Abstract
Aim
The placenta–brain axis reflects a developmental linkage where disrupted placental function is associated with impaired neurodevelopment later in life. Placental gene expression and the expression of epigenetic modifiers such as miRNAs may be tied to these impairments and are understudied.
Materials & methods
The expression levels of mRNAs (n = 37,268) and their targeting miRNAs (n = 2083) were assessed within placentas collected from the ELGAN study cohort (n = 386). The ELGAN adolescents were assessed for neurocognitive function at age 10 and the association with placental mRNA/miRNAs was determined.
Results
Placental mRNAs related to inflammatory and apoptotic processes are under miRNA control and associated with cognitive impairment at age 10.
Conclusion
Findings highlight key placenta epigenome–brain relationships that support the developmental origins of health and disease hypothesis.
Keywords: : cognition, epigenetics, epigenome, miRNA, mRNA, placenta
Plain language summary
Children born extremely preterm are at increased risk for neurodevelopmental impairments such as cerebral palsy, intellectual disability and autism. The biological processes that lead to these impairments likely begin before birth and involve altered placental function. In this study, the authors analyzed placental genomic and epigenomic data from children who were born extremely preterm in relation to cognitive assessments at 10 years of age. They examined the differences between the expression of placental genes and molecules that influence the expression of placental genes, comparing children who had impaired cognition at 10 years with children who did not. The results demonstrated elevated expression levels of genes involved in inflammatory processes and molecules that control the expression of these genes within the placentas of children who had impaired cognition at age 10.
Understanding the etiologic factors that underlie cognitive impairment in children is of great importance to public health. Individuals born prematurely are particularly at high risk for deficits in general cognitive ability (IQ) and executive function [1]. There is increasing evidence that in utero exposure to environmental chemicals and other stressors is associated with impaired neurodevelopment later in life [2]. The association between perinatal exposure to chemicals and stressors and impaired neurodevelopment is an exemplar of the developmental origins of health and disease (DOHaD) framework. This framework posits that the fetal environment contributes to long-term risk of noncommunicable health outcomes [3]. A target organ that may underlie the biological basis for the DOHaD framework is the placenta [4]. Disrupted placental functioning can alter the trajectory of fetal development, particularly in the brain [5]. This relationship between the placenta and fetal neurodevelopment is known as the placenta–brain axis [6].
The placenta is a multifunctional organ that mediates nutrient, gas and waste exchange between the mother and fetus during pregnancy. It also serves as a central regulator for healthy fetal development and a selective barrier limiting exposure to environmental toxicants [7]. Disrupted placental functioning during pregnancy is associated with adverse pregnancy and later in life health outcomes in offspring. These health outcomes include increased risk of hypertension, diabetes, obesity and adverse neurodevelopment [4,8,9]. Specifically, placental morphology and physiology, including epigenetic and gene expression changes, have been tied to later neurocognition [10–14].
Morphologic and physiologic disruptions in the placenta may be driven by transcriptomic alterations, which in turn can be mediated through epigenetic modifications [15]. Epigenetic modifiers include the methylation of CpG sites, the modification of histones, and altered miRNA expression [16]. The altered methylation of CpG sites within specific genomic locations can result in gene silencing or activation [17]. Previous research by the study authors has demonstrated links between placental CpG methylation and offspring neurocognition [12,18]. Other researchers have also found that the placental epigenome predicts neurodevelopmental outcomes, including aggressive behavior [12,18–20]. However, less is understood about the relationship between the expression of placental miRNAs, another critical epigenetic regulator, and later in life neurocognition.
MicroRNAs (miRNAs) are a class of short (∼22 nucleotides in length), noncoding RNAs that play an important role in the regulation of gene expression at the post-transcriptional level [21]. Gene expression is regulated by the binding of miRNAs to target sites within a target mRNA, where miRNA binding generally impedes translation, resulting in the suppression of protein synthesis [21]. Less often, miRNAs have been found to upregulate gene targets [22]. Several miRNAs display tissue-specific expression and are only expressed within the placenta [23,24]. The expression of miRNAs in the placenta has been tied to infant neurobehavior [25], supporting the potential for miRNAs in the placenta to impact neurobehavior later in life [26]. Few studies to date have evaluated the relationship between placental miRNA expression and later life neurocognition.
Understanding the role of sexual dimorphism is a vital facet of researching the association between placental -omics signatures and later life outcomes. Sex-based differences in infant and later in life health outcomes are well-described, as are sex-specific responses to maternal exposures to environmental toxicants [27]. Previous studies have identified sexually dimorphic expression of placental mRNAs and miRNAs, which highlight the sex-dependent genomic patterning in the placenta [26]. Further, sex-specific placental CpG methylation has been observed with striking patterns of hyper- or hypo-methylation, depending on the sex of the fetus [27]. While sex-based differences in the prevalence of cognitive impairment and other neurobehavioral outcomes are well characterized [28–30], sex-based differences in the placental epigenome as it relates to neurocognition has been understudied.
This study examined whether gene and miRNA expression levels in the placenta are associated with neurocognition at age 10. Placentas were collected at birth from the Extremely Low Gestational Age Newborn (ELGAN) study cohort. Within ELGANs, significant associations have been observed between postnatal circulating inflammatory-related proteins and adverse neurodevelopmental outcomes at 10 years [31]. Based on this, the authors hypothesized that mRNAs that encode for inflammatory-related proteins would be differentially expressed in the placenta in relation to cognitive impairment at age 10. They further hypothesized that miRNAs would control some of the differential expression observed in mRNAs. The results from this study may have implications for the application of the DOHaD framework to neurodevelopment.
Materials & methods
Study subject recruitment
The recruitment process for the ELGAN study has been described in detail elsewhere [32]. Between the years of 2002 and 2004, infants born prior to 28 weeks' gestational age were enrolled in 14 different hospitals in five states in the USA. The institutional review boards for all participating ELGAN sites approved of the study protocol. Key demographic and pregnancy data were collected via an interview and a review of the mothers' medical records.
The initial ELGAN study comprised 1249 mothers and 1506 participating infants. From these 1506 infants, a total of 1083 placentas were obtained. For the present study, a subset of 390 ELGAN placentas (n = 390) was assessed for the expression of miRNAs and mRNAs. For the final study, a subset of 21 samples was removed due to missing demographic data or failure to pass quality assessment/quality control steps. Thus, a total of 369 placentas were included in the final analyses. Such quality assessment/quality control steps are detailed below.
Placental sample collection
Placental collection for the ELGAN study has been described previously [32]. Briefly, placentas were collected on delivery, at which time the extra embryonic membranes were separated by retracting the amnion and exposing the underlying chorion. The final sample was taken from the base of the chorion, frozen in liquid nitrogen and stored at -80°C. The placental tissues were first processed on dry ice. To reduce any blood contamination, frozen tissue samples were sliced into approximately 0.025 g segments using a sterile dermal curette and washed in 1x phosphate-buffered saline (Fisher Scientific, MA, USA). The washed samples were then immediately snap frozen in homogenization tubes and placed back on dry ice to preserve sample integrity. Tissue segments were homogenized according to the manufacturer's guidelines (Qiagen, MD, USA) and processed samples were placed at -80°C until the time of extraction of nucleic acids.
Placental mRNA & miRNA extraction & quantification
The AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) was used to isolate RNA molecules ≥18 nucleotides. A LabChip (Perkin Elmer, MA, USA) instrument was used to determine RNA quality, generating RNA integrity numbers and DV200 values, which were in the acceptable range. The QuantSeq 3′ mRNA-Seq Library Prep Kit (Lexogen) was used to assess genome-wide mRNA expression, selected for its sensitivity to transcripts with reduced RNA integrity numbers [33]. Lexogen-derived protocols were used for library preparation, and the Illumina Hiseq 2500 was used to sequence single-end 50 bp reads of RNA. To prevent batch-to-batch artifacts, the Sciclone G3 (Perkin Elmer) was used for automated library preparation. The GENCODE database v30 and Salmon (version 0.11.3) were used to align the sequencing read counts and organize the data, respectively. A total of 37,268 unique human RNA transcripts, comprising both protein-coding and noncoding RNAs resulted from this analysis [34,35]. The Salmon alignment package was used because it represents a relatively recent advancement in quantifying transcript abundance from RNA-sequencing data, with advantages including the correction for fragment biases yielding improved abundance estimate accuracies [34]. This package has been tested specifically for utility in differential expression analyses through algorithms embedded in DESeq2, used here for the identification of transcripts associated with cognitive outcomes [34]. Furthermore, this method is well recognized within the transcriptomics field, is computationally efficient and has been successfully integrated into the authors' prior analyses leveraging the RNA-sequencing data in this cohort [26,36,37]. The aligned count data were used for all subsequent data processing and statistical analyses, detailed as follows.
The HTG EdgeSeq miRNA Whole Transcriptome Assay (HTG Molecular Diagnostics, AZ, USA) was used to assess genome-wide miRNA expression profiles for 2083 human miRNA transcripts. The counts of sequencing reads were aligned to miRBase v20 and organized using Parser (HTG Molecular Diagnostics) [38]. Raw and processed mRNA and miRNA count data have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus repository and are publicly available under Gene Expression Omnibus series GSE154829 [39].
Neurocognitive assessment of later life outcomes
Cognitive assessments of participants were conducted when they were 10 years of age and have been described previously [40]. General cognitive ability was assessed using the School-Age Differential Ability Scales-II (DAS-II) Verbal (verbal IQ) and Nonverbal Reasoning (nonverbal IQ) scales. The DAS-II and the Developmental NEuroPSYchological Assessment Second Edition (NEPSY-II) were used to assess attention and executive function. Two subtests from the DAS-II and five subsets from the NEPSY-II were used to assess executive function. DAS-II recall of digits backward and recall of sequential order measured verbal working memory. Five subtests from the NEPSY-II (auditory attention, auditory response set, inhibition, inhibition switching, and animal sorting) were used to assess attention, inhibition and set switching (cognitive flexibility). A latent profile analysis (LPA) of the nine measures was conducted based on the 873 children, yielding four classifications: ‘normal’ (34% of the sample), ‘low normal’ (41%), ‘moderately impaired’ (17%) and ‘severely impaired’ (8%) cognitive function. For this study, the LPA classifications were dichotomized into normal/low normal cognitive function (LPA score: 0) and moderate/severe cognitive impairment (LPA score: 1), termed ‘no cognitive impairment’ and ‘cognitive impairment’, respectively [18,41].
Statistical analysis of differential mRNA expression associated with later in life neurocognitive impairment
mRNA sequencing data were processed using R (v4.0.2) (cran.r-project.org/). Universally lowly expressed transcripts were filtered out from the count data. Specifically, for a gene to be included, >25% of the samples were required to be expressed at signals above the overall median signal intensity across all genes, as done in the authors' previous genome-wide mRNA and miRNA analyses [42–46]. This resulted in a total of 10,412 mRNA transcripts included in the analyses. Quality assessment/quality control was conducted as described elsewhere [26]. Three surrogate variables captured variability across the principal components of the mRNA and miRNA data (Supplementary Figure 1). The addition of these surrogate variables to the statistical model controls for batch effects and sample heterogeneity due to causes such as cell type proportion variation.
A causal directed acyclic graph approach was used to identify a minimal sufficient adjustment set of covariates to be included in the statistical model (Supplementary Figure 2). Directed acyclic graph analysis was conducted in Dagitty (v3.0) [47]. After considering collinearity, the authors retained the following variables for the final models: birth weight, infant sex, maternal age and health insurance [48–57]. Importantly, all of these variables have known relationships with later neurocognitive function in other cohort studies [12,58,59]. In addition to the confounders identified, three surrogate variables were included in each model. Therefore, the final model included cognitive impairment (independent variable), birth weight, infant sex, maternal age and health insurance (all confounders) as well as three surrogate variables.
mRNAs that were associated with cognitive impairment were identified using negative binomial generalized linear models within DESeq2 [60]. DESeq2 uses a shrinkage estimator for fold changes for differential expression analysis, which is well suited for the comparison of fold change across the wide dynamic range of RNA-sequencing experiments [61]. The Benjamini–Hochberg (BH) procedure was then used to adjust p-values, in order to account for multiple testing [62]. mRNAs that were defined as differentially expressed were those with a false discovery rate <10%, based on a BH-adjusted p-value, and fold change in expression of ≥ ± 1.3 (comparing samples from children with impaired/non-impaired neurocognition). The chromosomal locations of the mRNAs identified as showing differential expression in relation to cognitive impairment were established relative to the 30 GENCODE hg19 reference genome assembly.
Models were also run for males and females separately to test if there was a sex-dependent association between placental mRNA expression and later in life cognitive impairment.
Identification of miRNA gene targets & association between expression levels of miRNA–mRNAs
miRNA regulators of the cognitive impairment-associated mRNAs were identified using miRsystem [63]. miRsystem matches miRNAs of interest to up-to-date annotations and identifies the biological functions/pathways regulated by miRNAs determined through the functions of the predicted target genes [63,64]. Factors that influence the likelihood of miRNA–mRNA interactions, including binding site type and location, local adenine and uracil content, target site abundance, seed-pairing stability and supplementary pairing, are summarized by cumulative weighted context scores. The predicted gene targets were filtered to include only genes that were associated with neurocognitive function in the authors' statistical model, described above. The miRNA–mRNA pairs that remained were predicted to be miRNA–mRNA expression pairs. The predicted miRNA–mRNA expression pairs were queried for correlation between normalized counts. Pearson correlation coefficient (p < 0.1) was used as a cutoff to explore the correlations between the normalized counts of miRNA–mRNA expression pairs and to establish if the increased miRNA expression was associated with decreased mRNA levels or vice versa. Thus, three factors defined an expression pair: the mRNA was a predicted gene target of the miRNA, the mRNA was associated with cognitive impairment and a significant correlation existed between the mRNA and miRNA normalized counts. Taken together, the expression pairs represent miRNA–mRNA relationships in which the expression of the mRNA is predicted to be under miRNA control.
Results
Participant characteristics
In this study, the authors analyzed data from a total of 386 ELGANs. These subjects had information available on the expression levels of placental miRNAs and mRNAs as well as key demographic variables. The general characteristics of the ELGAN participants, including neurocognitive impairment classification at age 10, are provided in Table 1. In this subset, 87 of the placental samples were obtained from children with impaired neurocognition at age 10, and 299 were obtained from children without neurocognitive impairment at age 10. The majority of the mothers were between 21 and 35 years of age. The average gestational age for the infants was 182.5 days, or 26 weeks, with a gestational age range of 161–195 days. Based on self-reported racial characteristics, the cohort comprised 60.9% White mothers, 29% Black mothers and 9.06% other, representing the diversity within the participants. The majority of the participants did not report smoking during pregnancy (87.7%). While most of the mothers of children of normal neurocognition had received 16+ years of education (41.5%), 54% of the mothers of children with impaired neurocognition had received ≤12 years of education. Almost half of the mothers of children with impaired neurocognition received Medicaid insurance (49.4%), while the mothers of children with normal condition had mainly health maintenance organization or private insurance (65.9%).
Table 1. . Summary of ELGAN study demographics for the subcohort used in this analysis (n = 386), consisting of mothers who contributed placentas analyzed for miRNA and mRNA expression profiles.
| Overall (n = 386) |
Impaired neurocognition (n = 87) |
Normal neurocognition (n = 299) |
|
|---|---|---|---|
| N (%) or mean (SD) | N (%) or mean (SD) | N (%) or mean (SD) | |
| Gestational age (days) | 182.5 (165–195) | 180.5 (165–195) | 183.1 (165–195) |
| Maternal age category (years) | |||
| <21 | 44 (11.4) | 13 (14.9) | 31 (10.4) |
| 21–35 | 260 (67.4) | 60 (69.0) | 200 (66.9) |
| >35 | 82 (21.2) | 14 (16.1) | 68 (22.7) |
| Infant sex | |||
| Male | 203 (52.6) | 56 (64.4) | 147 (49.2) |
| Female | 183 (47.4) | 31 (35.6) | 152 (50.8) |
| Race | |||
| White | 235 (60.9) | 38 (43.7) | 197 (65.9) |
| Black | 112 (29.0) | 39 (44.8) | 73 (24.4) |
| Other | 35 (9.06) | 10 (11.5) | 25 (8.36) |
| Missing | 4 (1.04) | 0 (0) | 4 (1.34) |
| Ethnicity | |||
| Non-Hispanic | 354 (91.7) | 79 (90.8) | 275 (92.0) |
| Hispanic | 32 (8.3) | 8 (9.2) | 24 (8.0) |
| Smoking status | |||
| Yes | 41 (10.6) | 9 (10.3) | 32 (10.7) |
| No | 338 (87.6) | 76 (87.4) | 262 (87.6) |
| Missing | 7 (1.8) | 2 (2.3) | 5 (1.7) |
| Marital status (single) | |||
| Yes | 166 (43.0) | 46 (52.9) | 120 (40.1) |
| No | 220 (57) | 41 (47.1) | 179 (59.9) |
| Maternal years of education | |||
| ≤12 | 147 (38.1) | 47 (54.0) | 100 (33.4) |
| 13–15 | 84 (21.8) | 18 (20.7) | 66 (22.1) |
| 16+ | 144 (37.3) | 20 (23.0) | 124 (41.5) |
| Missing | 11 (2.8) | 2 (2.3) | 9 (3.0) |
| Maternal prepregnancy BMI | |||
| Underweight | 29 (7.5) | 7 (8.0) | 19 (6.3) |
| Normal | 199 (51.6) | 37 (42.6) | 162 (54.2) |
| Overweight | 67 (17.4) | 13 (14.9) | 54 (18.1) |
| Obese | 81 (21.0) | 27 (31.0) | 54 (18.1) |
| Missing | 13 (3.4) | 3 (3.5) | 10 (3.3) |
| Health insurance status | |||
| Health maintenance organization or private | 236 (61.1) | 39 (44.8) | 197 (65.9) |
| Self-pay | 5 (1.3) | 0 (0.0) | 5 (1.7) |
| Medicaid | 126 (32.6) | 43 (49.4) | 83 (27.8) |
| None | 5 (1.3) | 1 (1.2) | 4 (1.3) |
| Other | 6 (1.6) | 3 (3.4) | 3 (1.0) |
| Missing | 8 (2.1) | 1 (1.2) | 7 (2.3) |
| Birth weight category | |||
| <750 | 144 (37.3) | 44 (50.6) | 100 (33.4) |
| 751–1000 | 171 (44.3) | 33 (37.9) | 138 (46.2) |
| 1001–1250 | 63 (16.3) | 8 (9.2) | 55 (18.4) |
| <1250 | 8 (2.1) | 2 (2.3) | 6 (2.0) |
Participants with missing data were included in the percentage calculation.
SD: Standard deviation; BMI: Body mass index.
Identification of placental mRNAs with expression associated with cognitive impairment
A total of 48 mRNAs showed differential expression (BH-adjusted p < 0.1) in relation to later in life neurocognitive impairment (Figure 1). These analyses adjusted for maternal age, birth weight, infant sex and health insurance status in a nonsex-stratified approach. A secondary analysis compared the expression of these mRNAs in relation to chorioamnionitis and amnion inflammation and highlighted no statistical differences (Supplementary Table 1).
Figure 1. . Distribution of placental mRNAs with significant differential expression associated with impaired neurocognition (n = 48).
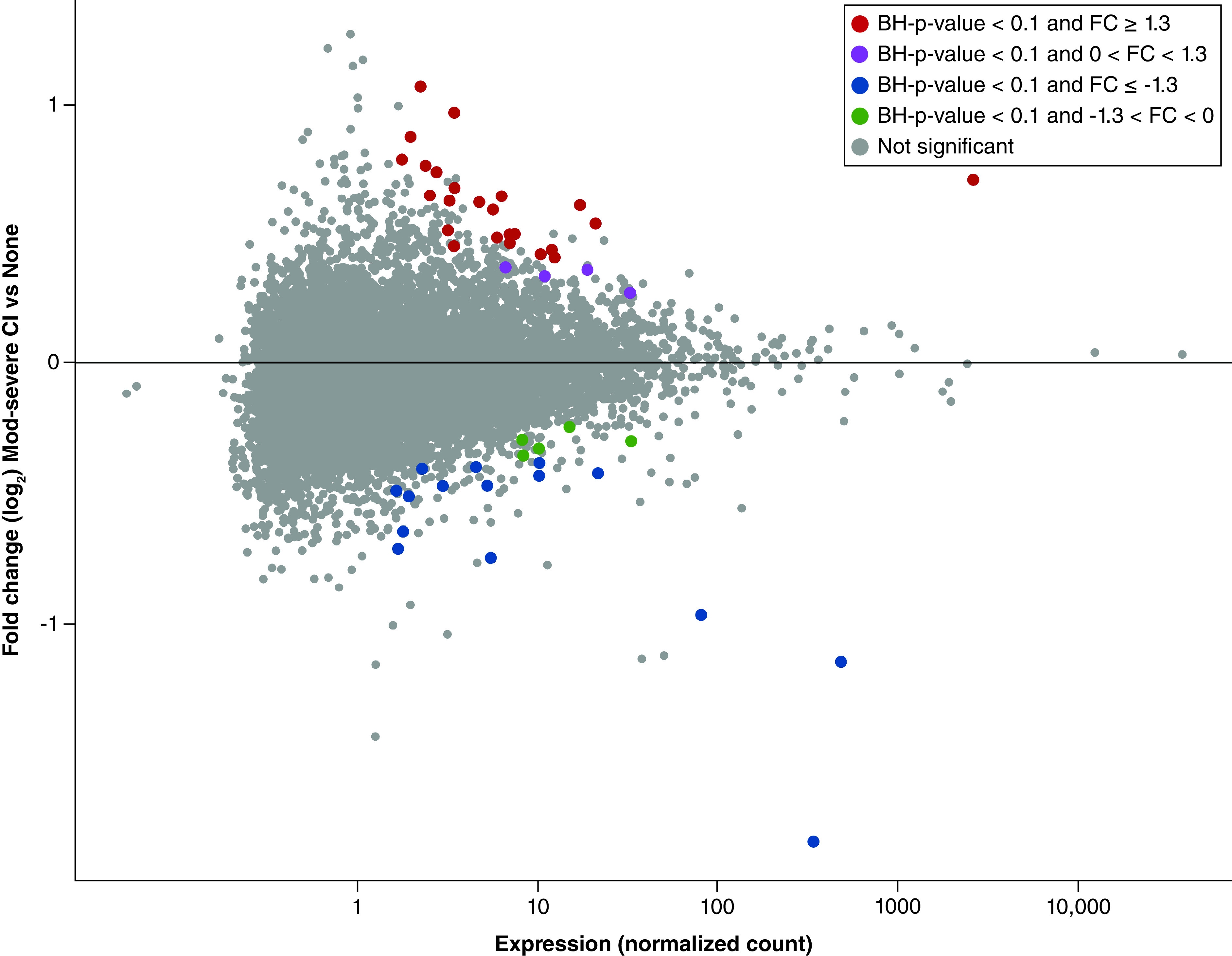
MA plots illustrate distributions for those identified in all placentas. Plots display the median normalized mRNA expression levels versus fold change in expression. FC values represent the ratio of mRNA expression in placentas derived from children with impaired neurocognition/normal.
BH: Benjamini-Hochberg; FC: Fold change.
A total of 28 (58.3%) mRNAs displayed increased expression in the placentas of subjects who exhibited cognitive impairment at age 10, and 20 (41.7%) mRNAs displayed decreased expression in the placentas of subjects who exhibited impaired cognitive function at age 10 (Supplementary Table 2). Sex-stratified findings are shown in Supplementary Table 3. The sex-stratified analysis demonstrated 0 significant differentially expressed genes among female-derived placentas and 37 genes that were differentially expressed among male-derived placentas (Supplementary Figure 3).
Among the 48 differentially expressed genes that were associated with neurocognitive impairment later in life, several were found to encode proteins that have roles in inflammation, apoptosis and/or immune response (Supplementary Table 2). The following displayed increased expression in relation to cognitive impairment: UCHL3, TMEM43, SRPK2, GDPD5, GNAI1, FN1, ITGB2 and PYGL. In contrast, the following displayed decreased expression in relation to cognitive impairment: RC3H1, ID2, CBX5, LOXL1, SDCBP, HBB and IQGAP1. RC3H1 and UCHL3 are among those known to be involved in inflammatory processes, and ID2 plays roles in both immune system regulation and inflammation. Genes that play apoptosis-associated roles include TMEM43, SRPK2 and GDPD5. Interestingly, several genes, including CBX5, RAB5A and GNAI1, demonstrate roles in both inflammation and apoptosis. A pathway analysis of the differentially expressed genes revealed a network involving FN1, ITGB2, LOXL1, SDCBP, PYGL, HBB and IQGAP1 relating to various functions involved in the inflammatory response (Figure 2).
Figure 2. . Pathway of inflammatory response-associated genes.
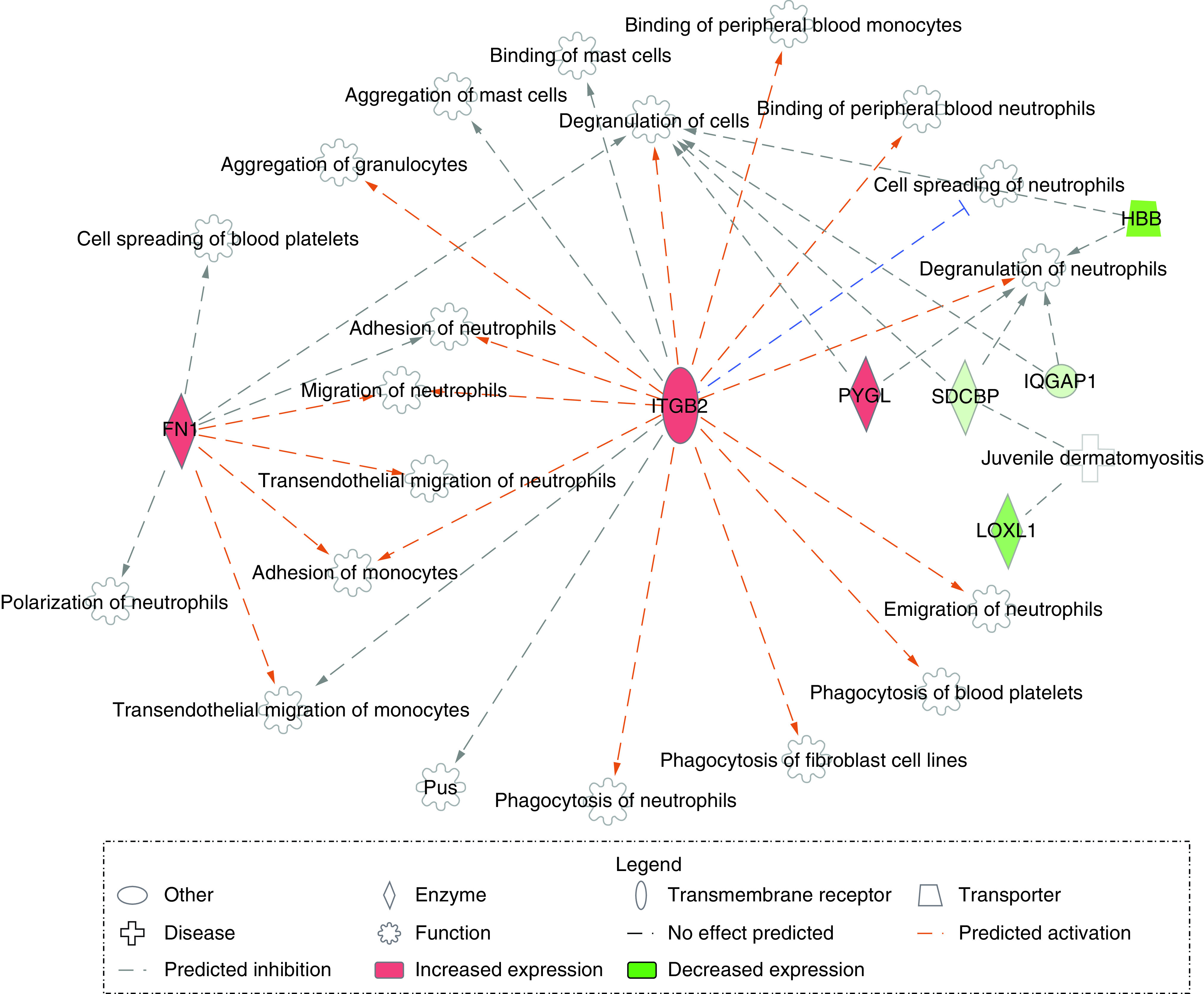
Genes are indicated in either red (increased predicted expression) or green (decreased predicted expression). Orange is indicative of predicted activation, and blue is indicative of predicted inhibition of the described function. Orange lines predict leads to activation, and blue lines predict leads to inhibition. Gray lines indicate there is no effect predicted.
Identification of miRNA–mRNA expression pairs
In addition to evaluating differentially expressed mRNAs, the authors set out to determine which mRNAs were potentially under miRNA control by identifying mRNA–miRNA expression pairs. Specifically, these data were analyzed in the context of the mRNAs that were associated with cognitive impairment. Among the 48 differentially expressed mRNAs, 37 (77.1%) were predicted to be controlled by 289 unique miRNAs. This generated a total of 1101 pairings overall, all of which were significantly correlated (p < 0.1; Supplementary Table 4). The general biological relationship between miRNA–mRNA expression pairs reflects that of a negative association, where increased miRNA expression is associated with decreased expression of the targeted mRNA (Figure 3). Thus, the authors investigated the roles of the miRNAs in the top 20 miRNA–mRNA expression pairs with the largest negative correlation coefficients, all of which were significant, with p < 0.1. Within these pairs, there were nine unique miRNAs: miR-765, miR-296-3p, miR-885-3p, miR-1207-5p, miR-671-5p, miR-936, miR-920, miR-877-5p, and miR-202-3p, representing nine targeted genes (Table 2). miR-765, miR-1207-5p, and miR-936 are related to the suppression of cellular proliferation and promotion of apoptosis. With regard to inflammation, miR-296-3p demonstrated differential expression in response to inflammatory signals and is associated with cellular invasion. Similarly, miR-885-3p and miR-920 increase in expression with increases in proinflammatory cytokine and the production of proinflammatory cytokines, respectively. Interestingly, miR-671-5p demonstrates roles in suppressing NF-κB activity and neuroinflammation, and miR-202-3p has been shown to reduce inflammatory factor expression and promote angiogenesis. The differentially expressed mRNAs represented by the miRNAs are all on different chromosomes, with the exception of RBM47 and TMEM192, which are both on chromosome 4. None of the chromosomes represented by these pairs are located on sex chromosomes.
Figure 3. . Six significantly correlated miRNA–mRNA expression pairings.
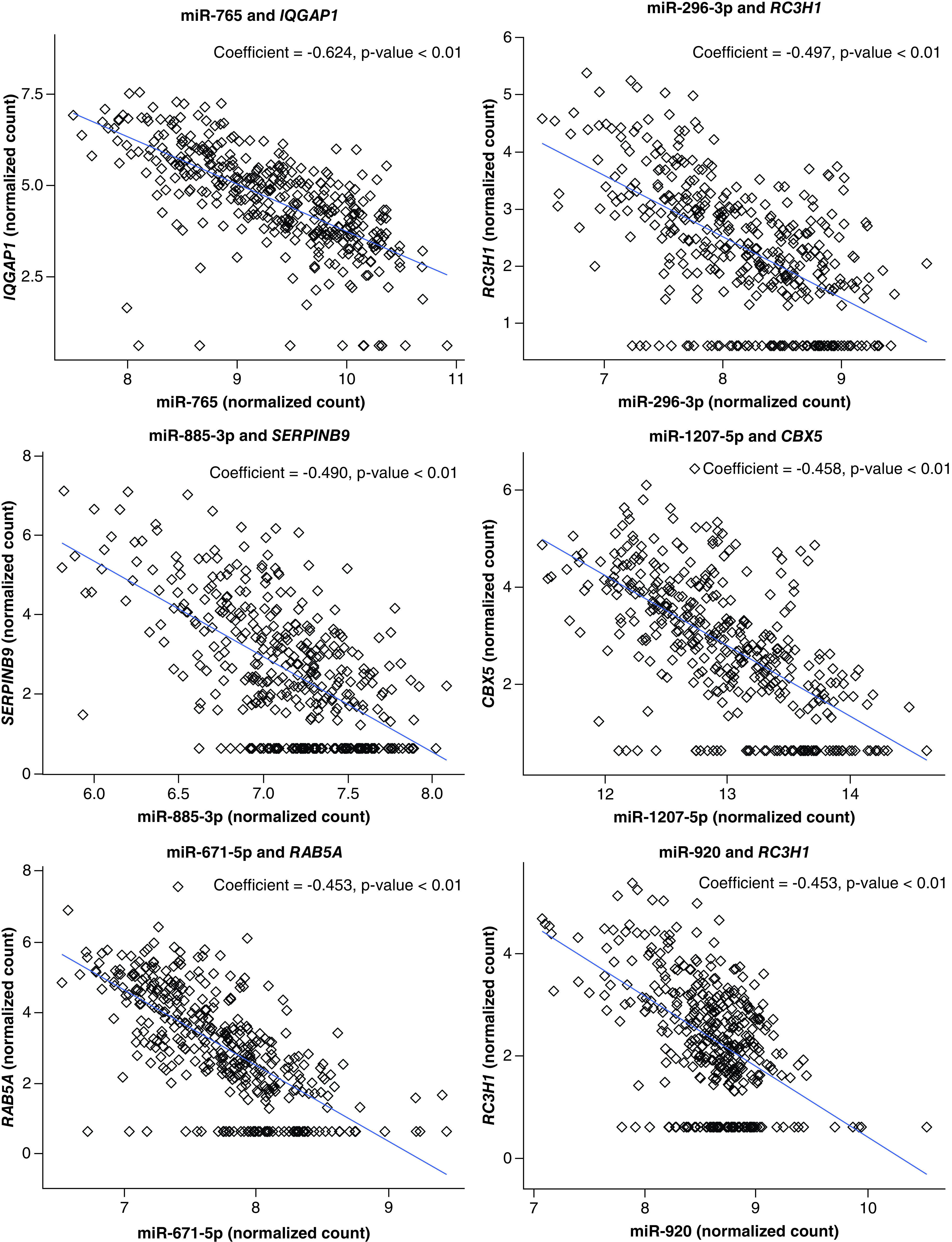
The x-axis represents the variance stabilized counts (‘normalized count’) of miRNAs. The y-axis reflects variance stabilized counts (‘normalized count’) of mRNAs. Correlation coefficients and p-values are at the top of the plot.
Table 2. . Summary of miRNAs from predicted miRNA–mRNA expression pairs (n = 20).
| miRNA | mRNA pair | Correlation coefficient | miRNA role |
|---|---|---|---|
| miR-765 | RAB5A | ‐0.590 | Apoptosis and suppressed cell proliferation |
| CBX5 | ‐0.580 | ||
| RBM47 | ‐0.535 | ||
| SERPINB9 | ‐0.510 | ||
| IQGAP1 | ‐0.624 | ||
| SDCBP | ‐0.492 | ||
| GDPD5 | ‐0.473 | ||
| miR-296-3p | IQGAP1 | ‐0.615 | Inflammation and cell invasion |
| RC3H1 | ‐0.497 | ||
| SERPINB9 | ‐0.495 | ||
| miR-885-3p | RC3H1 | ‐0.456 | Inflammation and enhanced NF-κB signaling |
| SERPINB9 | ‐0.490 | ||
| TMEM192 | ‐0.489 | ||
| miR-1207-5p | CBX5 | ‐0.458 | Inflammation |
| miR-671-5p | RAB5A | ‐0.453 | Suppressed NF-κB and neuroinflammation |
| miR-920 | RC3H1 | ‐0.453 | Production of proinflammatory cytokines |
| miR-936 | SDCBP | ‐0.428 | Apoptosis |
| RC3H1 | ‐0.447 | ||
| miR-877-5p | IQGAP1 | ‐0.439 | Reduced release of TNF-α, IL-1B and IL-6; suppressed inflammatory response; and apoptosis |
| miR-202-3p | RBM47 | ‐0.425 | Reduced inflammatory factor expression, angiogenesis and cancer cell apoptosis |
miRNAs chosen represent those observed within the first 20 pairs, based on correlation coefficient. Correlation between variance stabilized counts of miRNAs and mRNAs was calculated using the Pearson correlation coefficient. All expression pairs show p < 0.1.
Discussion
This study investigated the association between the expression levels of placental mRNAs and their targeting miRNAs as they relate to neurocognition later in life. These findings support a placenta epigenome–brain relationship in children born extremely preterm. With a specific focus on the placenta as a unique target tissue, the authors hypothesized that inflammation-related mRNAs would display higher expression in the placentas of children who went on to later in life cognitive impairment at age 10 years. This hypothesis is based on the knowledge that exposure to inflammation-related factors in utero disrupts fetal brain development [1,65]. Given the critical role of miRNAs as epigenetic regulators in the placenta, the authors also anticipated that some of the mRNAs expressed in the placenta would be under miRNA control.
Several of the mRNAs that were identified as having increased expressed in the placenta and as positively associated with cognitive impairment at age 10 are involved in proinflammatory signaling or apoptosis. Specifically, TMEM43 and UCHL3 are key molecular components of the NF-κB pathway [66–69]. While the magnitude of the gene expression changes is modest, the fact that there is an enrichment for these genes within common biological pathways, including the NF-κB pathway, is noteworthy. This pathway serves as a convergence point for proinflammatory responses and later in life health outcomes, as activation of the NF-κB pathway has been associated with brain damage in the newborn [15,70]. Accordingly, these findings are consistent with prior research indicating that inflammation early in life in ELGANs is associated with cognitive deficits [71–73], but they also support that the placenta is a target tissue for this genomic inflammation.
In support of the a priori hypothesis, the majority (88.9%) of the 48 cognitive function-associated mRNAs are predicted to be under epigenetic control of miRNAs. Additionally, in sex-stratified analysis, no significantly differentially expressed genes were identified among female-derived placentas, while 37 differentially expressed genes were identified among male-derived placentas. The genomic interactions observed in the analyses were defined based on three criteria: the miRNA is a predicted regulator of the gene target (mRNA), the expression levels of the miRNA and the targeted mRNA were associated with cognitive impairment and a significant correlation was observed between the mRNA and miRNA expression levels. Placental miRNAs are critical to mRNA expression as well as cognitive development [5,25,74]. Many of the identified miRNAs control inflammation or apoptotic processes in cells. Specifically, miR-765 and miR-936 have roles in apoptosis and the suppression of cell proliferation [75,76]. Studies have shown that miR-296-3p, miR-885-3p, and miR-920 demonstrate differential expression in response to inflammatory signals, as well as the increase in proinflammatory cytokine production [77–80]. These findings contribute to the growing literature showing that miRNAs in the placenta may serve as biomarkers of risk for impaired neurocognition later in life [5,25].
Several factors should be considered when interpreting the results of this study. First, the ELGAN cohort comprises children who were all born extremely prematurely, and thus the results may not be generalizable to non-preterm cohorts. To address this, future research should aim to replicate these analyses in term infants. Second, the present study focused solely on neurocognition as the health outcome of interest. However, preterm infants often experience comorbid neurodevelopmental impairments (e.g., depression, anxiety, autism, attention deficit disorder). While the ELGAN research team is investigating the associations between the placental genomic and epigenomic signatures and individual neurodevelopmental outcomes, such as social responsiveness and IQ [81], these studies have lower statistical power than the present analysis of cognitive impairment. Future placental programming studies should be designed to be powered to address comorbidity in neurodevelopmental outcomes. This study also has several unique strengths. It is among the first to demonstrate the placenta epigenome–brain axis as it relates to neurocognitive outcomes in children born preterm. While several studies have investigated the role that CpG methylation plays in fetal development, few studies have examined placental mRNAs and the role of miRNAs in controlling their expression. By connecting miRNAs to the genes they control, this study has further elucidated the relationship between the expression of inflammation-related genes, the epigenome and cognitive impairment later in life.
Conclusion
In summary, this study highlights the relationships among altered gene expression in the placenta in infants born preterm, gene targeting miRNAs, and cognitive impairment later in life. The identified genes play critical roles in inflammatory processes, which have established links to both preterm birth and later neurocognitive impairment [82–84]. This work provides a basis for further investigations of the role of miRNAs in the placenta epigenome–brain axis. The identified placental genes and miRNAs could serve as biomarkers for cognitive impairment risk in childhood.
Future perspective
This novel research is among the first to identify transcripts within the placenta that are under the epigenetic control of miRNAs and are associated with later life neurocognition. These transcriptomic differences in the placenta between individuals with and without impaired neurocognition at age 10 may be a result of exposure to harmful factors in the environment, including environmental toxicants. With further investigation, specific toxicants and stressors may also be identified that are potential drivers of these gene and miRNA changes. In the present study, placental genomic and epigenomic signatures were identified as biomarkers of susceptibility for neurodevelopmental later life health outcomes. Future research should investigate whether these biomarkers are associated with other later life neurocognitive outcomes, including autism spectrum disorder, depression and anxiety. Finally, replication of these results is needed through the study of separate children's cohorts, particularly those born at term. The identified biological genes and pathways in the placenta associated with neurocognitive impairments may be targets for interventions, including modulations within the maternal environment (i.e., chemical reduction, dietary modification and stress reduction).
Summary points.
The relationship between the placenta and fetal neurodevelopment is known as the placenta–brain axis.
Inflammatory signals during the neonatal period have been associated with damage to the developing fetal brain.
miRNAs are fundamental epigenetic regulators expressed in the placenta with critical roles during pregnancy.
miRNA expression in the placenta is associated with infant neurodevelopment.
Differences in placental mRNA and miRNA expression were evaluated in the ELGAN study (n = 386).
A total of 48 mRNAs were found to be differentially expressed, and 37 of the mRNAs (77%) were predicted to be under the control of 289 miRNAs.
Several miRNA-controlled mRNAs, including RAB5A, RC3H1, CBX5 and IQGAP1, have known roles in inflammatory processes.
These data provide insight into the molecular mechanisms by which the placenta is tied to cognitive outcomes.
Supplementary Material
Acknowledgments
The authors would like to thank the ELGAN Study Investigators and participants for their contributions to this research.
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/epi-2022-0061
Author contributions
M O'Shea and RC Fry conceived of the study design. All the authors interpreted the findings regarding associations with neurocognitive impairment in children born preterm. AN Freedman, LA Eaves, JE Rager, N Gavino-Lopez, L Smeester, J Bangma and HP Santos analyzed and interpreted the data-related mRNAs and miRNAs. KCK Kuban, RM Joseph, M O'Shea and RC Fry oversaw the analyses. AN Freedman drafted the manuscript, and all authors read, provided input and approved the final manuscript.
Financial & competing interests disclosure
This research was supported by grants from the following: National Institutes of Health (http://www.nih.gov): R01 ES019315, P42 ES005948; the Office of the NIH Director: UH3OD023348; the National Institute of Child Health and Human Development: R01HD092374, R03HD101413, UG3OD023348; the National Institute of Neurologic Disorders and Stroke: U01NS040069, U01NS040069, U01NS040069; the National Institute of Nursing Research: R01NR019245; and the National Institute for Occupational Safety and Health: T42/OH-008673. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Data sharing statement
All genomic data generated or analyzed in this study are included in this published article. mRNA- and miRNA-sequencing data have been deposited in Gene Expression Omnibus [GEO: GSE154829] (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154829).
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Tomlinson MS, Santos HP, Stewart JR et al. Neurocognitive and social-communicative function of children born very preterm at 10 years of age: associations with microorganisms recovered from the placenta parenchyma. J. Perinatol. 40(2), 306–315 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miguel PM, Pereira LO, Silveira PP, Meaney MJ. Early environmental influences on the development of children's brain structure and function. Dev. Med. Child Neurol. 61(10), 1127–1133 (2019). [DOI] [PubMed] [Google Scholar]; • Highlights links between in utero exposures and neurodevelopment.
- 3.O'Donnell KJ, Meaney MJ. Fetal origins of mental health: the developmental origins of health and disease hypothesis. Am. J. Psychiatry 174(4), 319–328 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Burton GJ, Fowden AL, Thornburg KL. Placental origins of chronic disease. Physiol. Rev. 96(4), 1509–1565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lester BM, Marsit CJ. Epigenetic mechanisms in the placenta related to infant neurodevelopment. Epigenomics 10(3), 321–333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenfeld CS. The placenta–brain-axis. J. Neurosci. Res. (2020) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gabory A, Roseboom TJ, Moore T, Moore LG, Junien C. Placental contribution to the origins of sexual dimorphism in health and diseases: sex chromosomes and epigenetics. Biol Sex Differ. 4(1), 5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Highlights the placenta–brain axis.
- 8.Barker DJ, Bull AR, Osmond C, Simmonds SJ. Fetal and placental size and risk of hypertension in adult life. BMJ (Clinical Research Ed.) 301(6746), 259–262 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ilekis JV, Tsilou E, Fisher S et al. Placental origins of adverse pregnancy outcomes: potential molecular targets: an executive workshop summary of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. Am. J. Obstet. Gynecol. 215(Suppl. 1), S1–S46 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redline RW, Minich N, Taylor HG, Hack M. Placental lesions as predictors of cerebral palsy and abnormal neurocognitive function at school age in extremely low birth weight infants (<1 kg). Pediatr. Dev. Pathol. 10(4), 282–292 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Nugent BM, Bale TL. The omniscient placenta: metabolic and epigenetic regulation of fetal programming. Front. Neuroendocrinol. 39, 28–37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tilley SK, Martin EM, Smeester L et al. Placental CpG methylation of infants born extremely preterm predicts cognitive impairment later in life. PlOS ONE 13(3), e0193271 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Describes CpG sites in the placenta from infants born preterm that are related to later-in-life neurocognition.
- 13.Boekelheide K, Blumberg B, Chapin RE et al. Predicting later-life outcomes of early-life exposures. Environ Health Perspect. 120(10), 1353–1361 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan W, Yue H, Ji X, Li G, Sang N. Prenatal NO2 exposure and neurodevelopmental disorders in offspring mice: transcriptomics reveals sex-dependent changes in cerebral gene expression. Environ. Int. 138, 105659 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Bangma JT, Hartwell H, Santos HP Jr, O'Shea TM, Fry RC. Placental programming, perinatal inflammation, and neurodevelopment impairment among those born extremely preterm. Pediatr. Res. (2020) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Illustrates the relationship between the NF-kB pathway and neurodevelopmental impairment in newborns.
- 16.Martin EM, Fry RC. Environmental influences on the epigenome: exposure-associated DNA methylation in human populations. Annu. Rev. Public Health 39, 309–333 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Jang HS, Shin WJ, Lee JE, Do JT. CpG and non-CpG methylation in epigenetic gene regulation and brain function. Genes (Basel) 8(6), 148 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meakin CJ, Martin EM, Santos HP Jr et al. Placental CpG methylation of HPA-axis genes is associated with cognitive impairment at age 10 among children born extremely preterm. Hormones and behavior 101, 29–35 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Dongen J, Hagenbeek FA, Suderman M et al. DNA methylation signatures of aggression and closely related constructs: a meta-analysis of epigenome-wide studies across the lifespan. Mol. Psychiatry (2021) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Dongen J, Nivard MG, Baselmans BM et al. Epigenome-wide association study of aggressive behavior. Twin Res. Hum. Genet. 18(6), 686–698 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Cai M, Kolluru GK, Ahmed A. Small molecule, big prospects: microRNA in pregnancy and its complications. J. Pregnancy 2017, 6972732 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaschetto LM. miRNA activation is an endogenous gene expression pathway. RNA Biol. 15(6), 826–828 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu Y, Sun J, Groome LJ, Wang Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am. J. Physiol. Endocrinol. Metab. 304(8), e836–e843 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gheorghe CP, Goyal R, Mittal A, Longo LD. Gene expression in the placenta: maternal stress and epigenetic responses. Int. J. Dev. Biol. 54(2-3), 507–523 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maccani MA, Padbury JF, Lester BM, Knopik VS, Marsit CJ. Placental miRNA expression profiles are associated with measures of infant neurobehavioral outcomes. Pediatr. Res. 74(3), 272–278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Demonstrates associations between placental miRNA expression and neurobehavioral outcomes in term infants.
- 26.Eaves LA, Phookphan P, Rager JE et al. A role for microRNAs in the epigenetic control of sexually dimorphic gene expression in the human placenta. Epigenomics 12(17), 1543–1558 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin E, Smeester L, Bommarito PA et al. Sexual epigenetic dimorphism in the human placenta: implications for susceptibility during the prenatal period. Epigenomics 9(3), 267–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baron-Cohen S, Lombardo MV, Auyeung B, Ashwin E, Chakrabarti B, Knickmeyer R. Why are autism spectrum conditions more prevalent in males? PLOS Biol. 9(6), e1001081 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frondas-Chauty A, Simon L, Branger B et al. Early growth and neurodevelopmental outcome in very preterm infants: impact of gender. Arch. Dis. Child. Fetal Neonatal Ed. 99(5), F366–F372 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Johnston MV, Hagberg H. Sex and the pathogenesis of cerebral palsy. Dev. Med. Child Neurol. 49(1), 74–78 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Kuban KCK, Heeren T, O'Shea TM et al. Among children born extremely preterm a higher level of circulating neurotrophins is associated with lower risk of cognitive impairment at school age. J. Pediatr. 201, 40–48 e44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Shea TM, Allred EN, Dammann O et al. The ELGAN study of the brain and related disorders in extremely low gestational age newborns. Early Hum. Dev. 85(11), 719–725 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.3′ mRNA-seq library prep ktt user guide (2020). www.lexogen.com/wp-content/uploads/2015/11/015UG009V0211_QuantSeq-Illumina.pdf
- 34.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14(4), 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrow J, Frankish A, Gonzalez JM et al. GENCODE: the reference human genome annotation for the ENCODE Project. Genome Res. 22(9), 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark J, Avula V, Ring C et al. Comparing the predictivity of human placental gene, microRNA, and CpG methylation signatures in relation to perinatal outcomes. Toxicol. Sci. 183(2), 269–284 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Payton A, Clark J, Eaves L et al. Placental genomic and epigenomic signatures associated with infant birth weight highlight mechanisms involved in collagen and growth factor signaling. Reprod. Toxicol. 96, 221–230 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozomara A, Griffiths-Jones S. miRbase: integrating microRNA annotation and deep-sequencing data. NAR 39, D152–D157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.NCBI. Gene expression omnibus. Placental Genomic and Epigenomic Signatures in Infants Born at Extremely Low Gestational Age. MD, USA: (2020). [Google Scholar]
- 40.Tilley SK, Joseph RM, Kuban KCK, Dammann OU, O'Shea TM, Fry RC. Genomic biomarkers of prenatal intrauterine inflammation in umbilical cord tissue predict later life neurological outcomes. PlOS ONE 12(5), e0176953 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heeren T, Joseph RM, Allred EN, O'Shea TM, Leviton A, Kuban KCK. Cognitive functioning at the age of 10 years among children born extremely preterm: a latent profile approach. Pediatr. Res. 82(4), 614–619 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rager JE, Auerbach SS, Chappell GA, Martin E, Thompson CM, Fry RC. Benchmark dose modeling estimates of the concentrations of inorganic arsenic that induce changes to the neonatal transcriptome, proteome, and epigenome in a pregnancy cohort. Chem. Res. Toxicol. 30(10), 1911–1920 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rager JE, Bailey KA, Smeester L et al. Prenatal arsenic exposure and the epigenome: altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ. Mol. Mutagen. 55(3), 196–2008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rager JE, Moeller BC, Miller SK et al. Formaldehyde-associated changes in microRNAs: tissue and temporal specificity in the rat nose, white blood cells, and bone marrow. Toxicol. Sci. 138(1), 36–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klaren WD, Ring C, Harris MA et al. Identifying attributes that influence in vitro-to-in vivo concordance by comparing in vitro Tox21 bioactivity versus in vivo drug matrix transcriptomic responses across 130 chemicals. Toxicol. Sci. 167(1), 157–171 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rager JE, Ring CL, Fry RC et al. High-throughput screening data interpretation in the context of in vivo transcriptomic responses to oral Cr(VI) exposure. Toxicol. Sci. 158(1), 199–212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Textor J, Van Der Zander B, Gilthorpe MS, Liśkiewicz M, Ellison GT. Robust causal inference using directed acyclic graphs: the R package ‘dagitty’. Int. J. Epidemiol. 45(6), 1887–1894 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Juracek J, Piler P, Janku P, Radova L, Slaby O. Identification of microRNA signatures in umbilical cord blood associated with maternal characteristics. PeerJ 7, e6981 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huan T, Chen G, Liu C et al. Age-associated microRNA expression in human peripheral blood is associated with all-cause mortality and age-related traits. Aging Cell 17(1), e12687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noren Hooten N, Fitzpatrick M, Wood WH 3rd et al. Age-related changes in microRNA levels in serum. Aging (Albany NY) 5(10), 725–740 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui C, Yang W, Shi J et al. Identification and analysis of human sex-biased microRNAs. Genomics Proteomics Bioinformatics 16(3), 200–211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loher P, Londin ER, Rigoutsos I. IsomiR expression profiles in human lymphoblastoid cell lines exhibit population and gender dependencies. Oncotarget 5(18), 8790–8802 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Telonis AG, Rigoutsos I. Race disparities in the contribution of miRNA Isoforms and tRNA-derived fragments to triple-negative breast cancer. Cancer Res. 78(5), 1140–1154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Telonis AG, Loher P, Jing Y, Londin E, Rigoutsos I. Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res. 43(19), 9158–9175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ostling H, Kruse R, Helenius G, Lodefalk M. Placental expression of microRNAs in infants born small for gestational age. Placenta 81, 46–53 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Rodosthenous RS, Burris HH, Sanders AP et al. Second trimester extracellular microRNAs in maternal blood and fetal growth: an exploratory study. Epigenetics 12(9), 804–810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Higashijima A, Miura K, Mishima H et al. Characterization of placenta-specific microRNAs in fetal growth restriction pregnancy. Prenat. Diagn. 33(3), 214–222 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Conradt E, Lester BM, Appleton AA, Armstrong DA, Marsit CJ. The roles of DNA methylation of NR3C1 and 11beta-HSD2 and exposure to maternal mood disorder in utero on newborn neurobehavior. Epigenetics 8(12), 1321–1329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bromer C, Marsit CJ, Armstrong DA, Padbury JF, Lester B. Genetic and epigenetic variation of the glucocorticoid receptor (NR3C1) in placenta and infant neurobehavior. Dev. Psychobiol. 55(7), 673–683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren X, Kuan PF. Negative binomial additive model for RNA-seq data analysis. BMC Bioinformatics 21(1), 171 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12), 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc.: series B (Methodological) 57(1), 289–300 (1995). [Google Scholar]
- 63.Lu TP, Lee CY, Tsai MH et al. miRSystem: an integrated system for characterizing enriched functions and pathways of microRNA targets. PLOS ONE 7(8), e42390 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.miRSystem (2016). http://mirsystem.cgm.ntu.edu.tw/
- 65.Hess JL, Radonjic NV, Patak J, Glatt SJ, Faraone SV. Autophagy, apoptosis, and neurodevelopmental genes might underlie selective brain region vulnerability in attention-deficit/hyperactivity disorder. Mol. Psychiatry (2020) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang C, Chen Q, Hamajima Y et al. Id2 regulates the proliferation of squamous cell carcinoma in vitro via the NF-kappaB/cyclin D1 pathway. Chin. J. Cancer 31(9), 430–439 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang C, Zhu Y, Zhou Z et al. TMEM43/LUMA is a key signaling component mediating EGFR-induced NF-kappaB activation and tumor progression. Oncogene 36(20), 2813–2823 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Zhang R, Li L, Chen L et al. MAP7 interacts with RC3H1 and cooperatively regulate cell-cycle progression of cervical cancer cells via activating the NF-kappaB signaling. Biochem. Biophys. Res. Commun. 527(1), 56–63 (2020). [DOI] [PubMed] [Google Scholar]
- 69.Murakawa Y, Hinz M, Mothes J et al. RC3H1 post-transcriptionally regulates A20 mRNA and modulates the activity of the IKK/NF-kappaB pathway. Nat. Commun. 6, 7367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leviton A, Gressens P, Wolkenhauer O, Dammann O. Systems approach to the study of brain damage in the very preterm newborn. Front. Syst. Neurosci. 9, 58 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang NM, Cowan M, Moonah SN, Petri WA Jr. The impact of systemic inflammation on neurodevelopment. Trends Mol. Med. 24(9), 794–804 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oldenburg KS, O'Shea TM, Fry RC. Genetic and epigenetic factors and early life inflammation as predictors of neurodevelopmental outcomes. Semin. Fetal Neonatal Med. 25(3), 101115 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tomlinson MS, Bommarito PA, Martin EM et al. Microorganisms in the human placenta are associated with altered CpG methylation of immune and inflammation-related genes. PLOS ONE 12(12), e0188664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lesseur C, Paquette AG, Marsit CJ. Epigenetic regulation of infant neurobehavioral outcomes. Med. Epigenet. 2(2), 71–79 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin J, Zhang D, Fan Y et al. Regulation of cancer stem cell self-renewal by HOXB9 antagonizes endoplasmic reticulum stress-induced melanoma cell apoptosis via the miR-765-FOXA2 axis. J. Invest. Dermatol. 138(7), 1609–1619 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Liu S, Gong Y, Xu XD et al. microRNA-936/ERBB4/AKT axis exhibits anticancer properties of gastric cancer through inhibition of cell proliferation, migration, and invasion. Kaohsiung J. Med. Sci. 37(2), 111–120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fayyad-Kazan H, Fayyad-Kazan M, Merimi M et al. The micronome of mesenchymal stromal cells is partially responsive to inflammation. Cell Biol. Int. 42(2), 254–260 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Zhang X, Gu H, Wang L, Huang F, Cai J. miR-885-3p is down-regulated in peripheral blood mononuclear cells from T1D patients and regulates the inflammatory response via targeting TLR4/NF-kappaB signaling. J. Gene Med. 22(1), e3145 (2020). [DOI] [PubMed] [Google Scholar]
- 79.Guo J, Zhao X, Liu Z et al. Transmissible gastroenteritis virus ORF3b up-regulates miR-885-3p to counteract TNF-alpha production via inhibiting NF-kappaB pathway. Vet. Microbiol. 261, 109189 (2021). [DOI] [PubMed] [Google Scholar]
- 80.Zhou W, Wang Y, Wu R, He Y, Su Q, Shi G. microRNA-488 and -920 regulate the production of proinflammatory cytokines in acute gouty arthritis. Arthritis Res. Ther. 19(1), 203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santos HP Jr, Bhattacharya A, Joseph RM et al. Evidence for the placenta–brain axis: multi-omic kernel aggregation predicts intellectual and social impairment in children born extremely preterm. Mol. Autism 11(1), 97 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Strauss JF 3rd, Romero R, Gomez-Lopez N et al. Spontaneous preterm birth: advances toward the discovery of genetic predisposition. Am. J. Obstet. Gynecol. 218(3), 294–314 e292 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bukowski R, Sadovsky Y, Goodarzi H et al. Onset of human preterm and term birth is related to unique inflammatory transcriptome profiles at the maternal fetal interface. PeerJ 5, e3685 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rangaraju S, Dammer EB, Raza SA et al. Identification and therapeutic modulation of a pro-inflammatory subset of disease-associated-microglia in Alzheimer's disease. Mol. Neurodegener. 13(1), 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


