Abstract
Although sequence analysis of Borrelia burgdorferi isolate B31 was recently declared “complete,” we found that cultures of this strain can contain a novel 9-kb circular plasmid, cp9-2. The newly described plasmid contains both sequence similarities with and differences from the previously identified B31 plasmid cp9-1 (formerly cp9). cp9-1 and cp9-2 each encode a unique allele of EppA, a putative membrane protein synthesized by B. burgdorferi during mammalian infection.
Natural isolates of the Lyme disease spirochete, Borrelia burgdorferi, contain large numbers of circular and linear extrachromosomal DNAs (2, 5, 9, 15, 25, 32, 46). Many of these plasmids have been found in every examined isolate, suggesting that they encode proteins essential for infection of the bacterium's warm-blooded and/or arthropod hosts. Consistent with this hypothesis, the majority of identified infection-associated proteins are encoded by extrachromosomal DNA species (for examples, see references 4, 20, 21, 23, 27, 35, and 38). Sustained cultivation in laboratory media results in loss of plasmids, with concomitant loss of ability to infect mammals (4, 17, 24, 29, 33, 47), further indications that plasmids encode virulence factors. In order to pinpoint genes for essential proteins, there have been several studies examining the correlation between bacterial infectivity and the presence or absence of certain plasmids (24, 29, 33, 36, 47; J. E. Purser and S. J. Norris, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. D-214, 2000).
The B. burgdorferi type strain B31 was isolated in 1981 from an infected Ixodes scapularis tick collected on Shelter Island, N.Y. (7, 18), and was cloned by limiting dilution (3). Since that time, different laboratories have independently maintained B31, and as a result, there are now many descendant cultures that differ in their plasmid content and infectivity. The chromosome and plasmids of an infectious B. burgdorferi isolate B31 culture, designated B31-MI, were recently sequenced by the Institute for Genomic Research (9, 15), data that can provide invaluable assistance to studies of the pathogenic mechanisms of Lyme disease spirochetes. As an example, it would appear that the plasmid contents of infectious and noninfectious B31 cultures can be compared using PCR primers or Southern blot probes designed from the published B31-MI sequences.
Bacteria of culture B31-MI contain nine different circular plasmids, which are named according to their approximate sizes in kilobases: cp9, cp26, and seven different cp32s (a group of largely homologous 30- to 32-kb circular plasmids [10, 43]). Our laboratories have extensively used B31 cultures with passage histories different from that of B31-MI (6, 10, 22, 30, 31, 38, 41, 44), and so we sought to determine the circular plasmid content of these other B31 bacteria. One such culture, designated herein B31-RML, is infectious to mice but is no longer clonal, since a small number of bacteria in this culture contain plasmid cp32-2, which is missing from the great majority of B31-RML bacteria (10). We also examined a second infectious B31 culture, B31-4a, grown from a single colony of B31-RML that had been plated in solid medium (10). Previous analyses demonstrated that bacteria in B31-RML contain cp26, all seven of the B31-MI cp32s plus two additional cp32s (cp32-2 and cp32-5), while B31-4a contains cp26 and eight cp32s (all except cp32-2) (9, 10). The presence of the other B31-MI circular plasmid, cp9, in these two B31 cultures was unknown, and so we sought to assess the presence of cp9 in B31-RML and B31-4a by PCR, using oligonucleotide primers complementary to DNA sequences on that plasmid.
B. burgdorferi B31-RML, B31-4a, and B31-MI plasmids were purified using Qiagen Midi kits (Qiagen, Valencia, Calif.) as previously described (13). Plasmid cp9 encodes EppA, a putative membrane protein synthesized by B. burgdorferi during mammalian infection (11). The published B31-MI cp9 sequence was used to design PCR oligonucleotide primer pairs complementary to DNA located 5′ and 3′ of eppA (EA-A and EA-B, respectively) and within eppA (EA-C and EA-D) (Table 1). PCR conditions consisted of 25 cycles of 94°C for 1 min, 45°C for 1 min, and 60°C for 2 min. Reaction products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. EA-A and EA-B, the oligonucleotide primer pair complementary to the eppA flanking sequences, yielded a PCR amplicon from both B31-RML and B31-MI (data not shown). However, PCR with EA-C and EA-D, the primer pair corresponding to internal eppA sequences, did not yield an appropriately sized product from B31-RML, although an amplicon was obtained from PCR of B31-MI DNA (data not shown). Variations of the PCR conditions, including lower annealing and elongation temperatures, failed to generate an amplicon from B31-RML DNA with primers EA-C and EA-D. No products were obtained from B31-4a using either oligonucleotide primer pair under any tested PCR condition.
TABLE 1.
Oligonucleotides used in this work
| Oligonucleotide | Sequence (5′ to 3′) |
|---|---|
| EA-A | AGCCCCTTTATCTCTATTATTGTTAATGGTCTTATTATTTGG |
| EA-B | CTCGCTTTATATAATGCTATCTATACGCCTCATAAGG |
| EA-C | AAAAGTGCTTTTTATGAGTCAAG |
| EA-D | TTTCTATTTTTAAAATTAATCTTTAGG |
| EA-E | TGCTAAAGCTCCAGAAATTAGGGG |
| EA-F | TAGCTTTTTCTCCATATTTTTCGG |
| EA-G | CTCCCCCCTAATTTCTGGAACACG |
| EA-H | TATGGAGAAGAGGCTAGGGAAAAG |
| 1 | GAACAAGGATTAATTCTTAAGTGG |
| 2 | GAACTTATAAAAACCTATTCTCTA |
| 3 | ACTATATTAAGTGTGTAGTAATAG |
| 4 | AATTTTGAGATTATTTGTGTATTG |
To answer the puzzle of why only one primer pair gave a PCR product from B31-RML, we cloned its EA-A–EA-B amplicon into the TA vector pCR2.1 (Invitrogen, San Diego, Calif.) and sequenced the insert. The identified sequence did not precisely match any in the Institute for Genomic Research B. burgdorferi B31 database (http://www.tigr.org/tdb/CMR/gbb /htmls/SeqSearch.html) and did not contain sequences complementary to oligonucleotides EA-C and EA-D. The B31-RML amplicon sequence contained an open reading frame that shared 72% identical nucleotides with the B31-MI eppA gene and encoded a protein predicted to share 63% identical amino acids with B31-MI EppA (Fig. 1). The DNA sequence flanking the B31-RML open reading frame was very similar to that flanking B31-MI eppA, indicating that a second B31 allele of eppA had been identified. We designate the previously reported B31 allele (11, 15) eppA1 and the new allele eppA2. Since PCR of B31-4a with EA-A and EA-B did not yield a product, it was concluded that this clonal culture did not contain eppA2.
FIG. 1.
Comparisons of eppA1 and eppA2 and their predicted proteins. (A) Nucleotides common to both eppA alleles are boxed and shaded. Predicted start and stop codons of each gene are indicated by boldface and asterisks. Additional DNA flanking eppA1 that corresponds with oligonucleotides EA-A and EA-B is also shown. (B) Amino acids predicted to be contained in both EppA1 and EppA2 are boxed and shaded.
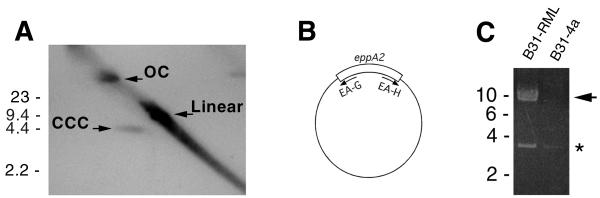
bapA, another B31 gene with similarity to eppA1, has been mapped to a 32-kb circular plasmid (42), and so we sought to identify the genomic location of eppA2. Two-dimensional Southern blotting was utilized to address whether eppA2 is located on a circular or a linear plasmid. B31-RML plasmid DNAs were electrophoresed at 1.3 V/cm for 12 h in a 0.35% agarose gel, whereupon the gel was rotated 90°, soaked in 15 μM chloroquine, and again electrophoresed for 12 h. Chloroquine has different effects on the supercoiling of linear and covalently closed circular DNAs, and so these two forms are readily distinguishable in two-dimensional gels (28). Different forms of a circular DNA species can typically be visualized on such two-dimensional gels, with linearized and open circular forms appearing on an axis, while the supercoiled form migrates off that axis. Linear plasmids, however, will resolve as a single band (14, 28, 39, 42). Separated DNAs were transferred to a Biotrans nylon membrane (ICN, Irvine, Calif.) and cross-linked in a Stratalinker 1800 (Stratagene, La Jolla, Calif.). A DNA probe was synthesized from the eppA2-containing recombinant plasmid by two rounds of PCR, using oligonucleotides EA-E and EA-F (Fig. 1 and Table 1) and conditions as previously described (42). The eppA2 probe was labeled with [α-32P]dATP (ICN) by random priming (Life Technologies, Gaithersburg, Md.) and hybridized with the nylon membrane overnight at 45°C, as previously described (22). The membrane was then washed at 55°C in 0.2× SSC (1× SSC is 0.15 M NaCl, 0.015 M sodium citrate, pH 7.0)–0.1% (wt/vol) sodium dodecyl sulfate at 55°C, and hybridized probe was visualized by autoradiography. The eppA2 probe gave a two-dimensional Southern blot pattern consistent with a circular plasmid (Fig. 2A), including hybridization with an approximately 9-kb linear DNA species that was presumably plasmid linearized by shearing during preparation.
FIG. 2.
(A) Two-dimensional Southern blotting of B31-RML plasmids hybridized with an eppA2-derived probe. Three forms of cp9-2 are evident and marked as CCC (covalently closed circular), OC (open circular), and Linear. (B) Diagram of cp9-2 indicating the locations of sequences complementary to oligonucleotides EA-G and EA-H, with 5′-to-3′ directions indicated by arrows. (C) Results of long-range PCR of B31-RML and B31-4a. The approximately 9-kb PCR product of cp9-2 is indicated by an arrow. A smaller PCR product was obtained from both cultures and is marked with an asterisk. Positions of DNA size standards (in kilobases) are indicated to the left of panels A and C.
The circular nature and size of the eppA2-containing plasmid were further addressed by PCR, using oligonucleotide primers directed away from eppA2 (Fig. 2B). This technique can amplify the remainder of a circular plasmid, a method used previously to confirm circularity of other borrelial plasmids (39, 40). B31-RML and B31-4a DNAs were subjected to PCR using oligonucleotides EA-G and EA-H (Fig. 2C and Table 1) with an Expand amplification system (Boehringer Mannheim, Indianapolis, Ind.). PCR conditions consisted of 10 cycles of 94°C for 10 s, 50°C for 30 s, and 68°C for 10 min, followed by 20 cycles with extension steps increasing by 20 s each cycle, as recommended by the manufacturer. Reaction products were separated by agarose gel electrophoresis and visualized with ethidium bromide, revealing the production of an approximately 9-kb amplicon from B31-RML, but not from B31-4a (Fig. 2C). A smaller, minor DNA product was also produced from both B31-RML and B31-4a (Fig. 2C), which was not a product of eppA2 extension, since B31-4a lacks this gene (see above).
Both two-dimensional Southern blotting and long-range PCR indicated that eppA2 is located on a circular plasmid having a size of approximately 9 kb, a size similar to that of the plasmid carrying eppA1 (11, 15). We therefore designate the eppA2-containing plasmid cp9-2 and rename the originally identified eppA1-carrying plasmid cp9-1.
Since some B. burgdorferi plasmids are known to encode virulence-associated proteins, it is of interest to identify which plasmids are essential for bacterial infectivity and which are dispensable. While an efficient method to analyze the plasmid content of infectious B31 cultures is by PCR with oligonucleotide primers derived from B31-MI plasmid sequences, a significant drawback to this approach was illustrated by our studies. PCR using oligonucleotide primers derived from the cp9-1 sequence can fail to amplify DNA from B31 cultures containing only cp9-2, which would lead one to incorrectly conclude that such bacteria lack an eppA-encoding plasmid. It is important for researchers to acknowledge that the reported sequence of the B31-MI genome does not represent all DNA species that can be carried by bacteria in B31 cultures and that plasmid content analyses of B31 cultures should not be based solely upon the sequences found in B31-MI.
To examine whether additional sequence similarities exist between cp9-1 and cp9-2, four oligonucleotide primers complementary to cp9-1 sequences were constructed: 1 and 2, flanking rev, and 3 and 4, flanking the paralog family 32 (ORF-3) gene (Table 1). PCR of B31-RML DNA failed to yield products with any combination of these primers. Likewise, no amplicons were produced from B31-RML DNA by PCR using cp9-1 primers paired with eppA2-specific primers. Control PCR of B31-MI with cp9-1 primer pairs yielded appropriately sized amplicons, as anticipated. We conclude that cp9-2 does not contain precise paralogs of the cp9-1 sequences comprising oligonucleotide 1, 2, 3, or 4.
Restriction mapping experiments also indicated differences between cp9-1 and cp9-2. The cp9-2 amplicon obtained with oligonucleotides EA-G and EA-H was digested with restriction endonucleases ApaLI, Bsu36I, EcoRI, EcoRV, and EcoO109 (New England Biolabs, Beverly, Mass.), enzymes that are predicted to cut cp9-1 (GenBank accession no. AE000791). Digested cp9-2 amplicons were separated by agarose gel electrophoresis and stained with ethidium bromide, and restriction fragment sizes were compared with those predicted for the non-eppA1 portion of cp9-1. cp9-2 contains single ApaLI, Bsu36I, and EcoRI sites, as does cp9-1, and digested fragments were of sizes similar to those predicted for cp9-1, indications of sequences held in common by both plasmids. In contrast, cp9-1 is predicted to contain two sites each for EcoRV and EcoO109, while the cp9-2 amplicon was digested only once by EcoO109, and not at all by EcoRV (data not shown).
As noted above, B31-4a, a clonal derivative of B31-RML, lacks eppA2 and, therefore, cp9-2. Experiments were performed to examine the frequency of cp9-2 in other clonal cultures derived from B31-RML. A mid-logarithmic-phase culture of B31-RML (approximately 107 bacteria/ml) was plated in solid medium, as previously described (19, 26). Visible colonies appeared after incubation for approximately 1 week at 37°C in a 5% CO2 atmosphere. Ten colonies were each inoculated into liquid medium, and bacteria were grown to mid-logarithmic phase. Bacteria from each culture were pelleted by centrifugation, washed twice with phosphate-buffered saline, resuspended in water, and lysed by boiling. An aliquot of each lysate was subjected to PCR with oligonucleotides EA-A and EA-B, using PCR conditions as described above. No amplicons were obtained from any of the 10 clonal cultures, while a product was obtained from a control PCR using uncloned B31-RML (data not shown). Since 11 separate clones of B31-RML lack cp9-2 (the 10 analyzed here plus B31-4a), it appears that a majority of bacteria in this infectious culture do not contain that plasmid. These data also indicate that bacteria lacking both cp9-1 and cp9-2 can be infectious, suggesting that neither of these plasmids contains genes essential for mammalian infection. Other researchers also found that B31 cultures lacking a 9-kb circular plasmid (it is unknown whether this was cp9-1 or cp9-2) could still infect hamsters, although the bacteria lacking such a plasmid were less infectious than those containing the plasmid (47). We are not aware of any studies that have compared the relative abilities of B31-MI, B31-RML, and B31-4a to infect animals. It is possible that the cp9s are somewhat redundant, since all the identified open reading frames on cp9-1 and cp9-2 have paralogs on other B31 plasmids that might compensate for loss of the 9-kb circular plasmids (9). For example, the B31 cp32-3 encodes BapA (42), which is homologous to EppA1 and EppA2 (45), and thus may perform functions similar to those proteins.
The similar sizes and presence of eppA genes on both cp9-1 and cp9-2 suggest relatedness between these two plasmids. Likewise, restriction mapping with ApaLI, Bsu36I, and EcoRI indicated the presence of other homologous DNA sequences on the two plasmids. However, EcoRV and EcoO109 restriction mapping and PCR analyses indicated that cp9-1 and cp9-2 contain many unique sequences, suggesting that cp9-2 is not simply cp9-1 with a variant eppA locus. The two identified B31 cp9s are similar in this regard to the cp32 plasmid family, whose members contain regions of highly conserved sequences interspersed with divergent sequences (9, 43). Perhaps not coincidentally, analysis of the cp9-1 DNA sequence indicated that it evolved from a cp32 (9). Clonal B. burgdorferi cultures that contain multiple different cp32s have been identified, indicating that many of those plasmids are compatible with each other (1, 10, 13, 42). We have yet to identify a clonal B31 culture that contains both cp9-1 and cp9-2, and so it is unknown whether the two cp9s are likewise compatible. We note, however, that B31 was reported as clonal when first described (3), and so the ancestral B31 organism presumably contained both cp9-1 and cp9-2.
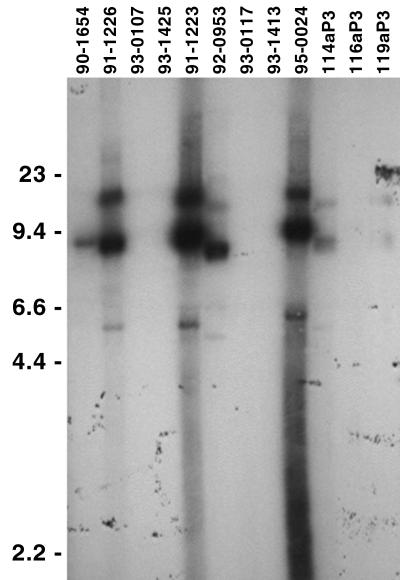
Circular plasmids of approximately 9 kb have also been observed for other B. burgdorferi isolates, although the presence of eppA genes on such plasmids is unknown (8, 10, 12, 16, 29, 32–34, 37, 48). To address that question, we used Southern blot analyses to examine 16 additional B. burgdorferi isolates, all cultured from human Lyme disease patients (22) (provided by M. Schriefer and K. Davis, Centers for Disease Control and Prevention, Ft. Collins, Colo.). Total genomic DNA was purified (22), separated by agarose gel electrophoresis, transferred to nylon membranes, incubated at 45°C overnight with the B31 eppA2-derived probe, and washed at 45°C in 2× SSC–0.1% sodium dodecyl sulfate. Autoradiography revealed that 7 of the 16 human isolates contained DNA that hybridized with the B31 eppA2 probe, all of which had the appearance of being circular DNAs of approximately 9 kb in size (Fig. 3 and data not shown). Southern blot signal strengths ranged from very intense (e.g., isolate 91-1223) to barely detectable (e.g., isolate 119aP3), which may indicate differences in sequences from that of B31 eppA2 or may be due to only a small fraction of the bacteria in some isolates carrying an eppA-containing plasmid (similar to B31-RML). The other nine examined isolates may have lacked cp9s or lost them during laboratory cultivation, as apparently happens frequently with the B31 cp9s (47). Alternatively, these other isolates may contain plasmids related to the B31 cp9-1 and cp9-2 but which lack an eppA locus, similar to the cp8.3 of isolate Ip90 (12). The widespread occurrence of eppA genes and 9-kb circular plasmids suggests that they contribute to the survival of B. burgdorferi in nature, and possibly to the pathogenesis of Lyme disease.
FIG. 3.
Southern blot analysis of B. burgdorferi isolated from human Lyme disease patients (22) using a B31 eppA2-derived probe. Positions of DNA size standards (in kilobases) are indicated to the left.
Nucleotide sequence accession number.
The sequence of B31 eppA2 has been deposited in GenBank and given accession no. AF213472.
Acknowledgments
We gratefully acknowledge support from the National Institutes of Health grant AI44254 and University of Kentucky Medical Center Research Fund grant 949 to B. Stevenson, and a Kentucky Opportunity Fellowship for Academic Excellence to J. Miller.
We thank M. Schriefer and K. Davis for providing B. burgdorferi isolates.
REFERENCES
- 1.Akins D R, Caimano M J, Yang X, Cerna F, Norgard M V, Radolf J D. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect Immun. 1999;67:1526–1532. doi: 10.1128/iai.67.3.1526-1532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbour A G. Plasmid analysis of Borrelia burgdorferi, the Lyme disease agent. J Clin Microbiol. 1988;26:475–478. doi: 10.1128/jcm.26.3.475-478.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbour A G, Burgdorfer W, Hayes S F, Peter O, Aeschlimann A. Isolation of a cultivable spirochete from Ixodes ricinus ticks of Switzerland. Curr Microbiol. 1983;8:123–126. [Google Scholar]
- 4.Barbour A G, Garon C F. The genes encoding major surface proteins of Borrelia burgdorferi are located on a plasmid. In: Benach J L, Bosler E M, editors. Lyme disease and related disorders. New York, N.Y: New York Academy of Sciences; 1988. pp. 144–153. [DOI] [PubMed] [Google Scholar]
- 5.Bergström S, Garon C F, Barbour A G, MacDougall J. Extrachromosomal elements of spirochetes. Res Microbiol. 1992;143:623–628. doi: 10.1016/0923-2508(92)90120-d. [DOI] [PubMed] [Google Scholar]
- 6.Bono J L, Tilly K, Stevenson B, Hogan D, Rosa P. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology. 1998;144:1033–1044. doi: 10.1099/00221287-144-4-1033. [DOI] [PubMed] [Google Scholar]
- 7.Burgdorfer W, Barbour A G, Hayes S F, Benach J L, Grunwaldt E, Davis J P. Lyme disease—a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 8.Carlyon J A, LaVoie C, Sung S-Y, Marconi R T. Analysis of the organization of multicopy linear- and circular-plasmid-carried open reading frames in Borrelia burgdorferi sensu lato isolates. Infect Immun. 1998;66:1149–1158. doi: 10.1128/iai.66.3.1149-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casjens S, Palmer N, van Vugt R, Huang W M, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson R J, Haft D, Hickey E, Gwinn M, White O, Fraser C. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516. doi: 10.1046/j.1365-2958.2000.01698.x. [DOI] [PubMed] [Google Scholar]
- 10.Casjens S, van Vugt R, Tilly K, Rosa P A, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J Bacteriol. 1997;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Champion C I, Blanco D R, Skare J T, Haake D A, Giladi M, Foley D, Miller J N, Lovett M A. A 9.0-kilobase-pair circular plasmid of Borrelia burgdorferi encodes an exported protein: evidence for expression only during infection. Infect Immun. 1994;62:2653–2661. doi: 10.1128/iai.62.7.2653-2661.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn J J, Buchstein S R, Butler L-L, Fisenne S, Polin D S, Lade B N, Luft B J. Complete nucleotide sequence of a circular plasmid from the Lyme disease spirochete, Borrelia burgdorferi. J Bacteriol. 1994;176:2706–2717. doi: 10.1128/jb.176.9.2706-2717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Hage N, Lieto L D, Stevenson B. Stability of erp loci during Borrelia burgdorferi infection: recombination is not required for chronic infection of immunocompetent mice. Infect Immun. 1999;67:3146–3150. doi: 10.1128/iai.67.6.3146-3150.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng S, Hodzic E, Stevenson B, Barthold S W. Humoral immunity to Borrelia burgdorferi N40 decorin binding proteins during infection of laboratory mice. Infect Immun. 1998;66:2827–2835. doi: 10.1128/iai.66.6.2827-2835.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidmann J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 16.Hyde F W, Johnson R C. Characterization of a circular plasmid from Borrelia burgdorferi, etiologic agent of Lyme disease. J Clin Microbiol. 1988;26:2203–2205. doi: 10.1128/jcm.26.10.2203-2205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson R C, Marek N, Kodner C. Infection of Syrian hamsters with Lyme disease spirochetes. J Clin Microbiol. 1984;20:1099–1101. doi: 10.1128/jcm.20.6.1099-1101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson R C, Schmid G P, Hyde F W, Steigerwalt A G, Brenner D J. Borrelia burgdorferi sp. nov.: etiologic agent of Lyme disease. Int J Syst Bacteriol. 1984;34:496–497. [Google Scholar]
- 19.Kurtti T J, Munderloh U G, Johnson R C, Ahlstrand G G. Colony formation and morphology in Borrelia burgdorferi. J Clin Microbiol. 1987;25:2054–2058. doi: 10.1128/jcm.25.11.2054-2058.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lam T T, Nguyen T-P K, Montgomery R R, Kantor F S, Fikrig E, Flavell R A. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect Immun. 1994;62:290–298. doi: 10.1128/iai.62.1.290-298.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marconi R T, Samuels D S, Garon C F. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J Bacteriol. 1993;175:926–932. doi: 10.1128/jb.175.4.926-932.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J C, El-Hage N, Babb K, Stevenson B. Borrelia burgdorferi B31 Erp proteins that are dominant immunoblot antigens of animals infected with isolate B31 are recognized by only a subset of human Lyme disease patient sera. J Clin Microbiol. 2000;38:1569–1574. doi: 10.1128/jcm.38.4.1569-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norris S J, Carter C J, Howell J K, Barbour A G. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect Immun. 1992;60:4662–4672. doi: 10.1128/iai.60.11.4662-4672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norris S J, Howell J K, Garza S A, Ferdows M S, Barbour A G. High- and low-infectivity phenotypes of clonal populations of in vitro-cultured Borrelia burgdorferi. Infect Immun. 1995;63:2206–2212. doi: 10.1128/iai.63.6.2206-2212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer N, Fraser C, Casjens S. Distribution of twelve linear extrachromosomal DNAs in natural isolates of the Lyme disease spirochetes. J Bacteriol. 2000;182:2476–2480. doi: 10.1128/jb.182.9.2476-2480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosa P, Samuels D S, Hogan D, Stevenson B, Casjens S, Tilly K. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J Bacteriol. 1996;178:5946–5953. doi: 10.1128/jb.178.20.5946-5953.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadziene A, Wilske B, Ferdows M S, Barbour A G. The cryptic ospC gene of Borrelia burgdorferi B31 is located on a circular plasmid. Infect Immun. 1993;61:2192–2195. doi: 10.1128/iai.61.5.2192-2195.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samuels D S, Garon C F. Coumermycin A1 inhibits growth and induces relaxation of supercoiled plasmids in Borrelia burgdorferi, the Lyme disease agent. Antimicrob Agents Chemother. 1993;37:46–50. doi: 10.1128/aac.37.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwan T G, Burgdorfer W, Garon C F. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect Immun. 1988;56:1831–1836. doi: 10.1128/iai.56.8.1831-1836.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwan T G, Piesman J. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J Clin Microbiol. 2000;38:382–388. doi: 10.1128/jcm.38.1.382-388.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwan T G, Schrumpf M E, Karstens R H, Clover J R, Wong J, Daugherty M, Struthers M, Rosa P A. Distribution and molecular analysis of Lyme disease spirochetes, Borrelia burgdorferi, isolated from ticks throughout California. J Clin Microbiol. 1993;31:3096–3108. doi: 10.1128/jcm.31.12.3096-3108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson W J, Garon C F, Schwan T G. Analysis of supercoiled circular plasmids in infectious and non-infectious Borrelia burgdorferi. Microb Pathog. 1990;8:109–118. doi: 10.1016/0882-4010(90)90075-2. [DOI] [PubMed] [Google Scholar]
- 34.Simpson W J, Garon C F, Schwan T G. Borrelia burgdorferi contains repeated DNA sequences that are species specific and plasmid associated. Infect Immun. 1990;58:847–853. doi: 10.1128/iai.58.4.847-853.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skare J T, Foley D M, Hernandez S R, Moore D C, Blanco D R, Miller J N, Lovett M A. Cloning and molecular characterization of plasmid-encoded antigens of Borrelia burgdorferi. Infect Immun. 1999;67:4407–4417. doi: 10.1128/iai.67.9.4407-4417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skare J T, Shang E S, Foley D M, Blanco D R, Champion C I, Mirzabekov T, Sokolov Y, Kagan B L, Miller J N, Lovett M A. Virulent strain associated outer membrane proteins of Borrelia burgdorferi. J Clin Investig. 1995;96:2380–2392. doi: 10.1172/JCI118295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stålhammar-Carlemalm M, Jenny E, Gern L, Aeschlimann A, Meyer J. Plasmid analysis and restriction fragment length polymorphisms of chromosomal DNA allow a distinction between Borrelia burgdorferi strains. Zentbl Bakteriol. 1990;274:28–39. doi: 10.1016/s0934-8840(11)80972-2. [DOI] [PubMed] [Google Scholar]
- 38.Stevenson B, Bono J L, Schwan T G, Rosa P. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect Immun. 1998;66:2648–2654. doi: 10.1128/iai.66.6.2648-2654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevenson B, Casjens S, van Vugt R, Porcella S F, Tilly K, Bono J L, Rosa P. Characterization of cp18, a naturally truncated member of the cp32 family of Borrelia burgdorferi plasmids. J Bacteriol. 1997;179:4285–4291. doi: 10.1128/jb.179.13.4285-4291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevenson B, Porcella S F, Oie K L, Fitzpatrick C A, Raffel S J, Lubke L, Schrumpf M E, Schwan T G. The relapsing fever spirochete Borrelia hermsii contains multiple, antigen-encoding circular plasmids that are homologous to the cp32 plasmids of Lyme disease spirochetes. Infect Immun. 2000;68:3900–3908. doi: 10.1128/iai.68.7.3900-3908.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevenson B, Schwan T G, Rosa P A. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevenson B, Tilly K, Rosa P A. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J Bacteriol. 1996;178:3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevenson, B., W. R. Zückert, and D. R. Akins. Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species. J. Mol. Microbiol. Biotechnol., in press. [PubMed]
- 44.Tilly K, Casjens S, Stevenson B, Bono J L, Samuels D S, Hogan D, Rosa P. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol Microbiol. 1997;25:361–373. doi: 10.1046/j.1365-2958.1997.4711838.x. [DOI] [PubMed] [Google Scholar]
- 45.Wallich R, Brenner C, Kramer M D, Simon M M. Molecular cloning and immunological characterization of a novel linear-plasmid-encoded gene, pG, of Borrelia burgdorferi expressed only in vivo. Infect Immun. 1995;63:3327–3335. doi: 10.1128/iai.63.9.3327-3335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y, Johnson R C. Analysis and comparison of plasmid profiles of Borrelia burgdorferi sensu lato strains. J Clin Microbiol. 1995;33:2679–2685. doi: 10.1128/jcm.33.10.2679-2685.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu Y, Kodner C, Coleman L, Johnson R C. Correlation of plasmids with infectivity of Borrelia burgdorferi sensu stricto type strain B31. Infect Immun. 1996;64:3870–3876. doi: 10.1128/iai.64.9.3870-3876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zückert W R, Meyer J. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J Bacteriol. 1996;178:2287–2298. doi: 10.1128/jb.178.8.2287-2298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]