Abstract
Objective
Amomum villosum (AV) is an herb whose dried fruit has been extensively used in modern medicine to treat digestive system diseases such as dysentery, vomiting and abdominal pain. This paper aims to supplement chloroplast (cp) genomic resources and to be used in phylogenetic studies and identification of AV related plants.
Methods
High-throughput sequencing technology was used to determine the complete sequence of the AV cp genome, and the sequence was then compared with three related species.
Results
The genome size of AV we obtained was 163,968 bp with an obvious tetrad structure. The AV cp genome was observed to contain 125 unique genes and 81 simple sequence repeat (SSRs) had been determined and the majority of which were adenine–thymine (AT)-rich. Comparative analysis of genome sequence of four ginger plants showed that the atpF, clpP and rpl32 genes are potential markers for identifying Amomum species. Phylogenetic analysis suggested that AV was closely related to A. kravanh and A. compactum.
Conclusion
These results have brought useful genetic resources for further identification researches, DNA barcoding, resolving taxonomy and understanding the evolutionary mode of Zingiberaceae cp genome.
Keywords: Amomum villosum Lour., chloroplast genome, phylogenetic analysis, Zingiberaceae
1. Introduction
The chloroplast (cp) is formed by endosymbiotic interactions between photosynthetic bacteria and non-photosynthetic hosts (Xiang et al., 2016). The cp plays important roles throughout the plant life cycle, including photosynthesis, metabolism, and starch, pigments, fatty acids, and amino acids biosynthesis (Daniell et al., 2016, Liu et al., 2018, Park et al., 2017c). Due to containing valuable information with highly conservative nature, the cp genome has been widely used in molecular markers, barcode identification, phylogenetic analysis and other fields (Wu et al., 2017, Provan et al., 2001, Park et al., 2017a, Park et al., 2017b). The cp genomes exhibit a circular quadripartite structure ranging from 120 to 160 kb in length, and these genomes typically consist of four parts: one long single-copy (LSC) region, one short single-copy (SSC) region, and two copies of a large inverted repeat (IR) region.
The cp genome exhibits a typical tetragonal structure with a length between 120 and 160 kb. These genomes are usually composed of four parts: a long single copy area (LSC), a short single copy area (SSC) and two reverse repeat regions (IR) (Choi and Park, 2015, Yu et al., 2017). The number of genome sequences was significantly increased because of the next-generation sequencing technologies development (Wu et al., 2017). As of February 2019, 2093 complete cp genomes of land plants had already been added to the GenBank, which enable us to gain insights into plant biological diversity, DNA barcoding, evolution, and population genetic analysis (Daniell et al., 2016, Benson et al., 2018, Baczkiewicz et al., 2017, Song et al., 2017).
Amomum villosum Lour. (AV, family Zingiberaceae) is a valuable herbaceous plant distributed in Southeast Asia (e.g., Burma, Laos, and especially in Southern China) (Wang et al., 2018, Li et al., 2010). Its medicinal parts are the ripe fruits or seed groups, which mainly contains volatile terpenes and has antibacterial, anti-ulcer, and anti-diarrhea activities (Huang et al., 2014, Xue et al., 2015, He et al., 2018, Chen et al., 2018). Due to its aroma and flavor, AV can also be used as culinary spices to prepare beverages, tea and some foods (Wang et al., 2018). The wide use of AV has increased demand for the fruits, which are mainly produced from cultivation. AV relies only on artificial pollination during the planting stage because of its flowers’ special structure, where the stigma of the pistil is higher than the stamen anther, making natural pollination difficult (He et al., 2014). Its yield is extremely low (36–60 kg/acre), with high market price of about 3000–5000 RMB/kg. Therefore, there are a number of adulterants and counterfeit AV in the market. Some related fruits or seeds are usually exchanged with AV, such as Amomum compactum Soland. ex Maton, Amomum kravanh Pierre ex Gagnep, Alpinia oxyphylla Miquel, and others, causing consumer health safety hazards and concerns (Wang et al., 2000). These factors have seriously affected the quality of AV. There is a high degree of complexity and diversity in morphology and internal structure, relying on traditional methods to identify them are more difficult. Although there have been some researches related on molecular identification and could provide some information about the taxonomy of AV and its related species (Wang et al., 2000, Wu et al., 2018, Pan et al., 2001, Zhang et al., 2018), few studies were performed on the genetic diversity of cp genome in AV. Therefore, there is a need to apply new methods to identify the AV and adulterants and abundant AV gene resources.
In this paper, the cp DNA structure of AV was determined and analyzed, including its essential organization, codon usage, and comparison of the entire genome. The phylogenetic tree was then constructed using the protein-coding genes of 11 plants in three genera of Zingiberales. Our results provide a complete AV cp genome, which is beneficial to phylogenetic research, breeding and identification of the plants related to AV.
2. Materials and methods
2.1. Leaf DNA extraction and sequencing
The fresh AV leaves were gathered from Yangchun City, which is located in Guangdong Province, China, well-known for producing genuine and high-quality AV. The samples were identified by Prof. Ping Ding (Guangzhou University of Chinese Medicine, Guangdong, China). Voucher specimens were deposited in the herbarium of Guangzhou University of Traditional Chinese Medicine, China. Total DNA was extracted from liquid nitrogen ground leaf powders using a Plant Genomic DNA Kit (Zhanchen Biotech Co., Guangzhou, China) (Wang et al., 2016). The quality and integrity of DNA samples were tested by NanoDrop 2000 spectrometer (Thermo Scientific, Waltham, MA, USA) and agarose gel electrophoresis (Saina et al., 2018). High-quality cpDNA was used to prepare 500 bp (insert size) pair-end DNA sequencing according to the manuscript library (Yang et al., 2018), and were sequenced by employing the Illumina Hiseq 4000 platform (Illumina Inc., San Diego, CA, USA).
2.2. Genome assembly
Briefly, filtering the sequencing reads based on quality value, and the bases which quality < 20 and error rate > 0.01 of 3′ downstream and 5′ upstream were cliped (Yang et al., 2018). First, pair-end sequencing reads were de novo assembled using SOAPdenovo 2 (http://soap.genomics.org.cn/soapdenovo.html) with multi-kmer (35–75). Second, all reported cp genome sequences of dicot were referenced, and all contigs were aligned using BLAST+ (National Center for Biotechnology Information, Bethesda MD, USA) by using the blastn method (Wang et al., 2016). Third, we assembled the contigs to the genome with overlap and read the pair-end relationship. Additionally, the polymerase chain reaction (PCR) was carried on to examine the assembly between LSC/SSC and IRs areas (Table S1).
2.3. Gene annotation and codon usage
The CPGAVAS (Chinese Academy of Medical Sciences, Beijing, China) and DOGMA (University of Texas at Austin, Austin, TX, USA) were used for preliminary gene annotation (Wu et al., 2017). The rRNA genes were confirmed using blastn with a nt database (Yang et al., 2018, Cheng et al., 2013, Iwasaki et al., 2013). The tRNAscan-SE v.2.0 (University of California Santa Cruz, CA, USA) software was used to verify the tRNA genes (Schattner et al., 2005) and the OGDRAW (the Max Planck Institute of Molecular Plant Physiology, Potsdam-Golm, Germany) program was used to generate a circular genome map. The final cp genome of AV was saved in GenBank with the accession number (pending upload). The GC contents and relative synonymous codon usage values (RSCU) of AV genome were analyzed using MEGA7 software to character codon usage (Kumar et al., 2016). The RSCU was the ratio between the actual observed value and the theoretical observed value of the codon (Gu et al., 2018).
2.4. Identification of long repetitive sequences and simple sequence repeats analysis
The size and position of long repeats in the cp genome of three species, including forward repeats, inverted repeats, palindrome repeats, and complement repeats were determined using REDuter software with following parameters: the minimum repeat length = 20 bp, sequence similarity > 90% and the Hamming distance = 3. The positions and types of simple sequence repeats (SSRs) within the AV genome were identified by the MISA software (Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Stadt Seeland, Germany). The microsatellites are some tandem repeats with one to six nucleotides distributed throughout the genome. The SSR thresholds were 10, 6, 5, 5, 5 and 5 for mononucleotide, dinucleotide, trinucleotide, tetanotide, pentotide and hexotide, respectively (Wang et al., 2016).
2.5. Analysis of synonymous and non-synonymous substitution rates
Using DnaSP 5 method (The University of Barcelona, Barcelona, Spain) to estimate the ratio of synonymous and (Ks) non-synonymous (Ka) substitution for protein-coding gene between the AV cp genomes and other two plants were estimated (Librado & Rozas, 2009). In order to evaluate the mutation rate of Ka/Ks, a Python script was carried out to extract the shared single protein coding exons and align them with MEGA7 (Wu et al., 2017).
2.6. Cgview comparison tool (CCT) map
The CCT method (University of Alberta, Alberta, Canada) was used to compare AV cp genome with other available plants of Zingiberaceae (Kyalo et al., 2018), and the result was shown as a circle. We signed genes by orthologous groups clusters, and the BLAST software was used to compare AV with other genomes. The distributions of AT were analyzed based on AT skewed as follows: AT skew = [A − T]/[A + T].
2.7. Phylogenetic analysis
A total of 77 common protein-coding genes from 12 species cp genomes were used to determine the AV phylogenetic location, including an outlier plant (T. latifolia). jModeltest 0.1.1 software (The University of Vigo, Vigo, Spain) was applied based on the Akaike information criterion (AIC) to analyze the model of GTR + G + I for the nucleotide sequence (Wang et al., 2016), followed by building the phylogenetic trees using RAxML 8.1.5 software with a rapid bootstrap analysis (1000 replicates) (Stamatakis, 2014). Phylobayes 4.1b was performed to Bayesian inference (BI) analysis with two chain max diff < 0.01 (Wang et al., 2016).
3. Results and discussion
3.1. Features of AV cp genome
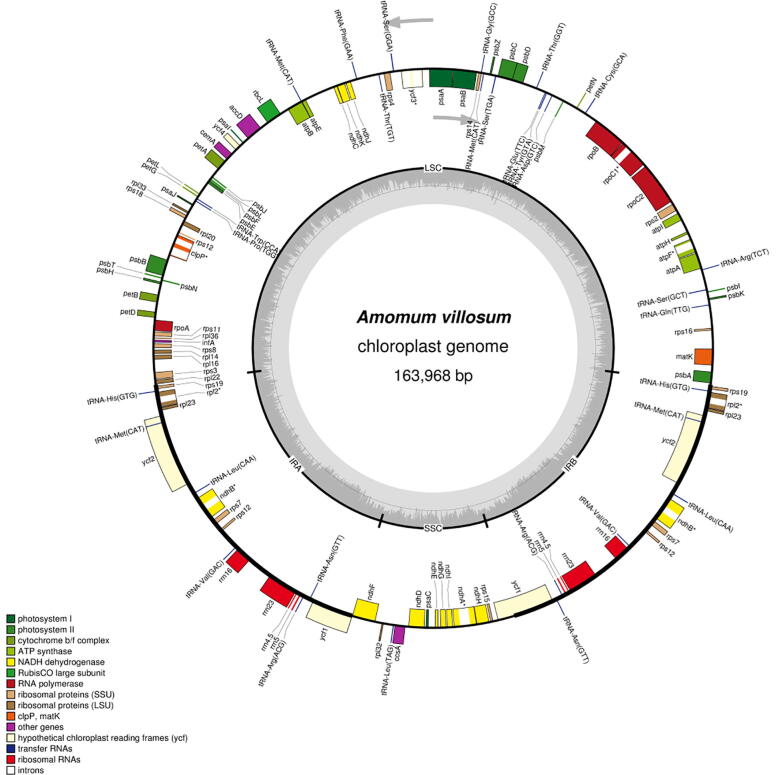
We determined the complete cp genome of AV, a typical quadripartite structure, to be 163,968 bp in size. The large (LSC; 88,798 bp) and short (SSC; 15,352 bp) single copy regions were splited by two inverted repeats (IRs; 29,909 bp) (Table 1 and Fig. 1). The overall GC content of the AV genome was 36.58%, with the IR regions possessing higher GC content (41.08%) than the LSC (33.72%) and SSC regions (29.99%) due to the reduction of AT nucleotides in the four duplicate rRNA genes. Within the protein-coding regions (CDS), the adenine–thymine (AT) content of the third-codon positions (71.2%) was higher than that of the first (55.47%) and second positions (62.57%). This preference for higher AT content in the third bit of codon is often used to distinguish cp DNA from nuclear DNA and mitochondrial DNA, similar to other reported cp genomes (Morton, 1994, Muse and Gaut, 1997, Yang et al., 2012). The cp genome was almost equally divided into non-coding (i.e., intergenic regions, pseudogenes, and introns) and coding regions (Wu et al., 2017).
Table 1.
Base composition of A. villosum (AV) chloroplast genome.
| A(U)/% | T/% | G/% | C/% | Length /bp | |
|---|---|---|---|---|---|
| LSC | 32.48 | 33.80 | 16.50 | 17.22 | 88,798 |
| SSC | 35.79 | 34.23 | 14.27 | 15.72 | 15,352 |
| IRA | 30.20 | 28.72 | 21.26 | 19.82 | 29,909 |
| IRB | 28.72 | 30.20 | 19.82 | 21.26 | 29,909 |
| Total | 31.69 | 32.26 | 18.29 | 18.29 | 163,968 |
| CDS | 31.51 | 31.57 | 19.74 | 17.18 | 83,271 |
| 1st position | 31.50 | 23.97 | 26.35 | 18.19 | 27,757 |
| 2st position | 30.20 | 32.37 | 17.38 | 20.05 | 27,757 |
| 3st position | 32.83 | 38.37 | 15.50 | 13.30 | 27,757 |
LSC: long single-copy; SSC: short single-copy; IR: inverted repeat; CDS: protein-coding regions; A: adenine; T: thymine; G: guanine; C: cytosine.
Fig. 1.
Complete chloroplast (cp) genome map of A. villosum (AV). Gene drawn inside circle is transcribed clockwise, while outside is just opposite. Color coding of genes is based on functional groups they belong to. Dark gray color of inner circle indicates GC content.
In total, the cp gene of AV has 125 unique functional genes, including 87 protein-coding genes (PCGs), 30 transfer RNA (tRNAs) genes, and eight ribosomal RNA (rRNAs) genes (Table 2). Of these, 18 genes were present as duplicates: six tRNAs, four rRNAs, and eight protein-coding genes (rps7, rps12, rps19, rpl2, rpl23, ndhA, ndhB, and ycf2). All eight rRNA were situated in IR regions (Liu et al., 2018, Liu et al., 2017, Zhou et al., 2018), which is consistent with numerous research results. The rps19 gene was found to be located in the IR and LSC boundary region, and ycf1 gene was mapped in the junction of IR and SSC.
Table 2.
Gene contents in A. villosum cp genome.
| Gene category | Gene groups | Gene names |
|---|---|---|
| Self-replication | Transfer RNAs | 30 tRNA genes |
| Ribosomal RNAs | rrn16 (×2), rrn23 (×2), rrn4.5 (×2), rrn5 (×2) | |
| Ribosomal proteins (SSU) | rps2, rps3, rps4, rps7 (×2), rps8, rps11, rps12 (×2), rps14, rps15, rps16, rps18, rps19 (×2) | |
| Ribosomal proteins (LSU) | rpl2 (×2), rpl14, rpl16, rpl20, rpl22, rpl23 (×2), rpl32, rpl33, rpl36 | |
| RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Photosynthesis | NADH-dehydrogenase | ndhA (×2), ndhB (×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ | |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Cytochrome b/f complex | petA, petB, petD, petG, petL, petN | |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| RubisCo Large subunit | rbcL | |
| Other | Translational initiation factor | infA |
| Maturase | matK | |
| Protease | clpP | |
| Envelope membrane protein | cemA | |
| Subunit of Acetyl-CoA-carboxylase | accD | |
| c-Type cytochrome synthesis gene | ccsA | |
| Hypothetical chloroplast reading frames (ycf) | ycf1, ycf2 (×2), ycf3, ycf4 | |
Note: (×2) Genes with two copies.
Introns play an inseparable role in regulating gene expression, and can increase the expression of foreign genes at a specific time and location, they therefore can be used as an important tool to improve the efficiency of transformation (Yi et al., 2012). Among the 125 functional genes, eight genes contained introns and most of them contained only one, whereas ycf3 and clpP harbored two, similar to other species (Chen et al., 2015, Curci et al., 2015). Among the eight intron genes, five protein-coding genes were mapped at the LSC, two at the IR, and only one gene at the SSC area (Table 3). In particular, the rps12 gene was a trans-spliced gene, in which 5′exon was located at the LSC area, while the 3′exon and intron were replicated in the IR area. The ndhA gene contained the longest intron region (1049 bp).
Table 3.
Genes with introns in AV cp genome, and lengths of exons and introns.
| Genes | Locations | Exon I /bp | Intron I /bp | Exon II /bp | Intron II /bp | Exon III /bp |
|---|---|---|---|---|---|---|
| atpF | LSC | 390 | 796 | 168 | ||
| rpoC1 | LSC | 1638 | 723 | 423 | ||
| ycf3 | LSC | 153 | 777 | 228 | 715 | 152 |
| clpP | LSC | 352 | 630 | 1300 | 841 | 69 |
| rpl2 | IR | 435 | 659 | 384 | ||
| ndhB | IR | 756 | 700 | 777 | ||
| ndhA | SSC | 540 | 1049 | 555 | ||
| rps12 | LSC/IR | 114 | – | 114 |
3.2. Codon usage
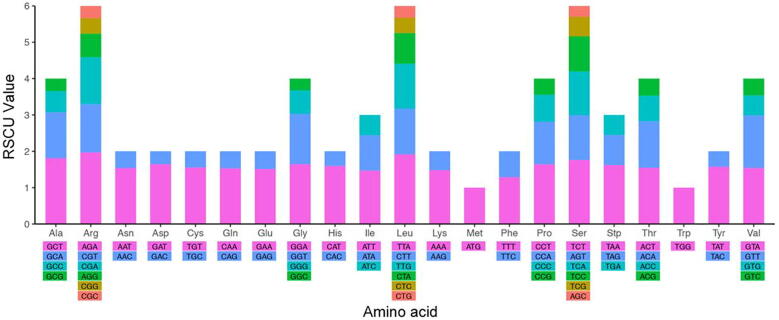
Codon usage biases have important ramifications for cellular function and reflect lineage specific translational systems, thus providing additional means for studying speciation and evolution at the molecular level (Gu et al., 2018, Plotkin and Kudla, 2011, Ikemura, 1981). Therefore, in this study, we examined the fency of codon usage in AV plastome (Fig. S1), as well as the RSCU value. As shown in Table S2, a total of 27,758 codons were involved in the protein-coding genes. Among these, leucine, isoleucine, and serine are the most frequent amino acids in AV, which encode in 2855 (10.28%), 2437 (8.77%), and 2182 (7.86%) codons, respectively.
RSCU represents a simple measure of the frequency of use of each codon encoding the same amino acid (Wu et al., 2017, Wang et al., 2016). It generally includes four types: lack of bias, low bias, moderate bias and high bias, and the corresponding RSCU ranges are < 1.0, 1.0 < RSCU < 1.2, 1.2 < RSCU < 1.3 and > 1.3, respectively (Liu et al., 2018b). In this study, there were 32 lack of bias codons (except tryptophan and methionine), two low bias codons, eight moderately biased codons, and 20 highly biased codons. Arginine, leucine and serine were each indicated by six synonymous codons with higher RSCU values (Fig. 2), which may be useful for protecting protein mutations given the function of the amino acids in biosynthesis (Park et al., 2017c, Wang et al., 2016, Zuo et al., 2017). The results showed that the RSCU was significantly biased except for tryptophan and methionine in AV. Notably, almost every amino acid has half of the codons ending in A or T (U), and the RSCU value is higher, while the remaining codons end in C or G with lower RSCU value, just like other plants reported (Gu et al., 2018, Raubeson et al., 2007, Delannoy et al., 2018), and is the main reason for the relative conservation of cp genes. Furthermore, the usage of the start codons AUG (encoding Met) and TGG (encoding Trp) exhibited no bias (RSCU = 1).
Fig. 2.
Relative synonymous codon usage (RSCU) of AV cp genome.
3.3. Repeat and simple sequence repeats analysis
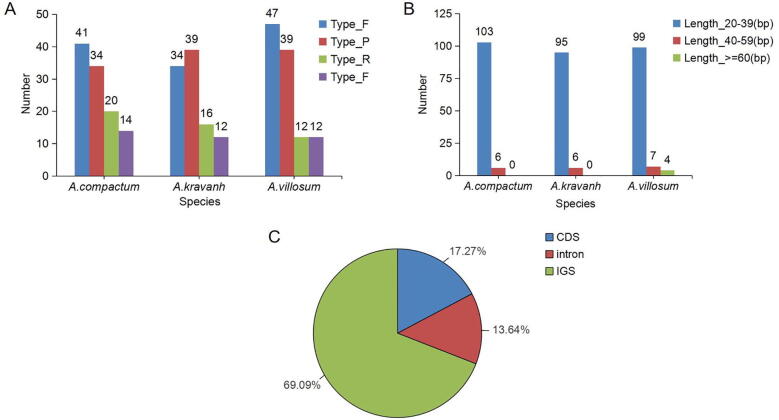
Repeated regions are of great significance in the evolutionary process, and can affect changes in genome structure like substitution and duplication, and they mostly occur in the sequences of intron and intergenic spacer (IGS) (Park, Kim, Yeo et al., 2017). In this study, we used REDuter to compare the forward (F), palindrome (P), reverse (R), and tandem (T) types (≥30 bp) of the three plants of Amomum, and found that the AV genome had the largest number of repeats (47F, 39P, 12 R, and 12 T types), followed by A. compactum (41F, 34P, 20 R and 14 T types), while A. kravanh had the least repeats. Among them, the F and P types accounted for the largest proportion with lengths mainly varied from 20 to 39 bp (Fig. 3A and B). The generation of F types is often related to the activity of transposons, which can lead to variations in genome structure, and is usually used as a marker for population relationship studies (Gu et al., 2018). There were 69.09% repeats located in the intergenic area, 17.27% in coding area, and 13.64% of the sites such as atpF and rps12 were located in the intron area (Fig. 3C).
Fig. 3.
Number of long repetitive repeats in three Amomum complete cp genomes. (A) Repeats number: F, forward; P, palindromic; R, reverse; T, tandem; (B) Repeats length range; (C) Repeats proportion in protein-coding (CDS), intergenic spacer (IGS), and intron regions.
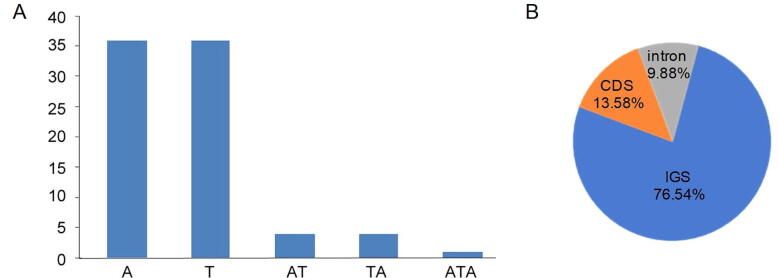
Simple sequence repeat (SSR), or microsatellite was extensively used in bioengineering, breeding and phylogenetic research as an effective technical method (Zhang et al., 2016). There were 81 SSRs have been identified, and mononucleotides had the largest number, accounting for 88.89% of the total SSRs, followed by dinucleotide SSRs (Fig. 4A). However, the tetra-, penta-, or hexa-nucleotide SSRs couldn’t be found according to our method. The mononucleotide repeats were mainly composed of A and T, which may lead to A/T richness in the angiosperm cp genomes (Liu et al., 2018). The results are similar to the statement, that is, most SSRs are consisted of short poly A (polyadenine) or poly T (polythymine) sequences. Approximately 13.58%, 9.88% and 76.54% SSRs were presented in protein-coding, introns, and intergenic areas of the AV, respectively (Fig. 4B). These findings are similar, as the distribution of SSR is unbalanced in the genome (Park et al., 2017).
Fig. 4.
Simple sequence repeats (SSRs) analysis in AV cp genome. (A) Frequency distribution of different classes of polymer in cp genome of AV and (B) SSRs frequency identified in intergenic spacer (IGS), protein-coding (CDS) and intron regions.
3.4. Analysis of synonymous and non-synonymous substitution rates
The non-synonymous (Ka) and synonymous (Ks) substitution ratio (denoted as Ka/Ks) is an important tool, mainly used to estimate the evolutionary pressures in specific groups of genes. Ratios > 1 indicate positive selection, values < 1 (especially if < 0.5) indicate negative selection, and values close to 1 indicate neutral selection (Wu et al., 2017). Here, we analyzed the Ka/Ks ratio of the 79 unique protein-coding genes in AV, A. compactum and A. kravanh genome (Table S3). Of these, most of the proteins possessed Ka/Ks ratios < 0.5, which suggests that most of protein-coding genes faced great pressure for purification and selection. In atpF and ycf1, the Ka/Ks values were > 1, which indicated a positive selection. In the genes involved in photosynthesis, for example, the atpF gene can encode a subunit of the H+-ATP synthase, thereby affecting the electron transport and photosynthetic phosphorylation in photosynthesis. These findings suggested that different levels of selective pressure in species may affect the function of the cp gene (Wu et al., 2017).
3.5. CG view comparison tool (CCT) map
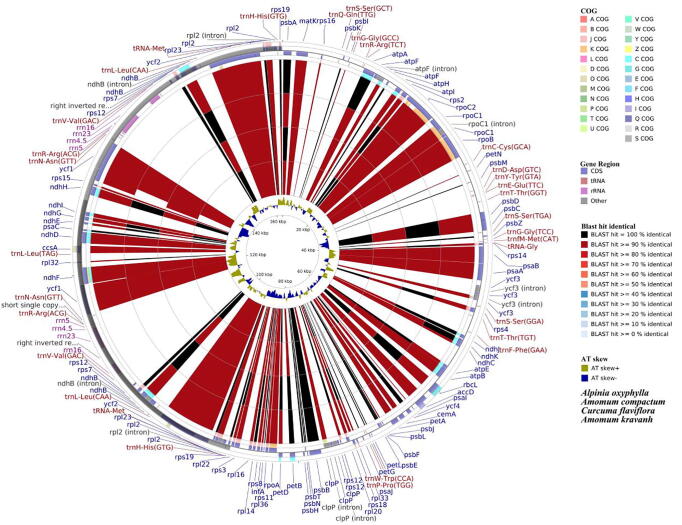
Four available cp genomes of Zingiberaceae species (A. compactum, A. kravanh, A. oxyphylla, and Curcuma flavifloraS Q. Tong) were selected for comparison with AV because the former is morphologically similar to AV (Fig. S2). The sequence identity between AV and other species' cp genomes was analyzed using CGView (University of Alberta, Alberta, Canada) with the annotated AV sequence as the reference. The closeness between plants generally reflects the similarity of gene sequences. The results revealed that the sequence similarity of A. compactum genome was the highest (>90%), followed by A. kravanh, similar to the cluster analysis results. The most similar regions were located in the IR area, and the SSC and LSC areas were quite different among these genomes involved in this study.
Differences in coding regions were smaller than non-coding regions, and the region with the greatest divergent lies in the intergenic area (Wu et al., 2017). The most divergent genes between AV and A. compactum were atpF, clpP, and rpl32, for which the blast identity values were 98.03, 98.87, 98.85, respectively (Fig. 5). The greatest difference in genes between AV and A. kravanh were atpF and rpl32, which were also considered to have high variability in other species (Yin et al., 2018). These three genes can be better used in the identification of Amomum family, even other species. Hence, the atpF, clpP, and rpl32 genes may be considered for development as molecular markers and barcoding to differentiate Amomum species. Among them, the atpF gene was also strongly positively selected. Above results bring a new insight for the development of molecular markers for Amomum family and even other species.
Fig. 5.
Genome comparison of four Zingiberaceae cp genomes to AV. Species involved are A. oxyphylla, A. compactum, C. flaviflora, and A. kravanh from outside. Four outermost rings represent protein-coding locations, while inner two rings indicated adenine–thymine (AT) skew. “AT skew +” indicates A > T, “AT skew –” indicates A < T.
3.6. Phylogenetic analysis
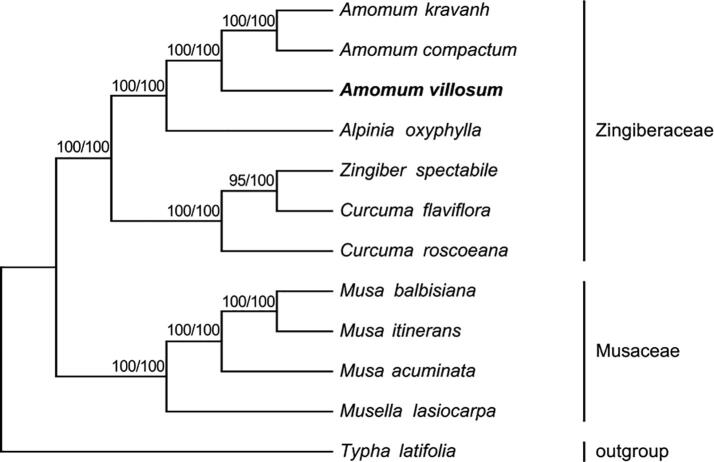
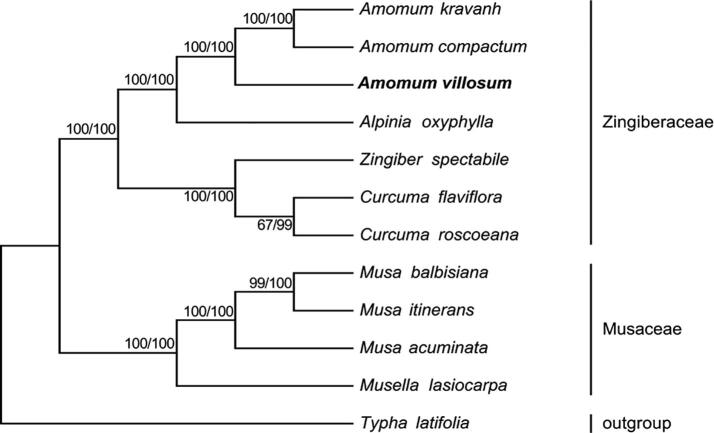
The cp sequences are often used in phylogenetics, evolution, and molecular systems studies (Liu et al., 2018). To determine the phylogenetic relationship of AV in Zingiberales, we used Maximum (ML) and Bayesian (BI) nucleic acid to analyze 77 protein-coding genes commonly found in 12 plants including AV. (Fig. 6). The results of ML and BI exhibited similar phylogenetic topologies. All nodes with a Bootstrap value of 100% were found using ML, and nine of them had observed bootstrap values ≥ 95% based on BI. Similarly, ML and BI protein analyses revealed that 8 of 9 nodes with bootstrap values of ≥ 99% (Fig. 7). Both nucleic acid and protein analyses showed that Amomum and Alpinia plants were sister groups. The four plants of AV, A. compactum, A. kravanh, and A. oxyphylla were grouped with 100% bootstrap values, and AV clustered more closely with A. compactum and A. kravanh than with A. oxyphylla.
Fig. 6.
Genetic relationship of AV based on maximum likelihood (ML) and bayesian inference (BI) nucleic acid analyses of 77 genes. First number represents BI bootstrap value of each branch, and the last one corresponds to ML. Phylogenetic tree was drawn using Typha latifolia as an outgroup. Position of AV is shown in boldface.
Fig. 7.
Genetic relationship of AV based on ML and BI protein analyses of 77 genes. First number represents BI bootstrap value of each branch, and the last one corresponds to the ML. Phylogenetic tree was drawn using Typha latifolia as an outgroup. Position of AV is shown in boldface.
The results of our cluster analysis are basically consistent with the phenotype-based clustering results reported in the literature (Zhang, 1994, Benedict et al., 2015). A. compactum and A. kravanh are plants from the same genus as AV, and are similar in the exterior shape and interior structure of their fruits. They are the most common counterfeits in many markets. A. oxyphylla is part of a related genus plants, and its seed groups are similar to AV's (Fig. 8) and it may be occasionally used as AV. The chemical constituents of the above four plants have considerable differences (Ding et al., 2004), and misuse may pose a threat to human health (Zhang, 1994). The results will facilitate the use of molecular markers to identify species of AV and other genera.
Fig. 8.

Appearance comparison of AV and its related fruits. A. compactum (A), A. kravanh (B), AV (C) and A. oxyphylla (D).
4. Conclusion
The AV entire cp genome was analyzed in this study, and the genome obtained had a quadruple structure. There were 81 SSRs in the AV genome, which will be used for further species identification. The ratio of Ka/Ks revealed that a large proportion of genes were in a state of strong purification selection. The fruits or seeds of A. kravanh, A. compactum and A. oxyphylla are usually substituted for AV due to highly similar morphological traits, and the phylogenetic tree fully supported AV as being closely related with A. kravanh and A. compactum, with a 100% bootstrap value. Comparison of cp genomes of the three plants indicated that the differences between them are very slight, while the atpF, clpP, and rpl32 genes are the most highly divergent regions, which will be developed as molecular markers that could discriminate the above related plants. The cp genome information of AV is an essential genetic resource that may facilitate the molecular identification of AV, and will lay a way for the breeding of a good cultivar of AV.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
This work was supported by the National Key R&D Program of China (2017YFC1701104) and Guangdong Province Applied Science and Technology R&D Special Fund Project (2015B020234002).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.chmed.2020.05.008.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Baczkiewicz A., Szczecinska M., Sawicki J., Stebel A., Buczkowska K. DNA barcoding, ecology and geography of the cryptic species of Aneura pinguis and their relationships with Aneura maxima and Aneura mirabilis (Metzgeriales, Marchantiophyta) PLoS ONE. 2017;12(12) doi: 10.1371/journal.pone.0188837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict J.C., Smith S.Y., Collinson M.E., Specht C.D., Marone F., Xiao X.H., Parkinson D.Y. Seed morphology and anatomy and its utility in recognizing subfamilies and tribes of Zingiberaceae. American Journal of Botany. 2015;102(11):1814–1841. doi: 10.3732/ajb.1500300. [DOI] [PubMed] [Google Scholar]
- Benson D.A., Karsch-Mizrachi I., Lipman D.J., Rapp B.A., Wheeler D.L. Genbank. Nucleic Acids Research. 2018;46(D1):D41–D47. doi: 10.1093/nar/gkx1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Zeng X., Ren G., Liu Z. CGAP: A new comprehensive platform for the comparative analysis of chloroplast genomes. BMC Bioinformatics. 2013;14:95. doi: 10.1186/1471-2105-14-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.H., Hao Z.D., Xu H.B., Yang L.M., Liu G.X., Sheng Y. The complete chloroplast genome sequence of the relict woody plant Metasequoia glyptostroboides Huet Cheng. Frontiers in Plant Science. 2015;6:447. doi: 10.3389/fpls.2015.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Ni W.Y., Yang C.X., Zhang T., Lu S.H., Zhao R.H., Mao X.J. Therapeutic effect of Amomum villosum on inflammatory bowel disease in rats. Frontiers in Pharmacology. 2018;9:639. doi: 10.3389/fphar.2018.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.S., Park S. The complete chloroplast genome sequence of Aster spathulifolius (Asteraceae); Genomic features and relationship with Asteraceae. Gene. 2015;572(2):214–221. doi: 10.1016/j.gene.2015.07.020. [DOI] [PubMed] [Google Scholar]
- Curci P.L., De P.D., Vendramin G.G., Sonnante G. Complete chloroplast genome of the multifunctional crop Globe Artichoke and comparison with other Asteraceae. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0120589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H., Lin C., Yu M., Chang W. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biology. 2016;17(1):134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy E., Fujii S., Colas des Francs-Small C., Brundrett M., Small I. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Molecular Biology and Evolution. 2018;28(7):2077–2086. doi: 10.1093/molbev/msr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding P., Fang Q., Xu H.H. GC fingerprint of components of Amomum villousm and its counterfeit species. West China Journal of Pharmaceutical Sciences. 2004;19(5):330–332. [Google Scholar]
- Gu C.H., Dong B., Xu L., Tembrock L.R., Zheng S.Y., Wu Z.Q. The complete chloroplast genome of Heimia myrtifolia and comparative analysis within Myrtales. Molecules. 2018;23(4):846. doi: 10.3390/molecules23040846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G.Z., Gao W., Su J., Li J.K., Tang L.Y. Morphological characteristics of the medicinal plant Amomum villosum flower organ. Chinese Bulletin of Botany. 2014;49(3):313–321. [Google Scholar]
- He X.Y., Wang H., Yang J.F., Deng K., Wang T. RNA sequencing on Amomum villosum Lour. induced by MeJA identifies the genes of WRKY and terpene synthases involved in terpene biosynthesis. Genome. 2018;61(2):91–102. doi: 10.1139/gen-2017-0142. [DOI] [PubMed] [Google Scholar]
- Huang Q., Duan Z., Yang J., Ma X., Zhan R., Xu H. SNP typing for germplasm identification of Amomum villosum lour. based on DNA barcoding markers. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0114940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: A proposal for a synonymous codon choice that is optimal for the E. coli translational system. Journal of Molecular Biology. 1981;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Iwasaki W., Fukunaga T., Isagozawa R., Yamada K., Maeda Y., Satoh T.P., Sado T. Mitofish and mitoannotator: A mitochondrial genome datebase of fish with an accurate and automatic annotation pipeline. Molecular Biology and Evolution. 2013;30(11):2531–2540. doi: 10.1093/molbev/mst141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. Mega7: Molecular evolutionary genetics analyses version 7.0 for bigger database. Molecular Biology and Evolution. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyalo C.M., Gichira A.W., Li Z.Z., Saina J.K., Malombe I., Hu G.W., Wang Q.F. Characterization and comparative analysis of the complete chloroplast genome of the critically endangered species Streptocarpus teitensis (Gesneriaceae) BioMed Research International. 2018;11:1507847. doi: 10.1155/2018/1507847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J. DanSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Li G., Chen A.J., Chen X.Y., Li X.L., Gao W.W. First report of Amomum villosum (cardamom) leaf lesion caused by Pyricularia costina in China. New Disease Reports. 2010;22:2. [Google Scholar]
- Liu X., Zhou B.Y., Yang H.Y., Li Y., Yang Q., Lu Y.Z., Gao Y. Sequencing and analysis of Chrysanthemum carinatum Schousb and Kalimeris indica. The complete chloroplast genomes reveal two inversions and rbcL as barcoding of the vegetable. Molecules. 2018;23(6):1358. doi: 10.3390/molecules23061358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yang H., Zhao B., Li T., Xiang B. The complete chloroplast genome sequence of the folk medicinal and vegetable plant purslane (Portulaca oleracea L.) European Journal of Horticultural Science. 2017;92(2):1–10. [Google Scholar]
- Liu X., Li Y., Yang H.Y., Zhou B.Y. Chloroplast genome of the folk medicine and vegetable plant Talinum paniculatum (Jacq.) Gaertn: Gene organization, comparative and phylogenetic analysis. Molecules. 2018;23(4):857. doi: 10.3390/molecules23040857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton B.R. Coden use and the rate of divergence of land plant chloroplast genes. Molecular Biology and Evolution. 1994;11(2):231–238. doi: 10.1093/oxfordjournals.molbev.a040105. [DOI] [PubMed] [Google Scholar]
- Muse S.V., Gaut B.S. Comparing patterns of nucleotide substitution rates among chloroplast loci using the related ratio test. Genetics. 1997;146(1):393–399. doi: 10.1093/genetics/146.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Huang F., Wang P., Zhou L., Cao L., Liang R. Identification of Amomum villosum, Amomum villosum var. xanthioides and Amomum longiligulare on ITS-1 sequence. Zhong Yao Cai. 2001;24(7):481–483. [PubMed] [Google Scholar]
- Park I., Yang S.Y., Choi G.Y., Kim W.J., Moon B.C. The complete chloroplast genome sequence of Aconitum pseudolaeve and Aconitum longecassidatum, and development of molecular markers for distinguishing species in the Aconitum subgenus Lycoctonum. Molecules. 2017;22(11):2012. doi: 10.3390/molecules22112012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I., Kim W.J., Yang S., Yeo S.M., Li H., Moon B.C. The complete chloroplast genome sequence of Aconitum coreanum and Aconitum carmichaelii and comparative analysis with other Aconitum species. PLoS One. 2017;12(9) doi: 10.1371/journal.pone.0184257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I., Kim W.J., Yeo S.M., Choi G., Kang Y.M., Piao R., Moon B.C. The complete chloroplast genome sequence of Fritillaria ussuriensis Maxim and Fritillaria cirrhosa D. Don, and comparative analysis with other Fritillaria species. Molecules. 2017;22(6):982. doi: 10.3390/molecules22060982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin J.B., Kudla G. Synonymous but not the same: The causes and consequences of codon bias. Nature Reviews Genetics. 2011;12(6):32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provan J., Powell W., Hollingsworth P.M. Chloroplast microsatellites: New tools for studies in plant ecology and evolution. Trends in Ecology & Evolution - Journal. 2001;16(3):142–147. doi: 10.1016/s0169-5347(00)02097-8. [DOI] [PubMed] [Google Scholar]
- Raubeson L.A., Peery R., Chumley T.W., Dziubek C., Fourcade H.M., Boore J.L., Jansen P.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics. 2007;8:174. doi: 10.1186/1471-2164-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saina J.K., Li Z.Z., Gichira A.W., Liao Y.Y. The complete chloroplast genome sequence of tree of Heaven (Ailanthus altissima (Mill.) (Sapindales: Simaroubaceae), an important pantropical tree. International Journal of Molecular Sciences. 2018;19(4):929. doi: 10.3390/ijms19040929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P., Brooks A.N., Lowe T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Research. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Chen Y., Lv J., Xu J., Zhu S., Li M., Chen N. Development of chloroplast genomic resources for Oryza species discrimination. Frontiers in Plant Science. 2017;8:1854. doi: 10.3389/fpls.2017.01854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. Raxml version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Ma D.M., Yang J.F., Deng K., Ji X.Y., Zhong L.T., Zhao H.Y. An integrative volatile terpenoid profiling and transcriptomics analysis for gene mining and functional characterization of AvBPPS and AvPS involved in the monoterpenoid biosynthesis in Amomum villosum. Frontiers in Plant Science. 2018;9:846. doi: 10.3389/fpls.2018.00846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Huang F., Zhou L., Cao L., Liang S., Xu H., Liu J. Analysis of Amomun villosum species and some adulterants of Zingiberaceae by RAPD. Journal of Chinese Medicinal Materials. 2000;23(2):71–74. [PubMed] [Google Scholar]
- Wang Y., Zhan D.F., Jia X., Mei W.L., Dai H.F., Chen X.T., Peng S.Q. Complete chloroplast genome sequence of Aquilaria sinensis (Lour.) Gilg and evolution analysis within the Malvales order. Frontiers Plant Science. 2016;7:280. doi: 10.3389/fpls.2016.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.L., Li Q., Xu J., Li X.W. Complete chloroplast genome of the medicinal plant Amomum compactum: Gene organization, comparative analysis, and phylogenetic relationships within Zingiberales. Chinese Medicine. 2018;13:10–21. doi: 10.1186/s13020-018-0164-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.L., Li Q., Hu Z.G., Li X.W., Chen S.L. The complete Amomum kravanh chloroplast genome sequence and phylogenetic analysis of the Commelinids. Molecules. 2017;22(11):1875. doi: 10.3390/molecules22111875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang B., Li X., Qian J., Wang L.Z., Ma L., Tian X., Wang Y. The complete chloroplast genome sequence of medicinal plant Swertia mussotii using the PacBio RS II platform. Molecules. 2016;21(8):1029. doi: 10.3390/molecules21081029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Yang D., Wang D., Xu X., Zhu L., Zhao Z. Solidification of floating organic drop liquid-phase microextraction cell fishing with gas chromatography-mass spectrometry for screening bioactive components from Amomum villosum Lour. Biomedical Chromatography. 2015;29(4):626–632. doi: 10.1002/bmc.3324. [DOI] [PubMed] [Google Scholar]
- Yang H.R., Xia J., Zhang J.E., Yang J.Z., Zhao H.H., Wang Q., Sun J.J. Characterization of the complete mitochondrial genome sequences of three Croakers (Perciformes, Sciaenidae) and novel insights into the phylogenetics. International Journal of Molecular Science. 2018;19(6):1741. doi: 10.3390/ijms19061741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Zhang X., Liu G., Yin Y., Chen K., Yun Q., Zhao D. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.) PLoS One. 2012;5(9) doi: 10.1371/journal.pone.0012762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi D.K., Lee H.L., Sun B.Y., Chung M.Y., Kim K.J. The complete chloroplast DNA sequence of Eleutherococcus senticosus (Araliaceae); comparative evolutionary analysis with other three asteroids. Molecules and Cells. 2012;33(5):497–508. doi: 10.1007/s10059-012-2281-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K.Q., Zhang Y., Li Y.J., Du F.K. Different, natural selection pressures on the atpF gene in evergreen sclerophyllous and deciduous oak species: Evidence from comparative analysis of the complete chloroplast genome of Quercus aquifolioides with other oak species. International Journal of Molecular Science. 2018;19(4):1042. doi: 10.3390/ijms19041042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.Q., Drew B.T., Yang J.B., Gao L.M., Li D.Z. Comparative chloroplast genomes of eleven Schima (Theaceae) species: Insights into DNA barcoding and phylogeny. PLoS One. 2017;12(6) doi: 10.1371/journal.pone.0178026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.F., Zhong Z.M., Huang S., Lai X.P. Identification and clustering analysis of Zingiberaceae with matK barcode. Lishizhen Medicine and Materia Medica Research. 2018;29(1):99–102. [Google Scholar]
- Zhang X.G. Identification of Amomum villosum and its confused varieties. Chinese Herbal Medicines. 1994;25(11):595–598. [Google Scholar]
- Zhang Y., Du L., Liu A., Chen J., Wu L., Hu W., Zhang W. The complete chloroplast genome sequences of five Epimedium species: Lights into phylogenetic and taxonomic analysis. Frontiers in Plant Science. 2016;7:306. doi: 10.3389/fpls.2016.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Cui Y., Chen X., Li Y., Xu Z., Duan B., Li Y. Complete chloroplast genomes of Papaver rhoeas and Papaver orientale: Molecular structures, comparative analysis, and phylogenetic analysis. Molecular. 2018;23(2):437. doi: 10.3390/molecules23020437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L., Shang A., Zhang S., Yu X., Ren Y., Yang M., Wang J. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: Genome comparative and taxonomic position analysis. PLoS One. 2017;12(2) doi: 10.1371/journal.pone.0171264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.