Abstract
Competence for genetic transformation in Streptococcus pneumoniae is coordinated by the competence-stimulating peptide (CSP), which induces a sudden and transient appearance of competence during exponential growth in vitro. Models of this quorum-sensing mechanism have proposed sequential expression of several regulatory genes followed by induction of target genes encoding DNA-processing-pathway proteins. Although many genes required for transformation are known to be expressed only in response to CSP, the relative timing of their expression has not been established. Overlapping expression patterns for the genes cinA and comD (G. Alloing, B. Martin, C. Granadel, and J. P. Claverys, Mol. Microbiol. 29:75–83, 1998) suggest that at least two distinct regulatory mechanisms may underlie the competence cycle. DNA microarrays were used to estimate mRNA levels for all known competence operons during induction of competence by CSP. The known competence regulatory operons, comAB, comCDE, and comX, exhibited a low or zero initial (uninduced) signal, strongly increased expression during the period between 5 and 12 min after CSP addition, and a decrease nearly to original values by 15 min after initiation of exposure to CSP. The remaining competence genes displayed a similar expression pattern, but with an additional delay of approximately 5 min. In a mutant defective in ComX, which may act as an alternate sigma factor to allow expression of the target competence genes, the same regulatory genes were induced, but the other competence genes were not. Finally, examination of the expression of 60 candidate sites not previously associated with competence identified eight additional loci that could be induced by CSP.
Competence for genetic transformation in Streptococcus pneumoniae depends on a system of coordinated gene regulation of significant complexity. The full extent of this regulation has not been determined, but the interactions and functions of all its components are beginning to be discerned. As currently understood, the number of genes involved (20 or more) and the number of different levels of control (two or more) are few enough that one might expect to describe the system completely. Yet, the complexity of the system is substantial enough that the gene-by-gene discovery and analysis approach is not entirely satisfactory. Thus, it represents a suitable target for exploiting the power of new methods for genome scale analysis of gene expression.
A central element in the regulation of pneumococcal competence is the competence-stimulating peptide (CSP), a pheromone coordinating the sudden and transient appearance of competence at some point during the exponential growth phase in vitro (8). Models of this quorum-sensing-dependent regulation of competence envision sequential expression of several regulatory genes, followed by that of a set of regulated genes encoding proteins of the DNA-processing pathway (3, 5, 9). Although many genes required for transformation are known to be expressed only during induction of competence, the extent to which such sequential activity is reflected in mRNA abundance changes has not been established. Current knowledge of competence gene regulation stems from several experimental methods, including pulse labeling, reporter gene fusion, Northern and Western blotting, oligonucleotide arrays, and genetic analysis. Metabolic pulse labeling of cellular proteins revealed (14) that an apparent global protein synthesis switch accompanies competence induction; two-dimensional polyacrylamide gel electrophoresis visualized at least 16 proteins from competent cells that were absent from noncompetent cultures (13). Additional relevant data has come from studies of transcriptional reporter gene fusions, which established that several genes are expressed specifically in competent cells, but detailed kinetics have been reported for only a few cases. The genes that have been shown to be expressed specifically during competence induction include celB, cflA, cglA, cglE, cinA, coiA, comA, comC, and comX (3, 11, 16, 19, 20). The hallmark of these genes is a very low basal expression state followed by a relatively large increase in expression during competence induction. The degree of expression change reported varies. For example, Campbell et al. (5) reported that cilA (ssbB), cilD (cglA), cilE (celA), and recA undergo inductions of 4- to 10-fold, while others (2, 11, 20) reported expression increases of more than 50-fold apparently for the same genes. Kinetic profiles have been reported for six genes by using a lacZ reporter; for all six, cinA (2), comC (3), comX (11), cgl, cel, and coi (20), β-galactosidase activity appeared a few (∼5 to 10) minutes after CSP treatment, accumulated during the ensuing ∼10 min while competence increased, and leveled off as competence reached a maximum. The reporter gene strategy is very sensitive to the onset of gene expression but is of limited value in establishing sequential expression or gene silencing unless a very unstable reporter is employed.
The first clear indication of sequential gene expression in competence regulation was a report of temporally distinct expression patterns for the genes cinA and comD (3). In Northern blots, the comD message rose from an undetectable level in uninduced cells to a maximum 5 min after CSP treatment and declined rapidly thereafter; in contrast, in the same cells, the cinA message reached its maximum at 10 min and declined soon thereafter. This differential timing suggested that at least two distinct expression patterns underlie the competence cycle. Genetic data and examination of apparent promoter structures have also indicated that competence genes are organized into at least two distinct regulons (11). The quorum-sensing genes, comABCDEX, depend on comE for activation but not on comX and apparently have canonical promoters. A larger set of genes, including many involved in DNA processing during transformation, depend on comE but also on comX for expression and have noncanonical promoters characterized by a conserved 8-bp −10 sequence, TACGAATA, termed the cinbox or combox (5, 20). Although the former have sometimes been conceptually classed as early genes and the latter as late genes, reflecting the dependence of the expression of the latter on the regulatory activities of the former, as mentioned above, experimental demonstration of a separation in the time of expression of these two sets of genes has been limited to very few specific cases.
Protein expression has been monitored by using antibody probes for the products of two competence genes. ComE is present at low levels in uninduced cells, increases strongly upon CSP treatment, and persists for at least one generation time after competence is lost (23), despite the early disappearance of the comE message mentioned above. A study of CSP secretion showed that, as expected, the level of this product of the comC gene rises dramatically in culture fluids soon after competence induction (9). In another report, oligonucleotide arrays were used to assay the expression of several competence genes, demonstrating that RNA levels for these genes were much higher in competent cells than in noncompetent cells (7). Finally, genetic analysis has shown that most of the competence-specific genes mentioned above are essential for the processes of genetic transformation, making the important linkage of expression data to biological significance.
In sum, these studies provide a picture of a system of genes tightly regulated for competence, with the final outcome of the CSP response a synchronous but brief synthesis of a new set of proteins. In general, however, the data available are of insufficient resolution to reveal the kinetics and temporal sequence of gene activation during the CSP response and of insufficient breadth to reveal the full set of genes involved.
High-density DNA microarrays have now been applied to a wide variety of systems to monitor gene expression in whole genomes, to detect DNA sequence polymorphisms, and to do comparative genomics (4, 6, 12, 21). In this paper, we describe the use of DNA microarrays to estimate the evolution of mRNA levels for all known competence operons during induction of competence in a culture exposed to CSP, show that several different expression patterns can be distinguished, and identify eight new CSP-inducible loci. The results illustrate the power of hybridization array technology as a quantitative tool to dissect gene expression patterns and to scan for new regulated genes.
MATERIALS AND METHODS
Bacterial strains, media, and DNA.
The pneumococcal recipient strains used in this study were CP1250 (hex bgl-1 str-r1 malM511) (20), CPM4 [hex bgl-1 str-r1 malM511 ΔcomX1::PcEm comX2′::pEVP3)::′comX2] (11), and CPM16 [hex bgl-1 str-r1 malM511 comX2′::pEVP3)::′comX2+] (11). A casein hydrolysate yeast extract medium (CAT) was used for both cell culture and transformation assays as described previously (11). Donor Novr DNA was from strain CP1500 (11).
Competence induction and RNA isolation.
Cells were grown to an optical density (550 nm) of 0.04 in CAT adjusted to 0.2% bovine serum albumin, 10 mM HCl, and 0.5 mM CaCl2, as described previously (11). The cultures were then split: the induced portion was supplemented with CSP to 200 ng/ml, while the control was untreated. CSP-1 (8) was obtained from Chiron Mimitopes (Raleigh, N.C.). Successive 100-ml culture samples were taken from the induced portion 1 min before (zero minute) and at specified intervals after the addition of CSP. For RNA isolation, the samples were mixed directly with 100 ml of hot acid phenol containing 0.1 M citrate buffer (pH 4.3) and 0.1% sodium dodecyl sulfate (SDS) and were mixed intermittently while they were standing in a boiling-water bath for 10 min. After being cooled in ice water and centrifuged at 8,000 × g for 20 min in the cold, the supernatants were extracted with 1 volume of acid phenol-chloroform (1:1) and then with chloroform at room temperature, with centrifugation for 15 min at 8,000 × g to separate phases. The final supernatant was adjusted to 0.3 M sodium acetate and mixed with 1 volume of isopropanol. After 20 min at 0°C, RNA was pelleted at 8,000 × g at 10°C for 15 min. The pellet was washed with 70% ethanol, dried, and dissolved in 5 ml of H2O. After the precipitation and washing steps were repeated, the pellets were finally dissolved in 500 μl of H2O and stored at −20°C. These samples were then treated with DNase I and purified over RNeasy columns (Qiagen) according to protocols supplied by the manufacturer. The typical yield, quantitated by measuring absorbance at 260 nm, was at least 200 μg of RNA.
Competence and LacZ assays.
Culture samples were exposed to 5 μg of Novr DNA/ml at 37°C for 5 min. After termination with DNase (50 μg/ml), incubation was continued for 40 min before plating. Appropriately diluted samples were plated to determine Novr recombinants as described previously (15). β-Galactosidase assays were done essentially as described previously (20) but using a direct lysis procedure in which culture samples were supplemented with 0.025% Triton X-100 and 10 mM EDTA, held at 37°C for 10 min, and then kept at 0°C until assay.
Open reading frame (ORF) amplification.
Genomic DNA from the type 4 strain of S. pneumoniae being sequenced at The Institute for Genomic Research (TIGR) (1) was used as a template. Oligonucleotide pairs (melting temperature, 55°C) were designed to represent internal portions of genes where possible so that the expected product was between 100 and 1,200 bp. Gene details are given in Table 1. Twenty nanograms of genomic DNA was used as a template for PCR using Perkin-Elmer Taq DNA polymerase in a total volume of 100 μl in 96-well microtiter plates; 0.2 μmol of each gene-specific primer/liter was used. The entire mixture was amplified for 35 PCR cycles as follows: 1 min at 94°C, 1 min at 55°C, 1 min at 72°C. Products were purified on Millipore MAFB NOB 96-well purification plates and eluted with 60 μl of MilliQ water. PCR products were assayed by agarose gel electrophoresis. The synthesis was repeated if the product was not a unique band of the expected size or was less than 10 μg.
TABLE 1.
Summary of DNA microarray target elements and gene expression patterns
| Gene namea | Combox positionb | PCR probe primersc
|
Amplicon size (bp) | Expression classd | Expression rangee | |
|---|---|---|---|---|---|---|
| Left | Right | |||||
| cclA | 164 | CACAGGTCTTCAATCGCTTTC | TCCGTTACGCTAAAGACGAGA | 361 | 3 | 50 |
| celA | 407 | GGAACGTAAACCAGAGCCTCA | CCAGCTCCACAAACACCTGTC | 296 | 3 | 50 |
| celA* | 77 | AATCATCGTCATCTGTACTGGTCT | AAAGTAATAGAAAACTCAGGTAAA | 660 | 3 | 14 |
| celA** | 642 | AGCTTAAAGACTATGTTACAGTGGATTAAG | GACAAGCACCGTCAAGGCAAAATTATCCAA | 960 | 3 | 250 |
| cflA | 544 | TCTGCTAACTGCTCCCCTTTC | TTTCGAACACCACTAGTTGTTGC | 419 | 3 | 330 |
| cflA* | 456 | CATAGATGTTTGTTTGGAGCTGTA | TCATAGACCAGCCTCCTTATTCAT | 850 | 3 | 140 |
| cglA | 171 | GTCGTCTTACCACTCCCAACC | TTGCAGCCGTTATCAGTCAC | 286 | 3 | 14 |
| cglA* | 71 | GCAGGATATCTATTTTGTCCCTAA | CCTCGGATACTCTTGGCGTGAATG | 660 | 3 | 200 |
| ciaH | NC | ACATGGTATGCGGATGACTTTAGT | CTCATGACTGGCATTTTCCACAAA | 660 | 1 | 1.8 |
| cinA | 316 | TATCTTTTTTGCCCTGCGACC | GCCTCTTCTTGACTGCTAGCC | 387 | 3 | 25 |
| coiA | 70 | ATAAACTTGAGAAGCAAGCATACA | TTTGATAATAAAGTTGTTGCCGGA | 660 | 3 | 59 |
| comA | NC | TGACTAAACTGCCACGTGAGC | TCGCAAAGAAGGACATAGGGA | 420 | 2 | 170 |
| comA* | NC | TATCGTCCGCAAGTGGATCAGATG | CTCCTGAGCGTAAGACAAGATTTG | 660 | 2 | 250 |
| comCD | NC | ATGAAAAACACAGTTAAATTGGAA | CCGTCACAACGAAAAAGAATGGGA | 531 | 2 | 10 |
| comD | NC | CCTTCTTTGCAATGAATTCTCATA | AGTTCAATTGGAAGCTTGGTAATC | 400 | 2 | 62 |
| comE | NC | GCTCAGCTCATTCGTCATTACAA | TGAGGAGAATAAAATCGCTGAGT | 400 | 2 | 200 |
| comX | NC | GGACTGGTAGACGATATTCCACG | TGAAAGAGATAATAATCATCTAGCCA | 191 | 2 | 170 |
| comX* | NC | ACCCTGAGAGAGGCTGGAGCCTCT | CTAATGGGTACGGATAGTAAACTC | 600 | 2 | 140 |
| dalA | 215 | AGAGGTTGTTCACCAGGTCCA | TGCGCATTTGTCGAAAGAGTT | 420 | 3 | 500 |
| dalA* | 215 | AGAGGTTGTTCACCAGGTCCA | GTAGAAAAAAGAAATGGAGTTATT | 660 | 3 | 110 |
| dnaK | NC | CTGACAAGATGGCAATGCAAC | CCGCACCCATAGCAACTACTT | 366 | 1 | 3.3 |
| endA | NC | CTACAGGGAGCTATTATATCAAGC | TCTGTGAAGCTGAGGGAACTAAAT | 660 | 1 | 1.7 |
| ftsH | NC | CCAGCACTATTCTTCCTGCAGAT | AGAACACCTGCTGGAATACGG | 376 | 1 | 1.5 |
| gyrA | NC | GCTGCCGCTCAACGTTATACC | GGATACCTGATTTCCCCATGA | 340 | 1 | 1.8 |
| lytA | 4798 | GCGGTTGAACTGATTGAAAGC | CGGTCTGCAAGCATATAGCC | 404 | 3 | 25 |
| mmsA | NC | TGCAGTTGCAGACGCTCAAGT | CTCACCCTTGGCAATGGTCTC | 410 | 1 | 2 |
| ply | NC | GGTCGCAACTACATTGTCACG | AAAATCCAGGAGATGTGTTTCA | 407 | 1 | 2.5 |
| recA | 1516 | CCTTGGCTCAGGTGGTTATCC | TTACGCATGGCCTGGCTCATC | 381 | 3 | 33 |
| recP | NC | TACTGGTTTTGCCCAAGCAGA | CGCCGTGTACAGCATTAGTTC | 412 | 1 | 3 |
| rpoC | NC | CCAATACCGACAAACCTTCGC | AACCTCGGTTACCACCACTCA | 391 | 1 | 1.8 |
| rpoD | NC | GTTTCCATTGCCAAACGCTAT | CTCACGCAAGACGATACGAGT | 465 | 3 | 7 |
| rpsF | NC | TATCATTCGTCCAAACATTGAAG | GTCAATTTTGACGATCATGTGAC | 262 | 1 | 1.5 |
| rRNA | NC | ATAGCCGACCTGAGAGGGTGA | TACAAGCCAGAGAGCCGCTTT | 463 | 1 | 1.1 |
| ssbA | NC | GCGTTATACCCCATCAAATGTAG | TGCTCCAAATGGATTTTCATTAC | 379 | 1 | 1.4 |
| ssbB | 21 | ATGTATAATAAAGTTATCTTGATT | TAATTCTTAAAATGGCAATTCTTC | 402 | 3 | 330 |
| tnpA | NC | AATCTCAGGAAGACGCGAAA | CAAGGAAATCATCGCCAAAC | 373 | 1 | 4.3 |
| tnpB | NC | TGAAAAAGAGGATGAACCTGCT | AGACAACTTTTCCCGTGTGCT | 380 | 1 | 2.4 |
| ccs1 | −8 | TCCGAATATAAAAGTGAACAA | TAACCAGCTGCCAAACCAGAA | 208 | 3 | 330 |
| ccs3 | 112 | ATCCTTGACAGCACTCGCACCGTAGAGACT | GAATGAATCTCTTGACGACGCACTTCGCCG | 949 | 1 | 1.7 |
| ccs4 | 18 | AATTCATGCTAACTAAGGAAGAAGTGAATG | AAAGCGAATACTGTCAATAGGATAACGAAT | 992 | 3 | 25 |
| ccs5 | 17 | GACAACAGAGCTAGGCACAGCAGTTGCGAG | CTAGCTCTTATAACAGGTTTGCTGAGTGCC | 800 | 1 | 1.6 |
| ccs10 | 0 | AGTCAGGCTGACGATATCCGTCCCACCACT | ACAGCGCTTGTTCTTGAAGATAAGGACTTG | 1,132 | 1 | 2 |
| ccs12 | 89 | CGTATAGGGTGGATCATCGTAAATTGTTTG | CTTTGTCTCAATACTCATGGTCTGCCTCGC | 1,000 | 3 | 33 |
| ccs15 | 84 | ATGAATTATCAGATATTGATGGCGGTCTCG | ACGGCCCTCAAGTTCTGAGCTGGCATCGAT | 1,230 | 3 | 500 |
| ccs16 | 137 | CATCTTATTTCAACTCACTATAGAAGGAGG | TTACAACAGTGCCTCAAATTCAGATACTGA | 955 | 3 | 100 |
| ccs18 | 434 | CTTGGCCAGTTGATTCGAGTTTGGTGATTT | TGTCATAAAACTACCTTCCGACCGCGAAAG | 952 | 1 | 1.1 |
| ccs19 | 106 | AAGCGTGAGACAGCGCATGCGGCTGGTTAC | CTATATCTCTAACTCACACTCAATCACTTG | 903 | 3 | 25 |
| ccs20 | 0 | GAAGAATATTACGAATATGGTCAACAACGT | CATAACCGCTCGCTGTAATTCCTCACGCGC | 1,100 | 3 | 4 |
| ccs23 | 209 | GGCTTTGAGTTTTATGATTGTTTCTTAGGT | GTCGCTCGTCATCGTCTCTTCGTAAGTCAT | 913 | 1 | 3.3 |
| ccs25 | 619 | ATAGACTTGTTTGCGCGTGGTGAGCCTCTG | CTAAATCCAATGAATCACAATGTCTCGCTT | 1,040 | 1 | 1.9 |
| ccs28 | 384 | CTGCACGCGCATCTGCTTAAAAGTAATGTC | CATGGAGTACGCGGAAACGAACGTAGTATT | 1,043 | 1 | 1.9 |
| ccs33 | 309 | GTTATATCTATTTTCATGGAATCACCTCAC | CTAGGTACTTGGTACTTCCCAGAAGCCGCT | 610 | 1 | 1.7 |
| ccs35 | 74 | AGAGGAACAAGTTATTACTTGAAGATGTCA | ACTATGGGCTCCATAGCTAGAGATGTTTCC | 1,020 | 3 | 1,000 |
| ccs36 | −46 | CAGCACCAATCATCAGCGAGCAGGCTCCTA | AATTAGATTGCCGGCGTAGAGGTTTTGTAA | 1,000 | 1 | 2.8 |
| ccs38 | 208 | CATTTAGAAATTGAATTGAAAACACTATTG | CATACTTTTGACAAATCGAACCAATTTTGA | 552 | 3 | 33 |
| ccs46 | 133 | GGATTGATGCTAGCAGCTGGTGATAGTGTC | ACTATGGGATCCATAGCTAGAGATGTTTCC | 960 | 3 | 250 |
| ccs50 | 39 | ATGTGGTCGTACTGCATAGCAAGGACAGGA | GGCTGATAGGGTACTTGGTCCTGCAACAAT | 1,170 | 3 | 8.3 |
| ccs56 | 83 | ATATTTCGTATAGGGTGGATCATCGTAAAT | TCAGATTGATATTCACTTACTCACAACAGA | 707 | 3 | 125 |
| ccs61 | 228 | GGATGAATAAAGGGATTTATCAGCATTTCT | CAATCAACTGGTTTGCTTGATTCCTAGATA | 615 | 1 | 1 |
| ccs62 | 147 | AACCAAGTTGTTCGTAGTCATTCTTGG | TAATTTTGATTTAGAGGAGAGTCGCCCGTA | 624 | 1 | 4 |
Candidate sites and recognized gene loci drawn from the TIGR 9 March 2000 release of S. pneumoniae genome data. The names assigned are from the literature, from strongly conserved homologues in Escherichia coli or Bacillus subtilis, or a serial number among candidate combox sites (ccsn). Candidates and amplicons for which no expression was detected are omitted. In cases where two or three amplicons were made for a single locus, they are distinguished by an asterisk and a double asterisk.
Number of base pairs between candidate combox and first probe primer; NC, not a combox gene. ccs1 and ccs15 are at the same candidate combox site. ccs35 and ccs46 are at the same candidate combox site. ccs12 and ccs56 contain insertion sequence-like repeated sequences.
Primers used to amplify target DNA for microarray elements.
Expression pattern after CSP treatment: 1, constant; 2, early induction; 3, late induction.
Expression range, calculated from CSP response data as maximal signal/minimal signal.
Arraying procedure and postprocessing.
PCR products (in 50% dimethylsulfoxide, 20 mM Tris-HCl, and 50 mM KCl, pH 6.5) were deposited onto 25- by 75-mm glass microscope slides (CMT-GAPS amino silane-coated slides; Corning) using a Molecular Dynamics Generation II array spotter. The humidity was maintained at ∼50% during printing. After being printed, the slides were air dried for 30 min and baked for 2 h at 80°C before the DNA was cross-linked to the surface by short-wavelength UV using a Stratagene Stratalinker and then were stored in a dessicator. For use, the slides were soaked for 2 h at 42°C in 50 ml of 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% SDS, and 1.0% bovine serum albumin, washed four times in MilliQ water and three times in isopropanol, and dried before hybridization probes were applied.
Three microarray designs were used in this work. A 40-gene array contained copies of 40 different genes, including all known competence operons as well as several genes known not to be induced at competence. A 68-gene array containing DNA for 60 candidates for new competence genes, chosen from genome data as described below, plus eight control genes was used to narrow the candidate pool. Finally, a combined 68-gene array including the known competence genes and a set of 31 positive candidates was used for collecting kinetic data. In all arrays, each gene-specific PCR product was present in at least 16 replicate spots.
Probe preparation.
Gene-specific primers were annealed to 2 μg of total RNA in a total volume of 11 μl by heating the reaction mixture to 70°C for 10 min and then quickly chilling it on ice. To this reaction mixture, 4 μl of First Strand reverse transcriptase buffer (250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2), 2 μl of 0.1 M dithiothreitol (Life Technologies), 1 μl of a deoxynucleoside triphosphate mixture (0.5 mM [each] dATP, dGTP, and dCTP and 0.25 mM dTTP; New England Biolabs Inc.), and 1 nmol of Cy3-dUTP or Cy5-dUTP (Amersham) were added. Incubations were performed in the dark. After incubation at room temperature for 5 min, 400 U of Superscript II reverse transcriptase (Life Technologies) was added, and incubation was continued at room temperature for an additional 5 min. The reaction mixtures were then placed at 42°C for 2 h, brought to 30 μl with water, and incubated at 100°C for 5 min. The RNA template was hydrolyzed by making the reaction mixtures 50 mM NaOH and incubating them at 37°C for 15 min. This reaction was then neutralized by making the solution 50 mM HCl and 100 mM Tris, pH 7.2. The probes were then purified over Pharmacia GFX columns and dried in a Speed-Vac to completion. In controls using random hexamer primers, ∼200 ng of cDNA was obtained from 2 μg of RNA template, with 30 pmol of dye nucleotide typically incorporated per μg of cDNA produced.
Hybridization and washes.
The dried probes were resuspended in a 40 μl volume (50% formamide, 5× SSC, 0.1% SDS, and 100 μg of salmon sperm DNA/ml). This mixture was heated for 5 min at 95°C and then added to a prehybridized slide under a coverslip. The slide was then placed at 42°C for 16 h in a sealed hybridization chamber humidified with 20 μl of 5× SSC. The arrays were then washed once at 55°C in a solution of 2× SSC and 0.1% SDS for 5 min, once in 0.1× SSC and 0.1% SDS for 5 min, and three times in 0.1× SSC for 2 min, followed by a final rinse in MilliQ water and drying.
Analysis.
The arrays were scanned on a Scanarrayer 3000 (General Scanning Inc.) with the excitation lasers at full power and a photomultiplier setting of 80%, with a separate scan for each fluorophore. The algorithm used to identify spots, calculate background, and quantitate fluorescent signals involved processing the entire image to allow a grid to be generated around each array element (V. Sharov, unpublished data; software available at http://webtest/softlab/). Local background levels were determined, and signals representing spots smaller than a user-specified size were discarded as noise. Only spots with a reference fluorescence signal at least three times the local background were accepted for quantitation. Spots above the background and size threshold located near the center of the grid were taken to be real and used to generate Cy3 and Cy5 signals as well as Cy5/Cy3 ratios. Finally, abberrant values were removed from the data set. Reasons for discarding array element readings were as follows: no signal (20%), weak signal (<3x background) (4.6%), slide problems (0.5%), and aberrant values (0.7%). The data for figures in this paper are posted at http://webtest/tdb/microarray.
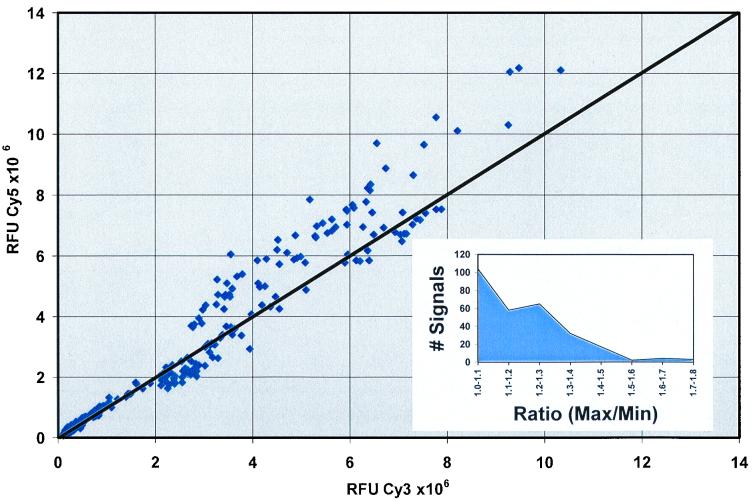
To define the operating limits of the DNA microarray as a quantitative tool for measuring bacterial RNA levels and to determine the total experimental error of the system, identical RNA samples were used to prepare cDNA with either Cy3 or Cy5 tags. These probes were mixed in a single hybridization reaction. After normalization, the ratio of signals (Cy3/Cy5) for each spot on the array was, as expected, near unity (Fig. 1); the deviation from this value can be considered to represent the sum total of experimental error for hybridization using the microarray in this context. While the source of a bias of about 20% toward Cy3 label for a few high-signal genes is unexplained, the histogram inset in Fig. 1 shows that the ratio of the two signals was less than 1.5 for 97% of the quantitated spots.
FIG. 1.
Correlation between relative message abundance measurements by hybridization with two independently labeled cDNA preparations. A scatter plot is shown of the pairs of Cy3 and Cy5 fluorescence values obtained by hybridization to a 40-gene microarray containing elements spotted at least 16 times each. cDNA was prepared from RNA (2 μg) isolated 10 min after exposure to CSP and was independently labeled with either Cy3 or Cy5 and subsequently combined for hybridization. The signal strength for Cy3 (550 nm) versus that for Cy5 (650 nm) is shown for each of 283 individual spots after normalization to yield 16S rRNA ratios of 1.0. The inset shows the frequency distribution of experimental error as the ratio of the larger signal divided by the smaller signal. RFU, relative fluorescence units.
Since many genes of interest are known not to be expressed in the absence of CSP exposure, we routinely used a reference probe in all experiments that represented an equal-mass mixture of RNA from the culture samples harvested between 0 and 15 min after CSP treatment. This standard eliminated statistical problems associated with using the low fluorescence values obtained from unstimulated cells as a denominator in comparisons. cDNA was prepared from this reference RNA with Cy5, while cDNA for individual kinetics samples was labeled with Cy3. Hybridizations were carried out by mixing the reference probe (Cy5) and an individual kinetics (Cy3) sample probe. Data normalization was accomplished by adjusting the Cy5 fluorescence data so that average Cy3/Cy5 ratios for 16S rRNA array elements were equal to 1.0. Genes with unchanging expression were then expected to exhibit a constant signal ratio of 1, while induced genes were expected to display initial values below 1 and peak induction values above 1. The results obtained substantiated the use of 16S rRNA as a normalization standard.
Computer scanning for potential comboxes.
A consensus sequence derived from Campbell et al. (5) [T(T/C)(T/C)(T/G) (7 to 11 nucleotides) T(A/C/G)CGAATA] was sought in the TIGR genome sequence of a type 4 strain of S. pneumoniae (http://www.tigr.org) using a perl script. Less stringent searches were also performed with TACGAATA, allowing for one mismatch anywhere in this consensus sequence. ORFs within 5 kb downstream of each consensus sequence were extracted by Glimmer (trained on S. pneumoniae sequences) and searched against a nonredundant amino acid sequence database using BlastP. To prepare a candidate array element, PCR primers were selected to represent the first one or two ORFs of significant size downstream of the consensus sequence or, in cases with no nearby large ORF, the first kilobase of downstream DNA.
RESULTS
CSP provokes two waves of gene expression.
To obtain a more comprehensive and direct picture of the events of competence induction in S. pneumoniae at the mRNA level, we constructed a DNA microarray containing internal-fragment probes for all the known competence operons, as well as other fragments representing genes not known to be necessary for transformation or to be differentially regulated during competence. The genes selected for inclusion in the microarray are listed in Table 1. RNA extracts were prepared from culture samples harvested at various times before, during, and after the appearance of competence following a dose of CSP sufficient to induce full competence. Parallel measurements of the same culture used for RNA extraction showed that competence for DNA uptake was maximal at 20 min and fell below 5% of maximum by 40 min, while the activity of a comA gene fusion reporter rose during the period from 8 to 20 min. Images obtained by scanning this microarray, illustrated in Fig. 2, showed that replicate spots displayed consistent fluorescence intensities while these intensities varied from gene to gene and, in some cases, with time after introduction of CSP.
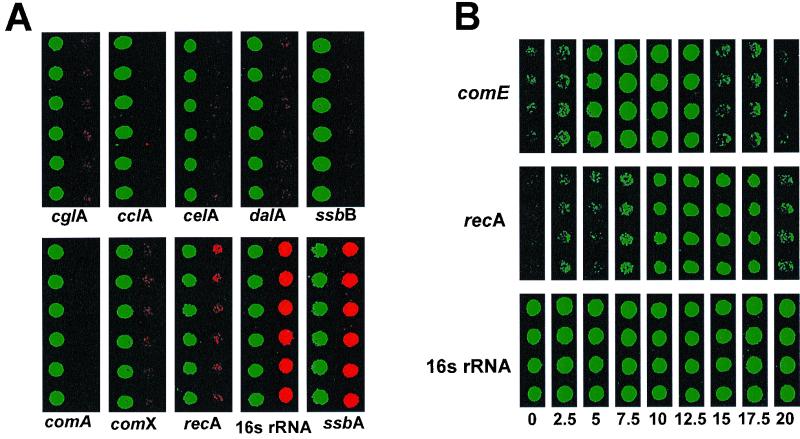
FIG. 2.
Hybridization images from representative sections of the 40-gene microarray. (A) Composite images of six replicate spots for the indicated competence genes and the control spots for ssbA and 16S rRNA. The left part of each box (Cy3; green) represents message abundance in cells 10 min post-CSP stimulation; the right side of each box (Cy5; red) represents message abundance in cells not exposed to CSP. (B) Differential temporal expression patterns of early and late competence-regulated genes. Quadruplicate spots for individual early (comE), late (recA), and control (16S rRNA) genes are shown after hybridization with cDNA prepared from RNA extracted at the indicated times (in minutes) after CSP treatment.
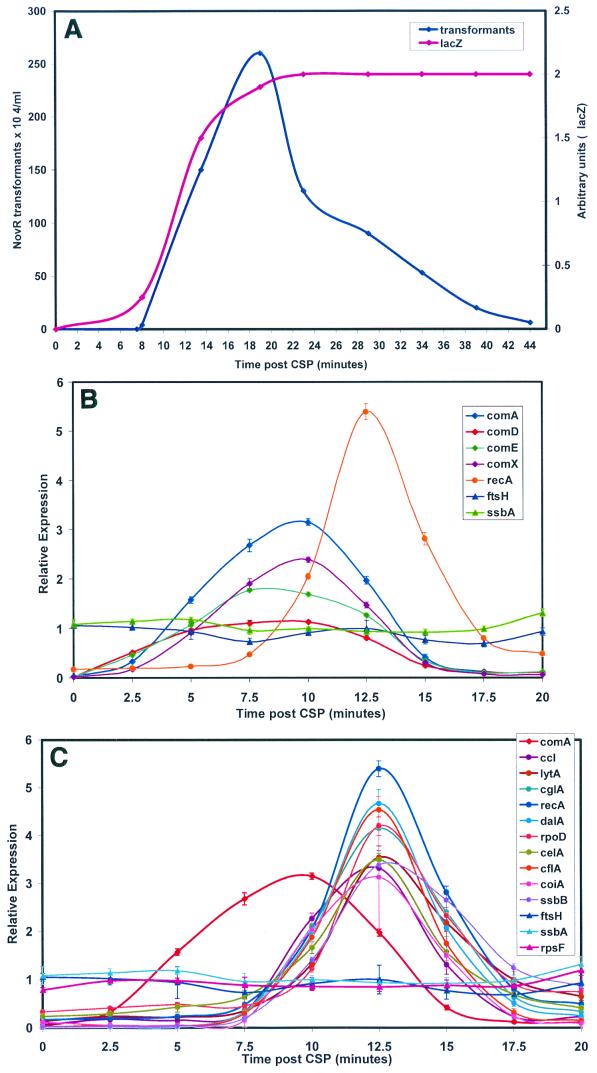
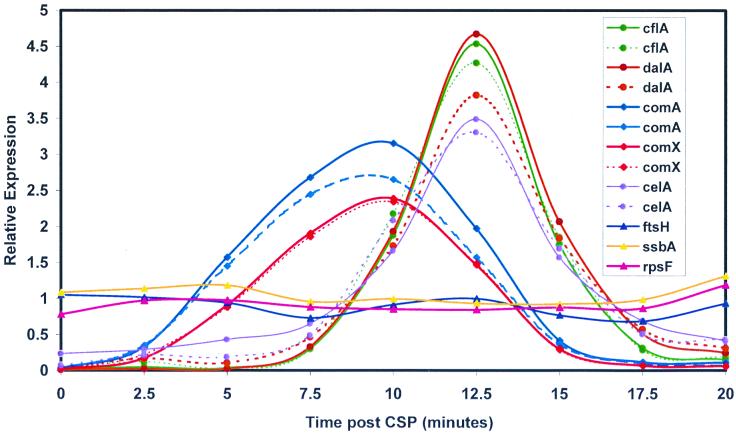
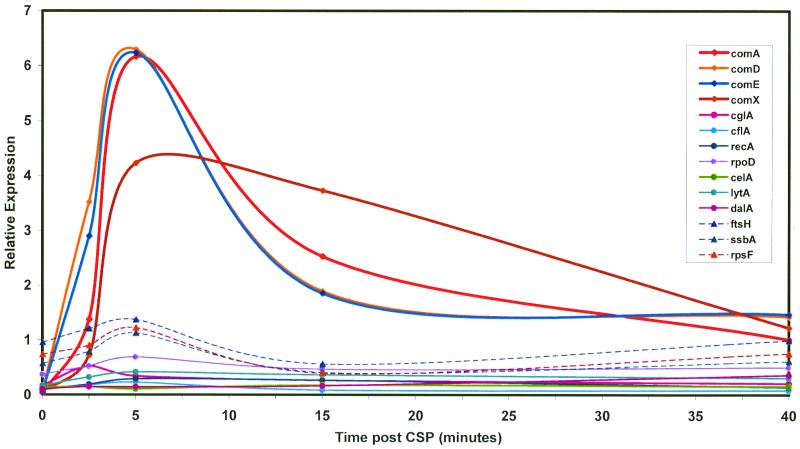
A standard reference probe was made by labeling an equal mixture of RNA samples harvested between 0 and 15 min after CSP exposure. This has the effect of normalizing the expression values so that variation in expression over the time course for each gene fluctuates around the average expression for a relevant portion of the time course and forestalls the inconvenience of vanishing denominators observed if RNA from uninduced cells is used as a reference probe. Measurements of RNA abundance obtained by quantitation of the complete DNA microarrays revealed three principal classes of expression pattern following treatment of noncompetent cultures with CSP (Fig. 3). For genes in the first class, mRNA levels remained constant (Cy3/Cy5 ratio, 1.0 ± 0.2). Expression of genes in the second class followed a clearly different course, exhibiting a low or zero initial (or uninduced) signal, strongly increased expression during the period between 2 and 15 min after CSP addition, with a maximum Cy3/Cy5 ratio in the range of 2.0 to 3.0 between 8 and 10 min, and a return to values well below 1.0 by 15 min (Fig. 3B). Genes in the third class displayed a qualitatively similar pattern of induction and decay of expression but with an additional delay of approximately 5 min before the onset of expression and a similarly tardy return toward preinduction levels (Fig. 3C). The two classes of induced expression patterns were distinguished reproducibly, as illustrated by Fig. 4, which compares the results obtained with two different sets of PCR products for selected genes.
FIG. 3.
Expression patterns of competence regulons following CSP treatment of strain CP1250. (A) Competence induction monitored as capacity for DNA uptake and recombination (Novr transformants) and expression from a comX::lacZ reporter fusion in a parallel culture of CPM16. (B) mRNA levels for class 1 and class 2 genes assayed by hybridization to the 40-gene microarray. The control genes not affected by CSP treatment are ftsH and ssbA. recA is a representative of the class 3 pattern. (C) mRNA levels for class 3 genes. comA is a representative of class 2 genes. The control genes shown are ftsH, ssbA, and rpsF. Cy3-labeled cDNA was prepared from RNA isolated at the indicated times after induction of strain CP1250 with CSP. The hybridization reference, prepared from an equal-mass mixture of six RNA preparations (0 to 15 min), was labeled with Cy5.
FIG. 4.
Reproducibility of expression patterns. Five CSP-inducible genes were each represented as two different PCR products on the 40-gene microarray as shown in Table 1; corresponding pairs of curves (solid and dashed) have the same color. The control genes shown (ssbA, ftsH, and rpsF) were represented twice on the microarray but with a single PCR product each. The cDNA probes were as for Fig. 3.
The genes in the first class were non-competence related, such as ply and gyrA, or genes with functions in transformation but previously reported not to display competence-specific expression, such as endA and mmsA. The genes in the other two classes were typically in operons known or suspected to be important for competence and to be regulated by CSP. Class 2, exhibiting the earliest induction of RNA, comprised genes in the quorum-sensing operons, comAB and comCDE, and the duplicate comX genes. Class 3 contained the nonregulatory competence genes, i.e., genes preceded by a combox. Inspection of class 3 genes revealed a subset that are known to function in noncompetent cells; these genes were strongly induced in response to CSP but also exhibited signals well above background in untreated cells. Included in this subclass were recA and lytA, which are known to be transcribed from both canonical and combox promoters (16), and rpoD, the gene for the principal sigma factor of RNA polymerase (discussed below).
comX is required for expression of the late competence genes and for prompt shutoff of early genes.
comX is a recently described gene that is required for competence and for the induced expression of four combox genes (11). As comX was not needed for induction of its own expression or that of the class 2 gene comA, and as its product, ComX, was found in association with RNA polymerase, it was proposed that comX encodes an alternate sigma factor required for induced expression of all combox genes. To test empirically the predicted generality of those results, a microarray hybridization experiment was carried out with a comX mutant. In this mutant, CPM4, expression patterns were indeed dramatically altered (Fig. 5): no class 3 genes were induced to a detectable degree, while quorum-sensing (class 2) genes were strongly induced, with their products rising to peak levels at 5 min. This result extends the previous observations to show that all known combox genes depend on comX while none of the quorum-sensing operons do. Furthermore, the induced expression of class 2 genes never returned to preinduction levels; expression at about one-fifth maximal levels continued for at least 40 min after CSP addition. Thus, CSP-induced expression of the comCDE, comAB, and comX loci is to some extent self-limiting, but its complete shutoff in wild-type cells depends largely on comX as well.
FIG. 5.
Effect of ComX deficiency on expression patterns of CSP-inducible genes. Strain CPM4 was treated with CSP at an optical density at 550 nm of 0.04. RNA extracts were prepared from samples taken 1 min before and 2.5, 5, 15, and 40 min after CSP addition. Cy3-labeled cDNA prepared from these RNA samples was analyzed by hybridization to the 40-gene microarray using a reference Cy5 cDNA mixture prepared from RNA isolated from wild-type cells at 0, 5, 10, 15, 20, 30, and 45 min after treatment with CSP.
Among 60 combox candidates surveyed with microarrays, 8 are strongly CSP regulated.
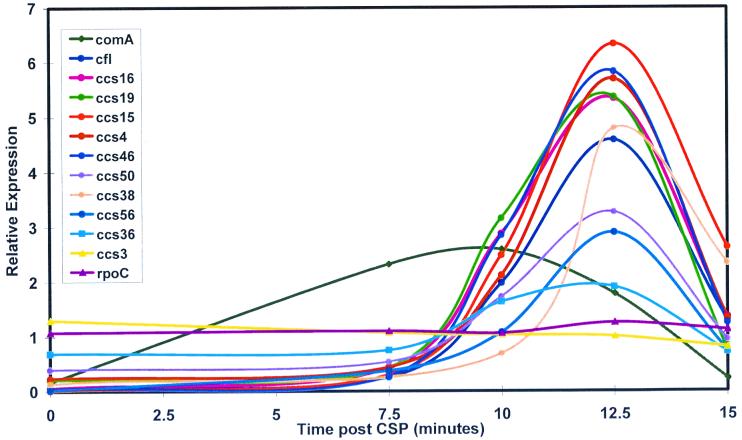
Using a partial sequence of the pneumococcal genome, Campbell et al. (5) identified six putative promoters matching TACGAATA with no more than one mismatch; they showed that five of them (cilA to -E) were both induced at competence and important for transformation. Pestova and Morrison (20) and Lee et al. (10) added coi and cfl to this list of competence-regulated loci. Using a more complete sequence data set, we identified 60 additional candidate sites which shared sequence elements with the Campbell combox and were located in apparent extragenic regions (Table 1). PCR amplification of DNA adjacent to these candidates allowed construction of a microarray for evaluating expression of sequences downstream of many potential combox sites in parallel. Probes derived from cells 10 min post-CSP treatment were used first to screen these candidate sites for new loci that appeared to be induced by CSP. On examining the detailed kinetics of the expression of these selected loci, it was found that eight exhibited threefold or more expression induction after CSP treatment (Fig. 6) and that these eight also depended on comX for their induction (data not shown). The expression of the eight newly identified CSP-inducible loci followed the same kinetics as that of the class 3 genes described above, with apparent induction amplitudes ranging to above 100× (Table 2). On the basis of searches of the DNA sequence downstream of these active combox sites, two categories of locus could be distinguished. In the larger group of loci (7), a conserved ORF was found a short distance (24 to 346 bp) downstream of each apparent combox. One of these hypothetical proteins appeared to be a dUTPase and another appeared to be a double-glycine type bacteriocin precursor, while five are similar to hypothetical proteins of unknown function in other bacterial species. In the second category was a single candidate combox site, ccs16, associated with no detectable nearby coding region oriented for transcription from the candidate combox. Targeted genetic analysis will be needed to define the extent of each CSP-inducible locus, to identify phenotypes associated with each induced region, and to learn if the induced expression is related to competence or to some other trait or is simply adventitious.
FIG. 6.
Expression patterns of candidate combox genes following CSP treatment. Hybridizations to a 68-gene microarray were performed with cDNA prepared from the same RNA extracts as in Fig. 3 and with the same hybridization reference as for Fig. 3. The profiles include controls, two uninducible candidates, and eight candidates with expression changes of threefold or more.
TABLE 2.
Characterization of new CSP-inducible sites
| Candidate site | CSP inductiona | Distance to ORFb | ORF categoryc |
|---|---|---|---|
| ccs1, ccs15 | 330 | 210 | GlyGly bacteriocin homologue; CAA90906 |
| ccs4 | 25 | 219 | Hypothetical protein of unknown function. |
| ccs16 | 100 | 683 | Hypothetical protein of unknown function; D90917 |
| ccs19 | 25 | 69 | dUTPase homologue; CAA72644 |
| ccs36 | 2.8 | 346 | Hypothetical protein of unknown function; B69843 |
| ccs38 | 33 | 195 | Hypothetical protein of unknown function; C69844 |
| ccs35, ccs46 | 250 | 58 | Hypothetical protein of unknown function; A41971 |
| ccs50 | 8.3 | 24 | Hypothetical protein of unknown function; B71466 |
Expression range, calculated from CSP response data as maximal signal/minimal signal.
Number of base pairs between candidate combox and first ORF shown here to be CSP inducible.
Characteristics of predicted product of downstream ORF and accession number of homologue.
DISCUSSION
It has been suggested that all operons marked by the combox motif are regulated in a common way (5, 11, 20). However, evidence supporting this view has not been comprehensive. The results displayed here show, for the first time, that messages from all eight known combox loci appear in parallel, with a constant delay of about 7 min after CSP treatment, and, further, that these RNAs disappear in parallel as well, declining from a maximum at 12 to 13 min to near zero by 20 min. Expression of each of the eight new CSP-inducible, comX-dependent loci described here follows the same pattern. This strictly parallel expression provides strong support for the hypothesis that the combox genes share a common regulatory mechanism. In addition, the fact that all 16 messages decay in parallel at the end of the CSP response suggests that a specific mechanism also coordinates the termination of competence.
A different temporal expression pattern was found for the competence regulatory genes, comA, -B, -C, -D, -E, and -X. (ComC, ComD, and ComE are hypothesized to have an autocatalytic function in quorum sensing which produces a burst of CSP and culminates in synchronous expression of com genes, but these interactions among the regulatory operons are presumably overwhelmed by the high dose of CSP used here.) Strongly induced by exposure to high levels of CSP, their messages began to accumulate immediately upon exposure to the pheromone. This immediate response distinguishes these genes from the combox genes and suggests a fundamentally different mode of regulation, which would be consistent both with their lack of combox consensus sites and with their comX independence. Early appearance of comX transcripts, independent of ComX function, is also consistent with the hypothesis (11) that activity of ComX as a sigma factor for specific transcription of the combox genes is the key to this distinction.
The sequence requirements for a functional combox are not completely defined. Since the 8-bp sequence occurs many times in the pneumococcal genome, it seems likely that there are other elements specifying authentic combox promoters. In Fig. 7, we show the sequences found upstream of the 16 apparent comboxes now linked to CSP-induced genes; all 8 bases of the −8 GAAT box consensus were highly conserved, and a TTTT element was often located 12 bases upstream of this box. However, as we found several cases of CSP-dependent expression near sites without the T4 consensus and as many sites with both consensus elements were not associated with CSP-inducible expression, it appears that some elements of the combox promoter remain to be identified.
FIG. 7.
Combox consensus sites. (A) Alignment of consensus sequences of CSP-inducible sites previously identified. TYG is induced by CSP but is not associated with a known protein product or function (5). (B) Alignment of candidate combox sites identified by pattern searches and validated as CSP inducible by expression measurements (Fig. 6). Regions (100 bp) spanning each consensus sequence were extracted and aligned with no gaps (consensus −10 and −25 elements are in boldface; mismatches to the consensus sequence are lowercase). When present, the first ATG codons of downstream ORFs are underlined.
The pattern of fluctuation in RNA abundance for competence genes shows that much, possibly most, of the regulation of these genes is probably transcriptional. However, detailed examination of the changes in expression of recA (16, 17) has suggested that posttranscriptional regulation plays a major role in limiting the level of at least the RecA protein. The protein pulse-labeling results obtained several years ago (13, 14) suggested that the CSP response entailed a brief cessation of synthesis of many proteins, while competence-specific proteins were made in abundance. Examination of the mRNA hybridization record for the noncombox genes sampled here shows no striking loss of their mRNA at any time during the CSP response. This suggests the possibility of some form of posttranscriptional regulation favoring the combox messages and raises the question of whether competence gene messages may carry especially efficient translation initiation signals.
While proteins responsible for the initiation of transcription of genes in both competence regulons have apparently been identified (e.g., ComE and ComX), essentially nothing is yet known about the mechanisms acting to achieve a rapid shutoff of this expression a few minutes after it begins, despite the continued presence of saturating amounts of the CSP signal. However, the present data show that the mechanism(s) affects mRNA levels severely, as all competence gene messages had largely disappeared by the time of maximal competence. Messages for the quorum-sensing genes (comABCDEX) declined before those of the combox genes. This timing and the early partial shutoff of the genes in a comX mutant both suggest that part of the control of early genes resulting in their earlier shutdown may be exerted by one of the early gene products. The proposal by Alloing et al. (3) that ComE acts both as a transcription-stimulatory protein and as an inhibitor in different phosphorylation states could account for this pattern. The surge in synthesis of the principal sigma factor after comX induction, if it proves to be reflected by a significant increase in sigma A activity, may also play a part in the reversal of competence gene induction. The mechanism of this brief hyperactivation of rpoD is unclear; it may depend on a comboxlike site upstream of rpoD, or it may be a regulatory reaction secondary to the effects of the burst of ComX synthesis.
Finally, it should be noted that the relation between competence and the combox regulon is complex. Thus, not all transformation genes are induced by CSP, since several genes with known roles in transformation but also with likely roles in other functions of DNA metabolism (endA, mmsA, hexA, and hexB) are constitutive. Conversely, while most CSP-induced genes are required for transformation, three have no known role (cinA, dinF, and lytA). lytA, the major autolysin gene, has long been known to be dispensable for transformation (22), casting doubt on any important role in competence or in cell wall remodeling for DNA uptake. However, it has also long been known that extracellular DNA is found in competent cultures (18), and the regulation of lytA does suggest a possible role of this gene in its release. Two other genes in the recA operon, cinA and dinF, are induced at competence but are dispensable for transformation (17). Third, although a few CSP-inducible genes are boosted only modestly in expression after CSP treatment, these increases can be crucial for the efficiency of DNA processing in transformation: in the case of recA, a modest increase in protein levels (4×) is responsible for a large proportion (95%) of the yield of recombinants (17). Thus, further experimental analysis of the new combox genes will be required to define their relation to competence or to other phenotypes that may depend on this quorum-sensing system. The number of new CSP-regulated genes, however, suggests the possibility that they reflect one or more phenotypes distinct from competence that depend on activation by this quorum-sensing mechanism.
The results described here exemplify the potential of microarrays in discovering new regulated genes and new regulatory patterns in bacterial systems when combined with genome sequence data and targeted experimental designs. Although several approaches to finding competence-regulated genes have been pursued over the past 5 to 10 years, including phenotypic mutant screens and reporter library screens, this small-scale microarray exploration has already doubled the number of target loci known to be regulated by the CSP quorum-sensing system. The DNA microarray provides a new tool worth using in parallel with other strategies, if not as a substitute for them: it relies on different assumptions, has different biases, and, perhaps most importantly, it is more readily driven to saturation. Would a larger microarray be useful for discovering additional genes in the competence regulons? The present data suggest that reproducible hybridization signal differences of twofold or more should be detectable with these methods. Since the expression signals for all of the genes already known to be in competence regulons varied in this study by at least five times this amount, it is apparent that additional genes regulated in this pattern would be readily detected when microarrays designed to assay expression of the entire genome become available.
ACKNOWLEDGMENTS
This work was supported in part by grants from the National Science Foundation (MCB-9722821, to D.A.M.) and from the Department of Energy (DEFC-0295-ER-61962, to S.P.).
REFERENCES
- 1.Aaberge I S, Eng J, Lermark G, Løvik M. Virulence of Streptococcus pneumoniae in mice: a standardized method for preparation and frozen storage of the experimental bacterial inoculum. Microb Pathog. 1995;18:141–152. doi: 10.1016/s0882-4010(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 2.Alloing G, Granadel C, Morrison D A, Claverys J P. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol Microbiol. 1996;21:471–478. doi: 10.1111/j.1365-2958.1996.tb02556.x. [DOI] [PubMed] [Google Scholar]
- 3.Alloing G, Martin B, Granadel C, Claverys J P. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum-sensing by the oligopeptide permease. Mol Microbiol. 1998;29:75–83. doi: 10.1046/j.1365-2958.1998.00904.x. [DOI] [PubMed] [Google Scholar]
- 4.Behr M A, Wilson M A, Gill W P, Salamon H, Skolnik G K, Rane S, Small P M. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 5.Campbell E A, Choi S Y, Masure H R. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol Microbiol. 1998;27:929–939. doi: 10.1046/j.1365-2958.1998.00737.x. [DOI] [PubMed] [Google Scholar]
- 6.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 7.de Saizieu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat Biotechnol. 1998;16:45–48. doi: 10.1038/nbt0198-45. [DOI] [PubMed] [Google Scholar]
- 8.Håvarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Håvarstein L S, Morrison D A. Quorum sensing and peptide pheromones in streptococcal competence for genetic transformation. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: ASM Press; 1999. pp. 9–46. [Google Scholar]
- 10.Lee M S, Dougherty B A, Madeo A C, Morrison D A. Construction and analysis of a library for random insertional mutagenesis in Streptococcus pneumoniae: use for recovery of mutants defective in genetic transformation and for identification of essential genes. Appl Environ Microbiol. 1999;65:1883–1890. doi: 10.1128/aem.65.5.1883-1890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee M S, Morrison D A. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol. 1999;181:5004–5016. doi: 10.1128/jb.181.16.5004-5016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lipshutz R J, Fodor S, Gingeras T, Lockhart D J. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21(Suppl. 1):20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 13.Morrison D A. Competence-specific protein synthesis in Streptococcus pneumoniae. In: Polsinelli M, Mazza G, editors. Transformation—1980. Oxford, United Kingdom: Cotswold Press Ltd.; 1981. pp. 39–53. [Google Scholar]
- 14.Morrison D A, Baker M F. Competence for genetic transformation in pneumococcus depends on synthesis of a small set of proteins. Nature. 1979;282:215–217. doi: 10.1038/282215a0. [DOI] [PubMed] [Google Scholar]
- 15.Morrison D A, Trombe M, Hayden M, Waszak G, Chen J. Isolation of transformation-deficient Streptococcus pneumoniae mutants defective in control of competence, using insertion-duplication mutagenesis with the erythromycin resistance determinant of pAMβ1. J Bacteriol. 1984;159:870–876. doi: 10.1128/jb.159.3.870-876.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mortier-Barriere I, de Saizieu A, Claverys J P, Martin B. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol Microbiol. 1998;27:159–170. doi: 10.1046/j.1365-2958.1998.00668.x. [DOI] [PubMed] [Google Scholar]
- 17.Mortier-Barrière I A. Contrôle génétique de la compétence chez la bactérie à Gram postif Streptococcus pneumoniae: étude de l'opéron tardif cinA. Ph.D. dissertation. Toulouse, France: Université Paul Sabatier; 1999. [Google Scholar]
- 18.Ottolenghi E, Hotchkiss R D. Release of genetic transforming agent from pneumococcal cultures during growth and disintegration. J Exp Med. 1962;116:491–519. doi: 10.1084/jem.116.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pestova E V, Håvarstein L S, Morrison D A. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 20.Pestova E V, Morrison D A. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J Bacteriol. 1998;180:2701–2710. doi: 10.1128/jb.180.10.2701-2710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richmond C S, Glasner J D, Mau R, Jin H, Blattner F R. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 1999;27:3821–3835. doi: 10.1093/nar/27.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez-Puelles J M, Ronda C, Garcia J L, Garcia P, Lopez R, Garcia E. Searching for autolysin functions. Characterization of a pneumococcal mutant deleted in the lytA gene. Eur J Biochem. 1986;158:289–293. doi: 10.1111/j.1432-1033.1986.tb09749.x. [DOI] [PubMed] [Google Scholar]
- 23.Ween O, Gaustad P, Håvarstein L S. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol Microbiol. 1999;33:817–827. doi: 10.1046/j.1365-2958.1999.01528.x. [DOI] [PubMed] [Google Scholar]