Abstract
A major phenotypic trait of the Mycobacterium avium complex is the ability to produce rough and smooth colony variants. The chemical basis of this morphological variation is the loss of an antigenic surface structure, termed glycopeptidolipid (GPL), by rough variants. Using M. avium serovar 2 strain 2151 as a model system, this laboratory previously reported that rough variants arise via the deletion of large genomic regions encoding GPL biosynthesis. One such deletion encompasses the gene cluster (ser2) responsible for production of the serovar 2 GPL haptenic oligosaccharide. In this study, nucleotide sequencing revealed that both ends of the ser2 gene cluster are flanked by a novel insertion sequence (IS1601) oriented as direct repeats. Detailed analyses of the site of deletion in the genome of M. avium 2151 Rg-1 demonstrated that a single copy of IS1601 remained and that the ser2 gene cluster was deleted by homologous recombination. This same deletion pattern was observed for 10 out of 15 rough colony variants tested. Additionally, these studies revealed that IS1601 contains portions of three independent insertion sequences. This report is the first to define the precise genetic basis of colony variation in Mycobacterium spp. and provides further evidence that homologous recombination between insertion sequence elements can be a primary determinant of genome plasticity in these bacteria.
Mycobacterium avium and Mycobacterium intracellulare comprise what is commonly referred to as the M. avium complex (MAC) (20). These bacteria are opportunistic pathogens and primary etiological agents of disseminated bacterial infections in patients in advanced stages of AIDS (13). MAC infections in these individuals are associated with an increased risk of death (8), and such infections are difficult to treat due to the innate resistance of the MAC to many common antimycobacterial drugs (13). A complicating factor in studying pathogenic mechanisms of the MAC is the high degree of genetic and phenotypic variability observed among strains (3, 28, 35). The most noted of these mutable phenotypic traits is the ability of MAC strains to produce three distinct colony morphologies: smooth transparent (SmT), smooth opaque (SmO), and rough (Rg) (16, 41). In vivo and in vitro models of infection demonstrate that SmT variants are highly virulent and SmO variants are avirulent (30, 31, 32, 38). However, Rg variants are reported to be either highly virulent or avirulent (30, 31, 32), suggesting that at least two forms of Rg morphological variants exist.
Early studies addressing the chemical basis of colony morphology in MAC strains revealed that Rg variants are devoid of the characteristic glycopeptidolipids (GPLs) (1) on the surface of SmO and SmT variants (2). More recently, detailed chemical analyses of M. avium serovar 2 strain 2151 demonstrated that two chemotypes of Rg variants are formed: one that is devoid of any vestige of the GPL (Rg-4), and one that lacks glycosylation but produces the lipopeptide core (Rg-1) (5). Additionally, the formation of these Rg chemotypes is associated with large genomic deletions (4). Specifically, Rg-1-type organisms arise through a deletion of the M. avium ser2 gene cluster, the genomic region encoding glycosylation of the serovar 2-specific GPL (4, 6, 29), whereas the deletion associated with Rg-4-type organisms maps to a region downstream of the ser2 gene cluster and presumably encompasses the genes encoding lipopeptide biosynthesis (4). Partial characterization of these deletions also led to the hypothesis that excision of the ser2 gene cluster (Rg-1) is mediated by recombination between homologous 2.8-kb ClaI fragments on either side of ser2 (4). However, the sequence of the 2.8-kb ClaI repeats and the precise mechanism of the deletion were not elucidated. Our current studies demonstrate that the ser2 gene cluster of M. avium 2151 SmO/SmT is flanked on either side with identical copies of a novel 4.3-kb insertion sequence (IS1601) oriented as direct repeats. PCR amplification and sequencing of the genomic site of the ser2 deletion in M. avium 2151 Rg-1 and 10 independently derived Rg colony variants indicate that the ser2 gene cluster of M. avium 2151 is usually deleted via homologous recombination between direct repeats of IS1601.
MATERIALS AND METHODS
Isolation of new rough colony variants, bacterial growth, and DNA isolation.
Additional Rg colony variants were isolated by spreading approximately 103 CFU of M. avium strain 2151 SmT and SmO (4) on plates (15 by 150 mm) of Middlebrook 7H11 agar (Difco Laboratories, Detroit, Mich.) containing 10% oleic acid–albumin–dextrose–catalase supplement (7H11-OADC) (10). After 2 weeks of incubation at 37°C, the plates were evaluated for the presence of Rg colony variants. Individual Rg colonies were picked and passed at least three times on 7H11-OADC plates to ensure morphological stability.
For DNA isolation, frozen stocks of M. avium serovar 2 strain 2151 Rg-1 (4) and 15 Rg colony variants were plated on 7H11-OADC medium. After 2 weeks of growth at 37°C, single colonies were used to inoculate Middlebrook 7H9 broth (Difco) containing 10% OADC. The 10-ml cultures were incubated with mild agitation for 1 week and scaled up to 150 ml of 7H9 broth, and at 2 weeks of growth the cells were harvested by centrifugation at 3,000 × g for 30 min. Genomic DNA was isolated from the cell pellets by minor modifications of the method of Belisle et al. (6). Briefly, the procedure involved cell lysis by treatment with lysozyme, proteinase K, and sodium dodecyl sulfate, followed by extraction with phenol-CHCl3-isoamyl alcohol (25:24:1) and CHCl3-isoamyl alcohol (24:1) prior to DNA precipitation. The DNA pellet was washed three times with 70% cold ethanol, dried, and dissolved in sterile H2O.
Escherichia coli strain DH5α (Life Technologies Gibco-BRL, Rockville, Md.) was used for propagation of all recombinant plasmids. Cells were grown with Luria-Bertani medium, and ampicillin (100 μg/ml) was added for the growth of recombinant E. coli clones. Transformation-competent cells of E. coli DH5α were generated as described by Sambrook et al. (37). Recombinant plasmids were isolated using the QIAprep Spin Miniprep kit (Qiagen, Valencia, Calif.).
DNA cloning.
All gel-purified DNA fragments were cloned into the corresponding restriction site of pBluescript II SK(−) (Stratagene, La Jolla, Calif.) unless otherwise noted. Recombinant plasmids pJTB232.1 and pJTB233.1 were derived from pJTB232 and pJTB233 (4), respectively, by subcloning the 2.8-kb ClaI fragments. For further sequencing up- and downstream of pJTB232.1, the 9.6-kb HindIII-DraI fragment of cosmid pJTB232 (Fig. 1) was cloned into the SmaI and HindIII sites of the vector to give plasmid pJTB232.2. To sequence the flanking region of pJTB233.1, the 7.9-kb HindIII fragment of cosmid pJTB230 (Fig. 1), which is similar to pJTB233 (4), was used to generate plasmid pJTB230.1. Subclones of pJTB232.2 and pJTB230.1 for DNA sequencing were generated with ClaI, EcoRI, EcoRV, SmaI, PstI, XhoI, and NotI (Life Technologies).
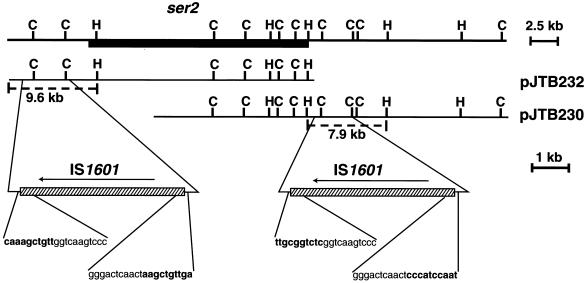
FIG. 1.
Organization of the ser2 gene cluster and flanking IS1601 sequences. The ser2 gene cluster is represented by the solid box, and IS1601 elements are represented by slashed boxes. Dashed lines depict the 9.6- and 7.9-kb fragments of pJTB232 and pJTB230 that were used as templates for the sequencing of IS1601-L (left copy) and IS1601-R (right copy), respectively. H, HindIII; C, ClaI.
PCR analyses.
Primers PL1 (5′CGCTGCGGCTATAGTCTTTAG3′), PR1 (5′CACGAGCACCCAGAAACATCA3′), PL2 (5′GCAATGCCGAAGGAACAGTCG3′), and PR2 (5′TACGGGCGCCATACACCCGGT3′) were synthesized by the Macromolecular Resources Facility at Colorado State University. All PCR amplifications were performed using a 2400 GeneAmp PCR System (Perkin-Elmer, Norwalk, Conn.) and Vent polymerase (New England Biolabs, Beverly, Mass.) or Taq polymerase (Life Technologies). The PCR program used with the above primers was 25 cycles of 94.0°C for 30 s, 63.0°C for 1.5 min, and 72.0°C for 1 min. Prior to the first cycle, the starting temperature of 94.0°C was held for 5 min, and at the end of the last cycle, a temperature of 72.0°C was held for 7 min. PCR amplification of the rtfA gene of M. avium was performed as previously described by Eckstein et al. (12). PCR products were resolved on a 0.8% agarose gel and stained with ethidium bromide.
DNA sequencing.
DNA sequencing of pJTB232.1, pJTB233.1, pJTB232.2, and pJTB230.1 (and their appropriate subclones) was performed with the pBluescript T3 and M13-20 primers. Custom primers were synthesized as necessary to resolve sequence ambiguities. The 4,451-bp PCR product obtained using primers PL1 and PR1 and M. avium 2151 Rg-1 genomic DNA was cloned into pCR-Blunt (Invitrogen, Carlsbad, Calif.), and the ends of this fragment were sequenced using the M13-20 and M13-20 reverse primers. Similarly, the 4,451-bp PCR products obtained from M. avium 2151 Rg-O2 and Rg-O7 were cloned into pGEM-T Easy (Promega, Madison, Wis.), and the ends of the fragments were sequenced using the T7 and SP6 primers. Sequencing of DNA was performed by the Macromolecular Resources Facility at Colorado State University or by the Department of Molecular, Cellular, and Developmental Biology at the University of Colorado in Boulder. DNA sequences, open reading frames (ORFs), and codon usage were determined with Sequencher 3.0 software (Gene Codes Corporation, Ann Arbor, Mich.) and FramePlot 2.3beta (http://www.nih.go.jp/∼jun/cgi-bin/frameplot-2.3b.pl) (22).
Nucleotide sequence accession number.
The sequence of IS1601 is available on GenBank at AF060182.
RESULTS
Two identical repetitive elements flank the ser2 gene cluster as direct repeats.
It was previously demonstrated using restriction endonuclease mapping and Southern blot hybridization that the ser2 gene cluster of M. avium 2151 SmO/SmT is flanked by 2.8-kb ClaI fragments and that the genomic region bracketed by these putative repetitive sequences had been deleted in the Rg-1 chemotype (4). To determine if these fragments were truly repetitive elements, the ClaI fragments were cloned, and DNA sequencing demonstrated that the left and right 2.8-kb ClaI fragments were identical. The complete sequence of these repetitive elements beyond the 2.8-kb ClaI fragments were determined by subcloning a 9.6-kb DraI-HindIII fragment containing the left 2.8-kb ClaI region and a 7.9-kb HindIII fragment containing the right 2.8-kb ClaI region from pJTB232 and pJTB230 (4), respectively (Fig. 1). The DNA sequence revealed that the full length of the repetitive element is 4,262 bp and that the left and right elements are identical. The complete sequencing of the ser2 gene cluster and flanking regions (T. M. Eckstein, M. L. Lambert, P. J. Brennan, J. T. Belisle, and J. M. Inamine, direct submission to GenBank, accession no. AF143772) showed that these elements are oriented as direct repeats in the genome (Fig. 1).
Repetitive element is a composite IS element designated IS1601. (i) Analysis of the ORFs.
The four ORFs (Fig. 2) of the repetitive element, designated IS1601, show a high degree of similarity to three separate IS families. ORF1 (nucleotides 126 to 1373) encodes a putative 45.3-kDa transposase, based on the high degree of homology (68% identity and 98% similarity) to the transposase of IS1512 from Mycobacterium gordonae (34) and to transposases of members of the IS256 family (11, 23, 33). The gene product of ORF2 (nucleotides 1632 to 2834) is predicted to be 44.2 kDa, and the amino acid sequence demonstrates significant homology (35% identity and 62% similarity) to the minicircle protein of IS117 (18) from Streptomyces coelicolor A3(2) that is a member of the IS110 family (26). Finally, the 13.0- and 18.7-kDa products predicted to be encoded by ORF3 (nucleotides 2886 to 3230) and ORF4 (nucleotides 3230 to 4147), respectively, are highly homologous to the transposase subunits of the IS3-like insertion sequences (IS987 and IS986) found in the Mycobacterium tuberculosis complex (19, 27).
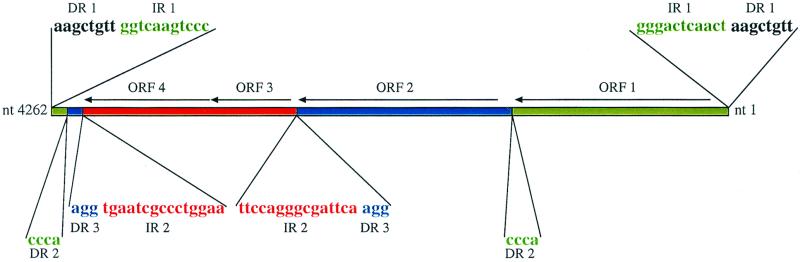
FIG. 2.
Organization of the ORFs of IS1601-L and the positions of the DRs and IRs that define parts A (green), B (blue), and C (red). The two copies of DR1 (shown in black) are found only in the left IS1601 (IS1601-L) shown in Fig. 1 and are not included in the 4,262-nucleotide (nt) position numbering.
(ii) Analysis of the direct and inverted repeats.
Supporting evidence that the 4,262-bp IS1601 is probably a composite of three different IS elements came from the localization of the direct repeats (DRs) and inverted repeats (IRs) within IS1601 with respect to the four ORFs (Fig. 2). By comparing these data with the genome sequence data from M. avium strain 104 provided by The Institute for Genomic Research (http://www.tigr.org) along with the DNA sequence of the ser2 gene cluster from strain 2151 (Eckstein et al., GenBank accession no. AF143772), three major observations were made. First, the 1,347-bp region of IS1601 (nucleotides 2832 to 4178; red area in Fig. 2) that contains ORF3 and ORF4 can be found as an independent IS element in strain 104 (http://www.tigr.org) and strain 2151. All of the independent copies of this 1,347-bp element possess terminal 15-bp IRs and appear to generate 3-bp DRs of the target sequence. The same organization of IRs and DRs is found within IS1601, and this 1,347-bp region, whose ends are defined by IR2 and DR3, is designated part C (red area in Fig. 2).
The second observation is that nucleotides 1 to 1373 and 4219 to 4262 of IS1601 (green areas in Fig. 2) together comprise the complete 1,417-bp independent IS element present in M. avium strain 104 (http://www.tigr.org). Moreover, the terminal 11-bp imprecise IRs at the ends of IS1601 (IR1 in Fig. 2) are the same as those of the 1,417-bp element in strain 104. The sequence from strain 104 indicates that the 1,417-bp element generated 8-bp DRs of the target sequence. An 8-bp duplication (DR1) was also found at both ends of the left copy of IS1601 (IS1601-L, Fig. 1 and 2), but not of the right copy (IS1601-R, Fig. 1). Thus, this 1,417-bp region, designated part A (green areas in Fig. 2), appears to be an independent IS element that was interrupted during the evolution of IS1601.
The third observation is that when parts A and C are subtracted from IS1601, the remaining 1,458 bp (nucleotides 1374 to 2831) and 36 bp (nucleotides 4179 to 4214) can form a 1,494-bp region (designated part B, blue areas in Fig. 2) containing ORF2 with a 4-bp DR at each end (designated DR2). Homology searches with the B region did not identify any similar sequences in the available M. avium strain 104 database, and there are no associated terminal IRs. Taken together, the physical relationship between these three regions suggests that IS1601 could have evolved from the insertion of B and C into A. As can be seen from Fig. 2, one possible scenario is that part C (red) is inserted into part B (blue), producing DR3, and this composite of B and C then is inserted into part A (green), generating DR2. Finally, the tripartite element containing A, B, and C (IS1601) is inserted into the ser2 region, giving rise to DR1.
Mechanism of deletion of the ser2 gene cluster.
The finding that IS1601 elements flank the ser2 gene cluster as direct repeats suggested that homologous recombination between the two copies of IS1601 might have mediated the deletion of the ser2 gene cluster to generate the Rg-1 mutant of M. avium strain 2151. However, there was also the possibility that the IS1601 elements and the intervening ser2 gene cluster formed a large composite transposon that was excised, as has been shown for the transpositional excision of λ prophage bracketed by direct repeats of IS1 (39). These two mechanisms could be readily discerned by the presence or absence of IS1601. If the deletion of the ser2 gene cluster had occurred via homologous recombination, then a single copy of IS1601 would remain at the site of deletion in the Rg-1 strain, while IS1601 would be absent from the deletion site if the ser2 genes had been excised by transposition (Fig. 3A).
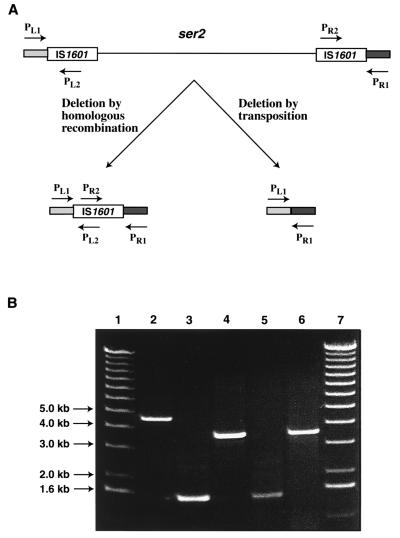
FIG. 3.
(A) Possible means of deletion of the ser2 gene cluster of M. avium 2151 SmT/SmO and the predicted organization of this genomic region in M. avium 2151 Rg-1 after deletion of the ser2 region. PL1, PL2, PR1, and PR2, and the corresponding arrows depict the locations of the PCR primers used to evaluate the deletion site in M. avium 2151 Rg-1. The sizes of the predicted PCR products are given in the text. (B) A 0.8% agarose gel of PCR products formed by amplification of M. avium 2151 Rg-1 genomic DNA and control DNAs using combinations of the primers PL1, PL2, PR1, and PR2. Lane 2, a 4.4-kb PCR product generated with primer pair PL1 and PR1 and Rg-1 genomic DNA. Lanes 3 and 5, the 1.5-kb PCR products generated using primer pair PL1 and PL2 with Rg-1 genomic DNA and pJTB232.2, respectively. Lanes 4 and 6, the 3.5-kb PCR products formed using primer pair PR1 and PR2 with Rg-1 genomic DNA and pJTB230.1, respectively. Lanes 1 and 7, molecular size markers.
A PCR strategy was devised to distinguish between these two mechanisms. Primers PL1 and PR1 were based on the sequences flanking IS1601-L and IS1601-R, respectively, and primers PL2 and PR2 were generated from internal sequences of IS1601 (Fig. 3A). Deletion of the ser2 gene cluster by transposition would produce a single PCR product of 189 bp (PL1 and PR1), whereas deletion by homologous recombination would result in products of 4,451 bp (PL1 and PR1), 1,496 bp (PL1 and PL2), and 3,508 bp (PR1 and PR2) with Rg-1 DNA. As shown in Fig. 3B, amplification of Rg-1 DNA resulted in PCR products of 4.4 kb (lane 2), 1.5 kb (lane 3), and 3.5 kb (lane 4). These results were compatible with the homologous recombination model.
To prove that IS1601 was indeed within the 4.4-kb PCR product derived with primers PL1 and PR1, the fragment was cloned and sequenced. This demonstrated that a single IS1601 element remained at the site of deletion in the Rg-1 chromosome. Moreover, the sequences flanking the left and right sides of this single copy of IS1601 were identical to those at the distal ends of IS1601-L and IS1601-R, respectively. These data, along with our previous biochemical characterization of M. avium Rg variants (5), provide definitive evidence that the deletion of a 21-kb genomic fragment containing the ser2 gene cluster that resulted in the Rg-1 morphotype was mediated by recombination between direct repeats of flanking IS1601 elements.
Screening of additional Rg mutants for the Rg-1 deletion.
In order to determine if the specific deletion observed for the Rg-1 morphotype was an isolated or rare event, 15 additional Rg colony variants (Rg-O1 to Rg-O10 and Rg-T1 to Rg-T5) were independently isolated from the M. avium 2151 SmO and SmT strains. Analysis by the PCR test described above showed that 9 of the 10 Rg isolates derived from the SmO morphotype and 1 of the 5 Rg isolates derived from the SmT morphotype yielded the 4.4-kb PCR fragment that is diagnostic for the Rg-1 deletion (Table 1). PCR analysis was also performed to determine the presence or absence of rtfA (12), a gene that is contained within the ser2 gene cluster and is thus a part of the genomic region that is lost in Rg-1. As expected, all of the newly isolated Rg variants that tested positive for the 4.4-kb PCR product lacked the rtfA gene, while four of the five that were negative for the Rg-1 deletion marker contained rtfA (Table 1). The exception was Rg-O8, a putative double-deletion mutant lacking both the 4.4-kb and rtfA PCR products, which is currently under investigation. The 4.4-kb PCR products from two of the Rg-1-like variants (Rg-O2 and Rg-O7) were selected at random, and the ends were sequenced. Both fragments had the same sequence as the 4.4-kb PCR product from Rg-1. Thus, these data demonstrate that recombination between these IS1601 elements and the resultant loss of the ser2 gene cluster are relatively common events. Additionally, the observation that not all Rg variants result from this type of deletion is in agreement with our earlier findings that an undefined deletion(s) outside of the ser2 gene cluster can also produce GPL-negative Rg mutants (4).
TABLE 1.
PCR amplification of the 4,451-bp Rg-1 deletion marker and the rtfA gene from Rg variants of M. avium strain 2151a
| Rough variants | 4,451-bp PCR product | rtfA gene |
|---|---|---|
| Rg-1b | + | − |
| Rg-4b | − | + |
| Rg-O1c | + | − |
| Rg-O2c | + | − |
| Rg-O3c | + | − |
| Rg-O4c | + | − |
| Rg-O5c | + | − |
| Rg-O6c | + | − |
| Rg-O7c | + | − |
| Rg-O8c | − | − |
| Rg-O9c | + | − |
| Rg-O10c | + | − |
| Rg-T1d | − | + |
| Rg-T2d | − | + |
| Rg-T3d | − | + |
| Rg-T4d | − | + |
| Rg-T5d | + | − |
The 4,451-bp PCR product was amplified using primers PL1 and PR1. rtfA was amplified using protocols described by Eckstein et al. (12).
Rg variants originally described by Belisle et al. (4).
Rg variants derived in this study from the SmO morphotype.
Rg variants derived in this study from the SmT morphotype.
DISCUSSION
Insertion elements have long been known to serve as substrates for homologous recombination. For example, the R100.1 plasmid of E. coli was observed to lose a 23-kb resistance determinant, and this deletion was mediated via RecA-dependent recombination between direct repeats of IS1 (9). Similarly, Ishiguro and Sato (21) demonstrated that spontaneous deletion of a citrate-utilizing gene cluster contained within a compound transposon (Tn3411) was mediated by homologous recombination between direct repeats of IS3411 elements at each end of Tn3411. However, deletion of this citrate-utilizing gene cluster was RecA independent. At present it is not known whether the 21-kb deletion observed in M. avium 2151 is RecA dependent or if recombination is mediated by an IS1601-encoded resolvase. However, since the predicted gene products of IS1601 do not show any homology to known resolvases, this latter mechanism would require a trans-acting resolvase.
The fact that the ser2 locus encodes a full complement of gene products for glycosylation of the serovar 2-specific GPL and is flanked by IS1601 elements suggested that this region of the M. avium genome might be a “biosynthetic island” and possibly a compound transposon. Sequencing of an 8.9-kb region of the ser2 gene cluster in M. avium subsp. paratuberculosis revealed a group of genes likely responsible for de novo synthesis of fucose and possessing a G+C content lower than that of the surrounding genome, and this led Tizard et al. (40) to speculate that this region represented a pathogenicity island. Pathogenicity islands generally are bordered by short direct repeats, possess other mobilization elements such as integrases, and in many cases possess a G+C content different from that of the surrounding chromosome (24). In comparison, the ser2 gene cluster (including the IS1601 elements) has a G+C content similar to that of the M. avium genome (Eckstein et al., GenBank submission), and the only direct repeat is the 4.3-kb IS1601. In addition, as noted by Tizard et al. (40), there is no direct evidence that the ser2 gene cluster contributes to the pathogenicity of the bacillus. In all, the ser2 gene cluster with its direct repeats of IS1601 does not fit the classical definition of a pathogenicity island, and, at least in the Rg-1-type mutants examined in this work, there is no evidence that it behaves like a compound transposon.
Genomic deletions in other Mycobacterium spp., in particular the M. tuberculosis complex, have been described. Mahairas et al. (25) isolated and characterized several regions of the genome of virulent Mycobacterium bovis (RD1, RD2, and RD3) that were absent from the genome of attenuated M. bovis strain BCG. Detailed sequence analyses of these regions and comparison to the deleted regions in M. bovis BCG demonstrated that a 24-bp imperfect direct repeat flanking the RD2 region was conserved at the site of deletion in BCG strains. This same type of organization was observed for the RD3 region; however, in this case the direct repeats were 12 bp in length. More recently, Gordon and colleagues (7, 17) have identified three deletions in M. tuberculosis H37Rv (RvD2, 7.9 kb; RvD3, 1 kb; and RvD4, 0.8 kb) that are mediated by homologous recombination between IS6110 elements. Similarly, Fang et al. (14) reported that deletions around the ilp locus of M. tuberculosis, identified as a hot spot for IS6110 insertions (19), were also produced by homologous recombination between copies of IS6110. Thus, deletion of the 21-kb ser2 gene cluster of M. avium 2151 via homologous recombination between IS elements is a further example of genome plasticity in Mycobacterium spp. and represents the first example where this genetic mechanism results in a gross morphological change in these bacteria.
A second interesting finding of this present work was that IS1601 appears to be a composite of three independent IS elements. The recently released sequence data for Streptomyces coelicolor A2(3) demonstrated a high concentration of IS elements in cosmid 3C8, with one of the IS110-like elements being disrupted by IS1648 (36). Similarly, Fang et al. (15) reported that the insertional hot spot for IS6110 in the ilp locus of M. tuberculosis is another IS element, IS1547. However, other copies of the disrupted IS110 or IS1547 have not been reported. In contrast, at least two identical copies of IS1601 were present in M. avium strain 2151, strongly suggesting that IS1601 is a true mobile genetic element. It was also noted that directly outside the imperfect inverted repeats of the IS1601-L were 8-bp DRs. At present it is not known which of the three putative transposases encoded on IS1601 is responsible for the movement of this IS element. However, members of the IS256 family typically produce 8-bp DRs (26), and part A of IS1601 (containing ORF1) is predicted to be a member of this IS family. Additionally, the 1,417-bp IS element present in the available genomic sequence of M. avium strain 104 (http://www.tigr.org) and analogous to part A of IS1601 is also associated with 8-bp DRs. In contrast, members of the IS110 family (part B of IS1601) typically do not generate DRs, and the transposase common to the IS3 family (part C of IS1601) produces short DRs of 3 to 5 bp (26). Given that the formation and the size of DRs are a function of the transposase (26), the transposition event responsible for the insertion of IS1601-L was probably mediated by the transposase encoded by ORF1 of IS1601. However, this may not be the only active transposase of IS1601, since IS1601-R lacks similar DRs. We are currently evaluating whether more copies of IS1601 are present in the M. avium genome and whether they mediated other deletions, or if they are specific to the gene cluster that is responsible for glycosylation of GPLs.
ACKNOWLEDGMENTS
This work was supported by grants (AI-18357 to Patrick J. Brennan and AI-41925 to Julia M. Inamine) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
We thank Jonathan Carlson, Fauzi Silbaq, and Alain Baulard for advice and thoughtful discussion. We also thank Nazir Barekzi for technical assistance. Preliminary sequence data for M. avium strain 104 were obtained from The Institute for Genomic Research website at http://www.tigr.org.
REFERENCES
- 1.Aspinall G O, Chatterjee D, Brennan P J. The variable surface glycolipids of mycobacteria: structures, synthesis of epitopes, and biological properties. Adv Carbohydr Chem Biochem. 1995;51:169–242. doi: 10.1016/s0065-2318(08)60194-8. [DOI] [PubMed] [Google Scholar]
- 2.Barrow W W, Brennan P J. Isolation in high frequency of rough variants of Mycobacterium intracellulare lacking C-mycoside glycopeptidolipid antigens. J Bacteriol. 1982;150:381–384. doi: 10.1128/jb.150.1.381-384.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belisle J T, Brennan P J. Molecular basis of colony morphology in Mycobacterium avium. Res Microbiol. 1994;145:237–242. doi: 10.1016/0923-2508(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 4.Belisle J T, Klaczkiewicz K, Brennan P J, Jacobs W R, Jr, Inamine J M. Rough morphological variants of Mycobacterium avium: characterization of genomic deletions resulting in the loss of glycopeptidolipid expression. J Biol Chem. 1993;268:10517–10523. [PubMed] [Google Scholar]
- 5.Belisle J T, McNeil M R, Chatterjee D, Inamine J M, Brennan P J. Expression of the core lipopeptide of glycopeptidolipid surface antigen in rough mutants of Mycobacterium avium. J Biol Chem. 1993;268:10510–10516. [PubMed] [Google Scholar]
- 6.Belisle J T, Pascopella L, Inamine J M, Brennan P J, Jacobs W R., Jr Isolation and expression of a gene cluster responsible for biosynthesis of the glycopeptidolipid antigens of Mycobacterium avium. J Bacteriol. 1991;173:6991–6997. doi: 10.1128/jb.173.21.6991-6997.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broch R, Philipp W J, Stavropoulos E, Colston M J, Cole S T, Gordon S V. Genomic analysis reveals variation between Mycobacterium tuberculosis H37Rv and attenuated M. tuberculosis H37Ra strain. Infect Immun. 1999;67:5768–5774. doi: 10.1128/iai.67.11.5768-5774.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaisson R E, Gallant J F, Keruly J C, Moore R D. Impact of opportunistic disease on survival in patients with HIV infection. AIDS. 1998;12:29–33. doi: 10.1097/00002030-199801000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Chandler M, Allet B, Gally E, Boy de la Tour E, Caro L. Involvement of IS1 in the dissociation of the r-determinant and RTF components of the plasmid R100.1. Mol Gen Genet. 1977;153:289–295. doi: 10.1007/BF00431594. [DOI] [PubMed] [Google Scholar]
- 10.Cohn M L, Waggoner R F, McClatchy J K. The 7H11 medium for the cultivation of mycobacteria. Am Rev Respir Dis. 1968;98:295–296. doi: 10.1164/arrd.1968.98.2.295. [DOI] [PubMed] [Google Scholar]
- 11.Collins D M, Stephens D M. Identification of an insertion sequence, IS1081, in Mycobacterium bovis. FEMS Microbiol Lett. 1991;67:11–15. doi: 10.1016/0378-1097(91)90435-d. [DOI] [PubMed] [Google Scholar]
- 12.Eckstein T M, Silbaq F S, Chatterjee D, Kelly N J, Brennan P J, Belisle J T. Identification and recombinant expression of a Mycobacterium avium rhamnosyltransferase gene (rtfA) involved in glycopeptidolipid biosynthesis. J Bacteriol. 1998;180:5567–5573. doi: 10.1128/jb.180.21.5567-5573.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellner J J, Goldberger M J, Parenti D M. Mycobacterium avium infection and AIDS: a therapeutic dilemma in rapid evolution. J Infect Dis. 1991;163:1326–1335. doi: 10.1093/infdis/163.6.1326. [DOI] [PubMed] [Google Scholar]
- 14.Fang Z, Doig C, Kenna D T, Smittipat N, Palittapongarnpim P, Watt B, Forbes K J. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J Bacteriol. 1999;181:1014–1020. doi: 10.1128/jb.181.3.1014-1020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang Z, Doig C, Morrison N, Watt B, Forbes K J. Characterization of IS1547, a new member of the IS900 family in the Mycobacterium tuberculosis complex, and its association with IS6110. J Bacteriol. 1999;181:1021–1024. doi: 10.1128/jb.181.3.1021-1024.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fregnan G B, Smith D W. Description of various colony forms of mycobacteria. J Bacteriol. 1962;83:819–827. doi: 10.1128/jb.83.4.819-827.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon S V, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole S T. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol Microbiol. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 18.Henderson D J, Lydiate D J, Hopwood D A. Structural and functional analysis of the mini-circle, a transposable element of Streptomyces coelicolor A3(2) Mol Microbiol. 1989;3:1307–1318. doi: 10.1111/j.1365-2958.1989.tb00112.x. [DOI] [PubMed] [Google Scholar]
- 19.Hermans P W, van Soolingen D, Mik E M, de Haas P E, Dale J W, van Embden J D. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inderlied C B, Kemper C A, Bermudez L E. The Mycobacterium avium complex. Clin Microbiol Rev. 1993;6:266–310. doi: 10.1128/cmr.6.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishiguro N, Sato G. Spontaneous deletion of citrate-utilizing ability promoted by insertion sequences. J Bacteriol. 1984;160:642–650. doi: 10.1128/jb.160.2.642-650.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishikama J, Hotta K. FramePlot: a new implementation of the frame analysis for prediction of protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol Lett. 1999;174:251–253. doi: 10.1111/j.1574-6968.1999.tb13576.x. [DOI] [PubMed] [Google Scholar]
- 23.Komeda H, Kobayashi M, Shimizu S. Characterization of the gene cluster of high-molecular-mass nitrile hydratase (H-NHase) induced by its reaction product in Rhodococcus rhodochrous J1. Proc Natl Acad Sci USA. 1996;93:4267–4272. doi: 10.1073/pnas.93.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C A. Pathogenicity islands and evolution of bacterial pathogens. Infect Agents Dis. 1996;5:1–7. [PubMed] [Google Scholar]
- 25.Mahairas G G, Sabo P J, Hickey M J, Singh D C, Stover C K. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J Bacteriol. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAdam R A, Hermans P W, van Soolingen D, Zainuddin Z F, Catty D, van Embden J D, Dale J W. Characterization of a Mycobacterium tuberculosis insertion sequence belonging to the IS3 family. Mol Microbiol. 1990;4:1607–1613. doi: 10.1111/j.1365-2958.1990.tb02073.x. [DOI] [PubMed] [Google Scholar]
- 28.McFadden J J, Butcher P D, Thompson J, Chiodini R, Hermon-Taylor J. The use of DNA probes identifying restriction-fragment-length polymorphisms to examine the Mycobacterium avium complex. Mol Microbiol. 1987;1:283–291. doi: 10.1111/j.1365-2958.1987.tb01934.x. [DOI] [PubMed] [Google Scholar]
- 29.Mills J A, McNeil M R, Belisle J T, Jacobs W R, Jr, Brennan P J. Loci of Mycobacterium avium ser2 gene cluster and their functions. J Bacteriol. 1994;176:4803–4808. doi: 10.1128/jb.176.16.4803-4808.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moehring J M, Solotorovsky M R. Relationship of colonial morphology to virulence for chickens of Mycobacterium avium and the nonphotochromogens. Am Rev Respir Dis. 1965;92:704–713. doi: 10.1164/arrd.1965.92.5.704. [DOI] [PubMed] [Google Scholar]
- 31.Olitzki A L, Davis C L, Schaefer W B, Cohn M L. Colony variants of avian-Battey group mycobacteria intracerebrally injected into mice. Pathol Microbiol. 1969;34:316–323. doi: 10.1159/000162176. [DOI] [PubMed] [Google Scholar]
- 32.Pedrosa J, Florido M, Kunze Z M, Castro A G, Portaels F, McFadden J, Silva M T, Appelberg R. Characterization of the virulence of Mycobacterium avium complex (MAC) isolates in mice. Clin Exp Immunol. 1994;98:210–216. doi: 10.1111/j.1365-2249.1994.tb06127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Picardeau M, Bull T J, Prod'hom G, Pozniak A L, Shanson D C, Vincent V. Comparison of a new insertion element, IS1407, with established molecular markers for the characterization of Mycobacterium celatum. Int J Syst Bacteriol. 1997;47:640–644. doi: 10.1099/00207713-47-3-640. [DOI] [PubMed] [Google Scholar]
- 34.Picardeau M, Bull T J, Vincent V. Identification and characterization of IS-like elements in Mycobacterium gordonae. FEMS Microbiol Lett. 1997;154:95–102. doi: 10.1111/j.1574-6968.1997.tb12629.x. [DOI] [PubMed] [Google Scholar]
- 35.Picken R N, Tsang A Y, Yang H L. Speciation of organisms within the Mycobacterium avium-Mycobacterium intracellulare-Mycobacterium scrofulaceum (MAIS) complex based on restriction fragment length polymorphisms. Mol Cell Probes. 1988;2:289–304. doi: 10.1016/0890-8508(88)90013-8. [DOI] [PubMed] [Google Scholar]
- 36.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb of Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schaefer W B, Davis C L, Cohn M L. Pathogenicity of transparent, opaque, and rough variants of Mycobacterium avium in chickens and mice. Am Rev Respir Dis. 1970;102:499–506. doi: 10.1164/arrd.1970.102.4.499. [DOI] [PubMed] [Google Scholar]
- 39.Shapiro J A, MacHattie L A. Integration and excision of prophage lambda mediated by the IS1 element. Cold Spring Harbor Symp Quant Biol. 1979;43(Pt. 2):1135–1142. doi: 10.1101/sqb.1979.043.01.127. [DOI] [PubMed] [Google Scholar]
- 40.Tizard M, Bull T, Millar D, Doran T, Martin H, Sumar N, Ford J, Hermon-Taylor J. A low G+C content genetic island in Mycobacterium avium subsp. paratuberculosis and M. avium subsp. silvaticum with homologous genes in Mycobacterium tuberculosis. Microbiology. 1998;144:3413–3423. doi: 10.1099/00221287-144-12-3413. [DOI] [PubMed] [Google Scholar]
- 41.Vestal A L, Kubica G P. Differential colonial characteristics of mycobacteria on Middlebrook and Cohn 7H10 agar-base medium. Am Rev Respir Dis. 1966;94:247–252. doi: 10.1164/arrd.1966.94.2.247. [DOI] [PubMed] [Google Scholar]