Abstract
Background
Crohn's disease (CD) is a chronic non-specific inflammatory bowel disease. Current CD therapeutics cannot fundamentally change the natural course of CD. Therefore, it is of great significance to find new treatment strategies for CD. Preclinical and clinical studies have shown that mesenchymal stromal cells (MSCs) are a promising therapeutic approach. However, the mechanism by which MSCs alleviate CD and how MSCs affect gut microbes are still unclear and need further elucidation.
Methods
We used 2,4,6-trinitrobenzenesulfonic acid (TNBS) to induce experimental colitis in mice and analysed the microbiota in faecal samples from the control group, the TNBS group and the TNBS + MSC group with faecal 16S rDNA sequencing. Subsequent analyses of alpha and beta diversity were all performed based on the rarified data. PICRUStII analysis was performed on the 16S rRNA gene sequences to infer the gut microbiome functions.
Results
MSC Treatment improved TNBS-induced colitis by increasing survival rates and relieving symptoms. A distinct bacterial signature was found in the TNBS group that differed from the TNBS + MSC group and controls. MSCs prevented gut microbiota dysbiosis, including increasing α-diversity and the amount of Bacteroidetes Firmicutes and Tenericutes at the phylum level and decreasing the amount of Proteobacteria at the phylum level. MSCs alleviated the increased activities of sulphur and riboflavin metabolism. Meanwhile some metabolic pathways such as biosynthesis of amino acids lysine biosynthesis sphingolipid metabolism and secondary bile acid biosynthesis were decreased in the TNBS group compared with the control group and the TNBS + MSC group
Conclusions
Overall, our findings preliminarily confirmed that colitis in mice is closely related to microbial and metabolic dysbiosis. MSC treatment could modulate the dysregulated metabolism pathways in mice with colitis, restoring the abnormal microbiota function to that of the normal control group. This study provides insight into specific intestinal microbiota and metabolism pathways linked with MSC treatment, suggesting a new approach to the treatment of CD.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13287-022-03118-1.
Keywords: Mesenchymal stem cells, Crohn's disease, TNBS, 16S rRNA gene sequences, Gut microbiota, Metabolism
Introduction
Crohn’s disease (CD) is a subtype of inflammatory bowel disease (IBD) characterized by chronic recurrent colonic mucosal inflammation. In CD, inflammation can be found throughout the gastrointestinal tract and it often extends deeper into the colonic tissue layer. IBD has become a global disease with an increasing incidence [1]. Developing countries are in the emergence stage, newly industrialized countries are in the acceleration of incidence stage, and western regions are in the compounding prevalence stage [2]. In western countries, although the incidence is stabilizing, the disease burden remains high, as the prevalence surpasses 0.3% [3].
Treatment for CD includes monoclonal antibody therapies, immunomodulators, and surgery, but the recurrence rate is high, and there is no effective cure [4]. Despite advances in IBD treatment, 30% of patients do not respond to current treatments, and 50% develop allergic reactions or become refractory over time [5]. IBD is a very complex and costly disease, and these data highlight the need for prevention research and health care system innovations.
In the last decade, MSCs have emerged as innovative and promising therapeutic options for many incurable diseases [6–8]. Further interest in MSCs has been attracted by the observation that they exhibit profound effects due to attractive features of plasticity, tropism for inflamed tissues, and high immunomodulatory potential. MSCs are nonhemopoietic cells capable of self-renewal and secreting antibacterial peptides and cytokines [9, 10]. In theory, they can be used as a potential treatment for CD. Clinical studies have suggested the potential benefits of MSC treatment for CD based on their immune regulatory functions [11, 12].
As it shares many features with human CD, the 2,4,6-trinitrobenzenesulfonic acid (TNBS)-induced colitis animal model has been used to study the mechanism and treatment of human CD. Preclinical animal experiments have confirmed that MSCs could significantly improve clinical abnormalities in TNBS-induced colitis mice and reduce lesion scores and inflammation in the gut [13]. However, the mechanism by which MSCs alleviate CD is still unclear and it needs further elucidation.
It is likely that CD is caused by a genetic predisposition combined with environmental triggers that shape the microbiome, such as diet and antibiotic exposure. It is caused by continuous inflammation in response to endogenous microbes in genetically predisposed individuals [14, 15]. The human gut microbiota, composed of trillions of individual microbes, has adapted to the uniquely diverse environments found in the intestine. Gut microbes are an essential part of the microbiota ecosystem. They outnumber human cells by tenfold [16]. In recent years, with the continuous progress of the theoretical research of gut microbiota and the gradual maturity of molecular technology, the relationship between CD and the gut microbiota is slowly being revealed. Jason lloyd-Price et al. [17] provided a comprehensive view of functional dysbiosis in the gut microbiome during IBD activity. The involvement of the gut microbiota in the maintenance of the gut ecosystem is twofold: it educates host immune cells and protects the host from pathogens. However, when the healthy microbial composition and function are disrupted (dysbiosis), the dysbiotic gut microbiota can trigger the initiation and development of various gastrointestinal diseases, including IBD [18].
Experiments on enteric microbiota from IBD patients have revealed that a decreased abundance of the Firmicutes and Bacteroidetes phyla with an increased abundance of Proteobacteria is significantly correlated with the severity of IBD, and genera of the Firmicutes phylum are reduced in IBD patients [19, 20]. Meanwhile, a network of bacteria-metabolite interactions has shown some mechanisms linked to disease activity in Crohn’s disease [21]. Thousands of candidate microbial proteins are likely to interact with the host immune system in IBD [22]. Researchers have found that MSC therapy may be associated with the gut microbiota in many diseases, including ulcerative colitis [23], acute liver injury [24], hypoxia-induced pulmonary hypertension [25], chronic hypoxia [26], and diabetes [27], which leads to gut dysbiosis. Whether the molecular mechanism of MSC therapy for CD is associated with the gut microbiota remains unknown.
Therefore, we hypothesised that treatment of colitis with MSCs could restore the microbiota composition and function in the TNBS-induced colitis model and reduce inflammation to a normal state. Then, we investigated the effects of MSC treatment on host disease status, gut microbiome composition and function using the 16S rRNA gene sequencing method in TNBS-induced colitis to evaluate how the gut microbiome responds to MSC treatment and contributes to colonic inflammatory etiopathogenesis.
Materials and methods
Mice
BALB/c mice were purchased from the Model Animal Research Center, Nanjing University (Nanjing, Jiangsu, China). All animals used for the in vivo studies were 8-week-old males. BALB/c mice were randomly allocated to each group. All animal protocols were reviewed and approved by the Sun Yat-sen University Institutional Animal Care and Use Committee.
Isolation and culture of MSCs
MSCs were isolated and expanded from human umbilical cords according to previously reported protocol [28]. Fresh human umbilical cords were obtained from newborns with parental consent and placed in PBS at 4 °C. The cord was washed twice with PBS to remove any remaining blood. The rinsed cords were cut into 10 mm3 pieces and placed in type I collagenase with hyaluronidase (0.1%) containing CaC12 (3 mM) for digestion at 37 °C for 4 h. The specimens were transferred to DMEM (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% foetal bovine serum (Pan Biotech, Aidenbach, Germany) at 37 °C in a humidified atmosphere with 5% CO2. The medium in the primary culture was changed 3 days later, and nonadherent cells were removed. After that the culture medium was refreshed every 4 days. Following the appearance of colonies of fibroblast-like cells after 14 days, the cells were trypsinized and transferred to a new flask for further expansion. We incubated the MSCs with 0.05% trypsin–EDTA(GIBCO, Invitrogen Inc., Carlsbad, CA, USA) at 37 °C and stored them at − 80 °C until analysis. For the collection of umbilical cords, the Human Ethics Committee of the Third Affiliated Hospital at Sun Yat-sen University approved this project. Written informed consent was obtained from all participants.
Flow cytometry
The characteristics of MSCs were identified by flow cytometry and the methods for identifying MSCs were described as previous methods [29]. CytoFLEX flow cytometers (Beckman Coulter) were used for flow cytometry, and CytoExpert software (Beckman Coulter) was used for data analysis. Anti-human CD90-FITC (Catalogue #IM1839U), anti-human CD19-FITC(Catalogue #A07768), anti-human CD11b-FITC(Catalogue #IM0530), anti-human HLA-DR-FITC(Catalogue #IM1638U), anti-human CD34-FITC (Catalogue #IM1870), anti-human CD45-FITC(Catalogue #AO7782), CD105-PE(Catalogue #B76299), and anti-human CD73-PE (Catalogue #B76299) (BECKMAN COULTER, Brea, CA, USA)were purchased from Beckman Coulter.
Differentiation assays
For osteogenic and adipogenic differentiation procedure of hUC-MSCs was described as previous method [30]. hUC-MSCs were seeded into 24-well plates for osteogenic differentiation and cultured for 12 h at a density of 6 × 104 cells per well. In the following days, the medium was changed to osteogenic differentiation medium (Biological Industries, Kibbutz Beit-Haemek, Israel) and it was refreshed every 3 days. Alizarin red solution (Biological Industries, Kibbutz Beit-Haemek, Israel) was used to stain the induced cells.
To differentiate hUC-MSCs into adipocytes, we seeded them in 24-well plates and cultured them for 12 h at a density of 6 × 104 cells per well. Following that the medium was replaced by adipogenic differentiation medium (Biological Industries, Kibbutz Beit-Haemek, Israel) for 21 days. The medium was then refreshed every 3 days. Oil Red O staining was used on the induced cells.
Experimental colitis induced by TNBS
For the colitis experiment, the backs of 8-week-old male BALB/c mice were smeared with 150 μl of presensitization solution (Sigma, St. Louis, MO, USA) 7 days before colitis was induced. We divided the mice into groups and fasted them (but still allowed them to drink freely) for 24 h. Our previous study described the procedures for inducing colitis and treating it with MSCs [31]. The parameters of body weight loss, diarrhoea, and survival were recorded daily for 7 days. Three days after TNBS injection (the peak of the disease), the length of the colon was measured from the caecum to the anus. The colons were examined for macroscopic damage based on the protocols described in our previous investigation [31].
Mesenchymal stem cell transplantation
MSC treatment was performed as described in a previous study [32]. The control mice received only 50% ethanol. MSCs were used for cell transplantation at passages 3–5. After instilling TNBS, the BALB/c mice were either intraperitoneally treated with the control medium (saline) or with 106 MSCs per mouse 2 h later.
Assessment of colitis severity
The animals were monitored for body weight loss, stool consistency, the presence of blood on the anus or in the stool and survival every day for a total of 7 days. The baseline data were collected before the instillation of TNBS. Disease activity and histologic scores were evaluated as previously described [33]. For disease activity, a scoring system incorporating the percentage of weight loss, stool consistency, and faecal occult blood test results was used [34, 35]. For histopathology analysis, a colon specimen from the middle part (1 cm from the anus to the caecum) was fixed in 10% buffered formalin phosphate and then embedded in paraffin. Sections were stained with haematoxylin and eosin, and inflammation was graded from 0 to 4 as follows, in a blinded fashion: 0, no signs of inflammation; 1, low leukocyte infiltration; 2, moderate leukocyte infiltration; 3, high leukocyte filtration, moderate fibrosis, high vascular density, thickening of the colon wall, moderate goblet cell loss and focal loss of crypts; and 4, transmural infiltrations, massive loss of goblet cells, extensive fibrosis, and diffuse loss of crypts.
Immunohistochemical staining
Following deparaffinization and rehydration, the tissue sections were placed in a repair box filled with citric acid (pH 6.0) antigen retrieval buffer and heated in a microwave oven to allow antigen retrieval. Then, the slides were transferred into an antigen retrieval solution and incubated with a primary antibody overnight at 4 °C. After incubation with a secondary antibody at room temperature for 1 h, 3,3-diaminobenzidine (Sigma, St Louis, MO) was used to develop the signal. After counterstaining with haematoxylin, optical microscopy was used to examine the sections. The primary antibodies included rabbit anti-human zonula occludens-1 (ZO-1, 1: 150) and rabbit anti-human occludin (1: 100). The antibodies were purchased from Wuhan Servicebio Technology Co., Ltd. The stained sections were read under a microscope and quantified using Image-Pro Plus (IPP) 6.0 software [36]. The quantified results of occludin and ZO-1 expression are presented as the mean density of 5 randomly selected fields from each group.
Collection and DNA extraction of faecal samples
The faeces of the mice were collected in sterile tubes and stored at – 80 °C until use. Microbial community genomic DNA was extracted from the samples using the E.Z.N.A.® soil DNA Kit(Omega Bio-Tek, Norcross, GA, USA) according to the manufacturer’s instructions. The concentration and purity of the DNA samples were determined using a NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE).
DNA extraction and PCR amplification
The hypervariable region V3-V4 of the bacterial 16S rRNA gene was amplified with the primer pairs 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R(5′-GGACTACHVGGGTWTCTAAT-3′) by an ABIGeneAmp®9700 PCR thermocycler (ABI, CA, USA). PCR amplification of the16S rRNA gene was performed as follows:initial denaturation at 95 °C for 3 min, followed by 27 cycles of denaturing at 95 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 45 s, and a single extension at 72 °C for 10 min, ending at 4 °C. The PCR mixtures contained 4 μL of 5 × TransStart FastPfu buffer, 2.5 mM dNTPs 2 μL, 0.8 μL of forward primer (5 μM), 0.8 μL of reverse primer (5 μM), 0.4 μL of TransStart FastPfu DNA Polymerase, 10 ng of template DNA, and ddH2O up to 20 μL. PCRs were performed in triplicate. The PCR product was extracted from a 2% agarose gel, purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) according to the manufacturer’s instructions and quantified using a Quantus™ Fluorometer (Promega, Madison, Wisconsin, USA).
Illumina MiSeq sequencing and processing of sequencing data
The purified amplicons were pooled equimolarly and sequenced on an Illumina MiSeq PE300/NovaSeq PE250 platform (Illumina, San Diego, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The raw reads were deposited into the NCBI Sequence Read Archive (SRA) database.
The raw 16S rRNA gene sequencing reads were demultiplexed, quality-filtered by fastp version 0.20.0 [37] and merged by FLASH version 1.2.7 [38] with the following criteria: (1) the 300 bp reads were truncated at any site receiving an average quality score of < 20 over a 50 bp sliding window, and the truncated reads shorter than 50 bp were discarded. Reads containing ambiguous characters were also discarded; (2) only overlapping sequences longer than 10 bp were assembled according to their overlapped sequence. The maximum mismatch ratio of the overlap region was 0.2. Reads that could not be assembled were discarded. (3) Samples were distinguished according to the barcode and primers, and the sequence direction was adjusted, with exact barcode matching and 2 nucleotide mismatches in primer matching.
Operational taxonomic units (OTUs) with a 97% similarity cut-off [39] were clustered using UPARSE version 7.1, and chimeric sequences were identified and removed. The taxonomy of each OTU representative sequence was analysed by Ribosomal Database Project (RDP) Classifier version 2.2 [40] against the 16S rRNA database (e.g., Silva v138) using a confidence threshold of 0.7.
The observed species, Sob, Shannon and Shannoneven are used to evaluate the complexity of species diversity. By performing linear discriminant analysis effect size (LEfSe), we identified the features contributing to the most variation between the control and treatment groups (linear discriminant analysis, LDA > 3). Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUStII) was further used for genome prediction of microbial communities in this study (Microbiota Sequencing) and then we performed functional categorization according to KEGG Orthology. STAMP3 was used for functional profiling [41].
Statistical analysis
Data in this study were analysed using IBM SPSS 21.0 software. All results are expressed as the mean ± SD. Tukey's post hoc multiple comparison tests were used to compare three groups, while Student's t tests were used for comparisons between only two groups. Analyses and graphs were generated using GraphPad Prism version 5.01. The relative abundance of bacterial groups at the phylum and genus levels between groups was tested by means of one-way nonparametric Analysis of Variance (ANOVA) (Kruskal–Wallis H test). P < 0.05 was considered significant. For the LEfSe test, the nonparametric factorial Kruskal–Wallis (KW) sum-rank test was used to detect significant differences in abundance and to identify the different groups. The Benjamini–Hochberg FDR method was used to correct significant P values associated with microbial clades identified by LEfSe. STAMP was used to assess the significance of P values associated with microbial function using one-way ANOVA followed by Tukey–Kramer post hoc analyses. The multiple comparisons were not corrected.
Results
Identification of hUC-MSC phenotypes
The hUC-MSCs showed a fibroblast-like morphology, expressed certain antigens (CD90, CD105 and CD73, ≥ 95% positive) (Additional file 1: Fig. S1a, g, h), and lacked haematopoietic lineage markers (CD11b, CD34, CD19, HLA-DR and CD45, ≤ 2% positive). The MSCs showed a spindle-shaped and fibroblast-like morphology (Additional file 1: Fig. S1i). After inducing osteogenesis and adipogenesis, mineral accumulation and bone nodule formation were identified by Alizarin red staining (Additional file 1: Fig. S1j), and the hUC-MSCs formed numerous neutral lipid droplets in the cytoplasm, as identified by Oil Red O staining (Additional file 1: Fig. S1k).
hUC-MSCs alleviate TNBS-induced colitis
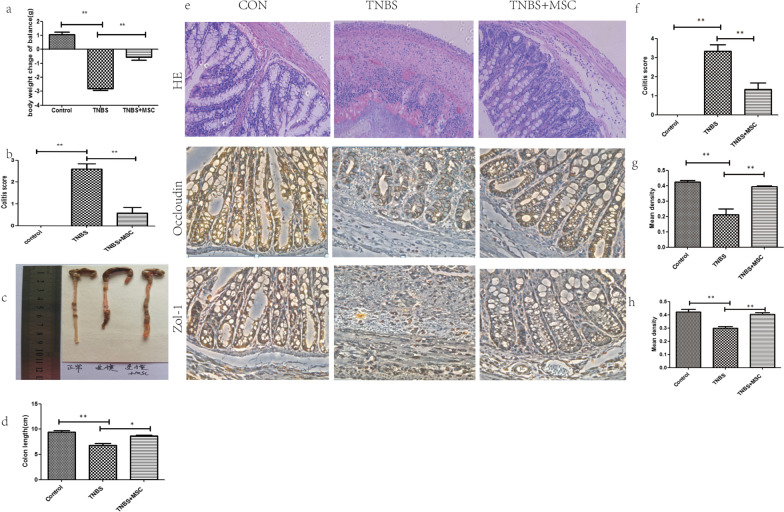
Mice treated with TNBS developed severe symptoms, including bloody diarrhoea, rectal prolapse, pancolitis, and sustained weight loss. Similar to the control mice, hUC-MSC treated mice had rapidly recovered body weight loss and milder inflammation. The disease activity index (DAI) score is an indicator of the severity of colitis and is based on results including weight loss, stool features and faecal occult blood. The DAI score for the TNBS group was significantly increased compared with that of the control group, while the DAI scores following treatment with MSC were significantly reduced compared with those of the TNBS group. (Fig. 1b). The colon tissue of the control group was normal in length, uniform in thickness, transparent and smooth, and light yellowish-white. The colon of the model group was thicker and shorter. Numerous ulcers and erosions were observed 2–4 cm from the anus. The colon injury in the hUC-MSC group was significantly reduced and mostly manifested as scattered congestion and erosion (Fig. 1c).
Fig. 1.
Human umbilical cord-derived mesenchymal stem cell (hUC-MSC) therapy mitigates TNBS-induced colitis. Colitis BALB/c mice were treated with hUC-MSCs (106 per mouse), while the control mice received only control medium (saline) 2 h following rectal administration of 2,4,6-trinitrobenzenesulfonic acid (TNBS). The animals were monitored every day for a total of 7 days for weight loss, the consistency of their stool, the presence of blood in their stool, disease activity and histologic scores, and the expression of mucosal barrier proteins. a Decreased weight loss; b alleviated colitis symptoms; c, d Colons were examined for their general form and length 3 days after TNBS intracolonic administration; e Histopathologic analysis (H&E staining and histological score). Inflammation was graded from 0 to 4; f Expression of the tight junction-related proteins occludin and ZO-1. The stained sections were read under a microscope and quantified using Image-Pro Plus (IPP) 6.0 software. The quantified results of occludin(g) and ZO-1(h) are presented as the mean density. The mean densities of 5 randomly selected fields in each group. Data are shown as the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and n.s. means not significant
Histopathological staining was used to evaluate the histological characteristics of the colon samples. The results indicated that mice maintained an integrated normal colonic structure in the control group, but mice with TNBS-induced colitis exhibited significant inflammatory cell infiltration, loss of crypts, destruction of the mucosal layer, and oedema (Fig. 1e). In contrast, TNBS-induced colitis in mice treated with hUC-MSCs exhibited mild inflammation. ZO-1 and occludin are important TJ proteins that maintain the integrity of the intestinal mucosal barrier and play an important role in maintaining intestinal permeability. Immunohistochemistry was used to determine the tissue distribution of occludin and ZO-1 in the colon of the mice. Immunohistochemistry showed positive staining for occludin and ZO-1 in the colon of the control group. However, the expression of these two proteins in the colon of the TNBS-induced colitis group was barely detected. TNBS-induced colitis in mice treated with hUC-MSCs resulted in mild occludin and ZO-1 levels (Fig. 1g, h).
Alpha and beta diversity among the three groups
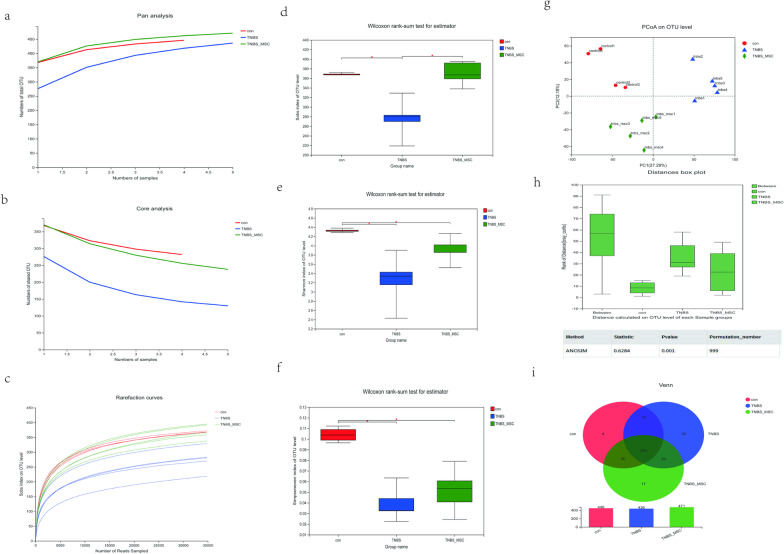
Pan/Core analysis was used to annotate the OTU species taxonomy and count the corresponding abundance information of each OTU annotation. When the species curve was flat, it indicated that the numbers of samples were sufficient for sequencing. Therefore, Pan/Core analysis can be used to evaluate whether the number of samples is sufficient for sequencing. Pan/Core analysis showed that the species curve was flat (Fig. 2a, b), indicating that the numbers of samples were sufficient for sequencing. Meanwhile, the rarefaction curve had already reached stable values in the current sequencing. The sequencing depth covered rare new phylotypes (Fig. 3c).
Fig. 2.
Changes in faecal microbial diversities among the control group (n = 4), TNBS group (n = 5) and TNBS + MSC group (n = 5). Faecal pellets were collected 3 days after MSC injection from the control TNBS and TNBS + MSC groups, and 16SrRNA sequences were determined as described in the "Methods". Alpha-diversity analysis and beta-diversity analysis among the three groups. a Pan genome analysis, b core genome analysis, c rank abundance curve of bacterial OTUs among the three groups. d–f The Sob, Shannon and Shannoneven indices were used to estimate the diversity of the faecal microbiota among the three groups (data expressed as the mean ± SD). g The plots shown were generated using principal coordinate analysis (PCoA). h Adonis shows that the difference between groups is significantly greater than that within groups (P < 0.001). i The Venn plot can be used to count the number of species that are common and unique among the three groups
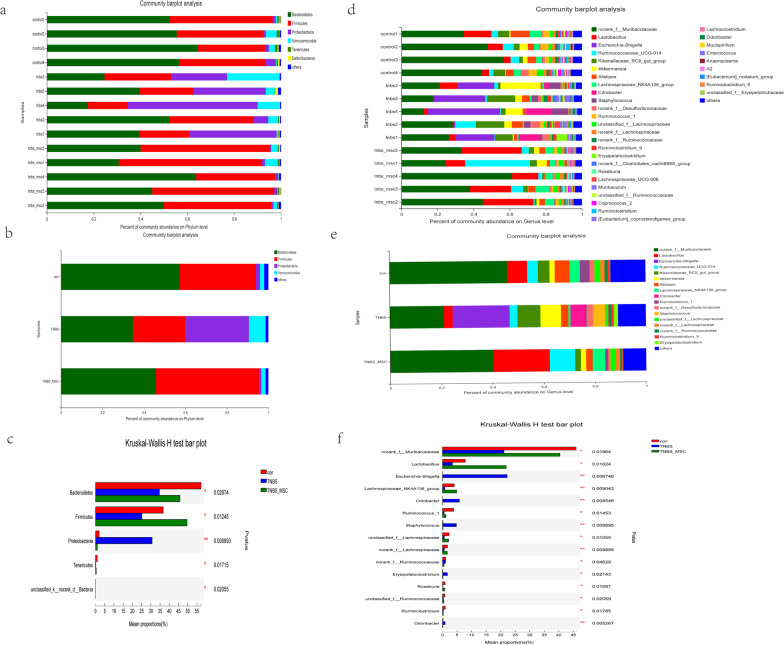
Fig. 3.
Structural comparison of the faecal microbiota among the control group (n = 4), TNBS group (n = 5) and TNBS + MSC group (n = 5). a Clustering of bacterial microbial composition at the phylum level in different samples. b The bacterial microbial composition in different experimental groups at the phylum levels. c The relative abundance of bacterial groups at the phylum level between groups tested by means of one-way nonparametric Analysis of Variance (ANOVA) (Kruskal–Wallis H test). *P < 0.05, **P < 0.01, ***P < 0.001. d Clustering of the bacterial microbial composition at the genus level in different samples. e The composition of bacterial microbial composition in different experimental groups at the genus level. f The relative abundance of bacterial groups at the genus level between groups tested by one-way nonparametric ANOVA(Kruskal–Wallis H test). *P < 0.05, **P < 0.01, ***P < 0.001
The Sob diversity index indices were significantly lower in the TNBS group than in the control and TNBS + MSC groups (P < 0.05 and P < 0.05, respectively) (Fig. 2d). Meanwhile, the Shannon and Shannoneven diversity index indices of the TNBS group were significantly lower than those of the control and TNBS + MSC groups (P < 0.05 and P < 0.05, respectively, Fig. 2e, f). A PCoA plot based on weighted UniFrac distance analysis was used to evaluate the beta diversity. As shown in Fig. 2g, an apparent clustering pattern was identified for the red, blue and green points, which represented the control group, the TNBS group and the TNBS + MSC group, respectively. Analysis of similarities (ANOSIM) suggested that the bacterial microflora composition difference between groups was greater than that within groups (P = 0.001) (Fig. 2h). Venn diagrams were constructed to evaluate the number and identity of the shared species among the control group, TNBS group and TNBS + MSC group. Venn diagram analysis revealed that the OTUs were shared by the three groups (Fig. 2i). Together, the results of the alpha and beta diversity analyses revealed significant microbiota variation in the gut microbiome among the control, TNBS and TNBS + MSC groups.
Gut microbiota differences among the three groups
Phylogenetic analysis of the gut microbiome from the phylum to the species level was performed on the samples using standard operational taxonomic unit (OTU) classification. A distinct bacterial signature was found in the TNBS group that differed from the TNBS + MSC group and controls. The TNBS group was particularly enriched with bacteria in the phylum Proteobacteria. At the phylum level, mice with colitis had decreased abundance of the phyla Bacteroidetes, Firmicutes and Tenericutes compared with control or MSC treated mice (Fig. 3a–c). A similar lineage pattern was seen at the genus level, as Escherichia-Shigella and Citrobacter were enriched in the TNBS group. At the genus level, mice with colitis had decreased abundance of Tenericutes, Lactobacillus, and Ruminococcus (Fig. 3d–f).
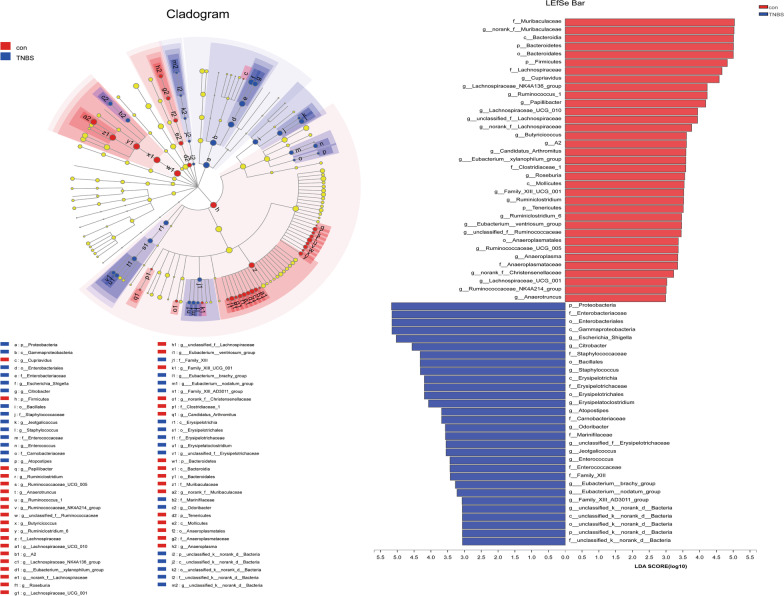
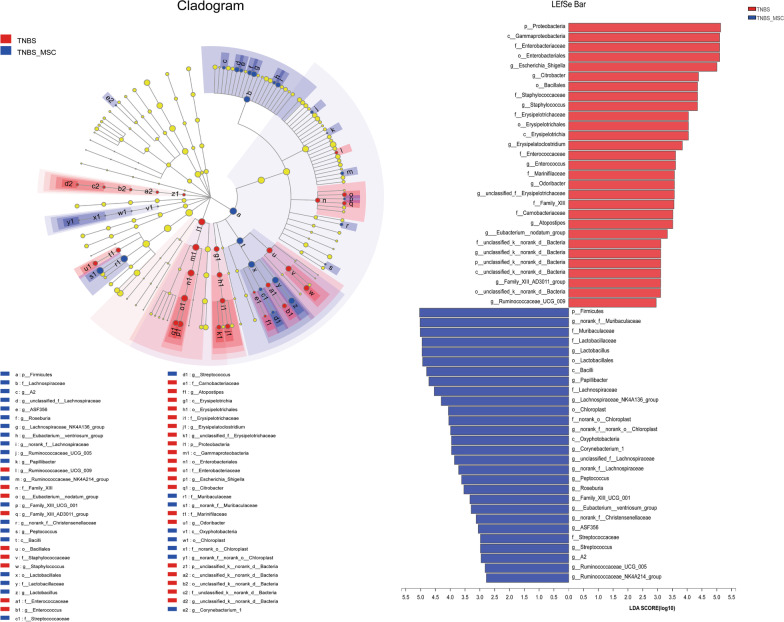
We used LEfSe to identify the specific bacterial phylotypes altered by TNBS administration and MSC treatment. To investigate the effects of experimental colitis on the microbiome, the differentiated taxa between the control group and the TNBS group were detected. The cladogram representative of the structure and its dominant bacteria is shown in Fig. 4. In comparisons of the control group versus the TNBS groups, linear discriminant analysis (LDA) scores and the corresponding cladogram showed that colitis mice were most enriched in the Proteobacteria phylum, which included the Gammaproteobacteria class, Enterobacteriaceae order, Enterobacteriales family, Escherichia_Shigella genus, and Citrobacter genus. However, the control group had higher enrichment of the Bacteroidetes phylum, which includes the Bacteroidia class, Bacteroidales order, Muribaculaceae family, and the norank_f_Muribaculaceae genus; and the Tenericutes phylum, which includes the Mollicutes class, Anaeroplasmatales order, Anaeroplasmataceae family, and the Anaerotruncus genus.
Fig. 4.
Differences in dominant microorganisms between the control and TNBS groups. In the comparisons of the control group versus the TNBS groups, colitis mice were enriched in the Proteobacteria phylum based on the LDA scores and cladogram. a LDA scores from LefSe analysis were performed on relative OTU abundances. Least discriminant analysis (LDA) effect size taxonomic cladogram comparing all samples categorized by control (red) and TNBS groups (blue). b Distribution histogram based on LDA (LDA score > 3). Significantly discriminant taxon nodes are coloured, and the branch areas are shaded according to the highest-ranked variety for that taxon. For each taxon detected, the corresponding node in the taxonomic cladogram is coloured according to the highest-ranked group for that taxon. If the taxon is not significantly differentially represented between sample groups, the corresponding node is coloured yellow
Moreover, to investigate the effects of the MSC on the microbiome, the differentiated taxon between the TNBS + MSC group and the TNBS group was assessed (Fig. 5). The TNBS group had higher enrichment of the Proteobacteria phylum, which includes the Gammaproteobacteria class, Enterobacteriaceae order, Enterobacteriales family, Escherichia_Shigella genus, and Citrobacter genus. However, the TNBS + MSC group had a higher enrichment of the Firmicutes phylum, which includes the Bacilli class, Lactobacillales order, Lactobacillaceae family, and Lactobacillus genus. Based on the results among the three groups, it was discovered that MSC treatment could restore the gut microbiome of mice with experimental colitis to a similar composition to the control group.
Fig. 5.
Differences in dominant microorganisms between the TNBS and TNBS + MSCgroups. The TNBS group had higher enrichment of the Proteobacteria phylum, while the TNBS + MSC group had higher enrichment of the Firmicutes phylum a LDA scores from LefSe analysis were obtained from the relative OTU abundances and the least discriminant analysis (LDA) effect size taxonomic cladogram comparing all samples categorized by TNBS (red) and the TNBS + MSC group (blue). b Distribution histogram based on LDA (LDA score > 3). Significantly discriminant taxon nodes are coloured and the branch areas are shaded according to the highest-ranked variety for that taxon. For each taxon detected, the corresponding node in the taxonomic cladogram is coloured according to the highest-ranked group for that taxon. If the taxon is not significantly differentially represented between the sample groups, the corresponding node is depicted in yellow
Microbial metabolic functions associated with MSC treatment in TNBS-induced colitis
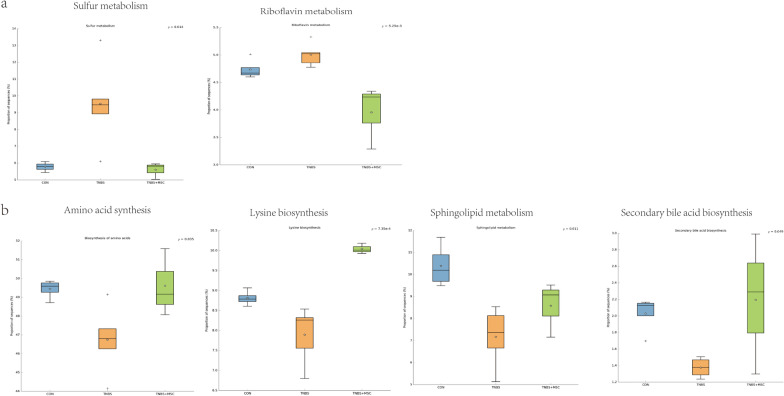
To investigate the gut microbiome functions related to TNBS administration and MSC treatment, we adopted PICRUStII to infer putative metagenomes from 16S rRNA gene profiles. STAMP was used to identify microbially relevant functions linked with the TNBS administration of MSCs. These pathways were further analysed at the KEGG level. This analysis permitted a comparison of the differences in the functional profiles among all groups and revealed pathways that were significantly different among the control, TNBS and TNBS + MSC groups (Fig. 6). Increased metabolism was present in the TNBS group compared with the control group and the TNBS + MSC group. As a result, MSC decreased the increased activities of sulphur metabolism and riboflavin metabolism (Fig. 6a). The increased levels of riboflavin and sulphur metabolism in the model group may indicate that the biosynthesis of compounds beneficial to oxidative stress has increased. An increase in those compounds may be due to a shift towards an inflammation-promoting microbiome. Some metabolic pathways were decreased in the TNBS group compared with the control group and the TNBS + MSC group, such as the biosynthesis of amino acids, lysine biosynthesis, sphingolipid metabolism, and secondary bile acid biosynthesis (Fig. 6b). The data show that amino acid, lysine, and sphingolipid metabolism, as well as secondary bile acid biosynthesis, were disrupted in the TNBS group. These metabolic changes were reversed by MSCs.
Fig. 6.
Gut microbiome functions inferred by PICRUStII from 16S rRNA gene sequences among the three groups (the control group, the TNBS group and the TNBS + MSC group). a The blue, golden yellow and green bar graphs indicate the inferred metabolism value of control group, TNBS group and TNBS + MSC group. KEGG orthology was used for functional categorization. STAMP3 was used for functional profiling. Elevated metabolic pathways after modelling. b Decreased metabolic pathways after modelling. A box plot shows the top quartile, median, and bottom quartile, while white stars indicate the average and a plus sign indicates the outlier. All differences were analysed using one-way ANOVA followed by the Tukey–Kramer post hoc test. The multiple comparisons were not corrected
Discussion
In the present study, we utilized a mouse model of TNBS-induced colitis. Mice were treated with MSCs to determine the beneficial effects of MSCs on the gut microbiota. First, we present evidence that MSCs ameliorate gut dysbiosis and impairment of the intestinal mucosal barrier. Furthermore, PICRUStII analysis data showed that amino acid, lysine, and sphingolipid metabolism, as well as secondary bile acid biosynthesis, were disrupted in the TNBS group. These metabolic changes can be reversed by MSCs.
The human body is colonized by a large number of microbes coexisting peacefully with their host. The most colonized site is the gastrointestinal tract. More than 70% of the microbes in the human body are found in the colon. The microorganism population is 10× larger than the total number of somatic and germ cells in the human body. Two bacterial phyla, making up more than 90% of the microbial cells, dominate the healthy adult intestine: Firmicutes and Bacteroidetes [42]. The microbiota acts as a barrier to pathogens, exerts important metabolic functions, and regulates the inflammatory response by stimulating the immune system [43]. The composition of the gut microbiota exhibits broad interindividual and intraindividual variability [44] and is thought to be a crucial determinant of host susceptibility to several diseases, including IBD. A key feature of IBD is alterations of the composition of the gut microbiota (known as dysbiosis) [45]; however, the precise role of dysbiosis in the disease remains poorly understood. Dysbiosis can destroy these mutualistic relationships and influence the host physiology, compromising human health status [46].
The pathogenesis of IBD in genetically susceptible hosts has been proposed to begin with a breakdown of the intestinal epithelial barrier, followed by a disproportionate immune response to the enteric microbiota, which results in a loss of intestinal homeostasis [47]. Such dysbiosis in the intestinal symbionts of IBD patients can be indicated by the low-level alpha diversity and it manifests as a decrease in many beneficial bacteria, including Firmicutes and Bacteroidetes, and an increased abundance of Proteobacteria [48, 49]. There were also significant changes in both the taxonomic structure and the functional composition of intestinal microbes in TNBS-induced colitis mice [50, 51]. Similar to previous clinical and animal research, our current results showed that TNBS treatment caused microbiota dysbiosis by reducing alpha diversity. At the phylum level, the TNBS group was characterized by enrichment in Proteobacteria and a reduction in Firmicutes and Bacteroidetes. The distribution of bacterial microbiota in TNBS-induced colitis mice was similar to that of CD patients.
Here, we demonstrated that MSC administration significantly ameliorated the loss of body weight and intestinal inflammation and decreased DAI in TNBS-induced colitis mice. After MSC administration, the phyla Bacteroidetes, Firmicutes and Tenericutes were upregulated, and the phylum Proteobacteria was downregulated. Linear discriminant analysis was performed to identify any key microbial biomarkers that might differentiate the disease and MSC treated states. In comparisons of the control versus TNBS groups, LDAs cores and the corresponding cladogram showed that colitis mice were most enriched in the Proteobacteria phylum, which included the Gammaproteobacteria class, Enterobacteriaceae order, Enterobacteriales family, Escherichia_Shigella genus, and Citrobacter genus. However, the control mice had higher enrichment of Muribaculaceae, Bacteroidetes, and Firmicutes. When comparing TNBS with the TNBS + MSC group, Firmicutes had the highest LDA score and enrichment in the MSC-treated mice.
Collectively, these data show that a combination of Proteobacteria phyla, which include the Gammaproteobacteria class, Enterobacteriaceae order, Enterobacteriales family, Escherichia_Shigella genus, and Citrobacter genus, could serve as potential biomarkers of colitis. Effective treatment with MSC was characterized by the enrichment of Firmicutes and Bacteroidetes. MSCs can modulate the gut microbiome during colitis in such a way that they prevent the overgrowth of pathogenic bacteria.
Researchers demonstrated that the microbiota of dextran sodium sulphate (DSS)-induced colitis mice was partially restored after treatment with MSCs [23]. DSS mice have symptoms similar to those of human ulcerative colitis, and TNBS mice have symptoms similar to those of human Crohn’s disease [52]. Our study has implications for understanding whether MSCs can affect gut microbiota in a Crohn's disease mouse model. In accordance with DSS-induced colitis, our study indicates that MSCs also restored the normal microbiota in TNBS-induced colitis mice.
The potential mechanism may be associated with the gut microbiota and host metabolism interaction, as the gut bacteria often target the host metabolism, which further drives immune activation and chronic inflammation [53]. Some pathways and genes were overrepresented and underrepresented in the TNBS group. Based on functional analysis, it is evident that the activities of sulphur and riboflavin metabolism increased while the biosynthesis of amino acids, lysine biosynthesis, sphingolipid metabolism, and secondary bile acid biosynthesis declined.
The TNBS group has an enhanced capacity for managing oxidative stress, a feature of the inflammatory environment, as indicated by incremental riboflavin and sulphur metabolism in the TNBS group compared with the control group and the TNBS + MSC group. Oxidative stress increases in response to inflammation, which allows the microbiota to maintain homeostasis. Thus, the increases in sulphur and riboflavin metabolism may reflect a mechanism by which the gut microbiome addresses the oxidative stress caused by inflammation.
IBD is also associated with a decrease in amino acid synthesis according to previous research [49]. Biosynthesis of amino acids and lysine biosynthesis were reduced in the TNBS group. Under states of inflammation, bacteria typically have a weak ability to produce their own nutrients [54]. The metabolism of amino acids is greatly perturbed. Genes involved in the metabolism and synthesis of nearly all amino acids (particularly lysine) declined in abundance.
Sphingolipid metabolism was decreased in the TNBS group. Sphingolipids play multiple roles in the healthy gut, including membrane structural components in intestinal cells and assigning molecules involved in cell fate decisions [55]. Previous research suggested that sphingolipid metabolism might be disrupted in IBD, causing an accumulation of compounds that promote inflammatory responses [56].
Secondary bile acid biosynthesis declined. Secondary bile acids (SBAs) are produced by bacteria from primary bile acids (PBAs), which have mainly anti-inflammatory effects. It has been found that IBD patients have disrupted BA metabolism, with faecal BA pools skewed towards lower SBAs than healthy controls [57], which was in agreement with our results.
Overall, the functional variation of microbiota among the control group, the TNBS group, and the TNBS + MSC group may indicate that MSC treatment could modulate the metabolism pathways in mice with colitis, restoring the abnormal microbiota function to a normal situation similar to the control group.
These results suggest that perturbations in the bacterial composition are associated with gut microbiome function. Overall, the functional variation of microbiota among the control group, the TNBS group, and the TNBS + MSC group may indicate that MSC treatment could modulate the metabolism pathways in mice with colitis, restoring the abnormal microbiota function to a normal situation as seen in the control group. Limitations in the current mouse experiments are that a mouse model may not fully mimic human Crohn’s disease, and the gut microbiota is different between humans and mice. Clinical studies in CD patients are a future requirement.
Conclusions
In summary, TNBS treatment induced colonic injury and an inflammatory response in mice. MSCs exerted a protective effect by repairing the intestinal mucosal barrier. MSCs notably restored the alpha diversity and reversed the alterations in the abundance of the gut microbiota in the mice treated with TNBS. MSC treatment could modulate the dysregulated metabolic pathways in mice with colitis. The mechanism might be associated with the gut microbiota, as MSCs improved gut microbiota communities in TNBS-induced murine colitis. To our knowledge, our study is the first to demonstrate MSC treatment and changes in the gut microbiota in a TNBS-induced colitis mouse model. These findings provide insight into specific intestinal microbiotas and metabolic pathways linked with MSC treatment, suggesting a new approach to treating CD. Thus, future perspectives include the following: clinical studies will need to be conducted to evaluate the efficacy and safety of MSCs and determine whether MSCs will normalize the gut microbiota in CD patients.
Supplementary Information
Additional file 1: Fig. S1 Surface marker expression and characterization of human umbilical cord-derived mesenchymal stromal cells (hUC-MSCs). hUC-MSCs at passage 3 were analysed using flow cytometry, and their osteogenic and adipogenic differentiation was confirmed using Alizarin red staining and Oil Red O, respectively. Flow cytometric analysis of surface markers (a. CD90, b. CD19, c. CD11b, d. HLA-DR, e. CD34, f. CD45, g. CD105, h. CD73), i. fibroblast-like morphology of MSCs, j. osteogenic differentiation (Alizarin red staining), k. Adipogenic differentiation (Oil Red Ostaining).
Acknowledgements
We thank all members of the Qi Zhang's laboratory for their support.
Abbreviations
- hUC-MSCs
Human umbilical cord-derived mesenchymal stem cells
- CD
Crohn’s disease
- IBD
Inflammatory bowel disease
- TNBS
2,4,6-Trinitrobenzenesulfonic acid
- OUT
Operational taxonomic unit
- ZO-1
Zonula occludens-1
- LEfSe
Linear discriminant analysis effect size
- LDA
Linear discriminant analysis
- PICRUSt
Phylogenetic investigation of communities by reconstruction of unobserved states
- DAI
Disease activity index
- IPP
Image-pro plus
- PCoA
Principal coordinates analysis
- PBAs
Primary bile acids
- SBAs
Secondary bile acids
- DSS
Dextran sodium sulphate
- RDP
Ribosomal database project
- ANOSIM
Analysis of similarities
- ANOVA
Analysis of variance
Author contributions
FY designed experiments, performed experiments, analysed data and wrote the paper. BBN, LL and XMZ analysed data and edited the paper. XYC, HZL and JXL collected human umbilical cord samples, performed experiments and edited the paper. QLL, XFZ, RXX and YZH performed animal experiments. FPH, QZ, XGZ and WJC designed the study, analysed data, and edited the paper. All authors read and approved the final manuscript.
Funding
This work was supported by project funded by National Natural Science Foundation of China (82260110, 81870449, 82170674, 51933011), China Postdoctoral Science Foundation (2019M653904XB), Natural Science Foundation of Xinjiang Uyghur Autonomous Region (2020D01C006), Tianshan Cedar Project Science and Technology Department of Xinjiang Uygur Autonomous Region (2018XS23), Science and Technology Projects in Guangdong Province (2018YJ007), Science and Technology Projects in Guangzhou (No 202102010310), The provincial and ministerial joint construction of the open project of the State Key Laboratory of High Incidence and Prevention in Central Asia (SKL-HIDCA-2020-10).
Availability of data and materials
All data and materials generated or used during the study are available from the corresponding authors upon reasonable request.
Declarations
Ethics approval and consent to participate
All experiments were performed in accordance with the institutional guidelines of Third Affiliated Hospital of Sun Yat-sen University. Use of animals in this study was approved by Institutional Animal Care and Use Committee of Sun Yat-sen University. Informed consent was obtained from all subjects and/or their legal guardian(s) under the ethical committee of the Third Affiliated Hospital of Sun Yat-sen University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fan Yang, Beibei Ni and Qiuli Liuare co-first authors.
Contributor Information
Qi Zhang, Email: zhangq27@mail.sysu.edu.cn.
Xiaoguang Zou, Email: zxgks@163.com.
Wenjie Chen, Email: chenwj5@mail.sysu.edu.cn.
References
- 1.Roda G, Chien Ng S, Kotze PG, et al. Crohn’s disease. Nat Rev Dis Primers. 2020;6(1):22. doi: 10.1038/s41572-020-0156-2. [DOI] [PubMed] [Google Scholar]
- 2.Kaplan GG, Windsor JW. The four epidemiological stages in the global evolution of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2021;18(1):56–66. doi: 10.1038/s41575-020-00360-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ng SC, Hi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet (London, England) 2017;390(10114):2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 4.Cushing K, Higgins PDR. Management of crohn disease: a review. JAMA. 2021;325(1):69–80. doi: 10.1001/jama.2020.18936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang JT. Pathophysiology of inflammatory bowel diseases. N Engl J Med. 2020;383(27):2652–2664. doi: 10.1056/NEJMra2002697. [DOI] [PubMed] [Google Scholar]
- 6.Abdelrazik H, Giordano E, Brodano GB, et al. Substantial overview on mesenchymal stem cell biological and physical properties as an opportunity in translational medicine. Int J Mol Sci. 2019;20(21):5386. doi: 10.3390/ijms20215386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Hao J, Hu Z, et al. Current status of clinical trials assessing mesenchymal stem cell therapy for graft versus host disease: a systematic review. Stem Cell Res Ther. 2022;13(1):93. doi: 10.1186/s13287-022-02751-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung JW, Chang WH, Bang OY, et al. Efficacy and safety of intravenous mesenchymal stem cells for ischemic stroke. Neurology. 2021;96(7):e1012–e1023. doi: 10.1212/WNL.0000000000011440. [DOI] [PubMed] [Google Scholar]
- 9.Alcayaga-Miranda F, Cuenca J, Khoury M. Antimicrobial activity of mesenchymal stem cells: current status and new perspectives of antimicrobial peptide-based therapies. Front Immunol. 2017;8:339. doi: 10.3389/fimmu.2017.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marx C, Gardner S, Harman RM, et al. Mesenchymal stromal cell-secreted CCL2 promotes antibacterial defense mechanisms through increased antimicrobial peptide expression in keratinocytes. Stem cells Transl Med. 2021;10(12):1666–16794. doi: 10.1002/sctm.21-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huldani H, Margiana R, Ahmad F, et al. Immunotherapy of inflammatory bowel disease (IBD) through mesenchymal stem cells. Int Immunopharmacol. 2022;107:108698. doi: 10.1016/j.intimp.2022.108698. [DOI] [PubMed] [Google Scholar]
- 12.Carvello M, Lightner A, Yamamoto T, et al. Mesenchymal stem cells for perianal Crohn's disease. Cells. 2019;8(7):764. doi: 10.3390/cells8070764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Santalla M, Garin MI. Improving the efficacy of mesenchymal stem/stromal-based therapy for treatment of inflammatory bowel diseases. Biomedicines. 2021;9(11):1507. doi: 10.3390/biomedicines9111507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Younis N, Zarif R, Mahfouz R. Inflammatory bowel disease: between genetics and microbiota. Mol Biol Rep. 2020;47(4):3053–3063. doi: 10.1007/s11033-020-05318-5. [DOI] [PubMed] [Google Scholar]
- 15.Henke MT, Kenny DJ, Cassilly CD, et al. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn's disease, produces an inflammatory polysaccharide. Proc Natl Acad Sci USA. 2019;116(26):12672–12677. doi: 10.1073/pnas.1904099116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Chen WD, Wang YD. The relationship between gut microbiota and inflammatory diseases the role of macrophages. Front Microbiol. 2020;11:1065. doi: 10.3389/fmicb.2020.01065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569(7758):655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagao-Kitamoto H, Kamada N. Host-microbial cross-talk in inflammatory bowel disease. Immune Netw. 2017;17(1):1–12. doi: 10.4110/in.2017.17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagao-Kitamoto H, Nobuhiko K, et al. Host-microbial Cross-talk in Inflammatory bowel disease. Immune Netw. 2017;17:1. doi: 10.4110/in.2017.17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu HM, Zhrou YL, Xu J, et al. Inhibition of PD-1 protects against TNBS-induced colitis via alteration of enteric microbiota. BioMed Res Int. 2021;2021(1):1–12. doi: 10.1155/2021/4192451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metwaly A, Dunkel A, Waldschmitt N, et al. Integrated microbiota and metabolite profiles link Crohn's disease to sulfur metabolism. Nat Commun. 2020;11(1):4322. doi: 10.1038/s41467-020-17956-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Bhosle A, Bae S, et al. Discovery of bioactive microbial gene products in inflammatory bowel disease. Nature. 2022;1:7. doi: 10.1038/s41586-022-04648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soontararak S, Chow L, Johnson V, et al. Mesenchymal stem cells (MSC) derived from induced pluripotent stem cells (iPSC) equivalent to adipose-derived MSC in promoting intestinal healing and microbiome normalization in mouse inflammatory bowel disease model. Stem Cells Transl Med. 2018;7(6):456–467. doi: 10.1002/sctm.17-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong X, Feng X, Liu J, et al. Characteristics of intestinal microecology during mesenchymal stem cell-based therapy for mouse acute liver injury. Stem Cells Int. 2019;2019:2403793. doi: 10.1155/2019/2403793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo L, Chen Q, Yang L, et al. MSCs therapy reverse the gut microbiota in hypoxia-induced pulmonary hypertension mice. Front Physiol. 2021;12:712139. doi: 10.3389/fphys.2021.712139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xing J, Ying Y, Mao C, et al. Hypoxia induces senescence of bone marrow mesenchymal stem cells via altered gut microbiota. Nat Commun. 2018;9(1):2020. doi: 10.1038/s41467-018-04453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lv W, Graves DT, He L, et al. Depletion of the diabetic gut microbiota resistance enhances stem cells therapy in type 1 diabetes mellitus. Theranostics. 2020;10(14):6500–6516. doi: 10.7150/thno.44113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandravanshi B, Bhonde RR. Human umbilical cord-derived stem cells: isolation, characterization, differentiation, and application in treating diabetes. Crit Rev Biomed Eng. 2018;46(5):399–412. doi: 10.1615/CritRevBiomedEng.2018027377. [DOI] [PubMed] [Google Scholar]
- 29.Del Angel-Mosqueda C, Gutiérrez-Puente Y, López-Lozano AP, et al. Epidermal growth factor enhances osteogenic differentiation of dental pulp stem cells in vitro. Head Face Med. 2015;11(1):29. doi: 10.1186/s13005-015-0086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Q, Chen X, Liu C, et al. Mesenchymal stem cells alleviate experimental immune-mediated liver injury via chitinase 3-like protein 1-mediated T cell suppression. Cell Death Dis. 2021;12(3):240. doi: 10.1038/s41419-021-03524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao Y, Lei J, Liu M, et al. Mesenchymal stromal cells mitigate experimental colitis via insulin-like growth factor binding protein 7-mediated immunosuppression. Mol Ther. 2016;24(10):1860–1872. doi: 10.1038/mt.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chao K, Zhang S, Qiu Y, et al. Human umbilical cord-derived mesenchymal stem cells protect against experimental colitis via CD5(+) B regulatory cells. Stem Cell Res Ther. 2016;7(1):109. doi: 10.1186/s13287-016-0376-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.González MA, Gonzalez-Rey E, Rico L, et al. Adipose-derived mesenchymal stem cells alleviate experimental colitis by inhibiting inflammatory and autoimmune responses. Gastroenterology. 2009;136(3):978–989. doi: 10.1053/j.gastro.2008.11.041. [DOI] [PubMed] [Google Scholar]
- 34.Wirtz S, Popp V, Kindermann M, et al. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc. 2017;12(7):1295–1309. doi: 10.1038/nprot.2017.044. [DOI] [PubMed] [Google Scholar]
- 35.Chao K, Zhang S, Qiu Y, et al. Correction to: human umbilical cord-derived mesenchymal stem cells protect against experimental colitis via CD5+ B regulatory cells. Stem Cell Res Ther. 2019;10(1):33. doi: 10.1186/s13287-019-1132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mei Y, Tang Z, Li Z, et al. Repeatability and reproducibility of quantitative corneal shape analysis after orthokeratology treatment using image-pro plus software. J Ophthalmol. 2016;2016:1732476. doi: 10.1155/2016/1732476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S, Zhou Y, Chen Y, et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics (Oxford, England) 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics (Oxford, England) 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 40.Wang Q, Garrity GM, Tiedje JM, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schippa S, Conte MP. Dysbiotic events in gut microbiota: impact on human health. Nutrients. 2014;6(12):5786–5805. doi: 10.3390/nu6125786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30:759–795. doi: 10.1146/annurev-immunol-020711-074937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carding S, Verbeke K, Vipond DT, et al. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. 2015;26:26191. doi: 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peloquin JM, Nguyen DD. The microbiota and inflammatory bowel disease: insights from animal models. Anaerobe. 2013;24:102–106. doi: 10.1016/j.anaerobe.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55(2):205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eom T, Kim YS, Choi CH, et al. Current understanding of microbiota- and dietary-therapies for treating inflammatory bowel disease. J Microbiol. 2018;56(3):189–198. doi: 10.1007/s12275-018-8049-8. [DOI] [PubMed] [Google Scholar]
- 50.Yang Y-W, Chen M-K, Yang B-Y, et al. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in mouse feces. Appl Environ Microbiol. 2015;81(19):6749–6756. doi: 10.1128/AEM.01906-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He Q, Li X, Liu C, et al. Dysbiosis of the fecal microbiota in the TNBS-induced Crohn's disease mouse model. Appl Microbiol Biotechnol. 2016;100(10):4485–4494. doi: 10.1007/s00253-015-7205-x. [DOI] [PubMed] [Google Scholar]
- 52.Shin K, Roh S-S. Comparative between DSS and TNBS-induced colitis on Sulfasalazine alone and combination with herbal medicine. 2017. [DOI] [PMC free article] [PubMed]
- 53.Han H, Yi B, Zhong R, et al. From gut microbiota to host appetite: gut microbiota-derived metabolites as key regulators. Microbiome. 2021;9(1):162–162. doi: 10.1186/s40168-021-01093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng H, Chen M, Li Y, et al. Modulation of Gut microbiome composition and function in experimental colitis treated with sulfasalazine. Front Microbiol. 2017;8:1703. doi: 10.3389/fmicb.2017.01703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abdel Hadi L, Di Vito C, Riboni L. Fostering inflammatory bowel disease: sphingolipid strategies to join forces. Mediators Inflamm. 2016;2016:3827684. doi: 10.1155/2016/3827684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fischbeck A, Leucht K, Frey-Wagner I, et al. Sphingomyelin induces cathepsin D-mediated apoptosis in intestinal epithelial cells and increases inflammation in DSS colitis. Gut. 2011;60(1):55–65. doi: 10.1136/gut.2009.201988. [DOI] [PubMed] [Google Scholar]
- 57.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol. 2019;4(2):293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1 Surface marker expression and characterization of human umbilical cord-derived mesenchymal stromal cells (hUC-MSCs). hUC-MSCs at passage 3 were analysed using flow cytometry, and their osteogenic and adipogenic differentiation was confirmed using Alizarin red staining and Oil Red O, respectively. Flow cytometric analysis of surface markers (a. CD90, b. CD19, c. CD11b, d. HLA-DR, e. CD34, f. CD45, g. CD105, h. CD73), i. fibroblast-like morphology of MSCs, j. osteogenic differentiation (Alizarin red staining), k. Adipogenic differentiation (Oil Red Ostaining).
Data Availability Statement
All data and materials generated or used during the study are available from the corresponding authors upon reasonable request.