Abstract
PseudobactinB10, the fluorescent siderophore produced by the rhizobacterium Pseudomonas strain B10, contains the hydroxamate ligand d-N5-hydroxyornithine (d-N5-OH-Orn). We cloned the l-Orn N5-oxygenase (psbA) gene from a genomic library of Pseudomonas strain B10 and demonstrated that PsbA is involved in the conversion of l-Orn to its N5-OH derivative. PsbA shows significant similarity to microbial ω-amino acid hydroxylases containing flavin adenine dinucleotide and NADP cofactor-binding sites and the FATGY signature of the putative substrate recognition pocket. The psbA gene is monocistronic, and its transcription is negatively controlled by iron. A site-specific psbA mutant of Pseudomonas strain B10 was biochemically complemented with the precursor l-N5-OH-Orn, suggesting that l-Orn is hydroxylated before conversion to the d isomer. The l-Orn N5-hydroxylase-defective mutants of Pseudomonas strain B10 and Pseudomonas aeruginosa PAO1 were much less effective than the parental strains in suppressing the growth of the phytopathogen Erwinia carotovora in iron-poor medium. The extent of in vitro inhibition of E. carotovora was strictly iron dependent and directly correlated with the amount of released siderophores. These data strengthen the role of fluorescent siderophores in biocontrol of deleterious rhizomicroorganisms.
Pseudobactins (synonym, pyoverdines), the fluorescent siderophores produced by group I pseudomonads, have been proposed to play a crucial role in the biological control of phytopathogenic microorganisms of the rhizosphere (reviewed in reference 26). Pseudobactins are thought to form stable complexes with soil iron, rendering this essential element nutritionally unavailable to deleterious rhizomicroorganisms. Pseudobactins are the most complex structures among microbial siderophores. They consist of a conserved dihydroxyquinoline derivative (the fluorescent chromophore) joined to a variable peptide which confers strict uptake specificities (8). As a general rule, the peptide backbone contains one or two hydroxamate ligands, in the form of d- or l-N5-OH-ornithine (d/l-N5-OH-Orn), which participate in Fe(III) coordination with the quinoline hydroxyls of the chromophore.
Pioneer studies with the rhizobacterium Pseudomonas strain B10 provided early evidence for the involvement of pseudobactinB10 in the disease-suppressing ability of the producing strain (14, 15, 16, 38, 41). PseudobactinB10 consists of a linear hexapeptide (l-Lys-d-threo-β-OH-Asp-l-Ala-d-allo-Thr-l- Ala-d-N5-OH-Orn) linked to the chromophore via an amide bond (39). In addition to pseudobactinB10, Pseudomonas strain B10 produces pseudobactin A, a nonfluorescent hydroxamatelike siderophore and a likely intermediate in pseudobactin biogenesis (40).
While the siderophore-mediated disease suppression correlates well with the elevated chemical stability and affinity for iron (Kf ≅ 1032) of pseudobactins (1, 5), the possibility exists that secondary iron-binding compounds from the same bacterium, namely, multiple pseudobactin forms (1, 40), ferribactins (21, 29), and salicylate-based siderophores (31, 43), may contribute to scavenge the limited iron pool in the rhizosphere.
Siderophore-deficient mutants proved to be valuable tools for determining the roles of individual iron chelators in biocontrol (reference 6 and references therein). However, mutations blocking siderophore biosynthetic pathways can lead to production of intermediates endowed with iron-binding activity (31, 37, 44) or overexpression of a secondary siderophore system(s) (31, 33, 37, 44). Moreover, mutations resulting in the siderophore-deficient phenotype can involve global regulatory genes which control a number of functions relevant to disease suppression besides iron uptake (4, 20, 30, 46). Synthesis of metabolites other than siderophores can also be altered due to intracellular iron depletion (4, 11, 19). Finally, at least for salicylic acid, a siderophore-independent activity in biocontrol has been demonstrated, due to the induction of systemic acquired resistance in plants (9). Because of these problems, we revisited the antagonistic role of pseudobactinB10 by a reverse approach. First, we identified a key enzyme of the biosynthetic pathway of the fluorescent siderophore; then we mutated the cognate gene and compared the antagonistic properties of wild-type and siderophore-deficient strains.
To gain insight into the genetic basis of pseudobactin (pseudobactinB10 and pseudobactin A) biogenesis, we cloned the l-Orn N5-hydroxylase gene of Pseudomonas strain B10. The bacterial strains and plasmids used are listed in Table 1. Eight pLAFR1-derived cosmids from a genomic library of Pseudomonas strain B10 (25) were individually screened for the ability to complement the pvdA mutation in the nonfluorescent, l-Orn N5-hydroxylase-deficient Pseudomonas aeruginosa mutant PALS124 (pvdA). Only pJLM1, carrying a DNA insert of approximately 23 kb (25), was able to restore the fluorescent phenotype in PALS124 grown on KB agar plates (13). Subcloning in the shuttle vector pUCP18 and deletion analysis localized the complementing DNA region within the 2.7-kb PstI-SphI insert of pCAΔSh. Plasmid pCAΔSh restored nearly wild-type production of both pyoverdine and hydroxamates in PALS124, as determined by spectrophotometric and chemical analyses of iron-poor (IFKB) culture supernatants (1, 18, 42, 44). As for wild-type Pseudomonas strain B10 and P. aeruginosa PAO1, synthesis of both pyoverdine and hydroxamates by PALS124 carrying either pJLM1 or pCAΔSh was under stringent iron control and was undetectable in cultures supplemented with 100 μM FeCl3 (data not shown).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and/or relevant characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| S17.1 | thi pro hsdR RP4-2-tet::Mu-Kan::Tn7 Tra+ Trir Strr | 42 |
| S17.1 λpir | S17.1-derived recA λpir phage lysogen | 27 |
| E. carotovora subsp. carotovora | Prototroph | L. Corazza (MAF Institute of Phytopathology, Rome, Italy) |
| P. aeruginosa | ||
| PAO1 (ATCC15692) | Prototroph | American Type Culture Collection |
| PALS124 | pvdA | 44 |
| PAAC1 | pvdA::tet site-specific mutant | 42 |
| Pseudomonas strain B10 | Prototroph | 25 |
| B10CA1 | psbA::cat site-specific mutant | This study |
| Plasmids | ||
| pLAFR1 | Broad-host-range cosmid vector derived from IncP1 plasmid pRK290; RK2 replicon λcos+ rlx lacZα Mob+ Tra− Tcr | 25 |
| pJLM1 | 23.2-kb EcoRI genomic fragment of Pseudomonas strain B10 ligated to pLAFR1 | 25 |
| pUCP18 | E. coli-Pseudomonas shuttle vector derived from pUC18; pMB1 and pRO1600 replicon lacZα bla Apr Cbr | 42 |
| pACYC184 | p15A replicon; Cmr Tcr | 7 |
| pJP5603 | R6K-based suicide vector | 27 |
| pJP1 | 4.1-kb psbA::cat fragment ligated to the PstI-HindIII sites of pJP5603 | This study |
| pCAΔSh | 2.7-kb PstI-SphI fragment of pJLM1 ligated to pUCP18 | This study |
Sequence analysis of the 2.7-kb DNA insert of pCAΔSh made it possible to identify an open reading frame of 1,335 bp. The putative gene, designated psbA for pseudobactin gene A, is predicted to encode a 49.8-kDa protein showing significant homology to microbial ω-amino acid hydroxylases. The PsbA protein shows the highest similarity to the l-Orn N5-oxygenase (PvdA1; 76% identity) of P. aeruginosa PAO1 (42) and to the homologous enzyme (PvdA2; 49% identity) from Burkholderia cepacia (33). Significant similarity (ca. 40% identity) was also found with the sid1 gene product, the l-Orn N5-oxygenase from Ustilago maydis (23), and with a putative hydroxylase (Hydro) from Aureobasidium pullulans (SwissProt accession no. U85909). Less similarity (ca. 30% identity) was found with the lysine N6-hydroxylases (IucD) of Escherichia coli (IucD1) and Shigella flexneri (IucD2) (reference 12 and SwissProt accession no. AAD44749, respectively), with the putrescine hydroxylases (AlcA) of Bordetella bronchiseptica (10), and with RhbE, a still-uncharacterized enzyme likely to be involved in hydroxamate biosynthesis in Sinorhizobium meliloti (SwissProt accession no. AAD09416).
The multiple alignment shown in Fig. 1 highlights three dominant areas of similarity along the whole sequence of ω-amino acid hydroxylases. The conserved N-terminal region (residues 2 to 34 relative to PsbA) is prevalently hydrophobic and contains the putative flavin adenine dinucleotide (FAD)-binding domain. The type 1 signature of the FAD-dependent oxidoreductases (motif GXGXXG; residues 16 to 21 of PsbA) shows a typical replacement of the last glycine with proline; this is a unique feature of hydroxylases involved in siderophore biosynthesis (34). Although such deviation is predicted to distort the α-helical structure within the secondary βαβ fingerprint of flavin-binding proteins (45), recent studies suggest that the deviated sequence is still compatible with the binding of FAD and association with the cytoplasmic membrane (35, 36). The existence of an additional dinucleotide (NADP)-binding site is inferred from the identification of a type 3 signature of the FAD-dependent oxidoreductases [GXGXX(G/A)] in the central portion of all the members of the group (residues 214 to 219 of PsbA) (34). The third signature region is located in the C-terminal half, corresponding to the L/FATGY motif (34). This element constitutes the core of a wider similarity region extending for 14 prevalently hydrophobic residues, starting with an aspartate and ending with a proline residue [D(X)3(L/F)ATGY(X)4(H/P)]. Although no specific function has been conclusively assigned to this element, its conservation in all ω-amino acid hydroxylases is consistent with the proposed role in substrate binding, with the (L/F)ATGY core providing the hydrophobic pocket and the highly conserved aspartate residue acting as proton-abstracting base to render the ω-amino group more reactive to flavin-dependent hydroxylation (34).
FIG. 1.
Multiple sequence alignment of microbial ω-amino acid hydroxylases. The deduced sequence of PsbA was used for BLAST database searches (National Center for Biotechnology Information), and the best-matching sequences were aligned by the BESTFIT and PILEUP programs (Genetics Computer Group [University of Wisconsin, Madison] package, version 10.0). IucD1, E. coli l-lysine N6-hydroxylase; IucD2, S. flexneri lysine N6-hydroxylase; AlcA, B. bronchiseptica putrescine hydroxylase; RhbE, S. meliloti hydroxylase; PvdA1, P. aeruginosa PAO1 l-Orn N5-oxygenase; PsbA, Pseudomonas strain B10 l-Orn N5-oxygenase; PvdA2, B. cepacia l-Orn N5-oxygenase; Hydro, A. pullulans putative hydroxylase; Sid1, U. maydis l-Orn N5-oxygenase. Residues common to the nine hydroxylases are shaded in dark gray and printed in white. Identical residues are shaded in medium gray, and similar residues are in light gray. Cysteine residues are printed in white and highlighted in black. Dominant areas of similarity are boxed. The two dinucleotide-binding domains were recognized by the MOTIF program (http://www.motif.genome.ad.jp). Putative transmembrane regions were localized by the PSORT (http://psort.nibb.ac.jp), TopPred2, and DAS (http://www.biokemi.su.se) programs. Secondary-structure predictions were inferred by the PredictProtein program (http://www.embl-heidelberg.de).
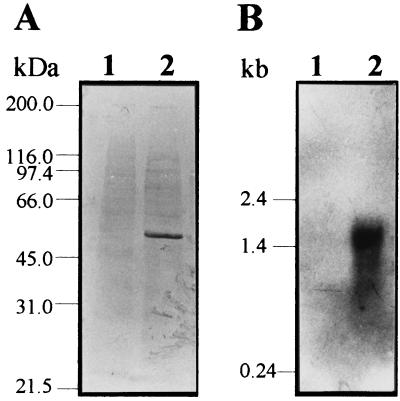
To investigate the regulation of psbA expression, Western blot analysis of Pseudomonas strain B10 cell lysates from iron-poor (IFKB) and iron-rich (IFKB plus 100 μM FeCl3) cultures was performed (18). Given the extensive similarity between PsbA and PvdA, an anti-PvdA mouse polyclonal serum (L. Putignani, A. Ciervo, and P. Visca, unpublished data) was used to detect the PsbA protein. The results show that anti-PvdA antibodies specifically recognize a 50-kDa protein band which is expressed under low-iron conditions but is undetectable in the iron-rich cell lysate (Fig. 2A).
FIG. 2.
Iron-regulated expression of the psbA gene. (A) Immunodetection of the 50-kDa PsbA protein in Pseudomonas strain B10 cell extracts probed with the anti-PvdA mouse antiserum. Lane 1, Pseudomonas strain B10 grown in IFKB medium supplemented with 100 μM FeCl3; lane 2, Pseudomonas strain B10 grown in IFKB medium. Reference molecular mass markers are shown on the left. (B) Northern blot analysis of the psbA transcript(s). Total RNA was isolated from Pseudomonas strain B10 grown in IFKB medium plus 100 μM FeCl3 (lane 1) or IFKB medium (lane 2). RNA samples (10 μg) were hybridized with a 530-bp SalI DNA probe, internal to the psbA coding sequence, which had been labeled with digoxigenin-11-dUTP (Boehringer Mannheim). The sizes of RNA standards are shown on the left.
To elucidate whether regulation of PsbA expression occurs at the transcriptional level, Northern blot analysis of the psbA messenger(s) was performed. Total cellular RNA was extracted from exponential cultures of Pseudomonas strain B10 grown under low-iron (IFKB) and high-iron (IFKB plus 100 μM FeCl3) conditions (17) and hybridized with a 530-bp psbA probe. A single band was observed with RNA from iron-deficient cells, while no signal was detected with iron-rich cells (Fig. 2B). The length of the psbA transcript(s) was estimated to be approximately 1,500 nucleotides, consistent with the existence of a potential stem-loop-like structure (ΔG ≅ −176 kJ/mol) spanning from bp 24 to 97 downstream of the psbA stop codon.
To confirm the implication of PsbA in the enzymatic conversion of l-Orn to l-N5-OH-Orn, a psbA mutant of Pseudomonas strain B10 was generated by gene disruption. The chloramphenicol resistance gene cassette (cat) of pACYC184 (7) was ligated to the unique BamHI site of psbA. The resulting psbA::cat element was cloned in the suicide vector pJP5603 (27), yielding pJP1. Conjugal transfer of pJP1 from E. coli S17.1 λpir (27) to Pseudomonas strain B10, followed by selection for chloramphenicol and kanamycin resistance (100 μg/ml each), made it possible to isolate the Pseudomonas strain B10CA1 mutant, carrying the insertionally disrupted psbA gene. pJP1 integration at the psbA locus was tested by Southern blot analysis of chromosomal DNA from B10CA1 and confirmed by the lack of PsbA expression in Western blot assays of iron-poor culture lysates probed with the anti-PvdA antiserum (data not shown).
Pseudomonas strain B10CA1 differed from wild-type Pseudomonas strain B10 in having impaired synthesis of fluorescent pigment and hydroxamate nitrogen in iron-poor (IFKB) medium. The hypothesis that it is an l-N5-OH-Orn auxotroph was confirmed by both genetic and biochemical complementation tests. In fact, B10CA1 produced detectable levels of fluorescent pigment and hydroxamate nitrogen following transformation with pJLM1 (psbA) or when fed with 400 μM l-N5-OH-Orn (44). Thus, PsbA is responsible for the enzymatic conversion of l-Orn to l-N5-OH-Orn, implying the existence in Pseudomonas strain B10 of an isomerase capable of turning l- to d-N5-OH-Orn during the assembly of the hexapeptide moiety of pseudobactin.
Though mutations of pseudobactin biosynthetic genes were previously reported to result in pleiotropic phenotypes (2), insertional inactivation of the l-Orn N5-hydroxylase gene in P. aeruginosa PAO1 and Pseudomonas strain B10 appeared to cause no secondary effects, except those which can be directly or indirectly predicted for a siderophore biosynthetic mutation (P. Visca and C. Ambrosi, unpublished data). However, unlike P. aeruginosa PAAC1 (pvdA), Pseudomonas strain B10CA1 (psbA) showed no reactivity on the CAS agar plates for siderophore detection (32) and failed to grow under severe iron deficiency, i.e., in IFKB supplemented with 600 μM 2,2′-dipyridyl. These phenotypic traits were ascribed to the absence in Pseudomonas strain B10 of additional siderophore systems capable of compensating for the defect in pseudobactin synthesis. In fact, thin-layer chromatography analysis of ethyl acetate extracts from IFKB culture supernatants of both wild-type and l-Orn N5-hydroxylase-defective mutants (43) did not detect either salicylate or pyochelin in culture extracts of Pseudomonas strains B10 and B10CA1, while both compounds were produced by P. aeruginosa PAO1 and PAAC1 (data not shown). This observation is consistent with the notion that fluorescent siderophore systems represent the primary strategy for iron assimilation in group I pseudomonads (24), a feature which explains the occurrence of natural strains unable to produce salicylate-based siderophores (22).
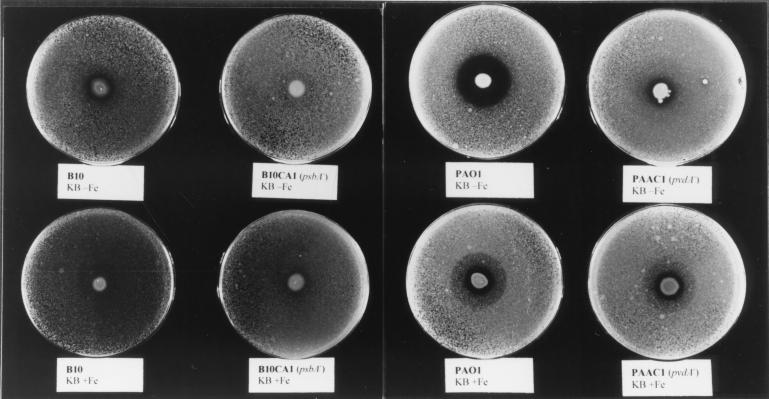
Pseudomonas strain B10CA1, due to its defect in the biosynthesis of both pseudobactin forms (i.e., pseudobactinB10 and pseudobactin A) and salicylate-based siderophores, was used as an ideal tool to verify the in vitro antagonistic activity of fluorescent siderophores against the soilborne pathogen Erwinia carotovora subsp. carotovora. This member of the family Enterobacteriaceae normally produces the catecholate siderophore chrysobactin, and occasionally aerobactin (3, 28). A previously described antibiosis assay (14) was used to compare the in vitro inhibitory activity of wild-type and l-Orn N5-hydroxylase-defective mutants of Pseudomonas strain B10 and P. aeruginosa PAO1. Inhibition of the test organism by Pseudomonas strain B10 occurred only in the iron-poor medium (KB) and not in the iron-rich medium (KB plus 100 μM FeCl3). Conversely, the pseudobactin-defective mutant B10CA1 did not exert any inhibitory activity independently of the iron concentration of the medium (Fig. 3). The antagonistic activity was more evident for wild-type P. aeruginosa PAO1, while it was strongly reduced for the pyoverdine-defective mutant PAAC1 (Fig. 3). UV light inspection of iron-poor plates disclosed that the fluorescent halo around PAO1 was much larger than that around B10. To correlate the inhibitory activity with siderophore production, the relationship between bacterial growth and release of fluorescent pigment was determined for P. aeruginosa PAO1 and Pseudomonas strain B10. The two strains showed comparable growth kinetics at 28°C but differed in siderophore biosynthetic capability, in that PAO1 produced threefold more fluorescent pigment than B10 after 24 h of growth in IFKB (data not shown). At this growth stage, PAO1 also released approximately 25 μM pyochelin and 50 μM salicylate into the medium, while B10 did not. These results are consistent with a dose-response effect for siderophore-dependent inhibition of E. carotovora.
FIG. 3.
Plate assay for siderophore-mediated antibiosis. KB agar plates were inoculated in the center with 10 μl of a suspension (≅106 CFU/ml) of the different Pseudomonas strains. The plates were incubated for 24 h at 28°C and then sprayed with a suspension of E. carotovora (≅106 CFU/ml) and examined after additional 24-h incubation. The inhibition of E. carotovora is indicated by the transparent halo surrounding the Pseudomonas patch. The strain and medium are indicated below each plate. KB-Fe, KB medium; KB+Fe, KB supplemented with 100 μM FeCl3.
Our experiments demonstrate that the ability of Pseudomonas strain B10 to produce pseudobactin is critical for in vitro antagonistic activity against E. carotovora. Transfer of Fe(III) from chrysobactin to fluorescent siderophores is expected on the basis of the unfavorable (2:1) iron-binding stoichiometry and the extremely low affinity for iron of chrysobactin compared with pseudobactin or pyoverdine (28). Given this assumption, the inability of chrysobactin to compete with pseudobactin for iron is the only plausible reason for the in vitro antagonistic activity of Pseudomonas strain B10. Likewise, the stronger antagonistic activity of P. aeruginosa PAO1 correlates well with its greater capability for iron withdrawal. The overexpression of salicylate and pyochelin iron uptake systems could be responsible for the residual antagonistic activity of the pvdA mutant under low-iron conditions (Fig. 3). Interestingly, both P. aeruginosa PAO1 and its pvdA derivative had residual inhibitory effects in iron-rich medium, meaning that additional antimicrobial metabolites, whose production is unaffected or increased by iron availability, may contribute to the in vitro antagonistic activity (4, 11, 26, 30). Thus, an efficient system for iron uptake is expected to be doubly advantageous for the success of fluorescent pseudomonads in biocontrol; not only would it suppress phytopathogens by scavenging the limited amounts of bioavailable iron in the rhizosphere, it would also provide the cell with sufficient iron to turn on the synthesis of secondary metabolites endowed with antimicrobial properties.
Nucleotide sequence accession number.
The sequence discussed in this paper has been deposited in the EMBL-GenBank database under accession no. AF230494.
Acknowledgments
We are grateful to J. Leong for the generous gift of cosmids carrying Pseudomonas strain B10 genomic DNA and for the encouragement to initiate this research. We also thank A. Petrucca for valuable technical assistance.
This work was supported by grants from “Istituto Pasteur-Fondazione Cenci Bolognetti” and MURST to N.O. and by the Ministry of Health to P.V.
REFERENCES
- 1.Abdallah M A. Pyoverdins and pseudobactins. In: Winkelman G, editor. CRC handbook of microbial iron chelates. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 139–153. [Google Scholar]
- 2.Adams C, Dowling D N, O'Sullivan D J, O'Gara F. Isolation of a gene (pbsC) required for siderophore biosynthesis in fluorescent Pseudomonassp. strain M114. Mol Gen Genet. 1994;243:515–524. doi: 10.1007/BF00284199. [DOI] [PubMed] [Google Scholar]
- 3.Barnes H H, Ishimaru C A. Purification of catechol siderophores by boronate affinity chromatography: identification of chrysobactin from Erwinia carotovora subsp. carotovora. BioMetals. 1999;12:83–87. doi: 10.1023/a:1009223615607. [DOI] [PubMed] [Google Scholar]
- 4.Blumer C, Haas D. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch Microbiol. 2000;173:170–177. doi: 10.1007/s002039900127. [DOI] [PubMed] [Google Scholar]
- 5.Buyer J S, Kratzke M G, Sikora L J. A method for detection of pseudobactin, the siderophore produced by a plant-growth-promoting Pseudomonasstrain, in the barley rhizosphere. Appl Environ Microbiol. 1993;59:677–681. doi: 10.1128/aem.59.3.677-681.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buysen S, Heungens K, Poppe J, Hofte M. Involvement of pyochelin and pyoverdin in suppression of Pythium-induced damping-off of tomato by Pseudomonas aeruginosa7NSK2. Appl Environ Microbiol. 1996;62:865–871. doi: 10.1128/aem.62.3.865-871.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelis P, Hohnadel D, Meyer J M. Evidence for different pyoverdin-mediated uptake systems among Pseudomonas aeruginosastrains. Infect Immun. 1989;57:3491–3497. doi: 10.1128/iai.57.11.3491-3497.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Meyer G, Capieau K, Audenaert K, Buchala A, Métraux J P, Höfte M. Nanogram amounts of salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa7NSK2 activate the systemic acquired resistance pathway in bean. Mol Plant-Microbe Interact. 1999;12:450–458. doi: 10.1094/MPMI.1999.12.5.450. [DOI] [PubMed] [Google Scholar]
- 10.Giardina P C, Foster L A, Toth S I, Roe B A, Dyer D W. Identification of alcA, a Bordetella bronchisepticagene necessary for alcaligin production. Gene. 1995;167:133–136. doi: 10.1016/0378-1119(95)00659-1. [DOI] [PubMed] [Google Scholar]
- 11.Gill P R, Jr, Warren G J. An iron-antagonized fungistatic agent that is not required for iron assimilation from a fluorescent rhizosphere pseudomonad. J Bacteriol. 1988;170:163–170. doi: 10.1128/jb.170.1.163-170.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrero M, de Lorenzo V, Neilands J B. Nucleotide sequence of the iucD gene of the pColV-K30 aerobactin operon and topology of its product studied with phoA and lacZgene fusions. J Bacteriol. 1988;170:56–64. doi: 10.1128/jb.170.1.56-64.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanine and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 14.Kloepper J W, Leong J, Teintze M, Schroth M N. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature (London) 1980;286:885–886. [Google Scholar]
- 15.Kloepper J W, Leong J, Teintze M, Schroth M N. Pseudomonassiderophores: a mechanism explaining disease suppressive soils. Curr Microbiol. 1980;4:317–320. [Google Scholar]
- 16.Kloepper J W, Schroth M N. Relationship of in vitroantibiosis of plant growth-promoting rhizobacteria to plant growth and the displacement of root microflora. Phytopathology. 1981;77:1020–1024. [Google Scholar]
- 17.Leoni L, Ciervo A, Orsi N, Visca P. Iron regulated transcription of the pvdA gene in Pseudomonas aeruginosa: effect of Fur and PvdS on promoter activity. J Bacteriol. 1996;178:2299–2313. doi: 10.1128/jb.178.8.2299-2313.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leoni L, Orsi N, de Lorenzo V, Visca P. Phylogenetic and functional analysis of PvdS, an iron-starvation sigma factor. J Bacteriol. 2000;182:1481–1491. doi: 10.1128/jb.182.6.1481-1491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Saux O, Robert-Baudouy J. Pyroglutamic acid and iron regulate the expression of the pcp gene in Pseudomonas fluorescensMFO. FEMS Microbiol Lett. 1997;155:209–215. doi: 10.1016/s0378-1097(97)00389-3. [DOI] [PubMed] [Google Scholar]
- 20.Liao C H, McCallus D E, Wells J M, Tzean S S, Kang G Y. The repB gene required for production of extracellular enzymes and fluorescent siderophores in Pseudomonas viridiflava is an analog of the gacA gene in Pseudomonas syringae. Can J Microbiol. 1996;42:177–182. doi: 10.1139/m96-026. [DOI] [PubMed] [Google Scholar]
- 21.Linget C, Azadi P, MacLeod J K, Dell A, Abdallah M A. Bacterial siderophores: the structures of the pyoverdins of Pseudomonas fluorescensATCC 13525. Tetrahedron Lett. 1992;33:1737–1740. [Google Scholar]
- 22.Maurhofer M, Reimmann C, Schmidli-Sacherer P, Heeb S, Haas D, Défago G. Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescensstrain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology. 1998;88:678–684. doi: 10.1094/PHYTO.1998.88.7.678. [DOI] [PubMed] [Google Scholar]
- 23.Mei B, Budde A D, Leong S A. sid1, a gene initiating siderophore biosynthesis in Ustilago maydis: molecular characterization, regulation by iron, and role in phytopathogenicity. Proc Natl Acad Sci USA. 1993;90:903–907. doi: 10.1073/pnas.90.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer J M. Exogenous siderophore-mediated iron uptake in Pseudomonas aeruginosa: possible involvement of porin OprF in iron translocation. J Gen Microbiol. 1992;138:951–958. doi: 10.1099/00221287-138-5-951. [DOI] [PubMed] [Google Scholar]
- 25.Moores J C, Magazin M, Ditta G S, Leong J. Cloning of genes involved in the biosynthesis of pseudobactin, a high affinity iron transport agent of a plant growth-promoting Pseudomonasstrain. J Bacteriol. 1984;157:53–58. doi: 10.1128/jb.157.1.53-58.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Sullivan D J, O'Gara F. Traits of fluorescent Pseudomonasspp. involved in suppression of plant root pathogens. Microbiol Rev. 1992;56:662–676. doi: 10.1128/mr.56.4.662-676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penfold R J, Pemberton J M. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene. 1992;118:145–146. doi: 10.1016/0378-1119(92)90263-o. [DOI] [PubMed] [Google Scholar]
- 28.Persmark M, Expert D, Neilands J B. Isolation, characterization, and synthesis of chrysobactin, a compound with siderophore activity from Erwinia chrysanthemi. J Biol Chem. 1989;264:3187–3193. [PubMed] [Google Scholar]
- 29.Philson S B, Llinas M. Siderochromes from Pseudomonas fluorescens. II. Structural homology as revealed by NMR spectroscopy. J Biol Chem. 1982;257:8086–8090. [PubMed] [Google Scholar]
- 30.Pierson L S, III, Wood D V, Pierson E A, Chancey S T. N-Acyl-homoserine lactone-mediated gene regulation in biological control by fluorescent pseudomonads: current knowledge and future work. Eur J Plant Pathol. 1998;104:1–9. [Google Scholar]
- 31.Reimmann C, Serino L, Beyeler M, Haas D. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF genes, are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology. 1998;144:3135–3148. doi: 10.1099/00221287-144-11-3135. [DOI] [PubMed] [Google Scholar]
- 32.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 33.Sokol P A, Darling P, Woods D E, Mahenthiralingam E, Kooi C. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding l-ornithine N5-oxygenase. Infect Immun. 1999;67:4443–4455. doi: 10.1128/iai.67.9.4443-4455.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stehr M, Diekmann H, Smau L, Seth O, Ghisla S, Singh M, Macheroux P. A hydrophobic sequence motif common to N-hydroxylating enzymes. Trends Biochem Sci. 1998;23:56–57. doi: 10.1016/s0968-0004(97)01166-3. [DOI] [PubMed] [Google Scholar]
- 35.Stehr M, Diekmann H, Smau L, Seth O, Ghisla S, Singh M, Macheroux P. A reply to Dick et al. Trends Biochem Sci. 1998;23:414–415. doi: 10.1016/s0968-0004(97)01166-3. [DOI] [PubMed] [Google Scholar]
- 36.Stehr M, Smau L, Singh M, Seth O, Macheroux P, Ghisla S, Diekmann H. Studies with lysine N6-hydroxylase. Effect of a mutation in the assumed FAD binding site on coenzyme affinities and on lysine hydroxylating activity. Biol Chem. 1999;380:47–54. doi: 10.1515/BC.1999.006. [DOI] [PubMed] [Google Scholar]
- 37.Stintzi A, Cornelis P, Hohnadel D, Meyer J M, Dean C, Poole K, Kourambas S, Krishnapillai V. Novel pyoverdin biosynthesis gene(s) of Pseudomonas aeruginosaPAO. Microbiology. 1996;142:1181–1190. doi: 10.1099/13500872-142-5-1181. [DOI] [PubMed] [Google Scholar]
- 38.Sunslow T V, Schroth M N. Rhizobacteria on sugar beets: effects of seed application and root colonization on yield. Phytopathology. 1982;72:199–206. [Google Scholar]
- 39.Teintze M, Hossain M B, Barnes C L, Leong J, van der Helm D. Structure of ferric pseudobactin, a siderophore from a plant-growth promoting Pseudomonas. Biochemistry. 1981;20:6446–6457. doi: 10.1021/bi00525a025. [DOI] [PubMed] [Google Scholar]
- 40.Teintze M, Leong J. Structure of pseudobactin A, a second siderophore from plant growth promoting PseudomonasB10. Biochemistry. 1981;20:6457–6462. doi: 10.1021/bi00525a026. [DOI] [PubMed] [Google Scholar]
- 41.Vandenbergh P A, Gonzales C F, Wright A M, Kunka B S. Iron-chelating compounds produced by soil pseudomonads: correlation with fungal growth inhibition. Appl Environ Microbiol. 1983;35:128–132. doi: 10.1128/aem.46.1.128-132.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visca P, Ciervo A, Orsi N. Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdin biosynthetic enzyme l-ornithine N5-oxygenase in Pseudomonas aeruginosa. J Bacteriol. 1994;176:1128–1140. doi: 10.1128/jb.176.4.1128-1140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Visca P, Ciervo A, Sanfilippo V, Orsi N. Iron-regulated salicylate synthesis by Pseudomonasspp. J Gen Microbiol. 1993;139:1995–2001. doi: 10.1099/00221287-139-9-1995. [DOI] [PubMed] [Google Scholar]
- 44.Visca P, Serino L, Orsi N. Isolation and characterization of Pseudomonas aeruginosamutants blocked in the synthesis of pyoverdin. J Bacteriol. 1992;174:5727–5731. doi: 10.1128/jb.174.17.5727-5731.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wierenga R K, Terpstra P, Hol W G J. Prediction of the occurrence of the ADP-binding βαβ fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 46.Whiteley M, Kimberly M L, Greenberg E P. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]