Abstract
In vitro oogenesis is key to elucidating the mechanism of human female germ‐cell development and its anomalies. Accordingly, pluripotent stem cells have been induced into primordial germ cell‐like cells and into oogonia with epigenetic reprogramming, yet further reconstitutions remain a challenge. Here, we demonstrate ex vivo reconstitution of fetal oocyte development in both humans and cynomolgus monkeys (Macaca fascicularis). With an optimized culture of fetal ovary reaggregates over three months, human and monkey oogonia enter and complete the first meiotic prophase to differentiate into diplotene oocytes that form primordial follicles, the source for oogenesis in adults. The cytological and transcriptomic progressions of fetal oocyte development in vitro closely recapitulate those in vivo. A comparison of single‐cell transcriptomes among humans, monkeys, and mice unravels primate‐specific and conserved programs driving fetal oocyte development, the former including a distinct transcriptomic transformation upon oogonia‐to‐oocyte transition and the latter including two active X chromosomes with little X‐chromosome upregulation. Our study provides a critical step forward for realizing human in vitro oogenesis and uncovers salient characteristics of fetal oocyte development in primates.
Keywords: ex vivo culture, fetal oocytes, humans, meiotic prophase, monkeys
Subject Categories: Development, Methods & Resources, Stem Cells & Regenerative Medicine
A novel fetal oocyte differentiation culture uncovers primate‐specific and evolutionarily conserved oogenesis programs.
Introduction
Germ cells differentiate either into oocytes or into spermatozoa, which unite to form zygotes with full developmental potential, thereby perpetuating and diversifying the genetic information across generations. An impairment in germ‐cell development leads to critical consequences, including infertility and genetic and epigenetic disorders of offspring. Understanding the mechanism of germ‐cell development is therefore a fundamental goal in both biology and medicine.
In mammals, germ cells arise as primordial germ cells (PGCs) early during embryonic development (Tang et al, 2016; Saitou & Hayashi, 2021). PGCs undergo migration and colonize the embryonic gonads, where they differentiate into oogonia in females or gonocytes in males. A key event that occurs in PGCs is epigenetic reprogramming, which leads to imprint erasure and X‐chromosome reactivation in females (Tang et al, 2016; Wen & Tang, 2019; Saitou & Hayashi, 2021). Consequently, oogonia and gonocytes show similar genetic and epigenetic properties until they start overt sexual differentiation, except that oogonia acquire two active X chromosomes, whereas gonocytes have one active X and one Y chromosome (Tang et al, 2016, Wen & Tang, 2019, Saitou & Hayashi, 2021) (see Conserved activities of X chromosomes section). In females, in response to cues from the embryonic ovaries, particularly from pre‐granulosa cells, oogonia differentiate into oocytes with an immediate entry into the first prophase of meiosis, and the oocytes that complete the meiotic prophase I are ensheathed by a single layer of granulosa cells to form primordial follicles, which serve as the source for oogenesis. In contrast, in males, in response to cues from the embryonic testes, particularly from Sertoli cells, gonocytes differentiate into pro‐spermatogonia and then into spermatogonia/spermatogonial stem cells (SSCs), which serve as the source for spermatogenesis (Griswold, 2016; Spiller et al, 2017; Wen & Tang, 2019; Saitou & Hayashi, 2021). The mechanism for mammalian germ‐cell development has been studied intensively using the mouse as a model organism, and based on such knowledge as well as the advancement of pluripotent stem cell (PSC) and reproductive technologies, the whole process of both female and male germ‐cell development in mice has been reconstituted in vitro using mouse PSCs (mPSCs) as a starting material (Hayashi et al, 2011, 2012; Hikabe et al, 2016; Ishikura et al, 2016, 2021), establishing the proof of concept of mammalian in vitro gametogenesis (IVG) research (Saitou & Hayashi, 2021).
The concept and strategy for mouse IVG have been progressively translated into the IVG research in other mammals, particularly humans (Irie et al, 2015; Sasaki et al, 2015; Yamashiro et al, 2018; Hwang et al, 2020), as human IVG serves as a foundation to explore the mechanism of human germ‐cell development, including the diseased states, which will provide salient information for disease etiologies and create novel possibilities in reproductive medicine (Saitou & Hayashi, 2021). With regard to the human oogenic pathway, human PSCs (hPSCs) are induced into human PGC‐like cells (hPGCLCs), which, upon aggregation culture with mouse embryonic ovarian somatic cells, differentiate into oogonia‐like cells with epigenetic reprogramming and then into early oocyte‐like cells at the outset of meiotic prophase I (Yamashiro et al, 2018, 2020). A key next step will therefore be to induce primordial follicles, that is, to differentiate oogonia‐ or early oocyte‐like cells into oocyte‐like cells that complete the meiotic prophase I and reach the diplotene stage by culturing them with human embryonic ovarian somatic cells or their in vitro counterparts induced from hPSCs (Yoshino et al, 2021). To attain such a goal, based on the history of mouse IVG research, we consider it an essential prerequisite to identify a condition that allows the in vitro maturation of human fetal ovaries containing mainly oogonia into those bearing primordial follicles.
Toward this end, we first used cynomolgus monkeys (cy: Macaca fascicularis), a primate closely related to humans and amenable for reproductive experimentations (Yamasaki et al, 2011), to screen for such an in vitro culture condition. We adopted ovary dissociation/reaggregation steps, as the dissociation of ovaries into single cells allows: (1) generation of their frozen stocks with an appropriate cell number, making a systematic exploration of their culture conditions feasible; (2) selective isolation/removal of specific cell populations by cell sorting; and (3) application to the reaggregation with oogonia‐like cells induced from PSCs. We then applied the condition identified with cynomolgus monkeys to humans to demonstrate that human fetal ovary reaggregates mainly with oogonia can also be matured into those with primordial follicles in vitro. We performed detailed cytological and single‐cell RNA sequence (scRNA‐seq) analysis not only to show that cy and human fetal oocyte development in vitro are an appropriate reconstitution of those in vivo, but also to identify evolutionarily conserved and primate‐specific pathways driving fetal oocyte development, including the progression of meiotic prophase I, among humans, monkeys, and mice. Our study creates a robust foundation for further promoting human IVG research as well as for exploring the mechanism of fetal oocyte development and meiotic prophase in diverse mammalian species.
Results
Cy fetal oocyte development
Cy PGCs are specified in the nascent amnion from around embryonic day (E) 11, migrate through the developing hindgut endoderm and mesentery, and colonize the incipient embryonic gonads from around E30 (Sasaki et al, 2016). In female embryos, embryonic gonads increase their size progressively and are specified into ovaries by around E40, whereas cyPGCs continue to colonize the developing gonads and also increase their numbers mitotically as oogonia (Sasaki et al, 2016, 2021). Thereafter, fetal oocyte development ensues in fetal ovaries (Fig 1A). To create a benchmark for the in vitro reconstitution of cy fetal oocyte development, we performed a careful characterization of cy fetal oocyte development in vivo from 8 weeks post‐fertilization (8 wpf: at E56–58) to 18 wpf (at E127 and 128) at two‐week intervals (Figs 1B and EV1A). The gestation period of cynomolgus monkeys is around 22 weeks.
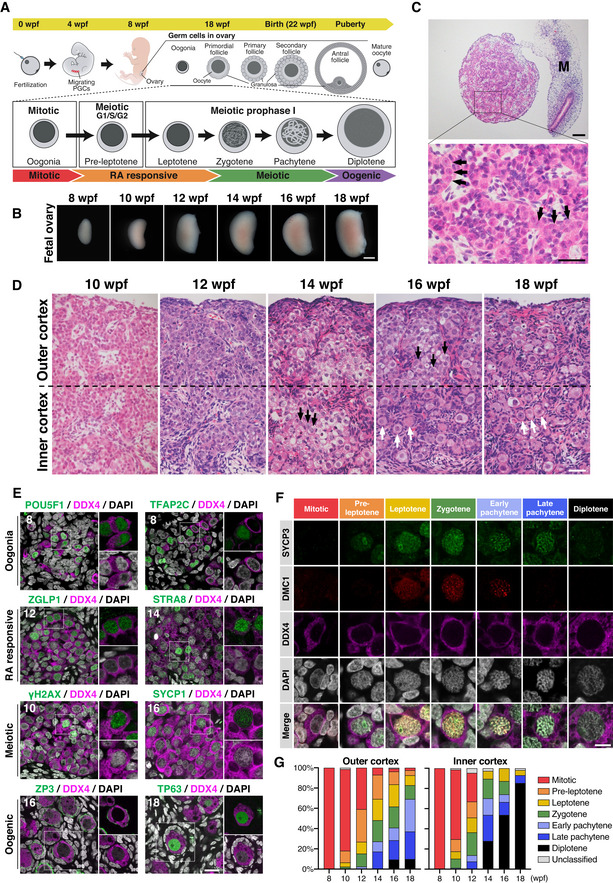
Figure 1. Fetal ovary development in cynomolgus monkeys.
-
AScheme of female germ‐cell development in cynomolgus monkeys. Wpf, weeks post‐fertilization; PGCs, primordial germ cells.
-
BGross appearances of developing cy ovaries at 8–18 wpf. Scale bar = 1 mm.
-
CHematoxylin and eosin (H&E) staining of a cy fetal ovary at 8 wpf. Arrows indicate oogonia. M, mesonephros. Scale bars = 100 μm (top), 40 μm (bottom).
-
DH&E staining of the ovarian cortex in cy fetuses at 10–18 wpf. The ovarian cortex was divided into the outer and inner cortex at the line bisecting the ovarian cortex. Black arrows, oocytes bearing a large nucleus with condensed, thread‐like chromatin; white arrows, primordial follicles. Scale bar = 50 μm.
-
EImmunofluorescence (IF) analysis for oogonia (POU5F1 and TFAP2C), RA responsive (ZGLP1 and STRA8), meiotic (γH2AX and SYCP1), and oogenic (ZP3 and TP63) markers during cy fetal ovary development. Representative images for each marker are shown. Germ cells were marked with DDX4 (magenta). Nuclear DAPI staining is shown in white. The upper‐right magnified images of each panel show the expression of key markers (green) co‐stained with DDX4 (magenta). The lower‐right magnified images of each panel show DDX4 expression (magenta) co‐stained with DAPI (white). The numbers written in the upper‐left corner indicate the stage (wpf) of the fetus. Scale bar = 20 μm.
-
FRepresentative images of germ cells immunostained for SYCP3 (green), DMC1 (red), and DDX4 (magenta) at each meiotic substage. Nuclear DAPI staining is shown in white. The substages of meiosis were defined based on the staining patterns of SYCP3, DMC1, and DAPI as described in Materials and Methods. Scale bar = 10 μm.
-
GPercentages of germ cells at each meiotic substage as defined in Fig 1F. The average percentages in the outer or inner ovarian cortex on each meiotic substage were calculated from at least three fetal ovaries [8 wpf (n = 4), 10/12/14/16/18 wpf (n = 3)]. The color‐coding is as indicated.
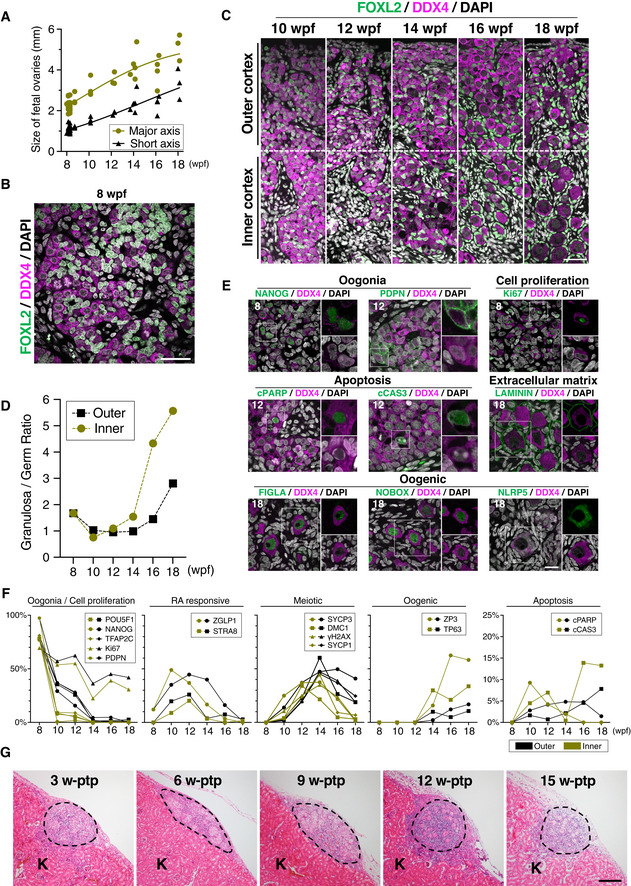
Figure EV1. Histological analyses of cy fetal ovaries in vivo and in xenotransplanted rOvaries.
-
ASize of developing cy fetuses and ovaries at 8–18 wpf [8/10/12/14/16/18 wpf (n = 26/3/3/5/4/3)]. Lines indicate LOESS curves fitted to the sample populations.
-
B, CIF of a cy fetal gonad at 8 wpf (B) and ovarian cortexes in cy fetuses at 10–18 wpf (C). The results of staining with FOXL2 (granulosa‐cell marker, green), DDX4 (germ‐cell marker, magenta), and DAPI (nucleus, white) are shown. The ovarian cortexes at 10–18 wpf were divided into an outer and inner cortex at the line bisecting the ovarian cortex. Scale bars = 40 μm (B) and 50 μm (C).
-
DThe granulosa/germ cell ratios at each developmental stage assessed from the IF data are shown [8–18 wpf (n = 1)].
-
EIF for oogonia (NANOG and PDPN), cell proliferation (Ki67), apoptosis (cleaved PARP and cleaved CASPASE 3), extracellular matrix (LAMININ), and oogenic (FIGLA, NOBOX, and NLRP5) markers during cy fetal ovary development. Representative images for each marker are shown. Germ cells were marked with DDX4 (magenta). Nuclear DAPI staining is shown in white. The upper‐right magnified images of each panel show the expression of key markers (green) co‐stained with DDX4 (magenta) except for NLRP5. The lower‐right magnified images of each panel show DDX4 expression (magenta) co‐stained with DAPI (white). The numbers written in the upper‐left corner indicate the stage (wpf) of the fetus. Scale bar = 20 μm.
-
FPercentages of cells positive for individual markers among DDX4+ germ cells from the IF of the cy fetal ovarian outer/inner cortex at 8–18 wpf. The mean values from more than three biological replicates are shown [8 wpf (n = 4), 10/12/14/16/18 wpf (n = 3)].
-
GH&E staining of transplanted cy rOvaries at 3‐/6‐/9‐/12‐/15‐week post‐transplantation (w‐ptp). Dashed lines indicate the transplanted cy rOvaries beneath the kidney capsule of KSN/Slc mice. K, mouse kidney. Scale bar = 200 μm.
Histological analysis
At 8 wpf, the ovaries exhibited a relatively uniform histology with abundant oogonia and no clear differentiation of ovarian cortex and medulla (Fig 1C). Oogonia were large in size (∼ 12 μm in diameter), bore a round nucleus with pale hematoxylin staining and a prominent eosinophilic nucleolus, and formed anastomosing cord‐like structures intercalated by relatively small cells with densely stained nuclei and a large nuclear–cytoplasmic ratio (Fig 1C). The interstitial stromal cells were evident, showed a spindle shape with a large nuclear–cytoplasmic ratio, and were embedded in loose connective tissues presumably synthesized by themselves (Fig 1C). Immunofluorescence (IF) analyses revealed that all oogonia expressed DDX4, an RNA‐binding protein expressed in oogonia and oocytes (Toyooka et al, 2000), whereas small, intercalated cells expressed FOXL2, a key transcription factor (TF) for granulosa‐cell development (Crisponi et al, 2001), demonstrating their granulosa‐cell identity (Fig EV1B). At 10 wpf, the ovaries grew in size with the acquisition of a kidney‐like shape, developing medulla consisting mainly of interstitial cells (Fig 1B). The cortex was populated with abundant oogonia, and the outer and inner cortexes, which extended over ∼ 450 μm, did not show overt histological differences (Figs 1D and EV1C). At 12 wpf, while oogonia were still abundant, germ cells with larger nuclei with condensed, thread‐like chromatin became apparent, particularly in the inner cortex (Figs 1D and EV1C), indicating that oogonia enter into the meiotic prophase I to differentiate into oocytes. At 14 wpf, oocytes at meiotic prophase I became dominant even in the outer cortex, and relatively large oocytes delineated by granulosa cells, that is, primordial follicles, began to be observed in the inner cortex (Figs 1D and EV1C). At 16 wpf, primordial follicles were observed in the outer cortex as well, and at 18 wpf, they became dominant both in the outer and in the inner cortexes (Figs 1D and EV1C). On the contrary, even at 18 wpf, oogonia‐like cells with no apparent sign of meiotic entry were observed in the outer cortex (Figs 1D and EV1C), demonstrating a substantial asynchrony in cy oocyte development. We measured the (pre‐)granulosa: germ‐cell ratio (FOXL2‐positive(+): DDX4+ cell ratio) to find that it was ∼ 2 at 8 wpf, dropped to and remained ∼ 1 at 10 and 12 wpf, and increased sharply after 14 wpf, particularly in the inner cortex, reaching ∼ 6 and ∼ 3 at 18 wpf in the inner and outer cortexes, respectively (Fig EV1D). This finding is consistent with the idea that germ cells increase their number by mitotic expansion of oogonia at least until 10 wpf and then their number decreases dramatically after 14 wpf, presumably due to apoptosis associated with meiotic prophase (see below), whereas granulosa cells maintain their number relatively constantly or proliferate slowly after their specification.
Key‐marker expression
An scRNA‐seq analysis classified human female fetal germ cells into four cell types: “mitotic” (oogonia), “retinoic acid (RA)‐responsive,” “meiotic,” and “oogenesis (hereafter, we call them “oogenic”)” (Li et al, 2017). Accordingly, we evaluated the cy fetal oocyte development with the expression of key markers of the four cell types. At 8 wpf, a vast majority of DDX4+ cells expressed markers for oogonia: Nearly all DDX4+ cells were positive for PDPN, and around 80% were positive for POU5F1, TFAP2C, and NANOG, whereas only a minority (∼ 10%) in the inner cortex expressed ZGLP1, a determinant for the oogenic fate and a marker for RA‐responsive cells (Figs 1E and EV1E and F) (Li et al, 2017; Nagaoka et al, 2020). At 10 wpf, despite their histological similarity to the cells at 8 wpf, germ cells expressing oogonia markers decreased dramatically, particularly in the inner cortex; on the contrary, those expressing markers for RA‐responsive cells increased (ZGLP1: ∼ 40–50%; STRA8: ∼ 10–20%), and some (∼ 10–20%) expressed SYCP3 (Figs 1E and EV1F), an axial component of the synaptonemal complex (SC) that initiates characteristic expression from the pre‐leptotene stage of meiosis (Yuan et al, 2000, 2002) (see below). While the oogonia markers, NANOG, POU5F1, and TFAP2C, showed a uniform distribution across oogonia nuclei, the RA‐responsive cell markers, ZGLP1 and STRA8, exhibited a granular distribution (Figs 1E and EV1E). At 12 wpf, germ cells expressing oogonia markers continued to decrease and those expressing markers for RA‐responsive and meiotic cells (SYCP3, DMC1, γH2AX, and SYCP1) continued to increase, and at 14 wpf, oogonia became minor (∼ 5%), meiotic cells became predominant (∼ 50%), and cells expressing oogenic markers (ZP3 and TP63) appeared in both the outer (∼ 10%) and the inner (∼ 30%) cortexes (Figs 1E and EV1F). γH2AX, a marker for double‐strand breaks (DSBs) upon meiotic recombination (Fernandez‐Capetillo et al, 2003), exhibited a dotted distribution in meiotic nuclei, while SYCP1, another axial component of SC (Meuwissen et al, 1992), showed a typical thread‐like distribution along synapsed homologous chromosomes in nuclei corresponding to the late zygotene or pachytene stage of the meiotic prophase I (see below). At 16 and 18 wpf, the oogenic cells (oocytes at the diplotene stage in the primordial follicles) (see below) were dominant in the inner cortex, and the meiotic cells were abundant in the outer cortex (Figs 1E and EV1F). In the oogenic cells, ZP3 was localized on the plasma membrane or in the extracellular space between oocytes and surrounding granulosa cells, indicating the formation of an incipient zona pellucida (ZP) (Fig 1E). TP63, FIGLA, and NOBOX, TFs critical for oogenesis (Liang et al, 1997; Suzumori et al, 2002; Suh et al, 2006), all showed a uniform localization in the nucleoplasm, except in the nucleolus, of the oocytes, whereas NLRP5, a maternal effect protein essential for early embryonic development (Tong et al, 2000), exhibited punctate localization throughout the cytoplasm (Figs 1E and EV1E). LAMININ, a marker for the basement membrane, delineated the basal side of the squamous granulosa cells composing primordial follicles, marking both single and sometimes multiple primordial follicles (Fig EV1E).
We next examined the expression of Ki67, a marker for cell proliferation (Gerdes et al, 1983), and the expressions of cleaved poly ADP‐ribose polymerase (cPARP) and CASPASE 3 (cCAS3), markers for apoptosis (Lazebnik et al, 1994; Tewari et al, 1995). At 8 wpf, a majority of oogonia (∼ 70%) expressed Ki67, which was localized mainly around the nucleolus (Fig EV1E and F). Although germ cells expressing oogonia markers decreased dramatically after 10 wpf, we observed Ki67+ cells throughout the stages we examined (∼ 40%) (Fig EV1F); these cells would have represented RA‐responsive and meiotic cells at an early stage of meiosis (see below). We did not detect germ cells with apoptotic markers at 8 wpf, but from 10 to 14 wpf with the onset and progression of meiotic prophase, and at 16 and 18 wpf with the formation of primordial follicles, ∼ 10% and ∼ 15% of germ cells, respectively, showed apoptotic markers (Fig EV1E and F). The apoptotic primordial follicles showed cCAS3, but not cPARP (Fig EV1F).
Progression of the meiotic prophase I
We determined the progression of the meiotic prophase I based on the chromatin architecture visualized by DAPI staining and the expression/localization of SYCP3 and DMC1, the latter being involved in single‐strand invasion for homologous recombination (Bishop et al, 1992; Fig 1F): Mitotic oogonia did not show overt signs of chromosome condensation, expressing neither SYCP3 nor DMC1. The pre‐leptotene oocytes initiated chromosome condensation and expressed SYCP3, which was localized around the nucleolus, but did not show DMC1. The leptotene oocytes exhibited further chromosome condensation and expressed SYCP3, which was localized both around the nucleolus and condensing chromosomes, in addition to DMC1, which was localized on condensing chromosomes. The zygotene oocytes exhibited signs of chromosome pairing and expressed SYCP3 and DMC1, which were localized on the paired chromosomes, with DMC1 showing multiple strongly stained foci. The early pachytene oocytes exhibited robust chromosome pairing with a clear axial structure, and expressed SYCP3 and DMC1, which showed co‐localization on multiple strongly stained foci, most likely crossover recombination sites of homologous chromosomes. The late pachytene oocytes were similar to the early pachytene oocytes, except that the former no longer expressed DMC1. The diplotene oocytes were larger in size, exhibited signs of diakinesis, and repressed both SYCP3 and DMC1. With this classification, a majority of germ cells were categorized as mitotic until 10 wpf, and it was demonstrated that the meiotic prophase I initiates in the inner cortex at 10 wpf and gradually proceeds during cy fetal development, with diplotene oocytes that complete the prophase appearing from 14 wpf onwards (Fig 1G). Collectively, these data update the classical observations in rhesus monkeys (Macaca Mulatta) (Baker, 1966) and create a cytological basis for the progression of cy fetal oocyte development, demonstrating that initiation of meiotic prophase I and overt morphological changes require around 2 weeks from the repression of oogonia markers and that the meiotic prophase I takes 4–6 weeks to complete.
Cy fetal oocyte development in xenotransplanted cy rOvaries
We next examined whether cy fetal oocyte development proceeds upon xenotransplantation of cy reaggregated ovaries (cy rOvaries) into immunodeficient mice (Fig 2A). We isolated cy fetal ovaries at 8 wpf, dissociated them into single cells (∼ 6.0 × 105 cells per ovary), reaggregated ∼ 50,000 cells (approximately 5,000 germ cells and 45,000 somatic cells) under a floating condition for 2 days to generate cy rOvaries, transplanted them under the surface epithelium of a kidney of immunodeficient mice, and analyzed the cy oocyte development at 3‐week intervals.
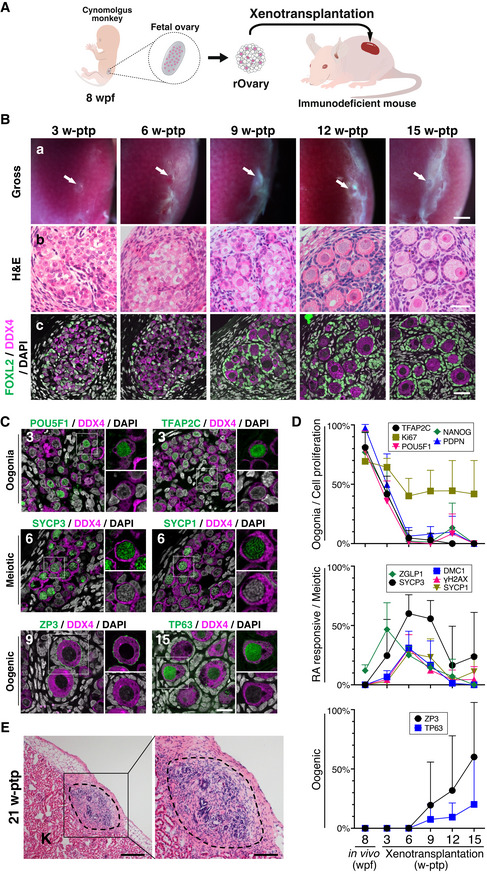
Figure 2. Derivation of cy ovarian follicles from fetal gonadal cells by xenotransplantation.
-
AScheme of the xenotransplantation experiment. The cy rOvaries generated from in vivo ovarian cells at 8 wpf were transplanted beneath the kidney capsule of immunodeficient KSN/Slc mice (see Materials and Methods).
-
BGross appearances (a), H&E staining (b), and IF for FOXL2/DDX4/DAPI (c) of transplanted cy rOvaries at 3‐/6‐/9‐/12‐/15‐week posttransplantation (w‐ptp). The primordial follicle‐like complexes surrounded with FOXL2+ granulosa cells were observed inside the rOvaries at 12‐ and 15‐w ptp. Arrows indicate the rOvaries. FOXL2, granulosa cell marker; DDX4, germ cell marker. Scale bars = 1 mm (a), 40 μm (b and c).
-
CIF for oogonia (POU5F1 and TFAP2C), meiotic (SYCP3 and SYCP1), and oogenic (ZP3, TP63) markers in the transplanted cy rOvaries. Germ cells and nuclei were marked with DDX4 (magenta) and DAPI (white), respectively. The upper‐right magnified images of each panel show the expression of each marker (green) co‐stained with DDX4 (magenta). The lower‐right magnified images of each panel show DDX4 expression (magenta) co‐stained with DAPI (white). The numbers written in the upper‐left corner indicate weeks after transplantation (w‐ptp). Scale bar = 20 μm.
-
DPercentages of cells positive for individual markers among DDX4+ germ cells from the IF of cy rOvaries at 3–15 w‐ptp [3 w‐ptp (n = 3), 6/9/12 w‐ptp (n = 4), 15 w‐ptp (n = 6)]. The average values with SDs are shown.
-
EH&E staining of transplanted cy rOvaries at 21‐w ptp. Dashed lines indicate the transplanted cy rOvaries beneath the kidney capsule of KSN/Slc mice. K, mouse kidney. Scale bars = 200 μm (left) and 100 μm (right).
At 3‐week post‐transplantation (3 w‐ptp), cy rOvaries bore an integrated structure, and although they did not exhibit overt outer/inner cortex stratification, germ cells reconstructed anastomosing cords intercalated with granulosa cells and interspersed with stromal cells (Figs 2B and EV1G). A majority of germ cells showed an immature morphology representing oogonia, while some exhibited larger nuclei with condensed chromatin, indicative of meiotic entry (Fig 2B). Consistent with these findings, ∼ 40% of germ cells expressed oogonia markers (NANOG, POU5F1, TFAP2C, and PDPN), while ∼ 50% expressed ZGLP1, ∼ 30% expressed SYCP3, and ∼ 10% expressed meiotic markers (DMC1, γH2AX, and SYCP1) (Fig 2C and D). At 6 w‐ptp, oocyte development progressed further in cy rOvaries, with a majority of germ cells showing the nuclear morphology in the meiotic prophase I, including that of the pachytene stage with synapsed chromosome pairs (< 10% positive for oogonia markers, ∼ 60% positive for SYCP3, and ∼ 30% positive for DMC1, γH2AX, and SYCP1), but there were no cells expressing the oogenic markers (Figs 2B−D and EV1G). From 9 w‐ptp onwards, there were differentiated primordial follicle‐like complexes with an oocyte expressing ZP3 and TP63 surrounded by a single layer of squamous granulosa cells expressing FOXL2, and by 15 w‐ptp, such complexes became dominant (Fig 2B−D). These findings demonstrate that, upon xenotransplantation into mice, cy fetal oocyte development and the meiotic prophase I proceed apparently normally, although the developmental progression seemed somewhat slower compared to that in vivo. We examined cy rOvaries at 21 w‐ptp and found that although they maintained a distinct structure, only granulosa and stromal cells survived, and essentially, all oocytes were degenerated (Fig 2E), indicating that further follicle growth does not proceed under the current xenotransplantation condition.
Reconstitution of cy fetal oocyte development in vitro
We next explored whether an in vitro culture would promote appropriate cy fetal oocyte development in cy rOvaries. We first examined an air–liquid interface culture that supports the development of both mouse embryonic ovaries and rOvaries (Hikabe et al, 2016; Morohaku et al, 2016). We generated cy rOvaries with ∼ 50,000 ovarian cells at 8 wpf, and cultured them on the Transwell‐COL membrane insert (Fig 3A). Initially, cy rOvaries on the Transwell‐COL membrane showed an integrated structure, but unlike in mouse rOvaries, some cells began to spread out from cy rOvaries from as early as 1 week of in vitro culture (1 w‐ivc), and at 3 w‐ivc, many cells had migrated out, leaving the cy rOvaries smaller and flattened (Figs 3B and EV2A). The cell spreading continued thereafter, and at 6 w‐ivc, the cy rOvaries were much diminished compared to their original sizes (Figs 3B and EV2A). Histological and IF analyses revealed that at 3 w‐ivc, cy rOvaries bore ovarian cords, but such areas were limited with a small number of germ cells, and at 6 w‐ivc, although FOXL2+ granulosa cells formed some clusters, there remained only a few germ cells with degenerating morphology (Fig 3B). We examined whether cy rOvaries can be maintained on a culture membrane coated with various extracellular matrix components, including collagen (type I, III, IV, V, VI), fibronectin, laminins, and their combinations (Fig EV2A). Under some conditions, such as on the membrane coated with collagen type I, III, IV, V, and VI, or iMatrix (laminin 511), cy rOvaries maintained their integrity up to 5 w‐ivc, but thereafter, they began to collapse, that is, some cells migrated out, and under all conditions examined, cy rOvaries were flattened out by 12 w‐ivc (Fig EV2A). These findings indicate that cy rOvaries require a condition distinct from that of mouse rOvaries for appropriate in vitro development.
Figure 3. In vitro culture system for inducing cy ovarian follicles from fetal gonadal cells.
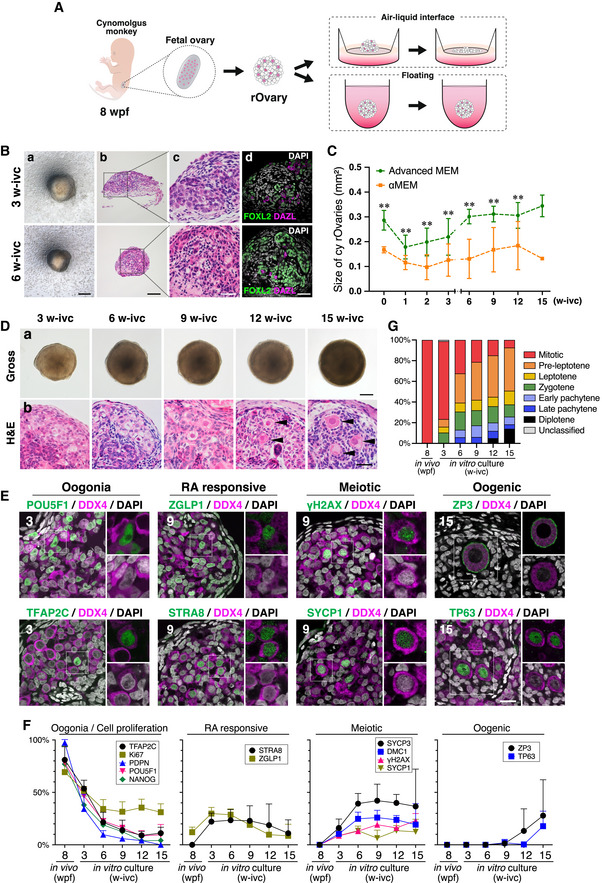
-
AScheme of the in vitro culture system for inducing cy ovarian follicles. The cy rOvaries generated from fetal gonadal cells at 8 wpf were cultured in either an air–liquid interface or a floating condition (see Materials and Methods).
-
BGross appearance (a), H&E staining (b and c), and IF for FOXL2/DAZL/DAPI (d) of cy rOvaries cultured in the air–liquid interface condition at 3 and 6 weeks of in vitro culture (w‐ivc). FOXL2, granulosa‐cell marker; DAZL, germ‐cell marker; DAPI, nucleus. Scale bars = 200 μm (a), 100 μm (b), and 40 μm (c and d).
-
CQuantitative analysis for the size of cy rOvaries cultured in Advanced MEM or αMEM as the basal media of P‐IVD medium. The mean values from more than three biological replicates are shown with SDs except for the rOvary in αMEM at 15 w‐ivc [0/1/2/3/6/9/12/15 w‐ivc (αMEM, n = 26/23/14/23/20/9/4/1; Advanced MEM, n = 14/14/14/14/9/8/7/4)]. **P < 0.01, Welch's T‐test.
-
DGross appearance (a) and H&E staining (b) images of cy rOvaries cultured with the P‐IVD medium at 3/6/9/12/15 w‐ivc. Arrowheads indicate primordial follicle‐like structures. Scale bars = 200 μm (a) and 40 μm (b).
-
EIF analyses of key markers as described in Fig 1E in the cultured cy rOvaries. Germ cells and nuclei were marked with DDX4 (magenta) and DAPI (white), respectively. The upper‐right magnified images of each panel show the expression of key markers (green) co‐stained with DDX4 (magenta). The lower‐right magnified images of each panel show DDX4 expression (magenta) co‐stained with DAPI (white). The numbers written in the upper‐left corner indicate the period of IVC (w‐ivc). Scale bar = 20 μm.
-
FPercentages of cells positive for individual markers among DDX4+ germ cells from the IF of cy rOvaries at 3–15 w‐ivc [3/6/9/12 w‐ivc (n = 3), 15 w‐ivc (n = 3 or 4)]. The average values with SDs are shown.
-
GPercentages of germ cells at each meiotic substage as defined in Fig 1F (see also Materials and Methods). The average percentages of each meiotic substage were determined from at least three cy rOvaries [3/6/9/12 w‐ivc (n = 3), 15 w‐ivc (n = 4)].
Figure EV2. Establishment of a novel in vitro culture system for cy oocyte differentiation.
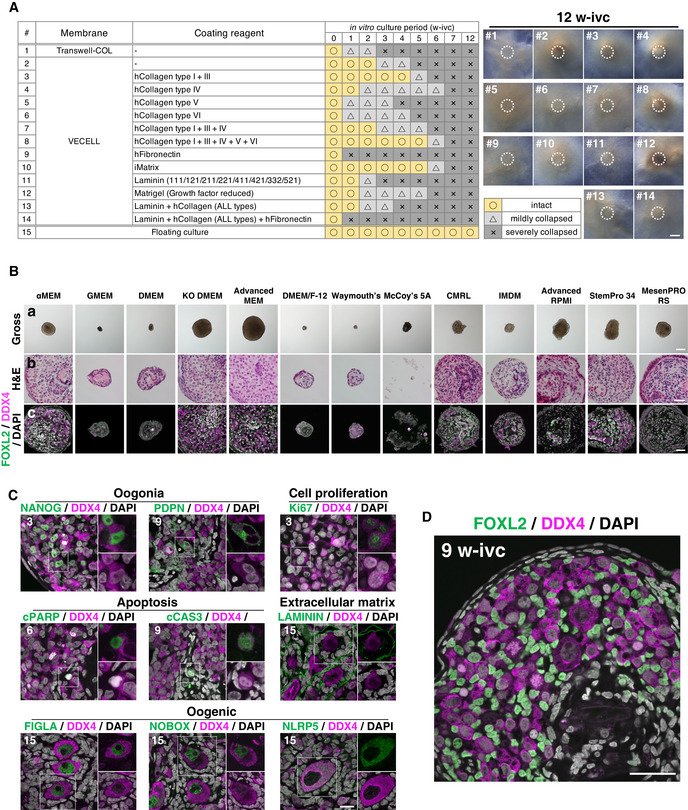
-
A(Left) Summary table of coating conditions tested for cy rOvaries. (Right) Representative images of cy rOvaries cultured on membranes at 12 w‐ivc. Dotted circles show the edge of cy rOvaries at the starting point of air–liquid interface culture. Scale bar = 500 μm.
-
BGross appearances (a), H&E staining (b), and IF for FOXL2/DDX4/DAPI (c) of cy rOvaries cultured in 13 different basal media at 6 w‐ivc. The rOvary cultured in Advanced MEM maintained its size, and DDX4+ germ and FOXL2+ granulosa cells were found within the rOvaries. Scale bars = 500 μm (a) and 40 μm (b and c).
-
CIF analyses of key markers as described in Fig EV1E in the cultured cy rOvaries. Germ cells and nuclei were marked with DDX4 (magenta) and DAPI (white), respectively. The upper‐right magnified images of each panel show the expression of key markers (green) co‐stained with DDX4 (magenta) except for NLRP5. The lower‐right magnified images of each panel show DDX4 expression (magenta) co‐stained with DAPI (white). The numbers written in the upper‐left corner indicate the period of IVC (w‐ivc). Scale bar = 20 μm.
-
DIF analysis for FOXL2 (granulosa cells)/DDX4 (germ cells) stained with DAPI (nucleus) in the cultured cy rOvary at 9 w‐ivc. Scale bar = 40 μm.
We therefore explored whether cy rOvaries can be maintained for the long‐term under a floating condition (Fig 3A). When grown by floating culture in αMEM+10% fetal bovine serum (FBS), a basal condition for mouse rOvary culture (Hikabe et al, 2016, Morohaku et al, 2016), at 6 w‐ivc, cy rOvaries showed an integrated appearance outlined by a few layers of squamous epithelial cells, although their central areas appeared somewhat necrotic, and a relatively large number of DDX4+ germ cells survived with intercalating FOXL2+ granulosa cells (Fig EV2B). We examined 13 different basal media including αMEM (αMEM, GMEM, DMEM, KO DMEM, Advanced MEM, DMEM/F‐12, Waymouth's, McCoy's 5A, CMRL, IMDM, Advanced RPMI, StemPro 34, MesenPRO RS) and found that Advanced MEM gave the best outcome regarding the size and histology of cy rOvaries at 6 w‐ivc (Fig EV2B). Cy rOvaries reduced their size at the outset of culture, possibly due to ongoing compaction of the aggregates, but they gradually regained their size after 1 w‐ivc (Fig 3C). The size of cy rOvaries cultured in Advanced MEM+10% FBS was consistently larger than that in αMEM+10% FBS during the 15‐week culture period (Fig 3C). We therefore analyzed cy fetal oocyte development in cy rOvaries cultured in Advanced MEM+10% FBS under the floating condition at 3‐week intervals (Fig 3D). At 3 w‐ivc, much as at 3 w‐ptp, although cy rOvaries did not show outer/inner cortex stratification, they exhibited anastomosing cord‐like structures with germ cells intercalated with granulosa cells, which were encapsulated by stromal cells (Appendix Fig S1). Most germ cells (∼ 50%) were oogonia expressing NANOG, POU5F1, TFAP2C, and PDPN, while ∼ 30% and ∼ 10% progressed to the RA‐responsive (ZGLP1+/STRA8+) and meiotic (DMC1+/γH2AX+/SYCP1+) stages, respectively (Figs 3E and F, and EV2C). Thereafter, oogonia decreased in number, while meiotic cells increased, albeit with slower kinetics compared to that in vivo and in xenotransplantation; nonetheless, oocytes expressing oogenic markers (ZP3, TP63, FIGLA, NOBOX, and NLRP5) in primordial follicle‐like structures differentiated from 12 w‐ivc onward (Figs 3E and F, and EV2C). The morphology of oogonia, and of RA‐responsive, meiotic, oogenic, and granulosa cells and the expression/subcellular localization of key markers in these cells in cy rOvaries in vitro were similar to those of the corresponding cell types in vivo (Figs 3E and EV2C and D). We examined the progression of the meiotic prophase using the criteria defined in Fig 1F, which revealed slow but steady progression of the meiotic prophase in cy rOvaries in vitro (Fig 3G). We noted that a relatively large fraction of germ cells remained in the pre‐leptotene stage from 6 w‐ivc to 15 w‐ivc (Fig 3G), suggesting that under the current condition, the pre‐leptotene to leptotene transition may be a rate‐limiting step. These findings demonstrate that cy fetal oocyte development proceeds in vitro, with mitotic oogonia differentiating into oocytes that complete the first meiotic prophase in the primordial follicle‐like structures over a period of 12 weeks.
Transcriptome dynamics for cy fetal ovary development in vivo, in xenotransplantation, and in vitro
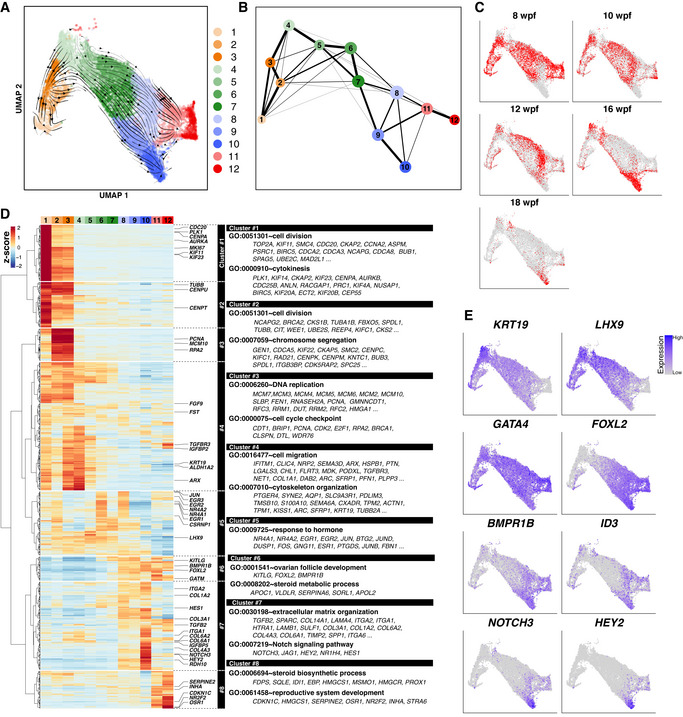
To define cy fetal ovary development in vivo as well as in xenotransplantation and under in vitro culture in a comprehensive manner, we performed single‐cell RNA‐sequence (scRNA‐seq) analysis of cy fetal ovaries and cy rOvaries (Datasets EV1 and EV2). First, we processed a total of 35,141 single cells isolated from cy fetal ovaries at 8, 10, 12, 16, and 18 wpf for single‐cell cDNA preparation using the 10X Chromium platform, and with sequencing, 29,127 cells passed key quality filters [number of genes detected (nGene), total unique molecular identifier (UMI) count, and % mitochondrial gene count] and processing with Scrublet [doublets/multiplets removal algorithm (Wolock et al, 2019)] for subsequent analysis (Dataset EV1). Batch‐effect removal from the principal component analysis (PCA) subspace by the fast mutual nearest neighbor (fastMNN) algorithm followed by graph‐based clustering using the Louvain algorithm classified these cells into five cell types: DDX4 + germ cells (2,283 cells), WT1 + pre‐granulosa cells (11,986 cells), TCF21 + stromal cells (14,028 cells), PECAM1 + endothelial cells (673 cells), and TYROBP + blood cells (157 cells) (Fig 4A−C). In all stages, larger numbers of genes were detected in germ cells than in somatic cells (Fig EV3A). We isolated germ cells and reanalyzed them with PCA, fastMNN, and Louvain clustering, which classified them into 11 clusters after manual removal of putative doublets/multiplets. Based on the expression of key markers, we annotated them as mitotic 1/2/3, pre‐leptotene 1/2/3, leptotene, zygotene, pachytene 1/2, and some of the pachytene 2 cells were manually classified as diplotene (Figs 4D and EV3B). The unclassified cells most likely represented apoptotic pre‐leptotene/leptotene cells, as they showed significantly lower levels of nUMI and nGene (Fig EV3C), and exhibited gene expression profiles highly correlated with those of pre‐leptotene/leptotene cells (see below for Fig 4I). The distributions of the cells annotated by scRNA‐seq across developmental stages were essentially consistent with those annotated by the IF analysis, except that there were only a few diplotene cells (Figs 4D and EV3B), which would at least in part be due to an incompatibility of diplotene cells with the 10X platform because of their large size. The numbers of genes detected and total UMI counts were relatively constant from mitotic to zygotene cells (on average, ∼ 7,000 and 50,000, respectively) but were somewhat decreased and more variable in pachytene cells (Fig EV3C).
Figure 4. Single‐cell RNA‐seq analyses of cy ovarian cells in vivo and in rOvaries.
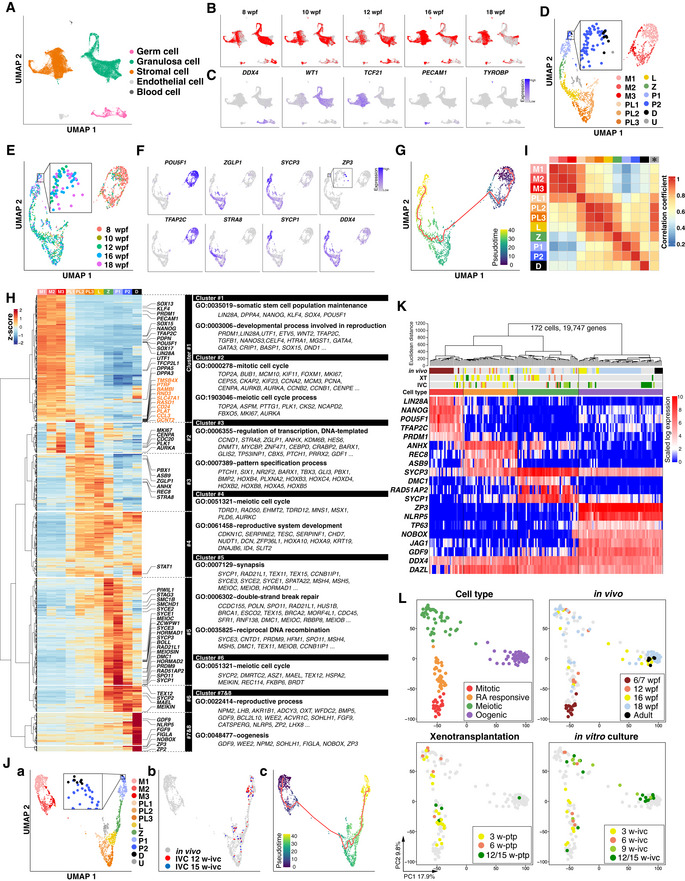
-
A–JAnalyses for cy germ cells by 10X scRNA‐seq. (A) Uniform manifold approximation and projection (UMAP) plot of cy fetal ovarian cells in vivo, colored by five computationally assigned major clusters based on the expression of cell‐type‐specific markers. The color‐coding is as indicated. (B) UMAP plot shown in Fig 4A, highlighting the cells for each fetal stage. (C) Feature plots for the expression of key marker genes for cy in vivo germ cells (DDX4), granulosa cells (WT1), stromal cells (TCF21), endothelial cells (PECAM1), and blood cells (TYROBP) overlaid on the UMAP plot in Fig 4A. (D) UMAP plot of cy in vivo germ cells shown with the clustering of meiotic substages. The color‐coding is as indicated. M, mitotic; PL, pre‐leptotene; L, leptotene; Z, zygotene; P, pachytene; D, diplotene; U, unclassified. (E) UMAP plot of cy in vivo germ cells colored by the fetal stage. The color‐coding is as indicated. (F) Feature plots for oogonia (POU5F1 and TFAP2C), RA‐responsive (ZGLP1 and STRA8), meiotic (SYCP3 and SYCP1), oogenic (ZP3), and germ‐cell (DDX4) markers during cy in vivo germ‐cell development. (G) UMAP plot of cy in vivo germ cells with the trajectory graph (red lines). Cells were colored by pseudotime. (H) (Left) Heatmap of the standardized expression of highly variable genes (HVGs, 1,481 genes) among in vivo cy germ‐cell clusters ordered by unsupervised hierarchical clustering (UHC); eight gene clusters were defined according to the UHC dendrogram. Key genes for female germ‐cell development are shown in black, and 10 genes transiently upregulated in the PL cluster are shown in orange. (Right) Representative genes and key gene ontology (GO) enrichments are shown. (I) Heatmap of the Pearson's correlation coefficients of the average expression levels of the 1,481 HVGs among the meiotic substages in cy in vivo germ cells. The asterisk indicates an unclassified cluster. The color‐coding is as indicated. (J) (a) UMAP plot for germ cells from cy in vivo fetal ovaries and in vitro cultured cy rOvaries, shown with the meiotic substages. The color‐coding is as indicated. (b) UMAP plot highlighting cells from in vitro cultured cy rOvaries at 12 and 15 w‐ivc. (c) UMAP plot with the trajectory graph (red lines). Cells were colored by pseudotime.
-
K, LAnalyses for cy germ cells by SC3‐seq. (K) UHC for cy germ cells in vivo and in rOvaries with all expressed genes (19,747 genes) and a heatmap of the expression levels of selected markers that differentially label mitotic, RA‐responsive, meiotic, and oogenic cells. Color bars under the dendrogram show the in vivo stage (1st bar), the period of xenotransplantation (2nd bar), the period of IVC (3rd bar), and the cell type (4th bar). The heatmap color‐coding is as indicated. The color‐coding of bars is as indicated in Fig 4L. (L) Principal component analysis (PCA) of cy germ cells in vivo and in rOvaries performed by SC3‐seq. The cells were plotted in the two‐dimensional plane defined by PC1 and PC2 values. The color‐coding is as indicated.
Figure EV3. 10X scRNA‐seq analysis of cy female oocyte development in vivo .
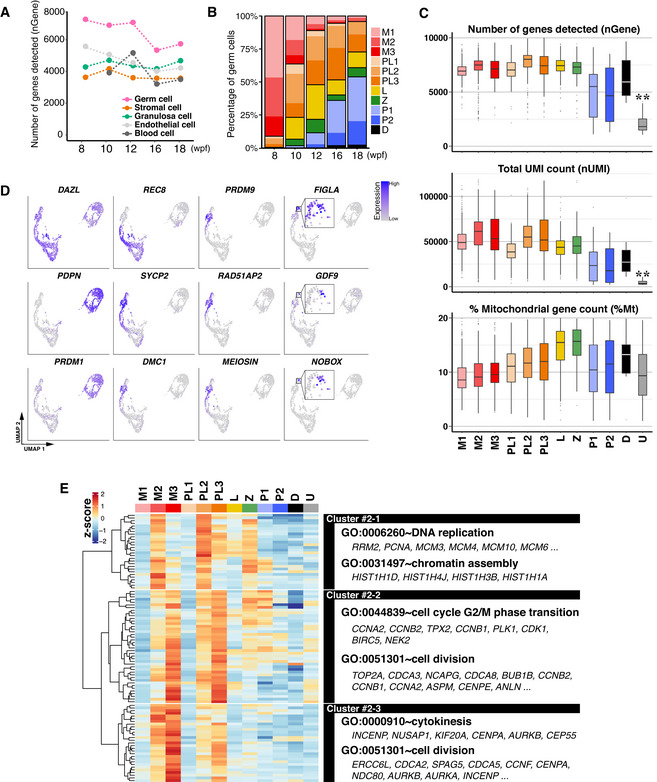
-
AThe average number of genes detected in five major cell types in cy in vivo fetal ovaries at 8–18 wpf.
-
BPercentages of germ cells at the indicated meiotic prophase substages defined in Fig 4D. The color‐coding is as indicated. M, mitotic; PL, pre‐leptotene; L, leptotene; Z, zygotene; P, pachytene; D, diplotene.
-
CThe detected gene number, UMI count, and percentage of mitochondrial genes in each germ‐cell cluster. The number of genes and UMI counts were significantly low in the “unclassified (U)” compared to any other germ‐cell cluster. **P < 0.01, Tukey–Kramer tests. In the boxplots, the central bands represent the median values; the lower/upper hinges represent the 25th/75th percentiles, respectively; the upper limits of the whiskers represent the largest values no further than 1.5 IQR (interquartile range) from the upper hinges; the lower limits of the whiskers represent the smallest values no further than 1.5 IQR from the lower hinges; the dots represent the outliers. The numbers of the cells used are 431/271/135/59/252/253/261/101/183/74/8/90 from 7 fetuses for M1/M2/M3/PL1/PL2/PL3/L/Z/P1/P2/D/U, respectively.
-
DFeature plots of key marker genes for oocyte development on the UMAP plot shown in Fig 4D.
- E
Mitotic 1/2/3 cells expressed key oogonia markers, including POU5F1, TFAP2C, PDPN, and PRDM1, and consisted predominantly of cells at 8 wpf, but included some cells from all developmental stages (Figs 4D and E, and F, and EV3B and D). Mitotic 1/2/3 cells were considered to represent cells in the G1, S, G2/M phases of the cell cycle, respectively (Fig EV3E). Pre‐leptotene 1 cells weakly expressed POU5F1 and ZGLP1, but did not show typical meiotic markers such as DMC1, PRDM9, and SYCP1, and consisted of cells from 8, 10, 12, and 16 wpf. They were in the G1 phase of the cell cycle (Figs 4D and F, and EV3D and E). Pre‐leptotene 2/3 cells expressed ZGLP1, REC8, and SYCP3, but were negative/very weak for DMC1, PRDM9, and SYCP1, consisted of cells from all developmental stages, and were in the S and G2 phases of the cell cycle, respectively (Figs 4D and F, and EV3D and E). Thus, pre‐leptotene 2 cells are those undergoing meiotic DNA replication. Leptotene and zygotene cells consisted of cells mainly after 12 wpf, and expressed REC8, SYCP2, and SYCP3 at a higher level than pre‐leptotene cells, with zygotene cells upregulating DMC1, PRDM9, and SYCP1 (Figs 4D and F, and EV3D). Pachytene 1/2 cells also consisted of cells mainly after 12 wpf, repressed ZGLP1 and REC8, and expressed SYCP1/2/3, DMC1, and PRDM9, with pachytene 2 cells initiating the downregulation of these genes and upregulation of FIGLA (Figs 4D and F, and EV3D). Diplotene cells (ZP3 + /GDF9 + /NOBOX + cells) consisted of cells at 16 and 18 wpf, downregulated meiotic markers, and expressed key genes for oocyte development, such as FIGLA and NLRP5 (Figs 4D and F, and EV3D). A pseudotime analysis for developmental trajectory by Monocle 3 (Cao et al, 2019) and a cell‐cycle phase prediction based on a widely used method (Tirosh et al, 2016) gave an outcome consistent with the above results (Fig 4G and Appendix Fig S2A−C).
We identified highly variable genes (HVGs) among the 11 clusters [top 2,000 HVGs minus genes with low‐expression levels (cluster average expression log (ssUMI+1) ≤ 0.5): 1,481 genes] (Dataset EV3). Unsupervised hierarchical clustering (UHC) classified such genes into 8 clusters (Fig 4H): the cluster 1 genes were expressed highly in mitotic 1/2/3 cells and downregulated acutely after the pre‐leptotene 1 stage, and were enriched in genes with gene ontology (GO) functional terms for “somatic stem cell population maintenance” and “developmental process involved in reproduction.” The cluster 2 genes showed an expression profile associated with cell‐cycle progression (expressed in mitotic 2/3, pre‐leptotene 2/3) and were indeed enriched for “mitotic cell cycle” and “meiotic cell cycle process” (Figs 4H and EV3E). The cluster 3 genes were expressed from pre‐leptotene 1 to zygotene and were enriched for “regulation of transcription, DNA‐templated,” and interestingly, “pattern specification process.” The cluster 4 genes were gradually upregulated from pre‐leptotene 1 onwards and were enriched for “meiotic cell cycle” and “reproductive system development.” The cluster 3 and 4 genes, particularly those expressed in pre‐leptotene 1/2, would include the oogenic fate determinant. The cluster 5 and 6 genes were expressed from leptotene to pachytene 2 (cluster 5) or diplotene (cluster 6), and included key genes for meiotic recombination [“synapsis/double‐strand break repair/reciprocal DNA recombination/meiotic cell cycle”]. The cluster 7 and 8 genes were strongly upregulated in diplotene and were enriched for “reproductive process” and “oogenesis.” The correlation analysis using the same genes revealed that the 11 clusters can be classified into 4 groups with higher correlations [mitotic 1‐to‐pre‐leptotene 1, pre‐leptotene 1‐to‐zygotene (corresponding to RA‐responsive), zygotene‐to‐pachytene 2 (corresponding to meiotic), pachytene 2‐to‐diplotene (corresponding to oogenic)], with 3 clusters (pre‐leptotene 1, zygotene, pachytene 2) playing a transitional role between the groups (Fig 4I), providing high‐resolution transcriptome categorization for primate fetal oocyte development. We confirmed that the pseudo‐temporal expression patterns of these HVGs as well as those of the HVGs defined based on the Moran's I statistic using the Monocle 3 package gave a consistent outcome (Appendix Fig S3A and B).
We generated 937 quality‐filtered single‐cell transcriptomes from cy rOvaries at 12 and 15 w‐ivc with 10X Chromium (73 germ cells, 702 (pre‐)granulosa cells, and 118 stromal cells) and compared them with those of cy fetal ovarian cells in vivo. PCA and fastMNN followed by Louvain clustering revealed that cy germ cells in cy rOvaries in vitro exhibited transcriptomes concordant with those of cy oocytes ranging from the pre‐leptotene to pachytene stage (Fig 4J). To gain further insights into the transcriptome dynamics associated with cy fetal oocyte development in cy rOvaries, we manually picked up single cells with germ‐cell appearance dissociated from cy rOvaries at 3/6/9/12/15 w‐ivc and w‐ptp as well as from cy ovaries at 12/16/18 wpf and in adult individuals, and performed scRNA‐seq analysis with single‐cell mRNA 3‐prime end sequencing (SC3‐seq), a highly sensitive scRNA‐seq method that allows a quantitative comparison of single‐cell gene expression profiles with a few (∼ 4–8) cells (Nakamura et al, 2015) (Fig 4K and L; Appendix Fig S4A, Dataset EV2) (the cells analyzed for the xenotransplantation experiment were small in number, in part due to a difficulty in dissociating transplanted cy rOvaries into single cells). UHC and PCA classified 172 quality‐filtered DDX4 + single cells (including 19 germ cells at 6/7 wpf in (Okamoto et al, 2021)) into four major groups corresponding to mitotic, RA‐responsive, meiotic, and oogenic cells, and in vivo, xenotransplanted, and in vitro cells were included in all groups, with intermingling transcriptome profiles within the groups (Fig 4K and L). The pseudo‐temporal expression patterns of the HVGs in Fig 4H during in vivo and in vitro oocyte development were consistent between the 10X scRNA‐seq and SC3‐seq datasets (Appendix Fig S3 and S4B). Accordingly, we found that the correlation coefficients between in vivo and in vitro RA‐responsive/meiotic/oogenic cells were 0.968/0.968/0.982, respectively (Appendix Fig S5A), and the differentially expressed genes (DEGs) between in vivo and in vitro RA‐responsive/meiotic/oogenic cells were few in number (Appendix Fig S5A, Dataset EV4). On the contrary, the DEGs included some key genes for oocyte development, including ZGLP1, REC8 (RA‐responsive), SOHLH1, NOBOX (meiotic), NLRP4, and ZP4 (oogenic) (Appendix Fig S5A, Dataset EV4), and their pseudo‐temporal expression profiles suggested that while the differential expression of SOHLH1, NOBOX (meiotic), NLRP4, and ZP4 (oogenic) may result from a difference in meiotic/oogenic stages in the compared cell populations, the differential expression of ZGLP1and REC8 (RA‐responsive) may reflect a significant difference (Appendix Fig S5B), which could explain why the pre‐leptotene to leptotene transition may be rate‐limiting under the current culture condition (Fig 3G). Collectively, these results led us to conclude that cy fetal germ cells undergo appropriate overall transcriptome maturation in cy rOvaries in vitro, while the upregulation of key genes for the pre‐leptotene to leptotene transition may not be optimal under the current condition.
We also examined the development of WT1 + cy pre‐granulosa cells in vivo and in vitro. Using the same analysis as for germ cells, cy (pre‐)granulosa cells in vivo were classified into 12 clusters after manual removal of putative doublets/multiplets (Fig EV4A). Analysis of the cluster distributions across developmental stages showed that most clusters (clusters 1, 2, 3, 4, 5, 6, 7, 9, 11, and 12) were present as early as 8 wpf, while clusters 8 and 10 emerged from 10 and 16 wpf onwards, respectively (Fig EV4A−C). We identified genes with highly variable expression among the 12 clusters [top 1,500 HVGs minus genes with low‐expression levels (cluster average expression log (ssUMI+1) ≤ 0.5): 788 genes] (Dataset EV5). UHC using these genes revealed that cells in clusters 2 and 3 were pre‐granulosa cells in the S phase of the cell cycle, while those in cluster 1 were in the G2/M phase; moreover, the cells in cluster 1 were abundant in the early stages (8/10/12 wpf) and, like the cells in cluster 4, were enriched with genes for “cell migration” and “cytoskeleton organization” (Fig EV4A−D). Cells in clusters 11 and 12 were also abundant in the early stages (8/10/12 wpf) and expressed genes for “steroid biosynthetic process” and “reproductive system development,” and thereby represented a unique pre‐granulosa cell subtype (Fig EV4A−D). The RNA velocity calculation (Bergen et al, 2020) and partition‐based graph abstraction (PAGA) (Wolf et al, 2019) suggested that pre‐granulosa cell differentiation proceeded from cluster 4 to 5 to 7 to 9 to 10 or from 4 to 5 to 6 to 8 to 10 (Fig EV4A and B), and cells in cluster 10, which emerged from 16 wpf onwards, were enriched with genes for “extracellular matrix organization” and “Notch signaling pathway,” and would therefore represent granulosa cells composing primordial follicles (Fig EV4D and E) (Li et al, 2017). Combined analysis of the transcriptomes of WT1 + pre‐granulosa cells in cy rOvaries in vitro revealed that the WT1 + cells at 12 and 15 w‐ivc formed two major clusters: one smaller cluster (cluster d) including a subset of granulosa cells at 16 wpf and one larger cluster (cluster l) close to/merged with a cluster of follicular granulosa cells at 16 and 18 wpf (cluster 10 in Fig EV4A), indicating that pre‐granulosa cells at 8 wpf underwent appropriate differentiation, including maturation into follicular granulosa cells, although the identity of cluster d, whose HVGs were enriched with genes for “extracellular matrix organization,” “angiogenesis,” and “cell–cell adhesion,” requires further investigations (Appendix Fig S6A−C). We noted, however, that follicular granulosa cells in vitro (cluster l) showed a signature for activated granulosa cells, including ID2/3/4 upregulation (i.e., activation of BMP signaling) and HEY2 upregulation and HEY1/HEYL downregulation (i.e., regulation of JAG1‐NOTCH2/3 signaling) (Appendix Fig S6D) (Vanorny & Mayo, 2017; Zhang et al, 2018), suggesting a possibility that granulosa‐cell development in culture proceeds to an activated primary follicle stage.
Figure EV4. 10X scRNA‐seq analysis for cy granulosa cells in vivo and in rOvaries.
-
A, BUMAP plot with RNA velocity (A) and PAGA graph (B) of cy in vivo granulosa cells as defined in Fig 4A. Granulosa cells were divided into 12 subclusters by Louvain clustering. The color‐coding is as indicated.
-
CUMAP plot shown in Fig EV4A, highlighting the cells for each fetal stage.
-
D(Left) Heatmap of the standardized expression of HVGs (712 genes) among in vivo cy granulosa‐cell subclusters ordered by UHC; eight gene clusters were defined according to the UHC dendrogram. (Right) Representative genes and key GO enrichments are shown.
-
EFeature plots of key marker genes for cy granulosa‐cell development on the UMAP plot shown in Fig EV4A.
Reconstitution of human fetal oocyte development in vitro
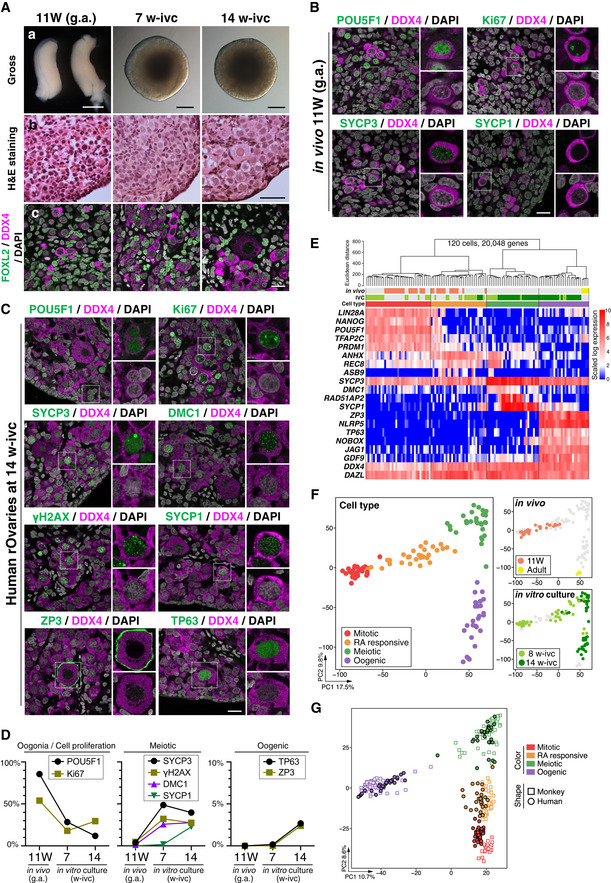
Having vestablished a condition for cy fetal oocyte development in vitro, we explored whether human fetal oocyte development can also be reconstituted in vitro. We were able to obtain two aborted human female fetuses at 11 weeks of gestation (11 W), and we isolated their gonads and processed them for histological analysis, human rOvary formation, and single‐cell transcriptome analysis. In addition, we isolated primordial follicles from a human adult ovary and processed their oocytes for single‐cell transcriptome analysis. All human samples were obtained upon appropriate informed consent (see Materials and Methods). Consistent with previous reports (Li et al, 2017), human fetal ovaries at 11 W were abundant with oogonia bearing a round nucleus with pale hematoxylin staining and a prominent nucleolus, but were also populated with cells with a larger nucleus with a more condensed chromatin appearance (Fig 5A). Accordingly, ∼ 85% of DDX4+ germ cells were POU5F1+ and a small fraction of them (∼ 3%) showed expression of early meiotic markers such as SYCP3, γH2AX, and DMC1, but no cells expressed SYCP1 (Fig 5B and D), demonstrating that most germ cells in human ovaries at 11 W are oogonia.
Figure 5. Derivation of human ovarian follicles from oogonia in vitro .
-
AGross appearance (a), H&E staining (b), and IF (c) for FOXL2/DDX4/DAPI of the human in vivo female gonads (11 W, left column) and the cultured rOvaries at 7 and 14 w‐ivc. FOXL2, granulosa‐cell marker; DDX4, germ‐cell marker; DAPI, nucleus. Scale bars = 1 mm (a, left), 200 μm (a, middle/right), 50 μm (b), and 20 μm (c). g.a., gestational age.
-
B, CIF for oogonia (POU5F1), cell proliferation (Ki67), meiotic (SYCP3, DMC1, γH2AX, and SYCP1), and oogenic (ZP3 and TP63) markers for human in vivo female gonad (11 W, B) and human in vitro cultured rOvaries at 14 w‐ivc (C). Germ cells and nuclei were marked with DDX4 (magenta) and DAPI (white), respectively. The upper‐right magnified images of each panel show the expression of key markers (green) co‐stained with DDX4 (magenta). The lower‐right magnified images of each panel show DDX4 expression (magenta) co‐stained with DAPI (white). Scale bars = 20 μm.
-
DPercentages of cells positive for individual markers among DDX4+ germ cells from the IF of human in vivo female gonads (11 W) and human cultured rOvaries at 7 and 14 w‐ivc. The mean values from two biological replicates are shown.
-
E, FAnalyses for human germ cells by SC3‐seq. (E) UHC for human germ cells in vivo and in rOvaries with all expressed genes (20,048 genes) and a heatmap of the expression levels of selected marker genes. Color bars under the dendrogram show the in vivo stage (1st bar), the period of IVC (2nd bar), and the cell type (3rd bar). The heatmap color‐coding is as indicated. The color coding in bars is as indicated in Fig 5F. (F) PCA of human germ cells in vivo and in rOvaries performed by SC3‐seq. The cells are plotted in the two‐dimensional plane defined by PC1 and PC2 values. The color‐coding is as indicated.
-
GPCA plot of all cy and human SC3‐seq data merged by canonical correlation analysis (CCA). Cell types were defined after CCA followed by Louvain clustering. The color and shape coding are as indicated.
Under the air–liquid interface condition, cells migrated out of human rOvaries (∼ 50,000 ovarian cells) and human rOvaries became flattened out and collapsed over an extended culture period; this was similar to the findings in cy rOvaries, but dissimilar to mouse rOvaries (Appendix Fig S7A). We therefore cultured human rOvaries under the floating condition defined for cy rOvaries and examined their properties at 7/8 and 14 w‐ivc. At 7 w‐ivc, human rOvaries showed an integrated structure, although like cy rOvaries, the central areas exhibited a necrotic appearance (Fig 5A). Histological sections revealed that the cortical areas were full of germ cells bearing oogonia‐like or early meiotic oocyte‐like morphology (Fig 5A). In good agreement with this result, ∼ 30% of DDX4+ germ cells were POU5F1+, ∼ 50% were SYCP3+, and ∼ 30% were γH2AX+/DMC1+, while only ∼ 2% were SYCP1+ (Fig 5C and D). At 14 w‐ivc, human rOvaries remained integrated and their cortexes were abundant with germ cells bearing a large nucleus with condensed, thread‐like chromatin and, remarkably, a number of primordial follicle‐like structures (Fig 5A). Consistent with these observations, the ratio of DDX4+/POU5F1+ cells decreased to ∼ 10%, while ∼ 30% became SYCP1+ and ∼ 3% were ZP3+/TP63+ (Fig 5C and D). The morphology of meiotic and primordial follicle‐like cells and the expression/subcellular localization of key markers in these cells were similar to those of corresponding in vivo cell types in cynomolgus monkeys and humans (Figs 1E and 5C; Baker, 1963).
Single‐cell transcriptome analysis by SC3‐seq of 120 quality‐filtered single cells (human fetal ovaries at 11 W: 33 cells; human adult ovaries: 4 cells; human rOvaries at 8 w‐ivc: 39 cells; human rOvaries at 14 w‐ivc: 44 cells) classified them into four cell types: mitotic, RA‐responsive, meiotic, and oogenic cells (Fig 5E and F, Dataset EV2). Consistent with the histological and IF analyses, most single cells picked up from dissociated human fetal ovaries at 11 W were mitotic oogonia expressing NANOG, POU5F1, TFAP2C, and PRDM1 (19/33) or RA‐responsive cells expressing ANHX, REC8, and ASB9 (14/33), whereas those from human rOvaries at 8 w‐ivc contained all four cell types (mitotic: 16/39; RA‐responsive: 13/39; meiotic: 7/39; oogenic: 3/39) and those at 14 w‐ivc were predominantly meiotic cells expressing DMC1, RAD51AP2, and SYCP1 or oogenic cells with ZP3, NLRP5, TP63, and NOBOX (mitotic: 0/44; RA‐responsive: 3/44; meiotic: 21/44; oogenic: 20/44; Fig 5E and F). Notably, the oogenic cells isolated from human rOvaries were clustered closely with adult oocytes in the primordial follicles (Fig 5E and F), indicating a proper progression of human oocyte development in human rOvaries in vitro. To substantiate this point further and complement the lack of human fetal ovary samples at later stages, we merged SC3‐seq data of human and cy germ cells by canonical correlation analysis (CCA) (Stuart et al, 2019) and performed PCA, which revealed a highly parallel transcriptomic progression of human oocytes in human rOvaries in vitro, and in cy oocytes in vivo and cy rOvaries in vitro (Fig 5G). In addition, we examined the pseudo‐temporal expression patterns of the human orthologues of cy fetal oocyte HVGs (Fig 4H) during human fetal oocyte development in vivo and in vitro, and found consistent gene expression changes between cy/h fetal oocyte development and between in vivo/in vitro human fetal oocyte development (Appendix Fig S4B and S7B). Taken together, these results indicated that human oocyte development can also be reconstituted in vitro, at least up to the oocytes that complete the meiotic prophase I in the primordial follicles.
Primate‐specific program for fetal oocyte development
In mammals, germ‐cell development proceeds with conserved cellular events, which include PGC specification from pluripotent embryonic precursors, epigenetic reprogramming, germ‐cell sex determination, and epigenetic programming coupled with (in females) or followed by (in males) meiotic divisions to generate sexually dimorphic haploid gametes. On the contrary, the precise cellular dynamics and molecular mechanisms underlying such conserved events appear to vary widely among species (Saitou & Hayashi, 2021). To gain insight into the primate‐specific program associated with fetal oocyte development, we went on to compare relevant transcriptome datasets among mice, monkeys, and humans. We merged our cynomolgus monkey and human datasets with four public datasets [one for humans (Chitiashvili et al, 2020) and three for mice (Niu & Spradling, 2020; Zhao et al, 2020; Ge et al, 2021)] using CCA, performed Louvain clustering, and defined the fetal oocyte developmental stage of the 15 clusters across the three species based on the expression of the relevant genes (mitotic 1–4, pre‐leptotene 1–3, leptotene 1–3, zygotene 1/2, pachytene 1/2, diplotene) (Fig 6A; Dataset EV1). The distributions of the cells with defined developmental stages in each developmental time point were in accord with previous observations (Fig 6B and C; Appendix Fig S8) (Chitiashvili et al, 2020) (Niu & Spradling, 2020, Zhao et al, 2020, Ge et al, 2021).
Figure 6. Cross‐species comparison analysis of fetal female germ‐cell development in vivo in humans, monkeys, and mice.
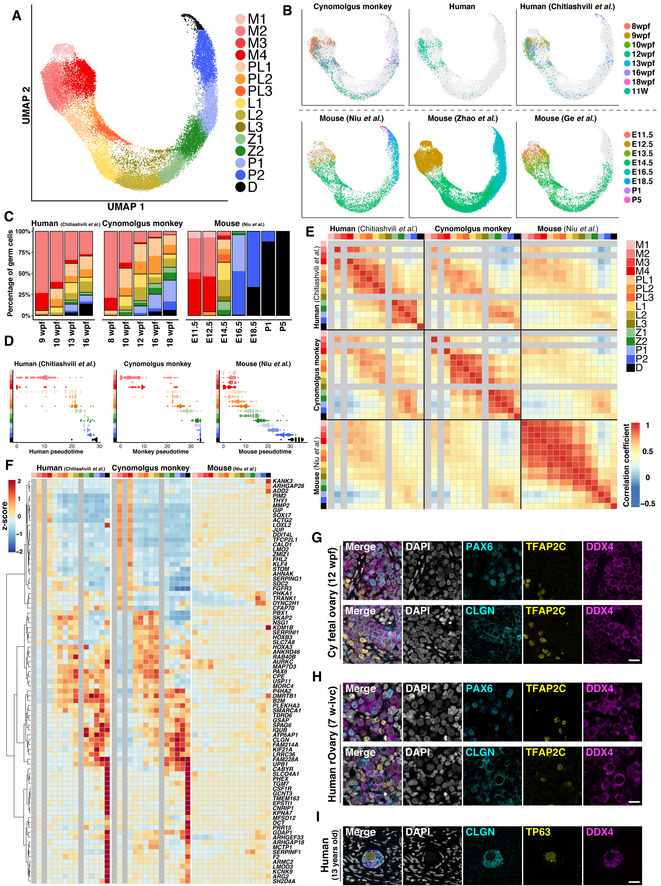
-
A–FAnalyses performed by 10X scRNA‐seq. (A) UMAP plot of germ cells from six 10X scRNA‐seq datasets (two for humans [this study and (Chitiashvili et al, 2020)], one for monkeys [this study], and three for mice [(Niu & Spradling, 2020, Zhao et al, 2020, Ge et al, 2021)]) integrated by CCA. Meiotic substages were defined after CCA followed by Louvain clustering. Colors indicate the computationally assigned meiotic substages defined by the expression levels of key meiotic markers conserved in all three species (see Materials and Methods). The color‐coding is as indicated. M, mitotic; PL, pre‐leptotene; L, leptotene; Z, zygotene; P, pachytene; D, diplotene. (B) The UMAP plot highlighting cells from each dataset. The color‐coding for each developmental stage is as indicated. (C) Percentages of germ cells belonging to each meiotic substage. The color‐coding for the germ‐cell stage is as indicated in Fig 6A. (D) Distribution of germ cells along the individually calculated species‐specific pseudotime trajectories (Monocle 3). The color‐coding for the germ‐cell stage is as indicated in Fig 6A. (E) Heatmap of the Pearson's correlation coefficients of the average expression levels of 237 HVGs (1–1–1 orthologues) among the meiotic substages in humans, monkeys, and mice (see Materials and Methods). The color‐coding for the germ‐cell stage is as indicated. (F) Heatmap of the standardized expression levels of selected genes using 10X scRNA‐seq data in all three species. Genes showing specific expression changes in humans and monkeys, but not in mice, during female germ‐cell development were manually selected from the list of DEGs acquired from the SC3‐seq analysis for cy/human in vitro cultured rOvaries (Dataset EV6). The heatmap color‐coding is as indicated. The color‐coding for the germ‐cell cluster is as indicated in Fig 6E.
-
G, HIf analyses of PAX6 and CLGN with TFAP2C (oogonia marker) and DDX4 (germ‐cell marker) in cy in vivo fetal ovaries at 12 wpf (G) and human cultured rOvaries at 7 w‐ivc (H). Nuclear DAPI staining is shown in white. Scale bars = 20 μm.
-
IIf analyses of CLGN with TP63 (oogenic marker) and DDX4 (germ‐cell marker) in a 13‐year‐old human oocyte of a primordial follicle. Nuclear DAPI staining is shown in white. Scale bar = 20 μm.
Pseudotime analysis by Monocle 3 revealed that in humans and monkeys, the mitotic to pre‐leptotene transition, that is, the entry into meiosis, produced relatively large transcriptomic alterations, whereas in all mouse datasets, such transition involved minor change, but the subsequent transitions gave rise to major alterations (Fig 6D; Appendix Fig S9A). We identified HVGs during fetal oocyte development in each species, among which 237 genes were orthologous and common to all three species. The correlation analysis of their expression in each developmental stage revealed a major distinction between the mitotic and pre‐leptotene cells in humans and monkeys, but not in mice (Fig 6E; Appendix Fig S9B). Furthermore, the expression properties of each cluster were relatively well conserved between humans and monkeys but were divergent in mice (Fig 6E). A previous study has shown that during the meiotic prophase for spermatogenesis in humans, monkeys, and mice, as meiosis progresses, the contribution to the overall expression of one‐to‐one‐to‐one (1–1–1) orthologous genes declines, whereas that of species‐specific genes increases in humans and mice (no clear conclusion was reached for monkeys due to the low level of gene annotation), arguing for species‐specific specialization for male meiotic prophase I (Lau et al, 2020). We performed the same analysis for female meiotic prophase I. In clear contrast to the findings in males, as meiosis progressed in females, the contribution to the overall expression of 1–1–1 orthologues gradually increased, whereas that of the species‐specific genes was consistently low (Appendix Fig S10). Thus, during oogenesis, in humans and monkeys, the mitotic‐to‐meiotic transition involves more drastic transcriptomic changes than in mice, and overall transcriptomic divergence among the three species derives from the distinct usage of orthologous genes.
We went on to identify primate‐specific genes that exhibit dynamic changes during fetal oocyte development. Based on the SC3‐seq analysis in Fig 5G, we identified differentially expressed 1–1 orthologues between the two successive cell types [log2 (normalized read counts +1) > 3, fold‐change > 2], extracted the common genes in humans and monkeys (397 genes), and examined their expression changes in the 10X datasets in fetal oocyte developmental stages as defined in Fig 6A (Dataset EV6). The expression profiles of 130 of the 397 genes were well conserved among the three species, and a notable trend was observed: Genes expressed in mitotic cells continued to be expressed in pre‐leptotene/leptotene cells, and those expressed in meiotic cells showed upregulation in relatively late stages in mice (Fig EV5A; Appendix Fig S11A, Dataset EV6). These genes included well‐known genes involved in fetal oocyte development, including PRDM1, TFAP2C, NANOS3 (mitotic), HORMAD1, REC8, STAG3 (leptotene∼), SYCP1, MEIOC, SPO11 (zygotene∼), NPM2, ZP2, NOBOX (pachytene 3/diplotene) (Fig EV5A; Appendix Fig S11A, Dataset EV6). On the contrary, 83 genes showed specific expression changes in humans and monkeys but not in mice, and these included, among other genes, THY1, SOX17, TFCP2L1 (mitotic), PBX1, PAX6, MORC4 (pre‐leptotene/leptotene), DMRTB1, CLGN, GSAP (leptotene∼), CSF1R, TMEM163, and LMOD3 (pachytene 2/diplotene) (Figs 6F and EV5B; Appendix Fig S11B, Dataset EV6). Among these, we verified the expression of PAX6, a homeobox TF involved in key cell‐fate specification processes including eye development (Walther & Gruss, 1991; Jordan et al, 1992), CLGN, a chaperone that is specifically expressed in the endoplasmic reticulum (ER) of spermatogenic cells and plays a key role in sperm binding to ZP (Ikawa et al, 1997), and LMOD3, an actin filament‐nucleating protein expressed in skeletal and cardiac muscle (Yuen et al, 2014). IF analysis revealed that PAX6 showed specific and strong expression in TFAP2C− germ cells at an early stage of the meiotic prophase in cynomolgus monkeys and human rOvaries (Fig 6G and H); CLGN was expressed and localized in the cytoplasm, most likely in the ER, in TFAP2C− germ cells in cynomolgus monkeys and human rOvaries, and in human juvenile primordial follicles (Fig 6G−I); and LMOD3 exhibited specific expression in primordial follicles in both cynomolgus monkeys and humans, with meshwork‐like localization in the cytoplasm (Fig EV5C). Interestingly, LMOD3 showed higher enrichment in the cytoplasm facing toward the ovarian medulla in primordial follicles in human juveniles (Fig EV5C).
Figure EV5. Cross‐species comparison analysis of oocyte development in humans, monkeys, and mice.
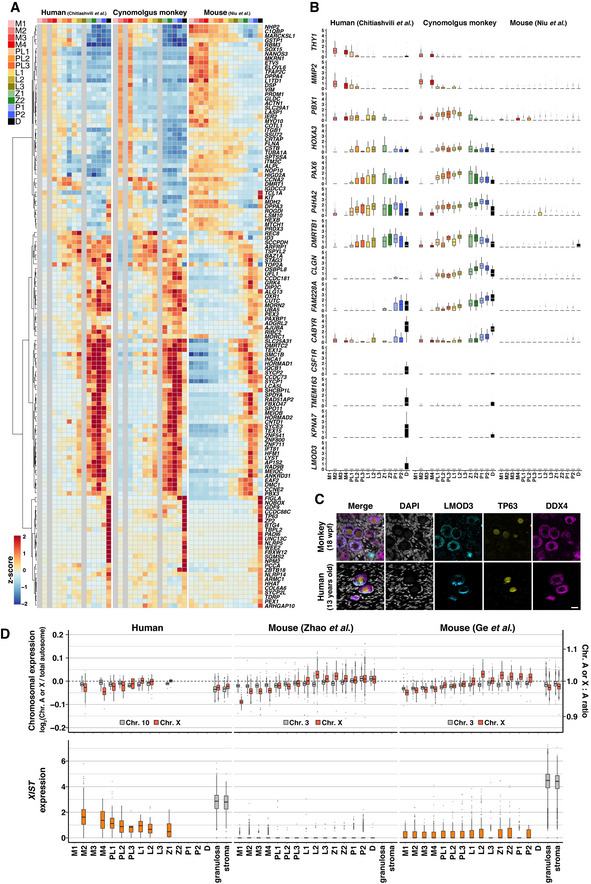
-
AHeatmap of the standardized expression levels of selected genes in all three species analyzed by 10X scRNA‐seq. Genes showing a conserved expression pattern in all three animal species during female germ‐cell development were manually selected from the list of DEGs acquired from the SC3‐seq analysis for cy/human in vitro cultured rOvaries (Dataset EV6). The color‐coding is as indicated.
-
BBox plots for the gene expression levels of selected markers showing primate‐specific expression dynamics during fetal germ‐cell development (see also Fig 6F). In the boxplots, the central bands represent the median values; the lower/upper hinges represent the 25th/75th percentiles, respectively; the upper limits of the whiskers represent the largest values no further than 1.5 IQR (interquartile range) from the upper hinges; the lower limits of the whiskers represent the smallest values no further than 1.5 IQR from the lower hinges; the dots represent the outliers. The numbers of the cells used are 0/606/0/117/133/76/6/72/68/2/12/14/45/16/47 from 4 fetuses for humans (Chitiashvili et al, 2020), 0/761/2/136/286/200/14/145/158/0/44/68/143/48/23 from 7 fetuses for cynomolgus monkeys (this study), and 26/261/21/232/259/149/209/450/422/134/232/163/339/630/298 for mice (Niu & Spradling, 2020) for M1/M2/M3/M4/PL1/PL2/PL3/L1/L2/L3/Z1/Z2/P1/P2/D, respectively. Cell clusters with more than two cells were analyzed.
-
CIF analyses of LEIOMODIN‐3 (LMOD3) with TP63 (oogenic marker) and DDX4 (germ‐cell marker) in cy in vivo fetal oocytes at 18 wpf (top) and 13‐year‐old human oocytes in primordial follicles (bottom). Nuclear DAPI staining is shown in white. Scale bar = 20 μm.
-
DThe autosome:A and X:A ratios (top) and XIST/Xist expression transitions (bottom) during female germ‐cell development in vivo in humans (this study) and mice (Zhao et al, 2020; Ge et al, 2021) analyzed by 10X scRNA‐seq. In the boxplots, the central bands represent the median values; the lower/upper hinges represent the 25th/75th percentiles, respectively; the upper limits of the whiskers represent the largest values no further than 1.5 IQR (interquartile range) from the upper hinges; the lower limits of the whiskers represent the smallest values no further than 1.5 IQR from the lower hinges; the dots represent the outliers. The numbers of the cells used are 0/275/0/40/81/40/5/20/31/0/6/1/1/2/2/3164/520 cells from 1 fetus for humans (this study), 643/1991/369/1819/306/179/490/1505/2051/523/1733/2529/2069/2863/102/0/0 for mice (Zhao et al, 2020), and 66/605/85/547/298/167/330/463/467/144/320/196/137/17/0/14962/5416 for mice (Ge et al, 2021) for M1/M2/M3/M4/PL1/PL2/PL3/L1/L2/L3/Z1/Z2/P1/P2/D/granulosa/stroma, respectively. Cell clusters with more than two cells were analyzed.
Conserved activities of X chromosomes
In mammals, females bear two X chromosomes (Xs), while males have one X and one Y chromosome. In females, one X is inactivated (Xi) in a random manner in early post‐implantation embryos (X‐chromosome inactivation: XCI) to compensate for the X‐linked gene dosage with males (Lyon, 1961; Zylicz & Heard, 2020). In somatic cells, one active X chromosome (Xa) is hyperactivated to compensate for the X chromosome: autosome (X:A) gene dosage (X‐chromosome upregulation: XCU) in both females and males (Ohno, 1967; Deng et al, 2014). On the contrary, in female germ cells, Xi is reactivated (X‐chromosome reactivation: XCR) (Monk & McLaren, 1981; Sugimoto & Abe, 2007; Guo et al, 2015; Tang et al, 2015; Vertesy et al, 2018; Chitiashvili et al, 2020; Okamoto et al, 2021), and it has been shown that germ cells exhibit a noncanonical X‐chromosome dosage compensation in mice and humans (Sangrithi et al, 2017), although another study reached a different conclusion (Chitiashvili et al, 2020).
We therefore sought to compare the X chromosome activities during fetal oocyte development in the three species. We first examined the X:A ratio of gene expression levels during mouse germ‐cell development based on bulk RNA‐seq data, which ensure high quantitative performance (Dataset EV7). In gonadal somatic cells, the X:A ratio was slightly below 1 (∼ 0.9), indicating that XCU occurs, if not fully, on a single Xa (Fig 7A; Dataset EV8). In female germ cells, the X:A ratio was below 1 (∼ 0.9) until E12.5 during their mitotic expansion, exceeded 1 (∼ 1.1) from E13.5 to E18.5 during the progression of meiotic prophase I, and was downregulated to ∼ 1 at postnatal day (P) 0 and 1 in the primordial follicles (Fig 7A; Dataset EV8). Given that both X chromosomes are progressively activated in germ cells by E14.5 (Sugimoto & Abe, 2007), this indicates a mild XCU during the meiotic prophase and its erasure in the primordial follicles. In male germ cells, the X:A ratio was ∼ 0.83 at E11.5, and it decreased to ∼ 0.65 at E12.5 and remained at ∼ 0.7 in the mitotic arrest phase (Fig 7A; Dataset EV8), indicating that XCU erasure also occurs in males. Interestingly, we detected a mild upregulation of Xist (X inactive specific transcript), the long noncoding RNA inducing a cascade of repressive events for XCI in mice (Brockdorff et al, 1992; Penny et al, 1996; Marahrens et al, 1997), in the primordial follicles, while it was essentially repressed in male germ cells (Fig 7A; Dataset EV8). The timing of the onset and erasure of a mild XCU in females was different from, but that of the XCU erasure in males was consistent with, a previous study (Sangrithi et al, 2017). We reanalyzed the datasets by Sangrithi et al (2017) using our pipeline and noted that the datasets by Sangrithi et al (2017) were somewhat variable in quality: For example, for gonadal somatic cells, the Chr3:A ratio ranged from ∼ 1.0 to 1.5, the X:A ratio ranged from ∼ 0.76 to 1.6, and the Xist expression levels ranged from ∼ 1.3 to 3.8 [log2(ssFPKM+1) value] (Appendix Fig S12). However, a relatively small number of replicates (2–5) was sampled for germ cells at each stage: For example, the X:A ratios of two datasets of germ cells at E12.5 were ∼ 0.9, while those of the other two were ∼ 1.2 (Appendix Fig S12).
Figure 7. X:A ratios and XIST expression during germ‐cell development in humans, monkeys, and mice.
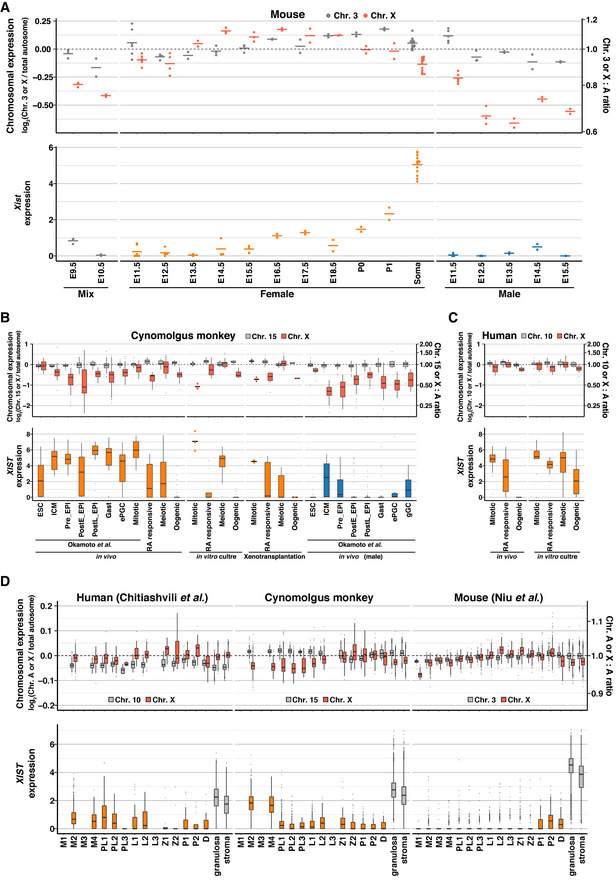
-
AThe Chr.3:A and X:A ratios (top), and Xist expression (bottom) in mouse fetal/neonatal gonadal cells analyzed by bulk RNA‐seq. Dot, each datum; bar, mean. Samples at E9.5/10.5 include germ cells from male and female embryos (mix). E, embryonic day; P, postnatal day; Soma, gonadal somatic cells.
-
BThe Chr.15:A and X:A ratios (top), and XIST expression (bottom) in cy female germ cells in vivo and in cultured/transplanted rOvaries analyzed by SC3‐seq. The Chr.15:A/X:A ratio transitions in the embryonic and hypoblast lineages reported in (Okamoto et al, 2021) are also shown. ESC, embryonic stem cell; ICM, inner cell mass; Pre_EPI, preimplantation epiblast; PostE_EPI, post‐implantation early epiblast; PostL_EPI, post‐implantation late epiblast; Gast, gastrulating cells; ePGC, early primordial germ cells. Data with small sample size (n ≦ 3) are indicated with dots (each datum) and bars (mean).
-
CThe Chr.10:A and X:A ratios (top), and XIST expression (bottom) in human female germ cells in vivo and in cultured rOvaries analyzed by SC3‐seq.
-
DThe autosome:A and X:A ratios (top), and XIST/Xist expression transitions (bottom) during female germ‐cell development in vivo in all three species analyzed by 10X scRNA‐seq.
Data information: In the boxplots, the central bands represent the median values; the lower/upper hinges represent the 25th/75th percentiles, respectively; the upper limits of the whiskers represent the largest values no further than 1.5 IQR (interquartile range) from the upper hinges; the lower limits of the whiskers represent the smallest values no further than 1.5 IQR from the lower hinges; the dots represent the outliers.
We next examined the X:A ratio during cy and human fetal oocyte development in vivo, in xenotransplantation, and in vitro based on the SC3‐seq datasets. During female cy embryonic development, XCI occurs coincidentally with XCU in somatic cells, and cy germ cells initiate XCR upon their specification and complete it by ∼ E50 (Okamoto et al, 2021). We found that in vivo, despite XCR, the X:A ratio was below 1 in mitotic and RA‐responsive cells (∼ 0.90 and ∼ 0.68, respectively), increased to ∼ 1 in meiotic cells, and then decreased below 1 (∼ 0.71) in oogenic cells (Fig 7B; Dataset EV8). XIST was expressed at a high level in mitotic germ cells, was repressed in RA‐responsive cells, exhibited an upregulation in meiotic cells, and was repressed again in oogenic cells (Fig 7B; Dataset EV8). The dynamics of the X:A ratio and XIST expression were similar in fetal oocyte development in xenotransplantation and in vitro as well as during human fetal oocyte development in vivo and in vitro (Fig 7B and C; Dataset EV8). We then analyzed the X:A ratio dynamics in the three species based on the 10X datasets, which revealed that although the dynamic range was narrower, the three species exhibited similar X:A ratio dynamics: The ratio was below 1 in mitotic and early meiotic (pre‐leptotene) oocytes, increased to ∼ 1 in late meiotic (zygotene/pachytene) oocytes, and decreased below 1 in oocytes in the primordial follicles (diplotene oocytes) (Figs 7D and EV5D; Dataset EV8). XIST showed a similar expression profile in humans and monkeys: It was expressed at a significant level in mitotic cells (similar to that in gonadal somatic cells) and then downregulated thereafter, whereas in mice, it was essentially repressed in mitotic and meiotic cells and showed a mild upregulation in diplotene oocytes (Figs 7D and EV5D; Dataset EV8). These findings are in agreement with the findings by Chitiashvili et al (2020), who reported the X:A dynamics during human fetal oocyte development (Chitiashvili et al, 2020), but are distinct from those of Sangrithi et al (2017), who reported a mild XCU in human mitotic oogonia (Sangrithi et al, 2017). Based on our collective data, we conclude that although there exist some levels of quantitative variability depending on the measurement methodology, on the whole, the X‐linked gene dosage is regulated in a similar manner in humans, monkeys, and mice, and fetal oocyte development proceeds with two Xa and with little, if any, XCU.
Discussion
In mammals, female embryonic germ‐cell development culminates in the generation of primordial follicles, which consist of oocytes arrested at the diplotene stage of the meiotic prophase I and a single layer of squamous granulosa cells that support subsequent oocyte growth and maturation (McGee & Hsueh, 2000; Edson et al, 2009). In humans, primordial follicles are a source for oogenesis that begins upon puberty and continues to provide mature oocytes across the nearly forty‐year reproductive lifecycle in females (McGee & Hsueh, 2000, Edson et al, 2009). Anomalies in primordial follicles or their exhaustion are a primary cause of infertility in females (McGee & Hsueh, 2000, Edson et al, 2009). Accordingly, induction of primordial follicles from hPSCs would be a major milestone of human IVG research. We have shown that both human and cy mitotic oogonia enter into and complete the meiotic prophase I to form primordial follicle‐like structures in rOvaries, providing a critical step toward realizing this milestone.
A notable finding is that, unlike mouse embryonic ovaries and rOvaries (Hikabe et al, 2016; Morohaku et al, 2016), both human and cy rOvaries failed to maintain their structural integrity on membrane inserts under air–liquid interface culture conditions, with ovarian cells, including germ cells, progressively spreading out on the membrane and rOvaries eventually flattening out (Figs 3B and EV2A and E). This difference would be a reflection of the different biophysical properties between mouse and primate ovarian somatic cells, the clarification of which requires further investigations. A key advantage of the air–liquid interface culture is the improved aeration and oxygen supply for cultured tissues, but it is unclear whether such a condition mimics embryonic ovary development and physiology in vivo. Indeed, it has been shown that the recruitment of primordial follicles requires a hypoxic condition (Shimamoto et al, 2019). Thus, the air–liquid interface culture that has been used successfully in mice may not be an essential requirement for mammalian rOvary culture in general.
Accordingly, we have shown that under a floating culture condition, both human and cy rOvaries maintained their structural integrity, increased their size, albeit to a mild degree, and underwent appropriate maturation, generating primordial follicle‐like structures over an extended culture period in vitro (humans: ∼ 14 weeks; monkeys: ∼ 12 weeks). The morphology, expression/subcellular localization of key markers, and transcriptome properties of meiotic cells and oocytes in primordial follicle‐like structures in human and cy rOvaries in vitro were very similar to those of the corresponding in vivo cell types in humans and cynomolgus monkeys (Figs 3, EV2, EV3, 4, EV4, 5), demonstrating an appropriate progression of fetal oocyte development in vitro. On the contrary, the proportion of oogonia that differentiated into oocytes in primordial follicle‐like structures appeared to be lower, with many germ cells arrested at the pre‐leptotene stage likely due to suboptimal upregulation of key genes for the pre‐leptotene to leptotene transition, and the time required for such differentiation was longer under the in vitro condition. In addition, we noted that granulosa cells that contributed to primordial follicle‐like structures acquired activated granulosa cell‐like properties as in the case of mouse rOvary culture, presumably due to a lack of appropriate rigidity/hydrostatic pressure in the surrounding tissue environments (Appendix Fig S6A and B and D) (Nagamatsu et al, 2019). Thus, based on our results using the presented strategy, it is important to further improve the human and cy rOvary culture condition to increase the efficiency, yield, and quality of primordial follicle‐like structures in a shortened, more physiologic time frame.
We have performed a detailed comparison of transcriptome dynamics during fetal oocyte development in humans, monkeys, and mice, which revealed a number of primate‐specific properties. One such property is that in humans and cynomolgus monkeys, the mitotic‐to‐pre‐leptotene transition involves a relatively large transcriptomic alteration, whereas in mice, it entails only a minor change (Fig 6D and E; Appendix Fig S9A and B). The transcriptome data for cy fetal oocyte development that we generated, which exhibited the highest quality among the data we used (Appendix Fig S8B), revealed that, upon such transition, most genes characterizing mitotic cells, including those associated with early PGC properties and pluripotency, were extinguished, and key oogenic genes identified in mice, such as ZGLP1 and STRA8 (Baltus et al, 2006; Nagaoka et al, 2020), initiated expression after the extinguishment of the oogonia genes in pre‐leptotene 1/2 cells (Fig 4H), suggesting that a primate‐specific mechanism exists that shuts off the oogonia program prior to the activation of oogenic/meiotic genes during the 2‐week mitotic‐to‐pre‐leptotene transition period. It is also interesting to note that both in humans and in cynomolgus monkeys, but not in mice, the activation of the oogenic program involves upregulation of many genes involved in “pattern specification process,” including PAX6, HES7, SIX1, TBX5, PBX1, and the HOX genes (Figs 4H and 6F, and EV5B), and the question of whether these genes play a key role in the oogenic/meiotic program warrants future investigations. In addition, we have identified many genes that exhibited dynamic expression during fetal oocyte development in a primate‐specific manner (Figs 6F−I and EV5B and C; Dataset EV6), and nonetheless found that a majority of genes contributing to fetal oocyte development, including those showing primate‐specific dynamic expression, are 1–1–1 orthologues but not species‐specific genes among humans, monkeys, and mice (Appendix Fig S10). This indicates that primate specificity is brought about by primate‐specific usage of orthologous genes, most likely through unique regulatory wiring acquired during primate evolution.
An appropriate X‐linked gene dosage is critical for fetal oocyte development both in vivo and in vitro (Ford et al, 1959; Singh & Carr, 1966; Carr et al, 1968; Burgoyne & Baker, 1985; Hamada et al, 2020), while how it is precisely regulated during mammalian oocyte development has not been well understood. One study has shown that, both in mice and in humans, XCR in the presence of XCU creates a state with excessive X‐linked gene dosage compensation (X:A ratio > 1) in mitotic oogonia and leptotene/zygotene oocytes, and subsequent erasure of XCU leads to a properly X‐linked gene‐dosage compensated state (X:A ratio ≅1) in pachytene/diplotene oocytes (Sangrithi et al, 2017), while another has documented that, in humans, mitotic oogonia and leptotene/zygotene oocytes exhibit an X:A ratio smaller than that in somatic cells due to X‐chromosome dampening (XCD) (presumable X:A ratio < 1), while pachytene oocytes show an X:A ratio greater than that in somatic cells due to XCD erasure (presumable X:A ratio > 1), and diplotene oocytes again realize an X:A ratio smaller than that in somatic cells, possibly through a distinct mechanism (presumable X:A ratio < 1) (Chitiashvili et al, 2020). The different conclusions by these studies could be due to the different technologies/methodologies used to determine the X:A ratio and/or the variable quality of the datasets (Appendix Fig S12). Using several different measurement technologies/methodologies (bulk RNA‐seq, SC3‐seq, 10X), we have shown that during fetal oocyte development, the X‐linked gene dosage is regulated in a similar manner in humans, monkeys, and mice, which is more consistent with the regulation of X‐linked gene dosage reported by Chitiashvili et al (2020) (Figs 7 and EV5D). On the contrary, throughout fetal oocyte development, the X‐linked gene dosage regulation occurred within a narrow range, with the X:A ratio fluctuating at around 0.8 to 1.2 with two Xa, and remaining consistently greater than that in male germ cells (∼ 0.7) (Figs 7 and EV5D). Whether the relatively minor X:A ratio change may be a passive consequence of a unique transcriptional regulation during the meiotic prophase or involve an active mechanism with functional relevance remains to be determined. In particular, the appropriate regulation of X‐chromosome activity poses an important challenge for human in vitro oogenesis from hPSCs, as the X chromosomes are vulnerable to epigenetic dysregulation in hPSCs and their differentiated progeny (Mekhoubad et al, 2012; Nazor et al, 2012; Patrat et al, 2020). In conclusion, we have not only demonstrated an appropriate in vitro maturation of fetal ovaries mainly with oogonia into those bearing primordial follicles both in humans and in monkeys, but have also elucidated evolutionarily conserved as well as primate‐specific programs that shape fetal oocyte development in humans, monkeys, and mice, providing a critical basis for human in vitro oogenesis research, including PSC‐based reconstitution of human oocyte development, and reproductive medicine.
Materials and Methods
Reagent Tools table
| Reagent/Resource | Reference or Source | Identifier or Catalog Number |
|---|---|---|
| Experimental Models | ||
| Slc:ICR (M. musculus) | Japan SLC | N/A |
| KSN/Slc (M. musculus) | Japan SLC | N/A |
| Vasa‐RFP Tg (M. musculus) | B6C3(Cg)‐Tg(Ddx4‐RFP,neo)1Tnc | RBRC03449 |
| Dppa3(Stella)‐EGFP Tg (M. musculus) | Patrat et al (2020), Seki et al (2007) | N/A |
| Cynomolgus monkey (M. fascicularis) | China / Philippines / Vietnam / Cambodia | N/A |
| Antibodies | ||
| Mouse anti‐AP‐2γ/TFAP2C, monoclonal | Santa Cruz Biotechnology | Cat # sc‐12762 |
| Rabbit anti‐Cleaved Caspase‐3, polyclonal | Cell Signaling Technology | Cat # #9661 |
| Rabbit anti‐Cleaved PARP, monoclonal | Cell Signaling Technology | Cat # #5625 |
| Rabbit anti‐CLGN, polyclonal | Atlas Antibodies | Cat # HPA048761 |
| Mouse anti‐DAZL, monoclonal | Santa Cruz Biotechnology | Cat # sc‐390929 |
| Mouse anti‐DDX4, monoclonal | Abcam | Cat # ab27591 |
| Rabbit anti‐DDX4, polyclonal | Abcam | Cat # ab13840 |
| Goat anti‐VASA/DDX4, polyclonal | R&D Systems | Cat # AF2030 |
| Mouse anti‐Dmc1, monoclonal | Santa Cruz Biotechnology | Cat # sc‐53269 |
| Rabbit anti‐FIGLA, polyclonal | Abcam | Cat # ab173725 |
| Goat anti‐FOXL2, polyclonal | Novus Biologicals | Cat # NB100‐1277 |
| Rabbit anti‐FOXL2, monoclonal | Abcam | Cat # ab246511 |
| Rabbit anti‐Ki67, polyclonal | Abcam | Cat # ab15580 |
| Rabbit anti‐Laminin, polyclonal | Abcam | Cat # ab11575 |
| Rabbit anti‐Laminin, polyclonal | Sigma‐Aldrich | Cat # L9393 |
| Rabbit anti‐Leiomodin‐3, polyclonal | Abcam | Cat # ab122305 |
| Rabbit anti‐NALP5/NLRP5, polyclonal | Abcam | Cat # ab185679 |
| Goat anti‐Nanog, polyclonal | R&D Systems | Cat # AF1997 |
| Mouse anti‐NOBOX, monoclonal | Novus Biologicals | Cat # NBP2‐46193 |
| Mouse anti‐p63/TP63, monoclonal | Abcam | Cat # ab735 |
| Rabbit anti‐PAX6, monoclonal | Abcam | Cat # ab195045 |
| Rat anti‐Podoplanin/PDPN, monoclolnal | BioLegend | Cat # 337001 |
| Mouse anti‐Oct3/4/POU5F1, monoclonal | Santa Cruz Biotechnology | Cat # sc‐5279 |
| Rabbit anti‐SCP1, polyclonal | Novus Biologicals | Cat # NB300‐228 |
| Rabbit anti‐SCP3, polyclonal | Abcam | Cat # ab15093 |
| Rabbit anti‐Stra8, polyclonal | Abcam | Cat # ab217380 |
| Rabbit anti‐ZGLP1, polyclonal | Atlas Antibodies | Cat # HPA049855 |
| Rabbit anti‐ZP3, polyclonal | Atlas Antibodies | Cat # HPA054061 |
| Rabbit anti‐gamma H2AX, polyclonal | Novus Biologicals | Cat # NB100‐2280 |
| Donkey anti‐Rat IgG (H+L) Highly Cross‐Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat # A21208 |
| Donkey anti‐Rabbit IgG (H+L) Highly Cross‐Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat # A21206 |
| Donkey anti‐Mouse IgG (H+L) Highly Cross‐Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat # A21202 |
| Donkey anti‐Goat IgG (H+L) Cross‐Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat # A11055 |
| Donkey anti‐Rabbit IgG (H+L) Highly Cross‐Adsorbed Secondary Antibody, Alexa Fluor 568 | Invitrogen | Cat # A10042 |
| Donkey anti‐Mouse IgG (H+L) Highly Cross‐Adsorbed Secondary Antibody, Alexa Fluor 568 | Invitrogen | Cat # A10037 |
| Donkey anti‐Goat IgG (H+L) Cross‐Adsorbed Secondary Antibody, Alexa Fluor 568 | Invitrogen | Cat # A11057 |
| Donkey anti‐Rabbit IgG (H+L) Highly Cross‐Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen | Cat # A31573 |
| Donkey anti‐Mouse IgG (H+L) Highly Cross‐Adsorbed Secondary Antibody, Alexa Fluor 647 | Invitrogen | Cat # A31571 |
| Donkey anti‐Goat IgG (H+L) Cross‐Adsorbed Secondary Antibody, Alexa Fluor 633 | Invitrogen | Cat # A21082 |
| Oligonucleotides and sequence‐based reagents | ||
| PCR primers | This study | Table EV1 |
| Chemicals, enzymes and other reagents | ||
| Leuplin for injection 1.88mg. | TAKEDA Pharmaceutical | N/A |
| uFSH injection | ASKA Pharmaceutical | N/A |
| Gonatropin | ASKA Animal Health | N/A |
| DMEM, high glucose, pyruvate, no glutamine | GIBCO | 10313021 |
| DMEM/F12 | GIBCO | 11330057 |
| MEM α, nucleosides | GIBCO | 12571063 |
| Advanced MEM | GIBCO | 12492013 |
| CMRL Medium, no glutamine | GIBCO | 11530037 |
| DMEM/F‐12, no glutamine | GIBCO | 21331020 |
| IMDM | GIBCO | 12440053 |
| KnockOut DMEM | GIBCO | 10829018 |
| McCoy's 5A (Modified) Medium | GIBCO | 16600082 |
| MesenPRO RS Medium | GIBCO | 12746012 |
| StemPro‐34 SFM (1X) | GIBCO | 10639011 |
| Waymouth's Medium | GIBCO | 11220035 |
| Glasgow's MEM (GMEM) | GIBCO | 11710035 |
| Advanced RPMI 1640 | GIBCO | 12633012 |
| Fetal bovine serum | GIBCO | 10437028 |
| BSA fraction V | GIBCO | 15260037 |
| Penicillin‐Streptomycin (5,000 U/ml) | GIBCO | 15070063 |
| MEM Non‐Essential Amino Acids Solution (100X) | GIBCO | 11140‐050 |
| Sodium Pyruvate (100 mM) | GIBCO | 11360070 |
| L‐Glutamine (200 mM) | GIBCO | 25030081 |
| L‐Ascorbic Acid | Sigma‐Aldrich | A4403 |
| 2‐Mercaptoethanol | GIBCO | 21985023 |
| Y‐27632 dihydrochloride | TOCRIS | 1254 |
| Trypsin‐EDTA (0.5%), no phenol red | GIBCO | 15400054 |
| TryPLE Express | GIBCO | 12604021 |
| Accumax | Innovative Cell Technologies | AM105 |
| Deoxyribonuclease I | MERCK | DN25 |
| CELLBANKER 1 plus | ZENOGEN PHARMA | CB021 |
| Human collagen type I | Millipore | CC050 |
| Human collagen type III | Corning | 354244 |
| Human collagen type IV | Corning | 354245 |
| Human collagen type V | Corning | 354246 |
| Human collagen type VI | Corning | 354261 |
| Human fibronectin | Corning | 354008 |
| iMatrix | Nippi | 892014 |
| Laminins (111/121/211/221/411/421/332/521) | VERITAS | BLA‐LNKT‐0201 |
| Matrigel Growth Factor Reduced | Corning | 356230 |
| Paraformaldehyde | Sigma‐Aldrich | 158127 |
| Hematoxylin | Fujifilm | 131‐09665 |
| Eosin | Fujifilm | 058‐00062 |
| OCT Compound | Sakura Finetek | 4583 |
| HistVT One | Nacalai Tesque | 06380‐76 |
| Bovine serum albumin | Sigma‐Aldrich | A7030 |
| Normal donkey serum | Jackson Immuno Research Laboratories | 017‐000‐121 |
| DAPI | Wako | 342‐07431 |
| VECTASHIELD | Vector Laboratories | H‐1000 |
| Software | ||
| FV10‐ASW | Olympus | N/A |
| FACSDiva | BD Biosciences | N/A |
| DAVID (v6.8; GO analysis) | N/A | https://david.ncifcrf.gov/tools.jsp |
| Cell Ranger (version 5.0.1) | N/A | https://github.com/10XGenomics/cellranger |
| R (version 3.6.3) | N/A | https://www.R‐project.org/ |
| Python (version 3.7.7) | N/A | https://www.python.org |
| Seurat (version 3.2.2; 10X analysis) | Stuart et al (2019) | https://github.com/satijalab/seurat |
| SeuratWrappers (Version 0.1.0) | N/A | https://github.com/satijalab/seurat‐wrappers/ |
| scran (version 1.14.1) | Lun et al (2016) | https://bioconductor.org/packages/release/bioc/html/scran.html |
| Monocle3 (version 0.2.1) | Cao et al (2019) | http://cole‐trapnell‐lab.github.io/monocle‐release/ |
| ComplexHeatmap (version 2.6.2) | Gu et al (2016) | https://github.com/jokergoo/ComplexHeatmap |
| LTLA/batchelor (version 1.2.1) | Haghverdi et al (2018) | https://rdrr.io/github/LTLA/batchelor/ |
| Scrublet (version 0.2.1) | Wolock et al (2019) | https://github.com/swolock/scrublet |
| scVelo (version 0.2.2) | Bergen et al (2020) | https://github.com/theislab/scvelo |
| Scanpy (version 1.6.0) | Wolf et al (2019) | https://github.com/theislab/scanpy |
| R (version 4.0.2) | N/A | https://www.R‐project.org/ |
| dendextend (version 1.15.1) | Galili et al (2015) | https://CRAN.R‐project.org/package=dendextend |
| amap (version 0.8‐18) | N/A | https://CRAN.R‐project.org/package=amap |
| pheatmap (version 1.0.12) | N/A | https://CRAN.R‐project.org/package=pheatmap |
| Seurat (version 4.0.4; SC3‐Seq analysis) | Stuart et al (2019) | https://github.com/satijalab/seurat |
| Other | ||
| Nunclon Sphera‐Treated U‐Shaped‐Bottom 96‐well plates | NUNC | 174925 |
| Transwell‐COL | Corning | 3495 |
| Preset VECELL | VECELL | PSVC12‐10 |
| QIAamp DNA Mini Kit | QIAGEN | 51304 |
| Power SYBR Green PCR Master Mix | Applied Biosystems | 4367659 |
| CFX384 Touch Real‐Time PCR Detection System | BIO‐RAD | N/A |
| DNA High Sensitivity Reagent Kit | PerkinElmer | CLS760672 |
| Chromium Single Cell 3′ GEM, Library & Gel Bead Kit v3 | 10X Genomics | 1000092 |
| Next GEM single cell 3' Library & Gel beads kit ver 3.1 | 10X Genomics | 1000128 |
| Chromium Single Cell B Chip Kit | 10X Genomics | 1000074 |
| Chroomium Next GEM single cell G chip kit | 10X Genomics | 1000120 |
| Mastercycler nexus | Eppendorf | 6331000033 |
| NucleoSpin RNA XS | Macherey‐Nagel | 740902 |
| NextSeq 500/550 | Illumina | N/A |
| NovaSeq 6000 | Illumina | N/A |
| NextSeq 500/550 High‐Output v2 Kit (75 cycles) | Illumina | FC‐404‐2005 |
| NovaSeq 6000 SP Reagent Kit (100 cycles) | Illumina | 20027464 |
| NovaSeq 6000 S1 Reagent Kit (100 cycles) | Illumina | 20012865 |
| Microtome | Leica | CX41 |
| Upright microscope | Olympus | BX53 |
| Inverted microscope | Olympus | IX71 |
| CCD camera | Olympus | DP70 |
| Cryostat | Leica | CM1850 |
Methods and Protocols
Animals
All animal experiments were performed with the ethical approvals of Kyoto University and Shiga University of Medical Science. ICR and immunodeficient KSN/Slc mice were purchased from SLC (Shizuoka, Japan). The C57BL/6N;mvh‐RFP Tg/Tg (VR) and Dppa3(Stella)‐EGFP Tg/Tg transgenic mice were established as described previously (Payer et al, 2006; Seki et al, 2007; Imamura et al, 2010; Miyauchi et al, 2017). Mice were housed in a specific pathogen‐free animal facility under a 14‐h light/10‐h dark cycle. For the animal experiments using cynomolgus monkeys, the procedures for oocyte collection, intracytoplasmic sperm injection, preimplantation embryo culture, and embryo transfer into foster mothers were as described previously (Yamasaki et al, 2011). Pregnancy was checked by transabdominal fetal echography at 4 wpf, and the blood plasma of the pregnant monkey was collected followed by the extraction of fetal cell‐free DNA. To minimize the number of cy fetuses used, cell‐free DNA in maternal plasma was purified by a QIAamp DNA Mini Kit (QIAGEN, 51304), and the sex of fetuses was determined by real‐time PCR targeted for SRY and DYS14 genes (Yasmin et al, 2015). For collecting the cy fetuses, cesarean sections were performed under isoflurane and ketamine‐based general anesthesia. The sex of the fetus was further confirmed by checking the gross appearance of its gonads or by PCR for UTX/UTY genes (Villesen & Fredsted, 2006). The light condition for monkeys was a 12‐h light/12‐h dark cycle. Temperature and humidity in the animal rooms were maintained at 25 ± 2°C and 50 ± 5%, respectively.
Collection of human embryo and ovary samples
Human fetal or adult ovaries were collected after informed consent from participants undergoing elective abortion or gynecological procedures. All experimental procedures were approved by the Institutional Review Board at Kyoto University (approval nos. G1047 and G1192). The sex of the fetus was determined by sex‐specific PCR for the ZFX/ZFY and SRY loci (Josef & Adam, 2003).
Histological analyses
For immunohistochemical analyses, fetal ovaries or rOvaries were fixed in 4% paraformaldehyde/PBS (Sigma‐Aldrich, 158127) at 4°C for 30 min or overnight, embedded in paraffin wax (Leica, 39601006) or OCT compound (Sakura Finetek, 4583), and sectioned at a thickness of 5 or 8 μm. After deparaffinization, the sections were pretreated with an antigen retrieval reagent, HistVT One (Nacalai Tesque, 06380–76), at 90°C for 40 min. Sections were treated with blocking solution (1% bovine serum albumin (Sigma‐Aldrich, A7030), 5% normal donkey serum (JIR, 017–000‐121), 0.1% Tween20, and 1× phosphate buffered saline) for 30 min at room temperature, and incubated with primary antibodies in blocking solution at 4°C overnight. The slides were further incubated with Alexa Fluor‐conjugated secondary antibodies. Fluorescence images were captured using a laser‐scanning confocal microscope (OLYMPUS, FV1000; ZEISS, LSM780). For the quantitative analysis of each marker, germ cells marked with DDX4 expression were analyzed. To define the “inner” and “outer” cortex for fetal ovaries at 10–18 wpf, the ovarian cortex was divided into two at the middle of the cortex. Hematoxylin and eosin staining were performed using the standard procedure.
Staging of meiosis in ovarian sections
Germ cells expressing DDX4 in fetal ovarian sections were classified into 7 stages by the pattern of nuclear staining of DAPI, SYCP3, and DMC1. The stages were defined as follows. Mitotic: cells without any SYCP3 and DMC1 signal. Pre‐leptotene: numerous short nuclear threads with low SYCP3 signal. Leptotene: numerous short nuclear threads with low SYCP3 and DMC1 signals. Zygotene: numerous long thin threads of SYCP3 staining with many DMC1 dots. Early pachytene: thick threads of SYCP3 staining with many DMC1 foci. Late pachytene: thick threads of SYCP3 staining with less than one DMC1 dot on each chromosome. Diplotene: numerous short nuclear threads with faint or no SYCP3 signal. Diplotene cells were distinguished from pre‐leptotene ones by their size and histological morphology.
Dissociation of cy/human fetal ovarian tissues and cy/human rOvaries
Fetal ovaries collected from cy fetuses at 8–18 wpf were carefully dissected out in chilled PBS. Fetal ovaries at 8–14 wpf were minced with scissors in 0.05% trypsin/EDTA/PBS solution (GIBCO, 15400–054), then incubated for 15 min at 37°C with gentle pipetting every 5 min. Dissociation medium [DMEM (GIBCO, 10313021) containing 10% FBS, 2 mM L‐glutamine (GIBCO, 25030081), 1X penicillin/streptomycin (GIBCO, 15070063), and 100 μg/mL DNaseI (MERCK, DN25)] was added for further dissociation. Dissociated cells were used for single‐cell RNA‐seq analyses, or resuspended in CELLBANKER 1 plus (ZENOGEN PHARMA, CB021) and stored at −80°C. Human fetal ovaries at 11‐week gestation were also dissociated and cryopreserved using the same procedures.
For fetal ovaries at 16 and 18 wpf, minced tissues were incubated in Accumax (Innovative Cell Technologies, AM105) for 40 min at 37°C with gentle pipetting every 5 min. Dissociation medium was added for further dissociation. For SC3‐seq analysis, cells were picked up under the inverted microscope (OLYMPUS, IX71) to exclude multiplet cells. For 10X experiments, cell suspensions were further treated with TryPLE Express (1×) (GIBCO, 12604021) for 5 min at 37°C to make the single‐cell suspension.
For dissociating cy/human rOvaries, tissues were incubated with TryPLE Express (1×) (GIBCO, 12604021) for 5 min. After adding dissociation medium, cells were processed for single‐cell analyses.
In vitro culture of cy/human rOvary under an air–liquid interface culture condition
-
i
The cy fetal ovarian cells (50,000 cells/well) at 8 wpf were plated on wells of Nunclon Sphera‐Treated U‐Shaped‐Bottom 96‐well plates (NUNC, 174925).
-
ii
The cells were cultured under a floating condition in αMEM medium containing 10% FBS, 2 mM L‐glutamine (GIBCO, 25030081), 1 × NEAA (GIBCO, 11140050), 1 mM Na pyruvate (GIBCO, 11360070), 1 × penicillin/streptomycin (GIBCO, 15070063), 100 μM 2‐mercaptoethanol (GIBCO, 21985023), and 10 μM Y‐27632 dihydrochloride (TOCRIS, 1254).
-
iii
After 2 days, using a glass capillary, the cy rOvaries were transferred onto a Transwell‐COL membrane insert (Corning, 3495) soaked in αMEM (GIBCO, 12571063) containing 10% FBS, 2 mM L‐glutamine (GIBCO, 25030081), 150 μM ascorbic acid (Sigma‐Aldrich, A4403), 1× penicillin/streptomycin (GIBCO, 15070063), and 55 μM 2‐mercaptoethanol (GIBCO, 21985023).
-
iv
Half the medium was changed every 2 days.
Cy/human rOvaries were cultured at 37°C under an atmosphere of 5% CO2 in air. The human rOvary was generated from human fetal ovarian cells at 11‐week gestation and cultured under the same conditions, except that Advanced MEM was used as the basal medium.
To assess the coating condition, Preset VECELL membranes (VECELL, PSVC12‐10) were coated with human collagen type I (Millipore, CC050), type III (Corning, 354244), type IV (Corning, 354245), type V (Corning, 354246), and type VI (Corning, 354261), human fibronectin (Corning, 354008), iMatrix (Nippi, 892014), laminins (111/121/211/221/411/421/332/521) (VERITAS, BLA‐LNKT‐0201), growth factor‐reduced Matrigel (Corning, 356230), and their combinations prior to IVC following the manufacturers' coating protocols.
In vitro culture of cy/human rOvaries under the floating condition
-
i
The cy fetal ovarian cells (50,000 cells/well) at 8 wpf were plated on wells of Nunclon Sphera‐Treated U‐Shaped‐Bottom 96‐well plates (NUNC, 174925).
-
ii
The cells were cultured under a floating condition in Advanced MEM (GIBCO, 12492013) containing 10% FBS, 2 mM L‐glutamine (GIBCO, 25030081), 1× NEAA (GIBCO, 11140050), 1 mM Na pyruvate (GIBCO, 11360070), 1× penicillin/streptomycin (GIBCO, 15070063), 100 μM 2‐mercaptoethanol (GIBCO, 21985023), and 10 μM Y‐27632 dihydrochloride (TOCRIS, 1254).
-
iii
After 2 days, the cy rOvary was transferred into another well of a U‐bottom culture plate using a glass capillary and cultured under a floating condition in Advanced MEM (GIBCO, 12492013) containing 10% FBS, 2 mM L‐glutamine (GIBCO, 25030081), 150 μM ascorbic acid (Sigma‐Aldrich, A4403), 1× penicillin/streptomycin (GIBCO, 15070063), and 55 μM 2‐mercaptoethanol (GIBCO, 21985023) [in vitro differentiation medium for primates: P‐IVD medium].
-
iv
Medium change was performed every 2 days by transferring the cy rOvary into another well with freshly prepared culture medium.
-
v
Cy rOvaries were analyzed at 3/6/9/12/15 w‐ivc (Day 23/44/65/86/107).
The human rOvary was generated from human fetal ovarian cells at 11 weeks of gestation and cultured under the same conditions.
Xenotransplantation experiment
Cy rOvaries generated by the culture method described above were transplanted under the kidney capsule of immunodeficient KSN/Slc mice. Transplantation procedures were performed under mixed anesthesia (midazolam, medetomidine, and butorphanol), and the cy rOvaries were placed beneath the kidney capsule by narrow glass capillaries. Histological and SC3‐seq analyses were performed at 3/6/9/12/15/21 w‐ptp.
Isolation of mouse gonadal/ovarian cells
To isolate mouse gonadal/ovarian cells at embryonic day (E) 11.5 for the bulk RNA‐seq analysis, VR females were crossed with the Dppa3(Stella)‐EGFP Tg/Tg males. Noon of the day when a copulation plug was identified was designated as E0.5. The sex of embryos at E11.5 was determined by sex‐specific PCR for the Ube1 locus. To isolate mouse gonadal/ovarian cells at E12.5–postnatal day (P)1, ICR females were crossed with the VR males, and the sex of embryos was determined by the gross appearance. The gonads/ovaries were dissociated into single cells by incubating with 0.05% trypsin–EDTA (GIBCO, 15400‐054) for 10 min at 37°C, and were quenched with an equal volume of DMEM containing 10% FBS. Large clumps of cells were removed using a nylon cell strainer (FALCON, 352350), and cells were centrifuged at 200 rcf for 5 min. The cell pellet was resuspended with DMEM/F12 medium (GIBCO, 1130–082) containing 0.1% BSA fraction V (Gibco, 15260037), and sorted into VR+ germ and VR− somatic cells with a fluorescence‐activated cell sorter (BD Biosciences, FACS Aria III).
10X genomics single‐cell RNA‐seq
Dissociated cells from cy fetal ovaries and cultured cy rOvaries were stained with DRAQ7 to distinguish dead cells. All DRAQ7− live cells were collected using a fluorescence‐activated cell sorter (BD Biosciences, FACS Aria III). Subsequent 10X scRNA‐seq library preparation (10X Genomics, Single Cell 3′ Kit v. 3 and v. 3.1) and sequencing on a NovaSeq 6000 platform (Illumina) were performed following the manufacturers' instructions. Raw files were processed with the “mkfastq” command in Cell Ranger (v. 5.0.1) and mapped to the cynomolgus monkey reference genome (MacFas5.0 for cynomolgus monkeys and GRCh38.p12 for humans).
Analysis for cy scRNA‐seq data (10X)
The gene‐barcode matrices were analyzed with the “Seurat” R package (v. 3.2.2) (Satija et al, 2015) following the online tutorials (https://satijalab.org/seurat/index.html). The low‐quality cells, putative doublets/multiplets, and putative stripped nuclei were excluded for downstream analyses (Dataset EV1). The raw counts for each cell were divided by their size factors calculated by the “scran” R package (v. 1.14.1), and then log‐transformed [log2(size‐scaled (ss) UMI + 1)] for further analyses. Doublet cells were removed using the “Scrublet” python package (v. 0.2.1) following the instructions reported in (Wolock et al, 2019). The threshold for doublet estimation was determined based on the bimodal distribution calculated by default parameters. Batch correction was then performed using the “RunFastMNN” function in the Seurat R package with the default setting. We used the Seurat “RunUMAP” function to characterize and visualize cell clusters. The resolution of the “FindClusters” function was set at 0.5 for the overall cell analyses and 0.8 for germ/granulosa cell analyses. Clusters were annotated by checking the expression pattern of previously characterized marker genes in human and mouse fetal ovarian cells. The sources and identification numbers of other R or Python packages are described in the Reagents and Tools table.
Analysis for cy germ cells (10X)
Cells in the DDX4 + germ‐cell cluster were further analyzed and divided into 13 clusters using the “FindClusters” function. After removing two small clusters consisting of putative doublets/multiplets that co‐expressed the markers of other cell types, 11 clusters were defined by the expression pattern of key germ‐cell markers as “mitotic 1/2/3,” “pre‐leptotene 1/2/3,” “leptotene,” “zygotene,” “pachytene 1/2,” and “unclassified.” Cells expressing GDF9/NOBOX/ZP3 were manually selected as another cluster defined as “Diplotene.” To identify highly variable genes (HVGs) during cy female germ‐cell development, we performed the “FindVariableFeatures (selection.method=“vst,” nfeatures = 2,000)” function in the Seurat R package across the 11 clusters (mitotic–diplotene) and detected 2,000 HVGs. Among them, excluding genes with low expression levels [cluster average of log(ssUMI+1) ≦ 0.5: 519 genes], 1,481 genes were used for heatmap visualization, GO enrichment analysis (see below), and calculation of the correlation coefficients between each cluster (Fig 4H and I; Dataset EV3).
Cell‐cycle scoring and annotation
Cell‐cycle annotation of fetal cy germ cells was performed using the “Cell‐Cycle Scoring” function of the Seurat package based on the scoring strategy described in Tirosh et al (2016). As our dataset includes oocytes in meiotic prophase I, we used orthologous genes from the annotation list for meiotic cells used in Shimada et al (2021) to score the meiotic prophase I. The CCNE2 gene was excluded due to its presence in both the G2/M and meiotic prophase I lists. Cell‐Cycle Scores for the S, G2/M, and meiotic prophase I phases were calculated using the “AddModuleScore” function, and cells without any of these annotations were assigned to the G1 phase.
Analysis for cy granulosa cells (10X)
Cells in the FOXL2 + granulosa‐cell cluster were further analyzed. The count matrices for spliced and unspliced transcripts were created by running the commands “count ‐‐include‐introns” and “count” in Cell Ranger (v. 5.0.1). The resolution of the “FindClusters” function was set at 0.8, and the cells were divided into 12 clusters after removing two small clusters consisting of putative doublets/multiplets that co‐expressed the markers of other cell types. RNA velocity analysis was performed using the “scvelo.pp.moments(n_pcs = 30, n_neighbors = 30)” and “scvelo.tl.velocity(mode = ‘stochastic’)” functions in the scVelo python package (v. 0.2.0) (Bergen et al, 2020). Partition‐based graph abstraction (PAGA) graph (Wolf et al, 2019) was computed on the clusters annotated in Fig EV4A using the “scanpy.tl.paga” function from the Scanpy (v. 1.2.2) in Python (v. 3.7.7) following the online tutorials (https://scanpy‐tutorials.readthedocs.io/en/latest/paga‐paul15.html), and the threshold for connection of clusters was set to 0.03. To identify HVGs among the 12 clusters, we used the “FindVariableFeatures (selection.method=“vst,” nfeatures = 1,500)” function in the Seurat R package and detected 1,500 HVGs. Among them, excluding genes with low expression levels [cluster average of log(ssUMI+1) ≦ 0.5: 712 genes], 788 genes were used for heatmap visualization and GO enrichment analysis (see below) (Fig EV4D, Dataset EV5). Cy granulosa cells from cultured rOvaries at 12/15 w‐ivc were also processed in the same manner.
SC3‐seq for cy/human germ cells
Dissociated germ cells from in vivo cy/human fetal ovaries and cy/human rOvaries were manually picked up using glass capillaries under a stereomicroscope (Leica, S8 APO). The following cDNA synthesis and amplification methods were previously described (Kurimoto et al, 2006, 2007; Nakamura et al, 2015, 2016). The quality of the amplified cDNA was measured by a SYBR real‐time PCR method targeting the PPIA, GAPDH, NANOG, DDX4, DAZL, SYCP3, SYCP1, ZP3, and FOXL2 genes. The primer sequences are listed in Table EV1. The SC3‐seq libraries were constructed as described previously (Nakamura et al, 2015, 2016; Okamoto et al, 2021), and the sequencing was performed on the Illumina NextSeq 500/550 platform with a NextSeq 500/550 high output kit v. 2 (75 cycles) (FC‐404‐2005; Illumina).
Bulk RNA‐seq analysis for mouse gonadal/ovarian cells
The mouse gonadal/ovarian cells were lysed and the total RNAs were purified using a NucleoSpin RNA XS (Takara Bio, U0902A) according to the manufacturer's instructions. 1 ng of total RNAs from each sample was used for the synthesis and amplification of cDNAs. The cDNA synthesis and library construction were as described previously (Nakamura et al, 2015; Ishikura et al, 2016; Okamoto et al, 2021). The sequencing was performed on the Illumina NextSeq 500/550 platform with a NextSeq 500/550 high output kit v. 2 (75 cycles) (FC‐404‐2005; Illumina).
Mapping reads of bulk RNA‐seq/SC3‐seq and conversion to gene expression levels
The genome sequence and transcript annotation (GRCm38.p6 for mice, GRCh38.p12 for humans, and MacFas5.0 for cynomolgus monkeys) were downloaded from the NCBI ftp site. Bulk RNA‐seq/SC3‐seq read only the 3′ end of the transcripts, so that the expression levels were calculated as genes (Entrez genes) but not mRNAs. Read trimming, mapping, and estimation of expression levels were performed as described previously (Ishikura et al, 2016; Nakamura et al, 2016).
Orthologous genes among three species
For the cross‐species comparison analyses for SC3‐seq and 10X scRNA‐seq data, we retrieved the lists of orthologous genes among three species from Ensembl BioMart (Ensembl 97: Jul 2019) as shown in Dataset EV9. One‐to‐one (1–1) orthologous genes between human and cy (16,734 genes) were used for the cross‐species integration of SC3‐seq data. One‐to‐one‐to‐one (1–1–1) orthologous genes across three species (15,007 genes) were used for the cross‐species integration of 10X data. As the orthologous gene list from Ensembl BioMart was relatively incomplete, we made a genomic coordinate comparison list by using the LiftOver tool as previously described (Sasaki et al, 2015; Nakamura et al, 2016) (Dataset EV9).
SC3‐seq data analysis for cy/human germ cells
The SC3‐seq data analysis was performed using the R software (v. 4.0.2). The read counts were normalized to 200,000 reads, and then log‐transformed (log2(normalized read counts+1)). We defined “all expressed genes” as genes whose log2(normalized read counts + 1) values were > 2 [greater than ∼ 10–20 copies per cell] in at least one sample (Nakamura et al, 2015). Unsupervised hierarchical clustering (UHC) was performed using the “hclust” function with Euclidean distances and Ward's method (ward.D2). The principal component analysis (PCA) was performed using the “prcomp” function without scaling.
For the cross‐species analysis between monkeys and humans, we used 1–1 orthologous genes between humans and monkeys, and processed data with the “Seurat” R package (v. 4.0.4). The cy SC3‐seq dataset was integrated with the human dataset using the Seurat CCA algorithm, that is, “FindIntegrationAnchors” (parameter “anchor.features” = 5,000) and “IntegrateData” functions with the default settings. Integrated germ cells were divided into four clusters using the Seurat “FindClusters” function (parameter “resolution” = 0.4). PCA was performed on the corrected count matrix obtained after adjusting for species differences. Gene expression levels were compared between two consecutive clusters, and DEGs were defined as those with greater than twofold change between clusters and an average gene expression level [log2(normalized read counts +1)] greater than 3 (Dataset EV6).
Gene ontology enrichment analysis
Gene ontology (GO) enrichment analysis for HVGs/DEGs was performed using the DAVID web tool (Huang da et al, 2009). As the annotation of Macaca fascicularis genes was relatively incomplete, human annotation corresponding to that of cynomolgus monkeys was used for GO analysis. Missing genes in the 1–1 orthologous gene list from Ensembl BioMart were complemented by the orthologous gene list produced with the LiftOver tool (Dataset EV9).
Single‐cell trajectory analysis for 10X/SC3‐seq datasets
The single‐cell trajectory analysis was performed using the “Monocle 3” R package (v. 0.2.1). We performed Moran's I statistical test using the “graph_test(neighbor_graph = “principal_graph”)” function to define the variable genes along the pseudotime trajectory.
Cross‐species analysis among three animal species using 10X public datasets
To perform the cross‐species analysis among humans, monkeys, and mice, we retrieved a 10X scRNA‐seq dataset for human in vivo fetal germ cells (Chitiashvili et al, 2020) and three datasets for mouse in vivo germ cells (Niu & Spradling, 2020; Zhao et al, 2020; Ge et al, 2021). Downloaded raw files were processed with the “mkfastq” command in Cell Ranger and mapped to the reference genome sequence (GRCm38.p6 for mice and GRCh38.p12 for humans). The low‐quality cells and putative doublets were excluded for downstream analyses. As the thresholds for these cells depend on the quality of each dataset (e.g., the reagent version and the reference genome used), the number of detected genes, UMIs, and mitochondrial genes in each dataset were finely assessed to define the thresholds (Dataset EV1). Each dataset was processed in the same manner as described above for the analysis of cy 10X scRNA‐seq data, and DDX4/Ddx4 + germ‐cell clusters were extracted for the following cross‐species comparison analysis. Our cy/human 10X scRNA‐seq datasets were integrated with the four public datasets. Each dataset was size‐scaled to 10,000 UMIs (ss10kUMI) and processed by using the CCA algorithm, that is, “FindIntegrationAnchors” (parameter “anchor.features” = 2,000) and “IntegrateData” functions in the “Seurat” R package (v. 3.2.2). PCA was performed on the corrected count matrix obtained after adjusting for species differences, and UMAP visualization was performed for 30 dimensions using the “RunUMAP” function. Integrated germ cells were divided into 15 clusters using the Seurat “FindClusters” function (parameter “resolution” = 0.8). Each cluster was defined by the expression pattern of key germ‐cell markers as “mitotic 1/2/3/4,” “pre‐leptotene 1/2/3,” “leptotene 1/2/3,” “zygotene 1/2,” “pachytene 1/2,” or “diplotene.” To confirm the accuracy of the merged clusters, we performed a pseudotime trajectory analysis on each dataset using the “Monocle 3” R package (v. 0.2.1). Among 1–1–1 orthologous genes complemented with the LiftOver tool, we obtained common HVGs (237 genes) across three datasets/species, and calculated Pearson's correlation coefficients of the average HVGs expression levels between the meiotic substages of the three animal species.
Conserved and primate‐specific variable genes during female meiosis
Among the DEGs identified by the cross‐species comparison analysis with SC3‐seq analysis (Dataset EV6) as described above, the commonly variable 1–1 orthologous genes were defined as the genes conserved across humans and monkeys. The clusters defined in the 10X cross‐species analysis were used to verify the average expression levels of these genes in 10X datasets from three animal species. Genes that varied identically in mice were selected manually as genes conserved among the three species (130 genes; Fig EV5A, Dataset EV6), and genes that showed no variation in expression in mice were selected as genes that varied specifically in primates (83 genes; Fig 6F; Dataset EV6).
Chromosomal expression ratio analysis
For bulk RNA‐seq/SC3‐seq data, the autosome (chromosome (chr.) 10 for humans, chr. 15 for monkeys, and chr. 3 for mice): total autosome ratio and the chr. X: total autosome ratio (X:A ratio) were calculated according to the method described previously (Okamoto et al, 2021). Briefly, 75th‐percentile log2(normalized read counts+1) values of the expressed genes [the maximum log2(normalized read counts + 1) values > 2, among the cells analyzed] on the autosomes and chr. X in individual samples/cells were calculated and used as representative expression values in the samples/cells. 19,307/811/728 genes on total autosome/Chr.10/Chr.X for humans, 20,576/849/841 genes on total autosome/Chr.15/Chr.X for monkeys, and 12,399/618/503 genes on total autosome/Chr.3/Chr.X for mice were used.
For 10X scRNA‐seq data, the average log2(ss10kUMI+1) values of all expressed genes, which were expressed in more than two cells, on the autosome (chr. 10 for humans, chr. 15 for monkeys, and chr. 3 for mice), all autosomes, and chr. X were used for the calculation of the autosome:A and X:A ratio. 17,190/682/725 genes on total autosome/Chr.10/Chr.X for humans (Chitiashvili et al, 2020), 18,066/736/791 genes on total autosome/Chr.15/Chr.X for monkeys, and 17,698/896/744 genes on total autosome/Chr.3/Chr.X for mice (Niu & Spradling, 2020) were used (see also Dataset EV8).
Reanalysis of the mouse bulk RNA‐seq by Sangrithi et al (2017)
The mouse RNA‐seq datasets by Sangrithi et al (2017), which were acquired by the Smart‐Seq2 platform, were mapped to GRCm38.p6 and reanalyzed using the bioinformatics pipeline for our bulk RNA‐seq datasets as described above. For chromosomal expression ratio analysis, 11,213/568/463 genes on total autosome/Chr.3/Chr.X were used.
Statistical analysis
All statistical analyses were performed using R software (v. 4.0.2). The Tukey–Kramer tests and T‐tests were performed with the “TukeyHSD” and “t.test” functions with the default settings.
Author contributions
Ken Mizuta: Conceptualization; data curation; formal analysis; funding acquisition; validation; investigation; visualization; methodology; writing – original draft; writing – review and editing. Yoshitaka Katou: Data curation; formal analysis; validation; investigation; methodology. Baku Nakakita: Data curation; investigation. Aoi Kishine: Data curation; investigation. Yoshiaki Nosaka: Data curation; investigation. Saki Saito: Data curation; investigation. Chizuru Iwatani: Data curation; investigation. Hideaki Tsuchiya: Data curation; investigation. Ikuo Kawamoto: Data curation; investigation. Masataka Nakaya: Data curation; investigation. Tomoyuki Tsukiyama: Data curation; investigation. Masahiro Nagano: Data curation; investigation. Yoji Kojima: Data curation; investigation. Tomonori Nakamura: Data curation; investigation. Yukihiro Yabuta: Data curation; investigation. Akihito Horie: Data curation; supervision; investigation. Masaki Mandai: Data curation; supervision; investigation. Hiroshi Ohta: Conceptualization; data curation; supervision; funding acquisition; validation; investigation; visualization; methodology; writing – original draft; project administration; writing – review and editing. Mitinori Saitou: Conceptualization; resources; supervision; funding acquisition; investigation; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
K.M., H.O., and M.S. are inventors on patent applications relating to ex vivo fetal ovary culture in humans and monkeys filed by Kyoto University. M.S. is an EMBO Associate Member.
Supporting information
Appendix S1
Expanded View Figures PDF
Table EV1
Dataset EV1
Dataset EV2
Dataset EV3
Dataset EV4
Dataset EV5
Dataset EV6
Dataset EV7
Dataset EV8
Dataset EV9
PDF+
Acknowledgments
We thank the members of our laboratory for their helpful input on this study, and Y. Nagai, N. Konishi, E. Tsutsumi, and M. Kawasaki of the Saitou Laboratory, S. Fujioka and M. Fujioka of the Ueno Laboratory, and T. Sato and M. Kabata of the Yamamoto Laboratory for their technical assistance. We are grateful to the animal care staff at the Research Center, Shiga University of Medical Science for their assistance with the animals, and to Single‐Cell Genome Information Analysis Core (SignAC) in ASHBi for the RNA sequence analysis. We also thank H. Watanabe, H. Ishikawa, and T. Yamaguchi of the Daigo Watanabe Clinic for their support with the human fetal tissue research. This work was supported by a Grant‐in‐Aid for JSPS Fellows from JSPS (19J22891), a Grant‐in‐Aid for Research Activity Start‐up from JSPS (21K20740) to K.M., a Grant‐in‐Aid for Scientific Research (B) from JSPS (19H03618) to H.O., and a Grant‐in‐Aid for Specially Promoted Research from JSPS (17H06098, 22H04920), a JST‐ERATO Grant (JPMJER1104), a Grant from HFSP (RGP0057/2018), and Grants from the Pythias Fund and Open Philanthropy Project (2018‐193685) to M.S.
The EMBO Journal (2022) 41: e110815.
Contributor Information
Hiroshi Ohta, Email: ohta@anat2.med.kyoto-u.ac.jp.
Mitinori Saitou, Email: saitou@anat2.med.kyoto-u.ac.jp.
Data availability
The accession numbers for the data in this study: GSE194266 (the GEO database, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE194266). The R and Python code to reproduce the analyses and figures are available on request.
References
- Baker TG (1963) A quantitative and cytological study of germ cells in human ovaries. Proc R Soc Lond B Biol Sci 158: 417–433 [DOI] [PubMed] [Google Scholar]
- Baker TG (1966) A quantitative and cytological study of oogenesis in the rhesus monkey. J Anat 100: 761–776 [PMC free article] [PubMed] [Google Scholar]
- Baltus AE, Menke DB, Hu YC, Goodheart ML, Carpenter AE, de Rooij DG, Page DC (2006) In germ cells of mouse embryonic ovaries, the decision to enter meiosis precedes premeiotic DNA replication. Nat Genet 38: 1430–1434 [DOI] [PubMed] [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ (2020) Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol 38: 1408–1414 [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N (1992) DMC1: A meiosis‐specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456 [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S (1992) The product of the mouse Xist gene is a 15 kb inactive X‐specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526 [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Baker TG (1985) Perinatal oocyte loss in XO mice and its implications for the aetiology of gonadal dysgenesis in XO women. J Reprod Fertil 75: 633–645 [DOI] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ et al (2019) The single‐cell transcriptional landscape of mammalian organogenesis. Nature 566: 496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr DH, Haggar RA, Hart AG (1968) Germ cells in the ovaries of XO female infants. Am J Clin Pathol 49: 521–526 [DOI] [PubMed] [Google Scholar]
- Chitiashvili T, Dror I, Kim R, Hsu FM, Chaudhari R, Pandolfi E, Chen D, Liebscher S, Schenke‐Layland K, Plath K et al (2020) Female human primordial germ cells display X‐chromosome dosage compensation despite the absence of X‐inactivation. Nat Cell Biol 22: 1436–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, Amati P, Bisceglia L, Zelante L, Nagaraja R, Porcu S et al (2001) The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet 27: 159–166 [DOI] [PubMed] [Google Scholar]
- Deng X, Berletch JB, Nguyen DK, Disteche CM (2014) X chromosome regulation: Diverse patterns in development, tissues and disease. Nat Rev Genet 15: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edson MA, Nagaraja AK, Matzuk MM (2009) The mammalian ovary from genesis to revelation. Endocr Rev 30: 624–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Capetillo O, Liebe B, Scherthan H, Nussenzweig A (2003) H2AX regulates meiotic telomere clustering. J Cell Biol 163: 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford CE, Jones KW, Polani PE, De Almeida JC, Briggs JH (1959) A sex‐chromosome anomaly in a case of gonadal dysgenesis (Turner's syndrome). Lancet 1: 711–713 [DOI] [PubMed] [Google Scholar]
- Galili T (2015) dendextend: an R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 31: 3718–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, Wang JJ, Zhang RQ, Tan SJ, Zhang FL, Liu WX, Li L, Sun XF, Cheng SF, Dyce PW et al (2021) Dissecting the initiation of female meiosis in the mouse at single‐cell resolution. Cell Mol Life Sci 78: 695–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J, Schwab U, Lemke H, Stein H (1983) Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer 31: 13–20 [DOI] [PubMed] [Google Scholar]
- Griswold MD (2016) Spermatogenesis: the commitment to meiosis. Physiol Rev 96: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, Schlesner M (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32: 2847–2849 [DOI] [PubMed] [Google Scholar]
- Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, Yong J, Hu Y, Wang X, Wei Y et al (2015) The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell 161: 1437–1452 [DOI] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, Marioni JC (2018) Batch effects in single‐cell RNA‐sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36: 421–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada N, Hamazaki N, Shimamoto S, Hikabe O, Nagamatsu G, Takada Y, Kato K, Hayashi K (2020) Germ cell‐intrinsic effects of sex chromosomes on early oocyte differentiation in mice. PLoS Genet 16: e1008676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M (2011) Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146: 519–532 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Ogushi S, Kurimoto K, Shimamoto S, Ohta H, Saitou M (2012) Offspring from oocytes derived from in vitro primordial germ cell‐like cells in mice. Science 338: 971–975 [DOI] [PubMed] [Google Scholar]
- Hikabe O, Hamazaki N, Nagamatsu G, Obata Y, Hirao Y, Hamada N, Shimamoto S, Imamura T, Nakashima K, Saitou M et al (2016) Reconstitution in vitro of the entire cycle of the mouse female germ line. Nature 539: 299–303 [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hwang YS, Suzuki S, Seita Y, Ito J, Sakata Y, Aso H, Sato K, Hermann BP, Sasaki K (2020) Reconstitution of prospermatogonial specification in vitro from human induced pluripotent stem cells. Nat Commun 11: 5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikawa M, Wada I, Kominami K, Watanabe D, Toshimori K, Nishimune Y, Okabe M (1997) The putative chaperone calmegin is required for sperm fertility. Nature 387: 607–611 [DOI] [PubMed] [Google Scholar]
- Imamura M, Aoi T, Tokumasu A, Mise N, Abe K, Yamanaka S, Noce T (2010) Induction of primordial germ cells from mouse induced pluripotent stem cells derived from adult hepatocytes. Mol Reprod Dev 77: 802–811 [DOI] [PubMed] [Google Scholar]
- Irie N, Weinberger L, Tang WW, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH, Surani MA (2015) SOX17 is a critical specifier of human primordial germ cell fate. Cell 160: 253–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikura Y, Yabuta Y, Ohta H, Hayashi K, Nakamura T, Okamoto I, Yamamoto T, Kurimoto K, Shirane K, Sasaki H et al (2016) In vitro derivation and propagation of Spermatogonial stem cell activity from mouse pluripotent stem cells. Cell Rep 17: 2789–2804 [DOI] [PubMed] [Google Scholar]
- Ishikura Y, Ohta H, Sato T, Murase Y, Yabuta Y, Kojima Y, Yamashiro C, Nakamura T, Yamamoto T, Ogawa T et al (2021) In vitro reconstitution of the whole male germ‐cell development from mouse pluripotent stem cells. Cell Stem Cell 28: 2167–2179.e9 [DOI] [PubMed] [Google Scholar]
- Jordan T, Hanson I, Zaletayev D, Hodgson S, Prosser J, Seawright A, Hastie N, van Heyningen V (1992) The human PAX6 gene is mutated in two patients with aniridia. Nat Genet 1: 328–332 [DOI] [PubMed] [Google Scholar]
- Josef B, Adam K (2003) Fast sex identification in wild mammals using PCR amplification of the Sry gene. Folia Zoologica 52: 269–274 [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Ono Y, Uno KD, Yamada RG, Ueda HR, Saitou M (2006) An improved single‐cell cDNA amplification method for efficient high‐density oligonucleotide microarray analysis. Nucleic Acids Res 34: e42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimoto K, Yabuta Y, Ohinata Y, Saitou M (2007) Global single‐cell cDNA amplification to provide a template for representative high‐density oligonucleotide microarray analysis. Nat Protoc 2: 739–752 [DOI] [PubMed] [Google Scholar]
- Lau X, Munusamy P, Ng MJ, Sangrithi M (2020) Single‐cell RNA sequencing of the cynomolgus macaque testis reveals conserved transcriptional profiles during mammalian spermatogenesis. Dev Cell 54:e7, 548, 566.e7 [DOI] [PubMed] [Google Scholar]
- Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC (1994) Cleavage of poly(ADP‐ribose) polymerase by a proteinase with properties like ICE. Nature 371: 346–347 [DOI] [PubMed] [Google Scholar]
- Li L, Dong J, Yan L, Yong J, Liu X, Hu Y, Fan X, Wu X, Guo H, Wang X et al (2017) Single‐cell RNA‐seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 20: 858–873.e4 [DOI] [PubMed] [Google Scholar]
- Liang L, Soyal SM, Dean J (1997) FIGalpha, a germ cell specific transcription factor involved in the coordinate expression of the zona pellucida genes. Development 124: 4939–4947 [DOI] [PubMed] [Google Scholar]
- Lun ATL, McCarthy DJ, Marioni JC (2016) A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5: 2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF (1961) Gene action in the X‐chromosome of the mouse (Mus musculus L.). Nature 190: 372–373 [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R (1997) Xist‐deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 11: 156–166 [DOI] [PubMed] [Google Scholar]
- McGee EA, Hsueh AJ (2000) Initial and cyclic recruitment of ovarian follicles. Endocr Rev 21: 200–214 [DOI] [PubMed] [Google Scholar]
- Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K (2012) Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell 10: 595–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen RL, Offenberg HH, Dietrich AJ, Riesewijk A, van Iersel M, Heyting C (1992) A coiled‐coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J 11: 5091–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi H, Ohta H, Nagaoka S, Nakaki F, Sasaki K, Hayashi K, Yabuta Y, Nakamura T, Yamamoto T, Saitou M (2017) Bone morphogenetic protein and retinoic acid synergistically specify female germ‐cell fate in mice. EMBO J 36: 3100–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M, McLaren A (1981) X‐chromosome activity in foetal germ cells of the mouse. J Embryol Exp Morphol 63: 75–84 [PubMed] [Google Scholar]
- Morohaku K, Tanimoto R, Sasaki K, Kawahara‐Miki R, Kono T, Hayashi K, Hirao Y, Obata Y (2016) Complete in vitro generation of fertile oocytes from mouse primordial germ cells. Proc Natl Acad Sci U S A 113: 9021–9026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamatsu G, Shimamoto S, Hamazaki N, Nishimura Y, Hayashi K (2019) Mechanical stress accompanied with nuclear rotation is involved in the dormant state of mouse oocytes. Sci Adv 5: eaav9960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka SI, Nakaki F, Miyauchi H, Nosaka Y, Yabuta Y, Kurimoto K, Hayashi K, Nakamura T, Yamamoto T, Saitou M (2020) ZGLP1 is a determinant for the oogenic fate in mice. Science 367: eaaw4115 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Yabuta Y, Okamoto I, Aramaki S, Yokobayashi S, Kurimoto K, Sekiguchi K, Nakagawa M, Yamamoto T, Saitou M (2015) SC3‐seq: A method for highly parallel and quantitative measurement of single‐cell gene expression. Nucleic Acids Res 43: e60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Okamoto I, Sasaki K, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Yamamoto T, Saitou M (2016) A developmental coordinate of pluripotency among mice, monkeys and humans. Nature 537: 57–62 [DOI] [PubMed] [Google Scholar]
- Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Muller FJ, Wang YC, Boscolo FS et al (2012) Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell 10: 620–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Spradling AC (2020) Two distinct pathways of pregranulosa cell differentiation support follicle formation in the mouse ovary. Proc Natl Acad Sci U S A 117: 20015–20026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S (1967) Sex chromosomes and sex‐linked genes. Berlin: Springer‐Verlag; [Google Scholar]
- Okamoto I, Nakamura T, Sasaki K, Yabuta Y, Iwatani C, Tsuchiya H, Nakamura SI, Ema M, Yamamoto T, Saitou M (2021) The X chromosome dosage compensation program during the development of cynomolgus monkeys. Science 374: eabd8887 [DOI] [PubMed] [Google Scholar]
- Patrat C, Ouimette JF, Rougeulle C (2020) X chromosome inactivation in human development. Development 147: dev183095 [DOI] [PubMed] [Google Scholar]
- Payer B, de Sousa C, Lopes SM, Barton SC, Lee C, Saitou M, Surani MA (2006) Generation of stella‐GFP transgenic mice: A novel tool to study germ cell development. Genesis 44: 75–83 [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N (1996) Requirement for Xist in X chromosome inactivation. Nature 379: 131–137 [DOI] [PubMed] [Google Scholar]
- Saitou M, Hayashi K (2021) Mammalian in vitro gametogenesis. Science 374: eaaz6830 [DOI] [PubMed] [Google Scholar]
- Sangrithi MN, Royo H, Mahadevaiah SK, Ojarikre O, Bhaw L, Sesay A, Peters AH, Stadler M, Turner JM (2017) Non‐canonical and sexually dimorphic X dosage compensation states in the mouse and human germline. Dev Cell 40:e3, 289, 301.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Yokobayashi S, Nakamura T, Okamoto I, Yabuta Y, Kurimoto K, Ohta H, Moritoki Y, Iwatani C, Tsuchiya H et al (2015) Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell 17: 178–194 [DOI] [PubMed] [Google Scholar]
- Sasaki K, Nakamura T, Okamoto I, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Shiraki N, Takakuwa T et al (2016) The germ cell fate of cynomolgus monkeys is specified in the nascent amnion. Dev Cell 39: 169–185 [DOI] [PubMed] [Google Scholar]
- Sasaki K, Oguchi A, Cheng K, Murakawa Y, Okamoto I, Ohta H, Yabuta Y, Iwatani C, Tsuchiya H, Yamamoto T et al (2021) The embryonic ontogeny of the gonadal somatic cells in mice and monkeys. Cell Rep 35: 109075 [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, Regev A (2015) Spatial reconstruction of single‐cell gene expression data. Nat Biotechnol 33: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M (2007) Cellular dynamics associated with the genome‐wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134: 2627–2638 [DOI] [PubMed] [Google Scholar]
- Shimada R, Koike H, Hirano T, Kato Y, Saga Y (2021) NANOS2 suppresses the cell cycle by repressing mTORC1 activators in embryonic male germ cells. iScience 24: 102890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto S, Nishimura Y, Nagamatsu G, Hamada N, Kita H, Hikabe O, Hamazaki N, Hayashi K (2019) Hypoxia induces the dormant state in oocytes through expression of Foxo3. Proc Natl Acad Sci U S A 116: 12321–12326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RP, Carr DH (1966) The anatomy and histology of XO human embryos and fetuses. Anat Rec 155: 369–383 [DOI] [PubMed] [Google Scholar]
- Spiller C, Koopman P, Bowles J (2017) Sex determination in the mammalian germline. Annu Rev Genet 51: 265–285 [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R (2019) Comprehensive integration of single‐cell data. Cell 177: 1888–1902.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M, Abe K (2007) X chromosome reactivation initiates in nascent primordial germ cells in mice. PLoS Genet 3: e116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh EK, Yang A, Kettenbach A, Bamberger C, Michaelis AH, Zhu Z, Elvin JA, Bronson RT, Crum CP, McKeon F (2006) p63 protects the female germ line during meiotic arrest. Nature 444: 624–628 [DOI] [PubMed] [Google Scholar]
- Suzumori N, Yan C, Matzuk MM, Rajkovic A (2002) Nobox is a homeobox‐encoding gene preferentially expressed in primordial and growing oocytes. Mech Dev 111: 137–141 [DOI] [PubMed] [Google Scholar]
- Tang WW, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, Surani MA (2015) A unique gene regulatory network resets the human germline epigenome for development. Cell 161: 1453–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WW, Kobayashi T, Irie N, Dietmann S, Surani MA (2016) Specification and epigenetic programming of the human germ line. Nat Rev Genet 17: 585–600 [DOI] [PubMed] [Google Scholar]
- Tewari M, Quan LT, O'Rourke K, Desnoyers S, Zeng Z, Beidler DR, Poirier GG, Salvesen GS, Dixit VM (1995) Yama/CPP32 beta, a mammalian homolog of CED‐3, is a CrmA‐inhibitable protease that cleaves the death substrate poly(ADP‐ribose) polymerase. Cell 81: 801–809 [DOI] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G et al (2016) Dissecting the multicellular ecosystem of metastatic melanoma by single‐cell RNA‐seq. Science 352: 189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, Dean J, Nelson LM (2000) Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet 26: 267–268 [DOI] [PubMed] [Google Scholar]
- Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T (2000) Expression and intracellular localization of mouse vasa‐homologue protein during germ cell development. Mech Dev 93: 139–149 [DOI] [PubMed] [Google Scholar]
- Vanorny DA, Mayo KE (2017) The role of notch signaling in the mammalian ovary. Reproduction 153: R187–R204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertesy A, Arindrarto W, Roost MS, Reinius B, Torrens‐Juaneda V, Bialecka M, Moustakas I, Ariyurek Y, Kuijk E, Mei H et al (2018) Parental haplotype‐specific single‐cell transcriptomics reveal incomplete epigenetic reprogramming in human female germ cells. Nat Commun 9: 1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villesen P, Fredsted T (2006) Fast and non‐invasive PCR sexing of primates: apes, Old World monkeys, New World monkeys and strepsirrhines. BMC Ecol 6: 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther C, Gruss P (1991) Pax‐6, a murine paired box gene, is expressed in the developing CNS. Development 113: 1435–1449 [DOI] [PubMed] [Google Scholar]
- Wen L, Tang F (2019) Human germline cell development: From the perspective of single‐cell sequencing. Mol Cell 76: 320–328 [DOI] [PubMed] [Google Scholar]
- Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Gottgens B, Rajewsky N, Simon L, Theis FJ (2019) PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol 20: 59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock SL, Lopez R, Klein AM (2019) Scrublet: computational identification of cell doublets in single‐cell transcriptomic data. Cell Syst 8: 281–291.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki J, Iwatani C, Tsuchiya H, Okahara J, Sankai T, Torii R (2011) Vitrification and transfer of cynomolgus monkey (Macaca fascicularis) embryos fertilized by intracytoplasmic sperm injection. Theriogenology 76: 33–38 [DOI] [PubMed] [Google Scholar]
- Yamashiro C, Sasaki K, Yabuta Y, Kojima Y, Nakamura T, Okamoto I, Yokobayashi S, Murase Y, Ishikura Y, Shirane K et al (2018) Generation of human oogonia from induced pluripotent stem cells in vitro . Science 362: 356–360 [DOI] [PubMed] [Google Scholar]
- Yamashiro C, Sasaki K, Yokobayashi S, Kojima Y, Saitou M (2020) Generation of human oogonia from induced pluripotent stem cells in culture. Nat Protoc 15: 1560–1583 [DOI] [PubMed] [Google Scholar]
- Yasmin L, Takano J, Nagai Y, Otsuki J, Sankai T (2015) Detection and quantification of male‐specific fetal DNA in the serum of pregnant cynomolgus monkeys (Macaca fascicularis). Comp Med 65: 70–76 [PMC free article] [PubMed] [Google Scholar]
- Yoshino T, Suzuki T, Nagamatsu G, Yabukami H, Ikegaya M, Kishima M, Kita H, Imamura T, Nakashima K, Nishinakamura R et al (2021) Generation of ovarian follicles from mouse pluripotent stem cells. Science 373: eabe0237 [DOI] [PubMed] [Google Scholar]
- Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Hoog C (2000) The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell 5: 73–83 [DOI] [PubMed] [Google Scholar]
- Yuan L, Liu JG, Hoja MR, Wilbertz J, Nordqvist K, Hoog C (2002) Female germ cell aneuploidy and embryo death in mice lacking the meiosis‐specific protein SCP3. Science 296: 1115–1118 [DOI] [PubMed] [Google Scholar]
- Yuen M, Sandaradura SA, Dowling JJ, Kostyukova AS, Moroz N, Quinlan KG, Lehtokari VL, Ravenscroft G, Todd EJ, Ceyhan‐Birsoy O et al (2014) Leiomodin‐3 dysfunction results in thin filament disorganization and nemaline myopathy. J Clin Invest 124: 4693–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yan Z, Qin Q, Nisenblat V, Chang HM, Yu Y, Wang T, Lu C, Yang M, Yang S et al (2018) Transcriptome landscape of human Folliculogenesis reveals oocyte and granulosa cell interactions. Mol Cell 72: 1021–1034.e4 [DOI] [PubMed] [Google Scholar]
- Zhao ZH, Ma JY, Meng TG, Wang ZB, Yue W, Zhou Q, Li S, Feng X, Hou Y, Schatten H et al (2020) Single‐cell RNA sequencing reveals the landscape of early female germ cell development. FASEB J 34: 12634–12645 [DOI] [PubMed] [Google Scholar]
- Zylicz JJ, Heard E (2020) Molecular mechanisms of facultative heterochromatin formation: an X‐chromosome perspective. Annu Rev Biochem 89: 255–282 [DOI] [PubMed] [Google Scholar]


