Abstract
The rfb region specifies the structure of lipopolysaccharide side chains that comprise the diverse gram-negative bacterial somatic (O) antigens. The rfb locus is adjacent to gnd, which is a polymorphic gene encoding 6-phosphogluconate dehydrogenase. To determine if rfb and gnd cotransfer, we sequenced gnd in five O55 and 13 O157 strains of Escherichia coli. E. coli O157:H7 has a gnd allele (allele A) that is only 82% identical to the gnd allele (allele D) of closely related E. coli O55:H7. In contrast, gnd alleles of E. coli O55 in distant lineages are >99.9% identical to gnd allele D. Though gnd alleles B and C in E. coli O157 that are distantly related to E. coli O157:H7 are more similar to allele A than to allele D, there are nucleotide differences at 4 to 6% of their sites. Alleles B and C can be found in E. coli O157 in different lineages, but we have found allele A only in E. coli O157 belonging to the DEC5 lineage. DNA 3′ to the O55 gnd allele in diverse E. coli lineages has sequences homologous to tnpA of the Salmonella enterica serovar Typhimurium IS200 element, E. coli Rhs elements (including an H-rpt gene), and portions of the O111 and O157 rfb regions. We conclude that rfb and gnd cotransferred into E. coli O55 and O157 in widely separated lineages and that recombination was responsible for recent antigenic shifts in the emergence of pathogenic E. coli O55 and O157.
The integration of foreign DNA into bacterial chromosomes has played an important role in the evolution of genomes and in the emergence of new pathogens (18, 22). These acquired segments confer upon the recipient cell the ability to express new phenotypes. For example, proteins of type III secretion systems that are encoded by genes in pathogenicity islands enable bacteria to export molecules that injure host epithelial cells (15), and O side chains (also termed somatic antigens) of bacterial lipopolysaccharide (LPS) that are specified by the rfb region are immunodominant surface molecules.
The rfb region is a complex locus; segment acquisition and recombination have played a major role in its evolution. This cluster, which typically varies in length between 8 and 14 kilobase pairs (kbp) and contains 8 to 14 genes, encodes the enzymes necessary for the synthesis of the O side chains that confer serogroup specificity. Many different O types of Escherichia coli and Salmonella enterica are associated with human disease, and the O side chain often induces bactericidal humoral immunity in infected hosts (13, 30, 31, 43). This LPS variability suggests that somatic antigens are under strong diversifying selection pressure to evade the host immune response.
Several lines of evidence indicate that somatic antigenic variation in E. coli and S. enterica is generated to a large extent by horizontal transfer and recombination of part or all of the rfb region (27, 30, 31). Closely related strains of E. coli can have different rfb genes encoding different O antigens (24). Conversely, distantly related organisms can have identical rfb genes that express the same O antigens (4, 35, 42, 46). Also, rfb genes usually have a low GC content compared to the total genomic DNA (30). This low GC content suggests that this DNA originated in a species other than E. coli.
How do recombination and diversifying selection at the rfb locus affect nearby genes? It has been suggested that the close proximity of the gnd locus to the rfb cluster underlies the extensive allelic diversity of 6-phosphogluconate dehydrogenase (6-PGD), the metabolic enzyme of the pentose phosphate shunt encoded by gnd. This concept teaches that new alleles of gnd, created either by point mutation or intragenic recombination, “hitchhike” to high frequency by diversifying selection favoring antigen variation at the adjacent rfb locus (27). In addition, local recombination events involving the rfb region and extending though the gnd locus could result in specific combinations of gnd alleles and rfb genes being cotransferred in nature.
E. coli O157:H7, a virulent food- and waterborne pathogen (39), and E. coli O55:H7, an enteropathogenic E. coli strain, are closely related members of the DEC5 lineage of diarrheagenic E. coli (44). Clonal analysis derived from multilocus enzyme electrophoresis (MLEE) suggests that E. coli O157:H7 evolved from a progenitor strain with serotype O55:H7 (11, 44). Furthermore, the nearly identical sequences of the H7 flagellin gene (32) and eae alleles (26) demonstrate the close relationship between E. coli O157:H7 and E. coli O55:H7. During the evolutionary descent of E. coli O157:H7 from its progenitor, an ancestral E. coli strain acquired an rfb region that specifies the O157 antigen (11), thereby replacing the O55 (23) with an O157 (29) LPS side chain. Also, as part of this evolution, E. coli O157:H7 acquired a different gnd allele, as evidenced by differences in the 6-PGD electromorph (44). These findings suggest that all or part of the gnd locus, in addition to rfb, was involved in this antigenic shift of the O side chain. Indeed, fewer than 200 bp separate gnd from the closest O157 rfb gene (35, 42), so cotransfer of these alleles within this lineage is quite plausible.
The purpose of the present study was to characterize gnd alleles in E. coli with specific LPS antigens in a variety of lineages, in order to gather sequence data in support of cotransfer as the mechanism of mobility of the rfb-gnd region. We also interrogated the region 3′ to gnd in selected E. coli in an attempt to determine the possible site and mechanism(s) of recombination. To accomplish these goals, we cloned and sequenced the gnd locus and neighboring DNA in a diverse collection of O55 and O157 strains, including E. coli within the DEC5 lineage, as well as in isolates with completely different chromosomal backgrounds.
MATERIALS AND METHODS
Wild-type bacteria.
Table 1 lists the wild-type bacteria from which gnd alleles were sequenced and the strains that were probed.
TABLE 1.
Wild-type E. coli strains used
| Lineage | Strain | Serotype
|
Source (reference or provider) | ECOR groupa | gnd allele | GenBank no.
|
||
|---|---|---|---|---|---|---|---|---|
| O | H | gnd | rfbE | |||||
| H7 (DEC5) | 86-24 | 157 | 7 | Washington State (40) | E | A | AF176356 | AF163327 |
| 85-07 | 157 | 7 | Washington State (40) | E | A | AF176359 | AF163328 | |
| 87-16 | 157 | 7 | Washington State (40) | E | A | AF176360 | AF163329 | |
| H8 | 157 | 7 | Colombia (S. Mattar via. S. Samadpour) | E | A | AF176357 | ||
| ADAL233 | 157 | 7 | Australia Government Analytical Laboratories (via S. Samadpour) | E | A | AF176358 | ||
| 2755 | 157 | − | Germany (L. Beutin) | E | A | AF176361 | AF163330 | |
| TB156A | 55 | 7 | Washington State (6) | E | ||||
| TB182A | 55 | 7 | Washington State (6) | E | D | AF176369 | ||
| DEC 5A-E | 55 | 7 | Pennsylvania State University (47) | E | ||||
| Non-H7 | 3004-89 | 157 | 3 | Centers for Disease Control (N. Stockbine) | A | B | AF176362 | AF163326 |
| G5933 | 157 | 12 | Centers for Disease Control (T. Barrett) | A | B | AF176363 | AF163331 | |
| 13A81 | 157 | 16 | Food and Drug Administration (S. Weagant) | A | C | AF176364 | AF163332 | |
| 3584-91 | 157 | —b | Centers for Disease Control (N. Stockbine) | D | C | AF176365 | AF163333 | |
| 3005-89 | 157 | 38 | Centers for Disease Control (N. Stockbine) | A | B | AF176366 | AF163334 | |
| DEC 7E | 157 | —b | Pennsylvania State University (47) | B1 | B | AF176367 | AF163335 | |
| 3260-92 | 157 | 16 | Centers for Disease Control (N. Stockbine) | A | B | AF176368 | AF163336 | |
| DEC 1A | 55 | 6 | Pennsylvania State University (47) | — | D | AF176370 | ||
| DEC 1B | 55 | 6 | Pennsylvania State University (47) | — | D | AF176371 | ||
| DEC 2A | 55 | 6 | Pennsylvania State University (47) | — | D | AF176372 | ||
| DEC 2B | 55 | 6 | Pennsylvania State University (47) | — | D | AF176373 | ||
ECOR group to which these strains belong by MLEE analysis. DEC1 and DEC2 E. coli O55:H6 strains do not fall within ECOR groups using MLEE analysis.
Nonmotile strains 3584-91 and DEC 7E have MLEE patterns identical to those of E. coli O157:H45 and O157:H43, respectively.
PCR.
Primers were purchased from Gibco-BRL (Gaithersburg, Md.). Primers A (5′CACGGATCCGATCACACCTGACAGGAGTA3′) and B (5′CCGGAATTCCGGGCAAAAAAAAGCCCGGTGCAA3′), with BamHI and EcoRI sites, respectively, were derived from published sequences (5) and amplify O157 gnd alleles. Primers C (5′CGGAATTCCGCGCTCAACATCGANAGCCGTGG3′) and D (5′CGGAATTCCGCCTGGATCAGGTTAGCCGG3′), with 5′ EcoRI sites, were chosen from consensus database gnd sequences to amplify a 1.3-kb fragment within the O55 gnd allele. Primer pairs E (5′CGGGGTACCCCGTAAGGGACCAGTTTCTTACCTGGG3′) (with a 5′ engineered KpnI site)-F (5′GCCCTATCTAGATAAAGG3′), G (5′AGTTAAAGCCTTCCGCGG3′)-H (5′TGCCCGCTACATCTCCTC3′), and I (5′GTTGTACTCTTCAGACGC3′)-J (5′TCGTCGCTTATGCGGTACAGAGCG3′) were selected from sites within the O55:H7 and O157:H7 gnd alleles to amplify circularized chromosomal DNA fragments spanning the 5′ and 3′ ends of the O55:H7 gnd and the 3′ end of the O157:H7 gnd, respectively (inverse PCR). Primers K (5′CCATCAGTAATAATGAAAAGGAAT3′) and L (5′ATCATTAGCTCCTCTTAAGATCGC3′), derived from the sequence of the products of inverse PCR with primer pairs E-F and G-H, respectively, produce panallelic amplifications of O55 gnd alleles. Primer pairs J-M (5′GCGTTCTTAAAGAGTCCTGC3′) and N (5′TGCCCGCTACATCTCCTC3′)-M amplify DNA spanning the 3′ ends of the O157:H7 and each of the O55 gnd alleles, respectively. Primers O (5′AAGATTGCGCTGAAGCCTTTG3′) and P (5′CATTGGCATCGTGTGGACAG3′) amplify sequences within rfbEEcO157:H7 (8) (also termed per [42]), which encodes RfbEEcO157:H7, which is a putative perosamine synthetase (4).
O55:H7 DNA was amplified with primers C and D using standard PCR conditions, Taq DNA polymerase, and a PTC-100 programmable thermal cycler (MJ Research, Watertown, Mass.). All other amplifications were performed using this cycler and the Expand long-template PCR system (Boehringer Mannheim, Indianapolis, Ind.), with supplied polymerases and BMB1 buffer, according to the manufacturer's instructions. For inverse PCR, O157:H7 and O55:H7 DNA were digested with BglII or SacII, respectively. Ligase was added to circularize the resulting fragments, and amplifications were performed with primer pair E-F, G-H, or I-J as described above. Ligases and restriction enzymes were purchased from Gibco-BRL, Boehringer Mannheim, New England Biolabs (Beverly, Mass.), or Promega (Madison, Wis.).
Cloning and sequencing.
Amplicons generated by primer pairs A-B or C-D were digested with BamHI and EcoRI or with EcoRI, respectively, and cloned into pSK+. All other cloning used the pGem-T Easy Vector (Promega). Inserts were sequenced in both directions with the Dye Terminator Cycle Sequencing or BigDye Terminator Cycle Sequencing Ready Reaction kits (Applied Biosystems, Foster City, Calif.) and an ABI 373 or 377 sequencer (Applied Biosystems). The entire length (1,407 bp) of gnd was sequenced in both directions in each strain in which this allele was cloned. In addition, the region 3′ to gnd was sequenced in E. coli O157:H7 strain 86-24 and E. coli O55:H7 strain TB182A. Sequences were aligned with the Genetics Computer Group program (University of Wisconsin). Searches were performed with a National Center for Biotechnology Information BLAST server (12).
MLEE and allelic relatedness.
Genetic distances and phylogenetic relationships between the clonal frames (chromosomal backgrounds) of strains were inferred by analyzing allelic variation at 20 or 38 enzyme loci, determined by MLEE (33). A neighbor-joining tree was used to infer allelic phylogeny. In the distance measures parameter model, distances on branches are expressed as the number of synonymous substitutions per 100 synonymous sites. Calculations were performed on MEGA (21).
Allelic breakpoints.
Intra-gnd regions of high and low homology and breakpoints between similar and dissimilar regions (i.e., putative recombination sites) were identified by a maximum chi-square method (36). This technique categorizes nucleotides surrounding each nucleotide in the gnd alleles as different or identical and calculates a 2 × 2 (identical versus different, left versus right) chi-square value for each position. Each sequence was compared to a reference sequence, and the point, kMAX, at which the chi-square statistic was maximum was determined. The sequence was then divided into two segments determined by the kMAX point, and a new maximum was found within each segment. This cycle was repeated four times so that 16 maxima were found. The significance of the kMAX values for the nested segments was tested by a Monte Carlo procedure, in which sites were placed randomly along the sequence 1,000 times and the null distribution of kMAX was tabulated. kMAX values exceeding values in the 5% tail of the null distribution were considered significant.
Southern hybridization.
Genomic DNA from E. coli HB101, E. coli O157:H7 strain 86-24, and each of the E. coli O55 strains in Table 1 was digested with EcoRV, separated in 1% agarose in 0.5× Tris-borate-EDTA (25), stained with ethidium bromide, photographed, denatured, and transferred to a nylon membrane (Micron Separations, Westboro, Mass.). Amplicons generated from the DNA of these E. coli O55 strains with primers M and N were digested with SacI and also analyzed by Southern hybridization. The immobilized DNA was probed with the cloned amplicon generated by primers M and N from O55:H7 DNA and labeled with the Megaprime DNA system (Amersham, Arlington Heights, Ill.) and [−α32P]dATP (New England Nuclear Research Products, Boston, Mass.).
RESULTS
Sequence analysis demonstrates that three alleles of gnd can be found in E. coli O157 strains belonging to diverse lineages. These alleles are unrelated to the gnd allele of E. coli O55:H7. The O55 gnd allele is conserved in O55 strains in diverse lineages, as is a region 3′ to gnd in O55 strains. These findings are discussed below.
E. coli O157:H7 gnd allele is conserved in all DEC5 E. coli O157 strains studied.
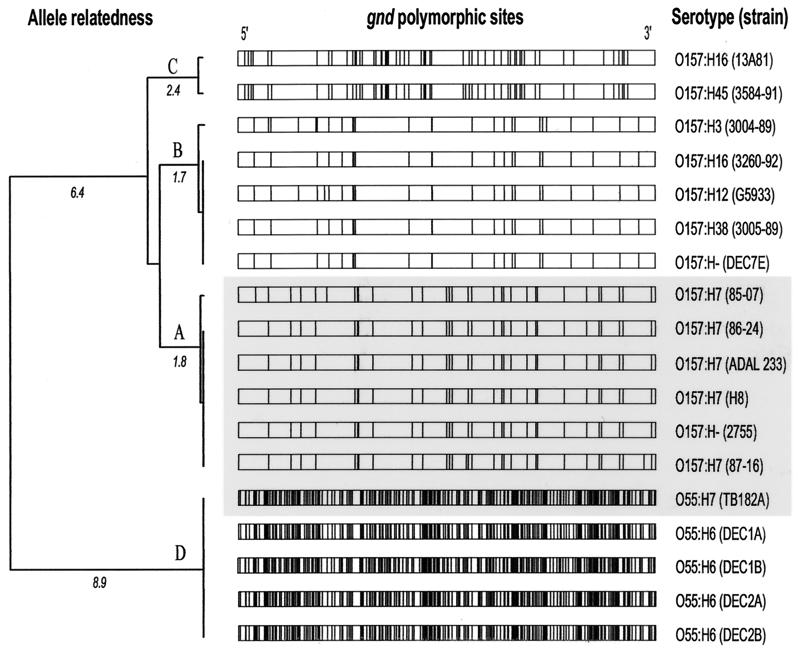
The 1,407 bp of the gnd locus in five E. coli O157:H7 strains and in a sorbitol-fermenting, Shiga toxin-producing E. coli O157:H− strain, each of which belongs to the DEC5 lineage, are nearly identical (Fig. 1 and Table 1). Among the six strains sequenced, which had been isolated from patients on four continents during two different decades, we could identify only two polymorphic sites in each of two E. coli strains. These two strains were isolated from Washington State patients in the 1980s.
FIG. 1.
Relationship between E. coli O55 and O157 gnd alleles. Horizontal bars represent gnd alleles. DEC5 lineage gnd alleles are highlighted in gray. Vertical lines within these bars denote polymorphisms compared to a consensus gnd sequence. The strains of origin of these alleles are noted to the right of each gnd. The evolutionary relationships inferred from gnd structure are represented by the dendrogram, which is derived from the percent nucleotide differences between gnd alleles. Phylogeny cannot be inferred because it is probable that the allelic structures resulted from recombination and not from point mutations.
Sequence variation among O157 strains.
Comparison of the 1,407 bp of the gnd locus of 13 O157 strains belonging to and outside the DEC5 lineage demonstrates 100 polymorphic nucleotide sites. A phylogenetic tree of the gnd sequences shows three branches or alleles, designated gnd alleles A, B, and C, defined for the purposes of this study as gnd genes composed of closely related sequences that differ at four or fewer nucleotide sites (Fig. 1 and Table 1). It is obvious that there are no regions in which polymorphisms are conserved between alleles A, B, and C (Fig. 1).
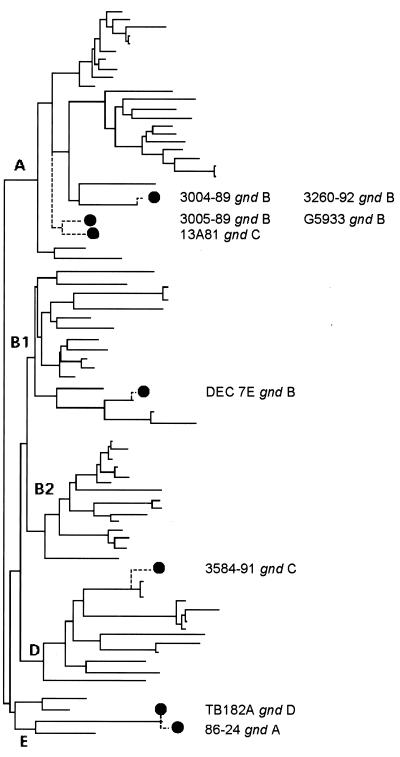
The gnd A allele is found only in DEC5 lineage O157 strains in this study, including strain E3406, an E. coli O157:H7 strain from the Pennsylvania State University E. coli collection (45). Allele A differs from alleles B and C at 4 and 6% of sites, respectively. The gnd B allele is found in a more diverse set of O157 strains, including those with H3, H12, H16, or H38 antigens. gnd allele B is also identified in strain 7E, a nonmotile E. coli O157 strain that has the same MLEE genotype as E. coli O157:H43 strains. Strain E8519 (also termed strain 851819), an E. coli O157:H− strain with an MLEE pattern resembling that of E. coli O157:H43 (46), also possesses gnd allele B. The gnd C allele is found in an O157:H16 strain and in nonmotile O157 strain 3584-91, which matches an E. coli O157:H45 strain in MLEE genotype analysis. E. coli strains in this study with gnd alleles B and C can be assigned to ECOR groups A and B1 and to groups A and D, respectively (Fig. 2), by clonal phylogenetic analysis (16, 45). The finding of gnd alleles B and C in E. coli O157 strains that belong to different lineages therefore supports the concept of cotransfer of gnd and rfb.
FIG. 2.
Relationship of E. coli O55 and O157 strains to ECOR strains. Dendrogram demonstrates the clonal relationships of the ECOR collection (from Herzer et al. [16], with additions) and the strains examined in this paper. Study strains are connected to the closest ECOR relative based on MLEE. DEC1 and -2 strains (serotype O55:H6) fall outside the ECOR tree. Solid lines are based on analysis of 38 enzyme loci. Dashed lines are based on the analysis of 20 enzyme loci.
Divergence at synonymous sites (ds) in gnd alleles (i.e., sites at which base pair changes do not affect the amino acid sequence of 6-PGD) suggests that the gnd A alleles are the most similar to one another (Table 2). Alleles B and C have approximately triple the variability of allele A. In contrast, the variability between alleles, as measured by ds, is over 30 times greater than the variability within alleles.
TABLE 2.
Nucleotide divergence between gnd sequences at synonymous and nonsynonymous sitesa
| Comparison (n) | Divergence
|
|
|---|---|---|
| Synonymous sites (dS × 100) | Nonsynonymous sites (dN × 100) | |
| O157 gnd allele A (7) | 0.09 ± 0.09 | 0.09 ± 0.05 |
| O157 gnd allele B (6) | 0.26 ± 0.19 | 0.10 ± 0.06 |
| O157 gnd allele C (2) | 0.29 ± 0.29 | 0.19 ± 0.13 |
| O55 gnd allele D (5) | 0.12 ± 0.12 | 0.04 ± 0.04 |
| O157 gnd alleles A, B, C (15) | 11.77 ± 1.35 | 0.28 ± 0.09 |
| O157 gnd allele A vs. ECOR 58 (2) | 9.52 ± 1.77 | 0.30 ± 0.17 |
| O157 gnd allele B vs. ECOR 29 (2) | 12.25 ± 2.03 | 0.30 ± 0.17 |
| O157 gnd allele C vs. ECOR 32 (2) | 15.47 ± 2.32 | 0.60 ± 0.25 |
| O157:H7 gnd allele D vs. O55:H7 allele D (2) | 130.52 ± 14.90 | 3.73 ± 0.60 |
The degree of divergence at silent sites within each allele group is approximately one-tenth that of the difference between that allele group and the closest gnd allele in the ECOR collection. The divergence between gnd alleles of E. coli O157:H7 and E. coli O55:H7 is over 1,400 times greater than the divergence between gnd A alleles.
Comparison of gnd alleles in O157:H7 and O55:H7 strains.
The background genotypes of O55:H7 and O157:H7 strains in the DEC5 lineages are closely related. However, gnd allele A and the O55:H7 gnd allele (allele D) are only 82% identical. Visual inspection demonstrates no region of conservation of polymorphisms between these two alleles (Fig. 1, shaded area). In fact the ds (130.5) exceeds the average divergence of homologous housekeeping genes of E. coli and S. enterica (34) (Table 2). Therefore, in the evolutionary separation of E. coli O157:H7 from E. coli O55:H7, all of gnd was exchanged, possibly via cotransfer with the adjacent rfb cluster.
Conservation of gnd allele D in diverse lineages.
E. coli O55 strains belonging to the DEC1 and DEC2 clonal groups possess gnd allele D, as found in DEC5 E. coli O55:H7 (Fig. 1). This conservation contrasts with the wide separation among the DEC1, -2, and -5 groups (Fig. 2). These data strongly suggest the cotransfer of gnd and the adjacent O55 rfb region.
Comparison of gnd alleles A, B, C, and D to gnd alleles from non-O157 and non-O55 strains.
We attempted to determine if part or all of gnd alleles A, B, C, and D was identical to gnd alleles in E. coli that express antigens other than O55 and O157. To do this, we compared alleles A, B, C, and D to 38 gnd sequences in public and internal databases, including those of 35 ECOR strains. Of the 1,407 nucleotides in gnd, 1,335 were analyzed. Alleles A, B, and C are most closely related to gnd alleles from ECOR strains 58 (serogroup O112), 29 (serogroup O150), and 32 (serogroup O7), respectively. There are differences at 33 (2.5%), 39 (2.9%), and 53 (4.0%) of the sites in these respective pairs. Each of the strains in which the related gnd alleles are found belongs to ECOR group B1. In contrast, gnd allele D is in a distinct branch of the phylogenetic tree and has only a distant relationship to the gnd allele of ECOR 4.
Evidence for intragenic recombination of alleles A and C.
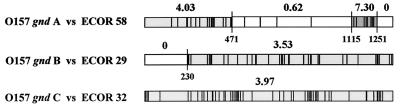
We next assessed the extent to which intragenic recombination contributed to the generation of allelic variation in gnd. To do this, we compared gnd alleles A, B, C, and D to the closest related gnd sequences in the ECOR database by the maximum chi-square method (Fig. 3). The comparison of gnd allele A to the ECOR 58 gnd indicates a mosaic structure, with breakpoints separating a central region of slight divergence from regions with more extensive divergence towards the ends of the gene. The comparison of the gnd allele B to ECOR 29 gnd identifies a single breakpoint at position 230 that separates segments of identical sequence from the remainder of the gene, the sequence of which is 3.5% divergent. Therefore, intragenic recombination has occurred in the evolutionary history of gnd alleles A and B. However, gnd allele C demonstrates no significant intragenic heterogeneity in its level of divergence from ECOR 32 gnd.
FIG. 3.
Mosaic structure of O157 gnd alleles. gnd alleles A, B, and C are compared to gnd alleles with the highest degree of structural similarity. Vertical lines denote sites differing from those in E. coli K-12. Significant kMAX points are marked with tall vertical lines, with corresponding numbers denoting the nucleotide positions. The chi-square values are kMAX 471 = 13.61, P < 0.009; kMAX 1115 = 10.91, P < 0.015; kMAX 1251 = 9.35, P < 0.026; kMAX 230 = 14.61, P < 0.005. The percent differences (in base pairs) between the sequences for each segment are noted in italics above the allele. For example, O157 gnd allele A and ECOR 58 are 4.0% different in segment 1 to 471 and 0.62% different in segment 471 to 1115. No mosaic structure is evident when O157 gnd allele C and ECOR 32 are compared.
rfbEEcO157:H7 sequences are conserved.
We assessed the extent of conservation of a central portion of the rfb region of E. coli O157:H7 to determine if any polymorphisms that might be found could shed light on the origin or mobility of this cluster. To do this, we sequenced the 456 nucleotides between primers O and P in 11 O157 strains. This segment was identical in the four DEC5 E. coli O157 strains tested and in E. coli O157:H3 strain 3004-89, H12 strain G5933, and H16 strain 3260-92. E. coli O157:H− strain DEC 7E has an A→G6651 substitution (nucleotide positions correspond to sites in GenBank submission AFO61251 [42]). An A between positions 6550 and 6557 of E. coli O157:H38 strain 3005-89 is deleted, resulting in a frameshift and a deduced gene product that is truncated compared to RfbEEcO157:H7. E. coli O157:H16 strain 13A81 and E. coli O157:H− strain 3584-91, the only two strains in this study that possess gnd allele C, each has G→C6511 and T→C6537 substitutions.
Sequence of the region 3′ of gnd in E. coli O55:H7.
Southern hybridization analysis of and restriction mapping of amplicons derived from O157:H7 and O55:H7 DNA suggest that the chromosome 3′ to gnd in these strains has unique as well as conserved regions (data not shown). Therefore, we analyzed the sequence downstream of gnd to attempt to find candidate sites of insertion of the rfb-gnd cluster into the O55:H7 and O157:H7 chromosomes.
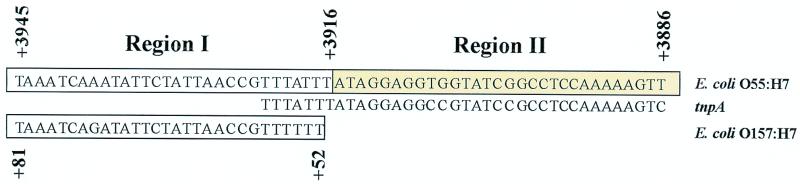
Figure 4 depicts elements of interest in the region 3′ to gnd in O55:H7 and O157:H7 DNA. Region I in each strain has 96% nucleotide identity and contains open reading frames (ORFs) that presumably encode UDP glucose-6-dehydrogenase (encoded by ugd) and an O antigen chain length-determining protein (encoded by wzz). The 3,915 and the 51 nucleotides 3′ to gnd in the O55:H7 and the O157:H7 strains, respectively, have no homology. These 3,915 O55:H7 nucleotides comprise region II in Fig. 4.
FIG. 4.
DNA surrounding gnd alleles of E. coli O55:H7 and E. coli O157:H7. DNA surrounding the O55:H7 and O157:H7 gnd alleles is depicted. Vertical numbers preceded by + represent distances in nucleotides from the 3′ end of gnd. Region I is 96% identical between the two strains. Region II includes segments homologous to tnpA of S. enterica serovar Typhimurium, an Rhs-like element including an H-rpt gene and flanking 11-mer inverted repeats, noncoding O157 rfb sequences, and O111 wbdJ and wbdK. Homologous proteins are noted directly above corresponding ORFs; the H-rpt corresponds to the ORF designated hrhEcO55:H7. Arrows represent the lateral borders of the 2.0- and 2.8-kb EcoRV fragments conserved among all E. coli O55 strains tested, as demonstrated in Fig. 6A. Asterisks represent locations of the 11-mer inverted repeats at the periphery of the Rhs-like element.
Region II has multiple components pertinent to genomic mobility. A segment of 1,129 nucleotides near its left border has extensive (96%) identity to DNA encoding an E. coli Rhs element (GenBank number L02370). This O55:H7 Rhs-like element includes an ORF that encodes a protein of 201 amino acids, 192 (96%) of which can be matched identically to amino acids encoded by ORF-H of RhsB, which encodes an H-rpt protein (49). This protein is depicted in Fig. 4 above its corresponding ORF. Eleven nucleotides (AGCTTGCCCTG) between positions +3799 and +3809, inclusive, and the nearly identical inversion (CAGGGAAGAT) of this 11-mer between positions +2655 and +2665 resemble inverted repeats flanking the H-rpt gene in other strains (49).
A segment of 7 region I and 107 region II nucleotides that straddle the O55 region I-II border has 92% identity to nucleotides at the 3′ end of tnpA of S. enterica serovar Typhimurium LT2 (Fig. 4 and 5). tnpA encodes IS200 transposase A (GenBank number AFO93749).
FIG. 5.
Sequence at border between regions I and II in E. coli O55:H7 and E. coli O157:H7. Common and unique regions of the O55 and O157 chromosome 3′ to gnd are provided, with homology to tnpA indicated. Vertical numbers preceded by + represent distance in nucleotides from the 3′ end of gnd in the respective strains.
Two contiguous region II ORFs have 75% identity to wbdJ and wbdK of the E. coli O111 rfb cluster. The deduced amino acid structures of the proteins that are encoded by these two genes are 67 and 80% identical to WbdJ and WbdK, respectively (2). Amino acid homologies at the peripheries of these ORFs exceed the corresponding nucleic acid homology (Fig. 4). Additionally, three segments within the O55 Rhs-like element have similarities (83 to 96% identity) to noncoding regions of the O157:H7 rfb cluster (35, 42).
Region 3′ to gnd is conserved in E. coli O55 belonging to diverse lineages.
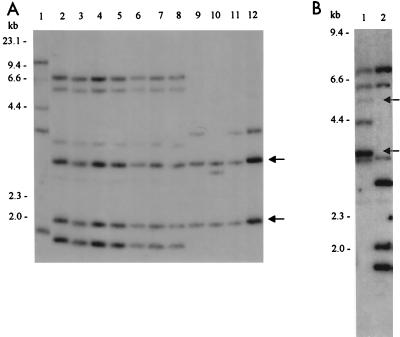
We attempted to determine if region II is conserved in 11 O55 strains belonging to the widely separated DEC lineages 1, 2, and 5. Primers M and N elicit 6.5-kb amplicons from the DNA of each of the 11 E. coli O55 strains listed in Table 1 but not from O157:H7 DNA (data not shown). These amplicons each contain a SacI site that corresponds in location to a SacI site in region II and hybridize to a probe consisting of the cloned amplicon generated by this primer pair (data not shown). This probe also detects in Southern hybridizations 2.0- and 2.8-kb EcoRV DNA fragments in each of these strains (Fig. 6A), which correspond to the DNA between the arrows in Fig. 4. These amplifications and hybridizations suggest cotransfer of the region 3′ to gnd, in addition to gnd and rfb, in O55 strains.
FIG. 6.
Conservation of the region 3′ to gnd in E. coli O55 strains in diverse lineages. EcoRV-digested bacterial DNA was electrophoresed and probed in Southern blots. The probe consists of the 3′ end of the O55:H7 gnd and regions I and II in Fig. 4. (A) DNA from E. coli HB101 (lane 1), DEC lineage 5 E. coli O55 H7 strains TB182A, TB156A, DEC 5A, 5B, 5C, 5D, and 5E (lanes 2 to 8, respectively), DEC lineage 1 E. coli O55:H6 strains 1A and 1B (lanes 9 and 10, respectively), and DEC lineage 2 E. coli O55:H6 strains 2A and 2B (lanes 11 and 12, respectively). Arrows indicate conserved EcoRV fragments corresponding to region II DNA between the arrows in Fig. 4. (B) DNA from E. coli O157:H7 (lane 1) and E. coli O55:H7 (lane 2). Arrows indicate O157:H7 fragments with predicted homology to the probe.
The 3.9- and 5.0-kb O157:H7 fragments detected by the probe generated by primers M and N (indicated by arrows in Fig. 6B) can be attributed to homology between known sequences in region I and the O157 rfb-like sequences that are in the probe fragment. The nature of the additionally detected fragments is not known but could signify IS200-tnpA-homologous elements in these isolates.
DISCUSSION
Our data shed light on the mechanisms of mobility of the rfb cluster of the E. coli chromosome and of the adjacent gnd allele. Specifically, the presence of identical gnd alleles in distantly separated lineages of E. coli O55 and of E. coli O157 provides evidence that strongly suggests the recent cotransfer of gnd and the adjacent O55 and O157 rfb clusters between unrelated E. coli strains in nature. Identical single-nucleotide polymorphisms in rfbEEcO157:H7 in E. coli O157 possessing gnd allele C in unrelated ECOR groups (46) lend additional credence to the theory that cotransfer is the mechanism of recent interbacterial movement of this region of the E. coli chromosome. However, we cannot exclude the possibility that recombination within gnd has occurred in some of the O157 strains studied or in a progenitor cell of these strains. Also, with respect to the gnd alleles described, we cannot assign donor or recipient status to any of the organisms in this study, as discussed below.
The panallelic discordance between the O55:H7 and the O157:H7 gnd alleles is consistent with cotransfer of gnd and rfb within the DEC5 lineage. However, because we have not found O157 gnd allele A in E. coli O157 outside the DEC5 lineage, we cannot state with certainty that this specific gnd allele and the adjacent O157 rfb region cotransferred into this or any other lineage. For example, intra-gnd recombination might well have occurred in a hypothetical ancestor to E. coli O157:H7 in its descent from E. coli O55:H7, resulting in gnd A.
It is important to note that despite the evidence we provide for cotransfer of gnd and rfb in the strains studied, it is apparent that intragenic recombination has also contributed to allelic variation. This finding is consistent with data from other E. coli and Salmonella strains (5, 10, 27). In particular, the corresponding breakpoints in the O157 gnd A and B alleles and the gnd alleles of E. coli O119 (ECOR 58) and E. coli O150 (ECOR 29), respectively, indicate that these genes are derived from recombination events of segments with different histories. However, it is not possible to infer the sequence of occurrence of this intragenic recombination when comparing two alleles with obviously common segments.
The association between the O157 rfb cluster and a limited number of gnd alleles might reflect the recent formation of this rfb cluster, such that the gnd alleles to which it is linked have not yet undergone extensive recombination. It is also possible that there is a selective advantage for E. coli O157 to carry gnd alleles A, B, and C rather than other gnd alleles.
We propose that the portion of the O55 chromosome that transfers between lineages, including the rfb cluster, gnd locus, and DNA 3′ to gnd including region II, be termed the E. coli O55 rfb-gnd conserved (O55 RGC) element, with yet to be defined borders. The mechanism of putative transfer of the O55 RGC element remains unknown, but several of its components warrant discussion. In particular, the H-rpt might be pertinent to rfb-gnd mobility. Transposition appears to be the mechanism of insertion of the Vibrio cholerae O139 rfb region (3, 7, 37, 38), and a construct of IS1358, an H-rpt protein gene homologue in the O139 rfb region, does transpose (9). Furthermore, the ISAS1 element of Aeromonas salmonicida (14) is an H-rpt homologue as well as a transposon. Also, H-rpt protein homologues have been proposed to play roles in rfb transfer in Salmonella (17, 48). Therefore, the H-rpt homologue gene of E. coli O55:H7, hrhEcO55:H7, encoding the O55 H-rpt protein depicted over the its corresponding ORF, might be necessary for the mobilization of this region. The location of the inverted repeat 11-mers is also interesting. In other E. coli strains studied, the 11-mer inversions are situated more closely to the H-rpt termini, whereas in E. coli O55, these inverted sequences are found at the peripheries of the Rhs-like element.
A short AT-rich region adjacent to a 3′ remnant of tnpA of IS200, a transposon which utilizes AT-rich integration sites, is also worthy of consideration as a region of insertion of the rfb-gnd region because it appears at the juncture between regions I and II. However, it is probable that the O55 RGC element includes region I, because the 4% discordance rate between the O55:H7 and the O157:H7 regions I is greater than would be expected had a common region I been present in a recent progenitor of these strains. Our data do not permit us to determine where the O55 RGC element or its components originated, the sequence in which the E. coli O55 strains studied acquired this region, and which, if any, of the E. coli O55 strains studied were donor strains for this element, nor do our data allow assignment of donor or recipient status to the cells containing the O157 alleles.
Additional region II components are noteworthy. Region II contains ORFs that encode proteins with homology to E. coli O111 WbdK and WbdJ; we have termed these O55 genes wbdKEcO55:H7 and wbdJEcO55:H7, respectively. The E. coli O111 WbdK is homologous to Yersinia pseudotuberculosis RfbH (20). RfbH is a CDP-4-keto-6-deoxy-d-glucose-3-dehydrase in the synthetic pathway of CDP-abequose, which is the d-isomer of colitose, a 3,6-dideoxyhexose that is a component of the O111 side chain. The O111 and Y. pseudotuberculosis O antigen synthetic pathways are believed to follow parallel sequences (2), and WbdK is postulated to be a pyridoxamine 5-phosphate-dependent dehydrase at a corresponding step leading to the synthesis of GDP-colitose (2). WbdJ is homologous to the E. coli K-12 fucose synthetase encoded by wcaG (fcl) (1), which converts GDP-4-keto-6-deoxymannose to GDP-l-fucose via GDP-4-keto-6-deoxygalactose. WbdJ has been proposed to function in an analogous role in the biosynthesis of the O111 LPS antigen by catalyzing the last two reactions in the cascade leading to the synthesis of GDP-colitose (41).
Colitose is an unusual residue among LPS sugars. However, this moiety is found in both O111 and O55 LPS side chains (2, 19, 23). Therefore, it is quite possible that wbdJEcO55:H7 and wbdKEcO55:H7 are necessary for the synthesis of the colitose component of the O55 LPS molecule.
The intercalation of a gene encoding an enzyme, 6-PGD, which is necessary for the viability of the bacterial cell, between loci that encode a specialized structure, the O antigen, which is presumably not necessary for viability, is surprising. However, the finding of O antigen biosynthesis genes 3′ to gnd is not unprecedented. Recently, Paton and Paton reported that wbnF, encoding a protein with homology to nucleotide sugar epimerases, is 3′ to gnd (28) in E. coli O113. It is presumed that wbnF is necessary for the synthesis of the O113 LPS antigen, because O113 rfb genes 5′ to gnd cannot confer the O113 phenotype upon E. coli K-12 without wbnF. The presence of genes necessary for the synthesis of the O antigen on both sides of gnd could, therefore, complicate long-range amplification of a complete O antigen-expressing cluster of genes. In particular, the use of primers within gnd, in combination with primers from the contralateral side of the rfb cluster (e.g., the JUMPstart sequence [42]), might not always amplify the full complement of genes necessary for the expression of the desired LPS antigen.
In contrast to the plausibility of the roles played by the proteins putatively encoded by wbdJEcO55:H7 and wbdKEcO55:H7 in the synthesis of the O55 LPS, the O55 RGC sequences that are homologous to the O157 rfb region are of less certain functional relevance. Specifically, O157 rfb-like sequences are in or near hrhEcO55:H7, and one of the 11-mer inverted repeats partially overlaps one of these O157-like sequences. In contrast, the sequences within the rfb region of E. coli O157:H7 to which these O55 sequences are homologous are not in ORFs. Therefore, this similarity is more likely to represent fortuitous homology biased by the still-limited number of rfb sequences in the database than the transfer of rfb components between the O55 and O157 chromosomes.
In summary, gnd has cotransferred with the adjacent rfb cluster into some or all of the E. coli O55 and O157 strains studied. Sequence analysis suggests that transposition might be the mechanism for this mobility in E. coli O55. O157 gnd alleles in multiple different lineages are stable and limited in number. There is no evidence that a recent common non-O157 ancestor contained any of the different O157 gnd alleles. However, portions of O157 gnd alleles A and B can be identified in the gnd alleles of other strains, demonstrating that intra-gnd recombination also contributed to the evolution of this region of the O157 chromosome. Nonetheless, in recent history, cotransfer appears to be the mechanism by which the O157 gnd allele B evolved. The mechanism(s) of rfb and gnd transfer warrants further elucidation.
ACKNOWLEDGMENTS
We thank Harry Yim for scientific advice, Steve Moseley for critical commentary on the manuscript, Charles Hill and Mary Berlyn for nomenclature suggestions, and Christine Merrikin and Kaye Green for secretarial assistance.
This research was supported by Public Health Service grants NIH R01 AI47499 (P.I.T.) and AI 42391 (T.S.W.) and USDA grant 96-01601 (P.I.T.).
REFERENCES
- 1.Andrianopolous K, Wang L, Reeves P. Identification of the fucose synthetase gene in the colanic acid gene cluster of Escherichia coli K-12. J Bacteriol. 1998;180:998–1001. doi: 10.1128/jb.180.4.998-1001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastin D A, Reeves P R. Sequence and analysis of the O antigen gene rfb cluster of Escherichia coli O111. Gene. 1995;164:17–23. doi: 10.1016/0378-1119(95)00459-j. [DOI] [PubMed] [Google Scholar]
- 3.Bik E M, Bunschoten A E, Gouw R D, Mooi F R. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilge S S, Vary J C, Jr, Dowell S F, Tarr P I. Role of the Escherichia coli O157:H7 O-side chain in adherence and analysis of an rfb locus. Infect Immun. 1996;64:4795–4801. doi: 10.1128/iai.64.11.4795-4801.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bisercic M, Feutrier J Y, Reeves P R. Nucleotide sequences of the gnd genes from nine natural isolates of Escherichia coli: evidence of intragenic recombination as a contributing factor in the evolution of the polymorphic gnd locus. J Bacteriol. 1991;173:3894–3900. doi: 10.1128/jb.173.12.3894-3900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bokete T N, Whittam T S, Wilson R A, Clausen C R, O'Callahan C M, Moseley S L, Fritsche T R, Tarr P I. Genetic and phenotypic analysis of Escherichia coli with enteropathogenic characteristics isolated from Seattle children. J Infect Dis. 1997;175:1382–1389. doi: 10.1086/516470. [DOI] [PubMed] [Google Scholar]
- 7.Comstock L E, Johnson J A, Michalski J M, Morris J G, Jr, Kaper J B. Cloning and sequence of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of Vibrio cholerae O1. Mol Microbiol. 1996;19:815–826. doi: 10.1046/j.1365-2958.1996.407928.x. [DOI] [PubMed] [Google Scholar]
- 8.Desmarchelier P M, Bilge S S, Fegan N, Mills L, Vary J C, Tarr P I. A PCR specific for Escherichia coli O157 based on the rfb locus encoding O157 lipopolysaccharide. J Clin Microbiol. 1998;36:1801–1804. doi: 10.1128/jcm.36.6.1801-1804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumontier S, Trieu-Cuot P, Berche P. Structural and functional characterization of IS1358 from Vibrio cholerae. J Bacteriol. 1998;180:6101–6106. doi: 10.1128/jb.180.23.6101-6106.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dykhuizen D E, Green L. Recombination in Escherichia coli and the definition of biological species. J Bacteriol. 1991;173:7257–7268. doi: 10.1128/jb.173.22.7257-7268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng P, Lampel K A, Karch H, Whittam T S. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998;177:1750–1753. doi: 10.1086/517438. [DOI] [PubMed] [Google Scholar]
- 12.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 13.Grossman N, Schmetz M A, Foulds J, Klima E N, Jimenez-Lucho V E, Leive L L, Joiner K A. Lipopolysaccharide size and distribution determine serum resistance in Salmonella montevideo. J Bacteriol. 1987;169:856–863. doi: 10.1128/jb.169.2.856-863.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustafson C E, Chu S, Trust T J. Mutagenesis of the paracrystalline surface protein array of Aeromonas salmonicida by endogenous insertion elements. J Mol Biol. 1994;237:452–463. doi: 10.1006/jmbi.1994.1247. [DOI] [PubMed] [Google Scholar]
- 15.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 16.Herzer P J, Inouye S, Inouye M, Whittam T S. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill C W, Sandt C H, Vlazny D A. Rhs elements of Escherichia coli: a family of genetic composites each encoding a large mosaic protein. Mol Microbiol. 1994;12:865–871. doi: 10.1111/j.1365-2958.1994.tb01074.x. [DOI] [PubMed] [Google Scholar]
- 18.Kaper J, Hacker J. Pathogenicity islands and other mobile virulence elements. Washington, D.C.: ASM Press; 1999. [Google Scholar]
- 19.Kenne L, Lindberg B, Soderholm E, Bundle D R, Griffith D W. Structural studies of the O-antigens from Salmonella greenside and Salmonella adelaide. Carbohydr Res. 1983;111:289–296. doi: 10.1016/0008-6215(83)88313-x. [DOI] [PubMed] [Google Scholar]
- 20.Kessler A, Haase A, Reeves P. Molecular analysis of the 3,6-dideoxyhexose pathway genes of Yersinia pseudotuberculosis serogroup IIA. J Bacteriol. 1993;175:1412–1422. doi: 10.1128/jb.175.5.1412-1422.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis, 1.0 ed. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 22.Lawrence J, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindberg B, Lindh F, Lonngren J. Structural studies of the O-specific side-chain of the lipopolysaccharide from Escherichia coli O55. Carbohydr Res. 1981;97:105–112. doi: 10.1016/s0008-6215(00)80528-5. [DOI] [PubMed] [Google Scholar]
- 24.Liu D, Reeves P R. Presence of different O antigen forms in three isolates of one clone of Escherichia coli. Genetics. 1994;138:6–10. [PMC free article] [PubMed] [Google Scholar]
- 25.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 26.McGraw E A, Li J, Selander R K, Whittam T S. Molecular evolution and mosaic structure of α, β, and γ intimins of pathogenic Escherichia coli. Mol Biol Evol. 1999;16:12–22. doi: 10.1093/oxfordjournals.molbev.a026032. [DOI] [PubMed] [Google Scholar]
- 27.Nelson K, Selander R K. Intergeneric transfer and recombination of the 6-phosphogluconate dehydrogenase gene (gnd) in enteric bacteria. Proc Natl Acad Sci USA. 1994;91:10227–10231. doi: 10.1073/pnas.91.21.10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paton A, Paton J. Molecular characterization of the locus encoding biosynthesis of the lipopolysaccharide O antigen of Escherichia coli serotype O113. Infect Immun. 1999;67:5930–5937. doi: 10.1128/iai.67.11.5930-5937.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry M B, MacLean L, Griffith D W. Structure of the O-chain polysaccharide of the phenol-phase soluble lipopolysaccharide of Escherichia coli O157:H7. Biochem Cell Biol. 1986;64:21–28. doi: 10.1139/o86-004. [DOI] [PubMed] [Google Scholar]
- 30.Reeves P. Role of O-antigen variation in the immune response. Trends Microbiol. 1995;3:381–386. doi: 10.1016/s0966-842x(00)88983-0. [DOI] [PubMed] [Google Scholar]
- 31.Reeves P R, Hobbs M, Valvano M, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C, Rick P. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 32.Reid S D, Selander R K, Whittam T S. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J Bacteriol. 1999;181:153–160. doi: 10.1128/jb.181.1.153-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharp P M. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J Mol Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu T, Yamasaki S, Tsukamoto T, Takeda Y. Analysis of the genes responsible for the O-antigen in enterohaemorrhagic Escherichia coli O157. Microb Pathog. 1999;26:235–247. doi: 10.1006/mpat.1998.0253. [DOI] [PubMed] [Google Scholar]
- 36.Smith J M. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 37.Stroeher U H, Jedani K E, Dredge B K, Morona R, Brown M H, Karageorgos L E, Albert M J, Manning P A. Genetic rearrangements in the rfb regions of Vibrio cholerae O1 and O139. Proc Natl Acad Sci USA. 1995;92:10374–10378. doi: 10.1073/pnas.92.22.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stroeher U H, Parasivam G, Dredge B K, Manning P A. Novel Vibrio cholerae O139 genes involved in lipopolysaccharide biosynthesis. J Bacteriol. 1997;179:2740–2747. doi: 10.1128/jb.179.8.2740-2747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarr P I. Escherichia coli O157:H7: clinical, diagnostic, and epidemiological aspects of human infection. Clin Infect Dis. 1995;20:1–8. doi: 10.1093/clinids/20.1.1. [DOI] [PubMed] [Google Scholar]
- 40.Tarr P I, Neill M A, Clausen C R, Newland J W, Neill R J, Moseley S L. Genotypic variation in pathogenic Escherichia coli O157:H7 isolated from patients in Washington. J Infect Dis. 1989;159:344–347. doi: 10.1093/infdis/159.2.344. [DOI] [PubMed] [Google Scholar]
- 41.Wang L, Curd H, Qu W, Reeves P R. Sequencing of Escherichia coli O111 O-antigen gene cluster and identification of O111-specific genes. J Clin Microbiol. 1998;36:3182–3187. doi: 10.1128/jcm.36.11.3182-3187.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang L, Reeves P R. Organization of Escherichia coli O157 O antigen gene cluster and identification of its specific genes. Infect Immun. 1998;66:3545–3551. doi: 10.1128/iai.66.8.3545-3551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;1:1–8. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 44.Whittam T S. Genetic population structure and pathogenicity in enteric bacteria. In: Baumberg S, Young J P W, Saunders S R, Wellington E M H, editors. Population genetics of bacteria. Cambridge, U.K: Cambridge University Press; 1995. pp. 217–245. [Google Scholar]
- 45.Whittam T S, Wachsmuth I K, Wilson R A. Genetic evidence of clonal descent of Escherichia coli O157:H7 associated with hemorrhagic colitis and hemolytic uremic syndrome. J Infect Dis. 1988;157:1124–1133. doi: 10.1093/infdis/157.6.1124. [DOI] [PubMed] [Google Scholar]
- 46.Whittam T S, Wilson R A. Genetic relationships among pathogenic Escherichia coli of serogroup O157. Infect Immun. 1988;56:2467–2473. doi: 10.1128/iai.56.9.2467-2473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whittam T S, Wolfe M L, Wachsmuth I K, Orskov F, Orskov I, Wilson R A. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiang S H, Hobbs M, Reeves P R. Molecular analysis of the rfb gene cluster of a group D2 Salmonella enterica strain: evidence for its origin from an insertion sequence-mediated recombination event between group E and D1 strains. J Bacteriol. 1994;176:4357–4365. doi: 10.1128/jb.176.14.4357-4365.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao S, Sandt C H, Feulner G, Vlazny D A, Gray J A, Hill C W. Rhs elements of Escherichia coli K-12: complex composites of shared and unique components that have different evolutionary histories. J Bacteriol. 1993;175:2799–2808. doi: 10.1128/jb.175.10.2799-2808.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]