogeneous associations with ovarian cancer risk by subtype (32). Hormonal therapy is less frequently used in ovarian than breast or endometrial cancer and its effectiveness remains unclear (33).
Unlike the dose-dependent effect of the adrenal-restrictive A allele observed in associations with endometrial and breast cancer genomic subtypes, the observed association with worse endometrial cancer outcomes was specific to homozygous adrenal-restrictive (AA) genotype, with AC and CC genotypes having similar outcomes to each other (Supplementary Figure 2, available online). In ER-positive breast cancer, worse outcomes have been observed with CC genotype (34). One could speculate a similar effect might exist in hormone-dependent subtypes of endometrial cancer. If this were the case, the worse survival conferred by the A allele’s association with hormone-independent endometrial cancer could be counteracted by worse survival with CC genotype within hormone-dependent endometrial cancer when all endometrial cancers are pooled together. It is important to interrogate survival by genotype within subtypes of endometrial and breast cancer in future studies. Additionally, treatment data were not available for the UK Biobank cohort in which the survival analysis was performed. Treatment, cancer subtype, and HSD3B1 genotype could potentially interact with each other, and this should be interrogated in cohorts in which all these data are available.
Previously, the adrenal-permissive HSD3B1(1245C), by enhancing production of androgens from adrenal precursors (DHEA → androstenedione → DHT), has been consistently shown to promote faster progression of castration-resistant prostate cancer with similar results obtained in at least 10 cohorts (6–14). Additionally, the homozygous adrenal-permissive genotype (which, in a breast cancer context, would instead lead to conversion of the increased androstenedione generated to estrogens) has been shown to be associated with ER-positive postmenopausal breast cancer in 3 cohorts (15) and with increased risk of distant metastatic recurrence and breast cancer–specific mortality in a separate cohort of ER-positive postmenopausal breast cancer patients (34). In the same 3 cohorts in which homozygous adrenal-permissive genotype was associated with ER-positive breast cancer, there were also trends toward the opposite association with ER-negative breast cancer (15). The basal subtype of breast cancer is largely triple-negative (ie, negative for ER, PR, and HER2 expression) (22) and comprises many, but not all, of the overall set of ER-negative tumors. Our results suggest the previously described trends toward an association with ER-negative breast cancer may be enhanced when looking specifically at the basal subtype.
The enzyme 3βHSD1 encoded by HSD3B1 is in the pathway to production of both potent androgens and estrogens from adrenally produced DHEA and also catalyzes the conversion of pregnenolone to progesterone (1). In addition to ER and PR, androgen receptor is also expressed in endometrial (35,36) and breast cancer (37,38). This suggests that the enhanced or limited production of androgens, estrogens, and progesterone because of HSD3B1 genotype could be relevant to associations between genotype and tumor subtype and/or outcomes.
The role of sex hormones in the different types of endometrial cancer is not fully resolved. For example, one study did not find differences in estradiol, progesterone, or testosterone levels between type I and type II endometrial cancer (39). On the other hand, CN-high endometrial cancer, composed mainly of serous as well as a subset of the high-grade endometrioid tumors that together would traditionally be considered type II, is characterized by lower levels of ER and PR expression as well as lower ER signaling in pathway analysis (21). The mechanisms underlying the association between adrenal-restrictive genotype and these tumors are a subject for further research. Previous studies identifying associations between HSD3B1 genotype and other phenotypes have not found that circulating sex steroid levels differ by genotype (15,40,41). This suggests that effects may be mediated at the local cell or tissue level rather than in circulation, as steroid levels in tissues can be different from those in circulation (42-44).
Intriguingly, the CN-high endometrial cancer subtype and basal breast cancer subtype, which both tend to have worse outcomes (21,22), are more common in Black women (45) (Supplementary Tables 2 and 3, available online), who are more likely to inherit HSD3B1(1245A). This raises the question of whether the higher rates of these tumor subtypes in Black women are due in part to higher A allele frequencies. In comparison to univariate, all participants regressions of subtype against genotype, regressions that were adjusted for genetic ancestry or restricted to participants of European ancestry had somewhat attenuated odds in a similar manner in endometrial and breast cancer, but the associations remained present (Figure 1). Although genetic ancestry was accounted for, the participant populations may still not have been completely homogenous in ancestry, and confounding because of different allele frequencies by ancestry may not have been completely eliminated. It is important to validate the results in additional cohorts, including cohorts with larger populations of non-European ancestry, in which statistical power is more limited because of the low C allele frequencies in such populations. In contrast to TCGA, the UK Biobank population is overwhelmingly, although not entirely, of European ancestry. Adjusting the UK Biobank survival results for the first 10 genetic principal components to account for potential confounding by ancestry only slightly attenuated the associations between survival and HSD3B1 genotype, which remained strong (Table 5).
In addition to validation of our results in additional cohorts, future studies should look at whether HSD3B1 genotype is associated with outcomes within different subtypes of endometrial and breast cancer and whether HSD3B1 genotype affects the response to hormonal therapy. One could speculate that the effect of genotype on outcomes might differ by subtype and hormone-dependent vs hormone-independent nature of the tumor. If HSD3B1 genotype does prove to predict response to hormonal therapy, it could be an important biomarker for determining who will benefit from what therapies. Taken together, the findings we report here along with previous findings in prostate (6–14) and breast cancer (15,34) suggest a role for adrenal-restrictive vs adrenal-permissive steroidogenesis, by way of rs1047303 genotype, in the development of and/or outcomes from at least 3 different commonly hormone-associated types of cancer: prostate, breast, and endometrial.
Funding
This work has been supported by R01CA261995, R01CA236780, R01CA172382, and R01CA249279 (to NS).
Notes
Role of the funder: Not applicable.
Disclosures: NS is a co-inventor on a Cleveland Clinic patent on HSD3B1. The other authors have no disclosures.
Author contributions: JMM: conceptualization, formal analysis, investigation, methodology, visualization, writing—original draft; RV: conceptualization, writing—review and editing; PSB: data curation, methodology, writing—review and editing; FRS: conceptualization, writing—review and editing; NS: conceptualization, funding acquisition, supervision, writing—review and editing.
Acknowledgements: This research has been conducted using data from UK Biobank, a major biomedical database. The authors additionally acknowledge Navin Sabharwal for helping with the initial setup of the UK Biobank analysis.
Prior presentations: McManus JM, Vargas R, Bazeley P, Sabharwal N, and Sharifi N. Association between adrenal-restrictive HSD3B1 inheritance and hormone independent subtypes of endometrial and breast cancer. American Association for Cancer Research 2022 Annual Meeting, New Orleans, LA, April 10, 2022.
Supplementary Material
Contributor Information
Jeffrey M McManus, Genitourinary Malignancies Research Center, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Roberto Vargas, Department of Gynecologic Oncology, Women’s Health Institute, Cleveland Clinic, Cleveland, OH, USA; Department of Translational Hematology and Oncology Research, Taussig Cancer Institute, Cleveland, OH, USA.
Peter S Bazeley, Department of Quantitative Health Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA.
Fredrick R Schumacher, Department of Population Health and Quantitative Sciences, Case Western Reserve University School of Medicine, Cleveland, OH, USA; Case Comprehensive Cancer Center, Case Western Reserve University School of Medicine, Cleveland, OH, USA.
Nima Sharifi, Genitourinary Malignancies Research Center, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA; Department of Hematology and Oncology, Taussig Cancer Institute, Cleveland Clinic, Cleveland, OH, USA; Department of Urology, Glickman Urological and Kidney Institute, Cleveland Clinic, Cleveland, OH, USA.
Data Availability
TCGA data underlying this article are available from the NCI’s Genomic Data Commons Data Portal (portal.gdc.cancer.gov) and GDC Legacy Archive (portal.gdc.cancer.gov/legacy-archive). UK Biobank data underlying this article are available from the UK Biobank (www.ukbiobank.ac.uk) after applying for access.
References
- 1. Sabharwal N, Sharifi N.. HSD3B1 genotypes conferring adrenal-restrictive and adrenal-permissive phenotypes in prostate cancer and beyond. Endocrinology. 2019;160(9):2180-2188. doi: 10.1210/en.2019-00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Auchus RJ, Sharifi N.. Sex hormones and prostate cancer. Annu Rev Med. 2020;71:33-45. doi: 10.1146/annurev-med-051418-060357. [DOI] [PubMed] [Google Scholar]
- 3. Sharifi N. Minireview: androgen metabolism in castration-resistant prostate cancer. Mol Endocrinol. 2013;27(5):708-714. doi: 10.1210/me.2013-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chang KH, Li R, Kuri B, et al. A gain-of-function mutation in DHT synthesis in castration-resistant prostate cancer. Cell. 2013;154(5):1074-1084. doi: 10.1016/j.cell.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.dbSNP. https://www.ncbi.nlm.nih.gov/snp/. Accessed June 4, 2021.
- 6. Hearn JWD, AbuAli G, Reichard CA, et al. HSD3B1 and resistance to androgen-deprivation therapy in prostate cancer: a retrospective, multicohort study. Lancet Oncol. 2016;17(10):1435-1444. doi: 10.1016/S1470-2045(16)30227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Agarwal N, Hahn AW, Gill DM, Farnham JM, Poole AI, Cannon-Albright L.. Independent validation of effect of HSD3B1 genotype on response to androgen-deprivation therapy in prostate cancer. JAMA Oncol. 2017;3(6):856. doi: 10.1001/jamaoncol.2017.0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shiota M, Narita S, Akamatsu S, et al. Association of missense polymorphism in HSD3B1 with outcomes among men with prostate cancer treated with androgen-deprivation therapy or abiraterone. JAMA Netw Open. 2019;2(2):e190115.doi: 10.1001/jamanetworkopen.2019.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garcia Gil S, Ramos Rodriguez R, Plata Bello A, et al. Relationship between mutations in the HSD3B1 gene and response time to androgen deprivation therapy in the treatment of prostate cancer. Presented at European Society of Oncology Pharmacy. Nantes, France; October 25-27, 2018.
- 10. Hearn JWD, Xie W, Nakabayashi M, et al. Association of HSD3B1 genotype with response to androgen-deprivation therapy for biochemical recurrence after radiotherapy for localized prostate cancer. JAMA Oncol. 2018;4(4):558-562. doi: 10.1001/jamaoncol.2017.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hearn JWD, Sweeney CJ, Almassi N, et al. HSD3B1 genotype and clinical outcomes in metastatic castration-sensitive prostate cancer. JAMA Oncol. 2020;6(4):e196496.doi: 10.1001/jamaoncol.2019.6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu C, Terbuch A, Dolling D, et al. Treatment with abiraterone and enzalutamide does not overcome poor outcome from metastatic castration-resistant prostate cancer in men with the germline homozygous HSD3B1 c.1245C genotype. Ann Oncol. 2020;31(9):1178-1185. doi: 10.1016/j.annonc.2020.04.473. [DOI] [PubMed] [Google Scholar]
- 13. Khalaf DJ, Aragón IM, Annala M, et al. ; for the PROREPAIR-B investigators. HSD3B1 (1245A>C) germline variant and clinical outcomes in metastatic castration-resistant prostate cancer patients treated with abiraterone and enzalutamide: results from two prospective studies. Ann Oncol. 2020;31(9):1186-1197. doi: 10.1016/j.annonc.2020.06.006. [DOI] [PubMed] [Google Scholar]
- 14. Prizment AE, McSweeney S, Pankratz N, et al. Prostate cancer mortality associated with aggregate polymorphisms in androgen-regulating genes: the Atherosclerosis Risk in the Communities (ARIC) study. Cancers (Basel). 2021;13(8):1958. doi: 10.3390/cancers13081958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kruse ML, Patel M, McManus J, et al. Adrenal-permissive HSD3B1 genetic inheritance and risk of estrogen-driven postmenopausal breast cancer. JCI Insight. 2021;6(20):e150403. doi: 10.1172/jci.insight.150403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Amant F, Moerman P, Neven P, Timmerman D, Van Limbergen E, Vergote I.. Endometrial cancer. Lancet. 2005;366(9484):491-505. doi: 10.1016/S0140-6736(05)67063-8. [DOI] [PubMed] [Google Scholar]
- 17. Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15(1):10-17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 18. Hecht JL, Mutter GL.. Molecular and pathologic aspects of endometrial carcinogenesis. J Clin Oncol. 2006;24(29):4783-4791. doi: 10.1200/jco.2006.06.7173. [DOI] [PubMed] [Google Scholar]
- 19. Murali R, Soslow RA, Weigelt B.. Classification of endometrial carcinoma: more than two types. Lancet Oncol. 2014;15(7):e268-e278. doi: 10.1016/S1470-2045(13)70591-6. [DOI] [PubMed] [Google Scholar]
- 20. Tomczak K, Czerwińska P, Wiznerowicz M.. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn). 2015;19(1A):A68-A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kandoth C, Schultz N, Cherniack AD, et al. ; for the Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67-73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61-70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sanchez-Vega F, Mina M, Armenia J, et al. ; for the Cancer Genome Atlas Research Network. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173(2):321-337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yuan J, Hu Z, Mahal BA, et al. Integrated analysis of genetic ancestry and genomic alterations across cancers. Cancer Cell. 2018;34(4):549-560.e9. doi: 10.1016/j.ccell.2018.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203-209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Galinsky KJ, Bhatia G, Loh P-R, et al. Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am J Hum Genet. 2016;98(3):456-472. doi: 10.1016/j.ajhg.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wasserstein RL, Lazar NA.. The ASA statement on p-values: context, process, and purpose. Am Stat. 2016;70(2):129-133. doi: 10.1080/00031305.2016.1154108. [DOI] [Google Scholar]
- 28. Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284-296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shimodaira M, Nakayama T, Sato N, et al. Association of HSD3B1 and HSD3B2 gene polymorphisms with essential hypertension, aldosterone level, and left ventricular structure. Eur J Endocrinol. 2010;163(4):671-680. doi: 10.1530/eje-10-0428. [DOI] [PubMed] [Google Scholar]
- 30. Yang XY, Wu WJ, Yang C, et al. Association of HSD17B3 and HSD3B1 polymorphisms with acne vulgaris in Southwestern Han Chinese. Dermatology. 2013;227(3):202-208. doi: 10.1159/000353581. [DOI] [PubMed] [Google Scholar]
- 31. Sieh W, Köbel M, Longacre TA, et al. Hormone-receptor expression and ovarian cancer survival: an Ovarian Tumor Tissue Analysis consortium study. Lancet Oncol. 2013;14(9):853-862. doi: 10.1016/S1470-2045(13)70253-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wentzensen N, Poole EM, Trabert B, et al. Ovarian cancer risk factors by histologic subtype: an analysis from the Ovarian Cancer Cohort Consortium. J Clin Oncol. 2016;34(24):2888-2898. doi: 10.1200/jco.2016.66.8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H, Liu Y, Wang Y, Zhao X, Qi X.. Hormone therapy for ovarian cancer: emphasis on mechanisms and applications (review). Oncol Rep. 2021;46(4):223.doi: 10.3892/or.2021.8174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Flanagan MR, Doody DR, Voutsinas J, et al. Association of HSD3B1 genotype and clinical outcomes in postmenopausal estrogen-receptor-positive breast cancer. Ann Surg Oncol. 2022. doi: 10.1245/s10434-022-12088-w. [DOI] [PubMed] [Google Scholar]
- 35. Hackenberg R, Schulz KD.. Androgen receptor mediated growth control of breast cancer and endometrial cancer modulated by antiandrogen- and androgen-like steroids. J Steroid Biochem Mol Biol. 1996;56(1-6 spec no):113-117. doi: 10.1016/0960-0760(95)00228-6. [DOI] [PubMed] [Google Scholar]
- 36. Tangen IL, Onyango TB, Kopperud R, et al. Androgen receptor as potential therapeutic target in metastatic endometrial cancer. Oncotarget. 2016;7(31):49289-49298. doi: 10.18632/oncotarget.10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ricciardelli C, Bianco-Miotto T, Jindal S, et al. The magnitude of androgen receptor positivity in breast cancer is critical for reliable prediction of disease outcome. Clin Cancer Res. 2018;24(10):2328-2341. doi: 10.1158/1078-0432.CCR-17-1199. [DOI] [PubMed] [Google Scholar]
- 38. Hickey TE, Selth LA, Chia KM, et al. The androgen receptor is a tumor suppressor in estrogen receptor-positive breast cancer. Nat Med. 2021;27(2):310-320. doi: 10.1038/s41591-020-01168-7. [DOI] [PubMed] [Google Scholar]
- 39. Wan J, Gao Y, Zeng K, et al. The levels of the sex hormones are not different between type 1 and type 2 endometrial cancer. Sci Rep. 2016;6(1):39744.doi: 10.1038/srep39744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zein J, Gaston B, Bazeley P, et al. HSD3B1 genotype identifies glucocorticoid responsiveness in severe asthma. Proc Natl Acad Sci USA. 2020;117(4):2187-2193. doi: 10.1073/pnas.1918819117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McManus JM, Sabharwal N, Bazeley P, Sharifi N.. Inheritance of a common androgen synthesis variant allele is associated with female COVID susceptibility in UK Biobank. Eur J Endocrinol. 2022;187(1):1-14. doi: 10.1530/eje-21-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stanczyk FZ, Mathews BW, Sherman ME.. Relationships of sex steroid hormone levels in benign and cancerous breast tissue and blood: a critical appraisal of current science. Steroids. 2015;99(pt A):91-102. doi: 10.1016/j.steroids.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 43. Neuzillet Y, Raynaud JP, Radulescu C, et al. Sexual steroids in serum and prostatic tissue of human non-cancerous prostate (STERPROSER trial). Prostate. 2017;77(15):1512-1519. doi: 10.1002/pros.23429. [DOI] [PubMed] [Google Scholar]
- 44. McManus JM, Bohn K, Alyamani M, Chung Y-M, Klein EA, Sharifi N.. Rapid and structure-specific cellular uptake of selected steroids. PLos One. 2019;14(10):e0224081.doi: 10.1371/journal.pone.0224081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wright JD, Fiorelli J, Schiff PB, et al. Racial disparities for uterine corpus tumors. Cancer. 2009;115(6):1276-1285. doi: 10.1002/cncr.24160 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TCGA data underlying this article are available from the NCI’s Genomic Data Commons Data Portal (portal.gdc.cancer.gov) and GDC Legacy Archive (portal.gdc.cancer.gov/legacy-archive). UK Biobank data underlying this article are available from the UK Biobank (www.ukbiobank.ac.uk) after applying for access.