Abstract
Objective
To evaluate the quality of Arnebiae Radix (AR) and Dictamni Cortex (DC) and study the efficacy of herbal extracts of these two herbs on the treatment of allergic contact dermatitis (ACD).
Methods
Qualitative and quantitative analysis of effective components was performed using High Performance Thin Layer Chromatography (HPTLC), High Performance Liquid Chromatography (HPLC), and HPLC-Quadrupole Time of Flight-Mass Spectrometry (HPLC-QTOF-MS). In vitro allergic ACD 3D model was established by incubating 3D reconstructed human epidermis (RHE) with skin sensitizer, potassium dichromate. A total of 65 gene expression that were associated with ACD, which included 24 antioxidant responsive element (ARE) and 41 SENS-IS genes were quantified by qRT-PCR. More than or equal to 10 ARE genes and 18 SENN-IS genes were induced by 1.3-fold, demonstrating the successful establishment of in vitro ACD model. Oil extracts of AR and DC were applied on the in vitro ACD model to study the efficacy.
Results
Batch 3 of AR and batch 2 of DC showed presence of all active ingredients with the highest concentrations. Active ingredients of the herbs were extracted using a special oil and formulated into herbal oil extracts. The herbal oil extracts were able to down regulate the induced genes in the in-vitro ACD skin model, bringing the tissue back to homeostatic status.
Conclusion
The oil extracts showed the potent efficacy of using AR and DC in ACD treatment. The combination study will be done to optimize the formulation ratio which will be developed into a topical cream.
Keywords: allergic contact dermatitis, Arnebiae Radix, Chinese medicine, Dictamni Cortex, 3D reconstructed epidermis, quality control
1. Introduction
Allergic contact dermatitis (ACD) is a type IV or delayed hypersensitivity reaction and occurs after exposure to allergens such as fragrance, cosmetics, chromate, paraphenylenediamine in hair dye and nickel (Ahlström et al., 2019). It is known to be the most prevalent form of immunotoxicity. ACD affects nearly 20% of total population with increasing trend, and the symptoms of allergic contact may last for as long as months before resolving completely (Alinaghi et al., 2019). Recent study reveals that in US, one in four people was seen by physician for at least one skin disease. Among the 24 common skin diseases, ACD is ranked as the 5th claims-based prevalence. Skin disease resulted in 75 billion US dollar direct health care costs each year (Lim et al., 2017). In EU, the attributable healthcare costs for patients with ACD is around €1000, some can go as high as €10 000 (Saetterstrøm et al., 2014). Treatment relies on long term usage of corticosteroids, non-steroid anti-inflammatory drugs, antibiotics and immunosuppressive agents. However, these agents are always associated with various adverse effects and high cost. Due to its complexity and suppression of the immune system, long-term usage of topical corticosteroids could lead to detrimental consequences. Consequences include dermal atrophy and steroid-induced acne vulgaris. Dermal atrophy will cause the affected skin to be more vulnerable to damage. There are also arguments to whether long-term usage may increase the probability of skin and lymphoma cancer (Gordon et al., 2018). Hence, there is a shift towards natural products, especially traditional Chinese medicine (TCM), for alleviating the symptoms and treatment of ACD. The desire for an effective treatment of ACD with minor side effects has remained unfulfilled.
At the initial stage of ACD, the allergen penetrates through the stratum corneum of the skin into the epidermis layer at the sensitization phase. This activates secretion of cytokines such as IL-1 α, IL-18 and TNF-α by the keratinocytes, resulting in epidermal Langerhans cells (LCs) forming a hapten-carrier protein complex (Lee et al., 2013). Maturation of these hapten-carrier LCs is done through the upregulation of surface maturation markers which include CXCR4, CD83 and CD86 and increased CXCL8 secretion. They continue to mature as they migrate to the dermis. There also be upregulation of CCR7 receptors. Together with CXCR4, the complex migrates to the draining lymphoid tissue to be presented to antigen-responsive T cells. The T cells proliferate and mature into effector and memory cells (Nakae et al., 2003).
Exposure to the same allergen causes activation of memory T cell, leading to IL-1, IL-2 and interferon-γ production. This is the elicitation phase where cytotoxic T cells are activated. These effector cells and their pro-inflammatory cytokines attack the epidermis. This causes inflammation and activation of adaptive immune responses, resulting in dermatitis. Suppressor cells secretes cytokines such as IL-4 and IL-10 after several days to stop the reaction. Cytotoxic T cells and Th1 cells are involved in ACD inflammation, however there is a possibility that Th2, Th17 and Th22 are involved as well. Nickel, a common allergen is known to induce the innate immune pathways of Th1, Th17 as well as Th22. Rubber and fragrances have shown to induce Th2 activity with minimal Th1 and Th17 association. Elevated levels of IL-9 expression, a pro-inflammatory mediator, was observed in ACD patients as the Th9 transcription factors such as IRF4 and ETS1 can cause production of IL-9 by Th9 cells (Jadali, 2019). IL-9 can also increase the activity of innate leukocytes such as mast cells and eosinophils, which cause ACD skin infiltration (Barbaud, 2020).
Arnebiae Radix (AR, Chinese name: Zicao) and Dictamni Cortex (DC, Chinese name: Baixianpi) have been individually formulated into various prescriptions in the treatment of patients with skin allergy for thousands of years with little reported side effect (Liang and Wang, 2020, Chen et al., 2021, Yang et al., 2017). The long period of traditional use and experiential research are evidence for their safety and effectiveness. This prompted us to propose a novel herbal extract for ACD treatment. The formula of this product is derived from historical evidence of practitioners and is built on knowledge of the guiding theories of TCM.
Any herbal product available at the pharmacy should meet the strict requirements of the applicable state in Pharmacopoeia. However, raw herb contains numerous chemical compounds with holistic efficacy rather than a single active compound. Hence, it is difficult to standardize the active ingredients contained in herbal remedies given varying climate, regional, and production conditions. The complexities challenge the current official quality control mode for which only one or two markers were selected for quantitative assay stated in pharmacopoeia. Therefore, in order to guarantee the efficacy of this herbal product, it is essential to establish comprehensive analytical methods. Qualitative and quantitative analysis of active ingredients using High Performance Thin Layer Chromatography (HPTLC), High Performance Liquid Chromatography (HPLC) and High Performance Liquid Chromatography-Quadrupole Time of Flight-Mass Spectrometry (HPLC-QTOF-MS) put emphasis on systematic characterization of a complex matrix to evaluate authenticity, superiority and stability of herbs (Kharbach et al., 2020).
The reconstructed human epidermis (RHE) model consists of fully differentiated 3D epidermal tissue grown from normal human keratinocytes in a chemically defined medium at the air liquid interface. RHE is an artificial epidermis which is reconstructed to be similar to the human skin in terms of their structure and functionality. RHE is constructed by growing keratinocytes and melanocytes cells, which forms the epidermal layer, onto an inert filter substrate. The dermis layer containing fibroblasts and collagen cells is constructed as well. The RHE is exposed to the atmospheric air after several days of culture to stimulate differentiation of basal keratinocytes. This differentiation will form the outer layer of the epidermis, comprising of the stratified epidermis and stratum corneum (Rodrigues neves & Gibbs, 2018). RHE models can be used to test for irritant chemicals in topical cosmetic or medicinal products. Organisation for Economic Co-operation and Development (OECD) has approved the use of RHE models for skin irritation and corrosion testing to replace animal testing (Cottrez et al., 2016, Cottrez et al., 2015). This 3D skin model has been validated and regulatory adopted by OECD.
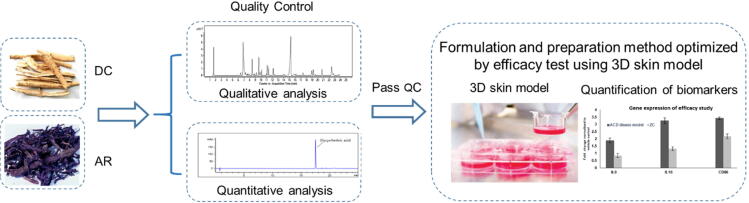
Potassium dichromate induced ACD has been validated based on SENS-IS and Redox/Detox (ARE) assay. A panel of 65 genes, belonging to the SENS-IS and Redox group was identified through data mining and by literature research. The SENS-IS group consists of 41 genes linked to inflammation, tissue repair, apoptosis, danger signals and cell migration. This would allow the complex cascade of events leading to activation of dendritic cells by a sensitizing chemical to be addressed (Cottrez et al., 2015). The Redox group contains a set of 24 genes that are linked to small Maf proteins. These proteins activate the ARE gene transcription, which provides contact allergens with the ability to modify and bind to cysteine amino acids of the Keap1-NRF2 complex. (Cottrez et al., 2015). This can lead to the overexpression of genes in the Redox group such as GSR, GSS, HMOX1, NQO1, TXN synthetase and reductase (Martin et al., 2011). The 24 ARE genes and 41 SENS-IS genes expression will be quantified using qRT-PCR. More than or equal to 10 ARE genes and 18 SENS-IS genes shall be induced by 1.3-fold to indicate successful establishment of an ACD model. Oil extracts of active ingredient of AR and DC will be applied on established ACD model. Efficacy will be evaluated through the 65 gene expression (Cottrez et al., 2015). The oil extracts should be able to downregulate the induced genes expressed in the ACD model and bring the tissue back to normal status. Fig. 1 shows the workflow of this project.
Fig. 1.
Schematic illustration of project workflow of this project.
2. Material and methods
2.1. Reagent and materials
All reference standards with purity higher than 98% were purchased from Biopurify Phytochemicals Ltd. LC-MS grade formic acid, acetonitrile, methanol, HPLC grade methanol was purchased from Merck. Ultrapure water was collected from Sartorius Stedim system. The Episkin Reconstructed Epidermis (13 days old, 1.07 cm2) was purchased from Episkin, L'Oréal France. RNeasy Fibrous Tissue Mini Kit was purchased from QIAGEN Singapore Pte Ltd. cDNA conversation kit (SuperScript™ IV VILO™ Master Mix with ezDNase™ Enzyme) was purchase from Life Technologies Holdings Pte Ltd. Customized Taqman Plate was purchased from ThermoFisher Scientific Applied Biosystems (ABI). TaqMan™ Fast Advanced Master Mix was purchased from ThermoFisher Scientific.
2.2. Quality control of AR
2.2.1. Identification of AR by HPTLC
To prepare reference standard solution, 5 mg of β-acetoxyisovalerylalkannin was weighed in 15 mL centrifuge tube followed by adding 2.5 mL of petroleum ether (60–80 °C). To prepare test solution, 1.0 g of the powdered AR was weighed into a 50 mL centrifuge tube, then 10 mL of petroleum ether (60–80 °C) was added. The mixture was sonicated for 30 min and centrifuged at 4000 g for 10 min. The supernatant was transferred to a 50 mL round bottomed flask. Solvent was evaporated to dryness using a vacuum rotary evaporator. The residue was dissolved in 1 mL of petroleum ether.
The separation was carried out using a CAMAG® HPTLC silica gel F254 plate. The HPTLC system contains a semi-auto sample dispenser, visualizer, development chambers, plate heater and immersion device. The reference standard solution and test solution were applied onto the plate using HPTLC semi-auto sample dispenser and solvent front was marked using pencil before developing. Developing solvent was prepared by mixing cyclohexane, ethyl acetate and formic acid (9:2:0.2, volume percentage) and placed into a twine trough glass chamber. Two filter papers were place on each trough to equilibrate for 30 min. After equilibration, HPTLC plate was placed in one trough. Development was carried out over a path of 7 cm. After the development, the plate was dried and examined under visible light.
2.2.2. Quantification of active ingredients in AR by HPLC assay
To prepare test solution, three batches of 0.5 g of powdered AR were weighed and placed into three 50 mL centrifuge tubes. Acetone (25 mL) was added into each 50 mL centrifuge tube. The mixture was sonicated for 30 min and thereafter centrifuged at 3000 g for 10 min. The supernatant was transferred into a 100 mL round bottom flask and the extraction was repeated for one more time. The supernatants were combined and evaporated to dryness using vacuum rotary evaporator. The residue was dissolved in HPLC grade methanol and transferred into a 10 mL volumetric flask. Methanol was used to make up to the mark on the volumetric flask. The solution was then filtered through a 0.22 µm filter membrane and injected into HPLC.
To prepare reference standard solution, 20 mg of β-acetoxyisovalerylalkannin was dissolved in 50 mL of methanol. Serial dilution was performed to prepare standard solution at concentrations of 400, 200, 100, 50, and 25 mg/L.
Separation was carried out on the Poroshell 120 Agilent Column EC-C18 (2.7 μm, 3.0 mm × 150 mm). The flow rate was set up at 0.3 mL/min. The mobile phase was a mixture of acetonitrile and 0.1% formic acid (70:30, volume percentage). The elution time was 30 min. Injection volume was 5 μL. Wavelength was set up at 516 nm.
2.2.3. Chemical profiling of AR by HPLC-QTOF-MS and HPLC fingerprinting
To prepare reference standard solution, 5 mg of acetylshikonin, β-acetoxyisovalerylalkannin, isobutyrylshikonin, β, β-dimethylacrylshikonin, isovalerylshikonin was dissolved in 5 mL of methanol, respectively.
To prepare test solution, 0.5 g of the powdered AR was weighed and placed in a 50 mL centrifuge tube, then 25 mL of acetone was added. The mixture was sonicated for 30 min and centrifuged at 3000 g for 10 min. The supernatant was transferred to 250 mL round-bottomed flask. The extraction was repeated for one more time. Supernatants were combined and the solvent was evaporated to dryness at reduced pressure in a rotary evaporator. Residue was dissolved in methanol and transferred the solution to a 10 mL volumetric flask and made up to the mark with methanol. The solution was directly filtered through 0.45 μm filter membrane and injected into HPLC. For HPLC-QTOF-MS injection, the test solution was diluted 2 folds and filtered through 0.22 μm filter membrane.
HPLC method for chemical profiling set up was same as HPLC assay (Section 2.2.2). For HPLC-QTOF-MS method set up, HPLC method was same as section 2.2.2. For QTOF-MS method set up, source parameters were optimized as followed, gas temperature, 170 °C; drying gas, 16 L/min; nebulizer, 35 psig; sheath gas temperature, 320 °C; sheath flow, 11 L/min and collision energy, 10 and 30. Auto MS/MS mode was chosen. Reference mass correction was set up at 301.9981. Ion polarity was set up as negative.
2.3. Quality control of DC
2.3.1. Identification of DC by HPTLC
To prepare reference standard solution, 3 mg of dictamnine was dissolved in 10 mL of methanol, 6 mg of fraxinellone was dissolved in 10 mL of methanol, and 6 mg of obacunone was dissolved in 10 mL methanol.
To prepare test solution, 2.0 g of the powdered DC and placed in a 50 mL centrifuge tube and 10 mL of methanol was added. The mixture was sonicated for 15 min and centrifuged at 4000 g for 10 min. Supernatant was filtered through 0.45 μm membrane.
The separation was carried out using a HPTLC silica gel F254 plate. Standard solution of dictamnine (2 μL), fraxinellone (5 μL) and obacunone (10 μL) was applied separately on the plate and 8 μL of test solution was applied to the plate using semi-auto sample dispenser. Solvent front was marked using pencil before developing. Developing solvent was prepared by mixing toluene, methanol and glacial acetic acid (10:0.5:0.1, volume percentage) and placed into a twine trough glass chamber. Two filter papers were place on each trough to equilibrate for 30 min. After equilibration, HPTLC plate was placed in one trough. Development was carried out over a path of 7 cm. After the development, the plate was removed from the chamber and dried in air. The plate was soaked in a chamber evenly with the spray reagent using CAMAG® chromatogram Immersion Device for 5 s and thereafter placed on the plate heater at temperature 105 °C until the spots or bands become visible. The plate was examined under UV light at 366 nm.
2.3.2. Quantification of active ingredients in DC by HPLC assay
To prepare reference standard solution, 5 mg of dictamnine, fraxinellone and obacunone was weighed and dissolved in 25 mL of HPLC grade ethanol as stock solutions, respectively. Serial dilution was performed to prepare standard solution at concentrations of 10, 5, 2.5, 1.25, 0.625, and 0.3125 mg/L for dictamnine; 50, 25, 12.5, 6.25, 3.125, and 1.5625 mg/L for fraxinelone; 200, 100, 50, 25, and 12.5 mg/L for obacunone.
To prepare test solution, three batches of 0.2 g of the powdered DC sample were weighed accurately into a 50 mL centrifuge tube, separately. Totally 10 mL of 70% ethanol was added to the powder. The mixture was sonicated for 15 min and centrifuged at 4000 g for 15 min. The supernatant was transferred to a 100 mL volumetric flask. The extraction was repeated two more times by using 10 mL and 5 mL 70% ethanol respectively. The supernatant was combined and topped up to the mark of volumetric flask with 70% ethanol. Supernatant was filtered through a 0.45 μm filter membrane and injected to HPLC.
Separation was performed using Poroshell 120 Agilent Column EC-C18 (2.7 μm, 3.0 mm × 150 mm). The flow rate was 0.3 mL/min. Injection volume was 5 μL. Wavelength was set up at 238 nm. Table S1 showed the gradient elution programme.
2.3.3. Chemical profiling of DC by HPLC-QTOF-MS and HPLC fingerprinting
To prepare reference standard solution, dictamnine, fraxinellone and obacunone are diluted from stock solution to 5, 25, and 100 mg/L, respectively. DC powder (1 g) was extracted using 10 mL of 70% ethanol by sonicating for 15 min, thereafter centrifuged at 4000 g for 5 min. The supernatant was diluted 2-fold and filtered through 0.22 μm membrane. The filtrate (2 μL) was injected into HPLC-QTOF-MS. The supernatant was filtered through 0.45 μm membrane and 5 μL of the filtrate was injected to HPLC. The wavelength for HPLC was set up at 254 nm. Table S2 showed the gradient elution programme set up for HPLC.
For QTOF-MS method, source parameters were optimized as followed, gas temperature, 170 °C; drying gas, 16 L/min; nebulizer, 35 psig; sheath gas temperature, 320 °C; sheath flow, 11 L/min and collision energy, 10, 20 and 30. Auto MS/MS mode was chosen. Reference mass correction was set up at 301.9981. Ion polarity was set up as positive.
2.4. HPTLC analysis of herbal oil extraction of AR and DC
AR and DC were soaked in a special oil for 4 weeks to produce herbal oil extract separately. For AR and DC test solution, 2 mL of herbal oil extract was place in a 15 mL centrifuge tube and 10 mL of analytical grade methanol was added. The mixture was vortexed for 1 min and then centrifuged at 5000 g for 10 min. A total of 5 mL of the supernatant was transferred to a 25 mL round bottom flask. It was evaporated using rotary evaporator, followed by reconstituting with 1 mL of petroleum ether. The reconstituted solution was filtered through 0.45 μL membrane. Reference standard solution preparation of AR is same as “Section 2.2.1″. Reference standard solution preparation of DC is same as “Section 2.3.1″.
2.5. 3D skin tissue culture and treatment
The Episkin RHE (13 days old, 1.07 cm2) were maintained in sterile 12-well culture plate containing 2 mL/well of maintenance medium supplied with the Episkin kit. The tissue was incubated at 37 °C with 5% CO2 overnight, upon receipt.
Potassium dichromate (20 mg) was dissolved in 2 mL of PBS. On day 14, the skin tissues were exposed to skin sensitizer. A total of 30 μL of potassium dichromate solution were applied onto the stratum corneum of the epidermis. The tissues were then incubated with the skin sensitizer for 15 min and washed with 25 mL of PBS per tissue. After washing, for negative control and ACD disease group, the tissues were tapped and dried on the sterilized gauze and placed into a new 12-well plate with 2 mL of assay medium in each well and further incubated at 37 °C, 5% CO2 for 6 h. For treatment group, 30 μL of herbal oil extracts was added evenly onto the stratum corneum of the epidermis and placed into a new 12-well plate with 2 mL of assay medium in each well and were further incubated at same condition as negative control. After 6 h incubation, skin tissue was washed with 25 mL of PBS and removed from the inserts with forceps and place in a 1.5 mL centrifuge tube with RNA protect tissue reagent and stored overnight at 4 °C.
2.6. RNA extraction
Total RNA was extracted using RNeasy Fibrous Tissue Mini Kit. Before RNA extraction, 10 μL of β-mercaptoethanol (β-ME) was added to 1 mL of Buffer RLT and can be stored at room temperature for up to 1 month. Lyophilized Dnase I was dissolved in 550 μL of the RNase-free water and can be stored at −20 °C for up to nine months.
Skin tissue was placed in a 1.5 mL RNase-free tube and 300 μL of Buffer RLT was added. Tissue was disrupted by a motor rod until the lysate is uniformly homogenized. A total of 590 μL of RNase-free water was added to the lysate, 10 μL of proteinase K solution was then added and mixed thoroughly by pipetting. The mixture was incubated at 55 °C for 10 min and thereafter centrifuged at 10 000 g for 3 min at 15–25 °C. A total of 900 μL of supernatant was transferred to a new 1.5 RNase free tube and 450 μL of ethanol (96–100%) was added and mixed well without centrifugation. The mixture (700 μL), including any precipitate, was transferred to a RNeasy Mini spin column, placed in a 2 mL collection tube and centrifuged at 10 000 g for 15 s at 15–25 °C. The flow-through was discarded and another 700 μL of the mixture was added and the process was repeated. Buffer RW1 (350 μL) was added to the RNeasy Mini spin column and centrifuged at 10 000 g for 15 s at 15–25 °C to wash the membrane. The flow-through was discarded. Thereafter, 10 μL of DNase I stock solution was added to 70 μL of Buffer RDD. The mixture (80 μL) was added directly to the RNeasy Mini spin column membrane and was incubated on the bench top for 15 min. After the incubation, 350 μL of Buffer RW1 was added to the RNeasy Mini spin column and centrifuged at 10 000 g for 15 s. The flow-through was discarded. Buffer RPE (500 μL) was then added to RNeasy Mini spin column and centrifuged at 10,000 g for 15 s. The flow-through was discarded. The process was repeated with 500 μL of Buffer RPE again. Finally, the RNeasy Mini spin column was placed in a new 2 mL collection tube (supplied) and spun at full speed for 1 min, to remove any residual buffer. The RNA was then eluted with 30 μL of RNase-free water to a new 1.5 mL centrifuge tube. Concentration and purity of RNA were tested using Nanodrop.
2.7. cDNA conversion
To prepare gDNA digestion reaction mix, 1 μL of 10 × ezDNase buffer, 1 μL of ezDNase enzyme, 2 μg template RNA was added into a PCR tube. RNase-free water was used to make up to 10 μL. The reaction mix was gently mixed and incubated at 37 °C for 2 min. After incubation, 4 μL of SuperScriptTM IV VILOTM Master Mix and 6 μL of nuclease-free water were added. The mixture was gently mixed and incubated at 25 °C for 10 min for primers to anneal, followed by incubation at 50 °C for 10 min for reverse transcription of RNA to cDNA. Then enzymes were inactivated at 85 °C for 5 min. The final cDNA product can be stored at −20 °C for up to one week.
2.8. qRT-PCR analysis
Quantitative RT-PCR was performed using customized 384 Taqman plate, preloaded with primers and probe. Table S3 showed the list of genes analyzed.Each gene was quantified in triplicates. The PCR reaction mix was prepared per reaction by adding 5 μL of TaqMan® Fast Advanced Master Mix (2X), 1 μL of cDNA template (1 pg to 100 ng in Nuclease free water) and 3.5 μL of nuclease free water. The PCR reaction mix is then added to each well on the customized Taqman plate. The plate was then sealed with optical adhesive film and centrifuged at 1000 g to bring the reaction mix to the bottom of the well. The QuantStudio 7 Real-time PCR instrument setup was UNG incubation at 50 °C for 2 min, followed by polymerase activation at 95 °C for 2 min. Afterwards, PCR (40 cycles), denature at 95 °C for 1 s and Anneal/extend at 60 °C for 20 s.
3. Results
3.1. HPTLC results of AR and DC
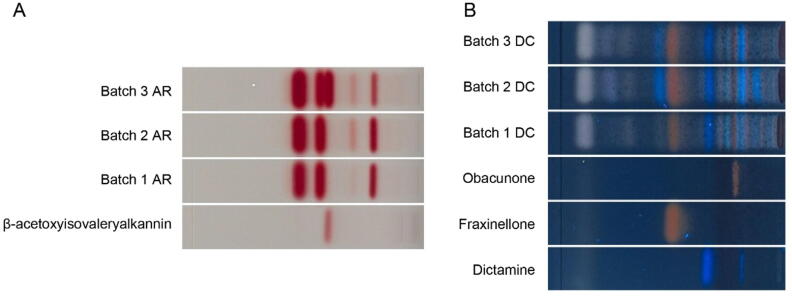
Fig. 2A showed that among three batches, β-acetoxyisovalerylalkannin, the active ingredient of AR, was only present in batch 3 while absent in other batches. Fig. 2B demonstrateed that dictamnine, fraxinellone and obacunone, the active ingredients of DC were present in all three batches.
Fig. 2.
HPTLC chromatogram of β-acetoxyisovalerylalkannin and AR (A) standards and DC (B).
3.2. Quantification of active ingredients in AR and DC using HPLC
The standard curve equation was y = 8.782x + 3.0083 and R2 = 1, where y is peak area and × is the concentration (mg/L). The mass percent concentration of β-acetylisovalerylalkanin in AR were not detected in batch 1 and 2. In batch 3, the weight of β-acetylisovalerylalkanin, to the weight of the herbal powder was 1.16%. It was higher than 0.1%, the limit set by the Hong Kong Materia Medica Pharmacopeia, thus passing the qualification. Hence batch 3 was used for chemical profiling analysis.
Standard curve equation of dictamnine is y = 241.45 x- 8.5353, R2 = 0.999, fraxinellone was y = 34.681 × – 5.0776, R2 = 0.999 and obacunone was y = 4.3397 × + 89.35, R2 = 0.999. All three batches passed the limits indicated in Hong Kong Materia Medica Pharmacopeia, for all active ingredients. For dictamnine, the mass percent concentration was 0.46%, 0.44% and 0.38% in batch 1, 2 and 3, respectively. For fraxinellone, the mass percent concentration was 2.36%, 1.86% and 0.63% in batch 1, 2 and 3, respectively. For obacunone, the mass percent concentration was 1.68%, 4.37% and 4.28% in batch 1, 2 and 3, respectively. However, comparison of all active ingredient, batch 2 showed the highest active ingredient content (mass percent concentration)in total. Since all passed the limits indicated, all three batches were used for chemical profiling analysis.
3.3. Chemical profiling of AR and DC using HPLC-QTOF-MS and HPLC
Five compounds were identified in AR (Table S4) using MS2 fragmentation. Four compounds were identified in DC (Table S5) using MS2 fragmentation.
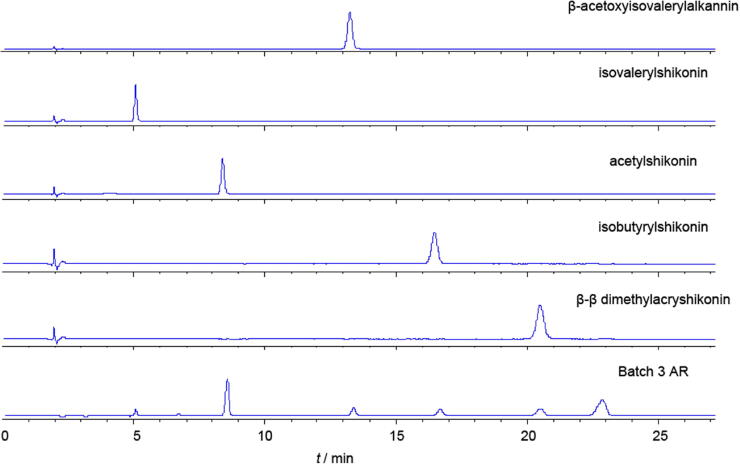
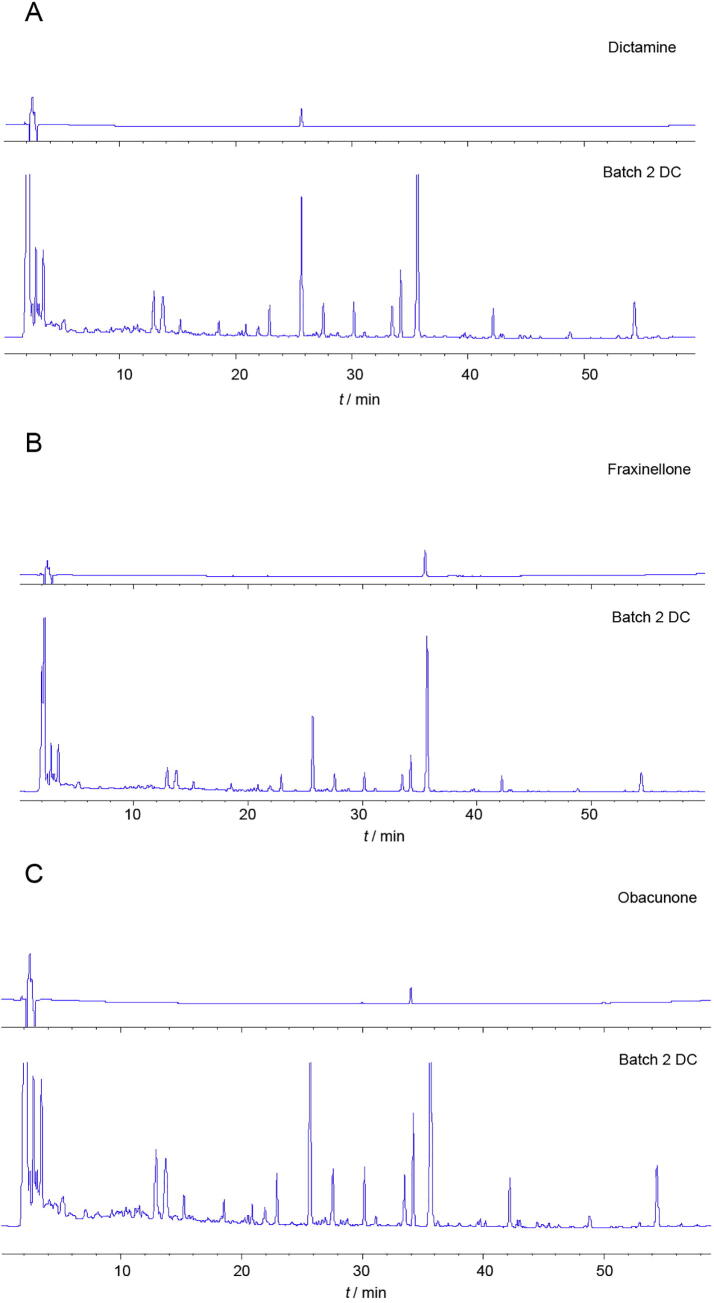
Fig. 3 showed that all of five compounds were present in batch 3 AR sample. Fig. 4 showed that dictamnine, fraxinellone, and obacunone were all present in batch 2 DC.
Fig. 3.
HPLC fingerprinting of five reference standards and AR (batch 3).
Fig. 4.
HPLC fingerprinting of (A) dictamine reference standard and DC (batch 2), (B) fraxinellone reference and standard DC (batch 2), (C) obacunone reference standard and DC (batch 2).
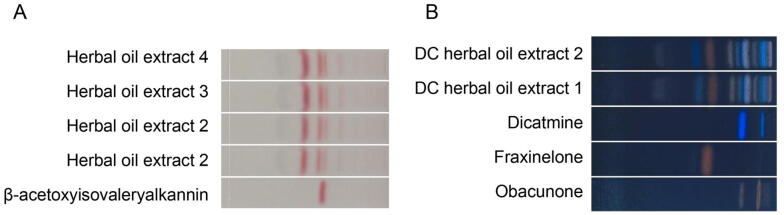
3.4. HPTLC analysis of herbal oil extraction of AR and DC
All active ingredients was found in the herbal oil extraction of AR and DC. Fig. 5A showed that β-acetoxyisovalerylalkannin was present in all herbal oil extracts of AR, while Fig. 5B showed that obacunone, fraxinellone, and dictamnine were present in all herbal extracts of DC.
Fig. 5.
HPLTC chromatogram of extracted AR in herbal oil (A) and extracted DC in herbal oil (B).
3.5. qRT-PCR result of induced gene for efficacy study
In vitro ACD disease model was developed by applying 1% potassium dichromate to the 3D skin cells. From Table 1, more than or equal to 18 SEN-IS and 10 ARE gene have been induced with >1.3-fold change in all three batches of qRT-PCR. After treatment of the ACD model by AR and DC, the number of induced gene decrease. Lower than 18 SENS-IS have been induced and lower than 10 ARE gene have been induced, demonstrating potent efficacy of AR and DC in the treatment of ACD.
Table 1.
qRT-PCR results of gene fold changes in ACD, AR and DC treatment groups.
| Groups | Batch 1 |
Batch 2 |
Batch 3 |
|||
|---|---|---|---|---|---|---|
| Gene number of SEN-IS with fold change > 1.3 | Gene number of ARE with fold change > 1.3 | Gene number of SEN-IS with fold change > 1.3 | Gene number of ARE with fold change > 1.3 | Gene number of SEN-IS with fold change > 1.3 | Gene number of ARE with fold change > 1.3 | |
| 1% Potassium dichromate induced ACD disease group | 20 | 16 | 20 | 16 | 18 | 10 |
| Treatment 1 AR | 8 | 10 | 5 | 11 | 17 | 3 |
| Treatment 2 DC | 10 | 7 | 9 | 7 | 10 | 3 |
4. Discussion
The choice of a RHE model as the in vitro test system is due to its similarity to human skin and conveniently allows the testing of chemicals under conditions close to the human use. For efficacy study, the first step was to establish an ACD disease model. In clinic, three skin sensitizers, potassium dichromate, formaldehyde and methyldibromo glutaronitrile are listed in the patch test, the diagnostic method used to determine which specific compound cause ACD in the patient (Fonacier & Noor, 2018). Previous experiments showed that the extreme skin sensitizer, potassium dichromate induced the most number of SENS-IS and ARE genes compared to the other two sensitizers. Hence, potassium dichromate was used to develop the ACD disease model. In our study, application of 1% potassium dichromate to the 3D RHE was able to induce > 1.3-fold change in>18 SEN-IS and 10 ARE gene, respectively. This demonstrated the successful establishment of an in-vitro 3D ACD model. Most of the genes that are induced by potassium dichromate are related to inflammation, cell stress and heat shock in SEN-IS category. For example, HSPB1 is one of the heat-shock protein family member and is involved in the initial pathway of skin sensitizer interactions with the epidermis in the ACD model (Scieglinska et al., 2019), while AKR1C2, GSTP1, ING1 and UGT1A1 are frequently induced in ARE group.
Due to the high stability of hyaluronic acid-collagen matrix, it is difficult to homogenize the skin tissue and extract high amounts of pure RNA. There are several available methods to extract RNA from skin such as using Qiazol reagent and the general RNeasy Mini Kit. Recently, researchers have demonstrated that the optimal method for extracting RNA from human skin tissue was to use RNeasy Fibrous Tissue Kit by Qiagen. The RLT lysis buffer combined with β-mercaptoethanol in this kit gives high yield of pure RNA (Reimann et al., 2019). The extracted RNA was quantified and purity was analyzed using Nanodrop. A260/A280 ratio of approximate 2 and A260/A230 ratio of 1.8–2.2 are generally accepted as pure RNA. A low A260/A280 ratio indicates the presence of protein, phenol or other contaminants and low A260/A230 means residual phenol, guanidine, carbohydrate and proteins are present in the sample. All RNA extracted from RHE in this project had an average concentration around 1 μg/μL with high purity.
For qRT-PCR analysis, SYBR green method was used in the Cottrez’s paper (Cottrez et al., 2015). When SYBR dye is added to sample, it immediately binds to double-stranded DNA. The target sequence is amplified by DNA polymerase, producing new copy of double-stranded DNA. SYBR dye will bind to each new copy of double-stranded DNA and the fluorescence intensity increases proportionally to the amount of PCR product. SYBR dye can be used to monitor the amplification of any double-stranded DNA. There is no requirement of using a probe thus reducing the running cost. However, the primary disadvantage is that it may generate false positive signals because it binds to any double-stranded DNA. This will result in low specificity. Therefore, it is essential to design primers that do not amplify non-target sequence and melt curve analysis should be performed (Cruz-Flores et al., 2019). In this project, we chose Taqman-based detection. Probes containing a fluorescent reporter dye on the 5′ end and a quencher dye on the 3′ end is designed to specifically anneal downstream from one of the primer sites. When the oligonucleotide probe is intact, the proximity of the quencher reduces the fluorescence emission by the reporter through fluorescence resonance energy transfer. When the probe binds to the specific target sequence, Taq DNA polymerase synthesis 5′ to 3′ new DNA strand and cleaves the probe. This cleavage removes the probe from the target sequence, separates reporter and quencher dye. The fluorescence emission is not inhibited by quencher anymore, hence, resulting in increase in fluorescence intensity that is proportional to the amount of amplicon produced (Zhou et al., 2017). Due to the advantage of Taqman has over SYBR, we chose customized Taqman plate with pre-loaded primers and probe loaded by automation. This will reduce human errors that can occur when adding primer individually into each 384 well.
Based on the qRT-PCR result, it demonstrated that both AR and DC can significantly decrease the induced gene expression and bring the skin tissue to homeostasis stage. In addition, DC shows slightly better efficacy than AR. Both DC and AR have been used in treating different types of skin allergy for thousand years in China. DC has strong anti-inflammatory effect. One recent case study showed that DC can alleviate the ACD caused by stainless steel implants for tibiofibular fracture. It is the top most common single herb prescribed for the treatment of patients with skin allergy in Taiwan (Weng et al, 2016).
5. Conclusion
The successful establishment of the in vitro 3D ACD model can be used as a platform for rapid screening of other herbs and active ingredients. It is a superior alternative to monolayer culture and it will also reduce the need for testing on animal models, while maintaining its experimental rigor. AR and DC have shown to be effective in reducing expression of the over expressed genes in the ACD model. This is a good starting point for the development of a topical cream to treat ACD. For future work, combination of AR and DC should be tested using the in-vitro 3D ACD model. The formulation ratio of these two herbs should be optimized and thereafter can be developed into a topical cream.
CRediT authorship contribution statement
Huan Li: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Visualization, Writing – review & editing. Esther Lim: Formal analysis, Project administration, Resources. Gladys Ang: Formal analysis, Project administration, Resources. Zhi-qing Lim: Formal analysis. Martin Hui Cai: Conceptualization, Formal analysis, Funding acquisition, Resources, Visualization. Jo-anne Loh: Formal analysis, Writing – review & editing. Celine Ng: Formal analysis, Funding acquisition, Project administration. Peijia Seetoh: Funding acquisition, Resources, Visualization. Edmund Tian: Funding acquisition, Resources, Visualization. Lay Beng Goh: Funding acquisition, Resources, Visualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The research was supported by 2019 Translational R&D and Innovation Fund Grant from Ministry of Education in Singapore.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.chmed.2021.10.006.
Contributor Information
Huan Li, Email: LI_Huan@TP.EDU.SG.
Edmund Tian, Email: Edmund_TIAN@TP.EDU.SG.
Lay Beng Goh, Email: GOH_Lay_Beng@TP.EDU.SG.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Ahlström M.G., Thyssen J.P., Wennervaldt M., Menné T., Johansen J.D. Nickel allergy and allergic contact dermatitis: A clinical review of immunology, epidemiology, exposure, and treatment. Contact Dermatitis. 2019;81(4):227–241. doi: 10.1111/cod.13327. [DOI] [PubMed] [Google Scholar]
- Alinaghi F., Bennike N.H., Egeberg A., Thyssen J.P., Johansen J.D. Prevalence of contact allergy in the general population: A systematic review and meta-analysis. Contact Dermatitis. 2019;80(2):77–85. doi: 10.1111/cod.13119. [DOI] [PubMed] [Google Scholar]
- Barbaud A. Mechanism and diagnosis of protein contact dermatitis. Current Opinion on Allergy and Clinical Immunology. 2020;20(2):117–121. doi: 10.1097/ACI.0000000000000621. [DOI] [PubMed] [Google Scholar]
- Chen Y., Xian Y.F., Loo S., Chan W.Y., Liu L., Lin Z.X. Anti-atopic dermatitis effects of dictamni cortex: Studies on in vitro and in vivo experimental models. Phytomedicine. 2021;82 doi: 10.1016/j.phymed.2020.153453. [DOI] [PubMed] [Google Scholar]
- Cottrez F., Boitel E., Auriault C., Aeby P., Groux H. Genes specifically modulated in sensitized skins allow the detection of sensitizers in a reconstructed human skin model. Development of the SENS-IS assay. Toxicol In Vitro. 2015;29(4):787–802. doi: 10.1016/j.tiv.2015.02.012. [DOI] [PubMed] [Google Scholar]
- Cottrez F., Boitel E., Ourlin J.C., Peiffer J.L., Fabre I., Henaoui I.S.…Groux H. SENS-IS, a 3D reconstituted epidermis based model for quantifying chemical sensitization potency: Reproducibility and predictivity results from an inter-laboratory study. Toxicol In Vitro. 2016;32:248–260. doi: 10.1016/j.tiv.2016.01.007. [DOI] [PubMed] [Google Scholar]
- Cruz-Flores R., Mai H.N., Dhar A.K. Multiplex SYBR Green and duplex TaqMan real-time PCR assays for the detection of Photorhabdus Insect-Related (Pir) toxin genes pirA and pirB. Molecular and Cell Probes. 2019;43:20–28. doi: 10.1016/j.mcp.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonacier L., Noor I. Contact dermatitis and patch testing for the allergist. Annals of Allergy Asthma & Immunology. 2018;120(6):592–598. doi: 10.1016/j.anai.2018.03.003. [DOI] [PubMed] [Google Scholar]
- Gordon W.C., López V.G., Bhattacharjee S., Gil D.R., Díaz J.A., de la Losa F.P.…Bazan N.G. A nonsteroidal novel formulation targeting inflammatory and pruritus-related mediators modulates experimental allergic contact dermatitis. Dermatologic Therapy (Heidelb) 2018;8(1):111–126. doi: 10.1007/s13555-018-0223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadali Z. Th9 cells as a new player in inflammatory skin disorders. Iranian Journal of Allergy Asthma and Immunology. 2019;18(2):120–130. [PubMed] [Google Scholar]
- Kharbach M., Marmouzi I., El Jemli M., Bouklouze A., Vander Heyden Y. Recent advances in untargeted and targeted approaches applied in herbal-extracts and essential-oils fingerprinting - A review. Journal of Pharmaceutical and Biomedical Analysis. 2020;177:112849. doi: 10.1016/j.jpba.2019.112849. [DOI] [PubMed] [Google Scholar]
- Lee H.Y., Stieger M., Yawalkar N., Kakeda M. Cytokines and chemokines in irritant contact dermatitis. Mediators of Inflammation. 2013;2013:1–7. doi: 10.1155/2013/916497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Wang S. Effectiveness of gromwell (Lithospermum erythrorhizon) oil in the treatment of toxic epidermal necrolysis in a patient with dermatomyositis. Dermatologic Therapy. 2020;33(6) doi: 10.1111/dth.14067. [DOI] [PubMed] [Google Scholar]
- Lim H.W., Collins S.A.B., Resneck J.S., Bolognia J.L., Hodge J.A., Rohrer T.A.…Moyano J.V. The burden of skin disease in the United States. Journal of the American Academy of Dermatology. 2017;76(5):958–972.e2. doi: 10.1016/j.jaad.2016.12.043. [DOI] [PubMed] [Google Scholar]
- Martin S.F., Esser P.R., Weber F.C., Jakob T., Freudenberg M.A., Schmidt M., Goebeler M. Mechanisms of chemical-induced innate immunity in allergic contact dermatitis. Allergy. 2011;66(9):1152–1163. doi: 10.1111/j.1398-9995.2011.02652.x. [DOI] [PubMed] [Google Scholar]
- Nakae, S., Komiyama, Y., Narumi, S., Sudo, K., Horai, R., Tagawa, Y., SEKIKAWA, K., Matsushima, K., Asano, M. & Iwakura, Y. (2003). IL-1-induced tumor necrosis factor-alpha elicits inflammatory cell infiltration in the skin by inducing IFN-gamma-inducible protein 10 in the elicitation phase of the contact hypersensitivity response. International Immunology, 15 (2), 251-60. [DOI] [PubMed]
- Reimann E., Abram K., Koks S., Kingo K., Fazeli A. Identification of an optimal method for extracting RNA from human skin biopsy, using domestic pig as a model system. Scientific Reports. 2019;9:20111. doi: 10.1038/s41598-019-56579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, neves C., Gibbs S. Progress on reconstructed human skin models for allergy research and identifying contact sensitizers. Current Topics in Microbiology and Immunology. 2018;430:103–129. doi: 10.1007/82_2018_88. [DOI] [PubMed] [Google Scholar]
- Saetterstrøm B., Olsen J., Johansen J.D. Cost-of-illness of patients with contact dermatitis in Denmark. Contact Dermatitis. 2014;71(3):154–161. doi: 10.1111/cod.12231. [DOI] [PubMed] [Google Scholar]
- Scieglinska Dorota, Krawczyk Zdzisław, Sojka Damian Robert, Gogler-Pigłowska Agnieszka. Heat shock proteins in the physiology and pathophysiology of epidermal keratinocytes. Cell Stress Chaperones. 2019;24(6):1027–1044. doi: 10.1007/s12192-019-01044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng S.W., Chen B.C., Wang Y.C., Liu C.K., Sun M.F., Chang C.M.…Yen H.R. Traditional Chinese medicine use among patients with psoriasis in Taiwan: A nationwide population-based study. Evidence-Based Complementary and Alternative Medicine. 2016;2016:1–13. doi: 10.1155/2016/3164105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Lee H.B., Kim S., Park Y.C., Kim K., Kim H. Decoction of Dictamnus dasycarpus Turcz. root bark ameliorates skin lesions and inhibits inflammatory reactions in mice with contact dermatitis. Pharmacognosy Magazine. 2017;13(51):483–487. doi: 10.4103/0973-1296.211034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.R., Zhang T.S., Song D.P., Huang T., Peng Q., Chen Y.J.…Tang Y.X. Comparison and evaluation of conventional RT-PCR, SYBR green I and TaqMan real-time RT-PCR assays for the detection of porcine epidemic diarrhea virus. Molecular and Cell Probes. 2017;33:36–41. doi: 10.1016/j.mcp.2017.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.