Abstract
Gene expression in the cyanobacterium Synechococcus elongatus PCC 7942 is under the control of a circadian oscillator, such that peaks and troughs of expression recur with a periodicity of about 24 h in the absence of environmental cues. This can be monitored easily as light production from luciferase gene fusions to S. elongatus promoters. All promoters seem to exhibit circadian oscillation of expression, but the phasing of peak and trough times differs among different genes. The majority of genes are designated class 1, with expression peaks near dusk or subjective dusk (the time corresponding to dusk in the absence of a diurnal cycle). A minority, of which purF is an example, have expression peaks approximately 12 h out of phase with class 1 genes. A screen of Tn5 mutants for those in which purF phasing is altered revealed a mutant that carries an insertion in the opcA gene, previously identified as essential for glucose-6-phosphate dehydrogenase function. However, a different enzymatic reporter and in vitro luciferase assays revealed that the expression pattern of the purF promoter is not altered by opcA inactivation, but rather the reduced flavin mononucleotide substrate of luciferase is limiting at the time of the natural circadian peak. The results suggest that OpcA is involved in temporally separated reductant-generating pathways in S. elongatus and that it has a role outside of its function in activating glucose-6-phosphate dehydrogenase. The opcA gene, expected to be cotranscribed with fbp and zwf, was shown to have its own class 2 promoter, whereas the fbp promoter was determined to be in class 1. Thus, opcA expression is likely to be constitutive by virtue of the activity of two promoters in nearly opposite circadian phases.
Cyanobacteria, like diverse eukaryotes, possess an internal timekeeping mechanism termed a circadian clock that controls oscillations of biological functions with an endogenous period of about 24 h (2, 6, 12, 14, 34). A circadian system is conventionally divided into three parts: a central oscillator that determines the circadian period, input pathways that synchronize the phasing of the oscillator with daily environmental cycles, and output pathways that confer circadian rhythmicity on the processes that are controlled by the clock. A locus of three genes, kaiA, kaiB, and kaiC, has been identified in the cyanobacterium Synechococcus elongatus PCC 7942 that is fundamentally important for circadian rhythmicity of gene expression (16). Mutations in any of these genes can alter the circadian period or cause arhythmicity of all genes that are examined.
Most or all of the genes in the S. elongatus genome are transcribed with a circadian rhythm. One demonstration of this was the insertion of promoterless Vibrio harveyi luciferase genes (luxAB) randomly throughout the genome. All bioluminescent colonies, indicating fusion of the reporter genes with a promoter, showed a circadian rhythm of light production (19). These and other data suggest that transcription is globally rhythmic in the organism. However, there is specialization in the phasing (timing of peaks and troughs), amplitude, and duration of the day with which individual genes are expressed. Most S. elongatus promoters drive luxAB with a peak of bioluminescence at dusk (in a light-dark cycle) or subjective dusk (in continuous light [LL]) and a trough at dawn or subjective dawn. By convention, subjective dusk is around 12 h, 36 h, and so on, and subjective dawn is around 24 h, 48 h, and so on, in LL after a 12-h pulse of darkness to entrain the clock. These dusk-peaking genes, including psbAI, psbAII, psbAIII, kaiA, kaiBC, and rrnA, are categorized as having class 1 expression patterns (16, 19). Even PconII, a synthetic promoter with a consensus Escherichia coli ς70 binding site, drives luxAB with a class 1 phase in S. elongatus (10, 35).
A minority of genes, among which only purF has been characterized (18), show the opposite phase of expression. Expression of these genes, grouped as class 2, peaks at dawn or subjective dawn and troughs at dusk or subjective dusk. purF encodes a key enzyme in the de novo purine biosynthesis pathway, glutamine phospho-ribosyl-pyrophosphate (PRPP) amidotransferase (amidophosphoribosyltransferase, EC 2.4.2.14). However, the purL gene, which is immediately upstream of purF and encodes a 5′-phospho-ribosyl N-formyl-glycinamidine (FGAM) synthetase (EC 6.3.5.3), another enzyme in the same pathway, belongs to class 1 (18). A potential advantage for this phasing of expression is that glutamine PRPP amidotransferase is oxygen sensitive, and the O2 concentration is low at dawn after a night of respiration, which consumes O2, the by-product of photosynthesis (18).
Mutagenesis has revealed that there are specific output pathways that affect subsets of the class 1 genes (17, 35). However, none of these affects circadian expression of the purF gene. To understand what decides the peaking of class 2 gene expression at dawn and how the temporal separation between class 1 and 2 gene expression is achieved, we attempted to identify mutations that would specifically eliminate the specialized phasing pattern of class 2 genes. We performed transposon mutagenesis on a bioluminescent reporter strain in which the purF promoter drives expression of the V. harveyi luxAB genes with a class 2 expression pattern and screened for mutants in which the phasing of the bioluminescence rhythm had reverted to class 1. The screen revealed a mutant that carries an insertion in the opcA gene, whose function was previously known only as being essential for function of glucose-6-phosphate dehydrogenase (G6PD), encoded by the upstream zwf gene (32, 33). G6PD is the first enzyme in the oxidative pentose phosphate pathway, the primary catabolic source of reductant in cyanobacteria (28).
Additional analysis revealed that the actual expression pattern of the purF promoter was unchanged in the opcA mutant, but that the reduced flavin mononucleotide (FMNH2) substrate of the luciferase was limiting at the time of the natural circadian peak. The opcA gene overlaps the upstream zwf gene by 1 bp, suggesting cotranscription (22). However, the phenotype of an opcA mutant is more severe than that of a zwf mutant, which cannot synthesize G6PD, indicating a broader function for OpcA than previously identified and lack of polarity of the zwf insertion on opcA. The opcA gene, whose product is important for reductant production at night in S. elongatus, has a class 2 promoter, independent of class 1 expression from the upstream fbp promoter.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
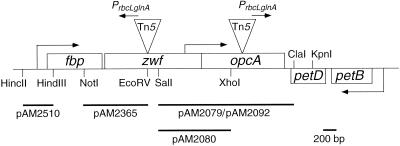
A list of all strains of cyanobacteria, Escherichia coli, and plasmids used in this work is presented in Table 1. Endpoints for restriction fragments used in reporter constructs are shown in Fig. 2. All reporter strains were derived from S. elongatus PCC 7942. This strain has previously been reported without a specific name as Synechococcus sp. strain PCC 7942. However, a pending update to Bergey's Manual of Determinative Bacteriology will support designation of the closely related strain PCC 6301 (13, 36) as the living neotype of S. elongatus (23, 24); PCC 7942 is also appropriately assigned to this species (R. Rippka, personal communication). S. elongatus strains were grown in liquid culture or on agar plates of modified BG-11 medium (BG-11M) (7) with appropriate antibiotics under continuous light at 30°C. E. coli strains were grown in liquid or on solid Luria-Bertani (LB) or Terrific broth (TB) medium (25) with appropriate antibiotics. Antibiotic concentrations in BG-11M were as follows: chloramphenicol (Cm), 2 μg/ml; spectinomycin (Sp) and streptomycin (Sm), 2 μg/ml each for selection of the omega cassette; and kanamycin (Km), 5 μg/ml. In LB or TB, concentrations were as follows: ampicillin (Ap), 100 μg/ml; Cm, 17 μg/ml; and Km, 50 μg/ml.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Characteristics or relevant genotype | Reference |
|---|---|---|
| E. coli | ||
| DH10B | Host for plasmids | |
| AM1037 | HB101 containing plasmid pAM1037, which carries a Tn5 derivative | 17 |
| AM1460 | HB101 containing plasmid pRK2013 to provide conjugal transfer functions | 9 |
| S. elongatus strains | ||
| PCC 7942 | Wild-type S. elongatus strain | |
| AMC393 | psbAI::luxAB fusion in NSII (Cmr) and psbAI::luxCDE fusion in NSI (Spr) | 1 |
| AMC395 | psbAI::luxCDE fusion in NSI (Spr) | |
| AMC408 | purF::luxAB fusion in NSII (Cmr) and psbAI::luxCDE fusion in NSI (Spr) | |
| AMC517 | Tn5 no. 91, uncharacterized class 2 reporter (Kmr and Spr) | |
| AMC530 | Tn5 no. 61, uncharacterized class 2 reporter (Kmr and Spr) | |
| AMC534 | AMC395 with opcA inactivated by Tn5 insertion | |
| AMC537 | psbAIII::luxAB fusion in NSI (Spr) and psbAI::luxCDE fusion in NSII (Cmr) | |
| AMC601 | purF::luc in NSII (Kmr) | |
| AMC617 | opcA::luxAB fusion (intact opcA gene) in NSII (Cmr) and psbAI::luxCDE fusion in NSI (Spr) | |
| AMC636 | opcA::luxAB fusion (N-terminal coding region only of opcA) in NSII (Cmr) and psbAI::luxCDE fusion in NSI (Spr) | |
| AMC661 | Derivative of AMC408 in which opcA has been inactivated by Tn5 insertion (reconstruction of the OP strain) | |
| AMC662 | Derivative of AMC636 in which opcA has been inactivated by Tn5 insertion | |
| AMC663 | purF::luxAB reporter strain, different from AMC408, in which zwf has been inactivated by Tn5 insertion | |
| AMC866 | fbp::luxAB fusion in NSII (Cmr) and psbAI::luxCDE fusion in NSI (Spr) | |
| Plasmids | ||
| pBGS18 | pUC18 with Apr gene replaced by Kmr gene | 31 |
| PIC20H and pIC20R | Cloning vectors | 21 |
| pAM1037 | Tn5 derivative pRL1058, further modified by insertion of a 0.7-kb fragment into the XbaI site to add outward-reading promoters from the Anabaena sp. strain PCC 7120 glnA and rbcL genes | 1 |
| pAM1406 | Cmr cassette inserted in EcoRV site of pIC20H | |
| pAM1580 | NSII vector with promoterless luxAB; Cmr selection for transformation of Synechococcus | |
| pAM2076 | Plasmid recovered by digestion of DNA from the OP mutant; Tn5 inserted in opcA | |
| pAM2079 | Intact opcA gene upstream of luxAB in pAM1580 | |
| pAM2080 | N-terminal coding region of opcA upstream of luxAB in pAM1580 | |
| pAM2084 | Cmr cassette inserted in opcA with the promoter of Cmr cassette convergent with the opcA promoter | |
| pAM2085 | Cmr cassette inserted in opcA with the promoter of Cmr cassette reading in the same direction as the opcA promoter | |
| pAM2092 | opcA SalI-ClaI fragment in pIC20R | |
| pAM2093 | EcoRI opcA fragment from pAM2092 inserted in the EcoRI site of pBGS18 | |
| pAM2095 | Kmr cassette in pIC20R with the promoter of Kmr cassette reading in the same direction as the lacZ promoter | |
| pAM2258 | PrbcLglnA removed from pAM2076 by digestion with XbaI followed by intramolecular ligation | |
| pAM2259 | Plasmid recovered by digestion of a low-bioluminescence mutant; Tn5 inserted in zwf | |
| pAM2260 | PrbcLglnA removed from pAM2259 by digestion with XbaI followed by intramolecular ligation | |
| pAM2510 | HincII-HindIII fragment of fbp upstream of luxAB in pAM1580 |
FIG. 2.
Physical and genetic map of the Synechococcus opcA locus. Genes are represented by labeled boxes. Triangles indicate the positions of insertions of Tn5 in mutants described in the text. Arrows show direction and approximate originations of transcription. Restriction sites are indicated for enzymes used in constructing relevant clones, and the inserts of wild-type DNA in specific plasmids are indicated by bars underneath the map, labeled with the plasmid names.
Transposon mutagenesis.
A modified Tn5 element that transposes in S. elongatus and confers Km resistance (Kmr) was introduced into S. elongatus on pAM1037, a derivative of pRL1058 (37), by triparental conjugation. Details of the procedure have been described elsewhere (1, 17).
Assay for bioluminescence rhythms.
Reporter strains were assayed for bioluminescence rhythms on a Packard TopCount luminometer (1, 17) in either LL or 12-h light–12-h dark cycles (LD). Luciferase reporter strains also carried a PpsbAI::luxCDE cassette to provide the long-chain aldehyde substrate for luciferase in vivo. Strains that carried purF fusions to the firefly luciferase gene (luc) were measured in the presence of firefly d-luciferin (Biosynth International, Naperville, Ill.) as described previously (17). Entrainment of circadian rhythms by temperature cycles in LL was performed with the following regimen while samples were in stackers on the TopCount: 12 h at 38°C, 12 h at 28°C, 12 h at 38°C, and then 28°C continuously. The cells perceive 12 h at 38°C as day and 12 h at 28°C as night (C. Inoue and T. Kondo, unpublished results). Except where indicated, figures show representative traces; in all cases in which different strains are compared, the wells chosen for comparison are comparable with respect to cell age, density, and position on the microtiter plate, so that quantitative values are meaningful.
Recovery of Tn5, sequencing of the flanking region, and removal of PrbcLglnA.
Restriction enzymes and most modifying enzymes were purchased from Promega and used as directed by the manufacturer. Genomic DNA from the S. elongatus mutant was isolated as described previously (8), digested with KpnI (KpnI does not cut within the Tn5), and religated by T4 DNA ligase. The ligation mixture was introduced into E. coli DH10B cells by electroporation, and Kmr E. coli colonies were selected on LB-agar plates. Plasmid DNA was prepared by standard methods (3). DNA flanking the transposon was sequenced on one strand by using a primer, AMO178, that is complementary to the N-terminal coding region of the transposase gene; this sequence should be adjacent to S. elongatus DNA after insertion of Tn5. The cycle sequencing method was used (dye terminator cycle sequencing ready reaction, ABI Prism; PE Applied Biosystems, Foster City, Calif.). PrbcLglnA was removed from the recovered plasmids from the opcA and zwf mutants (pAM2076 and pAM2259, respectively) by digestion with XbaI followed by intramolecular ligation to create pAM2258 and pAM2260, respectively.
Isolation and inactivation of opcA.
The intact opcA gene was isolated as a SalI-ClaI fragment (Fig. 2) from the plasmid that was recovered following Tn5 insertion into the zwf locus (pAM2259; U. Nair and S. Golden, unpublished results). The fragment was inserted into SalI- and ClaI-digested pIC20R to create pAM2092. The Kmr cassette from pAM2095 and the Cmr cassette from pAM1406 were isolated as XhoI-SalI fragments and inserted individually into the XhoI site of opcA in both orientations. The resultant four constructs were used to transform appropriate cyanobacterial reporter strains to inactivate opcA (11).
Complementation of opcA-inactivated mutants.
The opcA gene was excised from pAM2092 as an EcoRI fragment and inserted into EcoRI-digested pBGS18 to create pAM2093. opcA was then removed as an XbaI fragment and inserted at the NheI site of pAM1580 (1), a promoterless luxAB reporter vector for insertion at neutral site II (NSII) of the S. elongatus genome, creating pAM2079. In this construct, the entire opcA open reading frame (ORF) precedes luxA and luxB as a tricistronic operon. pAM2079 was digested with XhoI and religated to remove the carboxy-terminal coding region of opcA, resulting in pAM2080. In this construct, opcA upstream sequences drive expression of luxAB, but only a fragment of the OpcA polypeptide is encoded. pAM2079 and pAM2080 were used to transform AMC395, an S. elongatus strain that carries luxCDE driven by the psbAI promoter. These genes encode the synthetase responsible for biosynthesis of the luciferase long-chain aldehyde substrate.
In vitro luciferase assay and in vivo bioluminescence measurements.
Reporter strains were grown to an optical density at 750 nm of ca. 0.8 in flasks before they were split into two sets. One set was given a 12-h dark pulse and then 12-h light for the ZT12 group (Zeitgeber time is time relative to an entraining environmental signal). The other set was given 10-h light, 12-h darkness, and then 2-h light for the ZT2 group. The two groups were assayed at the same time. For in vivo assays, bioluminescence was measured from 200-μl samples of cultures after addition of 4 μl of decanal (n-decyl aldehyde; Sigma, St. Louis, Mo.) by a scintillation counter with coincidence disabled for photon counting (Beckman LS3801 liquid scintillation system). Strains with strongly expressed reporters were diluted before the assay. For in vitro assays, cells were broken with a French press, and luciferase activities were measured by the flavin injection assay (30, 38). Total protein concentrations were determined by the Lowry assay (20).
RESULTS
An opposite-phase mutant.
The purF::luxAB reporter strain AMC408 was mutagenized by the introduction of pAM1307, which carries a Tn5 transposon derivative. Exconjugants were transferred to agar pads in microtiter plates for screening of circadian phenotypes on a TopCount (Packard) cycling luminometer. Among approximately 3,000 screened colonies, a putative mutant was found, named OP, with low bioluminescence and an opposite phasing relative to the wild-type purF::luxAB bioluminescence rhythm (Fig. 1). The mutant phenotype was confirmed by several independent TopCount runs and also observed when the running conditions were 12-h light–12-h dark cycles (LD) (data not shown). Circadian rhythms of gene expression in S. elongatus can also be entrained by temperature cycles. The apparent phase change phenotype was also observed for the mutant when the circadian clock was entrained by a 38°C-28°C temperature cycle (data not shown).
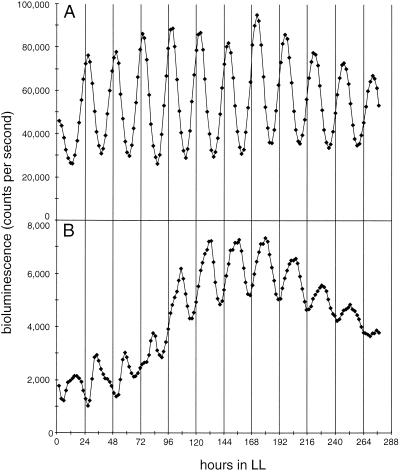
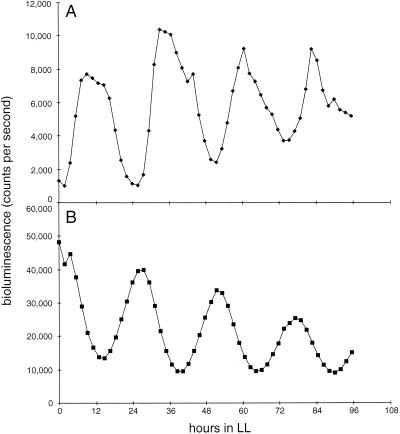
FIG. 1.
Circadian rhythms of bioluminescence from the purF::luxAB reporter in AMC408 (wild-type genetic background) (A) and the OP mutant (B). These representative traces demonstrate the altered phasing of peaks and troughs in OP as well as the characteristic decrease in overall bioluminescence level (peaks approximately 10% of the levels of those in a wild-type background). The x axis in these and all subsequent bioluminescence traces is hours in LL after a phase-synchronizing 12-h incubation in darkness. Bioluminescence values are counts per second measured from microtiter wells that contain reporter strains supported on agar cushions.
The Tn5 element and its flanking regions were recovered as a plasmid, pAM2076. This plasmid and a derivative, in which the outward-reading PrbcLglnA had been removed from the Tn5 element, were linearized and used to transform AMC408 by homologous recombination at the original insertion locus (1). All transformants showed the original OP phenotype (data not shown). This demonstrated that the mutant phenotype results from the insertion of Tn5 at the locus in OP rather than from a secondary mutation and that it did not depend on activity of PrbcLglnA. The S. elongatus DNA flanking Tn5 in pAM2076 was sequenced, revealing identity with the opcA gene previously sequenced from S. elongatus (GenBank accession numbers U33285 and X64768) (27). The transposon inserted at nucleotide 3124 in the GenBank DNA sequence corresponds to amino acid (aa) 133 of the 455-aa OpcA coding region (Fig. 2).
The biochemical activity of OpcA is unknown. However, it is required for the oxidative pentose phosphate (OPP) pathway (32, 33), specifically for the function of G6PD (525 aa) encoded by zwf. A Tn5 insertion into zwf, which is immediately upstream of opcA, was isolated independently in another screen (U. Nair and S. Golden, unpublished results) (Fig. 2). The insertion site corresponds to the codon for aa 197 of G6PD (GenBank accession numbers U33285 and X64768) (27). In the zwf mutant, PrbcLglnA reads upstream toward fbp (Fig. 2). The zwf insertion mutant was found initially in a psbAIII reporter background, AMC537. Tn5 and its flanking regions were recovered and used to transform both AMC537 (psbAIII::luxAB) and AMC408 (purF::luxAB). Bioluminescence from both reporter strains decreased, as previously observed for the original zwf insertion mutant in the AMC537 background (Fig. 3A). However, unlike the opcA insertion, zwf inactivation did not cause a phase change in the AMC408 background (data not shown). zwf and opcA overlap by one nucleotide and are assumed to be cotranscribed (26, 27). If OpcA were involved only in the OPP pathway and could only be cotranscribed with zwf, we would expect the OP phenotype when zwf is inactivated.
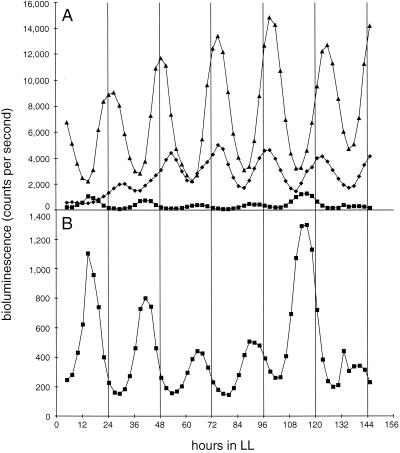
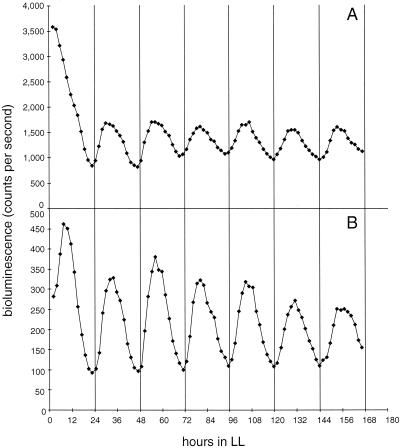
FIG. 3.
Bioluminescence traces from AMC408 and derivatives that carry Tn5 insertions in opcA or zwf that lack the outward-reading PrbcLglnA promoter. The fragment that carries PrbcLglnA was removed from the Tn5-bearing plasmids recovered from OP and AMC537(Tn5zwf). The resulting plasmids were then used to transform AMC408 to recreate Tn5 insertions in the zwf and opcA genes [AMC408(Tn5zwf) and AMC408(Tn5opcA), respectively]. (A) Bioluminescence traces from AMC408 (triangles), AMC408(Tn5zwf) (diamonds), and AMC408(Tn5opcA) (squares). (B) Expansion of the scale for the trace from AMC408(Tn5opcA) to demonstrate that the circadian rhythm persists with a phasing like that of OP in Fig. 1.
opcA is transcribed independently as a circadian class 2 gene.
To determine whether opcA is transcribed independently of zwf, two constructs were made in which all or part of the opcA gene, including upstream sequences internal to zwf, were used to drive promoterless luxAB genes. Two opcA::luxAB plasmids, pAM2079 and pAM2080, were used to transform AMC395, an S. elongatus strain that expresses the luxCDE genes to instruct in vivo synthesis of the long-chain aldehyde substrate of the luciferase (Table 1). In pAM2079, the entire opcA ORF is present upstream of luxAB, as are potential downstream terminator sequences from between opcA and petD (Fig. 2). The reporter fusion in pAM2080 carries only the first 439 bp of opcA (of 1,334 bp) upstream of luxAB (Fig. 2). Both plasmids target the reporters to a defined cloning site in the S. elongatus genome (NSII) (1). The resulting transformants were screened for circadian rhythms of bioluminescence, and all showed robust bioluminescence with a class 2 phase (Fig. 4A). This demonstrated that opcA has its own transcription signals in addition to expected cotranscription with zwf and that this independent transcription is in a class 2 phase. Assay of a fragment including zwf upstream sequences (Fig. 2) did not show promoter activity in this region (data not shown). The upstream region of fbp drives luxAB expression with a high-amplitude oscillation in class 1 (Fig. 4B). Thus, the independent transcription of opcA allows it to be expressed at a time when the remainder of the operon is quiescent.
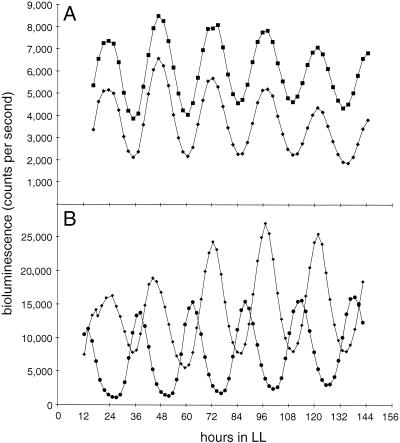
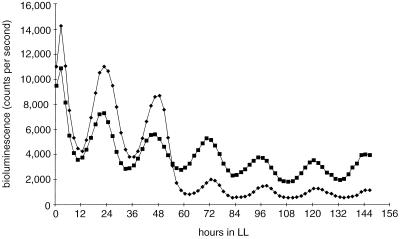
FIG. 4.
Rhythmic class 2 expression from luxAB fusions driven by the opcA upstream region. (A) In AMC617 (diamonds), luxAB is fused downstream of a complete opcA ORF and potential terminator sequences (see pAM2079 insert in Fig. 2). In AMC636 (squares), the reporter is fused after the first 439 bp of opcA (see pAM2080 insert in Fig. 2). The left endpoint of the regulatory region, within the zwf gene, is the same for both constructs (Fig. 2). The traces are mean averages from 12 samples of each strain. The mean peak levels (± SD) for the five complete oscillations are 5,403 ± 811 and 7,738 ± 548 for AMC617 and AMC636, respectively; trough values are 2,139 ± 155 and 4,290 ± 329, respectively. (B) Class 1 expression from the fbp upstream region in AMC866 (circles) compared with class 2 expression from opcA (diamonds). Average expression levels from the opcA reporter were approximately twice that from the fbp reporter.
Mutant phenotype caused by inactivation of opcA.
Mutants were constructed in which Kmr or Cmr cassettes were used to inactivate opcA in various reporter backgrounds to determine whether bioluminescence rhythms from other genes are affected. Each inactivating resistance cassette was inserted in both orientations into the XhoI site of opcA (Fig. 2). The two Kmr constructs were used to transform AMC408 (purF::luxAB, class 2). All transformants showed low bioluminescence and a class 1 phase, as first observed for the Tn5 mutant OP (data not shown). The two orientations of the Cmr cassette in opcA (pAM2084 and pAM2085) were used to transform two uncharacterized class 2 reporter strains, AMC517 and AMC530, obtained by random transposition of Tn5::luxAB throughout the S. elongatus genome (1; J. Shelton and S. Golden, unpublished results). All transformants showed low bioluminescence and class 1 phase (data not shown). These results indicate that the interruption of opcA can cause the mutant phenotype in other class 2 reporters.
We then tested whether opcA inactivation affects the pattern of opcA::luxAB expression. The opcA::luxAB reporter plasmids pAM2079 and pAM2080 were individually used to transform AMC534, a strain that carries the aldehyde biosynthesis genes and Tn5-inactivated opcA. Of the two sets of transformants, those from pAM2080 showed lower bioluminescence and a class 1 phase (Fig. 5A), indicating that the opcA bioluminescence rhythm, like that of purF, is affected by inactivation of opcA. However, the transformants from pAM2079 showed higher bioluminescence and a class 2 phase (Fig. 5B). This indicated that the opcA::luxAB construct in pAM2079, which carries the entire opcA ORF fused to luxAB, not only reported opcA expression but also complemented the mutant phenotype. In a wild-type (opcA+) background, pAM2080 transformants also exhibit class 2 phasing and the higher level of bioluminescence (Fig. 4A). These results demonstrate that the OP mutant phenotype—low bioluminescence and change of phase from class 2 to class 1—was caused by inactivation of opcA.
FIG. 5.
Phasing of bioluminescence from the opcA promoter is itself sensitive to opcA inactivation. A strain in which opcA is inactivated by Tn5 insertion was transformed with reporter plasmids that carry either an incomplete (A) or complete (B) copy of opcA fused to luxAB. When no intact opcA is present, the opcA::luxAB fusion shows the lower bioluminescence and reversed phasing of expression characteristic of the OP mutant (purF::luxAB reporter).
Inactivation of opcA affects reductant production at night and causes luciferase substrate limitation.
Several pieces of evidence suggested that the role of opcA in the OPP, an important pathway for generating reductant (NADPH) in the absence of photosynthesis, might be an important aspect of the phenotype. The Tn5 interruption of opcA was transferred to some class 1 reporters, including the psbAIII::luxAB reporter AMC537 (Fig. 6A). In general, the bioluminescence was lowered, more so at subjective dawn than at dusk; this caused an increase in the amplitude of the bioluminescence rhythm but did not change the phase (Fig. 6B). When opcA was inactivated in the psbAI::luxAB reporter AMC393 by transformation with pAM2076, bioluminescence from the transformants dropped dramatically during the first 3 days of monitoring (approximately 10 days after appearance of transformants), and the strains could not survive beyond that time. This early-death phenotype is not apparent in other reporter backgrounds or in an opcA+ psbAI::luxAB reporter background. An explanation for toxicity of the strong psbAI::luxAB expression in an opcA-null background might be consumption of limiting reductant by the luciferase enzyme. The promoter of the psbAI gene is the strongest we have measured in S. elongatus (among more than 20), and luciferase expression from this fusion is very high. The involvement of OpcA in the OPP pathway, which is the main catabolic source of reductant, made us suspect that inactivation of opcA caused limitation of FMNH2 substrate for the luciferase (4), especially at night.
FIG. 6.
Inactivation of opcA does not change the phasing of bioluminescence from a class 1 phase luxAB reporter. The Tn5 insertion in opcA was transferred to a psbAIII::luxAB reporter strain. Bioluminescence was monitored from the reporter in the wild-type background (AMC537) (A) and the opcA-inactivated background [AMC537(Tn5opcA)] (B).
If the mutant phenotype resulted from substrate limitation, we expected that the phasing of class 2 genes would be unaltered in an opcA mutant when measured by a reporter that does not depend on FMNH2. Strain AMC601 carries a translational fusion between purF and the firefly luciferase gene, luc. The luciferase encoded by luc, although it is an oxygenase like the luxAB product, does not use the reductant FMNH2 as a substrate, but rather requires ATP and a luciferin in addition to O2. We transformed AMC601 with pAM2084 and pAM2085 individually to inactivate opcA. All transformants tested showed the same bioluminescence phenotype as AMC601 (Fig. 7). This indicates that purF expression itself is not altered by loss of opcA and supports the likelihood that the OP phenotype is reporter specific, perhaps as a result of FMNH2 limitation.
FIG. 7.
Inactivation of opcA does not affect either the phasing or magnitude of purF expression when measured by a different reporter. A reporter was constructed in which the firefly luciferase gene, luc, is driven by the purF promoter. Phasing and overall magnitude of bioluminescence were similar from this gene fusion whether opcA was intact (AMC601, diamonds) or inactivated by insertion of a Cmr cassette [AMC601(CmopcA), squares].
To test directly for substrate limitation in the opcA background, we performed in vitro assays for luciferase activity from extracts of purF::luxAB and opcA::luxAB strains. Luciferase activity showed the wild-type pattern of higher activity at dawn and lower at dusk in both opcA and zwf mutants (AMC661 and AMC663, respectively) when exogenous FMNH2 and decanal were provided (Table 2). In vivo assays of opcA-inactivated strains still showed opposite phasing of bioluminescence from purF::luxAB (AMC661) and opcA::luxAB (AMC662) reporters relative to the wild-type controls even when decanal was added exogenously. The only untested substrate is O2, which is also a substrate for firefly luciferase. We did not see a mutant phenotype from the purF::luc reporter when opcA was inactivated. We conclude that the limitation of FMNH2 is solely responsible for the low bioluminescence and the phase change in the purF::luxAB reporter when opcA is absent. The FMNH2 level was especially low at dawn, such that the bioluminescence at dawn became lower than that at dusk, resulting in the apparent reversed phase.
TABLE 2.
Luciferase activities
| Strain | ZT time | In vitro
|
In vivo
|
||
|---|---|---|---|---|---|
| Activitya | Classb | Activityc | Class | ||
| AMC408 (purF::luxAB) opcA+ | ZT2 | 6.03 ± 1.00 | 2 | 7.90 ± 0.54 | 2 |
| ZT12 | 3.15 ± 1.00 | 3.44 ± 0.19 | |||
| AMC661 (purF::luxAB) opcA null | ZT2 | 5.60 ± 0.84 | 2 | 2.80 ± 0.86 | 1 |
| ZT12 | 2.54 ± 1.01 | 6.35 ± 1.84 | |||
| AMC636 (opcA::luxAB) opcA+ | ZT2 | 4.67 ± 1.02 | 2 | 13.02 ± 2.37 | 2 |
| ZT12 | 2.02 ± 0.95 | 4.58 ± 0.22 | |||
| AMC662 (opcA::luxAB) opcA null | ZT2 | 6.41 ± 0.60 | 2 | 19.58 ± 0.27 | 1 |
| ZT12 | 3.48 ± 0.13 | 44.33 ± 3.03 | |||
| AMC663 (purF::luxAB) zwf null | ZT2 | 5.87 ± 0.03 | 2 | NTd | |
| ZT12 | 2.88 ± 0.219 | NT | |||
Luminometer values (arbitrary units) per microgram of total protein ± standard deviation.
Class of expression; higher value at subjective dusk ZT12 (class 1) or dawn ZT2 (class 2).
Excess n-decyl aldehyde provided exogenously; values are 105 cpm ± standard deviation.
NT, not tested.
DISCUSSION
When the OP mutant data are reexamined with the knowledge that the opcA mutation causes a limitation in the reductant supply, the nature of the apparent phasing change of the purF::luxAB bioluminescence rhythm is evident. The absolute bioluminescence level at subjective dusk (when class 1 genes peak and purF troughs) is much less affected than the level at subjective dawn (when class 1 genes trough and class 2 genes peak). The same phenomenon is responsible for the increased amplitude of class 1 reporters when opcA is inactivated, as a result of lower bioluminescence during the trough at subjective dawn and a lesser effect on the subjective dusk peak. This asymmetry of the effect of decreased luciferase substrate suggests that the metabolic pathways for generating FMNH2 are not constitutive in S. elongatus but oscillate in a circadian fashion. A simplistic hypothesis is that photosynthetic activity is largely responsible for generating reductant during the day and subjective day, whereas respiratory pathways such as the OPP take over this role during subjective night. Surprisingly, firefly luciferase activity was not affected by inactivation of opcA, indicating that the cell has means of maintaining ATP concentrations when the OPP is disrupted.
A central question that remains regarding these data is why inactivation of opcA has a more severe phenotype than inactivation of zwf. The known function of OpcA is that it is required for G6PD activity. G6PD monomers are encoded by zwf. OpcA is involved in the oligomerization and activation of G6PD, as the subunits of the enzyme are monomeric, and there is little activity in an opcA mutant (33). Both zwf and opcA mutants appear to have defects in FMNH2 production, but only the inactivation of opcA lowered the substrate level enough at dawn to reverse the phase of bioluminescence. Our data suggest that OpcA has a role in a reductant-generating step beyond its involvement in activating G6PD. This could be another target within the OPP or in another metabolic pathway, but it is likely to be more active in the night or subjective night and under circadian control.
The opposite phasing of opcA and fbp expression is consistent with a role outside of OPP for OpcA; its independent transcription allows expression when the fbp and zwf genes are silent. Cotranscription of opcA with zwf may coordinate OpcA function in the OPP pathway, in which G6PD encoded by zwf is a crucial enzyme. Our data demonstrate that opcA can be transcribed independently and that its independent transcription is in class 2 phase, i.e., peaks at dawn. This, and the fact that the opcA mutant has such a low bioluminescence specifically at dawn, such that the original peak at dawn has become a trough, suggests that OpcA made at dawn is involved in a reductant production pathway at that time of day.
We have partially analyzed the purF promoter region (unpublished data). The identification of opcA as another class 2 gene affords the opportunity to compare potential regulatory elements. Functional analysis of these elements is in progress to determine the sequence information that is required for class 2 phasing of circadian expression.
The goal of the project was to identify mutants affected in generating class 2 expression, but these were not identified in our screen of approximately 5,000 Tn5 insertion mutants. There are several potential explanations, both technical and biological. One is that Tn5, thus far the only useful transposon for the organism, is simply a limited mutagen, which shows bias of insertion sites (5). Indeed, we have identified the zwf/opcA locus four more times in various screens with Tn5 mutagenesis and have yet to isolate an insertion in the known kaiABC locus that is required for circadian rhythmicity, even though loss of these genes is not deleterious under lab conditions.
Another more intriguing possibility is that there is a general mechanism underlying all class 2 gene expression and the classes of mutations we seek may be lethal. Temporal separation may be the original drive for the evolution of circadian clocks. Like purF, nitrogenase genes are expressed at night in unicellular diazatrophic cyanobacteria (12, 15, 29). The products of these genes are sensitive to O2, which is produced by photosynthesis during the (subjective) day. Phasing the expression of these genes at night may be advantageous, and changing all class 2 gene expression to class 1 may be disastrous. Support for this idea is provided by the cpmA mutant, in which some class 1 genes show dramatically altered phasing and the cells have linked growth and pigment defects (17). However, it is not clear whether the change in phasing itself is responsible for the poor fitness of the strain. Screens for conditional mutants may allow us to identify loci that are necessary for the generation of class 2 gene expression.
Prokaryotic circadian rhythms were first recognized in diazotrophic cyanobacteria because of temporal separation of the incompatible processes of oxygen-sensitive nitrogen fixation and oxygen-evolving photosynthesis (12). However, only gene expression has been examined as a circadian phenomenon in S. elongatus, the strain in which genetic analysis has identified components of the circadian clock. The current work provides an entry point to explore the extent and consequence of temporal regulation of metabolism in this genetically tractable strain.
ACKNOWLEDGMENTS
We are indebted to Thomas Baldwin, whose lab helped us carry out the in vitro luciferase assays. We thank Yao Ouyang and Carl Johnson for the purF::luc strain, Usha Nair for the zwf Tn5 insertion mutant, and Usha Nair, Ashok Gopalakrishnan, and Carl Strayer for assistance with analysis of circadian data.
This work was supported by National Science Foundation grants MCB-9513367 and MCB-9982852.
REFERENCES
- 1.Andersson C R, Tsinoremas N F, Shelton J, Lebedeva N V, Yarrow J, Min H, Golden S S. Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol. 2000;305:527–542. doi: 10.1016/s0076-6879(00)05511-7. [DOI] [PubMed] [Google Scholar]
- 2.Aschoff J. Temporal orientation: circadian clocks in animals and humans. Anim Behav. 1989;37:881–896. [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Assoc. and Wiley-Interscience; 1987. [Google Scholar]
- 4.Baldwin T O, Ziegler M M, Green V A, Thomas M D. Overexpression of bacterial luciferase and purification from recombinant sources. Methods Enzymol. 2000;305:135–152. doi: 10.1016/s0076-6879(00)05483-5. [DOI] [PubMed] [Google Scholar]
- 5.Berg D, Schmandt M, Lowe J. Specificity of transposon Tn5 insertion. Genetics. 1983;105:813–328. doi: 10.1093/genetics/105.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bünning E. The physiological clock. 3rd ed. New York, N.Y: Springer-Verlag; 1973. [Google Scholar]
- 7.Bustos S A, Golden S S. Expression of the psbDII gene in Synechococcus sp. strain PCC 7942 requires sequences downstream of the transcription start site. J Bacteriol. 1991;173:7525–7533. doi: 10.1128/jb.173.23.7525-7533.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bustos S A, Schaefer M R, Golden S S. Different and rapid responses of four cyanobacterial psbA transcripts to changes in light intensity. J Bacteriol. 1990;172:1998–2004. doi: 10.1128/jb.172.4.1998-2004.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen M F, Meeks J C, Cai Y A, Wolk C P. Transposon mutagenesis of heterocyst-forming filamentous cyanobacteria. Methods Enzymol. 1998;297:3–17. [Google Scholar]
- 10.Elledge S J, Sugiono P, Guarente L, Davis R W. Genetic selection for genes encoding sequence-specific DNA-binding proteins. Proc Natl Acad Sci USA. 1989;86:3689–3693. doi: 10.1073/pnas.86.10.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golden S S, Brusslan J, Haselkorn R. Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 1987;153:215–231. doi: 10.1016/0076-6879(87)53055-5. [DOI] [PubMed] [Google Scholar]
- 12.Golden S S, Ishiura M, Johnson C H, Kondo T. Cyanobacterial circadian rhythms. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:327–354. doi: 10.1146/annurev.arplant.48.1.327. [DOI] [PubMed] [Google Scholar]
- 13.Golden S S, Nalty M S, Cho D-C. Genetic relationship of two highly studied Synechococcus strains designated Anacystis nidulans. J Bacteriol. 1989;171:4707–4713. doi: 10.1128/jb.171.1.24-29.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamner K C, Takimoto A. Circadian rhythms and plant photoperiodism. Am Nat. 1964;98:295–322. [Google Scholar]
- 15.Huang T C, Lin R F, Chu M K, Chen H M. Organization and expression of nitrogen-fixation genes in the aerobic nitrogen-fixing unicellular cyanobacterium Synechococcus sp. strain RF-1. Microbiology. 1999;145:743–753. doi: 10.1099/13500872-145-3-743. [DOI] [PubMed] [Google Scholar]
- 16.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson C R, Tanabe A, Golden S S, Johnson C H, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 17.Katayama M, Tsinoremas N F, Kondo T, Golden S S. cpmA, a gene involved in an output pathway of the cyanobacterial circadian system. J Bacteriol. 1999;181:3516–3524. doi: 10.1128/jb.181.11.3516-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Tsinoremas N F, Golden S S, Kondo T, Johnson C H. Circadian expression of genes involved in the purine biosynthetic pathway of the cyanobacterium Synechococcus sp. strain PCC 7942. Mol Microbiol. 1996;20:1071–1081. doi: 10.1111/j.1365-2958.1996.tb02547.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Tsinoremas N F, Johnson C H, Lebedeva N V, Golden S S, Ishiura M, Kondo T. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 20.Markwell M A K, Haas S M, Bieber L L, Tolbert N E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 21.Marsh J L, Erfle M, Wykes E J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 22.Newman J, Karakaya H, Scanlan D J, Mann N H. A comparison of gene organization in the zwf region of the genomes of the cyanobacteria Synechococcus sp PCC 7942 and Anabaena sp PCC 7120. FEMS Microbiol Lett. 1995;133:187–193. doi: 10.1111/j.1574-6968.1995.tb07882.x. [DOI] [PubMed] [Google Scholar]
- 23.Rippka R, Cohen-Bazire G. The cyanobacteriales: a legitimate order based on the type strain Cyanobacterium stanieri? Ann Microbiol (Paris) 1983;134B:21–36. doi: 10.1016/s0769-2609(83)80094-5. [DOI] [PubMed] [Google Scholar]
- 24.Rippka R, Herdman M. Pasteur culture collection of cyanobacteria: catalogue and taxonomic handbook. I: catalogue of strains. Paris, France: Institut Pasteur; 1992. [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Scanlan D, Sundaram S, Newman J, Mann N, Carr N. Characterization of a zwf mutant of Synechococcus sp. strain PCC 7942. J Bacteriol. 1995;177:2550–2553. doi: 10.1128/jb.177.9.2550-2553.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scanlan D J, Newman J, Sebaihia M, Mann N H, Carr N G. Cloning and sequence analysis of the glucose-6-phosphate dehydrogenase gene from the cyanobacterium Synechococcus PCC 7942. Plant Mol Biol. 1992;19:877–880. doi: 10.1007/BF00027085. [DOI] [PubMed] [Google Scholar]
- 28.Schmetterer G. Cyanobacterial respiration. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 409–435. [Google Scholar]
- 29.Sherman L A, Meunier P, Colon-Lopez M S. Diurnal rhythms in metabolism: a day in the life of a unicellular, diazotrophic cyanobacterium. Photosynth Res. 1998;58:25–42. [Google Scholar]
- 30.Sinclair J F, Waddle J J, Waddill E F, Baldwin T O. Purified native subunits of bacterial luciferase are active in the bioluminescence reaction but fail to assemble into the alpha beta structure. Biochemistry. 1993;32:5036–5044. doi: 10.1021/bi00070a010. [DOI] [PubMed] [Google Scholar]
- 31.Spratt B G, Hedge P J, te Heesen S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 32.Summers M, Wallis J, Campbell E, Meeks J. Genetic evidence of a major role for glucose-6-phosphate dehydrogenase in nitrogen fixation and dark growth of the cyanobacterium Nostoc sp. strain ATCC 29133. J Bacteriol. 1995;177:6184–6194. doi: 10.1128/jb.177.21.6184-6194.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sundaram S, Karakaya H, Scanlan D, Mann N. Multiple oligomeric forms of glucose-6-phosphate dehydrogenase in cyanobacteria and the role of OpcA in the assembly process. Microbiology. 1998;144:1549–1556. doi: 10.1099/00221287-144-6-1549. [DOI] [PubMed] [Google Scholar]
- 34.Sweeney B M. Rhythmic phenomena in plants. 2nd ed. San Diego, Calif: Academic Press; 1987. [Google Scholar]
- 35.Tsinoremas N F, Ishiura M, Kondo T, Andersson C R, Tanaka K, Takahashi H, Johnson C H, Golden S S. A sigma factor that modifies the circadian expression of a subset of genes in cyanobacteria. EMBO J. 1996;15:2488–2495. [PMC free article] [PubMed] [Google Scholar]
- 36.Wilmotte A M R, Stam W T. Genetic relationships among cyanobacterial strains originally designated as ‘Anacystis nidulans’ and some other Synechococcus strains. J Gen Microbiol. 1984;130:2737–2740. [Google Scholar]
- 37.Wolk C P, Cai Y, Panoff J-M. Use of a transposon with luciferase as a reporter to identify environmentally responsive genes in a cyanobacterium. Proc Natl Acad Sci USA. 1991;88:5355–5359. doi: 10.1073/pnas.88.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziegler M M, Goldberg M E, Chaffotte A F, Baldwin T O. Refolding of luciferase subunits from urea and assembly of the active heterodimer. Evidence for folding intermediates that precede and follow the dimerization step on the pathway to the active form of the enzyme. J Biol Chem. 1993;268:10760–10765. [PubMed] [Google Scholar]