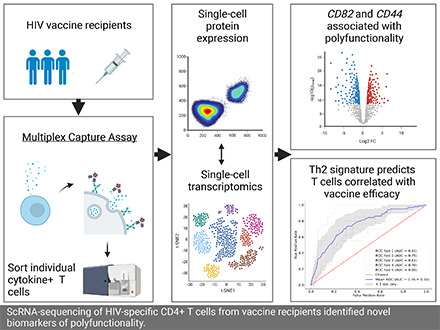
Abstract
Ag-specific T cells play a critical role in responding to viral infections. In the RV144 HIV vaccine clinical trial, a rare subset of HIV-specific polyfunctional CD4+ T cells correlated with reduced risk of HIV-1 infection. Polyfunctional T cells are a subset of Ag-specific T cells that are able to simultaneously produce multiple effector cytokines. Little is known about what differentiates polyfunctional T cells from other vaccine-elicited T cells in humans. Therefore, we developed a novel live-cell multiplexed cytokine capture assay to identify, isolate, and transcriptionally profile vaccine-specific polyfunctional CD4+ T cells. We applied these methods to samples from subjects who received the RV144 vaccine regimen, as part of the HVTN 097 clinical trial. We identified two surface receptors (CD44 and CD82) upregulated on polyfunctional T cells and a Th2-biased transcriptional signature (IL-4, IL-5, and IL-13) that predicted the envelope-specific polyfunctional CD4+ T cell profiles that had correlated with reduced risk of HIV infection in RV144. By linking single-cell transcriptional and functional profiles, we may be able to further define the potential contributions of polyfunctional T cells to effective vaccine-elicited immunity.
Visual Abstract
Key Points
A novel multiplexed assay isolates polyfunctional T cells for downstream analyses.
We identified surface markers upregulated on vaccine-specific polyfunctional CD4 T cells.
A signature predicts a CD4 T cell profile correlated with reduced risk of HIV infection.
Introduction
Almost 40 y since its identification as the causal agent of AIDS, HIV remains a significant public health challenge. Despite tremendous effort to develop a vaccine to prevent HIV infection, the pox-protein vaccine regimen that was tested for efficacy in the RV144 clinical trial remains the only vaccine, thus far, to have demonstrated vaccine-elicited protection from HIV acquisition, although with modest efficacy (1).
As a result of this ground-breaking achievement, a similar vaccine regimen was developed for use in sub-Saharan Africa that also used the ALVAC vector platform, but expressed a different HIV-1 envelope (env) gp120 insert, the subtype C zm96 strain (vCP2438), and the boost was coadministered with different bivalent gp120 proteins and different adjuvant, MF59 instead of alum. Lastly, the modified regimen included an additional vaccination at 1 y after enrollment to improve durability of the vaccine-elicited immunity (2). Notably, this subtype C–adapted RV144-similar pox-protein vaccine regimen was tested in a phase 2b-3 efficacy trial (HVTN 702) to evaluate HIV-1 prevention in populations at risk in sub-Saharan Africa and it failed to demonstrate any protection against HIV-1 acquisition (3). Thus, the original RV144 vaccine regimen continues to be the only vaccine to have demonstrated vaccine-mediated reduced risk of HIV infection. Thus, further investigation into the vaccine-induced correlates of protection identified in RV144, in particular, continues to be of significant interest to the field.
Utilizing an unbiased, Bayesian statistical approach, referred to as COMPASS, we previously determined that polyfunctional HIV env-specific CD4+ T cell responses correlated with a reduced risk of HIV infection in the RV144 trial (4). Polyfunctional T cells are able to simultaneously produce multiple functions or cytokines, and HIV-specific polyfunctional T cells have been repeatedly correlated with improved clinical outcomes in HIV infection (5, 6) and vaccination (7). In particular, two rare subsets of highly polyfunctional env-specific CD4+ T cell profiles, that is, 1) IL-4, IL-2, CD154, IFN-γ, and TNF-α, and 2) IL-4, IL-2, and CD154, correlated with reduced risk of HIV infection in Rv144 (odds ratio [OR]: 0.57, p = 0.006 and OR: 0.63, p = 0.013, respectively), whereas the overall magnitude of env-specific CD4+ T cell responses by IFN-γ and IL-2 expression did not (4). We sought to better understand the potential mechanisms by which these cells might contribute to prevention of HIV-1 infection. We hypothesized that these rare polyfunctional env-specific CD4+ T cells had unique functional properties that were either directly or indirectly correlated with their cytokine expression profiles.
Thus, we sought to determine whether these rare polyfunctional env-specific CD4 T cells had other characteristics that could explain their unique correlation with protection. Therefore, we performed single-cell transcriptional analyses of polyfunctional env-specific CD4+ T cells to gain a deeper understanding of the potential mechanism contributing to protective immune responses induced by the RV144 vaccine regimen. We sought to determine whether there were transcriptional signatures specific to these polyfunctional cells that were vital to vaccine efficacy compared with cells expressing one or two effector functions. However, the intracellular cytokine staining (ICS) methods traditionally used to identify polyfunctional T cell subsets require fixation and permeabilization, which damages the mRNA and precludes downstream transcriptional analysis. Alternatively, activation induced marker (AIM) assays have been used to identify and enrich Ag-specific T cells for transcriptional analyses (8–10). These assays rely on upregulation of surface markers that are preferentially upregulated after activation. However, the AIM assays do not allow for identification of polyfunctional T cells to be specifically interrogated. In addition, AIM markers have variable baseline expression that affects the specificity of detection of vaccine-specific cells, and upregulation of activation markers requires extended incubation times (up to 18 h), which can alter the transcriptional profiles and cause bystander activation of cells that are not vaccine specific.
Therefore, we developed a novel multiplexed cytokine capture assay to be able to identify and sort individual, viable, Ag-specific cytokine-expressing CD4+ T cells for single-cell RNA sequencing (scRNA-seq). The flow cytometry data were captured for each individual sorted T cell, enabling scRNA-seq data to be linked to the observed cytokine protein expression. The resulting analyses demonstrated that polyfunctional CD4+ T cells could be specifically detected among the vaccine-stimulated T cells by the multiplexed cytokine capture assay. It also led to the identification of novel surface-expressed activation markers, CD82 and CD44, which were significantly upregulated among polyfunctional CD4+ T cells. Lastly, we established that a Th2-biased transcriptional signature predicted the polyfunctional CD4+ T cell profiles that had correlated with reduced risk of HIV infection in the RV144 HIV efficacy trial, suggesting that the mechanism of vaccine-induced protection in RV144 was in part due to Th2 functions, likely through support of the humoral response.
Materials and Methods
HVTN 097 phase 1 clinical trial samples
HVTN 097 was a randomized, double-blind, placebo-controlled phase 1b clinical trial of the RV144 vaccine regimen and was conducted in South Africa (11). Vaccine recipients received ALVAC-HIV at months 0 and 1, followed by ALVAC-HIV plus AIDSVAX gp120B/E at months 3 and 6. PBMCs were collected at month 6.5 (2 wk after the fourth/last vaccination) for immunogenicity measurements, which were published previously (11). The study was approved by all appropriate Institutional Review Boards, and all participants provided written consent.
ICS assay
Flow cytometry was used to examine HIV-1–specific CD4+ T cell responses from cryopreserved PBMCs as previously described (11). Responses were evaluated to pools of overlapping peptides matched to the HIV-1 vaccine vector insert Ag, 92TH023-env, measured at 2 wk after final vaccination. Positivity of the ICS responses of individual cytokines or cytokine combinations was determined by a one-sided Fisher’s exact test applied to each peptide pool–specific response versus the negative control response with a discrete Bonferroni adjustment for the multiple comparisons due to testing against multiple peptide pools. Peptide pools with adjusted p values <0.00001 were considered positive. We selected samples for analysis by capture assay that had a positive response to the env peptide pool in the ICS assay during clinical trial testing.
Multiplexed cytokine capture assay
The multiplex cytokine capture assay is similar to the ICS assay by employing vaccine-specific peptides to induce the secretion of cytokines by vaccine-specific T cells. The catch reagents contain Abs specific for the cytokines of interest, which adhere to the cell via a proprietary mechanism (Miltenyi Biotec), likely an Ab specific for a surface cell marker expressed on leukocytes, such as CD45, to capture the cytokines as they are secreted by the cell. After the catch reagents have bound to the cell, the cells are diluted and incubated to allow for cytokine secretion from the activated T cells. The cytokines are captured by the catch reagents and tethered to the cell that released the cytokine. The captured cytokines can then be detected by flow cytometry using surface staining methods.
PBMCs were thawed quickly in warm RPMI 1640 media containing 10% FBS, l-glutamine, and penicillin-streptomycin (R10) and rested overnight. An aliquot of cells was stained in Guava ViaCount reagent and counted using a Guava easyCyte (EMD Millipore), then resuspended in R10. The cells were incubated with purified anti-CD40 mAb (Miltenyi Biotec) for 10 min before being stimulated (12). CD154-PE-Cy7 Ab was added along with 1 μg/ml anti-human CD28/CD49d costimulatory Abs and either 2 μg/ml vaccine-matched 92TH023-env peptide pool or with peptide diluent (DMSO) for 5 h at 37°C, 5% CO2. Cells were washed in cold wash buffer (1× PBS, 2 mM EDTA, 0.5% BSA) and then incubated with 10 µl per 106 cells each of IFN-γ catch reagent (Miltenyi Biotec), IL-2 catch reagent (Miltenyi Biotec), and IL-4 catch reagent (Miltenyi Biotec) in a final volume of 100 µl per 106 cells of wash buffer for 15 min on ice. Cells were washed with R10 media and then resuspended in a minimum of 1 ml per 106 cells (∼10 ml) of warm R10 media and incubated rotating at 37°C, 5% CO2 for 1 h. Samples were centrifuged and then washed with cold wash buffer and stained with Aqua viability dye for 10 min on ice. Samples were washed once with cold wash buffer. The cold buffer was decanted. Samples were stained with the Ab mixture containing lineage and phenotyping markers as well as cytokine detection Abs and incubated for 20 min on ice (Supplemental Table I). The samples were washed once more with cold wash buffer. Samples were resuspended in 800 µl of cold buffer and filtered immediately before sorting. Live CD14−CD19−CD56−CD8− CD4+ T lymphocytes that were either CD154+IFN-γ+IL-2+, CD154+IL-2+, or CD154+ were single-cell sorted with a FACSAria II cell sorter. Full functional profiles were determined postsorting using analysis of the individual cell-level flow cytometry data (acquired by indexing).
ICS assay including CD44 and CD82
The ICS assay was adapted to include surface staining for CD44 and CD82 (11). In brief, cryopreserved PBMCs from a healthy control were thawed quickly in warm R10, washed twice in R10, and rested overnight in a 37°C, 5% CO2. Cells were washed with R10 and resuspended at 5 × 106/ml in R10 plus 1 μg/ml anti-human CD28/CD49d costimulatory Abs and either 0.25 μg/ml staphylococcal enterotoxin B or with peptide diluent (DMSO), transferred to a 96-well round-bottom plate, and incubated for 6 h at 37°C, 5% CO2. Then, the cells were stained with the Live/Dead Blue viability dye in 1× PBS for 15 min and washed with FACS wash (1× PBS, 2% FBS). The cells were stained with Abs for CD44 and CD82 and incubated for 15 min (Supplemental Table I). The cells were washed with FACS wash and then resuspended with 100 μl of 1× FACS Lyse (BD Biosciences, no. 349202) for 10–15 min at room temperature. Then, 100 μl of wash buffer was added and cells were centrifuged, decanted, and resuspended with 200 μl of 1× FACS Perm II (BD Biosciences, no. 340973) and incubated for 10–15 min at room temperature. Wash buffer (100 μl) was added and cells were centrifuged, decanted, and resuspended in 50 μl of a mixture of the remaining Abs in FACS Wash (Supplemental Table I). Cells were incubated for 30 min at room temperature in the dark and then washed three times with FACS Wash and resuspended in 1% paraformaldehyde in 1× PBS. Cells were collected on an LSR II (BD Biosciences) and data were analyzed using FlowJo v9.9.4.
scRNA-seq
Samples were single-cell sorted into 96-well PCR plates containing 5 µl of DNA suspension buffer (Teknova) with 1% BSA (Sigma-Aldrich) per well. Plates were sealed, briefly vortexed, centrifuged at 800 × g for 1 min, and immediately transferred to dry ice. Plates were stored at −80°C for at least overnight before proceeding. The plates were thawed on ice and spun at 800 × g for 1 min to collect all liquid. We used a modified version of a previously published method to amplify the whole transcriptome (13). In brief, the reverse transcriptase that is used to make cDNA has a terminal transferase activity. Then, a “template‐switch” primer incorporates a second universal priming sequence yielding cDNA with two universal priming sequences.
In brief, reverse transcription occurred in two steps. The initial step was conducted in 5 µl of the lysed cell, 2.4 µM 3′ SMART CDS Primer II A (custom primer from IDT), 2.4 mM each 2′-deoxynucleoside 5′-triphosphate mix (GeneAmp), and 6.32 U of RNaseOUT (Invitrogen). The samples were incubated at room temperature for 1 min, 72°C for 3 min, and held on ice before beginning the second step. The next step was conducted with a SuperScript II reverse transcriptase kit (Invitrogen), using 7 µl of the step 1 product, 1× first-strand buffer, 2.5 mM DTT, 110 U of SuperScript II, and an additional 11 U of RNaseOUT, as well as 1 µM SMARTer II oligonucleotide (custom primer from IDT) and molecular-grade water to 11 µl. The samples were incubated at 42°C for 90 min, followed by a limited cycle amplification of 10 cycles at 50°C for 2 min and 42°C for 2 min, concluding with a final extension at 72°C for 15 min.
Whole-transcriptome amplification (WTA) immediately proceeded with the 11 µl of the reverse transcription product, using 1× KAPA HiFi HotStart ReadyMix (Roche) and 0.5 µM IS PCR Primer (IDT). Amplification parameters were 98°C for 3 min and 22 cycles of 98°C for 15 s, 67°C for 20 s, and 72°C for 6 min, followed by a final extension of 72°C for 5 min. The WTA product was cleaned using AMPure XP beads (Beckman Coulter). The PCR product was incubated at room temperature for 5 min with 0.8 vol of XP beads before being placed on a magnet. The beads were washed three times with 100 µl of 80% ethanol. After briefly drying, the beads were resuspended in 20 µl of 1× TE buffer (Sigma-Aldrich), and 18 µl of supernatant was transferred to a new 96-well PCR plate. Quality and quantitation were measured by TapeStation using a high-sensitivity D5000 ScreenTape (Agilent).
Samples with a concentration ≥200 pg/µl were carried forward for sequencing according to the Nextera XT DNA library preparation kit (Illumina) protocol. Samples were dual indexed using the Nextera index XT kit (Illumina) and a scheme that would allow for eight pools without overlapping indices. After pooling, the concentration of each pool was measured by a Qubit dsDNA HS assay kit (Invitrogen), and sizing was determined by fragment analyzer HS small fragment kit (Agilent). A smear analysis was conducted in the range of 150–1000 bp, and the average peak size along with the Qubit results were used to determine the molarity. Each pool was then diluted to a concentration of 2 nM. The samples were sequenced on the Illumina HiSeq 2500 using PE50 on a high-output eight-lane flow cell with one pool per lane.
Custom primer sequences
Custom primer sequences (5′ to 3′) were as follows: 3′ SMART CDS Primer II A (modified oligo(dT): AAGCAGTGGTATCAACGCAGAGTACT(30)VN; SMARTer II oligonucleotide (template switching oligonucleotide): AAGCAGTGGTATCAACGCAGAGTACATrGrGrG; IS PCR primer (WTA PCR primer): AAGCAGTGGTATCAACGCAGAGT.
Statistical analysis
scRNA-seq analysis
The fastq files were processed with kallisto v0.45.0 (14), and gene-level counts were generated from abundances scaled using the average transcript length, averaged over samples and to library size (lengthScaledTPM), using the tximport package v1.16.1 (15). Subsequent analysis was performed using a typical Bioconductor workflow (16). Briefly, the count matrix was imported of into R to create a SingleCellExperiment object. Cells with a library size of <30,000 reads were removed from the analysis. After quality control metric filtering, a total of 689 cells were kept for downstream analysis. The logNormCounts function from the scater package v1.16.2 (17) was used to generate log-transformed normalized expression values. Principal component analysis was performed using the top 4000 highly variable genes, and t-distributed stochastic linear embedding (18) and UMAP (uniform manifold approximation and projection) (L. McInnes, J. Healy, and J. Melville, manuscript posted on arXiv, DOI: 10.48550/arXiv.1802.03426) was subsequently performed using the top 30 principal components for data visualization. The top 30 principal components were also used to build a shared nearest neighbor (SNN) graph, and cell clusters were identified using the walktrap algorithm from the igraph package v1.2.5 (19). Differentially expressed genes between groups were identified using MAST v1.14.0 (20). Genes with a false discovery rate (FDR) of <0.05 and a fold change of >1.25 or less than −1.25 were considered significantly differentially expressed between groups.
Predicting cell-level functional phenotypes correlated with decreased HIV risk
We use tree boosting, as implemented in XGBoost (21), to build a nonlinear model predicting polyfunctionality from gene expression profiles. The log-transformed normalized expression values of 28,383 genes that were expressed in at least three cells were used to train a binary XGBoost classifier using the xgboost Python package v1.1.0 (21) with the following parameters: “objective”: “multi:softprob,” “num_class”: 2, “n_estimators”: 1000, “n_jobs”: 6, “random_state”: 0, “scale_pos_weight”: 5.51, “subsample”: 0.9. The classifier was evaluated using stratified 5-fold cross-validation and the area under the receiver operating characteristic curve metric. Shapley Additive Explanations (SHAP) values were calculated using the shap Python v0.37.0 (22).
Data availability
The accession number for the RNA-seq data reported in this study is Gene Expression Omnibus (GEO) GSE166945 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).
Results
Evaluation of vaccine-specific CD4+ T cells by multiplexed cytokine capture assay
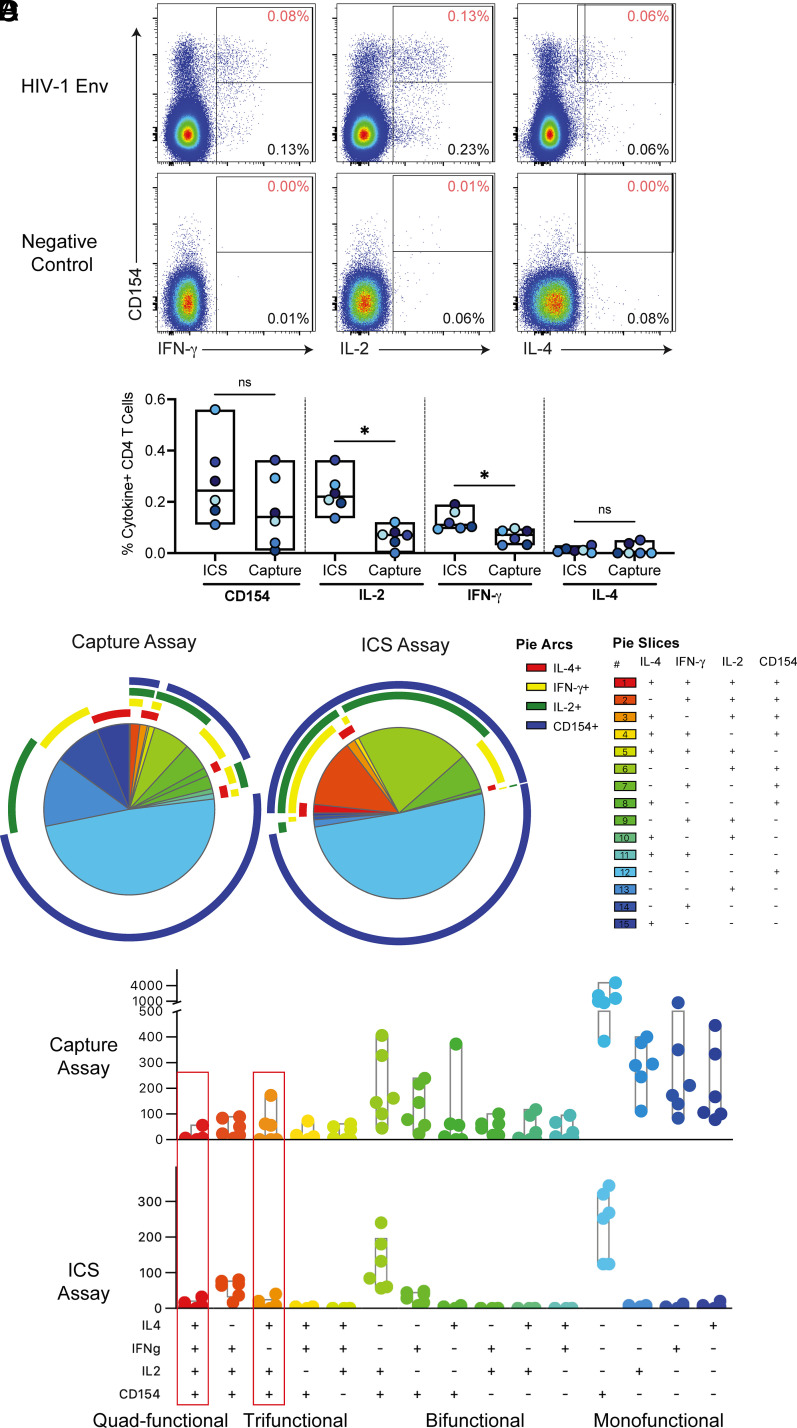
We sought to further interrogate the HIV env-specific CD4 T cells induced by the RV144 vaccine regimen; however, we did not have access to additional PBMC samples from RV144. Therefore, instead, we leveraged access to samples from a phase 1 clinical trial, HVTN 097, which tested the same pox-protein vaccine regimen as RV144. HVTN 097 was a phase 1b clinical trial that tested the prime-boost vaccine regimen that was used in the RV144 efficacy trial: ALVAC-gp140 with subtype B insert boosted with the coadministered bivalent subtype B/E gp120 proteins adjuvanted with alum, as described previously (1, 11). One of the primary goals of HVTN 097 was to confirm the immune profiles induced by the RV144 vaccine regimen, originally conducted in Thailand, in a different recipient population, South Africa. Indeed, the RV144 vaccine regimen tested in HVTN 097 induced env-specific polyfunctional CD4+ T cell responses, including the specific subsets IL-4, IL-2, CD154, IFN-γ, and TNF-α and IL-4, IL-2, and CD154 (4), which had correlated with reduced risk of infection in RV144 (11, 23).
To interrogate the transcriptional profiles of these rare subsets of cells, we developed a live-cell multiplexed cytokine capture assay to detect and sort vaccine-specific polyfunctional CD4+ T cells. To detect CD154 (CD40L), we preincubated the cells with Ab against CD40 to block uptake of its ligand, CD154, and then cocultured the cells with fluorescently labeled anti-CD154 Ab during the stimulation. Then, we multiplexed cytokine capture reagents to simultaneously detect secreted IFN-γ, IL-2, and IL-4. Owing to experimental limitations, we decided to not use TNF-α, as it was commonly coexpressed with IFN-γ among the polyfunctional T cell subsets and deemed not critical for distinguishing the env-specific CD4+ T cell “correlate” profiles: IFN-γ, TNF-α, IL-4, IL-2, and CD154 (OR: 0.57, p = 0.006) and IL-4, IL-2, and CD154 (OR: 0.63, p = 0.013) (4).
We selected HVTN 097 samples (n = 6) based on the presence of vaccine-induced env-specific CD4+ T cells as determined by IL-2+ and/or IFN-γ+ in the ICS assay (median 0.286% of CD4 T cells). The two polyfunctional correlate subsets encompass a very rare population. In this subset of samples, the env-specific T cells with either correlate profile were a median of 0.015% of CD4 T cells (range, 0.009–0.096%) and median of 2% (range, 0.81–7.73%) of the env-specific CD4 T cells, which was consistent with previous studies (11, 23).
Using the multiplexed cytokine capture assay, we single-cell sorted cytokine-positive env-specific CD4+ T cells from PBMCs from HVTN 097 vaccine recipients, 2 wk after the final HIV vaccination, for transcriptional profiling by scRNA-seq. In general, we were able to detect diverse functional env-specific CD4+ T cell responses (Fig. 1A, 1B). Although there was some monofunctional background in the individual cytokines in the capture assay, the background was low or nonexistent among bi-, tri-, or polyfunctional subsets (Fig. 1A). The magnitudes of env-specific CD154+ and IL-4+ CD4+ T cells were similar between the two assays with no significant differences, whereas frequencies of IL-2+ and IFN-γ+ CD4+ T cells were higher in the ICS assay compared with the capture assay (p = 0.03; (Fig. 1B). The higher IL-2+ and IFN-γ+ CD4 T cell frequencies in the ICS assay were likely due to the longer time during which secretory inhibitors allow cytokines to accumulate intracellularly for detection in the ICS assay (6 h). In contrast, in the multiplexed cytokine capture assay, the cytokine capture reagents are only added for the last hour of the incubation. Detection of secreted cytokines in the multiplexed cytokine capture assay is also limited by the number of the catch reagents bound to the surface of the cell.
FIGURE 1.
Comparison of env-specific CD4+ T cell responses postvaccination by multiplexed cytokine capture and ICS assays. (A) Example of multiplexed cytokine capture assay env and negative control stimulations of PBMCs from a HVTN 097 vaccine recipient. Data are gated on CD4+ T lymphocytes (live single, CD56−, CD19−, CD14−, CD3+, CD4+, and CD8−). The y-axis shows CD154 expression, and the x-axis shows IFN-γ (left), IL-2 (middle), and IL-4 (right). The percentage in the bottom right of each plot indicates the frequency of cytokine-positive events, and the percentage in the top right (in red) of each plot is the percentage of CD4 T cells that are coexpressing the indicated cytokine with CD154. (B) Background-subtracted percent of CD4 T cells expressing CD154, IFN-γ, IL-2, and IL-4 for each subject by multiplexed cytokine capture and ICS assays (n = 6). Significance was determined using a Wilcoxon rank test. *p < 0.05. (C) Pie charts show the median distribution of subsets among the env-specific CD4 T cells. Each pie slice indicates the proportion of env-specific CD4 T cells that express those cytokines (indicated in the legend) and is color-coded by the degree of polyfunctionality: monofunctional (blue), bifunctional (green), trifunctional (yellow/orange), and quad-functional (red). The pie arc colors allow visualization of the functions expressed by each slice. (D) Bar chart of the per subject counts of env-specific CD4+ T cell subsets by functional profile from ICS (out of a median of 556 total number of env-specific CD4+ T cells per subject) or multiplexed cytokine capture assay (out of a median of 1887 total number of env-specific CD4+ T cells per subject) and color-coded by the degree of polyfunctionality. The red boxes indicate the two subsets that correlated with reduced risk of HIV infection in the RV144 vaccine trial.
CD154 is a surface-expressed T cell activation marker that can be detected by intracellular staining as is done in the ICS assay. In the multiplexed cytokine capture assay, we used coculture with a fluorophore-labeled anti-CD154 detection Ab. Detection of CD154 by the two methods yielded similar frequencies and represented the single largest population of responding CD4+ T cells in both assays (Fig. 2B, 2C). In contrast, there were more monofunctional IFN-γ+, IL-2+, or IL-4+ CD4 T cells detected in the multiplexed cytokine capture assay compared with ICS (Fig. 1C, 1D). In general, there was a similar distribution of polyfunctional profiles among env-specific CD4+ T cells in the capture assay compared with the ICS assay (Fig. 1D). Indeed, the multiplexed cytokine capture assay had reliable detection of diverse polyfunctional vaccine-specific CD4+ T cells in each of the samples tested, including the two highly polyfunctional (three- and four-function) subsets that corresponded to the profiles that correlated with reduced risk of infection in RV144, that is, IFN-γ, IL-4, IL-2, and CD154; and IL-4, IL-2, and CD154 (Fig. 1D). Lastly, the multiplexed cytokine capture assay had negligible CD4+ T cells expressing two or more cytokines in the negative control stimulations, indicating the highly specific detection of polyfunctional T cells in the env-stimulated samples (Fig. 1A).
FIGURE 2.
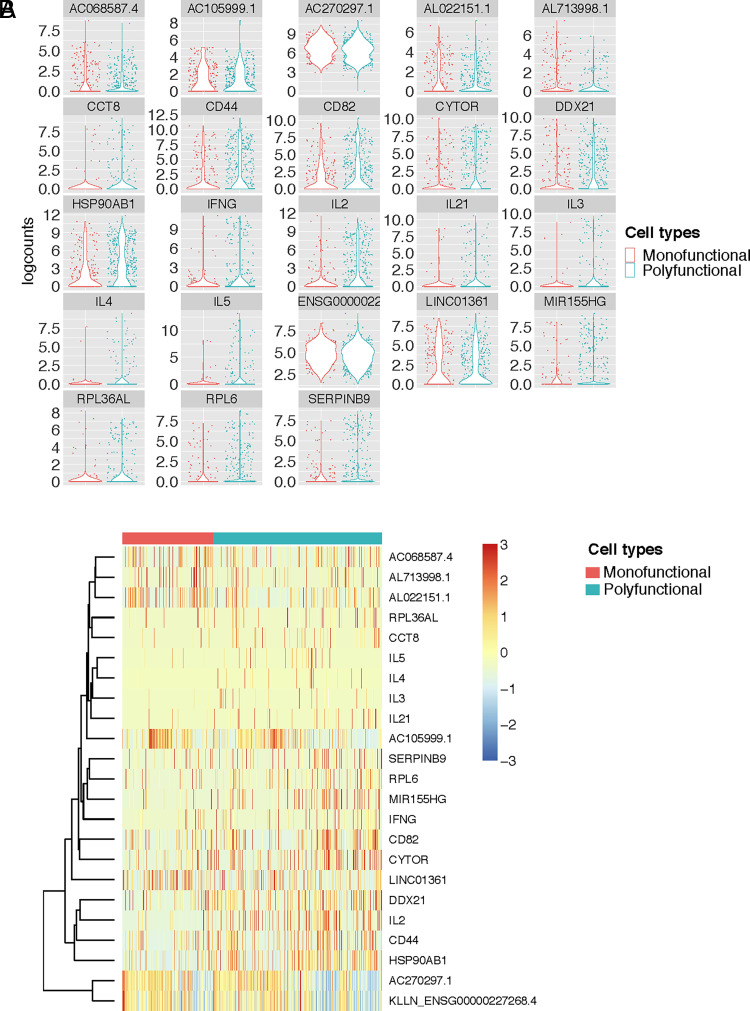
Individual genes that are differentially expressed by polyfunctional env-specific CD4 T cells. (A) Violin plots of log counts (log2-transformed normalized counts plus a pseudocount of one) of the genes that were significantly differentially expressed between monofunctional (producing only one cytokine/function) and polyfunctional (producing two or more cytokines/functions) env-specific CD4+ T cells. (B) Heatmap of Z scores (mean row-centered log2 counts per million) of the same differentially expressed genes clustered by polyfunctionality profile.
Single-cell gene expression analysis of vaccine-induced CD4+ T cells with diverse functional profiles
We used a combination of sort criteria in the multiplexed capture assay to single-cell index sort a diverse representation of cytokine-positive env-stimulated CD4+ T cells for transcriptional profiling by scRNA-seq. Because the polyfunctional CD4+ T cells are very rare, after sorting, we preferentially selected polyfunctional T cells in addition to a random selection of other cytokine-positive T cells for conducting scRNA-seq. We conducted reverse transcription using SMART primers, which incorporate a universal primer sequence to ensure sensitive and unbiased cDNA synthesis, and subsequently amplified the whole transcriptome (13).
We performed scRNA-seq on env-specific CD4+ T cells with diverse functional profiles from the postvaccination samples. The distribution of env-specific CD4+ T cells (n = 1423) for which scRNA-seq data were generated included representatives of all of the functional subsets identified by the flow cytometry data (Supplemental Fig. 1A). The largest subset of env-specific CD4+ T cells in the scRNA-seq dataset was characterized by IL-2 and CD40L protein coexpression. However, this was to be expected, as this cell subset was a dominant population among multifunctional subsets in both the ICS and multiplexed cytokine capture assay protein expression data by flow cytometry (Fig. 2D). Because of the sorting and sequencing strategy, polyfunctional CD4+ T cells, including the subsets that correlated with reduced risk of infection in RV144 as indicated by coexpression of IFN-γ, IL-4, IL-2, and CD154 and IL-4, IL-2, and CD154 (correlate), were enriched in the scRNA-seq dataset compared with the absolute frequencies by flow (Supplemental Fig. 1A, 1C, 1D).
We compared the detection of the individual cytokines IFN-γ, IL-4, IL-2, and CD154 by protein expression and transcript levels (Supplemental Fig. 1B). In all cases, cytokine transcript levels were significantly higher among CD4 T cells that were positive for that cytokine’s protein expression by flow cytometry (Supplemental Fig. 1B). This further supports the specificity of the detection of the cytokines by capture assay. We would not expect a perfect correlation between transcript and protein expression, as there are temporal differences in their expression with peak transcript levels generally preceding peak protein levels.
Using t-distributed stochastic linear embedding, we first looked to see whether dimension reduction analysis could identify gene expression clusters that corresponded to degree of functionality or protein expression of specific cytokines identified by the multiplexed cytokine capture assay (Supplemental Fig. 2). We overlaid the protein functional markers that were detected by flow cytometry on the cells clustered by gene expression. First, we looked by degree of polyfunctionality (or simply the number of functional markers coexpressed by each cell). The clusters defined by overall gene expression did not correspond to degree of polyfunctionality, except for a small distinct island at the top that was predominantly monofunctional. Then, we overlaid the protein expression of each cytokine/functional marker individually and found that once again they did not cluster by gene expression (Supplemental Fig. 2). The only exception was the one monofunctional island of cells that appeared to express only IFN-γ (Supplemental Fig. 2). Lastly, we assessed whether cells with correlate functional cytokine profiles would cluster together by gene expression, and we found that they did not (Supplemental Fig. 2). The lack of gene expression clusters corresponding with the functional profiles could be in part because it can be difficult to discern discrete clusters among a population with limited heterogeneity.
Polyfunctional vaccine-specific CD4+ T cells have increased expression of genes associated with T helper function
Thus, we looked at differential gene expression analysis using MAST to determine whether there were specific genes whose expression correlated with polyfunctionality. MAST is a generalized linear framework that accounts for the bimodality of scRNA-seq data and is suitable for identifying differentially expressed genes (20). MAST models cellular detection rate as a covariate and can control for differences in abundance due to extrinsic biological and technical effects (20). We compared the monofunctional versus polyfunctional (i.e., more than one cytokine/function) vaccine-specific CD4+ T cells and discovered 23 genes that were significantly differentially expressed. Interestingly, the top hits included a number of protein-coding genes upregulated among the polyfunctional T cells and directly linked to CD44 and play a role in cell adhesion and cell–cell interaction, and CD82 associates with CD4 and delivers costimulatory signals for the TCR/CD3 pathway. For CD4+ T cell function we used CD44, CD82, IFN-γ, IL-2, IL-3, IL-4, IL-5, and IL-21 (Fig. 2A, 2B). IFN-γ, IL-2, IL-3, IL-4, IL-5, and IL-21 are cytokines with antiviral or B cell activation, growth, and differentiation activity. Other transcripts that were significantly associated with polyfunctionality included multiple RNA- or protein-coding genes involved in regulation of translation (DDX21, RPL36AL, and RPL6), secretory pathway (CCT8), and cell cycle/proliferation (CYTOR, HSP90AB1, and MIR155HG) (Fig. 2A, 2B).
CD44 and CD82 were interesting, in particular, because they are both cell surface receptors that are upregulated by T cell activation and may be useful for identifying vaccine-induced polyfunctional T cells. To assess this, we evaluated CD44 and CD82 as part of an ICS assay utilizing PBMCs from healthy unvaccinated adult subjects and a polyclonal stimulant, staphylococcal endotoxin B. We found that CD44 and CD82 were each individually upregulated among the polyfunctional CD4 T cells as compared with the monofunctional CD4 T cells (Supplemental Fig. 3). In addition, 83% of polyfunctional CD4 T cells coexpressed both CD44 and CD82, in comparison with 60% of monofunctional CD4 T cells.
Th2-biased transcriptional signature predicts vaccine-specific CD4+ T cell profile correlated with reduced risk of HIV infection
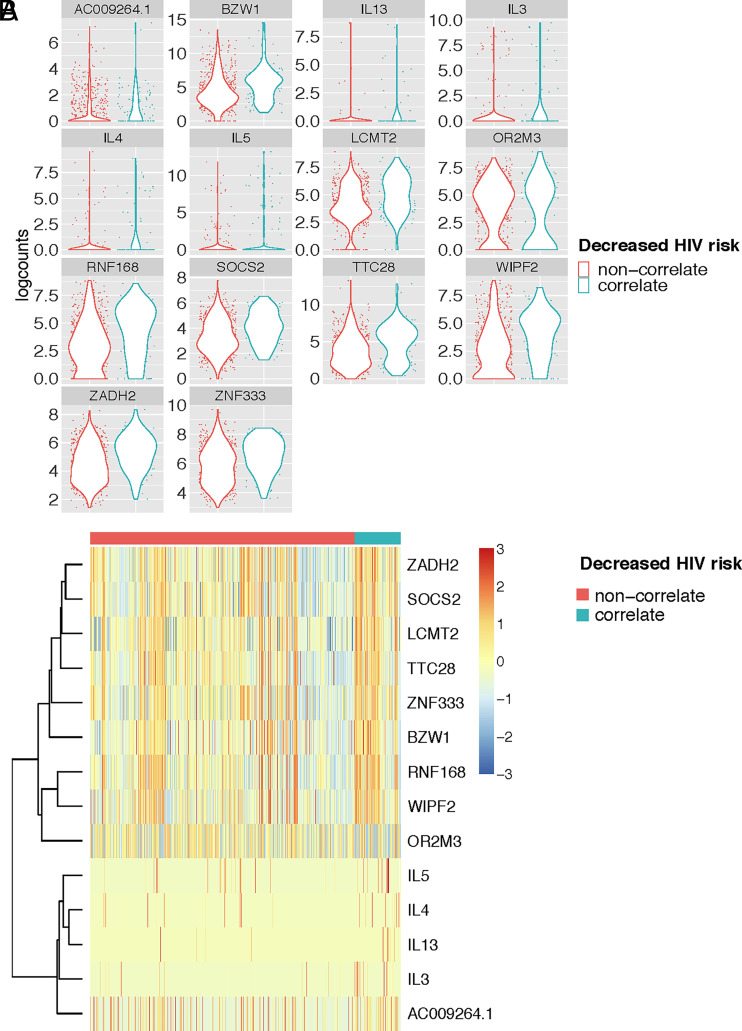
Next, we compared the gene expression profiles of the cell subsets that had been previously associated with reduced risk of HIV infection in RV144 (correlate) compared with all other subsets of cytokine-expressing env-specific CD4+ T cells (“non-correlate”) using MAST. We found 14 genes that were significantly differentially expressed among the env-specific CD4+ T cells with the correlate compared with the non-correlate functional profiles. In particular, we found that expression of IL-3, IL-4, IL-5, and IL-13 were significantly upregulated among the T cells with the correlate functional profile (Fig. 3A, 3B). Genes such as CD44, CD82, and IL-21 were not detected as differentially expressed in this analysis, either because the sample size was too small or because they were associated with polyfunctionality and not necessarily with the specific correlate phenotypes. Alternative analyses of these specific genes suggested that IL-21 was also associated with the correlate phenotype (FDR adjusted p value = 0.15), whereas CD44 and CD82 were not and were instead indicative of overall polyfunctionality (Supplemental Fig. 1C). Other transcripts that were significantly associated with the CD4+ T cells that exhibited the correlate functional profiles included protein-coding genes involved in regulation of JAK/STAT cell signaling pathway (SOCS2), transcription and/or translation (BZW1, LCMT2, and ZNF33), and cell cycle/proliferation (TTC28, WIPF2) (Fig. 3A, 3B).
FIGURE 3.
Individual genes that are differentially expressed by env-specific CD4 T cells expressing RV144 polyfunctional correlate profile compared with non-correlate env-specific CD4 T cells. (A) Violin plots of log counts of the scRNA sequences for the 14 significantly differentially expressed genes between the env-specific CD4+ T cells grouped by the cytokine protein expression profiles that correlated with reduced risk of HIV infection in RV144, labeled here as correlate (IL-4+, IL-2+, CD154+, and IFN-γ+ and IL-4+, IL-2+, and CD154+) or non-correlate. (B) Heatmap of Z scores of the differentially expressed genes clustered by T cells with correlate or non-correlate cytokine expression profile. Genes with FDR <0.05 and fold change of >1.25 or less than −1.25 were considered significantly differentially expressed between groups.
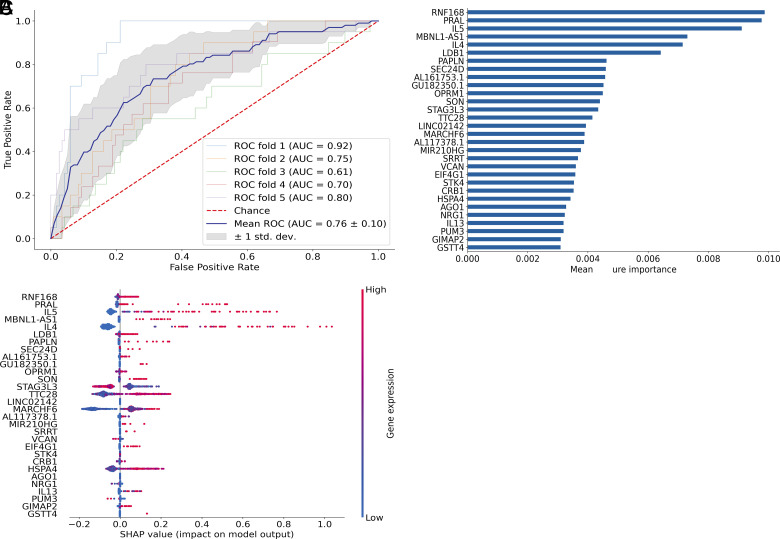
Building on this analysis, we asked whether there was a transcriptional signature that could predict the presence of the correlate functional profiles, using nonlinear boosting trees implemented in XGBoost (21). To avoid overfitting and more accurately evaluate model prediction performance, we conducted 5-fold cross-validation where the data were first split into five smaller sets. Subsequently, each unique set was retained as the test dataset, and a model was trained using the remaining four sets as training data and evaluated on the test set. The average area under the receiver operating characteristic curve for the model was 0.76 with a SD of 0.10 (Fig. 4A). We found that expression of IL-4, IL-5, and IL-13 were among the top 30 genes that contributed to the prediction of the T cells with the correlate profiles (Fig. 4B). We further calculated the SHAP values of the top 30 genes for every cell averaged across the five models to see which genes contributed to increased/decreased risk of infection (Fig. 4C). SHAP values indicate a gene’s impact on a change in the model output (22). As shown in (Fig. 4C, high expression of IL-5, IL-4, and, to a less extent, IL-13, drove the model predictions toward the functional profiles correlated with reduced risk of HIV infection. These data suggest that the expression of Th2 cytokines may be uniquely detected in these polyfunctional T cell subsets and that this mechanism may be directly or indirectly associated with an immune-mediated reduced risk of infection observed in RV144.
FIGURE 4.
Transcriptional signatures predicted rare env-specific polyfunctional CD4 T cell subsets that had correlated with reduced risk of HIV infection in the RV144 trial. XGBoost machine learning model was used to determine the genes that best predicted the two cytokine profiles associated with reduced risk of infection in Rv144 (correlate profile) versus other env-specific CD4+ T cell subsets (non-correlate profile). (A) Plot shows receiver operating characteristic (ROC) curves created from 5-fold cross-validation. The blue line depicts the mean across the five models. (B) Bar graph of the top 30 most important features based on mean feature importance across the five models. (C) SHAP values showing the contribution of each gene within each cell for the top 30 genes. Color indicates the level of gene expression in each cell for the corresponding gene.
Discussion
Thus far, RV144 remains the only vaccine trial to show at least partial efficacy at preventing HIV infection. In correlates analyses of RV144, two highly polyfunctional env-specific CD4+ T cell subsets correlated with reduced risk of HIV-1 infection (1, 4). Using PBMC samples from participants in the HVTN 097 clinical trial, who received the same vaccine regimen as RV144, we sorted env-specific CD4+ T cells using a novel multiplexed cytokine capture assay and conducted scRNA-seq to interrogate potential mechanisms underlying the contribution of env-specific polyfunctional CD4+ T cells to vaccine-elicited immunity in RV144. Importantly, we found that expression of Th2 cytokines was upregulated among vaccine-specific polyfunctional T cells and, specifically, in the CD4+ T cells that correlated with reduced risk of HIV infection in RV144. We also identified expression of the surface markers CD44 and CD82 as being significantly upregulated among polyfunctional env-specific CD4+ T cells. To our knowledge, this study represents the first time that individual polyfunctional vaccine-specific CD4+ T cells have been directly linked to their transcriptional profiles ex vivo and highlights potential mechanisms associated with different functional subsets of vaccine-specific CD4+ T cells.
The identification of the cell surface receptors CD44 and CD82 as being preferentially upregulated among polyfunctional CD4+ T cells suggests that they may serve as useful markers of polyfunctional T cells in future applications. In mice, CD44 is a canonical effector memory marker upregulated in response to Ag-specific activation and then maintained (24, 25). In humans, CD44 modulates migration and extravasation and is upregulated by T cell activation or exposure to inflammatory mediators (26). Similarly, CD82 is associated with adhesion and T cell activation (27). Thus, coexpression of CD44 and CD82 may be useful for identifying vaccine-specific polyfunctional CD4+ T cells and may correspond with increased trafficking. Although we were able to confirm upregulated protein expression on polyfunctional CD4 T cells with a polyclonal stimulation, in future studies we would like to confirm this in the context of vaccine-specific CD4 T cell responses.
In comparing the gene expression profiles of monofunctional and polyfunctional env-specific CD4+ T cells, we also identified an enrichment of genes that correspond to a number of cytokines: IFN-γ, IL-2, IL-3, IL-4, IL-5, and IL-21. Whereas IFN-γ, IL-2, and IL-4 correspond to proteins that were used to identify the polyfunctional T cells, IL-3, IL-5, and IL-21 were not part of the capture assay panel and suggest that these functions are preferentially upregulated specifically within the multifunctional CD4+ T cells. This is interesting, as these cytokines have been difficult to detect ex vivo among primary Ag-specific CD4+ T cells. In particular, secreted IL-21 has been previously detected after extended (40 h) in vitro stimulation and was associated with improved Ag-specific T helper function supporting both Ag-specific CD8+ T cells and B cells (28, 29). Similarly, the Th2 cytokines IL-5 and IL-13 have been difficult to detect by flow cytometry. The detection of IL-5 and IL-13 transcripts among IL-4–expressing T cells suggests that IL-4 may be a potential surrogate of additional Th2 cytokine activity.
Additionally, a Th2-biased transcriptional profile including expression of IL-4, IL-5, and IL-13, predicted the env-specific CD4+ T cells with the correlate phenotype. It has been previously demonstrated that the alum adjuvant tends to drive Th2-biased immune responses to vaccination (30). In contrast, MF59 tends to induce a more balanced ratio of Th1 and Th2 responses (31, 32). This difference in Th1 and Th2 bias was confirmed in a study where a SIV version of the ALVAC-gp140 vaccine regimen was adjuvanted MF59 or alum and compared side by side in the nonhuman primates model (33). The nonhuman primate study recapitulated the low level efficacy seen with the alum-adjuvanted vaccine regimen and predicted the lack of efficacy in the MF59-adjuvanted group. Moreover, they found increased Th1 CD4+ T cell responses in the MF59-adjuvanted group and Th2 transcriptional profiles associated with the alum-adjuvanted group (33). Thus, the data presented in this study are consistent with a Th2-biased immune response from the alum-adjuvanted vaccine regimen in RV144 and HVTN 097. In future studies, it would be interesting to compare this to env-specific CD4+ T cells induced by the MF59-containing HVTN 702 regimen.
There are multiple potential limitations of this study, including the selection of samples from the HVTN 097 study instead of the case control subset from the RV144 clinical trial. Although HVTN 097 tested the same vaccine regimen as RV144, it was conducted in a different study population. In addition, because HVTN 097 was not an efficacy trial, it was impossible to directly link the observed biomarkers with reduced risk of HIV infection. Another limitation is the study size. Although we were able to interrogate thousands of vaccine-specific CD4 T cells, those cells were isolated from only six independent subjects. The study is also limited by the fact that it was restricted to a single vaccine regimen. Future studies should evaluate whether these biomarkers of polyfunctionality and the correlate profiles are generalizable to other study populations and vaccine regimens. Lastly, it would be interesting to see the capture assay approach applied to detection of some of the cytokines that were detected in the transcriptomics, such as IL-3, IL-5, IL-13, and IL-21, to confirm that the transcriptional upregulation corresponded to increased cytokine secretion.
This study provides a proof of principle that the multiplex cytokine capture assay can be used to identify, isolate, and further interrogate vaccine-specific polyfunctional CD4+ T cells. The single-cell transcriptional profiling independently confirmed the flow cytometric detection of rare unique highly polyfunctional subsets among the vaccine-specific CD4+ T cells. The gene expression analyses also identified two cell surface receptors, CD44 and CD82, associated with T cell activation, which may be useful for enriching for polyfunctional Ag-specific CD4+ T cells by flow cytometry. Lastly, we identified a Th2-biased transcriptional profile that was specific to the subset of vaccine-specific CD4+ T cells that correlated with reduced risk of HIV infection in the RV144 vaccine trial. These results support the hypothesis that the env-specific polyfunctional CD4+ T cells induced by the RV144 vaccine regimen contributed to the reduction in HIV infection by modulating the Ab response through T cell help.
Supplementary Material
Acknowledgments
We thank the participants for volunteering their time and effort to participate in this study. We thank the HVTN 097 protocol leadership team, including Drs. Glenda Gray, Surita Roux, Nicole Grunenberg, Ying Huang, Edith Swann, and Georgia Tomaras, for oversight of the clinical trial. We thank the HVTN 097 trial site staff led by Drs. Linda-Gail Bekker, Lulu Nair, Gavin Churchyard, Craig Innes, and Fatima Laher for trial implementation and collecting the samples used in this study. At the Fred Hutchinson Cancer Research Center, we thank Dr. Laura Richert-Spuhler and Stephen Voght for assistance with the manuscript preparation.
This article is featured in Top Reads, p.
This work was supported by National Institute of Allergy and Infectious Disease/National Institutes of Health Grants U19AI128914 (to R.G. and M.J.M.) and UM1AI068618 (to M.J.M.), as well as by Vaccine and Infectious Disease Division Initiative awards from the Fred Hutchinson Cancer Research Center (to K.W.C., S.C.D.R., and R.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
R.G., S.C.D.R., and K.W.C. conceived the study. K.W.C., C.T., and A.S. planned and developed the methods, conducted the assays, and analyzed the data. A.S., Y.T., and R.A. conducted formal statistical analyses. K.W.C., Y.T., C.T., and A.S. drafted the original manuscript. R.G., S.C.D.R., M.J.M., and K.C. edited the manuscript. M.J.M., R.G., K.C., and S.C.D.R. secured funds and supervised the project. All authors read and approved the manuscript.
The RNA sequencing data presented in this article have been submitted to the Gene Expression Omnibus under accession number GSE166945.
The online version of this article contains supplemental material.
- AIM
- activation induced marker
- env
- envelope
- FDR
- false discovery rate
- ICS
- intracellular cytokine staining
- OR
- odds ratio
- R10
- RPMI 1640 media containing 10% FBS, l-glutamine, and penicillin-streptomycin
- scRNA-seq
- single-cell RNA sequencing
- SHAP
- Shapley Additive Explanations
- WTA
- whole-transcriptome amplification
Disclosures
R.G. has received consulting income from Takeda and declares ownership in Ozette Technologies as well as Modulus Therapeutics. R.A.A. is an employee and stockholder of Pfizer. The other authors have no financial conflicts of interest.
References
- 1. Rerks-Ngarm S., Pitisuttithum P., Nitayaphan S., Kaewkungwal J., Chiu J., Paris R., Premsri N., Namwat C., de Souza M., Adams E., et al. MOPH-TAVEG Investigators . 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361: 2209–2220. [DOI] [PubMed] [Google Scholar]
- 2. Bekker L. G., Moodie Z., Grunenberg N., Laher F., Tomaras G. D., Cohen K. W., Allen M., Malahleha M., Mngadi K., Daniels B., et al. HVTN 100 Protocol Team . 2018. Subtype C ALVAC-HIV and bivalent subtype C gp120/MF59 HIV-1 vaccine in low-risk, HIV-uninfected, South African adults: a phase 1/2 trial. Lancet HIV 5: e366–e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gray G. E., Bekker L. G., Laher F., Malahleha M., Allen M., Moodie Z., Grunenberg N., Huang Y., Grove D., Prigmore B., et al. HVTN 702 Study Team . 2021. Vaccine efficacy of ALVAC-HIV and bivalent subtype C gp120-MF59 in adults. N. Engl. J. Med. 384: 1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lin L., Finak G., Ushey K., Seshadri C., Hawn T. R., Frahm N., Scriba T. J., Mahomed H., Hanekom W., Bart P. A., et al. 2015. COMPASS identifies T-cell subsets correlated with clinical outcomes. Nat. Biotechnol. 33: 610–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Owen R. E., Heitman J. W., Hirschkorn D. F., Lanteri M. C., Biswas H. H., Martin J. N., Krone M. R., Deeks S. G., Norris P. J., NIAID Center for HIV/AIDS Vaccine Immunology . 2010. HIV+ elite controllers have low HIV-specific T-cell activation yet maintain strong, polyfunctional T-cell responses. AIDS 24: 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Betts M. R., Gray C. M., Cox J. H., Ferrari G.. 2006. Antigen-specific T-cell-mediated immunity after HIV-1 infection: implications for vaccine control of HIV development. Expert Rev. Vaccines 5: 505–516. [DOI] [PubMed] [Google Scholar]
- 7. Darrah P. A., Patel D. T., De Luca P. M., Lindsay R. W., Davey D. F., Flynn B. J., Hoff S. T., Andersen P., Reed S. G., Morris S. L., et al. 2007. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat. Med. 13: 843–850. [DOI] [PubMed] [Google Scholar]
- 8. Chattopadhyay P. K., Yu J., Roederer M.. 2005. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat. Med. 11: 1113–1117. [DOI] [PubMed] [Google Scholar]
- 9. Havenar-Daughton C., Reiss S. M., Carnathan D. G., Wu J. E., Kendric K., Torrents de la Peña A., Kasturi S. P., Dan J. M., Bothwell M., Sanders R. W., et al. 2016. Cytokine-independent detection of antigen-specific germinal center T follicular helper cells in immunized nonhuman primates using a live cell activation-induced marker technique. J. Immunol. 197: 994–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reiss S., Baxter A. E., Cirelli K. M., Dan J. M., Morou A., Daigneault A., Brassard N., Silvestri G., Routy J. P., Havenar-Daughton C., et al. 2017. Comparative analysis of activation induced marker (AIM) assays for sensitive identification of antigen-specific CD4 T cells. PLoS One 12: e0186998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gray G. E., Huang Y., Grunenberg N., Laher F., Roux S., Andersen-Nissen E., De Rosa S. C., Flach B., Randhawa A. K., Jensen R., et al. 2019. Immune correlates of the Thai RV144 HIV vaccine regimen in South Africa. Sci. Transl. Med. 11: eaax1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frentsch M., Arbach O., Kirchhoff D., Moewes B., Worm M., Rothe M., Scheffold A., Thiel A.. 2005. Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nat. Med. 11: 1118–1124. [DOI] [PubMed] [Google Scholar]
- 13. Trombetta J. J., Gennert D., Lu D., Satija R., Shalek A. K., Regev A.. 2014. Preparation of single-cell RNA-seq libraries for next generation sequencing. Curr. Protoc. Mol. Biol. 107: 4.22.1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bray N. L., Pimentel H., Melsted P., Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. [Published erratum appears in 2016 Nat. Biotechnol. 34: 888.] Nat. Biotechnol. 34: 525–527. [DOI] [PubMed] [Google Scholar]
- 15. Soneson C., Love M. I., Robinson M. D.. 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000 Res. 4: 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Amezquita R. A., Lun A. T. L., Becht E., Carey V. J., Carpp L. N., Geistlinger L., Marini F., Rue-Albrecht K., Risso D., Soneson C., et al. 2020. Orchestrating single-cell analysis with Bioconductor. [Published erratum appears in 2020 Nat. Methods 17: 242.] Nat. Methods 17: 137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCarthy D. J., Campbell K. R., Lun A. T., Wills Q. F.. 2017. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33: 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van der Maaten L., Hinton G.. 2008. Visualizing data using t-SNE. J. Mach. Learn. Res. 9: 2579–2605. [Google Scholar]
- 19. Csardi G., Nepusz T.. 2006. The igraph software package for complex network research. InterJournal Complex Systems 1695. [Google Scholar]
- 20. Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A. K., Slichter C. K., Miller H. W., McElrath M. J., Prlic M., et al. 2015. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16: 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen T., Guestrin C.. 2016. XGBoost: a scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. Association for Computing Machinery, San Francisco, CA, p. 785–794. [Google Scholar]
- 22. Lundberg S. M., Erion G., Chen H., DeGrave A., Prutkin J. M., Nair B., Katz R., Himmelfarb J., Bansal N., Lee S. I.. 2020. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao L. P., Fiore-Gartland A., Carpp L. N., Cohen K. W., Rouphael N., Fleurs L., Dintwe O., Zhao M., Moodie Z., Fong Y., et al. 2020. Landscapes of binding antibody and T-cell responses to pox-protein HIV vaccines in Thais and South Africans. PLoS One 15: e0226803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson P., Ruffell B.. 2009. CD44 and its role in inflammation and inflammatory diseases. Inflamm. Allergy Drug Targets 8: 208–220. [DOI] [PubMed] [Google Scholar]
- 25. Budd R. C., Cerottini J. C., Horvath C., Bron C., Pedrazzini T., Howe R. C., MacDonald H. R.. 1987. Distinction of virgin and memory T lymphocytes. Stable acquisition of the Pgp-1 glycoprotein concomitant with antigenic stimulation. J. Immunol. 138: 3120–3129. [PubMed] [Google Scholar]
- 26. Ariel A., Lider O., Brill A., Cahalon L., Savion N., Varon D., Hershkoviz R.. 2000. Induction of interactions between CD44 and hyaluronic acid by a short exposure of human T cells to diverse pro-inflammatory mediators. Immunology 100: 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shibagaki N., Hanada K., Yamaguchi S., Yamashita H., Shimada S., Hamada H.. 1998. Functional analysis of CD82 in the early phase of T cell activation: roles in cell adhesion and signal transduction. Eur. J. Immunol. 28: 1125–1133. [DOI] [PubMed] [Google Scholar]
- 28. Schultz B. T., Teigler J. E., Pissani F., Oster A. F., Kranias G., Alter G., Marovich M., Eller M. A., Dittmer U., Robb M. L., et al. 2016. Circulating HIV-specific interleukin-21+CD4+ T cells represent peripheral Tfh cells with antigen-dependent helper functions. Immunity 44: 167–178. [DOI] [PubMed] [Google Scholar]
- 29. Chevalier M. F., Jülg B., Pyo A., Flanders M., Ranasinghe S., Soghoian D. Z., Kwon D. S., Rychert J., Lian J., Muller M. I., et al. 2011. HIV-1-specific interleukin-21+ CD4+ T cell responses contribute to durable viral control through the modulation of HIV-specific CD8+ T cell function. J. Virol. 85: 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H., Willingham S. B., Ting J. P., Re F.. 2008. Cutting edge: inflammasome activation by alum and alum’s adjuvant effect are mediated by NLRP3. J. Immunol. 181: 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ott G., Barchfeld G. L., Chernoff D., Radhakrishnan R., van Hoogevest P., Van Nest G.. 1995. MF59. Design and evaluation of a safe and potent adjuvant for human vaccines. Pharm. Biotechnol. 6: 277–296. [DOI] [PubMed] [Google Scholar]
- 32. Galli G., Medini D., Borgogni E., Zedda L., Bardelli M., Malzone C., Nuti S., Tavarini S., Sammicheli C., Hilbert A. K., et al. 2009. Adjuvanted H5N1 vaccine induces early CD4+ T cell response that predicts long-term persistence of protective antibody levels. Proc. Natl. Acad. Sci. USA 106: 3877–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vaccari M., Gordon S. N., Fourati S., Schifanella L., Liyanage N. P., Cameron M., Keele B. F., Shen X., Tomaras G. D., Billings E., et al. 2016. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. [Published erratum appears in 2016 Nat. Med. 22: 1192.] Nat. Med. 22: 762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq data reported in this study is Gene Expression Omnibus (GEO) GSE166945 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).