Abstract
O-linked β-d-N-acetylglucosamine (O-GlcNAc) is an important post-translational modification of serine or threonine residues on thousands of proteins in the nucleus and cytoplasm of all animals and plants. In eukaryotes, only two conserved enzymes are involved in this process. O-GlcNAc transferase is responsible for adding O-GlcNAc to proteins, while O-GlcNAcase is responsible for removing it. Aberrant O-GlcNAcylation is associated with a variety of human diseases, such as diabetes, cancer, neurodegenerative diseases, and cardiovascular diseases. Numerous studies have confirmed that O-GlcNAcylation is involved in the occurrence and progression of cancers in multiple systems throughout the body. It is also involved in regulating multiple cancer hallmarks, such as metabolic reprogramming, proliferation, invasion, metastasis, and angiogenesis. In this review, we first describe the process of O-GlcNAcylation and the structure and function of O-GlcNAc cycling enzymes. In addition, we detail the occurrence of O-GlcNAc in various cancers and the role it plays. Finally, we discuss the potential of O-GlcNAc as a promising biomarker and novel therapeutic target for cancer diagnosis, treatment, and prognosis.
Keywords: O-GlcNAc, Post-translational modification, OGT, OGA, Biomarker, Cancer therapy
Background
O-linked β-d-N-acetylglucosamine (O-GlcNAc) is an important post-translational modification (PTM) comprising the reversible, highly dynamic, covalent attachment of β-N-GlcNAc to Ser/Thr residues on proteins, which was first reported by Hart in 1984 (Torres and Hart 1984). Unlike conventional complex glycans decorating the surface of cells, O-GlcNAc is a simple monosaccharide modification that mostly occurs inside cells, specifically in the nucleus or cytoplasm. With the development of technologies related to identification, site mapping, quantitation, and site-specific O-GlcNAc protein function determination of O-GlcNAc proteins (Ma and Hart 2014), studies have identified more than 16,000 proteins that are O-GlcNAcylated in 42 species (Wulff-Fuentes et al. 2021), including cytoskeletal proteins and their regulatory proteins (Arnold et al. 1996; Ding and Vandre 1996; Takahashi et al. 1999), nucleoporins (Davis and Blobel 1987), synaptic proteins (Luthi et al. 1991), heat shock proteins (Roquemore et al. 1996), tumor suppressor proteins, RNA polymerase II catalytic subunit (Kelly et al. 1993; Cervoni et al. 1997), as well as multiple transcription factors (Jackson and Tjian 1988; Yang et al. 2001; Iyer et al. 2003). These proteins are involved in all aspects of cellular function, including metabolism, signal transduction, transcriptional regulation, cell cycle control, protein trafficking, and regulation of the cell structure (Wells et al. 2001; Love and Hanover 2005; Zachara and Hart 2006; Slawson and Hart 2011).
O-GlcNAcylation has many unique characteristics compared to "classical" glycosylation. Chemically, O-GlcNAcylation only adds monosaccharides to the Ser/Thr residues of proteins and does not involve complex glycan structures. Spatially, O-GlcNAcylation mainly targets proteins in the nucleus, cytoplasm, and mitochondria rather than on the cell membrane (Hu et al. 2009). Temporally, similar to phosphorylation/dephosphorylation, O-GlcNAcylation achieves highly dynamic and fast-cycling PTM by reversibly adding and removing β-N-GlcNAc on protein Ser/Thr residues.
The diversity and importance of O-GlcNAc-modified proteins mean that a variety of human disorders are linked to aberrant O-GlcNAc, including diabetes (Issad et al. 2010; Ma and Hart 2013) and diabetic complications (Peterson and Hart 2016; Degrell et al. 2009; Akimoto et al. 2011), cardiovascular diseases (Chatham et al. 2008; Laczy et al. 2009; Fulop et al. 2007; Clark et al. 2003; Ngoh et al. 2009), cancer (Slawson and Hart 2011; Ma and Vosseller 2013; Fardini et al. 2013), neurodegenerative diseases (such as Alzheimer's disease) (Lazarus et al. 2009; Gong et al. 2012; Zhu et al. 2014), and immune system disorders (Golks and Guerini 2008; Golks et al. 2007). In recent years, because of the large proportion of cancer deaths among all deaths worldwide (Erratum: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. 2018), increasing numbers of studies have focused on the role of O-GlcNAcylation in many cancers, including their metabolism, proliferation, angiogenesis, and metastasis (Ferrer et al. 2016; Wu et al. 2020; Ma and Vosseller 2014). In this review, we first describe the general mechanism of O-GlcNAcylation and the structure of O-GlcNAc regulating enzymes and then focus on the important role played by O-GlcNAcylation in various cancers. Finally, we discuss the potential of O-GlcNAc as a cancer diagnostic and prognostic biomarker and therapeutic target.
The basic process of O-GlcNAcylation
Under euglycemic conditions, most glucose taken up by cells enters the glycolytic metabolic pathway; however, 3–5% of the glucose is still separated from the glycolytic pathway into the hexosamine biosynthetic pathway (HBP) (Marshall et al. 1991). In this process, an intermediate of glycolysis, fructose-6-phosphate, is converted to glucosamine 6-phosphate, catalyzed by glutamine fructose-6-phosphate amidotransferase (GFAT), and then the end product of the HBP, UDP-GlcNAc, is synthesized in the presence of many different metabolites, such as the fatty acid metabolite acetyl-coA and the nucleic acid metabolite UTP (Dennis et al. 2009). Eventually, UDP-GlcNAc acts as the sugar donor for O-GlcNAc. Under the action of O-GlcNAc modifying enzymes, β-N-GlcNAc is provided to complete the dynamic modification of countless intracellular proteins (Fig. 1).
Fig. 1.
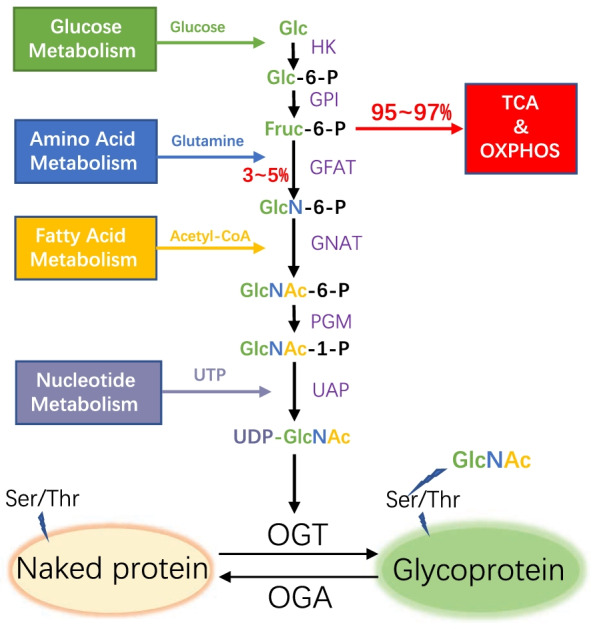
The process of hexosamine biosynthetic pathway (HBP) and O-GlcNAcylation. HBP integrates multiple metabolic pathways and the end product UDP-GlcNAc acts as a sugar donor for protein O-GlcNAcylation. HK hexokinase, GPI glucose-6-phosphate isomerase, GFAT glutamine:fructose-6-phosphate amidotransferase, GNAT glucosamine 6-phosphate N-acetyltransferase, PGM phosphoglucomutase, UAP UDP-N-acetylglucosamine pyrophosphorylase, TCA tricarboxylic acid cycle, OXPHOS oxidative phosphorylation
O-GlcNAc modifying enzymes
As an extensive PTM, the number of proteins modified by O-GlcNAc is similar to that of phosphorylation (Bond and Hanover 2015). However, unlike phosphorylation/dephosphorylation, only two highly conserved enzymes are involved in the addition and removal of O-GlcNAc (Hart 2019; Kreppel et al. 1997; Lubas et al. 1997). O-GlcNAc transferase (OGT) is responsible for connecting β-N-GlcNAc to the substrate protein, and β-N-acetylglucosaminidase (O-GlcNAcase, OGA) removes it. In contrast to this, there are more than 1000 kinases and more than 500 phosphatases involved in phosphorylation/dephosphorylation (Cohen 2000).
O-GlcNAc transferase (OGT)
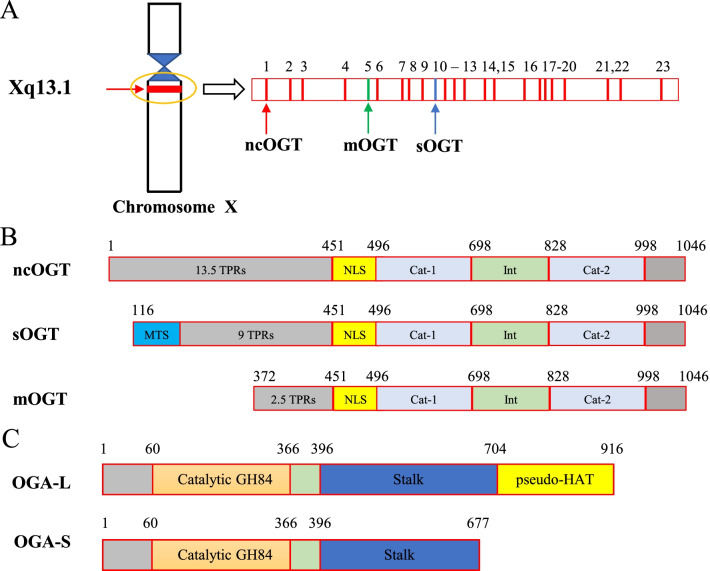
OGT is the only enzyme responsible for the addition of O-GlcNAc to the Ser/Thr residues of target proteins. In humans, the gene encoding OGT is located near the centromere of the X chromosome (Xq13.1) (Nolte and Muller 2002) and contains 23 exons and 21 introns. As a result of alternative splicing, there are three independent isoforms with different amino termini: nucleocytoplasmic OGT (ncOGT), mitochondrial OGT (mOGT), and short form OGT (sOGT) (Nolte and Muller 2002). Among them, ncOGT is the longest isoform and comprises 1046 amino acids, encoded by exons 1–4 and 6–23 (Hanover et al. 2003). The second longest, mOGT, is encoded by exons 5–23, and comprises 931 amino acids. mOGT localizes to the mitochondria, which is determined by its particular mitochondrial targeting sequence (MTS) at the N-terminus (Love et al. 2003). The shortest isoform, sOGT, is encoded by exons 10–23 and comprises 675 amino acids (Hanover et al. 2003) (Fig. 2A). OGT is widely expressed in all tissues, but varies in content, with the highest expression in pancreatic β-cells and brain, and the lowest in the liver and lung. (Weinstein et al. 2013; Alonso et al. 2014; Vander et al. 2009)
Fig. 2.
Schematic diagram of the gene and protein structure of O-GlcNAc modifying enzymes. Human OGT is composed of three regions and three splice forms are known. An N-terminal domain is formed from a series of TPRs and a C-terminal domain contains the glycosyltransferase catalytic domain. A Schematic of the genomic structure of the human OGT splice variants. B The linear domain organization is shown for the three OGT isoforms with the amino acids marking the boundary of each domain indicated. The TPRs are shown in light gray, the nuclear localization sequence (NLS) is in yellow, and the glycosyltransferase catalytic regions are in light blue (Cat-1and Cat-2). C The linear domain organization is shown for the two OGA isoforms with the amino acids marking the boundary of each domain indicated. The Catalytic GH84 is shown in orange, the pseudo-HAT is shown in yellow
From the crystal structure, OGT is composed of three regions: a tripeptide repeat (TPR) motif at the amino (N) terminal domain, a catalytic domain at the carboxyl (C) terminus, and a nuclear localization sequence (NLS) connecting these two domains (Kreppel et al. 1997; Lubas et al. 1997). The three isomers share the same C-terminal catalytic domain, which comprises an insertion domain separating two catalytic sheets. However, there is a structural difference at the N-terminus, which is manifested in the different number of TPR motifs: ncOGT contains 13.5 TPRs, mOGT contains 9, while sOGT contains only 2.5 TPRs (Ju 2020) (Fig. 2B).
Given that dysregulation of O-GlcNAcylation is associated with various diseases, tuning of OGT function is necessary and is achieved in three ways: First, the activity of OGT is influenced by the concentration of metabolites and enzymes involved in HBP, especially the precise regulation of the end product UDP-GlcNAc (Kreppel and Hart 1999); second, OGT itself is modified by various PTMs, such as phosphorylation, ubiquitination, (Peng et al. 2021) and S-Nitration (Ryu and Do 2011). Phosphorylation at Thr444 of OGT by AMP-activated protein kinase (AMPK) can alter its substrate selectivity and nuclear localization (Bullen et al. 2014); and glycogen synthase kinase-3β (GSK3β) can activate OGT by phosphorylating serine residues on OGT and regulates circadian rhythms (Kaasik et al. 2013). Interestingly, OGT can be O-GlcNAcylated (Fan et al. 2018; Griffin et al. 2016; Tai et al. 2004): O-GlcNAc at Ser389 affects its nuclear localization, whereas O-GlcNAc at Thr12 and Ser56 alters its substrate selectivity (Seo et al. 2016; Liu et al. 2019a). Finally, OGT can interact with other proteins thereby regulating its activity: under glucose deprivation, OGT can influence the activity of specific proteins by interacting with the stress kinase p38 (Cheung and Hart 2008); and peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), a transcriptional coactivator under hyperglycemic conditions, interacts with OGT to promote the activity of OGT on the transcription factor forkhead box O1 (FOXO1) (Housley et al. 2008, 2009).
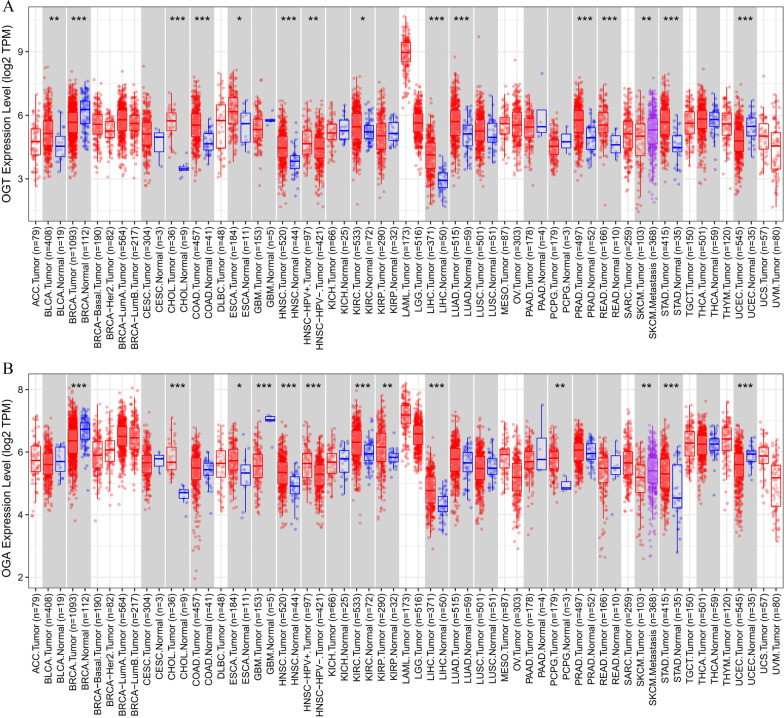
Abnormal OGT activity affects many downstream proteins. Therefore, the expression of OGT often changes in diseases states, especially in various cancers. To further understand the potential roles and clinical relevance of OGT in human cancers, our group investigated the OGT expression profiles in 33 major human cancer types in The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov) database (Weinstein et al. 2013; Blum et al. 2018; Roychowdhury and Chinnaiyan 2016), using Gene_DE module of the Tumor Immune Estimation Resource package (Li et al. 2020) (TIMER2.0, http://timer.cistrome.org). The results showed that: (1) Compared with that in adjacent normal tissues, OGT was expressed at markedly higher levels in bladder urothelial carcinoma (BLCA), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD). and (2) the expression level of OGT in breast invasive carcinoma (BRCA) and uterine corpus endometrial carcinoma (UCEC) was significantly lower than that in normal tissues (Fig. 3A).
Fig. 3.
Expression profile analyses of O-GlcNAc modifying enzymes across multiple cancers and normal tissues. Expression pattern of OGT (A) and OGA (B) in ACC, BLCA, BRCA, CESC, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LAML, LGG, LIHC, LUAD, LUSC, OV, PAAD, PCPG, PRAD, READ, SARC, SKCM, STAD, TGCT, THCA, THYM, UCEC, and UCS. TIMER2.0 was used to generate box plots profiling O-GlcNAc modifying enzymes expression patterns across multiple cancer types (TCGA tumor) and adjacent normal tissue samples (TCGA normal). Each dot represents the individual expression of a distinct tumor or normal sample. The statistical significance computed by the Wilcoxon test is annotated by the number of stars (*p-value < 0.05; **p-value < 0.01; ***p-value < 0.001). As displayed in gray columns when normal data is available
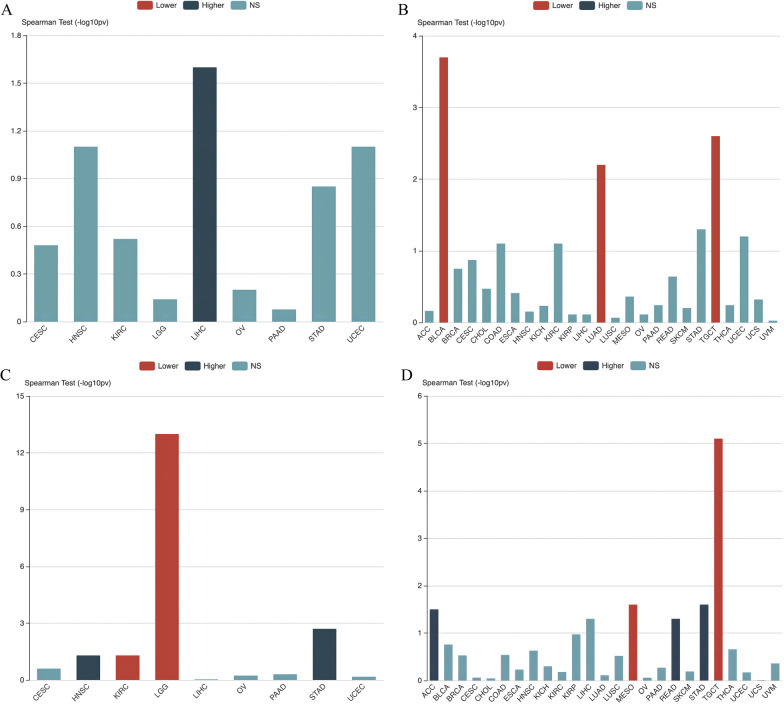
Furthermore, we analyzed the association between OGT expression and tumor-node-metastasis (TNM) stage, and the tumor grades across 33 cancer types using the Tumor and Immune System Interaction Database (TISIDB, http://cis.hku.hk/TISIDB) (Ru et al. (2019). The results showed that across 33 cancer types: (1) OGT expression was associated with higher grades only of LIHC (Fig. 4A); and (2) OGT expression was associated with lower tumor stages in BLCA, testicular germ cell tumors (TGCT), and LUAD (Fig. 4B) Overall, the expression of OGT is diverse in various cancers and is closely related to the grade and stage of tumors.
Fig. 4.
Association analyses between O-GlcNAc modifying enzymes and clinical features across multiple cancers. A Association between OGT expression and grade across human cancers. B Association between OGT expression and stage across human cancers. C Association between OGA expression and grade across human cancers. D Association between OGA expression and stage across human cancers. TISIDB was used to generate associations between the expression of OGT and OGA and pathological distribution across multiple cancers (TCGA tumor). Spearman test was used to calculate the associations, and a p-value < 0.05 was considered statistically significant
O-GlcNAcase (OGA)
O-GlcNAcase (OGA) is only enzyme which removes O-GlcNAc from the target proteins. In humans, OGA is encoded by the OGA gene on chromosome 10 (10q24.1–q24.3) and maps to a genetic loci involved in Alzheimer’s disease (Bertram et al. 2000; Kuwano et al. 2006). Human OGA contains 17 exons and encodes at least two (and possibly more) OGA isoforms of different lengths, caused by alternative splicing of the mRNA. To date, long form OGA (OGA-L) and short form OGA (OGA-S) have been well studied. OGA-L is the longest of the isomers and consists of 916 amino acids. OGA-S is a shorter isomer consisting of only 677 amino acids (Vocadlo 2012). OGA is a dual-domain protein, its N-terminus contains a member of the glycoside hydrolase family 84 (GH84) domain and is considered the catalytic domain (Hanover 2001) (Fig. 2C). The C-terminal domain is believed to have histone acetyltransferase (HAT) activity (Toleman et al. 2004), although this conclusion is still a matter of debate (Butkinaree et al. 2008; Alonso et al. 2014; Schultz and Pils 2002). Between the GH84 and HAT domains, there is an unstructured region in which Asp413 contains a cleavage site for caspase-3 and is processed during apoptosis (Butkinaree et al. 2008). Like OGT, OGA is expressed in all tissues, but varies in content (Gao et al. 2001). The highest expression is observed in the brain, lymph nodes, and spleen, while the lowest expression is observed in the pancreas and salivary glands.
Long-term difficulties in determining the OGA structure mean that the regulatory mechanism of OGA remains to be investigated. OGA is known to be modified by PTMs such as O-GlcNAcylation (Woo et al. 2018), phosphorylation (Dephoure et al. 2008; Olsen et al. 2010), and ubiquitination (Akimov et al. 2018). For example, OGA itself can be O-GlcNAcylated at Ser398, 399, 405, 410, and Thr415 as a substrate of OGT; however, the mechanism by which these PTMs regulate the function of OGA is largely unknown.
Similar to OGT, we used TCGA, TIMER2.0, and TISIDB to investigate the prognostic influence, pathological features, and clinical relevance of OGA expression in 33 major human cancer types (Weinstein et al. 2013; Blum et al. 2018; Roychowdhury and Chinnaiyan 2016). The results showed that, compared with that in adjacent normal tissues, (1) OGA expression is significantly higher in CHOL, ESCA, HNSC, KIRC, kidney renal papillary cell carcinoma (KIRP), LIHC, pheochromocytoma and paraganglioma (PCPG) and STAD. (Fig. 3B). (2) OGA expression was associated with higher grades of KIRC and brain lower grade glioma (LGG), but with lower grades of HNSC and STAD (Fig. 4C). (3) OGA expression was associated with higher tumor stages in mesothelioma (MESO) and TGCT, but with lower stages in ACC, READ, and STAD (Fig. 4D).
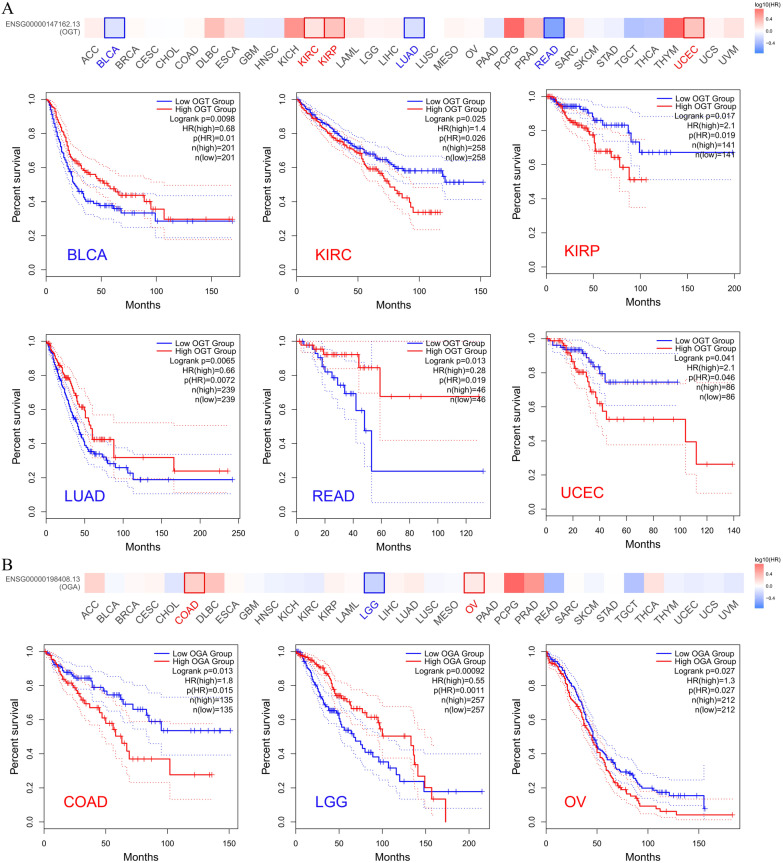
In addition, we further analyzed the relationship between the overall survival (OS) and the expression of OGT and OGA in 33 major human cancer types using the Gene Expression Profiling Interactive Analysis (GEPIA2, http://gepia2.cancer-pku.cn) package (Tang et al. 2019), the results showed that: (1) OGT expression was related to the OS of BLCA, KIRC, KIRP, LUAD, and READ, although the trends were inconsistent (Fig. 5A). (2) OGA expression was associated with the OS of COAD, LGG, and ovarian serous cystadenocarcinoma (OV), although the trend was inconsistent (Fig. 5B). In conclusion, the expression patterns of OGT and OGA are diverse and have both positive and negative effects on clinicopathological features, which indicates that the role of O-GlcNAc in cancer is very complex.
Fig. 5.
Correlation between OGT and OGA expression and survival prognosis of cancers in TCGA. We used the GEPIA2 tool to perform overall survival analyses of different tumors in TCGA by OGT (A) and OGA (B) expression. The survival map and Kaplan–Meier curves with positive results are given
O-GlcNAc and Cancer
An important characteristic of cancer is genomic instability and mutation (Hanahan and Weinberg 2011). In order to more intuitively understand the extent to which cancer is caused by the direct mutation of the O-GlcNAc site at Ser /Thr, we first selected the top ten most common mutated genes in the TCGA Pancancer Atlas Studies provided by cbioportal database (Cerami et al. 2012) (https://www.cbioportal.org/), they were TP53, TIN, MUC16, PIK3CA, CSMD3, RYR2, LRP1B, SYNE1, FLG, USH2A. Next, we sorted out their mutations at Ser/Thr, and then found their overlap sites according to the O-GlcNAc site information provided by the O-GlcNAc database (Wulff-Fuentes et al. 2021) (https://www.oglcnac.mcw.edu/). The results show that: (1) among the top ten most common mutations in TCGA Pancancer Atlas Studies, after excluding those proteins that are not included in the O-GlcANc database and have not been reported with the O-GlcANc site, there are four proteins whose mutation sites overlapped with the O-GlcNAc site, namely TP53, TTN, MUC16 and SYNE1. (2) Among these overlapping sites, the site S149 of TP53 is annotated as "likely oncogenic" while the role of overlapping sites of other genes is unknown (Table1).
Table 1.
The overlapping sites of O-GlcANc and cancer mutation
| Gene | Mutation proportion (%) | Protein | Uniprot ID | O-GlcNAc site | Cancer Type | Overlapping site | Protein Change | Annotation |
|---|---|---|---|---|---|---|---|---|
| TP53 | 37 | P53 | P04637 | S149 | Serous Ovarian Cancer | S149 | S149Ffs*32 | The TP53 S149Ffs*32 mutation is likely oncogenic |
| Prostate Adenocarcinoma | S149 | S149Pfs*21 | The TP53 S149Pfs*21 mutation is likely oncogenic | |||||
| Pancreatic Adenocarcinoma | S149 | S149Ffs*32 | The TP53 S149Ffs*32 mutation is likely oncogenic | |||||
| TTN | 30 | TITIN | Q8WZ42 | T671, T826, S1571, T3501, S4651, T4659, S7613, S10385, S10781, T12007, T14674, S28157, S28450, S33976 | Uterine Endometrioid Carcinoma | T671 | T671S | Unknown |
| Uterine Endometrioid Carcinoma | S1571 | S1571T | Unknown | |||||
| Cutaneous Melanoma | S1571 | S1571F | Unknown | |||||
| MUC16 | 19.3 | MUC16 | Q8WXI7 | S12117, S13054, T13833 | Uterine Endometrioid Carcinoma | S12117 | S12117N | Unknown |
| Renal Clear Cell Carcinoma | S13054 | S13054T | Unknown | |||||
| Renal Clear Cell Carcinoma | S13054 | S13054F | Unknown | |||||
| PIK3CA | 14.1 | PK3CA | P42336 | no data | ||||
| CSMD3 | 13.3 | CSMD3 | Q7Z407 | no data | ||||
| RYR2 | 13.2 | RYR2 | Q92736 | T1468 | No overlapping sites | |||
| LRP1B | 12.7 | LRP1B | Q9NZR2 | no data | ||||
| SYNE1 | 12.2 | SYNE1 | Q8NF91 | S1286, T1951, S3382, T4836, T7270 | Serous Ovarian Cancer | T1951 | T1951A | Unknown |
| Cutaneous Melanoma | S3382 | S3382N | Unknown | |||||
| FLG | 11.4 | FILA | P20930 | na | ||||
| USH2A | 11.2 | USH2A | O75445 | na |
no data: could not find human protein in O-GlcNAc database, na: no O-GlcNAc site reported
Another key hallmark of cancer is the reprogramming of energy metabolism in cancer cells (Hanahan and Weinberg 2011). The Warburg effect (Vander et al. 2009; Liberti and Locasale 2016) allows cancer cells to greatly increase glucose uptake and HBP flux. Therefore, as a downstream pathway of HBP, O-GlcNAcylation in cancer cells is largely affected by this particular mode of metabolism. Hyper-O-GlcNAcylation has been observed in almost all types of cancer (Ma and Vosseller 2014), including ovarian cancer (Queiroz et al. 2016), cervical cancer (Zeng et al. 2016), breast cancer (Caldwell et al. 2010), endometrial cancer (Krzeslak et al. 2012), liver cancer (Xu et al. 2017), colorectal cancer (Mi et al. 2011), cholangiocarcinoma (Phoomak et al. 2012), pancreatic cancer (Ma et al. 2013), gastric cancer (Jiang et al. 2016), esophageal squamous cell carcinoma (Qiao et al. 2012), bladder cancer, (Jin et al. 2020a) prostate cancer (Lynch et al. 2012), renal cell carcinoma (Wang et al. 2019), lung cancer (Lin et al. 2018), laryngeal cancer (Starska et al. 2015), thyroid papilloma (Li et al. 2021a), chronic and acute lymphoblastic leukemia (Shi et al. 2010; Zhang et al. 2017a) and glioblastoma (Ciraku et al. 2022). In addition to participating in metabolic reprogramming of cancer cells, O-GlcNAcylation also linked to various hallmarks of cancer, including cancer cell survival, proliferation, angiogenesis, invasion, metastasis, and epigenetics (Ma and Vosseller 2014). This part focuses on the role of O-GlcNAc in cancers from multiple systems (Table 2).
Table 2.
O-GlcNAc expression and roles in various cancers
| Cancer type | Research object | O-GlcNAc | OGT | OGA | Major findings | Clinical | References |
|---|---|---|---|---|---|---|---|
| Cervical cancer | HPV-related cervical tumors and human cervical cancer cell lines | Elevated | Elevated | no significant change | Elevated OGT activated the transcription of HPV E6/E7 and thus enhancing the oncogenic activity of HPV | Zeng et al. (2016) | |
| The human cervical cancer cell lines | Elevated | Elevated | Na | Elevated OGT not only increased the expression of E6/E7 oncoproteins but also promoted HCF-1-mediated transcriptional activity of the E6/E7 promoter | Xu et al. (2021) | ||
| The human cervical cell lines | Elevated | Elevated | Na | O-GlcNAcylation of NF-κB in cervical cancer promoted lung metastasis of cervical cancer by activating CXCR4 | Ali et al. (2017) | ||
| Breast cancer | Primary breast malignant tumors | Elevated | Elevated | Na | Reduction of O-GlcNAcylation inhibited the anchorage-independent growth of breast cancer cells | Champattanachai et al. (2013) | |
| Breast cancer cell lines | Elevated | Elevated | Na | O-GlcNAcylation enhanced the migration/invasion of breast cancer cells in vitro and lung metastasis in vivo | Gu et al. (2010) | ||
| Breast cancer cell lines | Elevated | Elevated | Na | Elevated O-GlcNAcylation and OGT levels contributed to cancer cell growth and invasion, | Caldwell et al. (2010) | ||
| Breast cancer cell lines | Elevated | Elevated | Na | Nutrient sensing pathway HBP connected with the SIRT1 deacetylase via O-GlcNAcylation to regulate cellular invasion via regulation of FOXM1 | Ferrer et al. (2017) | ||
| Breast cancer stem cells | Elevated | Elevated | Na | OGT played a key role in the regulation of breast CSCs in vitro and tumor initiation in vivo | Akella et al. (2020) | ||
| HR + /HER2- luminal breast cancer patient samples | Elevated | Na | Na | Hyper-O-GlcNAcylation was associated with poor 10-year DFS in patients with breast cancer | Poor survival | Kuo et al. (2021) | |
| Endometrial cancer | Endometrial cancer patient samples | Na | Elevated | Elevated | The OGT and OGA expression were significantly higher in tumors of a higher histological grade and associated with the depth of tumor invasion into the myometrium | Krzeslak et al. (2012) | |
| Endometrial cancer cell lines | Elevated | Na | Na | Hyper-O-GlcNAcylation promoted EMT in endometrial cancer cells | Jaskiewicz and Townson (2019) | ||
| OV | The human ovarian carcinoma cell lines | Elevated | Elevated | Na | O-GlcNAcylation decreased E-cadherin level, thereby inhibiting E-cadherin/catenin complex formation and reducing cell–cell adhesion, leading to cancer cell metastasis | Jin et al. (2013) | |
| The human ovarian carcinoma cell lines | Elevated | Elevated | Na | O-GlcNAcylation augments the motility of ovarian cancer cells via the RhoA/ROCK/MLC signaling pathway | Niu et al. (2017) | ||
| Liver cancer | HCC patient samples and cell lines | Elevated | Elevated | Na | O-GlcNAcylation of AGER increased its activity and stability to promote the development of HCC under high glucose conditions | Qiao et al. (2016a) | |
| HCC patient samples and cell lines | Elevated | Elevated | Na | O-GlcNAcylation of HDAC1 was overexpressed in HCC, and the progression of HCC can be inhibited by inhibiting the O-GlcNAcylation of HDAC1 | Zhu et al. (2016) | ||
| Livers of diabetic mice | Elevated | Na | Na | There is positive auto-regulatory feedback between O-GlcNAcylation and TRIB2, which might be critical for diabetes-associated liver cancer | Yao et al. (2016) | ||
| HCC patient samples and cell lines | Elevated | Elevated | Na | ACSL4 promoted HCC growth and survival by enhancing O-GlcNAcylation and activating mTOR signaling. Conversely, O-GlcNAcylation facilitated HCC growth via increasing ACSL4 expression and activating mTOR signaling | Wang et al. (2020) | ||
| NAFLD-HCC patient samples, and liver cancer cell lines | Na | Elevated | Na | OGT played an oncogenic role in NAFLD-associated HCC through regulating palmitic acid and inducing ER stress, consequently activating oncogenic JNK/c-Jun/AP-1 and NF-κB cascades | Xu et al. (2017) | ||
| Samples from patients with HCC recurrence after liver transplantation | Elevated | Elevated | Decreased | O-GlcNAcylation was significantly enhanced in the tumor tissues of patients who had suffered from HCC recurrence after LT compared with those who had not. Importantly, low expression of OGA was an independent prognostic factor for predicting tumor recurrence of HCC following LT, especially in patients with low AFP expression | Poor Survival | Zhu et al. (2012) | |
| HCC patient samples | Elevated | Na | Na | Increased O-GlcNAcylation of RACK1 is positively correlated with tumor growth, metastasis, and recurrence in patients with HCC | Duan et al. (2018) | ||
| Liver cancer patient samples and cell lines | Elevated | Na | Na | YAP was O-GlcNAcylated at Thr241 thereby antagonizing Hippo pathway-mediated phosphorylation of YAP, thus allowing YAP to promote liver tumorigenesis under diabetes-prone, high-glucose conditions | Zhang et al. (2017b) | ||
| CRC | CRC patient samples and the human colon tumor cell lines | Elevated | Elevated | no significant change | O-GlcNAcylation enhanced the anchorage-independent growth of colon cancer cells | Mi et al. (2011) | |
| CRC patient samples | Elevated | Elevated | no significant change | Abnormal O-GlcNAc-modified proteins, particularly annexin A2, may be novel biomarkers for CRC | Phueaouan et al. (2013) | ||
| CRC patient samples and the CRC cell lines | Elevated | Na | Na | O-GlcNAcylation at Thr236 of YY1 enhanced the expression of SLC22A15 and AANAT in cells and increase the protein stability of YY1 itself to exert its oncogenic effect | Zhu et al. (2019) | ||
| Human CRC cell lines | Elevated | Elevated | Na | Hyper-O-GlcNAcylation significantly contributed to tumor proliferation and metastasis and indicate a poor prognosis in patients with CRC | Poor survival | Wu et al. (2019) | |
| The murine colon carcinoma cells | Elevated | Na | Na | O-GlcNAcylation deregulated β-catenin and E-cadherin expression and activity in fibroblast cell lines and this might influence EMT and cell motility, which may further influence tumor development and metastasis | Harosh-Davidovich and Khalaila (2018) | ||
| Human colon cancer cells | Elevated | Elevated | Na | O-GlcNAcylation of XIAP at Ser406 is essential for its E3 ubiquitin ligase activity toward specifically OGT | Seo et al. (2020) | ||
| CRC patient samples, CRC cell lines | Elevated | Elevated | Na | ITGA5 overexpression accelerates the progression of CRC, which is closely associated with its enhanced O-GlcNAcylation | Yu et al. (2019) | ||
| PDAC | Human pancreatic cancer cells | Elevated | Elevated | Decreased | Hyper-O-GlcNAcylation played an important role in PDAC cells’ survival and constitutive NF-κB activity | Ma et al. (2013) | |
| PDAC cells | Elevated | Elevated | Elevated | OGA promotes OGT transcription through cooperation with the histone acetyltransferase p300 and transcription factor CCAAT/enhancer-binding protein β (C/EBPβ) | Qian et al. (2018) | ||
| The pancreatic cancer cell lines | Elevated | Elevated | Na | Triptolide-induced cell death in pancreatic cancer is mediated by alteration of O-GlcNAcylation of Sp1 | Banerjee et al. (2013) | ||
| GC | Primary GC patient samples | Elevated | Elevated | Na | O-GlcNAcylation was associated with the carcinogenesis and progression of GC | Poor survival | Jang and Kim (2016) |
| GC patient samples and cell lines | Elevated | Elevated | Na | Hyper-O-GlcNAcylation significantly promoted GC cell proliferation by modulating cell cycle-related proteins and ERK 1/2 signaling | Poor survival | Jiang et al. (2016) | |
| ESCC | ESCC patient samples | Elevated | Elevated | Na | Hyper-O-GlcNAcation stabilized proteins, leading to changes in cellular signal transduction and resulting in tumorigenesis and metastasis | Poor survival | Qiao et al. (2012) |
| ESCSs, ESCC cell lines | Na | Elevated | Na | OGT in exosomes from ECSCs protected ECSCs from CD8 + T cells through up-regulation of PD-1 | Yuan et al. (2021) | ||
| CHOL | CHOL patient samples | Elevated | Elevated | Decreased | Hyper-O-GlcNAcylation in CHOL tissues was associated with poor patient outcomes | Poor Survival | Phoomak et al. (2012) |
| PC | PC patient samples and cell lines | Elevated | Elevated | Na | OGT and O-GlcNAcylation were elevated in PC cells and required for growth, invasion, angiogenesis, and metastasis | Lynch et al. (2012) | |
| PC biopsy patient samples | Elevated | Na | Na | Hyper-O-GlcNAcylation was associated with decreased OS of patients | Poor Survival | Kamigaito et al. (2014) | |
| PC cell lines | Elevated | Elevated | Na | O-GlcNAcylation enhanced the malignancy of PC cells by inhibiting the formation of the E-cadherin/catenin/cytoskeleton complex | Gu et al. (2014) | ||
| PC patient samples | Na | Elevated | Na | Inhibition of OGT in PC cells resulted in slowing of the cell cycle and a reduction in DNA replication via a MYC-dependent pathway | Itkonen et al. (2013) | ||
| BC | Urine obtained from BC patients | Na | Elevated | Elevated | Analysis of urinary content of OGA and OGT mRNA was useful for bladder cancer diagnostics | Rozanski et al. (2012) | |
| BC patient samples and cell lines | Elevated | Elevated | Na | Hyper-O-GlcNAcylation enhanced oncogenic phenotypes and was involved in DNA damage response in BC | Wang et al. (2018) | ||
| BC patient samples and cell lines | Na | Elevated | Decreased | Knockdown of OGT inhibited cell proliferation, migration, invasion, and induce cell cycle arrest, while these effects were reversed when OGA is inhibited | Jin et al. (2020a) | ||
| RCC | RCC patient samples and cell lines | Elevated | Elevated | na | Hyper-O-GlcNAcylation was correlated with poor prognosis in RCC patients. OGT knockdown significantly suppressed RCC cell proliferation in vitro and in vivo | Poor survival | Wang et al. (2019) |
| Lung cancer | Lung cancer patient samples and cell lines | Elevated | Elevated | na | Hyper-O-GlcNAcylation increased the growth and invasion of lung cancer cells | Mi et al. (2011) | |
| LUAD patient samples | Elevated | Elevated | Elevated | High expression of OGT could independently predict poor survival outcomes in patients with stage I LUAD | Poor survival | Lin et al. (2018) | |
| Lung cancer patient samples and LUAD cell lines | Elevated | Elevated | na | O-GlcNAcylation promoted migration and invasion by activating IL-6/STAT3 signaling in lung cancer | Ge et al. (2021a) | ||
| SCLC | SCLC patient samples | Elevated | Elevated | Elevated | High OGT and OGA levels were associated with poor prognosis and could be considered new biomarkers of the invasive phenotype of tumor cells | Poor survival | Starska et al. (2015) |
| CLL | Blood from CLL patients, CLL cells | Elevated | Elevated | Na | Indolent and aggressive clinical behavior of CLL cells were correlated with higher and lower O-GlcNAcylation levels, respectively | Shi et al. (2010) | |
| AML | AML patient samples and cell lines | Na | Elevated | Na | Elevated OGT expression was significantly associated with poor OS in patients with AML. Inhibition of OGT inhibited AML cell proliferation and promoted AML cell apoptosis | Poor survival | He et al. (2021) |
| AML patient samples and cell lines | Elevated | Elevated | Na | Inhibition of HBP or OGT led to AML cell differentiation and apoptosis | Asthana et al. (2018) | ||
| ALL | Pre-B ALL patient samples and cell lines | Elevated | Elevated | Decreased | O-GlcNAcylation aggravated pre-B-ALL through regulation of glycolysis via the PI3K/Akt/c-Myc pathway | Zhang et al. (2017a) | |
| DLBC | DLBC patient samples and cell lines | Elevated | Elevated | Na | Elevated OGT levels were associated with poor survival of patients with DLBC. Targeting OGT in DLBC cells inhibited activation of O-GlcNAcylation and NF-κB | Poor survival | Pham et al. (2016) |
| TC | TC patient samples | Decreased | na | Elevated | OGA activity increased in TC in comparison to non-neoplastic lesions and adenomas | Krzeslak et al. (2010) | |
| Papillary thyroid cancer patient samples and cell lines | Elevated | Elevated | Na | O-GlcNAcylation of YAP at Ser109 dramatically inhibited its Ser127 phosphorylation, subsequently promoting tumor aggressiveness | Poor survival | Li et al. (2021a) | |
| GBM | GBM patient samples | Elevated | Elevated | Na | OGT regulates acetate-dependent acetyl-CoA and lipid production in GBM cells by regulating phosphorylation of ACSS2 by CDK5 | Ciraku et al. (2022) |
Cancers in the reproductive system
Cervical cancer
Determining the relationship between O-GlcNAc and cervical cancer, especially in human papilloma virus (HPV)-related cervical tumors, has attracted increased attention from researchers. Zeng et al. (Zeng et al. 2016) reported a significant upregulation of O-GlcNAcylation with increased OGT levels in HPV-induced cervical tumors, while OGA levels were not altered. Subsequently, some studies found that OGT could mediate O-GlcNAc modification of host cell factor C1 (HCF-1) to activate HPV E6/E7 transcription, thereby leading to immortalization and transformation of cells (Kim et al. 2016; Scheffner et al. 1990; Roman and Munger 2013). Furthermore, in addition to the observation that O-GlcNAcylation promotes tumorigenesis and metastasis by enhancing the oncogenic activity of HPV in vitro and in mouse models, O-GlcNAcylation was found to promote lung metastasis of cervical cancer by modifying nuclear factor kappa B (NF-κB) and thus activating C-X-C chemokine receptor 4 (CXCR4) (Ali et al. 2017).
Breast cancer
Many studies have focused on relationship between O-GlcNAcylation and the development and progression of breast cancer. Gu et al. (Gu et al. 2010) showed that O-GlcNAcylation in breast tumors was significantly increased compared with that in adjacent tissue. In addition, O-GlcNAcylation was more abundant in metastatic tissues compared with that in original tumor tissues. The authors also revealed that elevated O-GlcNAcylation could enhance the migration and invasion of breast cancer cells in vitro and lung metastasis in vivo, possibly by modifying P120 and β-catenin, thus inhibiting the binding of E-cadherin to P120 on the cell surface, thereby reducing intercellular adhesion. Caldwell et al. (Caldwell et al. 2010) showed that elevated O-GlcNAcylation and OGT promoted breast cancer cell growth and invasion, in part by regulating the oncogenic transcription factor forkhead box M1 (FOXM1) and multiple FOXM1-specific targets, such as s-phase kinase-associated protein 2 (SKP2) and cyclin-dependent protein kinase inhibitor (P27Kip1). Ferrer et al. (Ferrer et al. 2017) revealed that elevated O-GlcNAcylation promoted the invasion and metastasis of breast cancer by regulating the sirtuin 1 (SIRT1)/extracellular regulated kinase (ERK)/FOXM1 axis. Notably, Akella et al. (Akella et al. 2020) found that OGT/O-GlcNAc is essential and sufficient for maintaining breast cancer stem-like cells phenotype in breast cancer cells in vitro and plays a critical role in tumor-initiating potential in vivo. In addition, Kuo et al. (2021) showed that hyper-O-GlcNAcylation is associated with poor 10-year disease-free survival (DFS) in patients with breast cancer. These studies suggest that O-GlcNAcylation acts as a stimulating factor in the occurrence, invasion, metastasis, recurrence, and prognosis of breast cancer.
Endometrial cancer
Previous studies have determined the relationship between OGT and OGA mRNA expression levels and the clinical and pathological features of endometrial cancer (Krzeslak et al. 2012). The results showed that higher OGT and OGA levels were associated with higher histological grades and depth of tumor invasion into the myometrium. Research from Jaskiewicz et al. (Jaskiewicz and Townson 2019) showed that hyper-O-GlcNAcylation could promote cell proliferation, induce cytoskeletal reorganization, and induce epithelial-mesenchymal transition (EMT), thereby enhancing cell invasion. Interestingly, hypo-O-GlcNAcylation could also enhance the expression of the EMT-related gene WNT5B (encoding Wnt family member 5B), but decreased the overall proliferation and migration ability of the cells. These results suggested that breaking the O-GlcNAc cycle in endometrial cancer cells promotes EMT at the molecular and cellular levels, but only high O-GlcNAcylation causes proliferation, migration, and cytoskeletal reorganization of cells.
Ovarian cancer
Jin et al. (Jin et al. 2013) reported higher levels of O-GlcNAcylation and OGT mRNA in highly metastatic ovarian cancer HO-8910PM cells compared with low metastatic OVCAR3 cells and revealed the underlying molecular mechanism: hyper-O-GlcNAcylation decreased E-cadherin expression, thereby inhibiting E-cadherin/catenin complex formation, which reduced cell–cell adhesion, leading to cancer cell metastasis. Niu et al. (Niu et al. 2017) reported increased levels of O-GlcNAc and OGT in ovarian carcinoma cell lines and showed that O-GlcNAcylation of GTP-bound RhoA and modulator of VRAC current 1 (MLC1) phosphorylation might activate the RhoA/Rho associated coiled-coil containing protein kinase (ROCK)/MLC1 pathway to enhance ovarian cancer cell mobility, thus contributing to ovarian cancer cell migration and invasion.
Cancers in the digestive system
Liver cancer
Numerous studies have noted that O-GlcNAcylation is elevated in liver cancer, especially in hepatocellular carcinoma (HCC) (Qiao et al. 2016a; Zhu et al. 2016, 2012). Diabetes mellitus is an important risk factor for the development of liver cancer (Mukherjee et al. 2015); therefore, increasing numbers of studies have focused on the link between O-GlcNAcylation and high glucose stimulated liver cancer. Zhang et al. (Zhang et al. 2017b) reported that under high glucose condition, Yes-associated protein (YAP) can be O-GlcNAcylated at Thr241, thereby enhancing its expression, stability, and function, leading to the transformed phenotype of HCC cells. Importantly, they demonstrated that O-GlcNAcylation of YAP is essential during liver tumorigenesis induced by high glucose. Qiao et al. (Qiao et al. 2016a) found that the advanced glycosylation end product-specific receptor (AEGR) can increase its activity and stability through O-GlcNAcylation at Ser73, thus promoting the development of liver tumors under high glucose conditions.
O-GlcNAc can also modify many oncoproteins, thereby promoting liver tumorigenesis. Yao et al. (Yao et al. 2016) revealed that O-GlcNAcylation of tribbles pseudokinase 2 (TRIB2) enhances protein stability, which in turn promotes HBP and O-GlcNAcylation, thus maintaining the transformed phenotype of hepatoma cells. Another study showed that histone deacetylase-1 (HDAC1) is over O-GlcNAcylated in HCC and the progression of HCC can be reduced by inhibiting HDAC1 O-GlcNAcylation (Zhu et al. 2016). In non-alcoholic fatty liver disease-related hepatocellular carcinoma (NAFLD-HCC), researchers observed elevated OGT levels in patients with NAFLD-HCC and NAFLD-HCC cell lines and revealed that OGT could regulate lipid metabolism, thereby activating endoplasmic reticulum (ER) stress, and the JUN N-terminal kinase (JNK)/Jun proto-oncogene, AP-1 transcription factor subunit (c-Jun)/activator protein 1 (AP-1), and NF-κB cascades during the development of NAFLD-HCC (Xu et al. 2017); indeed, the latter has been shown to be a cancer-promoting factor in HCC (Han and Roman 2006). In addition, orthotopic HCC xenograft models indicated that OGT significantly promotes HCC lung metastasis by inhibiting E-cadherin and enhancing vimentin expression.
Liver transplantation (LT) is currently an effective way to treat patients with early HCC and cirrhosis. However, the frequent recurrence of tumors after LT still represents a great obstacle to the long-term survival of patients (Zheng et al. 2008). Zhu et al. (Zhu et al. 2012) observed significantly higher O-GlcNAcylation and lower OGA levels in tumor tissues from patients that suffered from HCC recurrence after LT compared with those who did not. Moreover, they revealed that elevated O-GlcNAcylation resulted in decreased E-cadherin and increased matrix metalloproteinase (MMP)1, MMP2, and MMP3 expression, thereby promoting migration and invasion. The study also identified lower OGA expression as an independent predictor for HCC tumor recurrence after LT, especially in patients with low alpha fetoprotein (AFP) expression. O-GlcNAcylation affects chemoresistance in HCC. Duan et al. (Duan et al. 2018) confirmed that O-GlcNAcylation of ribosome-activated C kinase 1 (RACK1) at Ser122 in HCC cells would lead to acquired chemoresistance and recurrence in patients. Taken together, these studies indicate that O-GlcNAcylation affects the occurrence, progression, and recurrence of HCC in a complex manner.
Colorectal cancer
Hyper-O-GlcNAcylation and elevated OGT levels were observed in colorectal cancer (CRC) tissues compared with those in adjacent normal tissues, while OGA expression showed no significant difference between tumor and normal tissues. (Mi et al. 2011; Phueaouan et al. 2013; Steenackers et al. 2016) Hyper-O-GlcNAcylation participates in the progression of CRC through multiple pathways. Recently, Zhu et al. (Zhu et al. 2019) showed that O-GlcNAcylation of transcription factor YIN-YANG-1 (YY1) at Thr236 could enhance the expression of solute carrier family 22 member 15 (SLC22A15) and aralkylamine N-acetyltransferase (AANAT) and increased its own protein stability, thereby exerting an oncogenic effect in CRC cells. Wu et al. (2019) found that RNA helicase p68/DEAD-box helicase 5 (DDX5) could be O-GlcNAcylated to enhance its stability, thus increasing the activation of the protein kinase B (AKT)/mechanistic target of rapamycin kinase (mTOR) signaling pathway, leading to the promotion of the malignant development of CRC. Lefebvre et al. (Olivier-Van et al. 2014, 2012) revealed that O-GlcNAcylation of β-catenin is the molecular event that links the glucose metabolism deregulation observed in metabolic disorders and the development of CRC. They observed that human colon tumors and colons from mice fed high-carbohydrate diets exhibited higher amounts of β-catenin and O-GlcNAc relative to healthy tissues and mice fed a standard diet, respectively. Subsequently, through analysis of β-catenin O-GlcNAcylation mutants, they identified Thr41 as the most crucial residue that controls the β-catenin degradation rate and found that β-catenin was O-GlcNAcylated at Thr41 thereby reducing its degradation and thus affecting the development of CRC. Harosh-Davidovich et al. (Harosh-Davidovich and Khalaila 2018) found that hyper-O-GlcNAcylation could not only enhance the expression of β-catenin and E-cadherin, but also increased the rate of β-catenin entry into the nucleus and enhanced its transcriptional activity, thereby promoting cell motility and tumorigenicity in CRC. Yu et al. (Yu et al. 2019) reported that O-GlcNAcylation of integrin α5 (ITGA5) would enhance its stability, thus promoting CRC cell proliferation and tumorigenesis, and reducing apoptosis. O-GlcNAcylation can also affect colon cancer stem cells (CCSCs). Guo et al. (Guo et al. 2017) demonstrated that O-GlcNAcylation in CRC could affect the epigenetic inheritance of CCSCs by regulating the transcription factor MYB proto-oncogene like 1 (MYBL1), thus regulating colon carcinogenesis.
Pancreatic cancer
In normal cells, OGT and OGA can regulate each other's activity and expression at the transcriptional and post-translational levels to maintain cellular O-GlcNAc levels in the "optimal region" (Yang and Qian 2017). However, studies have observed an increase in O-GlcNAcylation and OGT, and a decrease in OGA, in human pancreatic ductal adenocarcinoma (PDAC) (Ma et al. 2013). Qian et al. (Qian et al. 2018) revealed that this dysregulation was caused by the disruption of transcriptional regulation homeostasis in pancreatic cancer and proved that it was the abnormal activation of ERK signal transduction in cells that affected OGA-mediated OGT transcription, ultimately resulting in increased OGT expression (Wang et al. 1999; Liptay et al. 2003). O-GlcNAcylation was associated with immune evasion in pancreatic cancer. Shang et al. (Shang et al. 2021) observed an enhanced folate cycle, higher concentration of UDP-GlcNAc, and higher cMYC O-GlcNAcylation in patients with pancreatic cancer, which was proven to increase PDL1 (encoding programmed cell death 1 ligand 1) transcription, leading to immune escape. OGT can be used as a potential treatment target for pancreatic cancer. Triptolide, a diterpene epoxide from the Chinese plant Tripterygium wilfordii, targets OGT, resulting in downregulation of heat shock factor 1 (HSF1) and other heat shock proteins (HSPs), ultimately leading to tumor cell death (Banerjee et al. 2013). These studies suggested that dysregulation of O-GlcNAcylation is strongly associated with pancreatic cancer and could serve as a potential therapeutic target.
Gastric cancer
Previous studies have observed progressively higher OGT and O-GlcNAcylation levels as gastric cancer (GC) progresses: O-GlcNAcylation and OGT levels were higher in GC with the intestinal type, higher pathological and clinical stage, and more lymph node metastasis (Jang and Kim 2016). O-GlcNAcylation promotes GC progression through multiple pathways. Zhang et al. (Zhang and Chen 2016) revealed that O-GlcNAcylation enhanced GC cell invasion through the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/AKT pathway. Cheng et al. (Cheng et al. 2016a) reported that O-GlcNAcylation of intercellular scaffold protein guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 (GNB2L1) could promote its degradation, thereby blocking the inhibitory effect of GNB2L1 on gastric cancer cell migration, and eventually leading to metastases. Jiang et al. (Jiang et al. 2016) found that O-GlcNAcylation can regulate the cell cycle and ERK 1/2 pathway, thereby promoting the proliferation of GC cells. These studies further confirmed the adverse effect of hyper-O-GlcNAcylation in GC.
Esophageal cancer and cholangiocarcinoma
Qiao et al. (Qiao et al. 2012) reported higher O-GlcNAc and OGT levels in patients with esophageal squamous cell carcinoma (ESCC) compared with that in normal samples, they also revealed (Qiao et al. 2016b) that hyper-O-GlcNAcylation could promote tumorigenesis and metastasis of ESCC by raising the stability and expression of MMP9, and changing cellular signal transduction. Similar to pancreatic cancer, Yuan et al. (Yuan et al. 2021) reported that hyper-O-GlcNAcylation could improve the self-renewal capacity of esophageal cancer stem cells (ECSCs) and promote the high expression of programmed cell death 1 (PD1) in CD8 + T cells, resulting in immune escape. Higher O-GlcNAcylation and OGT levels, and lower OGA levels, were observed in cholangiocarcinoma compared with those in normal bile ducts, and this change led to poor prognosis of patients with cholangiocarcinoma (Phoomak et al. 2012).
Cancers in the urinary system
Prostate cancer
Lynch et al. (Lynch et al. 2012) first reported higher O-GlcNAcylation and OGT levels in prostate cancer (PC) tissues and cell lines compared with those in normal samples; they proved that this is related to the invasive phenotype and ability of PC cells and worse clinical outcomes. Furthermore, they reduced vascular endothelial growth factor (VEGF)-mediated angiogenesis of PC cells by targeting OGT. Gu et al. (Gu et al. 2014) revealed that hyper-O-GlcNAcylation promotes PC invasion by inhibiting the formation of the E-cadherin/catenin/cytoskeleton complex, rather than affecting CDH1 (encoding E-cadherin) mRNA levels.
Another study (Li et al. 2017) revealed that OGT could bind to the BMI1 proto-oncogene, polycomb ring finger (BMI-1)/protein regulator of cytokinesis 1 (PRC1) complex and modify BMI-1 at Ser255, thereby promoting its stability and oncogenic activity, resulting in inhibition of the tumor protein 53 (TP53), phosphatase and tensin homolog (PTEN), and cyclin dependent kinase Inhibitor (CDKN)1A/CDKN2A pathways. The importance of c-MYC for PC has been demonstrated (Hawksworth et al. 2010; Gurel et al. 2008). Slawson et al. (Slawson and Hart 2011) showed that O-GlcNAcylation of c-MYC at Thr58 could inhibit phosphorylation at nearby Ser62, thereby stimulating the growth of tumor cells. Itkonen et al. (Itkonen et al. 2013) found that inhibition of OGT in PC cells resulted in slowing of the cell cycle and a reduction in DNA replication via a MYC-dependent pathway, thereby reducing tumor growth. These studies all illustrated the important role played by O-GlcNAcylation in PC and the potential of OGT as a target to treat PC.
Bladder cancer and renal cell carcinoma
A previous study reported that the analysis of OGT and OGA content in urine might contribute to the diagnosis and grading of bladder cancer (BC) (Rozanski et al. 2012). OGT was found in most urine samples with BC, while OGT was not detected in the urine of healthy individuals. In addition, higher OGT and lower OGA levels were observed in the urine from patients with a higher tumor grade, although there were no differences in OGA levels in urine between healthy individuals and patients. Wang et al. (Wang et al. 2018) observed hyper-O-GlcNAcylation and increased OGT levels in BC tissues and cell lines compared with those in normal samples. In addition, they revealed that inhibiting OGT reduced BC cell proliferation and growth, triggered apoptosis, and led to cell cycle arrest, probably via increased autophagy. This was confirmed by a subsequent study (Jin et al. 2020b): Blockade of O-GlcNAcylation induced autophagy in BC cells through an mTOR-independent pathway. Jin et al. (Jin et al. 2020a) revealed that hyper-O-GlcNAcylation can promote the malignant phenotype of BC cells, while knockdown of OGT could reverse these effects. Recently, Chen et al. (Chen et al. 2021) reported similar results: Inhibiting OGT led to downregulation of the cell cycle-related protein nucleolar and spindle associated protein 1 (NUSAP1), thereby inhibiting the malignant progression of BC. Melatonin has previously been shown to inhibit the growth of BC (Reiter et al. 2017). Wu et al. (2021) further revealed the anti-prostate cancer mechanism of melatonin: Melatonin inhibits O-GlcNAcylation of cyclin-dependent kinase 5 (CDK5) at Thr246, thus promoting its degradation, and inhibiting the tumor-promoting effect of CDK5 on BC cells.
Wang et al. (Wang et al. 2019) first observed significantly higher OGT and O-GlcNAcylation levels in renal cell carcinoma (RCC) cell lines and tissues compared with those in normal samples, and found that hyper-O-GlcNAcylation was associated with higher grade and poor prognosis in patients. In addition, they revealed that knockdown of OGT in RCC cells could downregulate the epidermal growth factor receptor (EGFR) and PI3K/AKT pathways, thereby inhibiting the migration, invasion, and vascularization of RCC cells. Together, these studies demonstrated the potential of O-GlcNAcylation as a predictor and treatment target for BC and RCC.
Cancers in the respiratory system
Lung cancer and laryngeal cancer
Many studies have reported the role of O-GlcNAcylation in malignant tumors of the respiratory system. Mi et al. (Mi et al. 2011) not only observed elevated OGT and O-GlcNAcylation in lung cancer, but also indicated that hyper-O-GlcNAcylation would increase the growth and invasion of lung cancer cells. Lin et al. (Lin et al. 2018) showed a clinical relationship between OGT and lung cancer: Hyper-O-GlcNAcylation independently predicted a worse prognosis in patients with stage I lung adenocarcinoma. An inflammatory microenvironment is highly correlated with tumor initiation and malignant progression (Hanahan and Weinberg 2011; Pietila et al. 2016). Ge et al. (Ge et al. 2021a) confirmed that the inflammatory factor interleukin 6(IL-6), released by inflammatory cells or tumor cells, increased OGT expression in lung cancer cells though the NF-κB p65 signaling pathway, leading to migration and invasion.
Squamous cell laryngeal cancer (SCLC) is another respiratory malignancy. Researchers have observed hyper-O-GlcNAcylation and elevated OGT and OGA in SCLC samples compared with those in the normal laryngeal mucosa. In addition, this change is closely related to worse clinical outcomes of patients, including larger tumor size, higher pathological grade, more lymph node metastasis, and a higher recurrence rate.
Cancers in the hematopoietic system
Chronic lymphocytic leukemia
In addition to solid tumors, there are many reports about the role of O-GlcNAcylation in various blood cancers (Spaner 2021). Shi et al. (Shi et al. 2010) reported increased O-GlcNAcylation and OGT in chronic lymphocytic leukemia (CLL) cells compared with those in normal lymphocytes, and also revealed that this is caused by higher levels of the sugar donor UDP-GlcNAc. Notably, they found that hyper-O-GlcNAcylation appeared to be closely related to an indolent clinical course: Elevated O-GlcNAcylation can inhibit JNK signaling, weaken the response of CLL cells to proliferation signals, and hinder the division of CLL cells. Interestingly, the results of their study are somewhat contradictory to those in other cancers, because hyper-O-GlcNAcylation usually means more aggressive clinical behavior in solid tumors. However, similar to CLL, hyper-O-GlcNAcylation was also observed in chemosensitive ovarian cancer tissues compared with that in chemoresistant ovarian cancer tissues (Zhou et al. 2018). Another study explained the link between O-GlcNAcylation and the indolent clinical behavior of CLL (Lode et al. 2016; Yang et al. 2006): O-GlcNAcylation of p53 at Ser149 can inhibit cancer progression by stabilizing wild-type p53 and activating the intact p53 pathway. From a therapeutic aspect, Tomic et al. (Tomic et al. 2013) found that the anti-CLL activity of resveratrol (RSV) was associated with the reduction of O-GlcNAcylated proteins: RSV rapidly downregulated O-GlcNAcylation levels in CLL cells; the authors speculated that this might be a result of proteasomal activation.
Other hematological malignancies
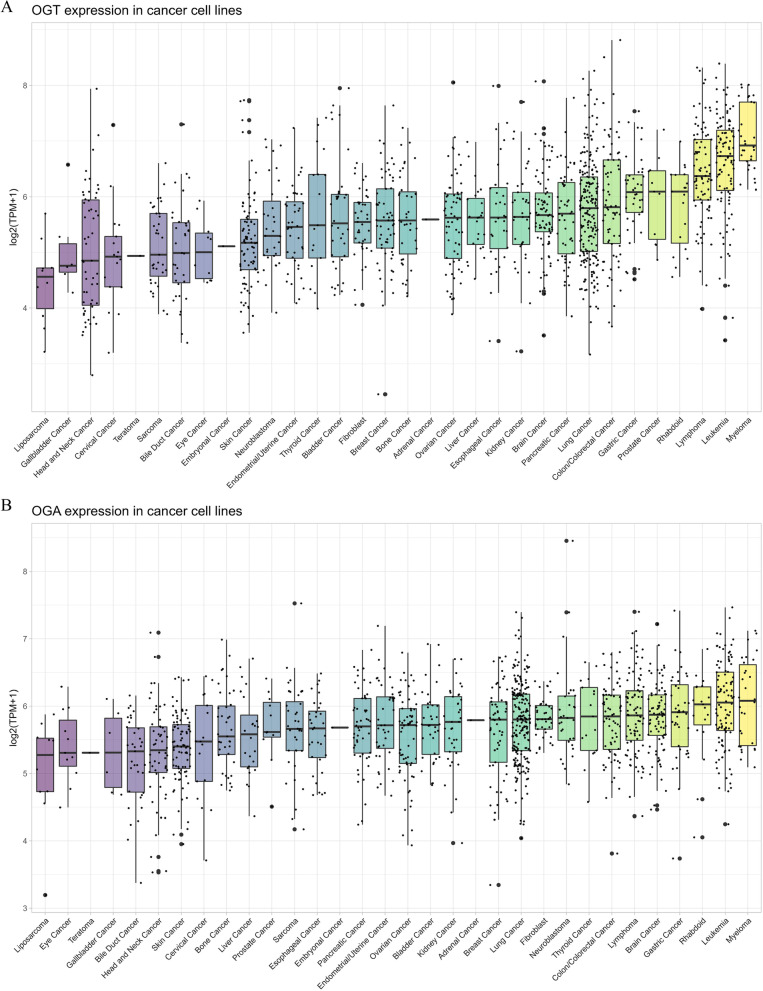
Besides CLL, abnormal O-GlcNAcylation has also been reported in myelodysplastic syndrome (MDS) (Li et al. 2021b), acute myeloid leukemia (AML) (He et al. 2021), acute lymphoblastic leukemia (ALL) (Zhang et al. 2017a), and diffuse large B-cell lymphoma (DLBC) (Pham et al. 2016). Interestingly, higher OGT and OGA expression levels were observed in tumor samples and cell lines from hematological malignancies compared with those in other solid tumors (Itkonen et al. 2021). In various cancer cell lines, the expression levels of OGT and OGA rank first and second in myeloma and leukemia, respectively (Fig. 6); in tumor samples from the TCGA database, the expression of OGT and OGA in LAML was much higher than that in other cancers (Fig. 3). Some studies have revealed the role of O-GlcNAcylation in blood cancers. Asthana et al. (Asthana et al. 2018) observed higher OGT and GFAT levels in primary AML cells and revealed that inhibiting the activity of HBP using the GFAT inhibitor 6-diazo-5-oxo-l-norleucine (DON), or using the OGT inhibitors OSMI-1 (OGT with a small molecule inhibitor) and BADGP (benzyl-2-acetamido-2-deoxy-α-d-galactopyranoside), can induce AML cell differentiation and apoptosis, but do not affect normal cells. Chemoresistance in AML is also associated with elevated O-GlcNAcylation. Liu et al. (Liu et al. 2018) reported that hyper-O-GlcNAcylation induced by chemotherapy drugs, such as doxorubicin (DOX) and camptothecin (CPT), can cause chemoresistance, thereby weakening the therapeutic effect, while targeting OGT enhanced the effect of chemotherapy. Zhang et al. (Zhang et al. 2017a) observed higher OGT and O-GlcNAcylation levels, and lower OGA levels, in pre-B acute lymphoblastic leukemia (pre-B-ALL) samples compared with those in healthy donors. Moreover, they demonstrated that hyper-O-GlcNAcylation aggravated pre-B-ALL by modulating glycolysis via the PI3K/AKT/c-MYC pathway. Interestingly, many previous studies have shown that elevated OGT is associated with tumor progression. However, Inoue et al. (Inoue et al. 2018) revealed the predictive and inhibitory effect of OGT in hematopoietic malignancies. They showed that additional sex combs like transcriptional regulator 1 (ASXL1) could be stabilized by O-GlcNAcylation at Ser199, allowing it to act as a hematopoietic malignant tumor suppressor. Furthermore, they showed that OGT has tumor suppressive activity in myeloid malignancies, especially in the presence of ASXL1 mutations (Abdel-Wahab et al. 2011; Bejar et al. 2011; Gelsi-Boyer et al. 2009; Yoshida et al. 2011). These results suggested that O-GlcNAcylation regulates hematopoietic malignancies in a complex way, and more studies are needed to clarify its role.
Fig. 6.
O-GlcNAc modifying enzymes expression in different cancer cell lines. Compared with other solid tumors, cancer cell lines of hematological malignancies showed higher expression levels of OGT and OGA. (A) OGT expression in different cancer cell lines. B OGA expression in different cancer cell lines. Data were downloaded from the DepMap-portal (https://depmap.org/portal). Each dot represents a cell line and the black bar graphs are median of expression in that tissue type
Cancers in the endocrine system
Thyroid cancers
Some studies have reported the role of O-GlcNAcylation in thyroid cancer (TC). Krześlak et al. (Krzeslak et al. 2010) observed the elevated OGA and decreased O-GlcNAcylation levels in TC samples compared with those in non-neoplastic lesions and adenocarcinoma. Yu et al. (Cheng et al. 2016b) revealed that hyper-O-GlcNAcylation improves TC cell viability and enhances its growth, migration, and invasion, thereby enhancing the malignant phenotype of TC. Krześlak et al. (Krzeslak et al. 2011) showed that elevated O-GlcNAcylation in thyroid anaplastic cancer cells could promote their proliferation through the insulin-like growth factor (IGF-1)-AKT-GSK3β-cyclin D1 pathway. Zhang et al. (Zhang et al. 2015) found that elevated O-GlcNAcylation could enhance the invasion of TC cells, in part through PI3K/AKT signaling. Recently, Li et al. (Li et al. 2021a) revealed that YAP, a core component of the Hippo pathway, was O-GlcNAcylated at Ser109, thereby inhibiting phosphorylation at Ser127. This allowed YAP to act as an oncogenic transcriptional coactivator to promote TC growth and metastasis, thereby influencing the recurrence-free survival and clinicopathological characteristics of patients with TC (Liu et al. 2017, 2019b). These studies suggested that high levels of O-GlcNAcylation are also characteristic of malignant TC, and targeting O-GlcNAcylation might be a potential strategy to treat TC.
Cancers in the nervous system
Brain cancer
Recently, the role of O-GlcNAcylation in brain tumors has attracted the attention of researchers. Lorela Ciraku et al. (Ciraku et al. 2022) reported that OGT and O-GlcNAcylation levels are elevated in glioblastoma (GBM) tissues and cells. In addition, they revealed that elevated O-GlcNAcylation in GBM cells regulates acetate-dependent acetyl-CoA and lipid production through the OGT/CDK5/acetyl-CoA synthetase 2 (ACSS2) pathway to adapt to lack of lipid availability in the brain environment. Specifically, elevated OGT in GBM cells increases the phosphorylation of ACSS2 at Ser267 in a CDK5-dependent manner, thereby reducing polyubiquitination and degradation to increase its stability. Chen et al. (Chen et al. 2022) observed the elevated O-GlcNAc levels in human samples of Sonic hedgehog (Shh)-subtype medulloblastoma and revealed that OGT in granule neuron precursors (GNPs) may contribute to medulloblastoma oncogenesis by activating the Shh signaling pathway via O-GlcNAcylation at S355 of GLI family zinc finger 2 (Gli2). These studies reveal the important role of O-GlcNAc signaling in brain cancer and deserve further exploration.
O-GlcNAc and cancer therapy
O-GlcNAcylation, as an emerging but ubiquitous protein PTM, has attracted great interest from researchers since it was reported in the early 1980s. To date, numerous publications have addressed its important functions, regulatory mechanisms, and links with human diseases. Although we lack detailed knowledge of the mechanisms, hyper-O-GlcNAcylation is a common feature in various cancers, and thus represents a promising potential target for cancer diagnosis and treatment.
O-GlcNAc as a biomarker for cancer diagnosis and prognosis
Many malignant tumors are characterized by difficult early diagnosis, poor prognosis, and easy recurrence, which poses a great challenge for their clinical treatment (Hanahan and Weinberg 2011). Therefore, identifying a reliable biomarker that can be used for early diagnosis, prognosis, and confirmation of recurrence has become an important research direction in cancer treatment. OGT and OGA expression levels have been assessed in patients and normal samples of major human cancers (Cancer Cell Line Encyclopedia Consortium 2015; UniProt Consortium 2019) (Fig. 3). The results showed that OGT and OGA expression levels changed in almost all cancers to varying degrees, suggesting that O-GlcNAcylation level and the levels of O-GlcNAc circulating enzymes might have significance for early cancer screening and prognosis. In fact, many studies have reported that hyper-O-GlcNAcylation often indicates poor prognosis of patients, shorter disease-free survival, and tumor recurrence: Kuo et al. (2021) found that high levels of O-GlcNAcylation was an important independent predictor of poor 10-year DFS for HR lysine demethylase and nuclear receptor corepressor (HR)+/human epidermal growth factor receptor 2 (HER2)-luminal breast cancer, and the predictive effect and potential can be greatly enhanced when combined with pyruvate kinase isoenzyme M2 (PKM2). Rozanski et al. (Rozanski et al. 2012) indicated that detecting OGA and OGT mRNA levels in urine might be helpful in the diagnosis of BC. Kamigaito et al. (Kamigaito et al. 2014) showed that OGT overexpression occurred in 39% of prostate cancer specimens from 56 patients who did not receive hormone therapy, and this OGT overexpression correlated significantly with reduced OS. Furthermore, Zhu et al. (Zhu et al. 2012) revealed that elevated O-GlcNAcylation and decreased OGA expression might indicate the recurrence of HCC after LT, and this predictive effect is more pronounced in patients with low AFP levels. Other studies indicated that the level of O-GlcNAcylation or OGT can be used as a biomarker to predict poor prognosis of LUAD, laryngeal carcinoma, CHOL, ESCC, GC, CRC, RCC, AML, DLBC, and TC (Table 2). These studies fully illustrate the great potential of O-GlcNAc as biomarkers for early diagnosis, prognosis, and confirmation of recurrence of cancer.
O-GlcNAc as target for cancer therapeutics
Given the adverse roles of dysregulated O-GlcNAcylation, especially hyper-O-GlcNAc, in cancers, such as promoting the growth, proliferation, invasion, and metastasis of cancer cells, inducing cancer angiogenesis, promoting the malignant phenotype of cancer, and promoting chemoresistance (Ma and Vosseller 2014), targeting O-GlcNAcylation is an attractive approach for clinical anticancer therapies.
Currently developed chemical drugs, therapeutic antibodies, or aptamers mainly regulate the O-GlcNAc pathway through two strategies to achieve therapeutic effects. The first strategy is to target the enzymes OGT and OGA directly for cancer therapy. The change in intracellular O-GlcNAcylation levels depends largely on change of these two enzymes, which leads to dysregulation of O-GlcNAcylation and the occurrence and progression of diseases. Thus, the adverse effects caused by dysregulated O-GlcNAcylation can be largely repaired or reversed by regulating the levels of OGT or OGA. Over the past few decades, investigators have developed various inhibitors against OGT, which belong to three categories: Substrate and product analogs (Dorfmueller et al. 2011), high-throughput screening (HTS)-derived inhibitors (Gross et al. 2005), and bisubstrate inhibitors (Borodkin et al. 2014) (Table 3). Alloxan was the first reported inhibitor of human OGT (Konrad et al. 2002). It is a uracil analog that can inhibit OGT activity, thus blocking hyper-O-GlcNAcylation in cells in a dose-dependent manner, presumably by binding to the uracil binding pocket or by covalent modification of cysteine residues (Konrad et al. 2002). However, reactive oxygen species (ROS) and free radicals produced by Alloxan during its involvement in extracellular redox processes can cause a large number of off-target effects and damage cellular structures, such as mitochondria and lysosomes (Zhang et al. 1992). Researchers have reported that UDP-S-GlcNAc, UDP-C-GlcNAc, and C-UDP act as OGT inhibitors (Dorfmueller et al. 2011). Notably, they are sub-millimolar inhibitors but are not cell-permeable. Gloster et al. (Gloster et al. 2011) reported that 5SGlcNAc and its analog Ac-5SGlcNAc could be recovered by cells and processed by the HBP to generate an efficient OGT inhibitor, UDP-5SGlcNAc. Importantly, some experiments have confirmed its anti-cancer roles in breast and prostate cancer, including inhibiting the proliferation, invasion, and angiogenesis of cancer cells. In addition, it can also promote metabolic stress and apoptosis by regulating the oncogenic proteins FOXM1, c-MYC, and hypoxia inducible factor 1 subunit alpha (HIF-1α) (Caldwell et al. 2010; Lynch et al. 2012; Itkonen et al. 2013; Ferrer et al. 2014; Sodi et al. 2015). However, Ac-5SGlcNAc affects the intracellular UDP-GlcNAc repertoire by hijacking the HBP pathway, thereby affecting N-glycosylation in cells and glycan synthesis outside the cell (Ortiz-Meoz et al. 2015).
Table 3.
The available inhibitors targeting OGT
| Categories | Compound | IC50 (μM) | Advantages | Disadvantages | References |
|---|---|---|---|---|---|
| Substrate and product analogs | Alloxan | 18 ± 1 | Cell-permeable | Potential off-target effects and general cellular toxicity | Konrad et al. (2002) |
| UDP- S -GlcNAc | 93 ± 15 | Sub-millimolar inhibitors | Lack of cell permeability | UniProt: a worldwide hub of protein knowledge (2019) | |
| UDP- C-GlcNAc | 41 ± 7 | Sub-millimolar inhibitors | Lack of cell permeability, a weak hOGT inhibitor | Dorfmueller et al. (2011) | |
| C-UDP | 9.0 ± 1.0 | Sub-millimolar inhibitors | Lack of cell permeability | Dorfmueller et al. (2011) | |
| UDP-5SGlcNAc | 5 | Cell-permeable | Affect N-glycosylation in cells and glycan synthesis outside the cells | Gloster et al. (2011) | |
| HTS-derived inhibitors | ST045849 | 53 ± 7 | Highly selective and cell-permeable | Potential off-target effects and cellular toxicity | Kamigaito et al. (2014) |
| OSMI-1 | 2.7 | Cell-permeable, not alter cell surface N- or O-linked glycans, on-target activity | Ortiz-Meoz et al. (2015) | ||
| Bisubstrate inhibitor | goblin1 | 18 | Can synergize with goblin2 to enhance inhibition | Lack of cell permeability | Borodkin et al. (2014) |
| goblin2 | 40 | Can synergize with goblin1 to enhance inhibition | Lack of cell permeability | Borodkin et al. (2014) |
The generation of another class of OGT inhibitors depends on high-throughput screening of large drug compound libraries. Using this approach, many OGT inhibitors have been developed (Gross et al. 2005), including the commercially available small molecule inhibitor ST045849. Itkonen et al. (Itkonen et al. 2016) confirmed the anti-cancer roles of ST045849: They found that inhibition of intracellular OGT activity in prostate cancer cells by ST045849 resulted in complete depletion of intracellular alanine, thereby inhibiting cancer cell viability and growth rate, and continuously induced the death of prostate cancer cells. OSMI-1 is another HTS-derived cell-permeable OGT inhibitor that inhibits O-GlcNAcylation, but does not alter the glycan structure on the cell surface (Ortiz-Meoz et al. 2015). OSMI-1 plays an anti-cancer role in various cancers: Lee et al. (2020) revealed that combined treatment using OSMI-1 and tumor necrosis factor (TNF)-related apoptosis inducing ligand (TRAIL) could synergistically enhance TRAIL-induced apoptosis through caspase-8 activation. In addition, OSMI-1 could induce apoptosis by blocking NF-κB signaling and activating the ER stress response, which enhanced the sensitivity of human colon cancer cells to TRAIL-induced cell death. OSMI-1 can also enhance the therapeutic effect of chemotherapeutic drugs. Lee et al. (2020) indicated the combination therapy of OSMI-1 and DOX significantly enhanced apoptosis and DOX-induced cell death of HCC cells by synergistically activating TP53 and mitochondrial B-cell CLL/lymphoma 2 (BCL2) pathways. Makwana et al. (Makwana et al. 2020) and Liu et al. (Liu et al. 2018) showed similar results in prostate cancer and breast cancer. In fact, more HTS-derived OGT inhibitors exist. Martin et al. (Martin et al. 2018) described the structure-based evolution of small molecule inhibitors of OGT and reported three cell-permeable compounds, later termed OSMI-2, OSMI-3, and OSMI-4. Among them, OSMI-2 acts as a rapid-acting OGT inhibitor in combination with anti-androgens to target MYC-dependent prostate cancer cells (Itkonen et al. 2019). Bisubstrate inhibitors against OGT mainly refer to two novel compounds, goblin1 and goblin2, which achieve selective inhibition by replacing the GlcNAc moiety of UDP-GlcNAc with a receptor peptide (Borodkin et al. 2014). However, there are still many limitations to the clinical application of such compounds, mainly because of their lack of cell permeability. Therefore, there is an urgent need for an OGT inhibitor with better specificity, potency, and cell permeability, both for laboratory studies and clinical cancer therapy.
The second strategy of using O-GlcNAcylation as a cancer therapeutic target is more targeted: changing the O-GlcNAc moieties on specific target proteins to achieve the therapeutic effect. This is quite attractive, because it can avoid the "accidental injury" caused by the alteration of global O-GlcNAcylation levels by targeting O-GlcNAc cycle enzymes. O-GlcNAcylation, as a widespread intracellular PTM, is involved in many aspects of the regulation of cellular life activities, and changes in its circulating enzymes could affect O-GlcNAcylation on thousands of proteins, many of which are not disease-related. Arbitrarily changing the O-GlcNAcylation of these proteins might have serious consequences. So far, a number of new technologies have emerged that can support the implementation of this idea, such as gene editing techniques, which eliminate the corresponding O-GlcNAcylation sites by introducing point mutations in proteins of interest; many laboratories have taken this approach when studying O-GlcNAcylation (Duan et al. 2018). In addition, aptamers, a single-stranded DNA or RNA that can specifically bind to cognate molecular targets, can be delivered into cells to specifically blocking O-GlcNAcylation on the corresponding sites of the target proteins (Zhu and Chen 2018). Furthermore, nanobodies have also been considered by researchers. Ramirez et al. (Ramirez et al. 2021) reported a nanobody-OGT fusion protein that can selectively increase O-GlcNAc levels of target proteins in cells, but does not disrupt the crosstalk or protein structure of PTMs. They also developed a nanobody-fused split OGA for selective removal of O-GlcNAc moieties from target proteins in cells and confirmed the effectiveness of the system by testing the target proteins (Ge et al. 2021b). Although there is no clinical report of targeting O-GlcNAcylation as a cancer therapy using nanobodies, we have reason to believe that with further research, this technique will show increased potential in the near future.
Concluding remarks and perspective
In summary, we reviewed the latest literature and silico analyses to show the links between dysregulation of O-GlcNAcylation and cancers, and highlighted the great prospects of O-GlcNAcylation as a cancer biomarker and therapeutic target. In the past few decades, O-GlcNAcylation has gone through a long process from its initial discovery and recognition as an important PTM to being considered as widely involved in all aspects of cellular life activities, even as a central hub of certain metabolic and signal transduction pathways. Although researchers have now realized that protein O-GlcNAcylation plays an important role in various human diseases, its detailed mechanisms in these diseases remains to be explored. Especially in cancer, determining how O-GlcNAcylation is involved in the regulation of numerous hallmarks of cancer, such as metabolic reprogramming, genomic instability, induction of angiogenesis, changes in the tumor microenvironment, and immune evasion, requires more exhaustive studies.
Hyper-O-GlcNAcylation occurs in most cancers, and can affect the growth, proliferation, invasion, metastasis, and chemoresistance in cancer cells. In addition, O-GlcNAcylation is involved in multiple signaling pathways and affects the expression of a variety of downstream molecules, such as HIF-1α, c-MYC, AMPK, mTOR, and NF-κB, all of which are main players in oncogenic pathways. Therefore, OGT or O-GlcNAcylation can be targeted for cancer therapy, by OGT inhibitors to regulate O-GlcNAc levels in cells, with the aim of treating cancer. This appears to be a promising approach, because many OGT inhibitors have shown tumor suppressive effects. However, there are still many obstacles to the entry of OGT inhibitors into clinical practice. First, we still need to study the physiological role of O-GlcNAcylation, because rashly changing the global O-GlcNAcylation in cells will cause changes in the O-GlcNAcylation level of many proteins unrelated to the disease, which might have serious consequences. Second, small molecule inhibitors that target OGT might present off-target toxicity, and most of them are not cell-permeable, thus the development of highly specific and cell-permeable OGT inhibitors is required. In addition, the anticancer effects of existing OGT inhibitors have not been tested in strict animal models and no kinetic and pharmacodynamic data have been reported. In other words, more preclinical data on OGT inhibitors are required. Furthermore, targeting a specific protein modified by O-GlcNAcylation is also a potential therapy, and currently emerging technologies, such as the CRISPR-Cas9 system, support the eliminate O-GlcNAcylation of proteins; however, these technologies are relatively new and the pathway to put them into clinical practice is long.
Acknowledgements
Not applicable.
Abbreviations
- O-GlcNAc
O-Linked β-d-N-acetylglucosamine
- PTM
Post-translational modification
- HBP
Hexosamine biosynthetic pathway
- GFAT
Glutamine fructose-6-phosphate amidotransferase
- OGT
O-GlcNAc transferase
- OGA
β-N-Acetylglucosaminidase
- ncOGT
Nucleocytoplasmic OGT
- mOGT
Mitochondrial
- sOGT
Short-form OGT
- MTS
Mitochondrial targeting sequence
- TPR
Tripeptide repeat
- NLS
Nuclear localization sequence
- AMPK
AMP-activated protein kinase
- GSK3β
Glycogen synthase kinase-3β
- PGC-1α
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- FOXO
Forkhead box O1
- TCGA
The Cancer Genome Atlas
- GEPIA
Gene expression profiling interactive analysis
- CHOL
Cholangiocarcinoma
- DLBC
Lymphoid neoplasm diffuse large B cell lymphoma
- LAML
Acute myeloid leukemia
- ACC
Adrenocortical carcinoma
- BRCA
Breast invasive carcinoma
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- COAD
Colon adenocarcinoma
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- OV
Ovarian serous cystadenocarcinoma
- PAAD
Pancreatic adenocarcinoma
- READ
Rectum adenocarcinoma
- SKCM
Skin cutaneous melanoma
- THCA
Thyroid carcinoma
- UCEC
Uterine corpus endometrial carcinoma
- UCS
Uterine carcinosarcoma
- TNM
Tumor-node-metastasis
- TISIDB
Tumor and Immune System Interaction Database
- LIHC
Liver hepatocellular carcinoma
- BLCA
Bladder urothelial carcinoma
- TGCT
Testicular germ cell tumors
- OGA-L
Long form OGA
- OGA-S
Short form OGA
- GH84
Glycoside hydrolase family 84
- HAT
Histone acetyltransferase
- PCPG
Pheochromocytoma and paraganglioma
- LGG
Brain lower grade glioma
- HNSC
Head and neck squamous cell carcinoma
- STAD
Stomach adenocarcinoma
- MESO
Mesothelioma
- OS
Overall survival
- BLCA
Bladder urothelial carcinoma
- HPV
Human papilloma virus
- HCF-1
Host cell factor C1
- NF-κB
Nuclear factor kappa B
- CXCR4
C-X-C chemokine receptor 4
- FOXM1
Forkhead box M1
- SKP2
S-phase kinase-associated protein 2
- P27Kip1
Cyclin-dependent protein kinase inhibitor
- SIRT1
Sirtuin 1
- ERK
Extracellular regulated kinase
- DFS
Disease-free survival
- EMT
Epithelial–mesenchymal transition
- WNT5B
Wnt family member 5B
- MLC1
Modulator of VRAC current 1
- ROCK
Rho associated coiled-coil containing protein kinase
- HCC
Hepatocellular carcinoma
- YAP
Yes-associated protein
- AEGR
The advanced glycosylation end product-specific receptor
- TRIB2
Tribbles pseudokinase 2
- HDAC1
Histone deacetylase-1
- NAFLD-HCC
Non-alcoholic fatty liver disease-related hepatocellular carcinoma
- ER
Endoplasmic reticulum
- JNK
JUN N-terminal kinase
- c-Jun
AP-1 transcription factor subunit
- AP-1
Activator protein 1
- LT
Liver transplantation
- MMP
Matrix metalloproteinase
- AFP
Alpha fetoprotein
- RACK1
Ribosome-activated C kinase 1
- CRC
Colorectal cancer
- YY1
YIN-YANG-1
- SLC22A15
Solute carrier family 22 member 15
- AANAT
Aralkylamine N-acetyltransferase
- DDX5
DEAD-box helicase 5
- AKT
Protein kinase B
- mTOR
Rapamycin kinase
- ITGA5
Integrin α5
- CCSCs
Colon cancer stem cells
- MYBL1
MYB proto-oncogene like 1
- PDAC
Pancreatic ductal adenocarcinoma
- PD-L1
Programmed cell death 1 ligand 1
- HSF1
Heat shock factor 1
- HSPs
Heat shock proteins
- GC
Gastric cancer
- PI3K
Phosphatidylinositol-4,5-bisphosphate 3-kinase
- G protein
Guanine nucleotide binding protein
- GNB2L1
Beta polypeptide 2-like 1
- ESCC
Esophageal squamous cell carcinoma
- ESCSs
Esophageal cancer stem cells
- PD1
Programmed cell death 1
- PC
Prostate cancer
- VEGF
Vascular endothelial growth factor
- BMI-1
Polycomb ring finger
- PRC1
Protein regulator of cytokinesis 1
- TP53
Tumor protein 53
- PTEN
Phosphatase and tensin homolog
- CDKN
Cyclin dependent kinase Inhibitor
- BC
Bladder cancer
- NUSAP1
Nucleolar and spindle associated protein 1
- CDK5
Cyclin-dependent kinase 5
- RCC
Renal cell carcinoma
- EGFR
Epidermal growth factor receptor
- IL-6
Interleukin 6
- SCLC
Squamous cell laryngeal cancer
- CLL
Chronic lymphocytic leukemia
- RSV
Resveratrol
- MDS
Myelodysplastic syndrome
- AML
Acute myeloid leukemia
- ALL
Acute lymphoblastic leukemia
- DON
6-Diazo-5-oxo-l-norleucine
- OSMI-1
OGT with a small molecule inhibitor
- BADGP
Benzyl-2-acetamido-2-deoxy-α-d-galactopyranoside
- DOX
Doxorubicin
- CPT
Camptothecin
- pre-B-ALL
Pre-B acute lymphoblastic leukemia
- ASXL1
Additional sex combs like transcriptional regulator 1
- TC
Thyroid cancer
- IGF
Insulin-like growth factor
- HR
HR lysine demethylase and nuclear receptor corepressor
- ACSS2
Acetyl-CoA synthetase 2
- Shh
Sonic hedgehog
- GNPs
Granule neuron precursors
- HER2
Human epidermal growth factor receptor 2
- PKM2
Pyruvate kinase isoenzyme M2
- HTS
High-throughput screening
- ROS
Reactive oxygen species
- TRAIL
Tumor necrosis factor (TNF)-related apoptosis inducing ligand
- BCL2
B-cell CLL/lymphoma 2
- HIF-1α
Hypoxia inducible factor subunit 1 alpha
Author contributions
XB and TL conceived this review. QL, XZ collected the literature. QL, XZ performed the bioinformatics analysis, data interpretation, and prepared the figures. QL, XZ drew the schematics and tables. QL, XZ wrote the manuscript. All authors read and approved the final manuscript version. QL, XZ contributed equally to the drafting process. XB, TL share senior authorship. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key Research and Development Program grant (2019YFC1316000 (TL)); National Natural Science Foundation of China (81871925 and 82071867 (XB), U20A20378 and 81830089 (TL)); the Key Research and Development Program of Zhejiang Province (2020C03117 (XB), 019C03019 (TL)); the Fundamental Research Funds for the Zhejiang Provincial Universities (2021XZZX031 (XB)); and the Major Scientific Project of Zhejiang Lab (2020ND8AD01).
Availability of data and materials
The datasets supporting the conclusions of this article are available in the TIMER database (http://timer.comp-genomics.org), the GEPIA2 database (http://gepia2.cancer-pku.cn), the TCGA database (http://cancergenome.nih.gov), the TISIDB database (http://cis.hku.hk/TISIDB), and the DepMap-portal database (https://depmap.org/portal).
Declarations
Ethics approval and consent to participate
As no human subjects were recruited for this study, obtaining consent is not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qingsong Lu and Xiaozhen Zhang have contributed equally.
Contributor Information
Tingbo Liang, Email: liangtingbo@zju.edu.cn.
Xueli Bai, Email: shirleybai@zju.edu.cn.
References
- Abdel-Wahab O, Patel J, Levine RL. Clinical implications of novel mutations in epigenetic modifiers in AML. Hematol Oncol Clin North Am. 2011;25(6):1119–1133. doi: 10.1016/j.hoc.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Akella NM, Le Minh G, Ciraku L, et al. O-GlcNAc transferase regulates cancer stem-like potential of breast cancer cells. Mol Cancer Res. 2020;18(4):585–598. doi: 10.1158/1541-7786.MCR-19-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimoto Y, Miura Y, Toda T, et al. Morphological changes in diabetic kidney are associated with increased O-GlcNAcylation of cytoskeletal proteins including alpha-actinin 4. Clin Proteomics. 2011;8(1):15. doi: 10.1186/1559-0275-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimov V, Barrio-Hernandez I, Hansen S, et al. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat Struct Mol Biol. 2018;25(7):631–640. doi: 10.1038/s41594-018-0084-y. [DOI] [PubMed] [Google Scholar]
- Ali A, Kim SH, Kim MJ, et al. O-GlcNAcylation of NF-kappaB promotes lung metastasis of cervical cancer cells via upregulation of CXCR4 expression. Mol Cells. 2017;40(7):476–484. doi: 10.14348/molcells.2017.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J, Schimpl M, van Aalten DM. O-GlcNAcase: promiscuous hexosaminidase or key regulator of O-GlcNAc signaling? J Biol Chem. 2014;289(50):34433–34439. doi: 10.1074/jbc.R114.609198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold CS, Johnson GV, Cole RN, et al. The microtubule-associated protein tau is extensively modified with O-linked N-acetylglucosamine. J Biol Chem. 1996;271(46):28741–28744. doi: 10.1074/jbc.271.46.28741. [DOI] [PubMed] [Google Scholar]
- Asthana A, Ramakrishnan P, Vicioso Y, Zhang K, Parameswaran R. Hexosamine biosynthetic pathway inhibition leads to AML cell differentiation and cell death. Mol Cancer Ther. 2018;17(10):2226–2237. doi: 10.1158/1535-7163.MCT-18-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Sangwan V, McGinn O, et al. Triptolide-induced cell death in pancreatic cancer is mediated by O-GlcNAc modification of transcription factor Sp1. J Biol Chem. 2013;288(47):33927–33938. doi: 10.1074/jbc.M113.500983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. J Clin Oncol. 2011;29(5):504–515. doi: 10.1200/JCO.2010.31.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Blacker D, Mullin K, et al. Evidence for genetic linkage of Alzheimer's disease to chromosome 10q. Science. 2000;290(5500):2302–2303. doi: 10.1126/science.290.5500.2302. [DOI] [PubMed] [Google Scholar]
- Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-analyzed tumors. Cell. 2018;173(2):530. doi: 10.1016/j.cell.2018.03.059. [DOI] [PubMed] [Google Scholar]
- Bond MR, Hanover JA. A little sugar goes a long way: the cell biology of O-GlcNAc. J Cell Biol. 2015;208(7):869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodkin VS, Schimpl M, Gundogdu M, et al. Bisubstrate UDP-peptide conjugates as human O-GlcNAc transferase inhibitors. Biochem J. 2014;457(3):497–502. doi: 10.1042/BJ20131272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen JW, Balsbaugh JL, Chanda D, et al. Cross-talk between two essential nutrient-sensitive enzymes: O-GlcNAc transferase (OGT) and AMP-activated protein kinase (AMPK) J Biol Chem. 2014;289(15):10592–10606. doi: 10.1074/jbc.M113.523068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkinaree C, Cheung WD, Park S, et al. Characterization of beta-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis. J Biol Chem. 2008;283(35):23557–23566. doi: 10.1074/jbc.M804116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell SA, Jackson SR, Shahriari KS, et al. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010;29(19):2831–2842. doi: 10.1038/onc.2010.41. [DOI] [PubMed] [Google Scholar]
- Cancer Cell Line Encyclopedia Consortium. Genomics of Drug Sensitivity in Cancer Consortium Pharmacogenomic agreement between two cancer cell line data sets. Nature. 2015;528(7580):84–7. doi: 10.1038/nature15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervoni L, Turano C, Ferraro A, et al. Glycosylation of RNA polymerase II from wheat germ. Febs Lett. 1997;417(2):227–230. doi: 10.1016/S0014-5793(97)01285-4. [DOI] [PubMed] [Google Scholar]
- Champattanachai V, Netsirisawan P, Chaiyawat P, et al. Proteomic analysis and abrogated expression of O-GlcNAcylated proteins associated with primary breast cancer. Proteomics. 2013;13(14):2088–2099. doi: 10.1002/pmic.201200126. [DOI] [PubMed] [Google Scholar]
- Chatham JC, Not LG, Fulop N, Marchase RB. Hexosamine biosynthesis and protein O-glycosylation: the first line of defense against stress, ischemia, and trauma. Shock. 2008;29(4):431–440. doi: 10.1097/SHK.0b013e3181598bad. [DOI] [PubMed] [Google Scholar]
- Chen Y, Liu J, Zhang W, et al. O-GlcNAcylation enhances NUSAP1 stability and promotes bladder cancer aggressiveness. Onco Targets Ther. 2021;14:445–454. doi: 10.2147/OTT.S258175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li Y, Song Z, et al. O-GlcNAcylation promotes cerebellum development and medulloblastoma oncogenesis via SHH signaling. Proc Natl Acad Sci USA. 2022;119(34):e2202821119. doi: 10.1073/pnas.2202821119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Ren J, Su L, et al. O-GlcNAcylation of the signaling scaffold protein, GNB2L1 promotes its degradation and increases metastasis of gastric tumours. Biochem Biophys Res Commun. 2016;478(4):1497–1502. doi: 10.1016/j.bbrc.2016.08.074. [DOI] [PubMed] [Google Scholar]
- Cheng YU, Li H, Li J, et al. O-GlcNAcylation enhances anaplastic thyroid carcinoma malignancy. Oncol Lett. 2016;12(1):572–578. doi: 10.3892/ol.2016.4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung WD, Hart GW. AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose deprivation. J Biol Chem. 2008;283(19):13009–13020. doi: 10.1074/jbc.M801222200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciraku L, Bacigalupa ZA, Ju J, et al. O-GlcNAc transferase regulates glioblastoma acetate metabolism via regulation of CDK5-dependent ACSS2 phosphorylation. Oncogene. 2022;41(14):2122–2136. doi: 10.1038/s41388-022-02237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark RJ, McDonough PM, Swanson E, et al. Diabetes and the accompanying hyperglycemia impairs cardiomyocyte calcium cycling through increased nuclear O-GlcNAcylation. J Biol Chem. 2003;278(45):44230–44237. doi: 10.1074/jbc.M303810200. [DOI] [PubMed] [Google Scholar]
- Cohen P. The regulation of protein function by multisite phosphorylation—a 25 year update. Trends Biochem Sci. 2000;25(12):596–601. doi: 10.1016/S0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc Natl Acad Sci USA. 1987;84(21):7552–7556. doi: 10.1073/pnas.84.21.7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Queiroz RM, Madan R, Chien J, Dias WB, Slawson C. Changes in O-linked N-acetylglucosamine (O-GlcNAc) homeostasis activate the p53 pathway in ovarian cancer cells. J Biol Chem. 2016;291(36):18897–18914. doi: 10.1074/jbc.M116.734533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrell P, Cseh J, Mohas M, et al. Evidence of O-linked N-acetylglucosamine in diabetic nephropathy. Life Sci. 2009;84(13–14):389–393. doi: 10.1016/j.lfs.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Dennis JW, Lau KS, Demetriou M, Nabi IR. Adaptive regulation at the cell surface by N-glycosylation. Traffic. 2009;10(11):1569–1578. doi: 10.1111/j.1600-0854.2009.00981.x. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105(31):10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding M, Vandre DD. High molecular weight microtubule-associated proteins contain O-linked-N-acetylglucosamine. J Biol Chem. 1996;271(21):12555–12561. doi: 10.1074/jbc.271.21.12555. [DOI] [PubMed] [Google Scholar]
- Dorfmueller HC, Borodkin VS, Blair DE, et al. Substrate and product analogues as human O-GlcNAc transferase inhibitors. Amino Acids. 2011;40(3):781–792. doi: 10.1007/s00726-010-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan F, Wu H, Jia D, et al. O-GlcNAcylation of RACK1 promotes hepatocellular carcinogenesis. J Hepatol. 2018;68(6):1191–1202. doi: 10.1016/j.jhep.2018.02.003. [DOI] [PubMed] [Google Scholar]
- Erratum: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2020;70(4):313. [DOI] [PubMed]
- Fan Q, Moen A, Anonsen JH, et al. O-GlcNAc site-mapping of liver X receptor-alpha and O-GlcNAc transferase. Biochem Biophys Res Commun. 2018;499(2):354–360. doi: 10.1016/j.bbrc.2018.03.164. [DOI] [PubMed] [Google Scholar]
- Fardini Y, Dehennaut V, Lefebvre T, Issad T. O-GlcNAcylation: a new cancer hallmark? Front Endocrinol (lausanne) 2013;4:99. doi: 10.3389/fendo.2013.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer CM, Lynch TP, Sodi VL, et al. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol Cell. 2014;54(5):820–831. doi: 10.1016/j.molcel.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer CM, Sodi VL, Reginato MJ. O-GlcNAcylation in cancer biology: linking metabolism and signaling. J Mol Biol. 2016;428(16):3282–3294. doi: 10.1016/j.jmb.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer CM, Lu TY, Bacigalupa ZA, et al. O-GlcNAcylation regulates breast cancer metastasis via SIRT1 modulation of FOXM1 pathway. Oncogene. 2017;36(4):559–569. doi: 10.1038/onc.2016.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop N, Marchase RB, Chatham JC. Role of protein O-linked N-acetyl-glucosamine in mediating cell function and survival in the cardiovascular system. Cardiovasc Res. 2007;73(2):288–297. doi: 10.1016/j.cardiores.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 2001;276(13):9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- Ge X, Peng X, Li M, et al. OGT regulated O-GlcNacylation promotes migration and invasion by activating IL-6/STAT3 signaling in NSCLC cells. Pathol Res Pract. 2021;225:153580. doi: 10.1016/j.prp.2021.153580. [DOI] [PubMed] [Google Scholar]
- Ge Y, Ramirez DH, Yang B, et al. Target protein deglycosylation in living cells by a nanobody-fused split O-GlcNAcase. Nat Chem Biol. 2021;17(5):593–600. doi: 10.1038/s41589-021-00757-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelsi-Boyer V, Trouplin V, Adelaide J, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145(6):788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- Gloster TM, Zandberg WF, Heinonen JE, et al. Hijacking a biosynthetic pathway yields a glycosyltransferase inhibitor within cells. Nat Chem Biol. 2011;7(3):174–181. doi: 10.1038/nchembio.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golks A, Guerini D. The O-linked N-acetylglucosamine modification in cellular signalling and the immune system'. Protein modifications: beyond the usual suspects' review series. Embo Rep. 2008;9(8):748–753. doi: 10.1038/embor.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golks A, Tran TT, Goetschy JF, Guerini D. Requirement for O-linked N-acetylglucosaminyltransferase in lymphocytes activation. Embo J. 2007;26(20):4368–4379. doi: 10.1038/sj.emboj.7601845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong CX, Liu F, Iqbal K. O-GlcNAc cycling modulates neurodegeneration. Proc Natl Acad Sci USA. 2012;109(43):17319–17320. doi: 10.1073/pnas.1215395109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin ME, Jensen EH, Mason DE, et al. Comprehensive mapping of O-GlcNAc modification sites using a chemically cleavable tag. Mol Biosyst. 2016;12(6):1756–1759. doi: 10.1039/C6MB00138F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross BJ, Kraybill BC, Walker S. Discovery of O-GlcNAc transferase inhibitors. J Am Chem Soc. 2005;127(42):14588–14589. doi: 10.1021/ja0555217. [DOI] [PubMed] [Google Scholar]
- Gu Y, Mi W, Ge Y, et al. GlcNAcylation plays an essential role in breast cancer metastasis. Cancer Res. 2010;70(15):6344–6351. doi: 10.1158/0008-5472.CAN-09-1887. [DOI] [PubMed] [Google Scholar]
- Gu Y, Gao J, Han C, et al. O-GlcNAcylation is increased in prostate cancer tissues and enhances malignancy of prostate cancer cells. Mol Med Rep. 2014;10(2):897–904. doi: 10.3892/mmr.2014.2269. [DOI] [PubMed] [Google Scholar]
- Guo H, Zhang B, Nairn AV, et al. O-Linked N-acetylglucosamine (O-GlcNAc) expression levels epigenetically regulate colon cancer tumorigenesis by affecting the cancer stem cell compartment via modulating expression of transcriptional factor MYBL1. J Biol Chem. 2017;292(10):4123–4137. doi: 10.1074/jbc.M116.763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurel B, Iwata T, Koh CM, et al. Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod Pathol. 2008;21(9):1156–1167. doi: 10.1038/modpathol.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SW, Roman J. Fibronectin induces cell proliferation and inhibits apoptosis in human bronchial epithelial cells: pro-oncogenic effects mediated by PI3-kinase and NF-kappa B. Oncogene. 2006;25(31):4341–4349. doi: 10.1038/sj.onc.1209460. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hanover JA. Glycan-dependent signaling: O-linked N-acetylglucosamine. FASEB J. 2001;15(11):1865–1876. doi: 10.1096/fj.01-0094rev. [DOI] [PubMed] [Google Scholar]
- Hanover JA, Yu S, Lubas WB, et al. Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch Biochem Biophys. 2003;409(2):287–297. doi: 10.1016/S0003-9861(02)00578-7. [DOI] [PubMed] [Google Scholar]
- Harosh-Davidovich SB, Khalaila I. O-GlcNAcylation affects beta-catenin and E-cadherin expression, cell motility and tumorigenicity of colorectal cancer. Exp Cell Res. 2018;364(1):42–49. doi: 10.1016/j.yexcr.2018.01.024. [DOI] [PubMed] [Google Scholar]
- Hart GW. Nutrient regulation of signaling and transcription. J Biol Chem. 2019;294(7):2211–2231. doi: 10.1074/jbc.AW119.003226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth D, Ravindranath L, Chen Y, et al. Overexpression of C-MYC oncogene in prostate cancer predicts biochemical recurrence. Prostate Cancer Prostatic Dis. 2010;13(4):311–315. doi: 10.1038/pcan.2010.31. [DOI] [PubMed] [Google Scholar]
- He N, Ma D, Tan Y, Liu M. Upregulation of O-GlcNAc transferase is involved in the pathogenesis of acute myeloid leukemia. Asia Pac J Clin Oncol. 2021. [DOI] [PubMed]
- Housley MP, Rodgers JT, Udeshi ND, et al. O-GlcNAc regulates FoxO activation in response to glucose. J Biol Chem. 2008;283(24):16283–16292. doi: 10.1074/jbc.M802240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housley MP, Udeshi ND, Rodgers JT, et al. A PGC-1alpha-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J Biol Chem. 2009;284(8):5148–5157. doi: 10.1074/jbc.M808890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Suarez J, Fricovsky E, et al. Increased enzymatic O-GlcNAcylation of mitochondrial proteins impairs mitochondrial function in cardiac myocytes exposed to high glucose. J Biol Chem. 2009;284(1):547–555. doi: 10.1074/jbc.M808518200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D, Fujino T, Sheridan P, et al. A novel ASXL1-OGT axis plays roles in H3K4 methylation and tumor suppression in myeloid malignancies. Leukemia. 2018;32(6):1327–1337. doi: 10.1038/s41375-018-0083-3. [DOI] [PubMed] [Google Scholar]
- Issad T, Masson E, Pagesy P. O-GlcNAc modification, insulin signaling and diabetic complications. Diabetes Metab. 2010;36(6 Pt 1):423–435. doi: 10.1016/j.diabet.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Itkonen HM, Minner S, Guldvik IJ, et al. O-GlcNAc transferase integrates metabolic pathways to regulate the stability of c-MYC in human prostate cancer cells. Cancer Res. 2013;73(16):5277–5287. doi: 10.1158/0008-5472.CAN-13-0549. [DOI] [PubMed] [Google Scholar]
- Itkonen HM, Gorad SS, Duveau DY, et al. Inhibition of O-GlcNAc transferase activity reprograms prostate cancer cell metabolism. Oncotarget. 2016;7(11):12464–12476. doi: 10.18632/oncotarget.7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkonen HM, Urbanucci A, Martin SE, et al. High OGT activity is essential for MYC-driven proliferation of prostate cancer cells. Theranostics. 2019;9(8):2183–2197. doi: 10.7150/thno.30834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkonen HM, Loda M, Mills IG. O-GlcNAc transferase—an auxiliary factor or a full-blown oncogene? Mol Cancer Res. 2021;19(4):555–564. doi: 10.1158/1541-7786.MCR-20-0926. [DOI] [PubMed] [Google Scholar]
- Iyer SP, Akimoto Y, Hart GW. Identification and cloning of a novel family of coiled-coil domain proteins that interact with O-GlcNAc transferase. J Biol Chem. 2003;278(7):5399–5409. doi: 10.1074/jbc.M209384200. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Tjian R. O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell. 1988;55(1):125–133. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- Jang TJ, Kim UJ. O-GlcNAcylation is associated with the development and progression of gastric carcinoma. Pathol Res Pract. 2016;212(7):622–630. doi: 10.1016/j.prp.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Jaskiewicz NM, Townson DH. Hyper-O-GlcNAcylation promotes epithelial–mesenchymal transition in endometrial cancer cells. Oncotarget. 2019;10(30):2899–2910. doi: 10.18632/oncotarget.26884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Qiu Z, Zhang S, et al. Elevated O-GlcNAcylation promotes gastric cancer cells proliferation by modulating cell cycle related proteins and ERK 1/2 signaling. Oncotarget. 2016;7(38):61390–61402. doi: 10.18632/oncotarget.11359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin FZ, Yu C, Zhao DZ, Wu MJ, Yang Z. A correlation between altered O-GlcNAcylation, migration and with changes in E-cadherin levels in ovarian cancer cells. Exp Cell Res. 2013;319(10):1482–1490. doi: 10.1016/j.yexcr.2013.03.013. [DOI] [PubMed] [Google Scholar]
- Jin L, Lu MH, Dai GC, et al. O-GlcNAcylation promotes malignant phenotypes of bladder cancer cells. Neoplasma. 2020;67(4):880–888. doi: 10.4149/neo_2020_191006N1009. [DOI] [PubMed] [Google Scholar]
- Jin L, Yuan F, Dai G, et al. Blockage of O-linked GlcNAcylation induces AMPK-dependent autophagy in bladder cancer cells. Cell Mol Biol Lett. 2020;25:17. doi: 10.1186/s11658-020-00208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju KE. O-GlcNAc transferase: structural characteristics, catalytic mechanism and small-molecule inhibitors. ChemBioChem. 2020;21(21):3026–3035. doi: 10.1002/cbic.202000194. [DOI] [PubMed] [Google Scholar]
- Kaasik K, Kivimae S, Allen JJ, et al. Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 2013;17(2):291–302. doi: 10.1016/j.cmet.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamigaito T, Okaneya T, Kawakubo M, et al. Overexpression of O-GlcNAc by prostate cancer cells is significantly associated with poor prognosis of patients. Prostate Cancer Prostatic Dis. 2014;17(1):18–22. doi: 10.1038/pcan.2013.56. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Dahmus ME, Hart GW. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J Biol Chem. 1993;268(14):10416–10424. doi: 10.1016/S0021-9258(18)82216-5. [DOI] [PubMed] [Google Scholar]
- Kim M, Kim YS, Kim H, et al. O-linked N-acetylglucosamine transferase promotes cervical cancer tumorigenesis through human papillomaviruses E6 and E7 oncogenes. Oncotarget. 2016;7(28):44596–44607. doi: 10.18632/oncotarget.10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad RJ, Zhang F, Hale JE, et al. Alloxan is an inhibitor of the enzyme O-linked N-acetylglucosamine transferase. Biochem Biophys Res Commun. 2002;293(1):207–212. doi: 10.1016/S0006-291X(02)00200-0. [DOI] [PubMed] [Google Scholar]
- Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem. 1999;274(45):32015–32022. doi: 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;272(14):9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- Krzeslak A, Pomorski L, Lipinska A. Elevation of nucleocytoplasmic beta-N-acetylglucosaminidase (O-GlcNAcase) activity in thyroid cancers. Int J Mol Med. 2010;25(4):643–648. doi: 10.3892/ijmm_00000387. [DOI] [PubMed] [Google Scholar]
- Krzeslak A, Jozwiak P, Lipinska A. Down-regulation of beta-N-acetyl-d-glucosaminidase increases Akt1 activity in thyroid anaplastic cancer cells. Oncol Rep. 2011;26(3):743–749. doi: 10.3892/or.2011.1333. [DOI] [PubMed] [Google Scholar]
- Krzeslak A, Wojcik-Krowiranda K, Forma E, Bienkiewicz A, Brys M. Expression of genes encoding for enzymes associated with O-GlcNAcylation in endometrial carcinomas: clinicopathologic correlations. Ginekol Pol. 2012;83(1):22–26. [PubMed] [Google Scholar]
- Kuo WL, Tseng LL, Chang CC et al. Prognostic significance of O-GlcNAc and PKM2 in hormone receptor-positive and HER2-nonenriched breast cancer. Diagnostics (Basel). 2021;11(8) [DOI] [PMC free article] [PubMed]
- Kuwano R, Miyashita A, Arai H, et al. Dynamin-binding protein gene on chromosome 10q is associated with late-onset Alzheimer's disease. Hum Mol Genet. 2006;15(13):2170–2182. doi: 10.1093/hmg/ddl142. [DOI] [PubMed] [Google Scholar]
- Laczy B, Hill BG, Wang K, et al. Protein O-GlcNAcylation: a new signaling paradigm for the cardiovascular system. Am J Physiol Heart Circ Physiol. 2009;296(1):H13–28. doi: 10.1152/ajpheart.01056.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus BD, Love DC, Hanover JA. O-GlcNAc cycling: implications for neurodegenerative disorders. Int J Biochem Cell Biol. 2009;41(11):2134–2146. doi: 10.1016/j.biocel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Kwon OS. O-GlcNAc transferase inhibitor synergistically enhances doxorubicin-induced apoptosis in HepG2 cells. Cancers (Basel). 2020;12(11). [DOI] [PMC free article] [PubMed]
- Lee SJ, Lee DE, Choi SY, Kwon OS. OSMI-1 Enhances TRAIL-Induced Apoptosis through ER Stress and NF-kappaB Signaling in Colon Cancer Cells. Int J Mol Sci. 2021;22(20). [DOI] [PMC free article] [PubMed]
- Li Y, Wang L, Liu J, et al. O-GlcNAcylation modulates Bmi-1 protein stability and potential oncogenic function in prostate cancer. Oncogene. 2017;36(45):6293–6305. doi: 10.1038/onc.2017.223. [DOI] [PubMed] [Google Scholar]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu Z, He J, et al. OGT regulated O-GlcNAcylation promotes papillary thyroid cancer malignancy via activating YAP. Oncogene. 2021;40(30):4859–4871. doi: 10.1038/s41388-021-01901-7. [DOI] [PubMed] [Google Scholar]
- Li HJ, Wang Y, Li BX, et al. Roles of ten-eleven translocation family proteins and their O-linked beta-N-acetylglucosaminylated forms in cancer development. Oncol Lett. 2021;21(1):1. doi: 10.3892/ol.2020.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41(3):211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Lin CH, Yeh YC, et al. High O-linked N-acetylglucosamine transferase expression predicts poor survival in patients with early stage lung adenocarcinoma. Oncotarget. 2018;9(57):31032–31044. doi: 10.18632/oncotarget.25772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liptay S, Weber CK, Ludwig L, et al. Mitogenic and antiapoptotic role of constitutive NF-kappaB/Rel activity in pancreatic cancer. Int J Cancer. 2003;105(6):735–746. doi: 10.1002/ijc.11081. [DOI] [PubMed] [Google Scholar]
- Liu Z, Zeng W, Wang S, et al. A potential role for the Hippo pathway protein, YAP, in controlling proliferation, cell cycle progression, and autophagy in BCPAP and KI thyroid papillary carcinoma cells. Am J Transl Res. 2017;9(7):3212–3223. [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Cao Y, Pan X, et al. O-GlcNAc elevation through activation of the hexosamine biosynthetic pathway enhances cancer cell chemoresistance. Cell Death Dis. 2018;9(5):485. doi: 10.1038/s41419-018-0522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li L, Ma C, et al. O-GlcNAcylation of Thr(12)/Ser(56) in short-form O-GlcNAc transferase (sOGT) regulates its substrate selectivity. J Biol Chem. 2019;294(45):16620–16633. doi: 10.1074/jbc.RA119.009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zeng W, Maimaiti Y, et al. High expression of yes-activated protein-1 in papillary thyroid carcinoma correlates with poor prognosis. Appl Immunohisto M m. 2019;27(1):59–64. doi: 10.1097/PAI.0000000000000544. [DOI] [PubMed] [Google Scholar]
- Lode L, Cymbalista F, Soussi T. Genetic profiling of CLL: a 'TP53 addict' perspective. Cell Death Dis. 2016;7:e2042. doi: 10.1038/cddis.2015.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the "O-GlcNAc code". Sci STKE. 2005;2005(312):re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- Love DC, Kochan J, Cathey RL, Shin SH, Hanover JA. Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J Cell Sci. 2003;116(Pt 4):647–654. doi: 10.1242/jcs.00246. [DOI] [PubMed] [Google Scholar]
- Lubas WA, Frank DW, Krause M, Hanover JA. O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J Biol Chem. 1997;272(14):9316–9324. doi: 10.1074/jbc.272.14.9316. [DOI] [PubMed] [Google Scholar]
- Luthi T, Haltiwanger RS, Greengard P, Bahler M. Synapsins contain O-linked N-acetylglucosamine. J Neurochem. 1991;56(5):1493–1498. doi: 10.1111/j.1471-4159.1991.tb02043.x. [DOI] [PubMed] [Google Scholar]
- Lynch TP, Ferrer CM, Jackson SR, et al. Critical role of O-Linked beta-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J Biol Chem. 2012;287(14):11070–11081. doi: 10.1074/jbc.M111.302547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Hart GW. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev Proteomics. 2013;10(4):365–380. doi: 10.1586/14789450.2013.820536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Hart GW. O-GlcNAc profiling: from proteins to proteomes. Clin Proteomics. 2014;11(1):8. doi: 10.1186/1559-0275-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Vosseller K. O-GlcNAc in cancer biology. Amino Acids. 2013;45(4):719–733. doi: 10.1007/s00726-013-1543-8. [DOI] [PubMed] [Google Scholar]
- Ma Z, Vosseller K. Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J Biol Chem. 2014;289(50):34457–34465. doi: 10.1074/jbc.R114.577718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Vocadlo DJ, Vosseller K. Hyper-O-GlcNAcylation is anti-apoptotic and maintains constitutive NF-kappaB activity in pancreatic cancer cells. J Biol Chem. 2013;288(21):15121–15130. doi: 10.1074/jbc.M113.470047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makwana V, Dukie AS, Rudrawar S. Investigating the impact of OGT inhibition on doxorubicin- and docetaxel-induced cytotoxicity in PC-3 and WPMY-1 cells. Int J Toxicol. 2020;39(6):586–593. doi: 10.1177/1091581820948433. [DOI] [PubMed] [Google Scholar]
- Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266(8):4706–4712. doi: 10.1016/S0021-9258(19)67706-9. [DOI] [PubMed] [Google Scholar]
- Martin S, Tan ZW, Itkonen HM, et al. Structure-based evolution of low nanomolar O-GlcNAc transferase inhibitors. J Am Chem Soc. 2018;140(42):13542–13545. doi: 10.1021/jacs.8b07328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi W, Gu Y, Han C, et al. O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim Biophys Acta. 2011;1812(4):514–519. doi: 10.1016/j.bbadis.2011.01.009. [DOI] [PubMed] [Google Scholar]
- Mukherjee B, Bhattacharya S, Chakraborty S, et al. Is type 2 diabetes mellitus a predisposal cause for developing hepatocellular carcinoma? Curr Diabetes Rev. 2015;11(2):64–70. doi: 10.2174/1573399811666150115110747. [DOI] [PubMed] [Google Scholar]
- Ngoh GA, Facundo HT, Hamid T, et al. Unique hexosaminidase reduces metabolic survival signal and sensitizes cardiac myocytes to hypoxia/reoxygenation injury. Circ Res. 2009;104(1):41–49. doi: 10.1161/CIRCRESAHA.108.189431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Xia Y, Wang J, Shi X. O-GlcNAcylation promotes migration and invasion in human ovarian cancer cells via the RhoA/ROCK/MLC pathway. Mol Med Rep. 2017;15(4):2083–2089. doi: 10.3892/mmr.2017.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte D, Muller U. Human O-GlcNAc transferase (OGT): genomic structure, analysis of splice variants, fine mapping in Xq13.1. Mamm Genome. 2002;13(1):62–64. doi: 10.1007/s00335-001-2108-9. [DOI] [PubMed] [Google Scholar]
- Olivier-Van SS, Guinez C, Mir AM, et al. The hexosamine biosynthetic pathway and O-GlcNAcylation drive the expression of beta-catenin and cell proliferation. Am J Physiol Endocrinol Metab. 2012;302(4):E417–E424. doi: 10.1152/ajpendo.00390.2011. [DOI] [PubMed] [Google Scholar]
- Olivier-Van SS, Dehennaut V, Buzy A, et al. O-GlcNAcylation stabilizes beta-catenin through direct competition with phosphorylation at threonine 41. Faseb J. 2014;28(8):3325–3338. doi: 10.1096/fj.13-243535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3(104):ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Ortiz-Meoz RF, Jiang J, Lazarus MB, et al. A small molecule that inhibits OGT activity in cells. Acs Chem Biol. 2015;10(6):1392–1397. doi: 10.1021/acschembio.5b00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K, Liu R, Jia C et al. Regulation of O-Linked N-acetyl glucosamine transferase (OGT) through E6 stimulation of the ubiquitin ligase activity of E6AP. Int J Mol Sci. 2021;22(19) [DOI] [PMC free article] [PubMed]
- Peterson SB, Hart GW. New insights: a role for O-GlcNAcylation in diabetic complications. Crit Rev Biochem Mol Biol. 2016;51(3):150–161. doi: 10.3109/10409238.2015.1135102. [DOI] [PubMed] [Google Scholar]
- Pham LV, Bryant JL, Mendez R, et al. Targeting the hexosamine biosynthetic pathway and O-linked N-acetylglucosamine cycling for therapeutic and imaging capabilities in diffuse large B-cell lymphoma. Oncotarget. 2016;7(49):80599–80611. doi: 10.18632/oncotarget.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoomak C, Silsirivanit A, Wongkham C, et al. Overexpression of O-GlcNAc-transferase associates with aggressiveness of mass-forming cholangiocarcinoma. Asian Pac J Cancer Prev. 2012;13(Suppl):101–105. [PubMed] [Google Scholar]
- Phueaouan T, Chaiyawat P, Netsirisawan P, et al. Aberrant O-GlcNAc-modified proteins expressed in primary colorectal cancer. Oncol Rep. 2013;30(6):2929–2936. doi: 10.3892/or.2013.2794. [DOI] [PubMed] [Google Scholar]
- Pietila M, Ivaska J, Mani SA. Whom to blame for metastasis, the epithelial-mesenchymal transition or the tumor microenvironment? Cancer Lett. 2016;380(1):359–368. doi: 10.1016/j.canlet.2015.12.033. [DOI] [PubMed] [Google Scholar]
- Qian K, Wang S, Fu M, et al. Transcriptional regulation of O-GlcNAc homeostasis is disrupted in pancreatic cancer. J Biol Chem. 2018;293(36):13989–14000. doi: 10.1074/jbc.RA118.004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Z, Dang C, Zhou B, et al. O-linked N-acetylglucosamine transferase (OGT) is overexpressed and promotes O-linked protein glycosylation in esophageal squamous cell carcinoma. J Biomed Res. 2012;26(4):268–273. doi: 10.7555/JBR.26.20110121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y, Zhang X, Zhang Y, et al. High glucose stimulates tumorigenesis in hepatocellular carcinoma cells through AGER-dependent O-GlcNAcylation of c-Jun. Diabetes. 2016;65(3):619–632. doi: 10.2337/db15-1057. [DOI] [PubMed] [Google Scholar]
- Qiao Z, Dang C, Zhou B, et al. Downregulation of O-linked N-acetylglucosamine transferase by RNA interference decreases MMP9 expression in human esophageal cancer cells. Oncol Lett. 2016;11(5):3317–3323. doi: 10.3892/ol.2016.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez DH, Ge Y, Woo CM. O-GlcNAc engineering on a target protein in cells with nanobody-OGT and nanobody-splitOGA. Curr Protoc. 2021;1(5):e117. doi: 10.1002/cpz1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter RJ, Rosales-Corral SA, Tan DX et al. Melatonin, a full service anti-cancer agent: inhibition of initiation, progression and metastasis. Int J Mol Sci. 2017;18(4) [DOI] [PMC free article] [PubMed]
- Roman A, Munger K. The papillomavirus E7 proteins. Virology. 2013;445(1–2):138–168. doi: 10.1016/j.virol.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roquemore EP, Chevrier MR, Cotter RJ, Hart GW. Dynamic O-GlcNAcylation of the small heat shock protein alpha B-crystallin. Biochemistry-Us. 1996;35(11):3578–3586. doi: 10.1021/bi951918j. [DOI] [PubMed] [Google Scholar]
- Roychowdhury S, Chinnaiyan AM. Translating cancer genomes and transcriptomes for precision oncology. CA Cancer J Clin. 2016;66(1):75–88. doi: 10.3322/caac.21329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozanski W, Krzeslak A, Forma E, et al. Prediction of bladder cancer based on urinary content of MGEA5 and OGT mRNA level. Clin Lab. 2012;58(5–6):579–583. [PubMed] [Google Scholar]
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- Ryu IH, Do SI. Denitrosylation of S-nitrosylated OGT is triggered in LPS-stimulated innate immune response. Biochem Biophys Res Commun. 2011;408(1):52–57. doi: 10.1016/j.bbrc.2011.03.115. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63(6):1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- Schultz J, Pils B. Prediction of structure and functional residues for O-GlcNAcase, a divergent homologue of acetyltransferases. Febs Lett. 2002;529(2–3):179–182. doi: 10.1016/S0014-5793(02)03322-7. [DOI] [PubMed] [Google Scholar]
- Seo HG, Kim HB, Kang MJ, et al. Identification of the nuclear localisation signal of O-GlcNAc transferase and its nuclear import regulation. Sci Rep. 2016;6:34614. doi: 10.1038/srep34614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HG, Kim HB, Yoon JY, et al. Mutual regulation between OGT and XIAP to control colon cancer cell growth and invasion. Cell Death Dis. 2020;11(9):815. doi: 10.1038/s41419-020-02999-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang M, Yang H, Yang R, et al. The folate cycle enzyme MTHFD2 induces cancer immune evasion through PD-L1 up-regulation. Nat Commun. 2021;12(1):1940. doi: 10.1038/s41467-021-22173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Tomic J, Wen F, et al. Aberrant O-GlcNAcylation characterizes chronic lymphocytic leukemia. Leukemia. 2010;24(9):1588–1598. doi: 10.1038/leu.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer. 2011;11(9):678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodi VL, Khaku S, Krutilina R, et al. mTOR/MYC axis regulates O-GlcNAc transferase expression and O-GlcNAcylation in breast cancer. Mol Cancer Res. 2015;13(5):923–933. doi: 10.1158/1541-7786.MCR-14-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaner DE. O-GlcNAcylation in chronic lymphocytic leukemia and other blood cancers. Front Immunol. 2021;12:772304. doi: 10.3389/fimmu.2021.772304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starska K, Forma E, Brzezinska-Blaszczyk E, et al. Gene and protein expression of O-GlcNAc-cycling enzymes in human laryngeal cancer. Clin Exp Med. 2015;15(4):455–468. doi: 10.1007/s10238-014-0318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenackers A, Olivier-Van SS, Baldini SF, et al. Silencing the nucleocytoplasmic O-GlcNAc transferase reduces proliferation, adhesion, and migration of cancer and fetal human colon cell lines. Front Endocrinol (lausanne) 2016;7:46. doi: 10.3389/fendo.2016.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai HC, Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC. Parallel identification of O-GlcNAc-modified proteins from cell lysates. J Am Chem Soc. 2004;126(34):10500–10501. doi: 10.1021/ja047872b. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Tsujioka Y, Yamada T, et al. Glycosylation of microtubule-associated protein tau in Alzheimer's disease brain. Acta Neuropathol. 1999;97(6):635–641. doi: 10.1007/s004010051040. [DOI] [PubMed] [Google Scholar]
- Tang Z, Kang B, Li C, Chen T, Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J Biol Chem. 2004;279(51):53665–53673. doi: 10.1074/jbc.M410406200. [DOI] [PubMed] [Google Scholar]
- Tomic J, McCaw L, Li Y, et al. Resveratrol has anti-leukemic activity associated with decreased O-GlcNAcylated proteins. Exp Hematol. 2013;41(8):675–686. doi: 10.1016/j.exphem.2013.04.004. [DOI] [PubMed] [Google Scholar]
- Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259(5):3308–3317. doi: 10.1016/S0021-9258(17)43295-9. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander HM, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vocadlo DJ. O-GlcNAc processing enzymes: catalytic mechanisms, substrate specificity, and enzyme regulation. Curr Opin Chem Biol. 2012;16(5–6):488–497. doi: 10.1016/j.cbpa.2012.10.021. [DOI] [PubMed] [Google Scholar]
- Wang W, Abbruzzese JL, Evans DB, et al. The nuclear factor-kappa B RelA transcription factor is constitutively activated in human pancreatic adenocarcinoma cells. Clin Cancer Res. 1999;5(1):119–127. [PubMed] [Google Scholar]
- Wang L, Chen S, Zhang Z, et al. Suppressed OGT expression inhibits cell proliferation while inducing cell apoptosis in bladder cancer. BMC Cancer. 2018;18(1):1141. doi: 10.1186/s12885-018-5033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Chen S, Zhang J, et al. Suppressed OGT expression inhibits cell proliferation and modulates EGFR expression in renal cell carcinoma. Cancer Manag Res. 2019;11:2215–2223. doi: 10.2147/CMAR.S190642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang Z, Yuan J, Wang J, Shen X. The positive feedback between ACSL4 expression and O-GlcNAcylation contributes to the growth and survival of hepatocellular carcinoma. Aging (albany NY) 2020;12(9):7786–7800. doi: 10.18632/aging.103092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein JN, Collisson EA, Mills GB, et al. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Hart GW. Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science. 2001;291(5512):2376–2378. doi: 10.1126/science.1058714. [DOI] [PubMed] [Google Scholar]
- Woo CM, Lund PJ, Huang AC, et al. Mapping and quantification of over 2000 O-linked glycopeptides in activated human T cells with isotope-targeted glycoproteomics (Isotag) Mol Cell Proteomics. 2018;17(4):764–775. doi: 10.1074/mcp.RA117.000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Jiang M, Han Y, et al. O-GlcNAcylation promotes colorectal cancer progression by regulating protein stability and potential catcinogenic function of DDX5. J Cell Mol Med. 2019;23(2):1354–1362. doi: 10.1111/jcmm.14038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Tan Z, Li H, et al. Melatonin reduces proliferation and promotes apoptosis of bladder cancer cells by suppressing O-GlcNAcylation of cyclin-dependent-like kinase 5. J Pineal Res. 2021;71(3):e12765. doi: 10.1111/jpi.12765. [DOI] [PubMed] [Google Scholar]
- Wu D, Jin J, Qiu Z, Liu D, Luo H. Functional analysis of O-GlcNAcylation in cancer metastasis. Front Oncol. 2020;10. [DOI] [PMC free article] [PubMed]
- Wulff-Fuentes E, Berendt RR, Massman L, et al. The human O-GlcNAcome database and meta-analysis. Sci Data. 2021;8(1):25. doi: 10.1038/s41597-021-00810-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Zhang X, Wu JL, et al. O-GlcNAc transferase promotes fatty liver-associated liver cancer through inducing palmitic acid and activating endoplasmic reticulum stress. J Hepatol. 2017;67(2):310–320. doi: 10.1016/j.jhep.2017.03.017. [DOI] [PubMed] [Google Scholar]
- Xu Z, Zhang Y, Ocansey D, Wang B, Mao F. Glycosylation in cervical cancer: new insights and clinical implications. Front Oncol. 2021;11:706862. doi: 10.3389/fonc.2021.706862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol. 2017;18(7):452–465. doi: 10.1038/nrm.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Su K, Roos MD, et al. O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc Natl Acad Sci USA. 2001;98(12):6611–6616. doi: 10.1073/pnas.111099998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WH, Kim JE, Nam HW, et al. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol. 2006;8(10):1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- Yao B, Xu Y, Wang J, et al. Reciprocal regulation between O-GlcNAcylation and tribbles pseudokinase 2 (TRIB2) maintains transformative phenotypes in liver cancer cells. Cell Signal. 2016;28(11):1703–1712. doi: 10.1016/j.cellsig.2016.08.003. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- Yu M, Chu S, Fei B, Fang X, Liu Z. O-GlcNAcylation of ITGA5 facilitates the occurrence and development of colorectal cancer. Exp Cell Res. 2019;382(2):111464. doi: 10.1016/j.yexcr.2019.06.009. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wang L, Ge D, et al. Exosomal O-GlcNAc transferase from esophageal carcinoma stem cell promotes cancer immunosuppression through up-regulation of PD-1 in CD8(+) T cells. Cancer Lett. 2021;500:98–106. doi: 10.1016/j.canlet.2020.12.012. [DOI] [PubMed] [Google Scholar]
- Zachara NE, Hart GW. Cell signaling, the essential role of O-GlcNAc! Biochim Biophys Acta. 2006;1761(5–6):599–617. doi: 10.1016/j.bbalip.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Zeng Q, Zhao RX, Chen J, et al. O-linked GlcNAcylation elevated by HPV E6 mediates viral oncogenesis. Proc Natl Acad Sci USA. 2016;113(33):9333–9338. doi: 10.1073/pnas.1606801113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Chen X. Potential role of O-GlcNAcylation and involvement of PI3K/Akt1 pathway in the expression of oncogenic phenotypes of gastric cancer cells in vitro. Biotechnol Appl Biochem. 2016;63(6):841–851. doi: 10.1002/bab.1441. [DOI] [PubMed] [Google Scholar]
- Zhang H, Gao G, Brunk UT. Extracellular reduction of alloxan results in oxygen radical-mediated attack on plasma and lysosomal membranes. APMIS. 1992;100(4):317–325. doi: 10.1111/j.1699-0463.1992.tb00878.x. [DOI] [PubMed] [Google Scholar]
- Zhang P, Wang C, Ma T, You S. O-GlcNAcylation enhances the invasion of thyroid anaplastic cancer cells partially by PI3K/Akt1 pathway. Onco Targets Ther. 2015;8:3305–3313. doi: 10.2147/OTT.S82845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Zhou P, Li X, et al. Bitterness in sugar: O-GlcNAcylation aggravates pre-B acute lymphocytic leukemia through glycolysis via the PI3K/Akt/c-Myc pathway. Am J Cancer Res. 2017;7(6):1337–1349. [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Qiao Y, Wu Q, et al. The essential role of YAP O-GlcNAcylation in high-glucose-stimulated liver tumorigenesis. Nat Commun. 2017;8:15280. doi: 10.1038/ncomms15280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SS, Xu X, Wu J, et al. Liver transplantation for hepatocellular carcinoma: Hangzhou experiences. Transplantation. 2008;85(12):1726–1732. doi: 10.1097/TP.0b013e31816b67e4. [DOI] [PubMed] [Google Scholar]
- Zhou F, Yang X, Zhao H, et al. Down-regulation of OGT promotes cisplatin resistance by inducing autophagy in ovarian cancer. Theranostics. 2018;8(19):5200–5212. doi: 10.7150/thno.27806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Chen X. Aptamer-based targeted therapy. Adv Drug Deliv Rev. 2018;134:65–78. doi: 10.1016/j.addr.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Zhou L, Yang Z, et al. O-GlcNAcylation plays a role in tumor recurrence of hepatocellular carcinoma following liver transplantation. Med Oncol. 2012;29(2):985–993. doi: 10.1007/s12032-011-9912-1. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Shan X, Yuzwa SA, Vocadlo DJ. The emerging link between O-GlcNAc and Alzheimer disease. J Biol Chem. 2014;289(50):34472–34481. doi: 10.1074/jbc.R114.601351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Tao T, Zhang D, et al. O-GlcNAcylation of histone deacetylases 1 in hepatocellular carcinoma promotes cancer progression. Glycobiology. 2016;26(8):820–833. doi: 10.1093/glycob/cww025. [DOI] [PubMed] [Google Scholar]
- Zhu G, Qian M, Lu L et al. O-GlcNAcylation of YY1 stimulates tumorigenesis in colorectal cancer cells by targeting SLC22A15 and AANAT. Carcinogenesis. 2019. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are available in the TIMER database (http://timer.comp-genomics.org), the GEPIA2 database (http://gepia2.cancer-pku.cn), the TCGA database (http://cancergenome.nih.gov), the TISIDB database (http://cis.hku.hk/TISIDB), and the DepMap-portal database (https://depmap.org/portal).