Abstract
Virginiae butanolide (VB)-BarA of Streptomyces virginiae is one of the newly discovered pairs of a butyrolactone autoregulator and a corresponding receptor protein of Streptomyces species and regulates the production of the antibiotic virginiamycin (VM) in S. virginiae. The gene vmsR was found to be situated 4.7 kbp upstream of the barA gene, which encodes the VB-specific receptor. The vmsR product was predicted to be a regulator of VM biosynthesis based on its high homology to some Streptomyces pathway-specific transcriptional regulators for the biosynthetic gene clusters of polyketide antibiotics, such as Streptomyces peucetius DnrI (47.5% identity, 84.3% similarity), which controls daunorubicin biosynthesis. A vmsR deletion mutant was created by homologous recombination. Neither virginiamycin M1 nor virginiamycin S was produced in the vmsR mutant, while amounts of VB and BarA similar to those produced in the wild-type strain were detected. Reverse transcription-PCR analyses confirmed that the vmsR deletion had no deleterious effects on the transcription of the vmsR-surrounding genes, indicating that VmsR is a positive regulator of VM biosynthesis in S. virginiae.
Streptomycetes are gram-positive filamentous bacteria that are well-known for producing a vast array of bioactive compounds, including more than 70% of commercially important antibiotics. The production of antibiotics by these organisms is regulated by a variety of physiological and nutritional conditions and is coordinated with processes of morphological differentiation, such as the formation of aerial mycelia and spores. Despite many years of research on antibiotics driven by their commercial importance, the overall regulatory pathway governing antibiotic production is still poorly understood. A detailed knowledge of the signal cascade and the genetic components involved in antibiotic production should permit construction of strains that can overproduce these commercially important compounds.
Antibiotic production and/or morphological differentiation is controlled in several Streptomyces species by low-molecular-weight compounds called butyrolactone autoregulators (32). To date, 11 butyrolactone autoregulators have been chemically identified and classified into three types based on minor structural differences in their C-2 side chains: (i) the virginiae butanolide (VB) type, containing a 6-α-hydroxy group (13, 31); (ii) the IM-2 type, containing a 6-β-hydroxy group (25, 27); and (iii) the A-factor type, containing a 6-keto group (16). Their effectiveness at nanomolar concentrations, as well as the presence in these species of specific receptor proteins (BarA as a VB-specific receptor in Streptomyces virginiae [9, 10, 21], FarA as an IM-2-specific receptor in Streptomyces lavendulae FRI-5 [23, 28], and ArpA as an A-factor-specific receptor in Streptomyces griseus [22]) as mediators of autoregulator signaling, implies that they should be regarded as Streptomyces hormones.
VB-BarA of S. virginiae has been among the most frequently studied pairs and is known to regulate the coordinated production of two structurally different compounds, virginiamycin M1 (VM1) and virginiamycin S (VS), a pair of antibiotics showing strong synergistic bactericidal activity. In vitro (11, 12) and in vivo (11, 17, 18) analyses have demonstrated that BarA in the absence of VB is a DNA-binding transcriptional repressor and that the release of DNA-bound BarA in the presence of VB from the promoter region of a target gene(s) will result, ultimately, in virginiamycin (VM) production. However, the VB signaling cascade beyond BarA is not clear at present.
In our previous study on a pleiotropic regulatory gene (barX) situated 259 bp upstream of the barA gene (8), a phenotypic defect in VM and VB production in the barX deletion mutant was correlated with the lack of transcription of a plausible pathway-specific regulatory gene (vmsR; formerly called orf2), which raised the possibility that the vmsR product might be responsible for the initiation of VM and/or VB production. To clarify the in vivo function of the VmsR protein, we created a vmsR deletion mutant of S. virginiae by homologous recombination and report here the results from phenotypic and transcriptional analyses on the vmsR mutant, which clearly indicate that VmsR is the regulator positively controlling the biosynthesis of both VM1 and VS in S. virginiae.
Bacterial strains, plasmids, growth conditions, and transformation.
S. virginiae (strain MAFF 10-06014; National Food Research Institute, Ministry of Agriculture, Forestry, and Fisheries, Tsukuba, Japan) was grown at 28°C in modified yeast extract-malt extract liquid medium (7) for protoplast formation, in tryptic soy broth (Oxoid, Basingstoke, Hampshire, United Kingdom) containing thiostrepton (5 μg ml−1) for plasmid preparation, on ISP2 agar medium (Difco, Detroit, Mich.) for spore formation, and in liquid f medium (33) for VM and VB production. S. virginiae was transformed as described previously (7). A plasmid, pSVR1, containing a 3.5-kbp EcoRI-BamHI fragment carrying barZ-vmsR-varM (8) (DDBJ accession no. AB035547) in the EcoRI-BamHI site of pUC18 was used as a template for PCR. DNA manipulations in Escherichia coli and S. virginiae were performed as described by Sambrook et al. (24) and Hopwood et al. (6), respectively.
Construction and identification of a vmsR deletion mutant.
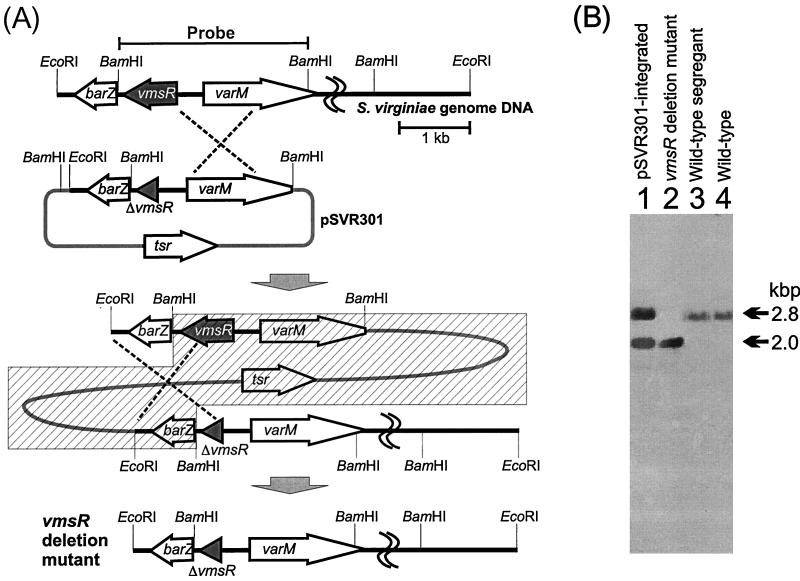
The vmsR gene product was predicted to be a regulator of VM biosynthesis based on its high homology to some Streptomyces pathway-specific transcriptional regulators of the biosynthetic gene clusters of polyketide antibiotics, such as Streptomyces peucetius DnrI (47.5% identity, 84.3% similarity), which controls daunorubicin biosynthesis (26); Streptomyces coelicolor A3(2) ActII-ORF4 (36.1% identity, 76.6% similarity), which controls actinorhodin biosynthesis (3); and S. coelicolor A3(2) RedD (28.9% identity, 67.3% similarity), which controls undecylprodigiosin biosynthesis (19). Alternatively, the VmsR protein might be responsible for the initiation of VB biosynthesis, because the phenotypic defect of VB production in the barX mutant correlated with the lack of vmsR transcription (8). In order to investigate these possibilities, a vmsR deletion mutant was created by homologous recombination within the S. virginiae genome (Fig. 1A).
FIG. 1.
Replacement of the S. virginiae vmsR gene with plasmid pSVR301 to form truncated vmsR by homologous recombination. (A) Restriction maps of pSVR301 and the genomic DNA of the S. virginiae wild-type strain. The varM promoter region was amplified by PCR with primers 5′-GGGGCCACTCGCGACTCCAACCGCCGC-3′ (a newly created recognition site for NruI is indicated by underlining) and 5′-GCAAGCGGCTCGCCGTACGGGAGCGCTG-3′ (a BsiWI recognition site is indicated by underlining) using pSVR1 as a template. The NruI-BsiWI-digested PCR fragment was then used to replace the original 1.0-kbp NruI-BsiWI fragment of pSVR1, resulting in the deletion of a 145-bp 5′ untranslated region and the N-terminal 698 bp of vmsR. From the resulting plasmid (pSVR201), a 2.8-kbp EcoRI-HindIII fragment containing barZ, the C-terminal 118 bp of vmsR, and the entire varM gene was cloned into the EcoRI-HindIII site of pUWL-KS to create plasmid pSVR301. A single crossover between pSVR301 and a homologous DNA in the genome gave the pSVR301-integrated strain, and a second crossover generated a vmsR-disrupted strain. Only one of the two possible first crossover events is shown. The hatched box indicates the region deleted by the second crossover. (B) Hybridization patterns of BamHI-digested genomic DNA of the pSVR301-integrated strain (lane 1), vmsR-disrupted strain (lane 2), wild-type segregant (lane 3), and the S. virginiae wild-type strain (lane 4). A 2.8-kbp BamHI fragment containing the vmsR gene was used as a probe. When hybridized with the probe, the vmsR-disrupted strain (lane 2) gave a 2.0-kbp fragment, as a result of the deletion of the vmsR gene.
Plasmid pSVR301 was constructed by ligating a 2.8-kbp EcoRI-HindIII fragment containing barZ-ΔvmsR-varM into the EcoRI-HindIII site of pUWL-KS (29). The S. virginiae wild-type strain was transformed with pSVR301, and a single crossover between pSVR301 and a homologous DNA in the S. virginiae genome gave the pSVR301-integrated strain. When we cultivated the pSVR301-integrated strain in liquid tryptic soy broth in the absence of thiostrepton, we obtained two types of strains: one is a vmsR deletion mutant, and the other is a regenerated wild-type strain (Fig. 1B). The integration of pSVR301 in the first crossover strain and its deletion in the second crossover strain were confirmed by Southern blot hybridization using a 2.8-kbp BamHI fragment encoding vmsR as a probe (Fig. 1B). We also confirmed the 802-bp deletion in the vmsR mutant by DNA sequence analysis. One of the resulting vmsR deletion mutants (strain DR1) was used for further investigation.
Phenotypes of the vmsR deletion mutant.
The amounts of VM produced were determined by reverse-phase high-pressure liquid chromatography (HPLC) using purified VM1 and VS as standards under the following conditions: column, Cosmosil 5C18 (4.6 by 100 mm; Nacalai Tesque, Kyoto, Japan); flow rate, 0.75 ml/min; temperature, 40°C; detection, UV radiation at 305 nm; elution, 5 min with 20% CH3CN containing 0.1% trifluoroacetic acid followed by the linear increase of CH3CN from 20 to 80% for 15 min. The amounts of VB were determined by measuring the VB-dependent production of VM (20). VB binding activity was assayed by the ammonium sulfate precipitation method (10) with 3H-labeled VB-C7 (54.6 Ci/mmol) in the presence and absence of 2,000-fold cold VB-C. When traits relating to VB-BarA in S. virginiae were measured (Table 1), all of them (VB production, VB binding activity, VM production, VM resistance, and morphology) were found to be identical, with high reproducibility, in the wild-type strain and the wild-type segregant (Table 1), indicating that no unexpected mutations relating to the VB-BarA signal transduction pathway took place during the pSVR301 integration into the wild-type strain or during the second crossover event generating either the wild-type segregant or the vmsR disruptant. The vmsR mutant, however, did not produce any VM, even after 48 h of cultivation, and was slightly more sensitive to VM1 and VS (Table 1). No differences in VB production, VB binding activity, growth rate, or morphology on either solid or liquid media were observed between the vmsR mutant and the wild-type strain (Table 1). The phenotypic defect of VM production in the vmsR mutant was complemented by introducing intact vmsR on pUWL-KS into the vmsR mutant (data not shown). These results suggested that VmsR does not participate in morphological control or VB biosynthesis in S. virginiae but does participate, directly or indirectly, in the regulation of VM biosynthesis.
TABLE 1.
Phenotypic comparison of wild-type S. virginiae, a vmsR disruptant, and a wild-type segregant
| Strain | VM productiona (μg/ml) | Resistance (MIC, μg/ml)
|
VB productionb (nM) | VB binding activityc (pmol/mg of protein) | Sporulation | |
|---|---|---|---|---|---|---|
| VM1 | VS | |||||
| Wild type | 86 | 300 | 300 | 390 | 0.44 | + |
| ΔvmsR | 0 | 200 | 200 | 390 | 0.44 | + |
| Wild-type segregant | 87 | 300 | 300 | 370 | 0.46 | + |
VM production was determined after 24 h of cultivation.
VB production was determined after 12 h of cultivation.
VB binding activity was determined for 12-h mycelia.
Analysis of antibiotic products.
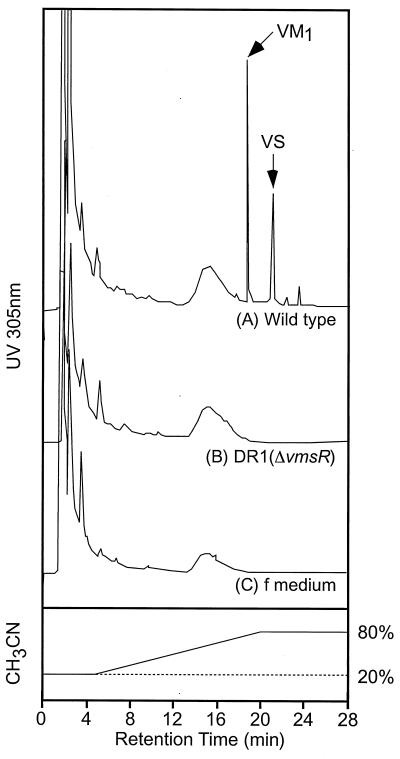
A further experiment was designed to determine the amounts of two components of VM, namely VM1 and VS, because the loss of either VM1 or VS production in the vmsR mutant would lead to great loss of antibiotic activity from the synergistic action of VM1 and VS. The 24-h culture supernatant was recovered after centrifugation, and the amounts of VM1 and VS were analyzed by HPLC. Under the HPLC conditions employed, VM1 and VS produced by the wild-type strain were detected easily at retention times of approximately 18 and 21 min, respectively (Fig. 2A), while the vmsR mutant did not show any signs of VM1 or VS (Fig. 2B), indicating clearly that the VmsR protein is essential for the production of both VM1 and VS.
FIG. 2.
Detection of two components of antibiotics, VM1 and VS, by C18 reverse-phase HPLC analysis. Authentic VM1 and VS were purified from the commercial animal-feed additive STAFAC 500 (Smith Kline-RIT, Rixensart, Belgium) by C18 reverse-phase HPLC (14).
Transcriptional patterns of the vmsR-surrounding genes in the vmsR mutant and the wild-type strain.
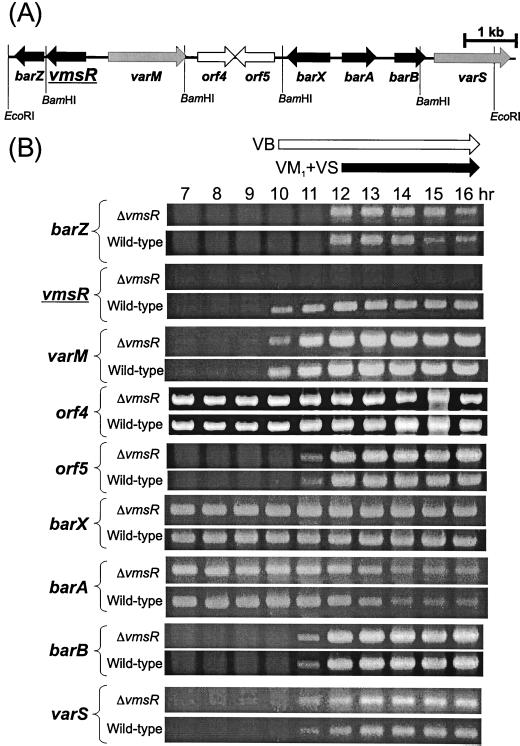
Reverse transcription (RT)-PCR of two genes immediately adjacent to vmsR, namely barZ transcribed in the same direction as vmsR and varM transcribed divergently from vmsR, revealed that transcription of these genes in the vmsR mutant is similar to that in the wild-type strain, indicating that the deletion in vmsR did not have a polar effect on barZ or a deleterious effect on varM. Therefore, the defect in VM biosynthesis in the vmsR mutant can be concluded to be due to the lack of functional VmsR protein rather than any undesired effect on the surrounding genes. Thus far, six other genes (orf4, orf5, barX, barA, barB, and varS) have been sequenced, and none of them showed any changes in transcription in the vmsR deletion mutant (Fig. 3), which suggests that they are not under the control of vmsR.
FIG. 3.
(A) Gene organization in the 10-kbp region containing the vmsR gene in S. virginiae. Solid arrows, gray arrows, and open arrows indicate regulator genes, resistance genes for VM, and genes of unknown function, respectively. (B) Transcriptional comparison of vmsR-surrounding genes in the vmsR disruptant and the wild-type strain by RT-PCR. RT-PCR conditions were previously described (8). Production of VB and VM1 plus VS is indicated by open and solid arrows, respectively. With the entire sample, RT-PCR was carried out without reverse transcriptase to confirm that the signals shown were derived from mRNA and not from contaminating genomic DNA. ΔvmsR, vmsR disruptant; Wild-type, S. virginiae MAFF 10-06014.
Based on the phenotypic analysis, the level of resistance to VM1 and VS was slightly reduced in the vmsR mutant compared to that in the wild-type strain (Table 1), although the transcription of the resistance genes (varS, which encodes the VS transporter and therefore confers VS resistance [14], and varM, which likely encodes the VM1 transporter [8]) was not impaired in the vmsR mutant (Fig. 3). In the case of varS, in addition to the VB-dependent derepression of barB-varS bicistronic transcription, varS monocistronic transcription was induced by the presence of VS (14), which should function to strengthen the VS resistance level. Similar regulation to enhance the VM1 resistance may be operative in the presence of VM1; the absence of VM1 and VS (i.e., not the absence of the VmsR protein per se) in the vmsR mutant seems to be the main reason for the slightly lowered resistance.
The amino acid sequence of VmsR is very similar to those of DnrI, ActII-ORF4, and RedD, all of which belong to an expanding family of Streptomyces antibiotic regulatory proteins (SARPs). SARPs are predicted to have a similar mechanism of transcriptional activation through binding to specific nucleotide sequences, and probable DNA recognition sites (helix α3 and a loop connecting two C-terminal β-strands [β6 and β7]) have been estimated on the basis of the crystal structure of OmpR (30). Alignment of the DNA-binding domains of OmpR with homologous regions of VmsR and SARPs reveals a number of highly conserved amino acids. Recently, as predicted by Wietzorrek and Bibb (30), the ActII-ORF4 protein has been confirmed to be a DNA-binding transcriptional activator by gel shift assays and DNase I footprinting (1). Thus, it seems highly probable that VmsR is a DNA-binding protein which acts as a transcriptional activator. It is currently unclear whether VmsR activates VM production by directly activating the transcription of the biosynthetic gene cluster for VM or via another regulator, because no biosynthetic genes for VM have been cloned from S. virginiae, although biosynthetic genes for the closely related antibiotic pristinamycin have been found in Streptomyces pristinaespiralis (2).
In this work, we obtained in vivo evidence that VmsR is a positive regulator of the biosynthesis of both VM1 and VS by constructing and analyzing the vmsR deletion mutant. However, it is not clear at present whether the transcription of vmsR is regulated directly by VB-BarA or indirectly via another regulator(s), such as the BarB protein. We are currently constructing a barB disruptant, the phenotypic and transcriptional analyses of which will clarify the transcriptional cascades among several regulators, such as BarA, VmsR, and BarB.
Acknowledgments
This work was supported in part by a grant from the Research for the Future Program of the Japan Society for the Promotion of Science (JSPS).
REFERENCES
- 1.Arias P, Fernández-Moreno M A, Malpartida F. Characterization of the pathway-specific positive transcriptional regulator for actinorhodin biosynthesis in Streptomyces coelicolor A3(2) as a DNA-binding protein. J Bacteriol. 1999;181:6958–6968. doi: 10.1128/jb.181.22.6958-6968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bamas-Jacques N, Lorenzon S, Lacroix P, de Swetschin C, Crouzet J. Cluster organization of the genes of Streptomyces pristinaespiralis involved in pristinamycin biosynthesis and resistance elucidated by pulsed-field gel electrophoresis. J Appl Microbiol. 1999;87:939–948. doi: 10.1046/j.1365-2672.1999.00955.x. [DOI] [PubMed] [Google Scholar]
- 3.Fernández-Moreno M A, Caballero J L, Hopwood D A, Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell. 1991;66:769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Jessee J, Bloom F R. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- 6.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 7.Kawachi R, Nihira T, Yamada Y. Development of a transformation system in Streptomyces virginiae. Actinomycetologica. 1997;11:46–53. [Google Scholar]
- 8.Kawachi R, Akashi T, Kamitani Y, Sy A, Wangchaisoonthorn U, Nihira T, Yamada Y. Identification of an AfsA homologue (BarX) from Streptomyces virginiae as a pleiotropic regulator controlling autoregulator biosynthesis, virginiamycin biosynthesis and virginiamycin M1 resistance. Mol Microbiol. 2000;35:302–313. doi: 10.1046/j.1365-2958.2000.01819.x. [DOI] [PubMed] [Google Scholar]
- 9.Kim H S, Nihira T, Tada H, Yanagimoto M, Yamada Y. Identification of binding protein of virginiae butanolide C, an autoregulator in virginiamycin production, from Streptomyces virginiae. J Antibiot. 1989;42:769–778. doi: 10.7164/antibiotics.42.769. [DOI] [PubMed] [Google Scholar]
- 10.Kim H S, Tada H, Nihira T, Yamada Y. Purification and characterization of virginiae butanolide C-binding protein, a possible pleiotropic signal-transducer in Streptomyces virginiae. J Antibiot. 1990;43:692–706. doi: 10.7164/antibiotics.43.692. [DOI] [PubMed] [Google Scholar]
- 11.Kinoshita H, Ipposhi H, Okamoto S, Nakano H, Nihira T, Yamada Y. Butyrolactone autoregulator receptor protein (BarA) as a transcriptional regulator in Streptomyces virginiae. J Bacteriol. 1997;179:6986–6993. doi: 10.1128/jb.179.22.6986-6993.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kinoshita H, Tsuji T, Ipposhi H, Nihira T, Yamada Y. Characterization of binding sequences for butyrolactone autoregulator receptors in streptomycetes. J Bacteriol. 1999;181:5075–5080. doi: 10.1128/jb.181.16.5075-5080.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo K, Higuchi Y, Sakuda S, Nihira T, Yamada Y. New virginiae butanolide from Streptomyces virginiae. J Antibiot. 1989;42:1873–1876. doi: 10.7164/antibiotics.42.1873. [DOI] [PubMed] [Google Scholar]
- 14.Lee C-K, Minami M, Sakuda S, Nihira T, Yamada Y. Stereospecific reduction of virginiamycin M1 as the virginiamycin resistance pathway in Streptomyces virginiae. Antimicrob Agents Chemother. 1996;40:595–601. doi: 10.1128/aac.40.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez-Hackert E, Stock A M. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure. 1997;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 16.Mori K. Revision of the absolute configuration of A-factor. J Antibiot. 1983;35:349–358. [Google Scholar]
- 17.Nakano H, Takehara E, Nihira T, Yamada Y. Gene replacement analysis of the Streptomyces virginiae barA gene encoding the butyrolactone autoregulator receptor reveals that BarA acts as a repressor in virginiamycin biosynthesis. J Bacteriol. 1998;180:3317–3322. doi: 10.1128/jb.180.13.3317-3322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano, N., C.-K. Lee, T. Nihira, and Y. Yamada. A null mutant of the Streptomyces virginiae barA gene encoding butyrolactone autoregulator receptor, and its pleiotropic and transcriptional analyses. J. Biosci. Biotechnol. 90:204–207. [DOI] [PubMed]
- 19.Narva K E, Feitelson J S. Nucleotide sequence and transcriptional analysis of the redD locus of Streptomyces coelicolor A3(2) J Bacteriol. 1990;172:326–333. doi: 10.1128/jb.172.1.326-333.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nihira T, Shimizu Y, Kim H S, Yamada Y. Structure-activity relationships of virginiae butanolide C, an inducer of virginiamycin production in Streptomyces virginiae. J Antibiot. 1988;41:1828–1837. doi: 10.7164/antibiotics.41.1828. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto S, Nakamura N, Nihira T, Yamada Y. Virginiae butanolide binding protein from Streptomyces virginiae. J Biol Chem. 1995;270:12319–12326. doi: 10.1074/jbc.270.20.12319. [DOI] [PubMed] [Google Scholar]
- 22.Onaka H, Ando N, Nihira T, Yamada Y, Beppu T, Horinouchi S. Cloning and characterization of the A-factor receptor gene from Streptomyces griseus. J Bacteriol. 1995;177:6083–6092. doi: 10.1128/jb.177.21.6083-6092.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruengjitchatchawalya M, Nihira T, Yamada Y. Purification and characterization of the IM-2-binding protein from Streptomyces sp. strain FRI-5. J Bacteriol. 1995;177:551–557. doi: 10.1128/jb.177.3.551-557.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Sato K, Nihira T, Sakuda S, Yanagimoto M, Yamada Y. Isolation and structure of a new butyrolactone autoregulator from Streptomyces sp. FRI-5. J Ferment Technol. 1989;68:170–173. [Google Scholar]
- 26.Stutzman-Engwall K J, Otten S L, Hutchinson C R. Regulation of secondary metabolism in Streptomyces spp. and overproduction of daunorubicin in Streptomyces peucetius. J Bacteriol. 1992;174:144–154. doi: 10.1128/jb.174.1.144-154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takano E, Nihira T, Hara Y, Jones J J, Gershater C J L, Yamada Y, Bibb M. Purification and structural determination of SCB1, a γ-butyrolactone that elicits antibiotic production in Streptomyces coelicolor A3(2) J Biol Chem. 2000;275:11010–11016. doi: 10.1074/jbc.275.15.11010. [DOI] [PubMed] [Google Scholar]
- 28.Waki M, Nihira T, Yamada Y. Cloning and characterization of the gene (farA) encoding the receptor for an extracellular regulatory factor (IM-2) from Streptomyces sp. strain FRI-5. J Bacteriol. 1997;179:5131–5137. doi: 10.1128/jb.179.16.5131-5137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wehmeier U F. New multifunctional Escherichia coli-Streptomyces shuttle vectors allowing blue-white screening on XGal plates. Gene. 1995;165:149–150. doi: 10.1016/0378-1119(95)00513-6. [DOI] [PubMed] [Google Scholar]
- 30.Wietzorrek A, Bibb M. A novel family of proteins that regulates antibiotic production in streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol Microbiol. 1997;25:1177–1184. doi: 10.1046/j.1365-2958.1997.5421903.x. [DOI] [PubMed] [Google Scholar]
- 31.Yamada Y, Sugamura K, Kondo K, Yanagimoto M, Okada H. The structure of inducing factors for virginiamycin production in Streptomyces virginiae. J Antibiot. 1987;40:496–504. doi: 10.7164/antibiotics.40.496. [DOI] [PubMed] [Google Scholar]
- 32.Yamada Y, Nihira T. Microbial hormones and microbial chemical ecology. In: Barton D H R, Nakanishi K, Meth-Cohn O, editors. Comprehensive natural products chemistry. Vol. 8. Oxford, United Kingdom: Elsevier Science Ltd.; 1999. pp. 377–413. [Google Scholar]
- 33.Yanagimoto M. Novel actions of inducer in staphylomycin production by Streptomyces virginiae. J Ferment Technol. 1983;61:443–448. [Google Scholar]
- 34.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]