Abstract
Stratifying patients according to disease severity has been a major hurdle during the COVID-19 pandemic. This usually requires evaluating the levels of several biomarkers, which may be cumbersome when rapid decisions are required. In this manuscript we show that a single nanoparticle aggregation test can be used to distinguish patients that require intensive care from those that have already been discharged from the intensive care unit (ICU). It consists of diluting a platelet-free plasma sample and then adding gold nanoparticles. The nanoparticles aggregate to a larger extent when the samples are obtained from a patient in the ICU. This changes the color of the colloidal suspension, which can be evaluated by measuring the pixel intensity of a photograph. Although the exact factor or combination of factors behind the different aggregation behavior is unknown, control experiments demonstrate that the presence of proteins in the samples is crucial for the test to work. Principal component analysis demonstrates that the test result is highly correlated to biomarkers of prognosis and inflammation that are commonly used to evaluate the severity of COVID-19 patients. The results shown here pave the way to develop nanoparticle aggregation assays that classify COVID-19 patients according to disease severity, which could be useful to de-escalate care safely and make a better use of hospital resources.
Abbreviations: Alb, albumin; AST, aspartate aminotransferaseALT, alanine aminotransferase; C1.75, protein concentration 1.75 × 10-4 g·dL-1; CPImin, protein concentration at PImin; Creat, creatinine; CRP, C-reactive protein; D-D, D-dimer; Ferr, ferritin; GGT, gamma-glutamyl transferase; Glu, glucose; Hb, hemoglobin; ICU, intensive care unit; INR, international normalized ratio (prothrombin time); LDH, lactate dehydrogenase; LSPR, localized surface plasmon resonance; MCV, mean corpuscular volume; Mono, monocytes; MPV, mean platelet volume; NIR, near-infrared; NLR, neutrophil-to-lymphocyte ratio; NTA, nanoparticle tracking analysis; PDW, platelet distribution width; PI, pixel intensity; PImin, minimum value of pixel intensity; PIdil, pixel intensity at plasma dilution 1:31250; PI1.75, pixel intensity at C1.75; PLR, platelet-to-lymphocyte ratio; RBC, red blood cells; RDW, red cell distribution width; TG, triglycerides; TotProt, total protein concentration; WBC, white blood cells
Keywords: Sepsis, SARS-CoV-2, Gold, Plasmonic, Colorimetric
Graphical Abstract
1. Introduction
Detection systems based on the aggregation of plasmonic nanoparticles have gained popularity due to the marked changes in color they produce, which can be evaluated by eye or with image processing software [1], [2]. In these detection systems, the specific detection of a target molecule requires highly controlled assay conditions in order avoid nanoparticle aggregation in the absence of the analyte. For example, it is well established that citrate-capped nanoparticles tend to aggregate in solutions containing highly concentrated ions or when buffered at low pH values [3]. The presence of biomolecules in these solutions may stabilize or destabilize the colloids depending on their concentration, and may also be modulated by other factors such as the pH or the presence of multivalent ions [4], [5]. For instance, proteins may aggregate nanoparticles or protect them from aggregation depending on their concentration and isoelectric point, among other factors [6], [7], [8]. These observations have paved the way to develop a new family of sensors that detect changes in the composition of human plasma, which contains proteins and other solutes, using nanoparticle aggregation assays ‘[9], [10]. For example, it has been shown that plasma samples from patients with acute COVID-19 have a different composition compared to samples from patients with less severe symptoms [11]. If these differences resulted in a different nanoparticle aggregation behavior, they could be used to identify patients that require critical care. While this approach would not reveal changes in the levels of specific biomarkers, being able to classify patients according to severity with a single measurement could improve their management and make a better use of hospital resources [12].
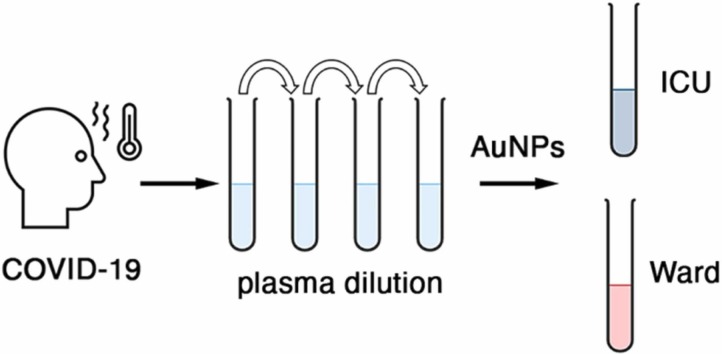
In this article we show that, in a narrow plasma dilution range, gold nanoparticles aggregate depending on disease severity when exposed to samples obtained from COVID-19 patients ( Fig. 1). Specifically, it will be shown that nanoparticles aggregate when the plasma was obtained from patients in the intensive care unit (ICU), but that, in the same conditions, do not aggregate so much when the plasma was obtained from patients that had been discharged from the ICU and were recovering in a hospital ward. Control experiments will demonstrate that plasma proteins are key to trigger the aggregation of the citrate-capped nanoparticles. Our method differs from previous approaches based on probing complex samples with colorimetric arrays of nanoparticles in that we use a single test to query the composition of the sample. Previously, a battery of nanoparticle aggregation conditions such as pH values, ionic strength, and surfactants, among others, were used to create an array of assays [13], [14], [15], [16], [17], [18], [19], [10]. This yielded a color pattern that was related to the composition of the sample. Instead, we dilute the sample so that the total protein concentration is the same, and then query if nanoparticles aggregate or not with a single test consisting of adding the diluted sample to the nanosensors. While our approach provides no information about the identity of specific proteins in the sample, the test output is a single value, which makes it easier to interpret the results and to correlate them with other clinical variables. It will be shown that the test results have an excellent correlation with prognosis biomarkers that have been shown to be dysregulated in severe COVID-19 patients. The ability to classify hospitalized patients according to disease severity and inflammatory status with a single colorimetric test could be useful to plan hospital resources as well as to guide the de-escalation of patient care, which have been key issues during the management of the COVID-19 pandemic.
Fig. 1.
Schematic representation of COVID-19 severity evaluation with plasma-induced nanoparticle aggregation. Probing a well-defined plasma dilution with gold nanoparticles generates a colorimetric signal associated to disease severity.
2. Material and methods
2.1. Nanoparticle synthesis
Citrate-capped gold nanoparticles with a diameter around 40 nm were synthetized using the Turkevich method [7]. Briefly, 49 mg gold(III) chloride trihydrate dissolved in 1 mL of Milli-Q water was added to 250 mL boiling Milli-Q water (final gold concentration 0.5 mM) and then sodium citrate was added to a final concentration of 0.75 mM for 15 min while vigorously stirring. The resulting gold nanoparticles were let to cool down at room temperature and then stored at 4 ºC until used.
2.2. Sample dilution protocol
Serial dilutions of plasma samples in Milli-Q water were obtained as follows. The first working dilution involved diluting plasma 104 times. This was obtained using a 10-fold dilution series starting with 5 μL of plasma. Samples were then further diluted in the range between 1:2500000 and 1:25000.
2.3. Nanoparticle assays
Nanoparticle assays were performed in 96-well plates (Thermo Scientific) previously blocked with 2 % PBS-BSA for 90 min at 37 ºC followed by rinsing 5 times with Milli-Q water. Each assay involved mixing 100 μL of nanoparticles with 100 μL of diluted plasma. The final nanoparticle concentration was 0.105 nM (Fig. S1). Pictures were taken 20–30 s afterwards using a homemade visible light transilluminator (Figs. S2 and S3). Pictures were taken with a dedicated 5MP Raspberry Pi camera module with Omnivision OV5647 CMOS sensor (model CMT-5MP-RP-OV5647-X010). The camera was controlled with a single board computer (Raspberry Pi Zero WH) and programmed to automatically upload images to a database at the push of a button. Extinction spectra were collected with a PowerWave HT plate reader (Biotek).
2.4. Plasma samples
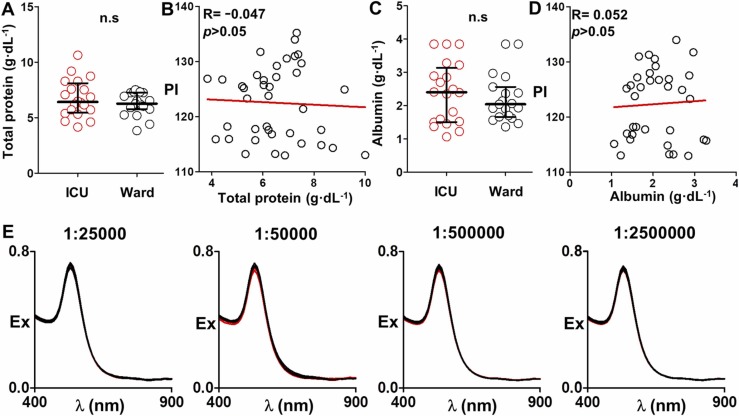
Blood samples from patients admitted to the ICU were collected in vacuum tubes containing ethylenediaminetetraacetic acid (EDTA) as anticoagulant. Twenty samples were collected from SARS-CoV-2-positive patients admitted to the ICU for less than 10 days, whereas 19 samples originated from patients already discharged from the ICU (ward patients). Gender distribution was similar across ICU and ward cohorts, with 30% and 36.8% females respectively (Fisher’s exact test p-value=0.74). Patients’ demographic and clinical variables at the time of sampling are provided in Table S1. All samples were collected after informed consent was obtained from the patient or a family member. The study was conducted according to the ethical guidelines of the 1975 Declaration of Helsinki (Ethics and Scientific Committee approval IB 4251/20 PI). Platelet-free plasma samples were obtained by following a double spin cycle. Briefly, whole blood samples were centrifuged at 1500 g for 10 min and then the resulting plasma samples were transferred to new tubes and centrifuged at 2500 g for 15 min. Samples were stored at − 80 °C in the Biobank IdISBa and CIBERES Pulmonary Biobank Consortium (integrated in the Spanish National Biobanks Network). In Fig. 5, the total protein concentration was determined in plasma samples diluted 1:2000 using a Bradford assay, whereas albumin levels in samples diluted 1:200,000 were determined with an ELISA test (reference RAB0603 purchased from Sigma-Aldrich).
Fig. 5.
Potential factors influencing nanoparticle aggregation after adding diluted plasma samples; (A) Distribution of total protein concentration values for ICU and ward patients; (B) Correlation plot comparing the total protein concentration and PIdil; (C) Distribution of albumin levels for ICU and ward patients; (D) Correlation plot comparing the albumin concentration and PIdil; (E) Extinction spectra of gold nanoparticles after adding 3 diluted ICU (red) and 3 ward (black) samples covering the whole dilution range shown in Fig. 3. Data in A and C are expressed as median with percentiles 25th and 75th. Mann-Whitey test proved non-significant (n.s.) statistical differences. Pdil; pixel intensity at plasma dilution 1:31250.
Experiments with deproteinized plasma samples were addressed after conducting the following deproteinization protocol. Briefly, plasma samples obtained from 3 ICU and 3 ward patients were diluted 1:100 in Milli-Q water (final volume of 500 μL). These pre-diluted samples were incubated in a bath at 90 ºC for 10 min and then centrifuged at 3400 g for 5 min. Next, 400 μL of the deproteinized supernatants was filtered using Amicon® Ultra-0.5 centrifugal 10 K filters (Millipore) following the manufacturers’ instructions.
2.5. Data analysis
The pixel intensity in images was evaluated by quantifying the “a” channel in the L*a*b color space in the region of interest with Adobe Photoshop. Statistical analysis was performed using GraphPad Prism software. The Mann-Whitney test was used to assess differences between colorimetric signals yielded by plasma samples from ICU and ward patients. A p value < 0.05 was considered statistically significant. Principal component analysis was done to evaluate relationships between patients’ clinical variables and colorimetric signals.
3. Results and discussion
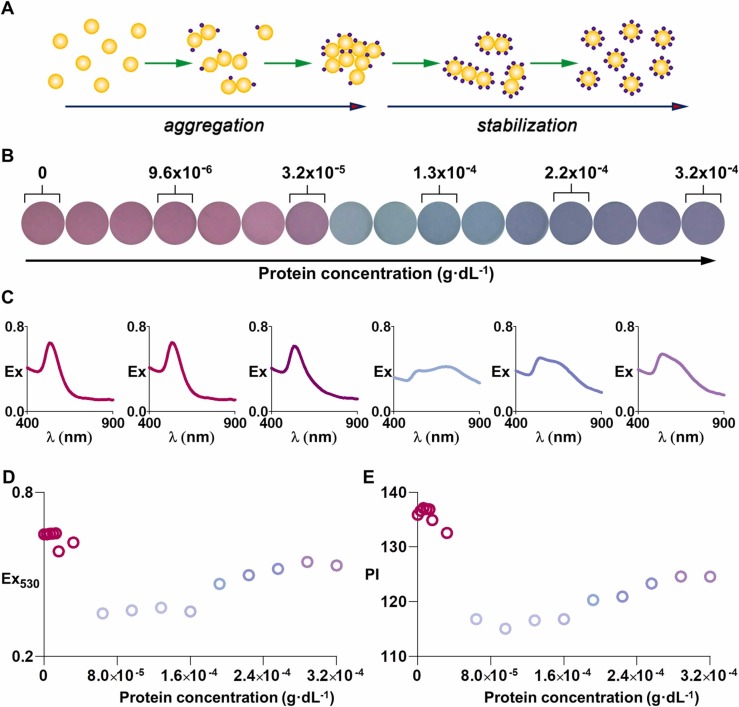
The aggregation of gold nanoparticles in a solution that contains proteins depends on the protein concentration, among other factors ( Fig. 2A). Therefore, we first endeavored to study the impact of the protein concentration on the aggregation of gold nanoparticles (40 nm diameter). To this end, we serially diluted plasma samples and then added nanoparticles at the same final concentration. Changes in the state of aggregation of the colloids were observed in the dilution range between 1:2,500,000 and 1:25,000. Fig. 2B shows a representative example of the results obtained from an ICU patient that had an initial total protein concentration value of 8 g·dL-1. In the proposed dilution range, when the concentration of proteins increases from 0 to 1.3 × 10-4 g·dL-1 the color of the colloidal suspension changes from mauve to blue and gray, which indicates that the nanoparticles aggregate (Fig. 2B). In Fig. 2C, extinction spectra taken in this concentration range show that the localized surface plasmon resonance (LSPR) at 530 nm decreases and a new LSPR appears at lower wavelengths, which demonstrates that the nanoparticles are clustering together. This was further supported using nanoparticle tracking analysis (NTA) (supporting Fig. S4). In this concentration range, protein adsorption disturbs the homogenous distribution of negative charges around the colloids and destabilizes them, as schematically shown in Fig. 2A. When the protein concentration increases further the color of the suspension slowly transitions to blue and purple (Fig. 2B). The LSPR at 530 increases and the near-infrared LSPR progressively disappears, which demonstrates that nanoparticles are less aggregated (Fig. 2C). In this concentration range a protein corona is generated that stabilizes the colloids as depicted in Fig. 2A. Figs. 2D and 2E show the evolution of the extinction value at 530 nm, as well as the pixel intensity measured from photographs taken with a raspberry pi camera. Both parameters decrease as the.
Fig. 2.
Representative example of plasma-induced nanoparticle aggregation generated with a sample from a critical patient; A) Schematic representation of the impact of adding proteins (purple dots) at different concentrations to gold nanoparticles (yellow dots); B) Photographs of the colloidal suspensions after adding fifteen decreasing dilutions of a sample from a critical patient with an initial total protein concentration value of 8 g·dL-1 (superscripts indicate the protein concentration in diluted samples prior to nanoparticle addition in g·dL-1) C) Extinction spectra of tests highlighted in panel B; D) Extinction at 530 nm (Ex530); and E) Pixel intensity (PI) as a function of the protein concentration prior to nanoparticle addition in diluted samples.
concentration of protein increases, and then slowly increase after achieving a minimum. These experiments demonstrate that the plasma-induced aggregation of gold nanoparticles depends on the degree of sample dilution, which changes the concentration of proteins and other solutes within it. Changes in nanoparticle aggregation can be monitored by measuring the pixel intensity of a photograph (densitometry), which in the future could be performed with a dedicated instrument or smartphone app [20], [21].
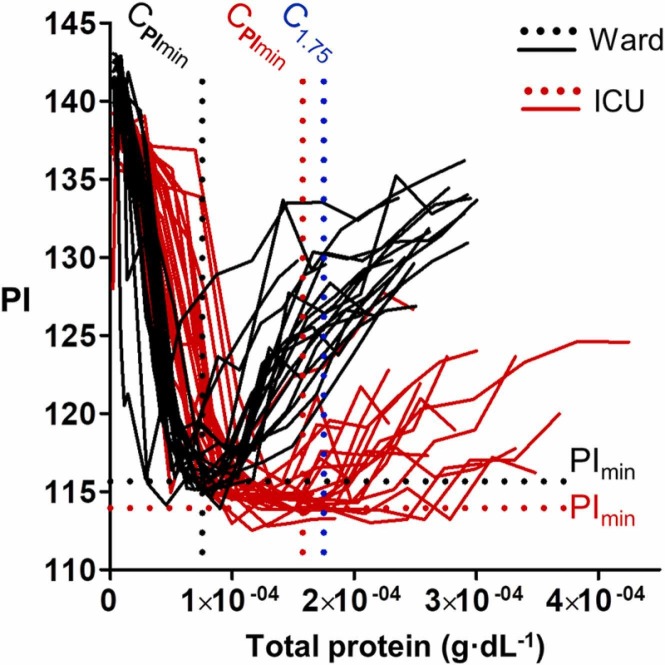
After studying the impact of protein concentration on nanoparticle aggregation, we sought to compare the aggregation pattern of nanoparticles after adding diluted plasma samples from patients. Fig. 3 shows the variation of the pixel intensity obtained from 20 ICU patients (red trace), as well as from 19 ward patients that were recovering after staying in the ICU (black trace). The same dilution protocol was applied to all samples. Results were then represented as a function of the actual protein concentration in each plasma sample, which was determined the same day that the sample was collected. In this Figure, the average minimum value of pixel intensity (PImin) is lower for ICU patients than for ward patients. This means that diluted plasma samples from ICU patients aggregate nanoparticles to a larger extent than plasma samples from ward patients. The average protein concentration at PImin (CPImin) is higher for ICU patients. This means that more concentrated proteins are required in order to achieve the highest level of aggregation compared to ward patients. Finally, the pixel intensity of ICU samples increases at a slower rate after PImin compared to samples from ward patients, which means that they stay highly aggregated in a larger range of protein concentrations. Due to this, the pixel intensity at a fixed total concentration value is lower for ICU patients than for ward patients in the range between 1 × 10-4 and 2 × 10-4 g·dL-1. For example, the pixel intensity from ICU patients differs from ward patients in most cases when the protein concentration is near 1.75 × 10-4 g·dL-1 (PI at C1.75). This.
Fig. 3.
Pixel intensity as a function of the protein concentration after adding diluted plasma samples from ICU (red) or ward patients (black) to gold nanoparticles. Dotted lines are a guide to eye. PImin: minimum value of pixel intensity; CPImin: protein concentration at PImin; C1.75: indicates that the protein concentration is 1.75 × 10-4 g·dL-1.
result indicates that a single aggregation assay at a fixed protein concentration could be enough to identify whether a patient requires critical care or not. This makes our approach different from previous methods based on analyzing the color of nanoparticle suspensions upon exposure to an array of aggregating stimuli [13], [14], [15], [16], [17], [18], [19], [10].
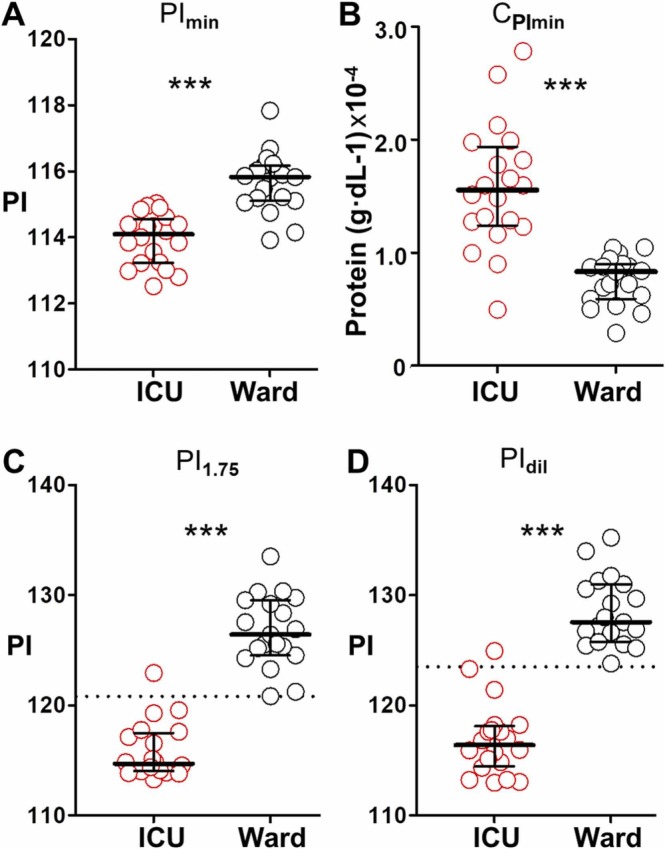
Fig. 4A-4C show the distribution of PImin, CPImin, and the PI at C1.75 (PI1.75) obtained from ICU and ward samples. Statistically relevant differences are observed in all cases. Nevertheless, the parameter that better distinguishes both populations is PI1.75 because test results overlap to a lesser extent than when plotting the other parameters. Indeed, only one ICU patient is miscategorized using PI1.75, in agreement with our previous observation that a single test at a fixed protein concentration (1.75 g·dL-1) can stratify patients according to severity. However, calculating PI1.75 would require measuring the total protein levels first followed by readjusting the dilution protocol in order to test plasma samples at the same final concentration, which would be cumbersome. As an alternative, we investigated if plasma samples could be diluted following the same proto-col and then analyze one dilution irrespective of the initial protein concentration. Fig. 4D shows that the PI at dilution 1:31250 (PIdil) also differentiates both sample collections except for the same ICU specimen that overlapped with the ward samples when plotting PI1.75. These experiments demonstrate that the color of a single test performed at a fixed dilution can be used to distinguish COVID-19 patients in the ICU from those that do not require intensive care anymore.
Fig. 4.
Comparison between different parameters proposed here to differentiate patient populations; (A) PImin (minimum value of pixel intensity); (B) CPImin (protein concentration at PImin); (C) PI1.75 (pixel intensity at C1.75); and (D) PIdil (pixel intensity at plasma dilution 1:31250) yielded by plasma samples from ICU (red) and ward (black) patients. The dotted lines represent two standard deviations above the mean of ICU patients. Data are expressed as median with percentiles 25th and 75th; *p-value was obtained with a Mann-Whitey test.
Next, we studied the origin of the differences in nanoparticle aggregation behavior observed in Fig. 2. We first checked whether changes in the initial protein concentration or in the concentration of the most abundant protein in plasma (albumin) could be the source of the observed differences. In Fig. 5A and 5B, ICU and ward patients do not have statistically different total protein levels, and there is no correlation between this parameter and PIdil. Similarly, there is no significant difference between the albumin levels of both patient populations, and there is no correlation between the albumin concentration and PIdil (Fig. 5C and 5D). These experiments demonstrate that, although the concentration of protein and albumin may influence the test results, they are not the only factor that determines the color of the test under the proposed conditions. It has been suggested that some drugs (e.g. aminoglycoside antibiotics) can trigger nanoparticle aggregation [22]. Critical patients often show higher lactic acid levels that could lead to small differences in pH [23], which is known to modulate nanoparticle aggregation [7], [24]. To demonstrate that proteins, and not other solutes in plasma, are the key to the observed changes in aggregation behavior, we repeated the same experiments with deproteinized plasma samples from 3 ICU and 3 ward patients. Under this condition, no aggregation was observed in the whole range of dilutions proposed, which demonstrates that proteins are essential in order to aggregate nanoparticles in Fig. 3 (Fig. 5E). Moreover, attempts to measure the pH of plasma samples diluted 1:25000 or more were not successful because the dilution factor was so high that the concentration of buffering molecules was negligible, and values fluctuated wildly. Nevertheless, no differences were observed when checking the mean pH after adding nanoparticles to 3 diluted ICU (4.05 ± 0.02) or ward (4.07 ± 0.03) samples. These experiments show that leftover citrate ions from the nanoparticle synthesis rather than intrinsic solutes in the highly diluted samples are responsible for the pH of the mixture. In summary these experiments demonstrate that, although solutes in the plasma may modulate the protein-induced nanoparticle aggregation, the biomolecules are essential to cluster the colloids together and yield the observed color changes.
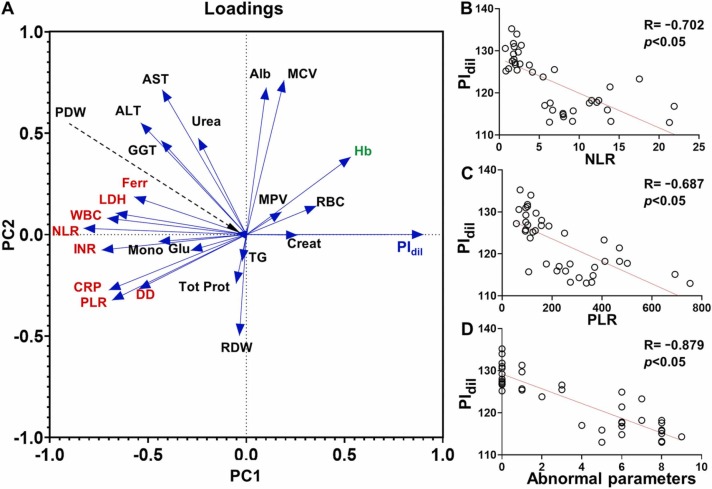
Fig. 6 shows a principal component analysis (PCA) comparing PIdil and clinical variables that are commonly used to evaluate disease severity and inflammatory status (see also Table S1). Fig. 6A shows two principal components clearly defined by hemostatic and inflammatory biomarkers (clustered in principal component 1, PC1), and albumin levels along with liver function (clustered in principal component 2, PC2). In this Figure, PIdil is highly correlated with PC1 (loading value of 0.901) and its correlation with PC2 is negligible (loading value of 0.001), which reinforces the idea that albumin is not a relevant clustering factor in the distribution of colorimetric data when comparing ICU and ward patients. This PCA demonstrates a well-defined negative correlation between PIdil and those clinical variables with which it forms nearly 180º angles (red-colored biomarkers in Fig. 6A). The strongest correlation is an inverse relationship between PIdil and the neutrophil-to-lymphocyte ratio (NLR, Fig. 6B) and the platelet-to-lymphocyte ratio (PLR, Fig. 6C). Indeed, high values of NLR and PLR have been associated to deterioration in COVID-19 patients, which agrees well with the low PIdil value found in ICU patients [25], [26]. Other strongly correlated variables are biomarkers of inflammation and poor prognosis such as C-reactive protein (CRP), D-dimer (DD), ferritin (Ferr), and lactate dehydrogenase (LDH), which have also been shown to be more elevated in patients with severe or acute COVID-19 [27], [28], [29], [30]. Coagulation biomarkers such as the prothrombin time (International Normalized Ratio, INR) were also inversely correlated with PIdil, which agrees well with reports on highly elevated INR levels in patients with severe COVID-19 [31]. Finally, PIdil values correlate positively with hemoglobin levels (Hb, green-colored biomarker in Fig. 6A), which have also been reported to be dysregulated in patients with severe COVID-19 [32]. These data analyses support the idea that our test is detecting patients that require intensive care, which could be useful to stratify them according to disease severity. We then hypothesized that PIdil values could be related to the number of dysregulated prognosis biomarkers that showed a strong correlation in Fig. 6A. To study this, we measured the number of these clinical variables that were out of normal range for ICU and ward patients (see Table S1) and plotted them against PIdil. In Fig. 6D there is a strong linear correlation between the number of dysregulated biomarkers and the test results, which further supports the idea that PIdil values are useful to determine disease severity. It should be noted that many patients in the ICU were younger than those in recovering in the ward, and therefore that the observed changes are not generated by higher inflammation levels originated by age (Table S1).
Fig. 6.
Correlation studies between PIdil and clinical variables; (A) Principal component analysis; (B-D) Correlation plots comparing PIdil with the neutrophil-to-lymphocyte ratio (NLR, B), the platelet-to-lymphocyte ratio (PLR, C) and the number of biomarkers out of normal range (within selected biomarkers highlighted in red and green in (A), D). PIdil; pixel intensity at plasma dilution 1:31250.
4. Conclusions
In conclusion, we have shown that diluted plasma samples from COVID-19 patients in the ICU aggregate gold nanoparticles differently than samples obtained from patients recovering in a hospital ward. In other words, the color of the colloidal suspension is different after adding plasma diluted to the same the extent from the proposed populations. The presence of proteins in the samples is crucial to trigger nanoparticle aggregation, although the exact factor or combination of factors responsible for clustering the colloids is unknown. The test results correlate well with the levels of biomarkers commonly used to evaluate severity and prognosis in COVID-19. Indeed, there is a linear correlation between the number of dysregulated biomarkers and the test outcome, which reinforces the idea that the color of the test is associated to disease severity. Future experiments will aim at studying whether the test can differentiate other severe infections that may require intensive care (e.g., cases of bacterial sepsis or septic shock) [33]. Larger populations will also be recruited to improve the statistical analysis and determine cut-off values. Using a dedicated tabletop instrument for plasma separation, liquid handling and test photographing along with software for automated color quantification could reduce the test variability and produce results within minutes, which would make our approach useful for making rapid decisions about patient stratification in hospitals.
Author Contributions
The manuscript was written through contributions of all authors. / All authors have given approval to the final version of the manuscript.
CRediT authorship contribution statement
Giulia Santopolo: Methodology, Data curation, Validation. Antonio Clemente: Conceptualization, Data curation, Formal analysis, Supervision, Methodology, Writing - review & editing. Marta Gonzalez-Freire: Data curation, Resources.Steven M. Russell: Methodology, Investigation. Andreu Vaquer: Methodology, Investigation. Enrique Barón: Methodology, Investigation. María Aranda: Investigation, Resources. Antonia Socias: Investigation, Resources. Alberto del Castillo: Investigation, Resources. Marcio Borges: Investigation, Resources Roberto de la Rica: Conceptualization, Funding acquisition; Investigation, Project administration; Resources; Supervision, Writing - original draft; Writing - review & editing.
Declaration of Competing Interest
There are no conflicts of interest.
Acknowledgment
R.R and M.B. acknowledge funding from Instituto de Salud Carlos III through the project PI20/00538 (Co-funded by European Regional Development Fund/European Social Fund "A way to make Europe"/"Investing in your future"). R.R., A.C., and G.S. acknowledge a Radix, Folium and Talent Junior fellowship, respectively, from IdISBa/Impost turisme sos-tenible/Agència d’Estratègia Turística de les Illes Bale-ars/Govern de les Illes Balears. M. G.-F. acknowledges a contract from the Miguel Servet Program (MS19/00201), Instituto de Salud Carlos III (ISCIII). E.B. acknowledges funding from the Instituto de Salud Carlos III (Sara Borrell contract). We want to particularly acknowledge the patients and the Biobank IdISBa and CIBERES Pulmonary Biobank Consortium (PT17/0015/0001) member of the Spanish National Biobanks Network financed by the Carlos III Health Institute.
Biographies
Giulia Santopolo obtained her Bachelor’s degree in Chemistry and Chemical Technology and her Master’s degree in Clinical, Forensic Chemistry and Doping Control from the University of Turin in 2015 and 2019, respectively. She is a member of the Multidisciplinary Sepsis Group (Balearic Islands Health Research Institute, IdISBa) and is currently developing her Ph.D. on diagnostic applications of plasmonic nanosensors.
Dr. Antonio Clemente obtained his MSc degree on Microbiology in 2008 and his Ph.D. degree on Immunology from the Autonomous University of Barcelona in 2014. During 1-year postdoctoral position at the August Pi Sunyer Biomedical Research Institute (IDIBAPS) he studied nanotechnology applications to immunological disorders therapies. Currently, he is working at the Health Research Institute of the Balearic Islands (IdISBa) with a 2-year post-doc contract (FOLIUM). His research focuses into biosensor diagnosis of bacterial infections.
Dr. Marta Gonzalez-Freire completed his Ph.D. at the European University of Madrid in Madrid, Spain. After that she started a postdoctoral fellowship at the National Institutes of Health (NIH), USA, for almost 6 years. She has experience in epidemiology studies and biology of aging and inflammation. She is currently a Miguel Servet Investigator, a tenure track position, at the Balearic Islands Health Research Institute, IdISBa since January 2020.
Steven M. Russell is a researcher in computer vision and machine learning. His work applies real-time image processing to biosensing. He is also interested in the 3D modeling of biochemical interactions and in developing paper-based microfluidic devices.
Andreu Vaquer obtained his degree in Chemistry from Balearic Islands University in 2017. He is currently on the first year of the M.Sc. Science and Chemical technology at the University of the Balearic Islands. He is a member of the Multidisciplinary Sepsis Group (Balearic Islands Health Research Institute, IdISBa) since April 2019. His research interest focuses into paper microfluidic devices and nanoparticle functionalization.
Dr. Enrique Barón obtained his MSc degree on Advanced Chemistry in 2012 and his Ph.D. degree on Analytical Chemistry from the University of Barcelona in January 2016. During his Ph.D. he studied the environmental behavior of different halogenated flame retardants. Currently, he is working at the Health Research Institute of the Balearic Islands (IdISBa) with a 3-year post-doc contract (Sara Borrell). His research focuses into the development of biosensors for the detection of different disease biomarkers.
María Aranda Perez obtained her degree in Medicine from Universidad Complutense de Madrid in 2007. She specialized in Critical Care Medicine at HU Son Llàtzer hospital in 2015 and from 2015 she is working at HU Son Llàtzer as intensivist and is member of Sepsis Unit from its creation.
Antonia Socias obtained her degree in Medicine from Universitat Autonoma de Barcelona in 2000. She specialized in Critical Care Medicine at Vall d'Hebron hospital and from 2006 she is working at HU Son Llàtzer as intensivist and is member of Sepsis Unit from its creation. She is currently a Ph.D. student in Toxicology.
Alberto del Castillo obtained his degree in Medicine from University of Salamanca in 2002 and completed the Intensive Care specialty at the Hospital Dr.Peset (Valencia) in 2008. He is currently working in the Intensive Care Unit of the University Hospital Son Llàtzer. He is a member of the Multidisciplinary Sepsis Group (Balearic Islands Health Research Institute, IdISBa). His research interest focuses into sepsis.
Marcio Borges Coordinator of Multidisciplinary Sepsis Unit. Intensive Care Unit. Son Llàtzer University Hospital. Associated Professor in Infectious Diseases by Balearic Islands University (UIB). Coordinator of the Multidisciplinary Sepsis Group (Balearic Islands Health Research Institute, IdISBa) since January 2017. Director of Sepsis Area by Iberian and Panamerican Federation of Intensive Care. Spanish´s Director of Sepsis Code endorsed by Health Ministry of Spain.
Dr. Roberto de la Rica completed his Ph.D. at the National Center for Microelectronics in Barcelona, Spain. He has worked at the City University of New York, at the University of Twente and at Imperial College London. He has experience in nanolithography, synthesis of nanomaterials, biosensors and bio-enabled nanofabrication. He is the coordinator of the technological division of the Multidisciplinary Sepsis Group (Balearic Islands Health Research Institute, IdISBa) since January 2019.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.snb.2022.132638.
Appendix A. Supplementary material
Supplementary material
.
Data availability
Data will be made available on request.
References
- 1.Ran B., Zheng W., Dong M., Xianyu Y., Chen Y., Wu J., Qian Z., Jiang X. Peptide-mediated controllable cross-linking of gold nanoparticles for immunoassays with tunable detection range. Anal. Chem. 2018;90:8234–8240. doi: 10.1021/acs.analchem.8b01760. [DOI] [PubMed] [Google Scholar]
- 2.Chen P., Liu X., Goyal G., Tran N.T., Shing Ho J.C., Wang Y., Aili D., Liedberg B. Nanoplasmonic sensing from the human vision perspective. Anal. Chem. 2018;90:4916–4924. doi: 10.1021/acs.analchem.8b00597. [DOI] [PubMed] [Google Scholar]
- 3.Paterson S., de la Rica R. Solution-based nanosensors for in-field detection with the naked eye. Analyst. 2015;140:3308–3317. doi: 10.1039/C4AN02297A. [DOI] [PubMed] [Google Scholar]
- 4.Huang R.H., Nayeem N., He Y., Morales J., Graham D., Klajn R., Contel M., O’Brien S., Ulijn R.V. Self-complementary zwitterionic peptides direct nanoparticle assembly and enable enzymatic selection of endocytic pathways. Adv. Mater. 2022;34 doi: 10.1002/adma.202104962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vesga M.J., McKechnie D., Laing S., Kearns H., Faulds K., Johnston K., Sefcik J. Effect of glycine on aggregation of citrate-functionalised gold nanoparticles and SERS measurements. Colloids Surf. A Physicochem. Eng. Asp. 2021;621 doi: 10.1016/j.colsurfa.2021.126523. [DOI] [Google Scholar]
- 6.Ho Y.T., Ain Azman N., Loh F.W.Y., Ong G.K.T., Engudar G., Kriz S.A., Kah J.C.Y. Protein corona formed from different blood plasma proteins affects the colloidal stability of nanoparticles differently. Bioconjug. Chem. 2018;29:3923–3934. doi: 10.1021/acs.bioconjchem.8b00743. [DOI] [PubMed] [Google Scholar]
- 7.Santopolo G., Clemente A., Aranda M., Socias A., del Castillo A., Chica A., Borges M., de la Rica R. Colorimetric detection of sepsis-derived hyperdegranulation with plasmonic nanosensors. ACS Sens. 2021;6:4443–4450. doi: 10.1021/acssensors.1c01884. [DOI] [PubMed] [Google Scholar]
- 8.Santopolo G., Doménech-Sánchez A., Russell S.M., de la Rica R. Ultrafast and ultrasensitive naked-eye detection of urease-positive bacteria with plasmonic nanosensors. ACS Sens. 2019;4:961–967. doi: 10.1021/acssensors.9b00063. [DOI] [PubMed] [Google Scholar]
- 9.Papafilippou L., Claxton A., Dark P., Kostarelos K., Hadjidemetriou M. Protein corona fingerprinting to differentiate sepsis from non-infectious systemic inflammation. Nanoscale. 2020;12:10240–10253. doi: 10.1039/d0nr02788j. [DOI] [PubMed] [Google Scholar]
- 10.Bordbar M.M., Samadinia H., Sheini A., Aboonajmi J., Hashemi P., Khoshsafar H., Halabian R., Khanmohammadi A., Nobakht M.Gh.BF., Sharghi H., Ghanei M., Bagheri H. Visual diagnosis of COVID-19 disease based on serum metabolites using a paper-based electronic tongue. Anal Chim Acta. 2022;1226 doi: 10.1016/j.aca.2022.340286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell S.M., Alba-Patiño A., Barón E., Borges M., Gonzalez-Freire M., De La Rica R. Biosensors for managing the COVID-19 cytokine storm: challenges ahead. ACS Sensors. 2020;5:1506–1513. doi: 10.1021/acssensors.0c00979. [DOI] [PubMed] [Google Scholar]
- 13.Bigdeli A., Ghasemi F., Golmohammadi H., Abbasi-Moayed S., Nejad M.A.F., Fahimi-Kashani N., Jafarinejad S., Shahrajabian M., Hormozi-Nezhad M.R. Nanoparticle-based optical sensor arrays. Nanoscale. 2017;9:16546–16563. doi: 10.1039/C7NR03311G. [DOI] [PubMed] [Google Scholar]
- 14.Li Z., Askim J.R., Suslick K.S. The optoelectronic nose: colorimetric and fluorometric sensor arrays. Chem. Rev. 2019;119:231–292. doi: 10.1021/acs.chemrev.8b00226. [DOI] [PubMed] [Google Scholar]
- 15.Xi H., He W., Liu Q., Chen Z. Protein discrimination using a colorimetric sensor array based on gold nanoparticle aggregation induced by cationic polymer. ACS Sustain. Chem. Eng. 2018;6:10751–10757. doi: 10.1021/acssuschemeng.8b02063. [DOI] [Google Scholar]
- 16.Mao J., Lu Y., Chang N., Yang J., Zhang S., Liu Y. Multidimensional colorimetric sensor array for discrimination of proteins. Biosens. Bioelectron. 2016;86:56–61. doi: 10.1016/j.bios.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 17.Yu H., Long D., Huang W. Organic antifreeze discrimination by pattern recognition using nanoparticle array. Sens. Actuators, B Chem. 2018;264:164–168. doi: 10.1016/j.snb.2018.02.180. [DOI] [Google Scholar]
- 18.Sun J., Lu Y., He L., Pang J., Yang F., Liu Y. A colorimetric sensor array for protein discrimination based on carbon nanodots-induced reversible aggregation of AuNP with GSH as a regulator. Sens. Actuators, B Chem. 2019;296 doi: 10.1016/j.snb.2019.126677. [DOI] [Google Scholar]
- 19.Rogowski J.L., Verma M.S., Gu F.X. Discrimination of proteins using an array of surfactant-stabilized gold nanoparticles. Langmuir. 2016;32:7621–7629. doi: 10.1021/acs.langmuir.6b01339. [DOI] [PubMed] [Google Scholar]
- 20.Russell S.M., Alba-Patiño A., Vaquer A., Clemente A., de la Rica R. Improving the quantification of colorimetric signals in paper-based immunosensors with an open-source reader. Sensors. 2022;22:1880. doi: 10.3390/s22051880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L.J., Chang Y.C., Sun R., Li L. A multichannel smartphone optical biosensor for high-throughput point-of-care diagnostics. Biosens. Bioelectron. 2017;87:686–692. doi: 10.1016/j.bios.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 22.Zheng T., Li Sip Y.Y., Leong M.B., Huo Q. Linear self-assembly formation between gold nanoparticles and aminoglycoside antibiotics. Colloids Surf. B. Biointerfaces. 2018;164:185–191. doi: 10.1016/j.colsurfb.2018.01.027. [DOI] [PubMed] [Google Scholar]
- 23.Maciel A.T., Noritomi D.T., Park M. Metabolic acidosis in sepsis. Endocr. Metab. Immune Disord. Drug Targets. 2010;10:252–257. doi: 10.2174/187153010791936900. [DOI] [PubMed] [Google Scholar]
- 24.Santopolo G., Rojo-Molinero E., Clemente A., Borges M., Oliver A., de la Rica R. Bedside detection of carbapenemase-producing pathogens with plasmonic nanosensors. Sens. Actuators, B Chem. 2021;329 doi: 10.1016/j.snb.2020.129059. [DOI] [Google Scholar]
- 25.Zeng Z.-Y., Feng S.-D., Chen G.-P., Wu J.-N. Predictive value of the neutrophil to lymphocyte ratio for disease deterioration and serious adverse outcomes in patients with COVID-19: a prospective cohort study. BMC Infect. Dis. 2021;21:80. doi: 10.1186/s12879-021-05796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkar S., Kannan S., Khanna P., Singh A.K. Role of platelet-to-lymphocyte count ratio (PLR), as a prognostic indicator in COVID-19: a systematic review and meta-analysis. J. Med. Virol. 2022;94:211–221. doi: 10.1002/jmv.27297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stringer D., Braude P., Myint P.K., Evans L., Collins J.T., Verduri A., Quinn T.J., Vilches-Moraga A., Stechman M.J., Pearce L., Moug S., McCarthy K., Hewitt J., Carter B. The role of C-reactive protein as a prognostic marker in COVID-19. Int. J. Epidemiol. 2021;50:420–429. doi: 10.1093/ije/dyab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.H. Zhan, H. Chen, C. Liu, L. Cheng, S. Yan, H. Li, Y. Li, Diagnostic Value of D-Dimer in COVID-19: A Meta-Analysis and Meta-Regression., Clin. Appl. Thromb. Off. J. Int. Acad. Clin. Appl. Thromb. 27 (2021) 10760296211010976. https://doi.org/10.1177/10760296211010976. [DOI] [PMC free article] [PubMed]
- 29.Kaushal K., Kaur H., Sarma P., Bhattacharyya A., Sharma D.J., Prajapat M., Pathak M., Kothari A., Kumar S., Rana S., Kaur M., Prakash A., Mirza A.A., Panda P.K., Vivekanandan S., Omar B.J., Medhi B., Naithani M. Serum ferritin as a predictive biomarker in COVID-19. A systematic review, meta-analysis and meta-regression analysis. J. Crit. Care. 2022;67:172–181. doi: 10.1016/j.jcrc.2021.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martha J.W., Wibowo A., Pranata R. Prognostic value of elevated lactate dehydrogenase in patients with COVID-19: a systematic review and meta-analysis. Postgrad. Med. J. 2022;98:422–427. doi: 10.1136/postgradmedj-2020-139542. [DOI] [PubMed] [Google Scholar]
- 31.Zinellu A., Paliogiannis P., Carru C., Mangoni A.A. INR and COVID-19 severity and mortality: a systematic review with meta-analysis and meta-regression. Adv. Med. Sci. 2021;66:372–380. doi: 10.1016/j.advms.2021.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayneris-Perxachs J., Russo M.F., Ramos R., de Hollanda A., Arxé A.A., Rottoli M., Arnoriaga-Rodríguez M., Comas-Cufí M., Bartoletti M., Verrastro O., Gudiol C., Fages E., Giménez M., de A., Gil G., Bernante P., Tinahones F., Carratalà J., Pagotto U., Hernández-Aguado I., Fernández-Aranda F., Meira F., Castro Guardiola A., Mingrone G., Fernández-Real J.M., Obesity-T2DM Covid19 Study Group M.R., Fumaña S., Jimenez-Murcia M., Eatxandi A., Rombauts G., Abelenda-Alonso C., Guidone D., Anello G., Giannetti E., Ortega I., Conget C., Viñals C., Hernández-Aguado J.-M., Sirvent R., Orriols M., Fernández-Balsells W.Ricart. Blood hemoglobin substantially modulates the impact of gender, morbid obesity, and hyperglycemia on COVID-19 death risk: a multicenter study in Italy and Spain. Front. Endocrinol. (Lausanne). 2021;12 doi: 10.3389/fendo.2021.741248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheini A. A point-of-care testing sensor based on fluorescent nanoclusters for rapid detection of septicemia in children. Sens. Actuators, B Chem. 2021;328 doi: 10.1016/j.snb.2020.129029. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.