Abstract
Background
Onco-immunogenic molecule CD155 is overexpressed in various tumor microenvironments (TME) including in colorectal cancer (CRC). Tumor-associated macrophages (TAMs) are the most abundant immune cells in CRC TME and play a vital role in CRC progression and metastasis. Most studies have focused on investigating the role of CRC cell-specific CD155 on CRC progression, while the contribution of TAMs-specific CD155 is still unknown. Here, we sought to investigate the expression pattern of CD155 in CRC TAMs and its role in tumor immunity and progression.
Methods
CD155 expression patterns in CRC TAMs and macrophages in paratumor or adjacent normal tissue were analyzed in 50 patients with CRC using flow cytometry and in 141 patients with CRC using immunohistochemistry. The correlation of CD155 expression level in TAMs with M1 and M2 phenotypic transition was analyzed. The role of macrophage-specific CD155 in CRC progression and tumor immune response was investigated in vitro and in vivo. We further analyzed the effect of CRC cells on the regulation of CD155 expression in macrophages.
Results
CRC TAMs from clinical samples showed robustly higher expression of CD155 than macrophages from paratumor and adjacent normal tissues. The CD155 expression level was higher in TAMs of CRC at III/IV stages compared with the I/II stages and was negatively associated with the survival of patients with CRC. CD155+ TAMs showed an M2 phenotype and higher expression of interleukin (IL)-10 and transforming growth factor (TGF)-β. CD155+ macrophages promoted CRC cell migration, invasion, and tumor growth supporting the findings from the clinical tissue analysis. This effect was mainly regulated by TGF-β-induced STAT3 activation-mediated release of matrix metalloproteinases (MMP)2 and MMP9 in CRC cells. CD155–⁄– bone marrow transplantation in wild-type mice, as well as CD155– macrophages treatment, promoted the antitumor immune response in the mice ectopic CRC model. Additionally, CRC cells released IL-4 to trigger CD155 expression in macrophages indicating the regulatory role of CRC cells in the development of CD155+ TAMs.
Conclusions
These findings indicated that CD155+ TAMs are responsible for the M2-phenotype transition, immunosuppression, and tumor progression in CRC. The specific localization of CD155+ TAMs in CRC tissue could turn into a potential therapeutic target for CRC treatment.
Keywords: tumor microenvironment, tumor escape, immunity
Background
Colorectal cancer (CRC) is the third most diagnosed cancer (10.0%) and the second leading cause of cancer death (9.4%) worldwide.1 2 Data from 2020 showed >1.9 million new CRC cases and 935,000 deaths, representing approximately 1 in 10 cancer deaths. In recent years, the development of effective cancer screening and preventive measures has successfully improved the outcome of localized CRC treatment. However, distant metastasis to vital organs and postoperative recurrence are the major causes of CRC-related death.3 The tumor microenvironment (TME) consisting of various non-cancerous cells, such as regulatory T cells, tumor-associated macrophages (TAMs), endothelial cells, etc, plays an important role in the antitumor immune response, angiogenesis, cancer progression, and metastasis.4 5 Recent studies have revealed that CD155 is overexpressed in the TME of various cancers, such as lung cancer, melanoma, and colon cancer.6–8 CD155 was originally identified as a poliovirus receptor expressed on the surface of hematopoietic and non-hematopoietic cells.9 The CD155 expression level is associated with the pathophysiology and therapeutic efficacy in various cancers, including CRC.10 11 However, the cell types expressing CD155 in the CRC TME and their role in the pathophysiology of CRC are still unknown.
TAMs are the most abundant immune cells in the TME and play a vital role in the progression and immunogenicity of various cancers including CRC.12 13 The functional heterogeneities of TAMs are associated with phenotypic subsets, that is, tumor-inhibiting M1-like and tumor-promoting M2-like macrophages.14 15 Under normal circumstances, the biological function of M2 macrophages is to inhibit inflammation and promote tissue repair.16 M2 TAMs enhance cancer cell invasion, regulate T cell function, and promote tumor progression.17 Moreover, M2 macrophages present in TAMs suppress the antitumor immune response of T cells and disrupt immune cell interactions resulting in an immunosuppressive CRC TME.18 19 A recent study has confirmed higher CD155 messenger RNA expression in TAMs.20 Similarly, CD155 is overexpressed in CRC tissue and prevents the apoptosis of cancer cells.21 However, the CD155 expression pattern in CRC TAMs and its role in CRC pathophysiology should still be unraveled.
In this study, we aimed to investigate: (1) the protein expression pattern of CD155 in CRC TAMs using clinical tissue samples from CRC patients, (2) the role of CD155 expression levels in macrophages on M1 and M2 polarization in the TME, and (3) the role of CD155 expression levels in macrophages on the pathophysiology of CRC.
Methods
Patients with CRC and tissues
This study recruited 50 patients with histologically proven CRC who underwent surgery at the Third Affiliated Hospital of Sun Yat-sen University between 2019 and 2021. CRC tissue, paratumor intestinal tissue (within 2 cm from the tumor edge), and adjacent normal intestinal tissue (2 cm away from the tumor edge) were collected. A blood sample (8 mL) from each patient was collected in an EDTA vial. Patients’ demographic, clinical information, and histopathological data were presented in online supplemental table 1. Patients with preoperative antitumor treatments (neoadjuvant radiotherapy or chemotherapy) were excluded. All patient specimens were obtained with written informed consent. In addition, paraffin sections of samples from a total of 141 patients with complete clinical data who were diagnosed with CRC at Third Affiliated Hospital of Sun Yat-sen University between 2009 and 2012 were used to detect CD155 expression on TAMs with immunohistochemistry, and the clinicopathological parameters were presented in online supplemental table 1.
jitc-2021-004219supp001.pdf (185.8KB, pdf)
Processing of human samples
A standard Ficoll (Biosharp, China) procedure was utilized to obtain peripheral blood mononuclear cells (PBMCs).22 Tumor-infiltrating mononuclear cells were isolated as described previously.23 Briefly, freshly resected human colorectal tissue samples (within 2 hours) were minced by scissors into 2–4 mm diameter pieces and digested in 2.5 mg/mL collagenase IV and 2.5 mg/mL DNase containing RPMI 1640 (Gibco) media at 37°C for 1 hour. The enzymatic reaction was stopped by adding phosphate buffered saline (PBS) to a double volume of the sample. Afterward, the homogenate was filtered through a 70 µm cell strainer and centrifuged at 1500 rpm for 5 min. To isolate tumor-infiltrating mononuclear cells, 30% and 70% Percoll (Cytiva, USA) was utilized during centrifugation at 2500 rpm for 20 min. Next, the tumor-infiltrating mononuclear cells were washed with PBS and resuspended in RPMI 1640 medium to obtain a single-cell suspension. The remaining methods can be seen in the online supplemental material.
jitc-2021-004219supp002.pdf (3.9MB, pdf)
Statistical analysis
Data were presented as means±SD. Quantitative data were analyzed using t-test or one-way analysis of variance. For flow cytometry data, FlowJo software (Tree Star) was utilized for every individual sample. The cut-off value of CD155 integrated optical density was determined by the median value of CD155 density in macrophages, and Kaplan-Meier survival curves were plotted using patient survival data and tested by log-rank test. SPSS V.22.0 (Chicago, USA) and Prism V.7.0 (California, USA) software were adopted for data analysis. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 and ns, not significant. P value <0.05 was considered to indicate a statistically significant difference.
Results
TAMs in CRC tissues showed higher expression of CD155 and exhibited an M2-phenotype
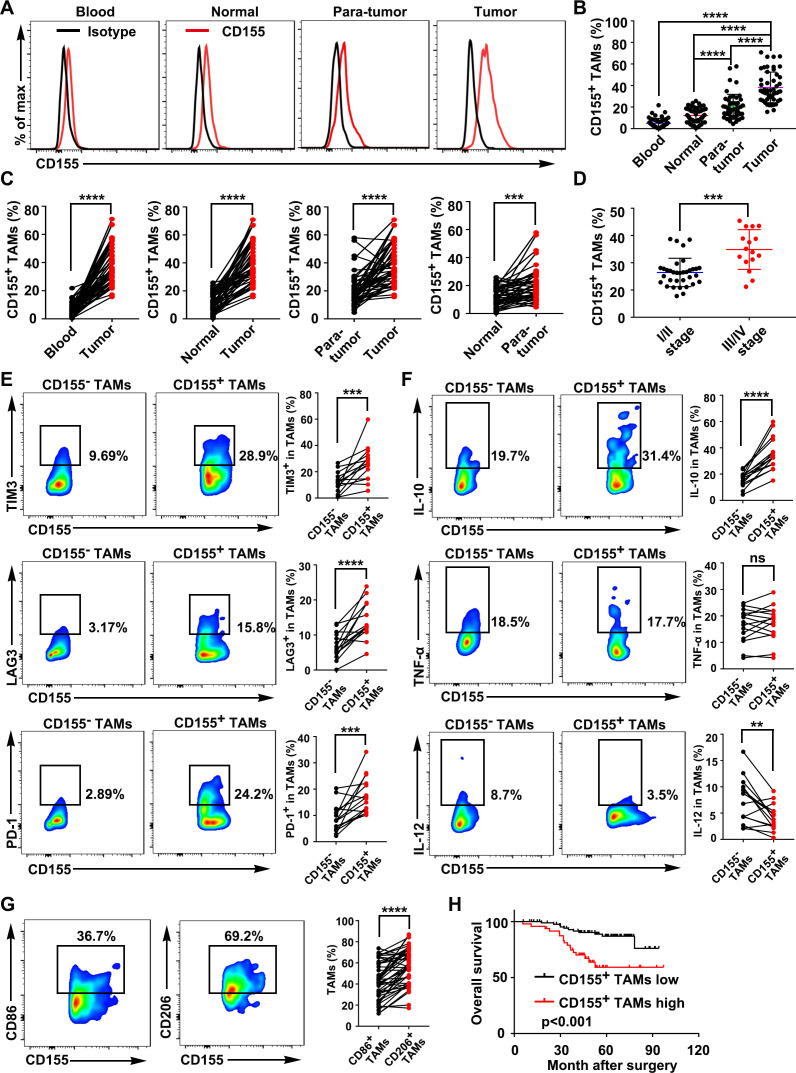
The CD155 expression in macrophages was determined with flow cytometry in paired blood samples, normal tissues, paratumor tissues, and tumor tissues from 50 patients with CRC. Gating strategies to investigate the proportion of various immune cells in samples were presented in online supplemental figure S1. The results showed that intratumoral macrophages expressed a higher level of CD155 than non-tumor tissue macrophages and peripheral macrophages (figure 1A–C, online supplemental figure S2A). Tumor tissue showed a higher number of CD155+ TAMs than the paratumor tissue. Similarly, paratumor tissues showed a higher number of CD155+ TAMs than the adjacent normal intestinal tissue. In addition, immunofluorescence and immunohistochemistry staining exhibited higher numbers of CD155+ TAMs in CRC tissue than paired adjacent normal intestinal tissue (online supplemental figure S2B–G). The number of macrophages (CD68+ cells) in the TME was robustly higher than in adjacent normal tissue (online supplemental figure S2C-G). The percentage of CD155+ TAMs in tumor tissue was higher in patients with CRC with disease stages III/IV than in those with disease stages I/II, indicating the clinical relevance of intratumoral CD155+ TAMs in CRC (figure 1D). The phenotype of CD155+ TAMs was further analyzed based on the levels of cell surface inhibitory receptors, cytokines, and macrophage polarization markers. CD155+ TAMs from CRC tissue showed higher expression of cell surface inhibitory receptors T-cell immunoglobulin and mucin domain 3 (TIM-3), lymphocyte-activation gene 3 (LAG-3), and programmed cell death protein- 1 (PD-1) (figure 1E). The expression of the M2 phenotype markers interleukin (IL)-10 and transforming growth factor (TGF)-β were elevated and the M1 phenotype marker IL-12 was reduced in CD155+ TAMs compared with CD155– TAMs in human CRC tissues (figure 1F, online supplemental figure S2E, H). However, the inflammatory marker tumor necrosis factor-α expression pattern was similar in CD155+ TAMs and CD155– TAMs (figure 1F). We further analyzed the typical M1 and M2 cell surface markers in TAMs, and the M2 marker CD206 was more highly expressed on CD155+ TAMs compared with the M1 marker CD86 (figure 1G).
Figure 1.
CD155+ TAMs were predominant in CRC tissue and showed an M2 macrophage phenotype. (A) Representative FACS images showed the expression patterns of CD155 on macrophages from the paired blood samples, normal tissues, paratumor tissues, and tumor tissues of patients with CRC (gated on CD68+ cells). (B) Quantitative analysis of CD155+ macrophages from the FACS analysis (n=50). (C) Percentage of CD155+ macrophages presented in paired blood samples and tumors, paired normal tissues and tumors, paired paratumor tissues and tumor tissues, and paired normal tissues and paratumor tissues of patients with CRC (n=50). (D) Percentage of CD155+ TAMs in CRC tissue of patients with tumor stages I/II and III/IV. (E) Expression pattern of TIM-3, LAG-3, and PD-1 on CD155– and CD155+ TAMs presented in CRC tissues (n=15). (F) Expression pattern of IL-10, TNF-α, and IL-12 in CD155– and CD155+ TAMs presented in CRC tissues (n=15). (G) Expression pattern of the M1 phenotype marker CD86 and the M2 phenotype marker CD206 in CD155+ TAMs (n=50). (H) Kaplan-Meier analysis of overall survival according to low and high CD155 expression in 141 patients with CRC. Data were presented as mean±SD. A significant difference between the groups, **p<0.01, ***p<0.001, and ****p<0.0001. CRC, colorectal cancer; FACS, fluorescence activated cell sorter; IL, interleukin; LAG-3, lymphocyte-activation gene 3; ns, no significant difference; PD-1, programmed cell death protein-1; TAM, tumor-associated macrophages; TIM-3, T-cell immunoglobulin and mucin domain 3; TNF, tumor necrosis factor.
CD155+ TAMs were negatively associated with the survival of patients with CRC
CD8+ T cells are the most powerful effectors in the anticancer immune response. The percentage of CD155+ TAMs was negatively correlated with CD8+ T cells in CRC tissues (online supplemental figure S3). Among the 50 patients with CRC, 3 patients diagnosed with lymph node metastasis were used for further study. Metastasized lymph nodes showed a higher number of CD155+ TAMs than the primary tumor (online supplemental figure S4). These results indicated that CD155+ TAMs selectively accumulate in the CRC tissues and lymph node metastasized tumors with a protumor M2-like phenotype creating favorable conditions for cancer progression. According to the median value of CD155 density in TAMs, patients with CRC who underwent surgery with complete follow-up data were divided into a CD155+ TAMs high group (n=48, online supplemental table 1) and a CD155+ TAMs low group (n=93, online supplemental table 1), and the readouts showed a negative association between the frequency of CD155+ TAMs and the prognosis of patients with CRC (figure 1H, online supplemental figure S5).
CD155 level in macrophages regulates their phenotypic and functional transition
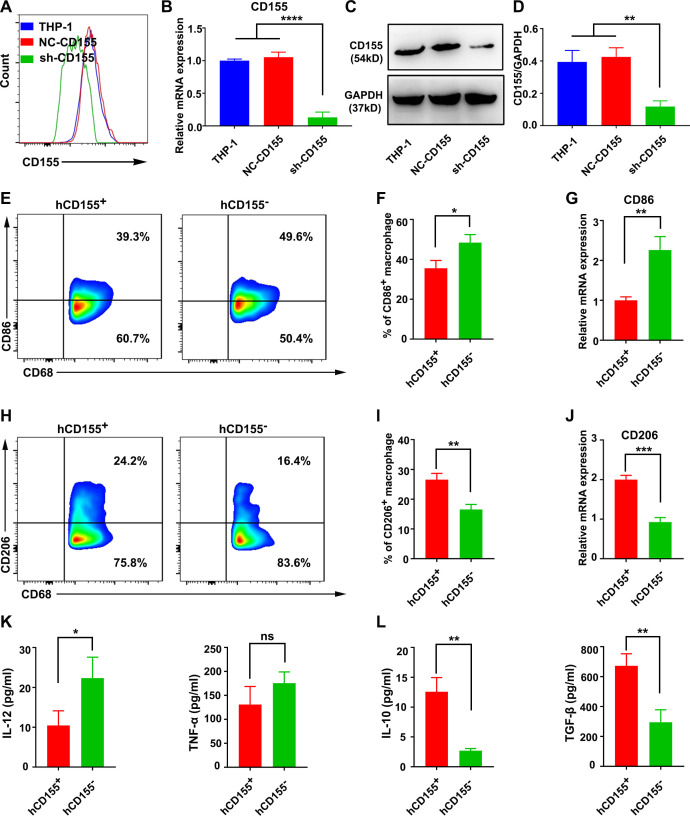
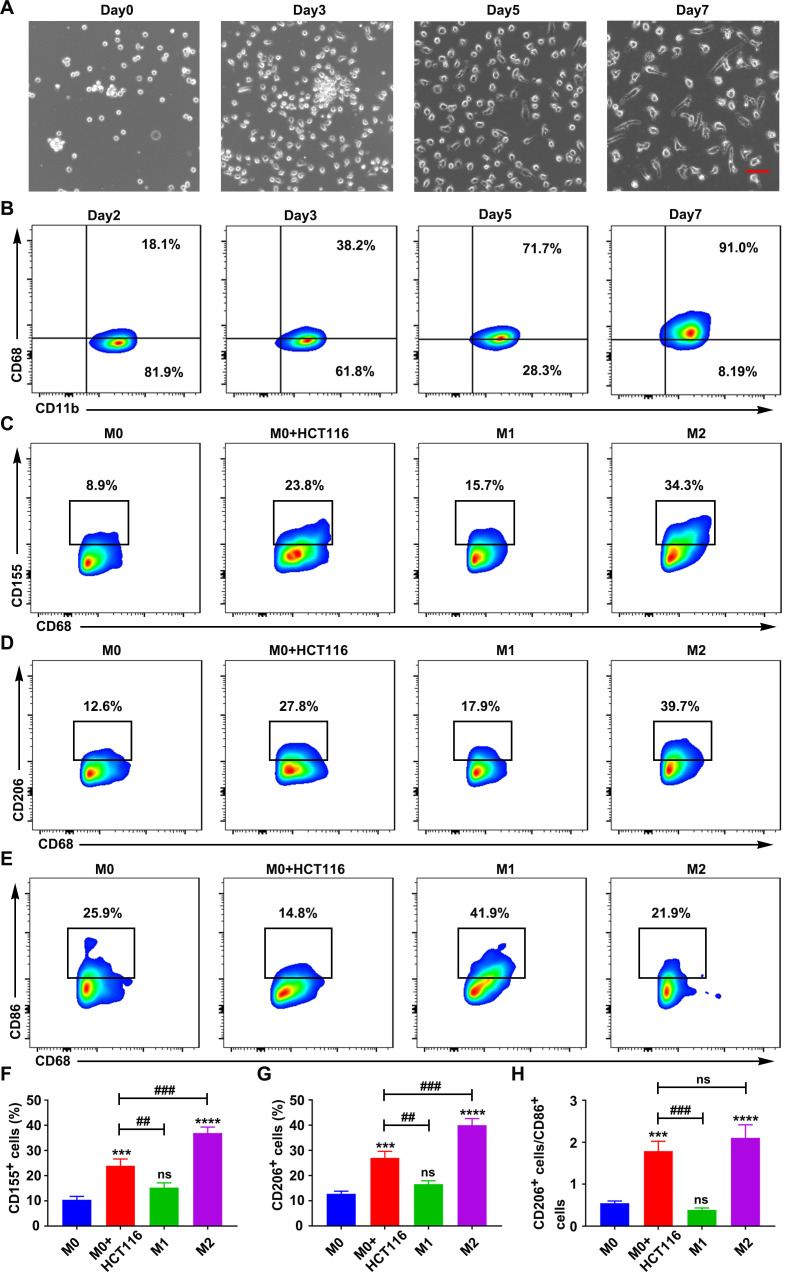
We analyzed the biological function of M2 polarized CD155+ macrophages in vitro using the human monocyte cell line THP-1 and the mouse macrophage cell line RAW264.7. The THP-1 cells were differentiated into macrophages by incubation with 100 ng/mL 12-myristate 13-acetate (PMA, Sigma, USA) in RPMI 1640 medium for 24 hours, and the CD68 expression was determined by fluorescence activated cell sorter (FACS) (online supplemental figure S6A, B). THP-1 and RAW264.7-derived macrophages showed higher expression of CD155 than the human PBMCs-derived and mouse bone marrow-derived macrophages, respectively (online supplemental figure S6C, D). This could be due to the tumor origin of THP-1 and RAW264.7 cells. Therefore, in this study, we used THP1 and RAW264.7-derived macrophages as models of CD155 overexpressing macrophages (CD155+) for in vitro studies. To create CD155 knockdown macrophages (CD155−), THP-1 and RAW264.7 cells were transfected with the sh-CD155 expression virus (online supplemental figure S6E, F). Then, puromycin was utilized to select resistant colonies, that is, 2 µg/mL for THP-1 cells and 6 µg/mL for RAW264.7 cells (online supplemental figure S6G, H). The expression of CD155 was downregulated in stable transformants of THP1 (figure 2A–D) and RAW264.7 cells (online supplemental figure S7A–D) as confirmed by flow cytometry, real time quantitative PCR (RT-qPCR), and Western blot analysis.
Figure 2.
CD155 level in macrophages determined phenotypic transition and function. Expression pattern of CD155 in THP-1 cells, NC-CD155 transfected THP-1 (hCD155+), and sh-CD155 transfected THP-1 (hCD155–) cells analyzed by FACS (A), RT-qPCR (B), and Western blot analysis (C and D). (E–G) CD86 expression patterns in LPS+IFN-γ-treated hCD155+ and hCD155– macrophages. (H–J) CD206 expression patterns in IL-4+IL-13-treated hCD155+ and hCD155– macrophages (gated on CD68+ cells). (K) IL-12 and TNF-α protein expression patterns in LPS/IFN-γ-treated hCD155+ and hCD155– macrophages. (L) IL-10 and TGF-β protein expression patterns in IL-4/IL-13-treated hCD155+ and hCD155– macrophages. Data were presented as mean±SD, n=3. A significant difference between the groups, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. FACS, fluorescence activated cell sorter; IL, interleukin; IFN, interferon; LPS, lipopolysaccharide; mRNA, messenger RNA; ns, no significant difference; RT-qPCR, real time quantitative PCR; TGF, transforming growth factor; TNF, tumor necrosis factor.
After treatment with lipopolysaccharide (LPS)+interferon (IFN)-γ, CD155– human macrophages showed a higher M1 polarization concomitant with increased CD86 expression compared with CD155+ macrophages (figure 2E–G). After treatment with IL-4+IL-13, CD155– human macrophages showed a reduced M2 polarization commitment with reduced CD206 expression (figure 2H–J). A similar trend of CD86 and CD206 expression was observed in M1 and M2-induced CD155– mouse macrophages compared with CD155+ macrophages (online supplemental figure S7E, F). The M1 marker IL-12 was enhanced in M1-induced CD155– human macrophages compared with CD155+ human macrophages (figure 2K). The protein expression levels of M2 markers IL-10 and TGF-β were reduced in M2-induced CD155– human macrophages compared with CD155+ human macrophages (figure 2L). These results suggested the role of CD155 levels in the phenotypic transition and function of macrophages.
CD155+ macrophages promoted the migration and invasion of CRC cells
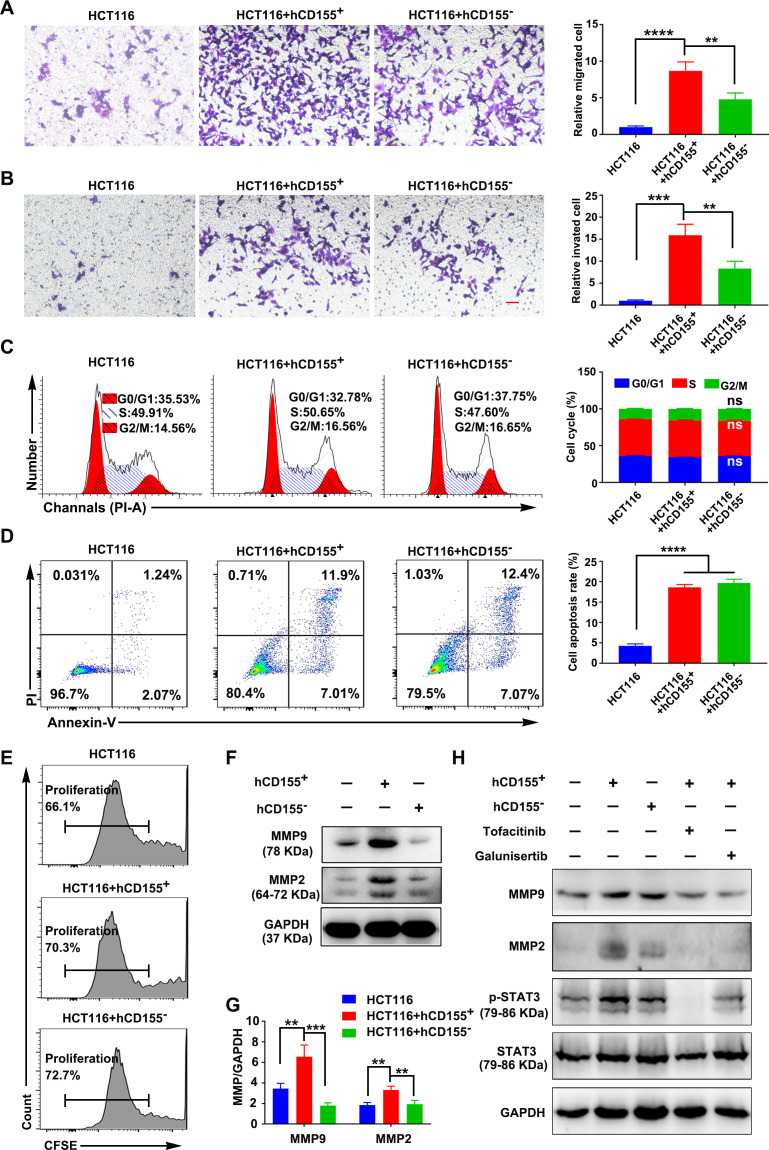
Macrophages and CRC cells co-culture showed that CD155– human and mouse macrophages inhibited CRC cell migration and invasion compared with the respective CD155+ macrophages (figure 3A, B, online supplemental figure S8A, B). The CD155– macrophages did not affect the CRC cell cycle and apoptosis compared with CD155+ macrophages (figure 3C, D, online supplemental figure S8C, D). CD155+ macrophages did not affect the CRC cell proliferation rate (figure 3E, online supplemental figures S8E and 9A, B). These results indicated the regulatory role of the CD155 expression level in macrophages on CRC cell migration and invasion.
Figure 3.
CD155+ macrophages (THP-1-derived) enhanced the migration and invasion of human CRC cells. The migration (A), invasion (B), cell cycle status (C), apoptosis (D), and proliferation rate (E) of CRC cells (HTC116) during co-cultured with hCD155+ or hCD155– macrophages. Scale bar: 100 µm. (F and G) MMPs expression in CRC cells during co-cultured with hCD155+ or hCD155– macrophages. (H) Expression pattern of MMPs and pSTAT3/STAT3 in CRC cells during co-cultured with hCD155+ or hCD155– macrophages with or without inhibition of STAT3 or TGF-β signaling. Data were presented as mean±SD, n=3. A significant difference between the groups, **p<0.01, ***p<0.001, and ****p<0.0001. ns, no significant difference. Tofacitinib (2.5 µM): JAK/STAT3 signaling inhibitor, galunisertib (10 µM): TGF-β signaling inhibitor. CRC, colorectal cancer; MMP, matrix metalloproteinases; TGF, transforming growth factor.
CD155+ macrophages induced MMPs expression and activated pSTAT3 signaling in CRC cells
The migratory and invasive potential of CRC cells is dependent on matrix metalloproteinases (MMPs) anchored on the cell surface.24 Thus, we speculated that MMPs may play a role in macrophage-mediated CRC invasion and migration. CRC cells co-cultured with CD155– macrophages showed reduced expression of MMP2 and MMP9 compared with the CRC cells co-cultured with CD155+ macrophages (figure 3F, G, online supplemental figure S8F, G).
STAT3 signaling mediated CRC cell migration and invasion.25 26 CRC cells co-cultured with CD155– macrophages showed reduced expression of pSTAT3 compared with the CRC cells co-cultured with CD155+ macrophages. Inhibition of STAT3 signaling by tofacitinib in CRC cells during co-culture with CD155+ macrophages reduced the expression of p-STAT3, MMP2, and MMP9 in CRC cells (figure 3H, online supplemental figures S8H and S9C, D).
CD155+ macrophage-produced TGF-β promoted the migration and invasion of CRC cells possibly via activation of the STAT3/MMPs signaling cascade
TGF-β was overexpressed in M2-induced CD155+ macrophages compared with CD155– macrophages (figure 2L). TGF-β, an M2 macrophage phenotype marker, induces cancer cell invasion.27 28 We further analyzed the possible role of CD155+ M2 macrophage-produced TGF-β on CRC cell migration and invasion. Inhibition of TGF-β signaling by galunisertib in CRC cells during co-culture with CD155+ macrophages reduced the expression of pSTAT3, MMP2, and MMP9 in CRC cells (figure 3H, online supplemental figures S8H and S9C, D), as well as the migration and invasion of CRC cells (online supplemental figure S10). Interestingly, the inhibition of STAT3 signaling during CRC cells and CD155+ macrophage co-culture dramatically reduced the migration and invasion of CRC cells (online supplemental figure S10). Taken together, we speculated that CD155+ macrophages participated in CRC progression via TGF-β-mediated activation of the STAT3/MMPs cascade in CRC cells.
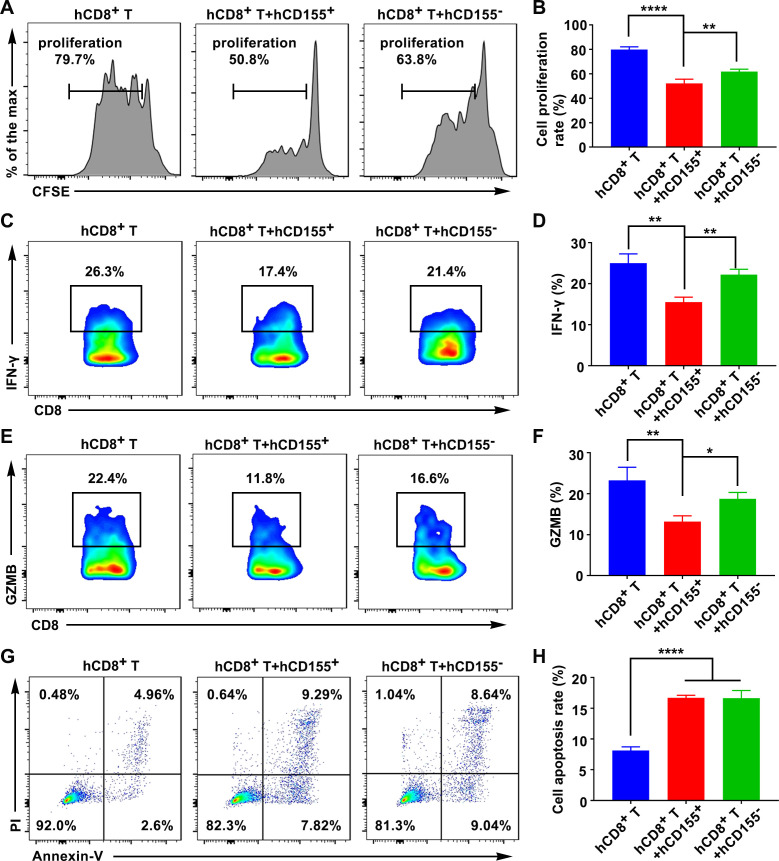
CD155+ macrophages suppressed CD8+ T-cell proliferation and function
CD8+ T cells are the most powerful effectors in the antitumor immune response. To investigate whether CD155+ macrophages regulated CD8+ T-cell function in the TME, CD8+ T cells were isolated using immunomagnetic beads from healthy subjects or normal mice (online supplemental figure S11A–D). CD155+ or CD155– macrophages were co-cultured with anti-CD3/CD28-stimulated CD8+ T cells for 3 days. CD155+ macrophages inhibited and CD155– macrophages enhanced the proliferation of cytotoxic CD8+ T cells (figure 4A, B, online supplemental figure S12A, B). IFN-γ and granzyme B (GZMB) are polyfunctional phenotype markers of CD8+ T cells. CD155+ macrophages inhibited the expression of IFN-γ and GZMB in cytotoxic CD8+ T cells compared with CD155– macrophages (figure 4C–F, online supplemental figure S12C–F). The high and low levels of CD155 in macrophages were not related to CD8+ T-cell apoptosis (figure 4G, H, online supplemental figure S12G, H).
Figure 4.
CD155 level in human macrophages regulated CD8+ T-cell proliferation and function. (A and B) The proliferation rate of CD8+ T cells during co-cultured with hCD155+ or hCD155– macrophages. Expression pattern of IFN-γ (C and D), and GZMB (E and F), in CD8+ T cells during co-cultured with hCD155+ or hCD155– macrophages (gated on CD8+ cells). (G and H) Apoptosis rate of CD8+ T cells during co-cultured with hCD155+ or hCD155– macrophages. Data were presented as mean±SD, n=3. A significant difference between the groups, *p<0.05, **p<0.01, and ****p<0.0001. GZMB, granzyme B; IFN, interferon; ns, no significant difference.
CRC cells induced CD155 expression in macrophages
The phenotypic transition of macrophages was analyzed during co-culture with CRC cells. CD14+ monocytes from human peripheral blood and mouse bone marrow cells were induced to differentiate into macrophages (figure 5A, B, online supplemental figure S11E, F and S13A, B). CD155 expression on human and mouse primary macrophages was relatively low and was not affected during the period of macrophage transition (online supplemental figure S14). The expression patterns of CD155, CD206, and CD86 were analyzed in macrophages co-cultured with CRC cells, M1-induced macrophages, and M2-induced macrophages. Macrophages co-cultured with CRC and M2-induced macrophages alone showed higher expression of CD155 than M1-induced macrophages (figure 5C, F, online supplemental figure S13C, F). M2-induced macrophages showed the highest expression of CD206 compared with macrophages co-cultured with CRC cells and M1-induced macrophages (figure 5D, G, online supplemental figure S13D, G). Macrophages co-cultured with CRC cells and M2-induced macrophages alone showed a similar pattern of CD206:CD86 expression ratio (figure 5E, H, online supplemental figure S13E, H). These results indicated that CRC cells increased the expression of CD155 in macrophages and the M2:M1 ratio creating a favorable TME for cancer growth. IL-4 and IL-13 are the key inducers of M2-like TAM polarization in the TME.29 30 Our results showed higher expression of IL-4 in CRC cells than in normal intestinal epithelial cells (online supplemental figure S15A, C). There was no obvious difference in IL-13 expression in CRC cells and normal intestinal epithelial cells (online supplemental figure S15B, D). Moreover, CD155 expression decreased in primary macrophages co-cultured with CRC cells with anti-IL-4 antibody treatment compared with the macrophages co-cultured with CRC cells without anti-IL-4 antibody treatment (online supplemental figure S15E–H). These results suggested that IL-4 secreted by CRC cells may trigger CD155+ macrophage expansion with a protumor M2-like phenotype.
Figure 5.
CRC cells triggered CD155 expression in human macrophages. (A) Microscopic images of differentiating macrophages from human PBMC-derived monocytes. Scale bar: 50 µm. (B) CD68 expression pattern during macrophagic differentiation of monocytes. CD155 expression (C), CD206 expression (D), and CD86 expression (E), in macrophages during co-cultured with CRC cells, M1 (LPS+IFN-γ-treated) and M2 (IL-4+IL-13-treated) polarization (gated on CD68+ cells). (F–H) Statistical analysis of CD155 expression, CD206 expression, and the ratio of CD206 to CD86 in macrophages during co-cultured with CRC cells, M1 (LPS+IFN-γ-treated) and M2 (IL-4+IL-13-treated) polarization. Data were presented as mean±SD, n=3. A significant difference between the groups, ##p<0.01 and ###p<0.001, and compared with M0 group ***p<0.001 and ****p<0.0001. CRC, colorectal cancer; IFN, interferon; IL, interleukin; LPS, lipopolysaccharide; ns, no significant difference; PBMC, peripheral blood mononuclear cell.
Bone marrow transplantation from CD155–⁄– mice inhibited ectopic CRC tumor growth in wild-type mice
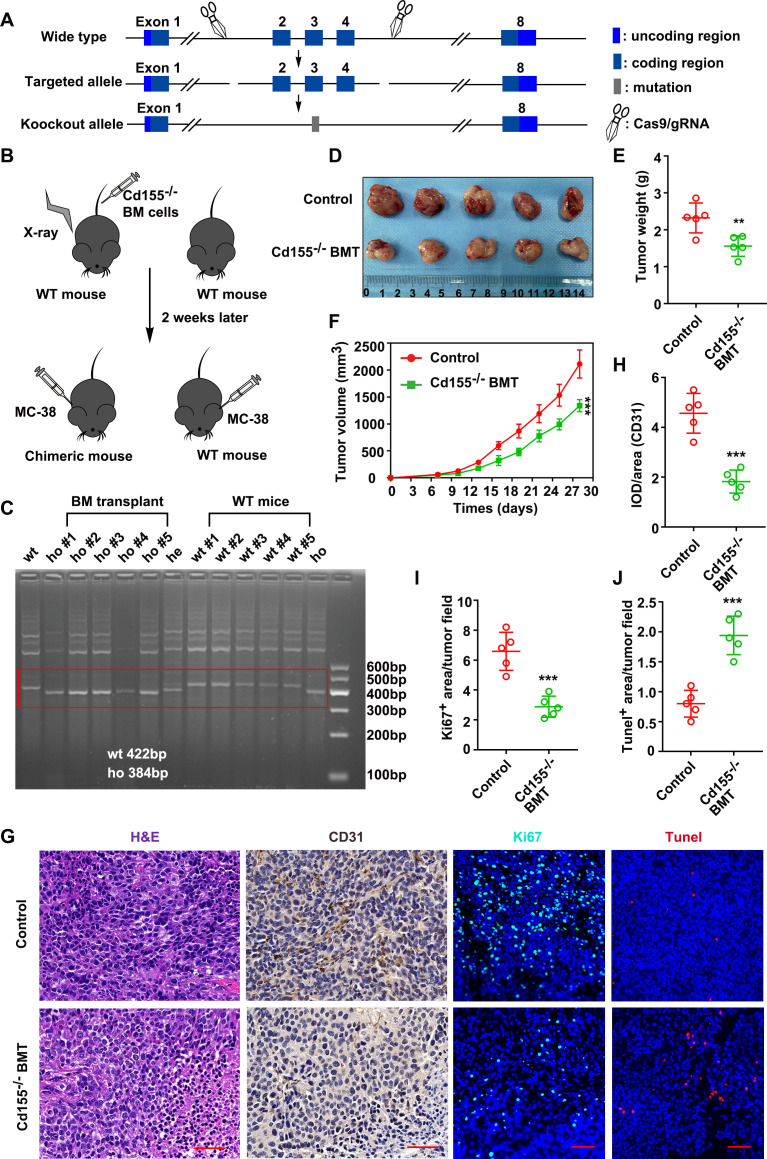
To explore the protumor effect of CD155+ macrophages in vivo, two subcutaneous CRC models (CT26 cells in BALB/c and MC-38 cells in C57BL/6 mice) were established. Cd155–⁄– mice were purchased from Shanghai Model Organisms (Shanghai, China), and a bone marrow transplantation (BMT) from these null mice into irradiated wild-type mice (C57BL/6) was performed to populate a BM with CD155 null monocytes (figure 6A–C, online supplemental figure S16A, B). Then, the subcutaneous tumor model was established with MC-38 CRC cells. online supplemental figures S17,18a Cd155–⁄– BMT mice inhibited tumor growth compared with the wild-type mice as indicated by the tumor size, weight, and volume (figure 6D–F). Tumor cells from the Cd155–⁄– BMT group showed lower tumor vessels and proliferation, and a higher cell apoptosis rate than those from the wild-type group as indicated by CD31, Ki-67, and Tunel staining (figure 6G–J). These results indicated the direct role of CD155+ macrophages on CRC progression.
Figure 6.
CD155–⁄– BMT inhibited tumor growth in the mouse ectopic CRC model. (A) Schematic illustration of CD155 knockout. (B) Cd155–⁄– BM cells were transplanted into irradiated wild-type mice to populate a BM with CD155 null monocytes. (C) Mouse blood genotypes were verified by agarose gel electrophoresis. (D) Macroscopic images of tumor tissues from wide type and CD155–⁄– BMT groups. Quantitative analysis of tumor weight (E), and tumor volume (F). (G) Representative microscopic images of tumor tissue sections showed tumor morphology (H&E staining), blood vessels (CD31 IHC), cell proliferation (Ki-67 IHC), and apoptosis (TUNEL). Scale bar: 50 µm. (H–J) Quantitation of blood vessels, proliferation rate, and apoptosis rate in tumor tissue sections. Data were presented as mean±SD, n=5. A significant difference between the groups, **p<0.01 and ***p<0.001. BM, bone marrow; BMT, bone marrow transplantation; CRC, colorectal cancer; IHC, immunohistochemistry; WT, wild-type.
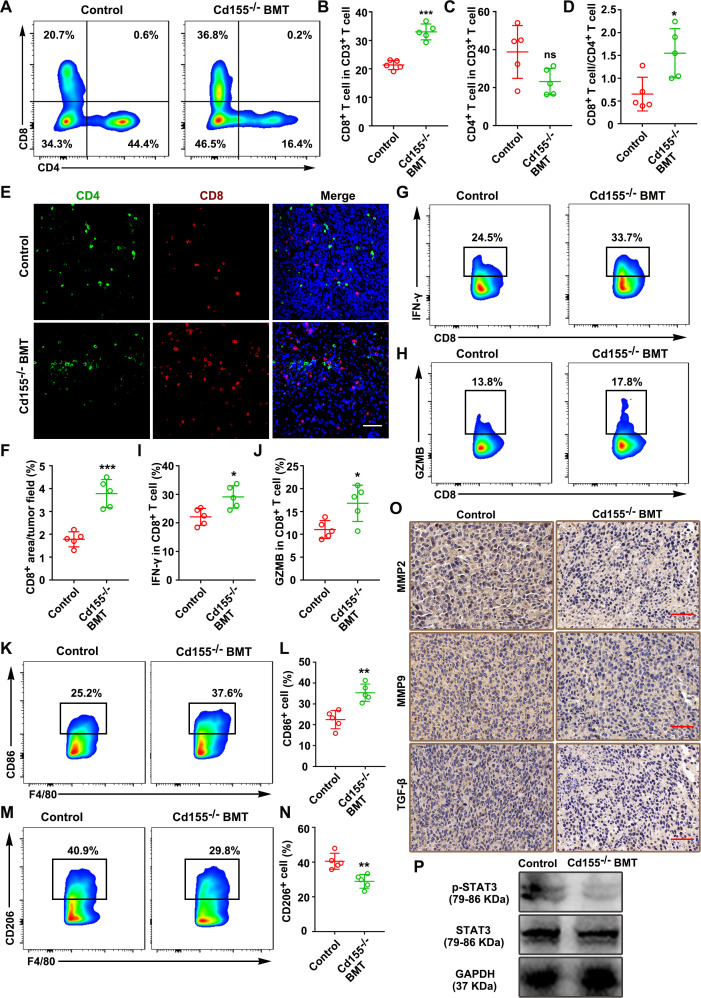
To better understand the tumor growth-promoting effect of CD155+ macrophages, we analyzed the T-cell subpopulations in tumor tissues. CD155+ TAMs expression was relatively low in the tumor tissue from the Cd155–⁄– BMT group compared with the wild-type group (online supplemental figure S16A, B). The Cd155–⁄– BMT group showed a higher percentage of CD8+ T cells than the wild-type group (figure 7A, B). However, there was no obvious difference in the percentage of CD4+ T cells (figure 7C). Interestingly, the CD8+/CD4+ ratio was increased in the Cd155–⁄– BMT group compared with the wild-type group (figure 7D). Immunofluorescence staining of tumor sections showed increased infiltration of CD8+ T cells in tumor tissues of the Cd155–⁄– BMT group compared with that of the wild-type group (figure 7E, F). IFN-γ and GZMB expression was higher in the Cd155–⁄– BMT group than wild-type group (figure 7G–J). Taken together, these results indicated that the increased infiltration of CD8+ T cells could be a factor for tumor growth inhibition in the Cd155–⁄– BMT group.
Figure 7.
CD155–⁄– BMT promoted the antitumor immune response in the mouse ectopic CRC model. Number of CD8+ and CD4+ T cells (A–C), and CD8+/CD4+ T-cell ratio (D), in mouse tumor tissues examined by flow cytometry (gated on CD3+ cells). (E and F) Representative immunofluorescence images of CD4 and CD8 expressing cells in tumor tissues. Scale bar: 50 µm. (G–J) Expression pattern of IFN-γ and GZMB in CD8+ T cells of tumor tissue. CD86 (K and L) and CD206 expression patterns (M and N) of TAMs in tumor tissues (gated on F4/80+ cells). (O) Representative microscopic images of MMP2 (IHC), MMP9 (IHC), and TGF-β (IHC) in tumor tissues. Scale bar: 50 µm. (P) Expression pattern of pSTAT3/STAT3 in tumor tissues. Data were presented as mean±SD, n=5. A significant difference between the groups, *p<0.05, **p<0.01, and ***p<0.001. BMT, bone marrow transplantation; CRC, colorectal cancer; GZMB, granzyme B; IFN, interferon; IHC, immunohistochemistry; MMP, matrix metalloproteinases; ns, no significant difference; TAM, tumor-associated macrophages.
In addition, the proportion of M1-like TAMs (CD11b+F4/80+CD86+) was increased in the Cd155–⁄– BMT group (figure 7K, L), and the proportion of M2-like TAMs (CD11b+F4/80+CD206+) was decreased in the Cd155–⁄– BMT group (figure 7M, N) compared with the wild-type group. Besides, the MMPs, TGF-β, and p-STAT3/STAT3 expression levels were decreased in the Cd155–⁄– BMT group compared with the wild-type group (figure 7O, P, online supplemental figure S16C–F). The original macrophages in BALB/c mice were depleted by clodronate liposome injection intraperitoneally two times a week (online supplemental figures S17,18a). Similar trends in the results from Cd155–⁄– BMT mice group were observed in macrophage ablated BALB/c mice treated with CD155– macrophage (online supplemental figures S18 and S19). Collectively, our results suggested that CD155+ TAMs are responsible for tumor cell progression possibly via suppression of the adaptive anticancer immune response.
Discussion
CD155 is an onco-immunogenic molecule associated with tumor growth, progression, and metastasis.7 Previous studies have reported the overexpression of CD155 in the CRC TME.11 21 TAMs are the most abundant immune cells in the CRC TME and play a vital role in CRC progression and metastasis.30 However, the CD155 expression levels in CRC TAMs and the role of CRC TAMs-specific CD155 in cancer progression and the anticancer immune response have not been reported yet. We found overexpression of CD155 in CRC TAMs compared with macrophages in paratumor and adjacent normal tissues. The CD155 level was higher in TAMs of CRC at III/IV stages than that at I/II stages, and the CD155+ TAMs were negatively associated with the survival of patients which indicates a positive correlation of CD155 expression levels in TAMs with CRC progression. In addition, CD155+ TAMs showed an M2 phenotype and enhanced CRC cell migration, invasion, and tumor growth. Moreover, CD155+ TAMs inhibited CD8+ T-cell proliferation and function as well as the CD8+/CD4+ T-cell ratio indicating its role in cancer-induced immunosuppression. On the other hand, CRC cells enhanced CD155 expression in macrophages indicating the stimulatory role of CRC cells in the development of CD155+ TAMs. Taken together, our results indicate that CD155+ TAMs are responsible for the M2-phenotype transition, immunosuppression, and tumor progression in CRC.
CD155 expression was robustly higher in RAW264.7 and THP-1-derived macrophages than in primary macrophages derived from mouse BM cells and human monocytes, respectively. Therefore, CD155 was knocked down in THP-1 and RAW264.7 cells by using a set of distinct CD155 shRNAs. TIM3, LAG3, and PD-1 are the key immune checkpoint molecules in immune cells of various cancers including CRC.31 CD155+ CRC TAMs showed higher expression of TIM3, LAG3, and PD-1 than CD155– CRC TAMs. A previous study has reported the TAMs-specific overexpression of PD-1 and its inhibitory role in phagocytosis and tumor immunity.32 This is the first study to report the expression pattern of TIM3, LAG3, and PD-1 in CD155+ TAMs indicating the possible role of CD155+ TAMs in CRC immunity.
In vitro, LPS+IFN-γ treatment in CD155+ macrophages resulted in a reduced expression of CD86 and IL-12 compared with that of CD155– macrophages. In contrast, IL-4+IL-13 treatment in CD155+ macrophages resulted in a higher expression of CD206, IL-10, and TGF-β compared with that of CD155– macrophages. The results from clinical CRC tissues also indicated higher expression of the M2-specific cytokine TGF-β in CRC tissues. Taken together, CD155+ macrophages showed a higher tendency to polarize toward M2 macrophages. M2 macrophages in the TME trigger CRC progression and metastasis.33 However, the role of CD155+ TAMs in CRC progression and metastasis has not yet been investigated. Our in vitro study showed that CD155+ macrophages exhibit an M2 phenotype and promote CRC cell migration and invasion. Furthermore, Cd155–⁄– BMT robustly inhibited tumor growth in the CRC ectopic model compared with wild-type mice, supporting the findings from the clinical study.
CRC TME showed higher expression of CD155 and TGF-β, and CD155+ macrophages showed robustly higher expression of TGF-β than the CD155– macrophages. TAMs have been reported to regulate pancreatic ductal adenocarcinoma progression via TGF-β signaling.34 Moreover, TGF-β signaling activates STAT3 to promote cancer invasion and metastasis.35 CD155+ macrophages and CRC cells co-culture activated STAT3 in CRC cells. STAT3 activation regulated MMP2 and MMP9 expression, tumor invasion, and metastasis.36 Overexpressed MMPs in the TME regulated matrix remodeling, angiogenesis, cell signaling, and migration.37 MMP2 and MMP9 overexpressed in the serum and TME of CRC and promoted CRC progression, angiogenesis, and metastasis.38 39 In this study, CD155+ macrophages induced MMP2 and MMP9 production in CRC cells. Inhibition of STAT3 signaling prevented CD155+ macrophage-induced MMP2/MMP9 production, migration, and invasion of CRC cells. Moreover, inhibition of TGF-β signaling inhibited STAT3 activation, MMP2/MMP9 expression, migration, and invasion of CRC cells. Taken together, CD155+ macrophages promoted CRC progression and invasion via the TGF-β-mediated STAT3/MMPs axis.
CD8+ T cells are the most powerful effectors in the anticancer immune response. TAMs promoted CD8+ T-cell dysfunction and tumor growth.40 M2 macrophage-released IL-10 in the TME inhibits the CD8+ T cell-dependent response to chemotherapy.41 In contrast, M1 macrophages release IL-12 which enhances the proliferation and cytotoxic activity of CD8+ T cells.42 In the current study, CD155+ TAMs showed robustly higher expression of IL-10 and lower expression of IL-12 than CD155– TAMs. The CD155 expression level in TAMs is inversely correlated with the number of CD8+ T cells in the CRC TME. A high CD8+/CD4+ T-cell ratio is associated with a favorable prognosis in various cancers including CRC.43 44 The number of CD155+ TAMs inversely correlated with the CD8+/CD4+ T-cell ratio in the CRC TME. In co-culture, CD155+ macrophages inhibited CD8+ T cells’ proliferation and the expression of IFN-γ and GZMB compared with CD155– macrophages. Similarly, Cd155–⁄– BMT increased the CD8+ T-cell level, and CD8+/CD4+ T cells’ ratio in the CRC ectopic model compared with wild-type mice. In addition, IFN-γ and GZMB levels were also increased in the Cd155–⁄– BMT group compared with the wild-type group. Our results indicated the possible inhibitory role of CD155+ TAM-released higher levels of IL-10 and TGF-β, and reduced level of IL-12 on CD8+ T-cell proliferation and function causing anti-CRC immune suppression.
Cancer cells created a favorable TME for their growth and progression by modulating the function of immune cells.45 CRC cells have been reported to promote exosome-mediated M2 macrophage polarization.46 In this study, CD155 expression was relatively low and stable during the monocyte–macrophage transition. Macrophages co-cultured with CRC cells showed higher CD155 expression and CD206/CD86 expression ratio. CRC cell-produced IL-4 increased the M2/M1 TAMs ratio.47 When primary macrophages are co-cultured with CRC cells with anti-IL-4 antibody treatment, the increased CD155 expression can be neutralized. Thus, increased M2/M1 ratio and CD155 expression in macrophages co-cultured with CRC cells might be the effect of CRC-released IL-4. Further in vivo studies blocking IL-4 signaling is necessary to validate this hypothesis.
This study extensively analyzed the expression pattern of CD155 in CRC TAMs and investigated its role in CRC progression and immunosuppression using clinical samples, in vitro studies, and mouse ectopic CRC models. However, we did not investigate the molecular mechanism of CD155-mediated macrophage polarization, which is the key limitation of this study.
In conclusion, CD155+ TAMs were robustly higher in the TME than in the peripheral blood and adjacent normal tissue of patients with CRC. CD155 overexpression in TAMs correlated with M2 macrophage polarization, CD8+ T-cell proliferation and function, and tumor progression. In vitro and in vivo studies showed the stimulatory role of CD155+ macrophages in CRC cell migration/invasion and tumor growth but an inhibitory role in tumor immunity. CRC cells seemed to stimulate CD155 expression in macrophages and M2 polarization, indicating the possible role of CD155 in the crosstalk of macrophages and cancer cells within the CRC TME. Taken together, CD155+ TAMs are responsible for M2-phenotype transition, immunosuppression, and tumor progression in CRC. The specific localization of CD155+ TAMs in CRC tissue and paratumor tissue could turn into a potential therapeutic target for CRC treatment.
Footnotes
XZ and RL contributed equally.
Contributors: Conception and design: XZ, RL, HW, BW. Collection of samples: ZZ, TC, YH, JL, RL, DD, SH, JS. Data analysis and interpretation: XZ, RL, DD, BW. Manuscript writing: all authors. Review: JLP and BW. BW is responsible for the overall content as guarantor. Final approval of manuscript: all authors.
Funding: This work was supported by the Science and Technology Planning Project of Guangdong Province (2021A0505030020, and 2017B020227009), China Postdoctoral Science Foundation (2021M703721).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by the Ethical Committee of Third Affiliated Hospital of Sun Yat-sen University (reference number: 2021-02-012-01). Participants gave informed consent to participate in the study before taking part.
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, Sierra MS, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017;66:683–91. 10.1136/gutjnl-2015-310912 [DOI] [PubMed] [Google Scholar]
- 3.Brenner H, Kloor M, Pox CP. Colorectal cancer. The Lancet 2014;383:1490–502. 10.1016/S0140-6736(13)61649-9 [DOI] [PubMed] [Google Scholar]
- 4.Belli C, Trapani D, Viale G, et al. Targeting the microenvironment in solid tumors. Cancer Treat Rev 2018;65:22–32. 10.1016/j.ctrv.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 5.Mu Q, Najafi M. Modulation of the tumor microenvironment (Tme) by melatonin. Eur J Pharmacol 2021;907:174365. 10.1016/j.ejphar.2021.174365 [DOI] [PubMed] [Google Scholar]
- 6.Masson D, Jarry A, Baury B, et al. Overexpression of the CD155 gene in human colorectal carcinoma. Gut 2001;49:236–40. 10.1136/gut.49.2.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lepletier A, Madore J, O'Donnell JS, et al. Tumor CD155 expression is associated with resistance to anti-PD1 immunotherapy in metastatic melanoma. Clin Cancer Res 2020;26:3671–81. 10.1158/1078-0432.CCR-19-3925 [DOI] [PubMed] [Google Scholar]
- 8.Gao J, Zheng Q, Shao Y, et al. Cd155 downregulation synergizes with adriamycin to induce breast cancer cell apoptosis. Apoptosis 2018;23:512–20. 10.1007/s10495-018-1473-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendelsohn CL, Wimmer E, Racaniello VR. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 1989;56:855–65. 10.1016/0092-8674(89)90690-9 [DOI] [PubMed] [Google Scholar]
- 10.Okumura G, Iguchi-Manaka A, Murata R, et al. Tumor-Derived soluble CD155 inhibits DNAM-1-mediated antitumor activity of natural killer cells. J Exp Med 2020;217:20191290. 10.1084/jem.20191290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin Z-B, Long P, Zhao Z, et al. Long Noncoding RNA KCNQ1OT1 is a Prognostic Biomarker and mediates CD8+ T cell exhaustion by regulating CD155 Expression in Colorectal Cancer. Int J Biol Sci 2021;17:1757–68. 10.7150/ijbs.59001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol 2012;33:119–26. 10.1016/j.it.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sunakawa Y, Stintzing S, Cao S, et al. Variations in genes regulating tumor-associated macrophages (TAMs) to predict outcomes of bevacizumab-based treatment in patients with metastatic colorectal cancer: results from tribe and FIRE3 trials. Ann Oncol 2015;26:2450–6. 10.1093/annonc/mdv474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ning W-R, Jiang D, Liu X-C, et al. Carbonic anhydrase XII mediates the survival and prometastatic functions of macrophages in human hepatocellular carcinoma. J Clin Invest 2022;132:153100. 10.1172/JCI153110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamerkar S, Leng C, Burenkova O, et al. Exosome-Mediated genetic reprogramming of tumor-associated macrophages by exoASO-STAT6 leads to potent monotherapy antitumor activity. Sci Adv 2022;8:7002. 10.1126/sciadv.abj7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature 2013;496:445–55. 10.1038/nature12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seoane S, Martinez-Ordoñez A, Eiro N, et al. Pou1F1 transcription factor promotes breast cancer metastasis via recruitment and polarization of macrophages. J Pathol 2019;249:381–94. 10.1002/path.5324 [DOI] [PubMed] [Google Scholar]
- 18.Fang W, Zhou T, Shi H, et al. Progranulin induces immune escape in breast cancer via up-regulating PD-L1 expression on tumor-associated macrophages (TAMs) and promoting CD8+ T cell exclusion. J Exp Clin Cancer Res 2021;40:4. 10.1186/s13046-020-01786-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez-Garcia A, Lynn RC, Poussin M, et al. Car-T cell-mediated depletion of immunosuppressive tumor-associated macrophages promotes endogenous antitumor immunity and augments adoptive immunotherapy. Nat Commun 2021;12:877. 10.1038/s41467-021-20893-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKay ZP, Brown MC, Gromeier M. Aryl hydrocarbon receptor signaling controls CD155 expression on macrophages and mediates tumor immunosuppression. J Immunol 2021;206:1385–94. 10.4049/jimmunol.2000792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Q, Wang B, Gao J, et al. Cd155 knockdown promotes apoptosis via AKT/Bcl-2/Bax in colon cancer cells. J Cell Mol Med 2018;22:131–40. 10.1111/jcmm.13301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawada Y, Honda T, Nakamizo S, et al. Resolvin E1 attenuates murine psoriatic dermatitis. Sci Rep 2018;8:11873. 10.1038/s41598-018-30373-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark NM, Bos PD. Tumor-Associated macrophage isolation and in vivo analysis of their tumor-promoting activity. Methods Mol Biol 2019;1884:151–60. 10.1007/978-1-4939-8885-3_10 [DOI] [PubMed] [Google Scholar]
- 24.Björklund M, Heikkilä P, Koivunen E. Peptide inhibition of catalytic and noncatalytic activities of matrix metalloproteinase-9 blocks tumor cell migration and invasion. J Biol Chem 2004;279:29589–97. 10.1074/jbc.M401601200 [DOI] [PubMed] [Google Scholar]
- 25.Rokavec M, Öner MG, Li H, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest 2014;124:1853–67. 10.1172/JCI73531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park JH, van Wyk H, McMillan DC, et al. Signal transduction and activator of transcription-3 (STAT3) in patients with colorectal cancer: associations with the phenotypic features of the tumor and host. Clin Cancer Res 2017;23:1698–709. 10.1158/1078-0432.CCR-16-1416 [DOI] [PubMed] [Google Scholar]
- 27.Derynck R, Turley SJ, Akhurst RJ. Tgfβ biology in cancer progression and immunotherapy. Nat Rev Clin Oncol 2021;18:9–34. 10.1038/s41571-020-0403-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calon A, Espinet E, Palomo-Ponce S, et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 2012;22:571–84. 10.1016/j.ccr.2012.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foucher ED, Ghigo C, Chouaib S, et al. Pancreatic ductal adenocarcinoma: a strong imbalance of good and bad immunological cops in the tumor microenvironment. Front Immunol 2018;9:1044. 10.3389/fimmu.2018.01044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian W, Lei N, Zhou J, et al. Extracellular vesicles in ovarian cancer chemoresistance, metastasis, and immune evasion. Cell Death Dis 2022;13:64. 10.1038/s41419-022-04510-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasidharan Nair V, Toor SM, Taha RZ, et al. Dna methylation and repressive histones in the promoters of PD-1, CTLA-4, Tim-3, LAG-3, TIGIT, PD-L1, and galectin-9 genes in human colorectal cancer. Clin Epigenetics 2018;10:104. 10.1186/s13148-018-0539-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gordon SR, Maute RL, Dulken BW, et al. Pd-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 2017;545:495–9. 10.1038/nature22396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu C, Fan L, Lin Y, et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes 2021;13:1980347. 10.1080/19490976.2021.1980347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ottaviani S, Stebbing J, Frampton AE, et al. TGF-β induces miR-100 and miR-125b but blocks let-7a through LIN28B controlling PDAC progression. Nat Commun 2018;9:9. 10.1038/s41467-018-03962-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Wu C, Zhang C, et al. TGF-β-induced STAT3 overexpression promotes human head and neck squamous cell carcinoma invasion and metastasis through malat1/miR-30a interactions. Cancer Lett 2018;436:52–62. 10.1016/j.canlet.2018.08.009 [DOI] [PubMed] [Google Scholar]
- 36.Pakala SB, Rayala SK, Wang R-A, et al. Mta1 promotes STAT3 transcription and pulmonary metastasis in breast cancer. Cancer Res 2013;73:3761–70. 10.1158/0008-5472.CAN-12-3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 2010;141:52–67. 10.1016/j.cell.2010.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mochizuki S, Ao T, Sugiura T, et al. Expression and function of a disintegrin and metalloproteinases in cancer-associated fibroblasts of colorectal cancer. Digestion 2020;101:18–24. 10.1159/000504087 [DOI] [PubMed] [Google Scholar]
- 39.Zucker S, Vacirca J. Role of matrix metalloproteinases (MMPs) in colorectal cancer. Cancer Metastasis Rev 2004;23:101–17. 10.1023/a:1025867130437 [DOI] [PubMed] [Google Scholar]
- 40.Dong L, Chen C, Zhang Y, et al. The loss of RNA N6-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8+ T cell dysfunction and tumor growth. Cancer Cell 2021;39:945–57. 10.1016/j.ccell.2021.04.016 [DOI] [PubMed] [Google Scholar]
- 41.Ruffell B, Chang-Strachan D, Chan V, et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell 2014;26:623–37. 10.1016/j.ccell.2014.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian Y, Qiao S, Dai Y, et al. Molecular-Targeted immunotherapeutic strategy for melanoma via dual-targeting nanoparticles delivering small interfering RNA to tumor-associated macrophages. ACS Nano 2017;11:9536–49. 10.1021/acsnano.7b05465 [DOI] [PubMed] [Google Scholar]
- 43.Kimura H, Matsui Y, Ishikawa A, et al. Randomized controlled phase III trial of adjuvant chemoimmunotherapy with activated cytotoxic T cells and dendritic cells from regional lymph nodes of patients with lung cancer. Cancer Immunol Immunother 2018;67:1231–8. 10.1007/s00262-018-2180-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato E, Olson SH, Ahn J, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci U S A 2005;102:18538–43. 10.1073/pnas.0509182102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hara T, Chanoch-Myers R, Mathewson ND, et al. Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell 2021;39:779–92. 10.1016/j.ccell.2021.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao S, Mi Y, Guan B, et al. Tumor-Derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol 2020;13:156. 10.1186/s13045-020-00991-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin X, Wang S, Sun M, et al. miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J Hematol Oncol 2019;12:20. 10.1186/s13045-019-0708-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-004219supp001.pdf (185.8KB, pdf)
jitc-2021-004219supp002.pdf (3.9MB, pdf)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.