Abstract
Objective
To develop a powerful integrated strategy based on liquid chromatography coupled with mass spectrometry (LC-MS) systems for the comprehensive characterization and quantification of multiple components of herbal medicines.
Methods
Firstly, different mobile phase additives, analysis time, and MS acquisition modes were orthogonally tested with liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (LC-QTOF/MS) in order to detect as many components of Gelsemium elegans as possible with high peak intensity. Secondly, several data mining strategies, including database searching, diagnostic ion filtering and neutral loss filtering, were utilized to perform chemical profiling. Subsequently, this study focused on the quantification and validation of the performance of a liquid chromatography-triple mass spectrometry (LC-QqQ/MS) assay based on derivative multiple reaction monitoring (DeMRM).
Results
A total of 147 components from G. elegans were characterized, among them 116 nontarget components were reported for the first time. A sensitive and reproducible LC-QqQ/MS method was successfully developed and validated for the simultaneous relative quantification of 41 components of G. elegans. This LC-QqQ/MS method was then applied to compare the contents of components in the roots, stems and leaves.
Conclusion
The present integrated strategy would significantly contribute to chemical studies on herbal medicine, and its utility could be extended to other research fields, such as metabolomics, quality control, and pharmacokinetics.
Keywords: Gelsemium elegans (Gardn. & Champ.) Benth, herbal medicine, LC-MS, mass spectrometry
1. Introduction
There is rising global interest in herbal medicines as a promising source for new drug discovery and development (Shi et al., 2018, Wang et al., 2018). The use of herbal medicines in the clinic also requires the elucidation of as many components as possible to ensure improved quality control and clinical outcomes. A significant feature of herbal medicine is the number of chemical components, as each herbal medicine contains hundreds or even thousands of different components, depending on the number of genes. These components are small molecules (<1500 Da) among many different chemical substance classes, which can be present in many different concentrations (Yan et al., 2013, Mustafa et al., 2018, Tan et al., 2018). Due to their high diversity, the comprehensive characterization and quantification of multiple components of herbal medicines remain a great challenge and bottleneck.
Currently, liquid chromatography coupled with various types of mass spectrometers, especially hybrid mass spectrometers, has been increasingly accepted as the predominant platform for the global analysis of complex herbal medicines. Among these mass spectrometry techniques, time-of-flight (TOF) mass spectrometry and liquid chromatography coupled with hybrid tandem mass spectrometry (LC-QTOF/MS) are expected to be the most powerful tools for structurally characterizing complex components of herbal medicines due to the high resolution of both precursor and product ions with these methods (Lei et al., 2018, Zhang et al., 2017b, Zuo et al., 2018). Both types of information are important and reliable for identifying components. Previously, most reported methodologies have been limited to target components and mainly depended on the use of reference components and/or comparison with literature data (Huo et al., 2018, Li et al., 2015a). Considering that the reference components that can be obtained are limited and most components contained in herbal medicines are unknown (nontarget components), these methods are apparently insufficient for the comprehensive detection and characterization of the complicated components of herbal medicines.
The characterization of nontarget components of herbal medicines is never an easy task (Chingin, Makarov, Denisov, Rebrov, & Zubarev, 2014; Xue et al., 2016; Samanipour, Reid, Bæk, & Thomas, 2018). Great challenges remain in optimizing the detection of herbal medicines and processing and mining data from the complex information obtained. Recently, corresponding strategies based on diagnostic fragment ions, key ion filtering, and mass defect filtering have been developed to characterize nontarget components from complex mixtures (Hao et al., 2008, Cai et al., 2014, Zhang et al., 2016, Shi et al., 2017). However, it is worth noting that these studies mainly focused on qualitative aspects (Chen et al., 2016, Ma et al., 2016). Moreover, few studies on quantifying the amounts of multiple components or monitoring the chemical variations that are frequently present in different parts of herbal medicines during different seasons have been conducted due to the high diversity of components and lack of standards (Li et al., 2015b, Yang et al., 2013). Therefore, the development of more comprehensive and effective strategies for the simultaneous characterization of nontarget components and quantification of multiple components of herbal medicines in the absence of reference substances is imperative.
To address this challenge, the present study provides an integrated strategy to simultaneously identify target and nontarget components and monitors the concentrations of multiple components of herbal medicines. The general flowchart for this integral strategy is shown in Fig. 1. First, different mobile phase additives, analysis time and MS acquisition modes were orthogonally tested to detect as many components of herbal medicines as possible with high peak intensities based on LC-QTOF/MS. Then, various data mining techniques, including database searching, diagnostic ion filtering and neutral loss filtering, were used to characterize nontarget components under the optimized conditions. Furthermore, to determine the relative concentrations of the characterized components, we quantified and validated the performance of an LC-QqQ/MS method based on derivative multiple reaction monitoring (DeMRM). A single calibration curve was developed to calculate the relative contents of multiple components without authentic standards for each component.
Fig. 1.
Workflow of the integrated strategy toward comprehensive characterization of nontarget components and derivative multiple reaction monitoring for multiple components quantification of herbal medicine using LC-QTOF/MS and LC-QqQ/MS.
The proposed strategy was demonstrated on the herbal medicine Gelsemium elegans (Gardn. & Champ.) Benth, a species of flowering plant in Loganiaceae family, is known as a toxic plant. In China, it is known as Gouwen, Dachayao or Duanchangcao (Ornduff, 1970) and has been used as a traditional Chinese medicine (TCM) for the treatment of rheumatoid arthritis, neuropathic pain, spasticity, skin ulcers and cancer for many years. The bioactive components of G. elegans have attracted much attention from chemists, pharmacologists and toxicologists in recent years due to their multiple biological effects, such as anti-inflammatory, immunomodulating, analgesic, anxiolytic, antitumor, and neuropathic pain-relieving properties (Ling et al., 2014, Liu et al., 2013, Meyer et al., 2013, Xu et al., 2012a, Xu et al., 2012b, Zhang et al., 2015b). To date, a total of 121 alkaloids, 25 iridoids and a number of other components from a wide spectrum of secondary metabolite classes have been isolated from G. elegans and characterized (Jin et al., 2014, Liu et al., 2017b, Yamada et al., 2011, Zhang et al., 2017a). Previous studies have focused mainly on the isolation and purification of these components. Only our laboratory has recently published an analytical strategy for the characterization and structural analysis of target components from G. elegans by using LC-QTOF/MS based on the use of accurate mass databases combined with MS/MS spectra (Liu et al., 2017b, Xiao et al., 2017, Yang et al., 2018a). However, many nontarget components in G. elegans have not been characterized. There are few reports on quantification methods for G. elegans formulations, and the number of analytes quantified in these reports is at most three (Hu et al., 2017, Liang et al., 2010, Wang et al., 2018, Yang et al., 2018b, Zhang et al., 2015a). We developed a practical and reliable high-performance liquid chromatography-ultraviolet detector (HPLC-UV) method for fingerprint analysis using two major components, gelsemine and koumine. The results showed that at least seven relatively major components present in G. elegans may be useful for its quality control (Liu et al., 2017a). Using the methodology established in the study, a total of 147 components were characterized from G. elegans, and among these components, 116 nontarget components were reported for the first time. The simultaneous quantification of 41 components of G. elegans was achieved using DeMRM mode.
2. Materials and methods
2.1. Chemicals and materials
The reference components gelsemine (>98%), koumine (>98%), koumidine (>98%) and gelsenicine (>98%) were purchased from Shanghai Jiwei Biochemical Technology Co., Ltd. (Shanghai, China). HPLC-grade acetonitrile and methanol were purchased from Merck (Darmstadt, Germany). Ammonium formate, ammonium acetate, formic acid and acetic acid were purchased from SIGMA-ALDRICH (USA). Deionized water was purified by a Milli-Q water purification system (Bedford, MA, USA). The other chemicals used were of analytical grade. All reference components were weighed, dissolved in methanol and then diluted to appropriate concentrations for LC-QTOF/MS and LC-MS/MS analysis.
2.2. Plant materials and sample preparation
A total of nine samples including G. elegans roots, stems and leaves from three different periods were collected from Guangxi Province in China (see Supporting information, Table S1). The samples were authenticated by the author Qi Tang and stored at Hunan Key Laboratory of Traditional Chinese Veterinary Medicine. The samples were dried and then ground into powder. According to our optimized method, a 1 g aliquot of the powder was extracted twice by ultrasonication with 80% ethanol (1:25) for 0.5 h at 60 °C. The extraction solution was combined for filtration, and 1 mL of the filtered solution was evaporated and dissolved in 1 mL of acetonitrile-ammonium acetate (1:4, volume percent). Then, the solution was filtered through a 0.22 μm membrane before use. A 5 μL aliquot was injected for analysis.
2.3. LC-QTOF/MS conditions
Analysis was performed on an Agilent series 1290 Infinity HPLC instrument coupled with an Agilent 6530 Q-TOF mass spectrometer (Agilent Technologies, Santa Clara, CA, USA). Samples were separated on a Waters C18 column (3.5 μm, 4.6 mm × 150 mm). The flow rate was 0.3 mL/min, and the column temperature was maintained at 30 °C. To detect as many chemical components as possible with high peak intensities, an L9 (3) orthogonal array was used to examine the influence of difference mobile phase additives, gradient analysis times and MS acquisition mode (see Supporting information, Table S2). The gradient elution for 40 min was as follows: 0–2 min, 10% B; 2–7 min, 10% B to 15% B; 7–20 min, 15% B to 35% B; 20–30 min, 35% B to 90% B; 30–33 min, 90% B; 33.01–40 min, 10% B. The gradient elution for 50 min was as follows: 0–2 min, 10% B; 2–7 min, 10% B to 15% B; 7–30 min, 15% B to 45% B; 30–40 min, 45% B to 90% B; 40–43 min, 90% B; 43.01–50 min, 10% B. The gradient elution for 60 min was as follows: 0–2 min, 10% B; 2–7 min, 10% B to 15% B; 7–40 min, 15% B to 55% B; 40–50 min, 55% B to 90% B; 50–53 min, 90% B; 53.01–60 min, 10% B.
Mass spectrometric detection was performed in positive electrospray ionization (ESI) mode. The operating parameters were as follows: fragmentor voltage, 30 V; capillary voltage, 3500 V; dry gas temperature, 300 °C; sheath gas temperature, 350 °C; dry gas (N2) flow rate, 9 L/min; sheath gas flow rate, 11 L/min; nebulizer, 35 psi; VCap, 4000; nozzle voltage, 1000 V; fragmentor, 175; Skimmer1, 65. Data were acquired in a mass range of m/z 50–1000. Auto MS/MS mode was utilized to obtain abundant structural information without knowledge of the sample. The collision energy (CE) was set at 10, 20, and 30 V.
The instrument performed internal mass calibration automatically using an automated calibrant delivery system. The calibration solution contained internal reference masses of m/z 121.0508 and 922.0098 in positive mode. All of the data acquisition was controlled by Agilent MassHunter workstation software (version B.01.03 Build 1.3.157.0 2).
2.4. LC-QqQ/MS and MS/MS conditions
Analysis was performed on an Agilent series 1290 Infinity HPLC instrument coupled with an Agilent 6460 QqQ/MS mass spectrometer (Agilent Technologies, Santa Clara, CA, USA). Samples were separated on a Waters C18 column (3.5 μm, 4.6 mm × 150 mm). The flow rate was 0.3 mL/min, and the column temperature was maintained at 30 °C.
Auto MS/MS analysis was performed in both scan mode and multiple reaction monitoring (MRM) mode. The MS/MS switching time and scan speed were set at < 2 ms and 5200 amu/s, respectively. The operating parameters were set as follows: scan range, m/z 50–1000; capillary temperature, 350 °C; capillary voltage, 3000 V; dry gas (N2) flow rate, 12 L/min; nebulizer pressure, 40 psi. Full mass spectra were recorded at a mass resolving power of 0.7 Da (full width at half maximum).
2.5. Relative quantitation of multiple components
In the present study, DeMRM was used to obtain the mass responses of targeted analytes, and a koumidine calibration curve was employed instead of authentic standards to determine the concentrations of multiple components of G. elegans. The method was first validated with four representative components (gelsemine, koumine, koumidine and gelsenicine) in terms of linearity, limits of detection and quantification (LOD and LOQ), and intraday and interday precision. LOD and LOQ were determined at a signal-to-noise ratio of approximately 3 and 10, respectively. Intraday precision was determined by analyzing six replicates of each representative component at 10 ng/mL and 100 ng/mLl within one day, while interday precision was assessed on three consecutive days. Then, the DeMRM method was carried out with six G. elegans samples to demonstrate that the whole method was reliable and acceptable for multiple component quantification. Six samples were prepared separately according to the above method and analyzed to measure repeatability. To assess stability, the samples were exposed to ambient temperature and then analyzed at 0, 2, 4, 8, and 24 h. The relative concentrations of multiple components of G. elegans were calculated from the koumidine calibration curve.
3. Results and discussion
3.1. Optimization of LC-QTOF/MS conditions
This study examined different mobile phase additives, analysis times and MS acquisition modes, which could help to significantly improve the information obtained regarding the content of nontarget components in herbal medicines
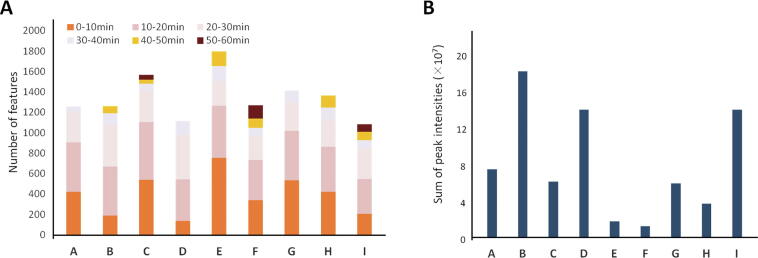
The results of the detection of all components in G. elegans are shown in Fig. 2. These results showed that there were great differences in the number of G. elegans components detected under different test conditions. As shown in Fig. 2A, among the experimental groups, the number of components detected in the D and I groups was the smallest, and the number of components detected in the C and E groups was the highest. Relative to the number detected with the other two mobile phases, the total number of compounds detected in groups A, B and C with a 0.1% formic acid–water mobile phase was significantly large. The number of compounds detected in the corresponding groups C, F, and I with a consistent analysis time of 60 min was not significantly different from that detected in groups B, E, and H, corresponding to an analysis time of 50 min. As shown in Fig. 2A, the component peaks were mainly concentrated in the first 30 min of analysis, so the number of components did not change by extending the analysis time over 50 min. Fig. 2B showed that among the experimental groups, the peak intensities of the components in G. elegans in groups B, D, and I were the highest, and the intensities of the components in groups E and F were the lowest. These results suggested that compared to the other two acquisition modes, the auto MS/MS mode not only had higher peak intensities but also was more efficient. What’s more, compared with ammonium acetate and ammonium formate as additives, an aqueous solution of 0.1% formic acid provided higher peak intensities. Thus, we recommend the application of a 50 min gradient, MS/MS mode and 0.1% formic acid to ensure better results.
Fig. 2.
Effect of mobile phases, analysis time and mass spectrometry modes on number of detected features (A) and sum of intensities (B) for G. elegans components. A, 0.1% FA, 40 min of gradient separation and MS mode; B, 0.1% FA, 50 min of gradient separation and auto MS/MS mode; C, 0.1% FA, 60 min of gradient separation and target MS/MS mode; D, 5 mmol/L NH4Ac, 40 min of Gradient separation and auto MS/MS mode; E, 5 mmol/L NH4Ac, 50 min of gradient separation and target MS/MS mode; F, 0.1% 5 mmol/L NH4Ac, 60 min of Gradient separation and MS mode; G, 10 mmol/L NH4F, 40 min of Gradient separation and target MS/MS mode; H, 10 mmol/L NH4F, 50 min of gradient separation and MS mode; I, 10 mmol/L NH4F, 60 min of gradient separation and auto MS/MS mode.
3.2. Characterization of target components in G. elegans
The optimal LC-QTOF/MS conditions were used to acquire information about G. elegans sample No. S1. The first step of this analytical strategy was to extract components from raw acquisition data with the “Find by Auto MS/MS” function. The mass match tolerance was set to 0.05 m/z, and the peak abundance was set to 1000. Following this step, the target components were matched with personal database searching and characterized according to our published analytical approach (Liu et al., 2017b).
The precise mass of precursor ions (within ± 5 ppm) found to match data in the MS database of Gelsemium would give the exact element compositions and the possible known structure of the components. Structural characterization of the components was achieved on the basis of determining the accurate mass and fragmentation behavior of the product ions. A total of 31 components were matched with the personal database and characterized. Thirteen of them were gelsedine-type alkaloids, which was the largest class in G. elegans. They were components 28, 40, 75, 83, 94, 100, 109, 110, 118, 124, 126, 131 and 134. Components 99, 117, 119, 128 and 143 were target sarpagine-type alkaloids obtained after matching. The [M + H]+ ions of components 133 and 137 were m/z 355.2025 and 371.1974, respectively, which corresponded to humantenine-type alkaloids. Components 53 and 95 had masses of m/z 325.1917 and 307.1812, respectively, and were characterized as koumine-type alkaloids. Components 63 and 72 were target gelsemine-type alkaloids obtained after matching. Four kinds of iridoid components were detected by matching with Gelsemium database: 30, 57, 91 and 92. Only two phenolic acids were detected in G. elegans after matching with the MS database.
3.3. Characterization of nontarget components in G. elegans
The second step was to characterize the nontarget components when a component fails to match the information in our personal database. Based on the fragmentation of the target components, common fragment ions and neutral loss ions could be classified into families. According to this idea, some post-acquisition data mining procedures, including key ion filtering, diagnostic ion filtering, neutral loss filtering and online database (Metlin and HMDB public database) searching, were performed in this study (Qiao et al., 2016). The structure of the nontarget components could be characterized by accurate MS/MS spectra and fragmentation comparisons. As a result, a total of 116 nontarget components were characterized. Table 1 summarized the exact mass, fragment ion, and retention time for the characterized components of Gelsemium elegans. They could be divided into seven groups according to their structural types and MS/MS fragmentation pathways.
Table 1.
Retention time, accurate mass, fragment ions of target and non-target compounds.
| No. | t/R(min) | [M + H]+ | Fragment ions | Formula | Tentative characterization | Classification |
|---|---|---|---|---|---|---|
| 1 | 5.4 | 150.0921 | 132.0807,117.0574,77.0383 | C9H11NO | Unknown | Amino acid |
| 2 | 5.5 | 521.2131 | 490.1948,459.1777,441.1624,403.1448,359.1604,328.1423,297.1249,279.1138,269.1286,256.0980,108.0806 | C25H32N2O10 | Unknown | Gelsedine-type |
| 3 | 5.6 | 537.2074 | 506.1900,489.1864,475.1775,457.1617,375.1521,344.1369 | C25H32N2O11 | Unknown | Gelsedine-type |
| 4 | 5.8 | 229.1551 | 170.0785,142.0858,132.0788,114.0550,96.0812,70.0659,58.0667 | C11H20N2O3 | Pro Leu or Pro Ile | Amino acid |
| 5 | 6.0 | 236.1323 | 218.1163,150.0921,132.0811,117.0593,87.0449 | C13H17NO3 | Unknown | Amino acid |
| 6 | 6.4 | 195.0626 | 177.0483,149.0596,121.0621,91.0552,77.0366 | C10H10O4 | Ferulic acid | Phenolic acids |
| 7 | 6.4 | 213.0735 | 195.0627,177.0517,165.0518,149.0568,131.0466,121.0621,103.0519,91.0523,77.0370,69.0317,57.0323 | C10H12O5 | Gelsemide | Iridoids |
| 8 | 6.4 | 231.0841 | 216.1015,195.0604,177.0518,149.0568,121.0625,107.0465,93.0672,91.0521,77.0365,57.0321 | C10H14O6 | Geleganoid A/GRIR-1 | Iridoids |
| 9 | 6.6 | 229.1536 | 195.0628,177.0517,142.0854,121.0626,70.0645 | C11H20N2O3 | Pro Leu or Pro Ile | Amino acid |
| 10 | 6.8 | 199.0949 | 135.0774,121.0622,117.0865,109.0641,91.0533,79.0540,67.0542,55.0540 | C10H14O4 | 9-DeoxyGRIR-2 | Iridoids |
| 11 | 6.8 | 217.1057 | 163.0705,135.0786,109.0640,91.0536,79.0538,67.0535,55.0175 | C10H16O5 | 7-Hydroxygelsemiol/9-Hydroxygelsemiol | Iridoids |
| 12 | 6.8 | 239.0877 | 177.0541 | C12H14O5 | Hydroxyl of ferulic acid ethyl ester | Phenolic acids |
| 13 | 7.0 | 408.1505 | 213.0748,195.0648,151.0738 | C17H21N5O7 | Unknown | Nucleoside |
| 14 | 7.3 | 187.0959 | 109.0639,105.0698,95.0854,91.0542,81.0701,79.0544,77.0385,67.0553 | C9H14O4 | 7-Deoxygeleganoid D/9-Deoxygeleganoid D | Iridoids |
| 15 | 7.7 | 217.1073 | 135.0802,117.0701,109.0650,91.0540,79.0549,67.0547,55.0186 | C10H16O5 | Isomer of 7-Hydroxygelsemiol/9-Hydroxygelsemiol | Iridoids |
| 16 | 7.8 | 239.0866 | 177.0534,84.0794,58.0655 | C12H14O5 | Hydroxyl of ferulic acid ethyl ester | Phenolic acids |
| 17 | 8.2 | 422.1295 | 325.0941,277.0637,213.0759,151.0761,133.0635 | C17H19N5O8 | Unknown | Nucleoside |
| 18 | 8.3 | 236.1285 | 150.0906,132.0791,117.0553,87.0447 | C13H17NO3 | Unknown | Amino acid |
| 19 | 8.6 | 213.0759 | 195.0636,177.0543,149.0583,131.0477,121.0630,91.0546,77.0398,57.0344 | C10H12O5 | Isomer of GEIR-1 | Iridoids |
| 20 | 8.8 | 187.0968 | 123.0804,105.0708,95.0855,91.0564,81.0691,77.0396,67.0550,55.0558 | C9H14O4 | 7-Deoxygeleganoid D /9-Deoxygeleganoid D | Iridoids |
| 21 | 8.9 | 217.1059 | 135.0848,117.0683,91.0559,81.0576 | C10H16O5 | Isomer of 7-Hydroxygelsemiol/9-Hydroxygelsemiol | Iridoids |
| 22 | 8.9 | 407.1820 | 376.1611,345.1455,331.1300,166.0879 | C20H26N2O7 | Tihydroxygelsemicine | Gelsedine-type |
| 23 | 9.2 | 422.1300 | 395.1598,213.0757,195.0638,175.0232,151.0750,129.0183,73.0295 | C17H19N5O8 | Unknown | Nucleoside |
| 24 | 9.4 | 187.0966 | 123.0786,109.0650,105.0707,95.0852,91.0536,81.0698,77.0397,67.0548,55.0548 | C9H14O4 | Isomer of 7-Deoxygeleganoid D /9-Deoxygeleganoid D | Iridoids |
| 25 | 9.6 | 199.0937 | 135.0776,117.0675,115.0515,105.0667,91.0521,79.0522,67.0519,55.0162 | C10H14O4 | 9-Deoxygeleganoid F | Iridoids |
| 26 | 9.6 | 217.1045 | 163.0717,135.0781,117.0676,109.0624,105.0669,91.0521,79.0525, 67.0526,55.0164 | C10H16O5 | 7-Hydroxygelsemiol/9-Hydroxygelsemiol | Iridoids |
| 27 | 9.6 | 239.0855 | 177.0447,105.0674 | C12H14O5 | Hydroxyl of ferulic acid ethyl ester | Phenolic acids |
| 28 | 9.8 | 359.1578 | 328.1395,311.1366,297.1212,279.1105,269.1259,256.0960,228.0718,124.0740,108.0791,96.0790 | C19H22N2O5 | 11,14-Dihydroxygelsenicine | Gelsedine-type |
| 29 | 9.9 | 195.0642 | 177.0546,121.0623.91.0534,77.0375 | C10H10O4 | Isomer of ferulic acid | Phenolic acids |
| 30 | 9.9 | 213.0754 | 195.0644,177.0511,149.0579,121.0626,91.0532,79.0529,77.0372,57.0326,55.0171 | C10H12O5 | GEIR-1 | Iridoids |
| 31 | 10.1 | 375.1553 | 344.1359,327.1335,313.1175,295.1072,279.1139,267.1137,124.0756 | C19H22N2O6 | 11,14,15-Trihydroxygelsenicine | Gelsedine-type |
| 32 | 10.7 | 187.0969 | 133.0695,123.0822,109.0638,105.0706,95.0847,91.0544,81.0702,79.0539,77.0386,67.0542,55.0541 | C9H14O4 | Isomer of 7-Deoxygeleganoid D /9-Deoxygeleganoid D | Iridoids |
| 33 | 10.7 | 417.1649 | 386.1485,355.1260,166.0854 | C21H24N2O7 | Isomer of 14-Acetoxy-dihydroxygelsenicine | Gelsedine-type |
| 34 | 10.8 | 213.0738 | 177.0499,149.0581,121.0628,103.0522,91.0529,77.0374,67.0533,55.0532 | C10H12O5 | Isomer of gelsemide | Iridoids |
| 35 | 10.9 | 395.1603 | 364.1396,347.1355,333.1212,307.1060,277.0950,246.1106,228.0630,132.0789 | C22H22N2O5 | Unknown | Gelsedine-type |
| 36 | 11.4 | 373.1768 | 342.1581,327.1357,311.141,271.1448,180.1024,122.0965,108.0810 | C20H24N2O5 | 14,15-Dihydroxyrankinidine | Humantenine-type |
| 37 | 11.5 | 339.1707 | 293.1616,236.1063,195.0677 | C20H22N2O3 | Gelsemine oxide | Gelsemine-type |
| 38 | 11.6 | 408.1510 | 213.0765,195.0636,177.0518,167.0732,149.0606 | C17H21N5O7 | Unknown | Nucleoside |
| 39 | 11.8 | 325.1913 | 307.1808,281.1644,238.1268,220.1179,194.0997,150.0908,130.06481,70.0655 | C20H24N2O2 | 19-(S)-Hydroxydihydrokoumine/19-(R)-Hydroxydihydrokoumine | Coumine-type |
| 40 | 11.9 | 329.1499 | 311.1387,269.1300,168.1021,148.0381,122.0949,108.0809 | C18H20N2O4 | Nb-Methylgelsedilam | Gelsedine-type |
| 41 | 11.9 | 391.1869 | 360.1639,329.1497,311.1414,168.1018 | C20H26N2O6 | Dihydroxy gelsemicine | Gelsedine-type |
| 42 | 12.2 | 361.1763 | 330.1564,301.1184,283.1101,162.0545,84.0812 | C19H24N2O5 | 11,14-Dihydroxy-dihydrogelsenicine | Gelsedine-type |
| 43 | 12.3 | 422.1293 | 213.0759,195.0642,177.0538,149.0578,121.0653 | C17H19N5O8 | Unknown | Nucleoside |
| 44 | 12.4 | 323.1757 | 293.1646,279.1496,198.0908,138.0907,70.0606 | C20H22N2O2 | Isomer of gelsemine | Gelsemine-type |
| 45 | 12.4 | 377.1717 | 346.1527,317.1131 ,299.1021,271.1068,232.1196,203.0812,138.0552 | C19H24N2O6 | 11,14,15-Trihydroxy-dihydro gelsenicine | Gelsedine-type |
| 46 | 12.7 | 339.1709 | 323.1703,252.1005,70.0657 | C20H22N2O3 | Gelsemine oxide | Gelsemine-type |
| 47 | 12.9 | 311.1760 | 293.1654,252.1387 | C19H22N2O2 | Hydroxy of koumidine | Sarpagine-type |
| 48 | 12.9 | 385.1769 | 354.1602,339.1356,311.1336,134.0956,122.0995 | C21H24N2O5 | 15-Hydroxyhumantenox-enine | Humantenine-type |
| 49 | 13.2 | 571.2292 | 540.2101,509.1940,481.1919,359.1604,328.1422,297.1245, | C29H34N2O10 | Diydroxyl of gelseiridone/gelseganine D | Gelsedine-type |
| 50 | 13.4 | 527.2389 | 508.2282,496.2177,479.2188,339.1347 | C28H34N2O8 | Unknown | Gelsedine-type |
| 51 | 13.5 | 341.1866 | 323.1742,311.1156,297.1663,238.1224 | C20H24N2O3 | 19S-Hydroxydihydrogelsemine | Gelsemine-type |
| 52 | 13.5 | 391.1872 | 360.1683,329.1499,281.1284,238.0866,148.0396 | C20H26N2O6 | Dihydroxy gelsemicine | Gelsedine-type |
| 53 | 13.7 | 325.1916 | 307.1810,281.1661,238.1226,220.1116,194.0995,70.0660 | C20H24N2O2 | 19-(S)-Hydroxydihydrokoumine/19-(R)-Hydroxydihydrokoumine | Koumine-type |
| 54 | 13.9 | 323.1750 | 307.1801,238.1236,220.1118,194.1006,150.0913,122.0960 | C20H22N2O2 | Koumine N-oxide | Koumine-type |
| 55 | 14.1 | 422.1300 | 197.0816,179.0702,161.0589,153.0889,135.0796,108.0656 | C17H19N5O8 | Nucleoside | |
| 56 | 14.2 | 339.1712 | 325.1546,297.1603,252.1007,210.0910,70.0657 | C20H22N2O3 | Gelsemine oxide | Gelsemine-type |
| 57 | 14.4 | 197.0817 | 151.0752,133.0639,115.0534,105.0703,91.0547,81.0705,79.0543,77.0383,67.0543 | C10H12O4 | 7-Deoxygelsemide /9-Deoxygelsemide | Iridoids |
| 58 | 14.4 | 375.1558 | 344.1735,326.1625,313.1450,311.1388,299.1051,298.1672,122.0965,108.0809 | C19H22N2O6 | 11,14,15-Trihydroxygelsenicine | Gelsedine-type |
| 59 | 14.5 | 321.1604 | 291.1520,236.1068,210.0932,178.0846,136.0753 | C20H20N2O2 | Gelebolines A | Other types of alkaloids |
| 60 | 14.7 | 327.1711 | 311.1756,297.1581,291.1470,238.1229,135.0806 | C19H22N2O3 | 3-Hydroxykoumidine Oxide | Sarpagine-type |
| 61 | 14.8 | 527.2396 | 509,2273,496.2207,479.2195,468.2315,339.1342 | C28H34N2O8 | Unknown | Gelsedine-type |
| 62 | 14.9 | 355.1031 | 328.1448,309.1591,163.0393,145.0286,135.0442,117.0338,89.0400 | C16H18O9 | 1-O-Caffeoylquinic acid/4-O-Caffeoylquinic acid | Phenolic acids |
| 63 | 15.1 | 341.1869 | 323.1759,311.1751,297.1605,281.1452,238.1221,158.0595,70.0656 | C20H24N2O3 | 19R-Hydroxydihydrogelsemine | Gelsemine-type |
| 64 | 15.2 | 197.0807 | 179.0702,151.0745,133.0646,115.0539,105.0697,91.0544,81.0699,79.0547,77.0385,67.0550 | C10H12O4 | 7-Deoxygelsemide /9-Deoxygelsemide | Iridoids |
| 65 | 15.2 | 373.1759 | 342.1570,325.1550,311.1394,293.1285,108.0808 | C20H24N2O5 | 11,14-Dihydroxyrankinidine | Humantenine-type |
| 66 | 15.3 | 375.1919 | 357.1798,326.1625,311.1388,297.1584,283.1448,198.1100,122.0965,108.0809 | C20H26N2O5 | 14,15-dihydroxy-19,20dihydrorankinidine | Humantenine-type |
| 67 | 15.4 | 387.1913 | 369.1850,356.1697,341.1448,325.1548,311.1734,194.1165,138.0899 | C21H26N2O5 | Dihydrogelegamine A | Humantenine-type |
| 68 | 15.5 | 394.1343 | 341.0486,295.0510,197.0804,179.0705,161.0593,153.0910,135.0804,108.0658 | C16H19N5O7 | Unknown | Nucleoside |
| 69 | 15.6 | 343.1647 | 312.1465,295.1437,281.1281,264.1019,240.1012,212.0746,159.0673,108.080714,71.0734 | C19H22N2O4 | 11-Hydroxygelsenicine | Gelsedine-type |
| 70 | 15.6 | 505.2169 | 474.1980,443.1818,425.1636,339.1511,371.1530,341.0567,325.1311,297.1600,240.1194 | C25H32N2O9 | Unknown | Gelsedine-type |
| 71 | 15.9 | 359.1606 | 328.1422,285.1235,95.0732 | C19H22N2O5 | Hydroxyl of gelseziridine | Gelsedine-type |
| 72 | 16.2 | 323.1753 | 293.1627,262.1222,236.1072,70.0658 | C20H22N2O2 | Gelsemine | Gelsemine-type |
| 73 | 16.2 | 424.1452 | 3397.0429,341.00501,197.0804,179.0698,161.0959,135.0805 | C17H21N5O8 | Unknown | Phenolic acids |
| 74 | 16.7 | 359.1600 | 328.1420,311.1234,297.1238,279.1125,251.1168,225.1020 | C19H22N2O5 | 14,15-Dihydroxygelsenicine | Gelsedine-type |
| 75 | 16.8 | 343.1641 | 312.1473,281.1289,263.1181,253.1336,240.1033,212.0764,139.0990,124.0758,108.0811 | C19H22N2O4 | 14-Hydroxygelsenicine | Gelsedine-type |
| 76 | 17.0 | 406.1351 | 378.1419,343.1626,197.0796,179.0695,161.0591,153.0888,135.0792,107.0848,81.0729 | C17H19N5O7 | Unknown | Phenolic acids |
| 77 | 17.2 | 339.1707 | 323.1508,312.1470,295.1451,281.1279,251.1339,108.0812 | C20H22N2O3 | Gelsemine N-oxide | Gelsemine-type |
| 78 | 17.3 | 409.1756 | 378.1576,361.1555,347.1380,162.0904 | C23H24N2O5 | Unknown | Gelsedine-type |
| 79 | 17.4 | 375.1911 | 344.1728,313.1547,295.1430,253.1331,168.1019 | C20H26N2O5 | 11-Hydroxygelsemicine | Gelsedine-type |
| 80 | 17.4 | 423.1558 | 392.1369,375.1340,361.1186,335.1069,305.0927,188.0714,176.0707,151.0633,107.0731 | C23H22N2O6 | Unknown | Gelsedine-type |
| 81 | 17.6 | 379.1655 | 348.1469,331.1440,317.1286,291.1147,289.1335,261.1034,212.0706,132.0807,107.0734 | C22H22N2O4 | Unknown | Gelsedine-type |
| 82 | 17.8 | 345.1818 | 314.1617,285.1230,267.1147,168.1011 | C19H24N2O4 | 14-Hydroxygelsedine | Gelsedine-type |
| 83 | 17.8 | 401.1712 | 370.1526,339.1340,311.1398,295.1131,166.0864,154.0849 | C21H24N2O6 | 14-Acetoxy-15-hydroxygelsenicine | Gelsedine-type |
| 84 | 17.9 | 361.1758 | 330.1571,301.1181,283.1078,271.1045,138.0544 | C19H24N2O5 | Isomer of 11,14-Dihydroxy-dihydro gelsenicine | Gelsedine-type |
| 85 | 18.0 | 307.1802 | 293.1654,277.1697,251.1539,220.1113,138.0905,108.0806 | C20H22N2O | Isomer of koumine | Koumine-type |
| 86 | 18.0 | 343.1651 | 312.1460,295.1495,281.1281,165.1330,108.0806,95.0730 | C19H22N2O4 | Hydroxyl of gelsenicine | Gelsedine-type |
| 87 | 18.0 | 449.1078 | 431.0937,413.0829,395.0740,377.0659,353.0643,329.0654,299.0547 | C21H20O11 | Orientine/iso-orientine | Flavone |
| 88 | 18.1 | 305.1653 | 277.1680,220.1117,162.0904,130.0650,120.0803,70.0655 | C20H20N2O | Dehydrokoumine | Koumine-type |
| 89 | 18.2 | 311.1755 | 293.1656,269.1647,251.1546,138.0918,108.0816 | C19H22N2O2 | Hydroxy of koumidine | Sarpagine-type |
| 90 | 18.2 | 428.2539 | 398.1746,339.0895,299.0594,118.0864 | C24H33N3O4 | Unknown | Oil |
| 91 | 18.3 | 183.1009 | 137.0908,119.0842,107.0851,93.071,91.0541,77.0387,67.0545 | C10H14O3 | GSIR-1 | Iridoids |
| 92 | 18.3 | 201.1118 | 147.0788,135.0788,119.0845,107.0843,93.0691,91.0536,81.0692,79.0538,77.0382, 67.0541,55.0180 | C10H16O4 | Gelsemiol | Iridoids |
| 93 | 18.3 | 369.1802 | 338.1611,323.1733,307.1432,178.1215,122.0952 | C21H24N2O4 | Gelsevirine N-oxide | Gelsemine-type |
| 94 | 18.5 | 373.1767 | 342.1570,313.1564,311.1394,293.1285,283.1432,270.1129,189.0778,108.0808 | C20H24N2O5 | GS-2 | Gelsedine-type |
| 95 | 18.7 | 307.1810 | 277.1701,233.1229,220.1122,176.1074,130.0654,70.0658 | C20H22N2O | Koumine | Koumine-type |
| 96 | 19.0 | 359.1611 | 328.1391,311.1351,283.1451,271.1076,254.0819,190.0723,150.0907,138.0909 | C19H22N2O5 | 19,20-Dihydroxygelsenicine | Gelsedine-type |
| 97 | 19.0 | 449.1084 | 413.0855,383.0752,353.0650,329.0650,299.0547,209.1642 | C21H20O11 | Orientine/iso-orientine | Flavone |
| 98 | 19.4 | 371.1966 | 340.1781,313.1542,311.1404,295.1437,277.1331 | C21H26N2O4 | 15-hydroxyhumantenine | Humantenine-type |
| 99 | 19.5 | 295.1802 | 277.1697,222.1269,156.0802,144.0806,138.0908,108.0807 | C19H22N2O | Koumidine | Sarpagine-type |
| 100 | 19.5 | 375.1916 | 344.1723,313.1540,299.1388,265.1323,257.1268,198.1108,132.0441 | C20H26N2O5 | Hydroxylation of Gelsemicine | Gelsedine-type |
| 101 | 19.6 | 359.1606 | 328.1405,311.1387,299.1414,281.0969,185.0702 | C19H22N2O5 | 14,19-Dihydroxygelsenicine | Gelsedine-type |
| 102 | 19.8 | 417.1655 | 386.1469,368.1498,341.1498,329.1128,323.13838,311.1122,283.1073,194.0804 | C21H24N2O7 | 14-Acetoxy-dihydroxygelsenicine | Gelsedine-type |
| 103 | 19.9 | 391.1868 | 360.1680,331.1283,329.1478,313.1176,217.0959,138.0546 | C20H26N2O6 | Dihydroxy gelsemicine | Gelsedine-type |
| 104 | 20.1 | 369.1816 | 325.1542,311.1405,295.1418 | C21H24N2O4 | Humantenoxenine | Humantenine-type |
| 105 | 20.2 | 433.1975 | 402.1784,371.1601,343.1628,311.1386,283.1443,150.0915 | C22H28N2O7 | 11-Hydroxy-14-acetoxygelselegine | Gelsedine-type |
| 106 | 20.3 | 371.1977 | 340.1774,325.1548,323.1762,138.0913 | C21H26N2O4 | 19(R)-Hydroxydihydrogelsevirine/19(S)-hydroxydihydrogelsevirine | Gelsemine-type |
| 107 | 20.9 | 371.1973 | 340.1772,323.1750,212.0715,122.0961 | C21H26N2O4 | 19(R)-Hydroxydihydrogelsevirine/19(S)-hydroxydihydrogelsevirine | Gelsemine-type |
| 108 | 21.1 | 295.1816 | 277.1700,247.1239,156.0806,144.0812,138.0918,120.0813,108.0814 | C19H22N2O | Isomer of koumidine | Sarpagine-type |
| 109 | 21.1 | 405.2028 | 374.1823,343.1652,329.1502,325.1525 | C21H28N2O6 | 11-Methoxy-19-(R)-hydroxygelselegine | Gelsedine-type |
| 110 | 21.3 | 327.1720 | 296.1534,265.1357,225.1055,108.0825,95.0747,71.0750 | C19H22N2O3 | Gelsenicine | Gelsedine-type |
| 111 | 21.6 | 371.1966 | 340.1782,325.1552,311.1410,178.1229,122.0965 | C21H26N2O4 | 6-hydroxyhumantenine | Humantenine-type |
| 112 | 21.9 | 357.1809 | 326.1632,311.1397,297.1263,269.1285,178.1228,164.1073,122.0967,108.0816 | C20H24N2O4 | 14-Hydroxyrankinidine | Humantenine-type |
| 113 | 22.1 | 353.1865 | 323.1722,322.1675,291.1491,164.1067,108.0809 | C21H24N2O3 | Gelsevirine | Gelsemine-type |
| 114 | 22.5 | 343.1663 | 312.1480,281.1265,255.1127,238.0867,210.0915,174.0783,136.0785,118.0653 | C19H22N2O4 | Hydroxyl of gelsenicine | Gelsedine-type |
| 115 | 22.6 | 325.1914 | 307.1796,281.1658,243,1500,158.0617,136.1124 | C21H26N2O2 | Gardnerine | Sarpagine-type |
| 116 | 22.8 | 363.1712 | 332.1515,301.1133,261.1032,212.0702,144.0803,121.0855 | C22H22N2O3 | Unknown | Gelsedine-type |
| 117 | 22.9 | 311.1761 | 293.1674,267.1490,249.1398,229.1338,158.0605,138.0900,122.0964,108.0815 | C19H22N2O2 | 3-Hydroxykoumidine | Sarpagine-type |
| 118 | 22.9 | 357.1811 | 326.1625,297.1428,295.1442,278.1185,254.1174,213.0919,108.0813,71.0740 | C20H24N2O4 | 4,20-Dehydrogelsemicine | Gelsedine-type |
| 119 | 22.9 | 383.1970 | 365.1857,341.1872,321.1592,180.1017,172.0753,138.0913 | C22H26N2O4 | Gelsempervine A | Sarpagine-type |
| 120 | 23.7 | 339.1709 | 308.1521,277.1339,225.1019,176.1067,148.1119,114.0918 | C21H26N2O2 | Nb-Demethylgelsevirine | Gelsemine-type |
| 121 | 23.7 | 533.2496 | 515.2388,502.2278,484.2206,467.2177,453.2030,381.1824,353.1859,339.1698 | C27H36N2O9 | Unknown | Gelsedine-type |
| 122 | 23.9 | 309.1963 | 291.1840,265.1713,220.1113,178.1226,122.0960 | C20H24N2O | Dihydrokoumine | Koumine-type |
| 123 | 23.9 | 353.1870 | 322.1677,295.1721,291.1488,239.1178,121.0883 | C21H24N2O3 | 19-(Z)-Akuammidine | Sarpagine-type |
| 124 | 23.9 | 359.1964 | 328.1778,297.1602,279.1484,222.0943,182.1161 | C20H26N2O4 | Gelsemicine | Gelsedine-type |
| 125 | 23.9 | 417.2016 | 399.1909,376.1818,368.1717,357.1786,326.1627,298.1686,269.1284,163.0985 | C22H28N2O6 | Unknown | Gelsedine-type |
| 126 | 24.1 | 385.1764 | 354.1573,323.1407,311.1390,295.1444,263.1179,237.1045,121.0880,108.0812,95.0732 | C21H24N2O5 | 14-Acetoxygelsenicine | Gelsedine-type |
| 127 | 24.3 | 517.1342 | 488.1965,443.1667,163.0386,145.0282,117.0338 | C25H24O12 | 1,3-dicaffeoylquinic acid | Phenolic acids |
| 128 | 24.9 | 369.1812 | 323.1412,309.1539,307.1439,265.1079,172.1063,148.1113,122.0964,107.0742 | C21H24N2O4 | 19E-16-epi-Voacarpine | Sarpagine-type |
| 129 | 25.0 | 329.1859 | 298.1671,269.1279,257.1167,152.1059,84.0808 | C19H22N2O3 | Gelsedine | Gelsedine-type |
| 130 | 25.1 | 417.2016 | 386.1832,355.1651,343.1652,295.1433,225.1015,150.0907 | C22H28N2O6 | 14-Acetoxygelselegine | Gelsedine-type |
| 131 | 25.5 | 389.2069 | 358.1881,327.1703,309.1577,284.1405 | C21H28N2O5 | 11-Methoxygelselegine | Gelsedine-type |
| 132 | 25.7 | 357.2166 | 326.1984,311.1773,298.2032,239.1178,181.1452,124.1115 | C21H28N2O3 | 19,20-Dihydrohumantenine | Humantenine-type |
| 133 | 25.9 | 355.204 | 325.1911,324.18556,311.1703,310.1658,309.1625,178.1253122.0992 | C21H26N2O3 | Humantenine | Humantenine-type |
| 134 | 25.9 | 429.2018 | 398.1839,385.1536,353.1862,339.1695,222.1110,166.0849,122.0946 | C23H28N2O6 | Gelseoxazolidinine | Gelsedine-type |
| 135 | 26.1 | 309.1616 | 295.1466,281.1322,138.0939,132.0478,120.0813,108.0847 | C19H20N2O2 | Oxokoumidine | Sarpagine-type |
| 136 | 26.1 | 341.1896 | 311.1749,310.1712,295.1483,281.1355,178.1266,164.1109,122.1003,108.0850,96.0849 | C20H24N2O3 | Rankinidine (Gelsemamides) | Humantenine-type |
| 137 | 27.4 | 371.1951 | 340.1764,325.1531,311.1396,164.1055,122.0946,108.0797 | C21H26N2O4 | 11-hydroxyhumantenine | Humantenine-type |
| 138 | 27.5 | 385.2134 | 354.1920,339.1716,311.1720,178.1228,122.0972 | C22H28N2O4 | 11-Methoxyhumantenine | Humantenine-type |
| 139 | 27.8 | 357.2162 | 326.1968,311.1726,297.1587,181.1431,124.1223 | C21H28N2O3 | Isomer of 19,20-Dihydrohumantenine | Humantenine-type |
| 140 | 28.9 | 373.2127 | 342.1929,327.1680,313.1549 | C21H28N2O4 | Acetyl of 14-Hydroxygelsedine | Gelsedine-type |
| 141 | 29.7 | 325.1911 | 294.1728, 279.1519, 164.1066 | C20H22N2O2 | Na-Desmethoxyhumante-nine | Humantenine-type |
| 142 | 30.2 | 273.1388 | 257.1064,254.1068 | C19H16N2 | Sempervirine | Yohimbane |
| 143 | 30.6 | 339.2068 | 308.1862,293.1644,279.1541,178.1221,164.1068,148.1117,136.1116,122.0961,108.0815 | C21H26N2O2 | Na-Methoxy-19(Z)anhydrovobasinediol | Sarpagine-type |
| 144 | 33.6 | 343.1654 | 312.1450,281.1317,265.1067,255.1137,238.0862,210.0917,174.0872,118.0860 | C19H22N2O4 | Hydroxyl of gelsenicine | Gelsedine-type |
| 145 | 36.0 | 343.1655 | 312.1448,295.1285,281.1294,265.1071,255.1113,238.0862,210.0991,174.0773,136.0750 | C19H22N2O4 | Hydroxyl of gelsenicine | Gelsedine-type |
| 146 | 40.0 | 343.1184 | 327.0868,313.0706,299.0907,282.0881 | C19H18O6 | Unknown | Flavone |
| 147 | 40.7 | 279.1598 | 265.0186,219.1034,149.0232,121.0285,93.0342,57.0706 | C16H22O4 | Phthalic acid dibutyl ester | Oil |
3.3.1. Characterization of gelsedine-type alkaloids
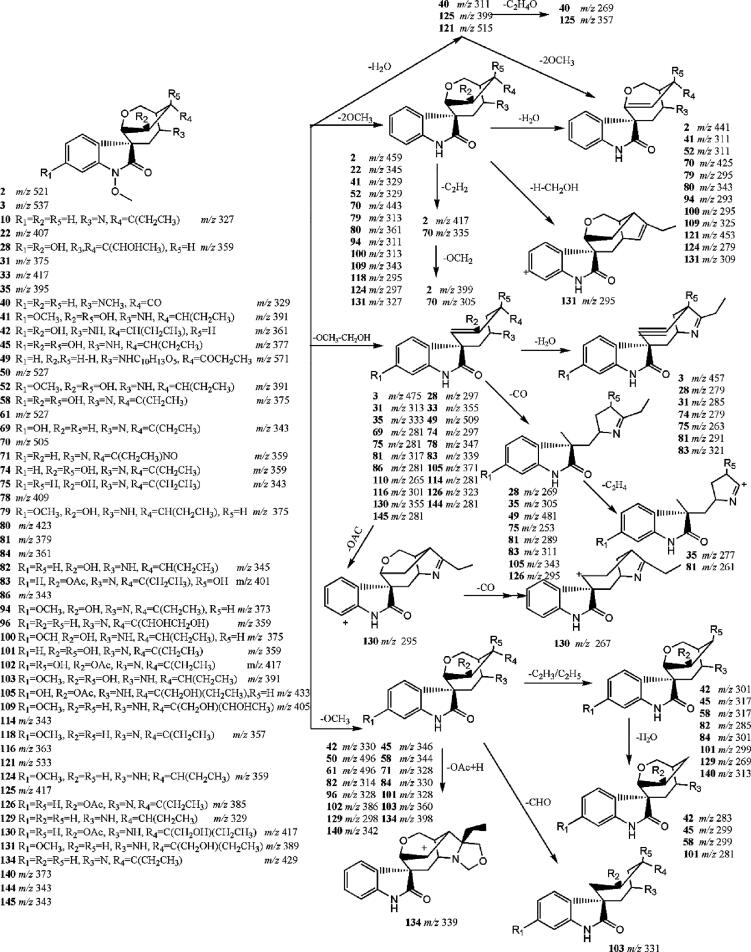
A total of 52 components were recognized as gelsedine-type alkaloids. Based on the fragmentation behavior of these components, they contained a Na-methoxy group. On the one hand, there was also a methoxy group on the C-11 position for some components; on the other hand, some of the components had a CH2OH group at this position. Therefore, the main fragmentation pattern of components could be a neutral loss of 62 Da (two OCH3 or OCH3 plus CH2OH). The neutral loss chromatogram (pNLC) of m/z 62 was shown in Fig. 3. For example, components 74 and 124 displayed [M + H]+ ions at m/z 359.1600 and 359.1964, respectively. Both of them showed a fragment loss of 62 Da. However, for component 74, a fragment of m/z 279 (M + H-80 Da) was lost, indicating that component 74 had more than one hydroxyl group; moreover, component 74 was an isomer of 28. Therefore, components 74 and 124 were characterized as 14,15-dihydroxygelsenicine and gelsemicine, respectively.
Fig. 3.
EIC spectrum (A), MS/MS spectrum (B) of component 94 (GS-2), pNLC spectrum of gelsedine type alkaloids with neutral loss ion at m/z 62 (C) and pNLC spectrum of G. elegans compounds (D).
The fragment ions of component 58 were m/z 344, 327, and 267, which were each 16 Da higher than the corresponding fragment ions m/z 328, 311, and 251 of component 96, respectively. Therefore, component 58 was the hydroxylated product of component 96. Components 58 and 45 provided the same fragment ion at m/z 299 by loss of a C2H3 or C2H5 group, respectively, indicating that component 45 was a reduction product of component 58. Components 41 and 52 were 16 Da and 32 Da higher in molecular mass than components 100 and 124, respectively, suggesting that they were dihydroxylation products of gelsemicine. The fragmentation pathways of the gelsedine-type alkaloids were summarized in Fig. 4.
Fig. 4.
Mass spectral fragmentation pathways of gelsedine-type alkaloids.
3.3.2. Characterization of sarpagine-type alkaloids
Compared with the target components, component 99 was characterized as koumidine. Furthermore, component 108 was an isomer of 99. According to the fragmentation pathway of koumidine (99), component 117 was characterized as a hydroxylated derivative of component 99. Components 89 and 117 were characterized as 3-hydroxykoumidine and hydroxylated koumidine, respectively. Component 60 was the N-oxide form of 3-hydroxykoumidine. The sarpagine-type components could first lose the group at the Na position, C-3 position or C-16 position. The diagnostic ion of these alkaloids was m/z 138. Components 123 and 113 had the same molecular formula, but component 113 had a diagnostic ion at m/z 323. The most abundant fragment ion of component 123 was at m/z 295. Therefore, component 123 was characterized as 19-(Z)-akuammidine. The fragmentation pathway of the sarpagine-type alkaloids was shown in Fig. 5.
Fig. 5.
Mass spectral fragmentation pathways of sapargine-type alkaloids.
3.3.3. Characterization of humantenine-type alkaloids
The key filter ions of humantenine-type alkaloids were m/z 311.17 (components 67, 132, 133, 136, 138 and 139) and 311.14 (components 36, 48, 65, 66, 98, 104, 111, 112 and 137). The results indicated that humantenine-type alkaloid components could be found by filtering m/z 311 in the extracted-ion chromatogram (EIC) MS/MS spectrum. The EIC MS/MS spectrum of m/z 311 was shown in Fig. 6. These components could lose H2O, hydroxymethyl, methoxy, methyl, or methylene groups to form the fragment ion at m/z 311. For example, component 112 could lose OCH3 and CH3 groups to form m/z 311, and component 137 could lose CH2 after losing 46 Da (OCH3 plus CH3) to form m/z 311. The molecular formulas of compounds 98, 111, and 137 were calculated as C21H26N2O4 based on their measured accurate mass of m/z 371.196, which suggested that components 98, 111, and 137 were isomers. Components 36 and 65 were 16 Da higher in molecular weight than component 112, which showed they were formed by dihydroxylation of component 112. As a result, components 112 and 137 were characterized as 14-hydroxyrankinidine and 11-hydroxyhumantenine, respectively. Through online database searching, component 104 was tentatively characterized as humantenoxenine, which belonged to the humantenine-type alkaloid family. The fragmentation pathways of the humantenine-type alkaloids were shown in Fig. 6.
Fig. 6.
EIC spectrum (A) and MS/MS spectrum (B) of component 133 (humantenine); MS/MS spectrum extracted by extracting diagnostic ions at m/z 311.17, and 311.14 (C). Mass spectral fragmentation pathways of humantenine-type alkaloids (D).
3.3.4. Characterization of gelsemine-type alkaloids
The precursor ions of 11 gelsemine-type alkaloids, including compounds 46, 44, 51, 56, 63, 72, 77, 93, 106, 107, and 113 were screened out by using the diagnostic ion at m/z 323. The fragmentation pathways of the gelsemine-type alkaloids were shown in Fig. S1. These components would first lose the group at the Na position, the methyl at the Nb position and the group at the C-19 position to form the diagnostic ion at m/z 323. For example, components 106 and 107 could lose OCH3 (31 Da) to form the product ion at m/z 340 and then could lose OH (17 Da) at the C-19 position to form m/z 323. Based on the fragmentation data and accurate mass values, components 106 and 107 were a pair of isomers and were characterized as 19(R)-hydroxydihydrogelsevirine or 19(S)-hydroxydihydrogelsevirine, respectively. Component 120 was 14 Da lower in molecular weight than component 113 (loss of a methyl group at the Nb position), and component 93 was 16 Da higher in molecular weight than component 113 (Nb position). Thus, components 120, 113, and 93 were characterized as Nb-demethylgelsevirine, gelsevirine and gelsevirine N-oxide. Furthermore, components 37, 46, 56, and 77 were the oxidized form of components 44 or 72.
3.3.5. Characterization of koumine- type alkaloids
A total of seven components, compounds 39, 53, 54, 85, 88, 95, and 122 were classified as koumine-type alkaloids since they all yielded a diagnostic ion at m/z 220. Koumine-type alkaloids could gradually lose the groups at the C-19 position, Nb position, C-16 and C17 positions. The fragmentation pathways of the koumine-type alkaloids are shown in Fig. S2. By comparison to a koumine standard, component 95 was characterized as koumine. Component 95 was 2 Da higher in molecular weight than component 88, and components 95 and 88 had the same fragment ions, which showed that component 88 was the dehydrogenation product of component 95. Therefore, component 88 was named dehydrokoumine. Components 39 and 53 also had the same fragment ions and were characterized as 19-(S)-hydroxydihydrokoumine and 19-(R)-hydroxydihydrokoumine by literature searching (Kitajima, Kobayashi, Kogure, & Takayama, 2010), respectively.
3.3.6. Characterization of iridoids
Iridoids were filtered by a neutral loss of 46 Da (CH2O2) and a diagnostic ion at m/z 91. This filtering could be applied to EIC MS/MS and pNLC spectra due to the structure of iridoids. For example, components 10 and 25 had the same formula, but component 25 could lose 82 Da (2H2O plus CH2O2), whereas component 10 could lose only 64 Da (H2O plus CH2O2), which proved that compared to component 10, component 25 had an additional hydroxyl group. Components 11 and 26 were 16 Da higher in molecular weight than component 92 and were characterized as 7-hydroxygelsemiol or 9-hydroxygelsemiol, respectively. Through the combination of target ion and database searching, the nontargeted components were characterized quickly. The fragmentation pathways of the iridoids are shown in Fig. S3.
3.3.7. Characterization of phenolic acids
Only two types of phenolic acids were detected in G. elegans after matching with the MS database, and their structures were determined based on the MS/MS spectra. Component 62 produced a sodium-adduct molecular ion [M + Na]+ at m/z 377.0866 and a protonated molecular ion [M + H]+ at 355.1024, and the enriched fragment ion at m/z 163 formed by losing glucuronic acid from the protonated molecule. The ion at m/z 145.0291 was formed by the loss of a molecule of H2O (18 Da) from m/z 163.0394. The minimum ion at m/z 89.0403 was generated by the neutral loss of two CO molecules (28 Da) at m/z 145.0291. The structure of the phenolic acids was based on a benzene ring and numerous hydroxyl groups, so its fragment ions were formed by losses of H2O and CO. Component 6 was established as ferulic acid, and it was proposed as an isomer of component 29. Component 62 was characterized tentatively as 1-O-caffeoylquinic acid or 4-O-caffeoylquinic acid. Through an online database search, components 12, 16, 27, 73, and 127 were classified as phenolic acids. In addition, components 12, 16, and 27 were hydroxylated derivatives of ethyl ferulic acid.
3.4. MRM transitions derived from MS2
With a combination of LC-QTOF/MS and LC-QqQ/MS, we developed a quantitative approach for analysis in the absence of standards. The present LC-MS/MS strategy enabled the separation of 41 components in the method validation protocol. Many parameters must be optimized during the development of a DeMRM method, which is always laborious and time-consuming. Therefore, a simple standard operation procedure was recommended in the present study.
First, the precursor ions and corresponding product ions of each component were obtained by LC-QTOF/MS and were used to form an ion pair; in general, each precursor ion had 2–3 product ions. Second, higher precursor ion and product ion responses were obtained through adequate optimization of instrumental parameters by repeatedly testing four standards (gelsemine, koumine, koumidine and gelsenicine) in reference multiple reaction monitoring (RMRM) mode. Finally, multiple components of G. elegans were optimized in terms of ion pairs and CE in LC-QqQ/MS (Table 2).
Table 2.
Monitor ion pairs, CE, segment, repeatability and stability of multiple compounds in G. elegans samples.
| No. | Analytes | t/R (min) | Ion pair | CE/eV | Seg | Repeatability (mg/g ± RSD%) | Stability/RSD(%) |
|---|---|---|---|---|---|---|---|
| 1 | 7-Deoxygelsemide or 9-deoxygelsemide | 6.2 | 197.1 → 105.1 | 30 | 2 | 0.001 ± 3.31 | 3.02 |
| 2 | 11,14-Dihydroxygelsenicine | 11.4 | 359.2 → 108.1 | 30 | 3 | 0.086 ± 2.88 | 1.63 |
| 3 | 14,15-Dihydroxygelsenicine | 11.5 | 359.2 → 328.1 | 30 | 3 | 0.066 ± 2.96 | 2.56 |
| 4 | Unknown (375) | 15.6 | 375.2 → 311.1 | 30 | 4 | 0.029 ± 1.42 | 1.98 |
| 5 | Gelsemine | 16.5 | 323.2 → 70.1 | 33 | 4 | 3.787 ± 0.94 | 1.58 |
| 6 | 11-Hydroxygelsenicine | 16.5 | 343.2 → 281.1 | 30 | 4 | 0.017 ± 1.27 | 3.42 |
| 7 | Gelsemicine | 17.1 | 359.2 → 108.1 | 30 | 4 | 0.002 ± 2.79 | 2.12 |
| 8 | GSIR-1 | 17.4 | 183.1 → 91.1 | 30 | 4 | 0.001 ± 2.76 | 4.68 |
| 9 | 14-Hydroxygelsemicine or other hydroxylation of gelsemicine | 17.5 | 375.2 → 313.2 | 35 | 4 | 0.237 ± 2.36 | 1.56 |
| 10 | 14-Hydroxygelsedine | 17.8 | 345.2 → 285.1 | 30 | 4 | 0.034 ± 2.13 | 2.46 |
| 11 | 14-Hydroxygelsenicine | 18.0 | 343.2 → 108.1 | 30 | 5 | 0.730 ± 3.92 | 1.63 |
| 12 | Koumine | 19.1 | 307.2 → 180.0 | 53 | 5 | 0.830 ± 1.83 | 2.07 |
| 13 | 14-Dehydroxygelsefuranidine or other dehydroxylgelsefuranidine (2) | 19.4 | 405.2 → 343.1 | 30 | 5 | 0.071 ± 2.70 | 1.86 |
| 14 | 11-methoxy-14-hydroxygelsenicine | 19.5 | 373.2 → 108.1 | 30 | 5 | 0.142 ± 2.16 | 2.69 |
| 15 | Unknown (295) | 19.8 | 295.2 → 138.1 | 30 | 5 | 0.058 ± 1.78 | 3.03 |
| 16 | Hydroxyl of gelsedine | 19.9 | 345.2 → 285.1 | 30 | 5 | 0.001 ± 4.46 | 4.20 |
| 17 | Gelsemoxonine | 20.1 | 359.2 → 311.1 | 30 | 6 | 0.942 ± 3.38 | 2.10 |
| 18 | One of other 5 hit compounds | 21.1 | 371.2 → 323.1 | 30 | 6 | 0.067 ± 2.84 | 1.64 |
| 19 | 14-Dehydroxygelsefuranidine or other dehydroxylgelsefuranidine (1) | 21.1 | 405.2 → 374.2 | 30 | 6 | 0.049 ± 2.92 | 4.24 |
| 20 | Koumidine | 21.2 | 295.2 → 144.1 | 30 | 6 | 0.042 ± 2.01 | 2.06 |
| 21 | 14-Acetoxygelsedilam or other acetoxyl of gelsedilam (1) | 21.4 | 373.2 → 342.2 | 30 | 6 | 0.005 ± 4.13 | 2.06 |
| 22 | 11-Hydroxyhumantenine | 21.5 | 371.2 → 325.2 | 30 | 6 | 0.312 ± 1.77 | 2.26 |
| 23 | Gelsevirine | 22.0 | 353.2 → 291.2 | 30 | 7 | 0.191 ± 2.30 | 1.39 |
| 24 | Gelsenicine | 22.2 | 327.2 → 265.1 | 30 | 7 | 0.958 ± 1.75 | 1.41 |
| 25 | 12β-hydroxy-pregn-4,16-diene-3,20-dione | 22.2 | 329.2 → 97.1 | 30 | 7 | 0.002 ± 4.96 | 3.00 |
| 26 | Koumicine | 23.8 | 353.2 → 166.1 | 30 | 7 | 0.342 ± 3.81 | 1.70 |
| 27 | Gelsedine or Nb-Methylgelsedilam | 23.4 | 329.2 → 298.2 | 30 | 7 | 0.004 ± 1.54 | 2.01 |
| 28 | Gelseoxazolidinine | 23.9 | 429.2 → 339.2 | 30 | 7 | 0.003 ± 2.53 | 2.99 |
| 29 | 16-epi-voacarpine or gelsevirine N-oxide | 24.0 | 369.2 → 166.1 | 30 | 7 | 0.823 ± 1.20 | 1.84 |
| 30 | Humantenoxenine | 24.3 | 369.2 → 108.1 | 30 | 7 | 0.009 ± 3.48 | 3.31 |
| 31 | Iso-Gelsedine or Nb-Methylgelsedilam | 24.7 | 329.2 → 269.3 | 30 | 7 | 0.014 ± 4.55 | 2.48 |
| 32 | 1-O-Caffeoylquinic acid or 4-O-Caffeoylquinic acid | 25.0 | 355.1 → 135.0 | 30 | 8 | 0.117 ± 1.24 | 1.39 |
| 33 | Gelseziridine | 25.7 | 343.2 → 108.1 | 30 | 8 | 0.001 ± 2.57 | 2.27 |
| 34 | Na-Desmethoxyhumantenine | 25.8 | 325.2 → 136.1 | 35 | 8 | 0.020 ± 2.63 | 1.55 |
| 35 | 14-Acetoxygelsedilam or other acetoxyl of gelsedilam (2) | 26.6 | 373.2 → 342.2 | 30 | 8 | 0.033 ± 2.14 | 1.95 |
| 36 | 14-Acetoxygelsedilam or other acetoxyl of gelsedilam (3) | 27.3 | 373.2 → 342.2 | 30 | 8 | 0.033 ± 2.48 | 2.57 |
| 37 | 19R-Hydroxydihydrogelsevirine or 19S-Hydroxydihydrogelsevirine | 26.5 | 371.2 → 164.1 | 30 | 8 | 4.526 ± 4.24 | 4.18 |
| 38 | 11-Methoxyhumantenine | 26.8 | 385.2 → 339.2 | 30 | 8 | 0.061 ± 4.33 | 4.53 |
| 39 | iso-12β-Hydroxy-5α-pregn-16-ene-3,20-dione | 31.2 | 331.2 → 109.1 | 30 | 11 | 0.003 ± 4.42 | 4.02 |
| 40 | Gelse-norursane E | 31.7 | 471.2 → 217.1 | 30 | 10 | 0.008 ± 4.28 | 3.99 |
| 41 | 12β-Hydroxy-5α-pregn-16-ene-3,20-dione | 31.8 | 331.2 → 97.1 | 30 | 11 | 0.038 ± 4.24 | 4.18 |
To avoid interference with some low-concentration components, ion pairs were set to several segments to improve sensitivity. The multiple components need to be optimized in terms of the analysis time period by adjusting the LC conditions to ensure good peak shape without tailing and drift. In addition, it is observed that compounds with similar structures are often assigned similar MRM parameters and transitions. Therefore, we developed a DeMRM method to monitor multiple components in herbal medicines, even those present at a trace level. In this study, G. elegans was selected as an example to demonstrate our approach. The retention times, monitored ion pairs, and related voltage parameters of multiple components in G. elegans were shown in Table 2.
3.5. Method validation of proposed method
Table S3 summarized the validation results of the four representative components (gelsemine, koumine, koumidine and gelsenicine) for RMRM. The correlation coefficients of the four compounds were higher than 0.99 in the concentration range of 10–200 ng/mL. The LODs of gelsemine, koumine, koumidine and gelsenicine were 2.5, 2, 5 and 1.5 ng/mL, respectively. The LOQs of gelsemine, koumine, koumidine and gelsenicine were 5, 5, 10 and 5 ng/mL, respectively.
The DeMRM method was examined in terms of specificity, accuracy, and stability. As presented in Fig. S4, no interfering peaks were observed at the retention times of the 41 components in the DeMRM chromatograms of G. elegans samples. Most componets had no the phenomenon of trailing, incomplete peak of the sample and the retention time of the sample was suitable. For example, the retention time of gelsemine was 16.5 min, and that of koumidine was 22.2 min, which indicated the specificity of this analytical method. Table 2 summarizes the relative concentrations of 41 components in the G. elegans sample and relative standard deviations (RSDs) of the concentrations. The intraday and interday precision were expressed as the RSD. The RSDs of the four representative components at the two tested concentrations were all within 10%. Moreover, all the RSDs of the DeMRM method were within the accepted variable limits. The results support that the DeMRM method has reasonable accuracy and stability and is applicable to the quantitative analysis of complex herbal medicines.
3.6. Sample analysis
The validated DeMRM method was subsequently applied to determine the relative concentrations of multiple components in different tissues of G. elegans during different periods. The quantitative performance of the DeMRM method was examined by comparing the experimental values of the four representative components obtained by DeMRM and RMRM. The DeMRM results were expressed as the relative content of herb based on a koumidine calibration curve, while the RMRM results were converted to the herb contents by calculating the absolute amounts of the components in the herb. Taking koumine in the root as an example, as shown in Fig. 7 (Root), the RMRM results indicated that the koumine content obtained from chemical standards was the highest in December, followed by September, and was the lowest in November, which is the result of RMRM. In Table 3, the relative contents of koumine obtained from the koumidine calibration curves were 1.376, 1.089, and 1.772 mg/g in September, November, and December, respectively, which indicated that the results obtained by DeMRM were consistent with the above trends observed by RMRM. The same trend proved that the results of the relative quantification of 41 components calculated by DeMRM in G. elegans were reliable. Although some deviation exists, the approach proposed herein still offered a direct and rapid method for semiquantitative determination with reasonable accuracy in cases when authentic standards are not available and/or the absolute quantity is not needed.
Fig. 7.
Differences contents of four standards between roots (A), stems (B) and leaves (C) from G. elegans.
Table 3.
Multi-compounds contents in Gelsemium elegans samples.
| No. | Analytes | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7-Deoxygelsemide or 9-Deoxygelsemide | – | – | 0.004 | – | – | 0.001 | – | – | 0.001 |
| 2 | 11,14-Dihydroxygelsenicine | 0.043 | 0.041 | 0.114 | 0.012 | 0.058 | 0.193 | 0.009 | 0.148 | 0.472 |
| 3 | 14,15-Dihydroxygelsenicine | 0.032 | 0.032 | 0.087 | 0.009 | 0.045 | 0.148 | 0.009 | 0.114 | 0.365 |
| 4 | Unknown (375) | 0.037 | 0.007 | 0.005 | 0.032 | 0.015 | 0.015 | 0.043 | 0.015 | 0.009 |
| 5 | Gelsemine | 3.036 | 4.145 | 4.679 | 2.208 | 3.014 | 3.565 | 3.956 | 4.678 | 3.19 |
| 6 | 11-Hydroxygelsenicine | 0.048 | 0.032 | 0.08 | 0.008 | 0.039 | 0.009 | 0.002 | 0.003 | 0.006 |
| 7 | Gelsemicine | – | – | – | 0.001 | 0.001 | 0.002 | 0.003 | 0.004 | 0.004 |
| 8 | GSIR-1 | 0.001 | 0.001 | 0.003 | – | – | 0.001 | – | – | 0.001 |
| 9 | 14-Hydroxygelsemicine or other hydroxylation of gelsemicine | 0.424 | 0.145 | 0.129 | 0.273 | 0.187 | 0.13 | 0.417 | 0.136 | 0.088 |
| 10 | 14-Hydroxygelsedine | 0.048 | 0.005 | 0.007 | 0.066 | 0.005 | 0.004 | 0.132 | 0.008 | 0.005 |
| 11 | 14-Hydroxygelsenicine | 0.383 | 0.171 | 0.576 | 0.704 | 0.407 | 0.552 | 0.951 | 1.198 | 0.925 |
| 12 | Koumine | 1.376 | 0.644 | 0.693 | 1.089 | 0.562 | 0.51 | 1.772 | 0.333 | 0.197 |
| 13 | 14-Dehydroxygelsefuranidine or other dehydroxylgelsefuranidine (2) | 0.076 | 0.037 | 0.106 | 0.064 | 0.041 | 0.143 | 0.117 | 0.03 | 0.097 |
| 14 | 11-Methoxy-14-hydroxygelsenicine | 0.064 | 0.02 | 0.099 | 0.133 | 0.069 | 0.091 | 0.224 | 0.182 | 0.169 |
| 15 | Unknown (295) | 0.077 | 0.039 | 0.023 | 0.071 | 0.027 | 0.016 | 0.152 | 0.025 | 0.008 |
| 16 | Hydroxyl of gelsedine | 0.001 | – | – | – | – | – | 0.003 | – | – |
| 17 | Gelsemoxonine | 0.158 | 0.04 | 0.455 | 0.167 | 0.207 | 1.994 | 0.617 | 1.066 | 3.185 |
| 18 | One of other five hit compounds | 0.08 | 0.028 | 0.065 | 0.069 | 0.022 | 0.071 | 0.184 | 0.017 | 0.039 |
| 19 | 14-Dehydroxygelsefuranidine or other dehydroxylgelsefuranidine (1) | 0.059 | 0.013 | 0.034 | 0.06 | 0.01 | 0.046 | 0.111 | 0.019 | 0.03 |
| 20 | koumidine | 0.022 | 0.04 | 0.02 | 0.028 | 0.028 | 0.022 | 0.107 | 0.028 | 0.012 |
| 21 | 14-Acetoxygelsedilam or other Acetoxyl of gelsedilam (1) | 0.01 | 0.01 | 0.008 | 0.001 | 0.005 | 0.003 | 0.004 | 0.003 | 0.003 |
| 22 | 11-Hydroxyhumantenine | 0.987 | 1.069 | 0.963 | 0.186 | 0.541 | 0.185 | 0.077 | 0.097 | 0.1 |
| 23 | Gelsevirine | 0.257 | 0.108 | 0.062 | 0.389 | 0.08 | 0.02 | 0.631 | 0.03 | 0.007 |
| 24 | Gelsenicine | 0.944 | 0.228 | 0.823 | 1.085 | 0.805 | 0.55 | 1.508 | 0.901 | 0.488 |
| 25 | 12β-Hydroxy-pregn-4,16-diene-3,20-dione | 0.002 | – | 0.001 | 0.002 | 0.001 | 0.001 | 0.002 | 0.001 | 0.001 |
| 26 | Koumicine | 0.326 | 0.237 | 0.349 | 0.219 | 0.217 | 0.446 | 0.441 | 0.199 | 0.327 |
| 27 | Gelsedine or Nb-Methylgelsedilam | 0.007 | 0.001 | 0.002 | 0.006 | 0.002 | 0.001 | 0.01 | 0.001 | 0.001 |
| 28 | Gelseoxazolidinine | 0.015 | 0.005 | 0.01 | 0.002 | – | 0.003 | 0.002 | – | 0.001 |
| 29 | 16-epi-Voacarpine or gelsevirine N-oxide | 0.853 | 0.514 | 0.812 | 0.57 | 0.645 | 1.13 | 0.934 | 0.817 | 1.033 |
| 30 | Humantenoxenine | 0.006 | 0.008 | 0.02 | 0.003 | 0.004 | 0.013 | 0.009 | 0.009 | 0.011 |
| 31 | Iso-Gelsedine or Nb-Methylgelsedilam | 0.012 | 0.002 | 0.002 | 0.042 | 0.001 | 0 | 0.042 | 0.001 | – |
| 32 | 1-O-Caffeoylquinic acid or 4-O-Caffeoylquinic acid | 0.176 | 0.044 | 0.072 | 0.165 | 0.059 | 0.071 | 0.268 | 0.064 | 0.045 |
| 33 | Gelseziridine | 0.001 | – | – | 0.003 | – | 0.001 | 0.004 | – | – |
| 34 | Na-Desmethoxyhumantenine | 0.028 | 0.005 | 0.012 | 0.028 | 0.01 | 0.014 | 0.046 | 0.012 | 0.009 |
| 35 | 14-Acetoxygelsedilam or other Acetoxyl of gelsedilam (2) | 0.039 | 0.008 | 0.019 | 0.068 | 0.019 | 0.02 | 0.069 | 0.019 | 0.019 |
| 36 | 14-Acetoxygelsedilam or other Acetoxyl of gelsedilam (3) | 0.028 | 0.009 | 0.023 | 0.087 | 0.017 | 0.019 | 0.09 | 0.018 | 0.018 |
| 37 | 19R-Hydroxydihydrogelsevirine or 19S-Hydroxydihydrogelsevirine | 3.436 | 1.261 | 1.335 | 9.918 | 2.4 | 2.677 | 10.567 | 1.584 | 2.629 |
| 38 | 11-Methoxyhumantenine | 0.105 | 0.08 | 0.04 | 0.081 | 0.034 | 0.034 | 0.272 | 0.016 | 0.011 |
| 39 | iso-12β-Hydroxy-5α-pregn-16-ene-3,20-dione | 0.004 | 0.001 | 0.004 | 0.002 | – | 0.006 | 0.008 | – | 0.005 |
| 40 | Gelse-norursane E | 0.012 | 0.001 | 0.002 | 0.006 | 0.001 | 0.007 | 0.036 | 0.001 | 0.003 |
Note: -, nondetected.
4. Conclusion
The present work contributed to the development of a powerful integrated strategy based on liquid chromatography coupled with mass spectrometry (LC-MS) systems. The results demonstrated the significant advantages of this strategy over other strategies. First, the number of components detected with high peak intensity was successfully maximized by comprehensively optimizing the LC-QTOF/MS method, and 31 target components were characterized through matching analysis with our established personal Gelsemium database. Second, various data mining techniques, including database searching, diagnostic ion filtering and neutral loss filtering, were implemented to fully and systematically clarify the structure of various chemical components in G. elegans. A total of 147 components were characterized from G. elegans, and among them, 116 nontarget components were reported for the first time. A sensitive and reproducible LC-QqQ/MS method was successfully developed and validated for the simultaneous relative quantification of 41 components of G. elegans. This method was effective in identifying a variety of nontarget components and provided a technical reference for the characterization of other chemical components. The present integrated strategy would significantly contribute to chemical studies on herbal medicine, and its utility could be extended to other research fields, such as metabolomics, quality control and pharmacokinetics.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by National Key R&D Program of Intergovernmental Key Projects (Grant No: 2018YFE0101700) and National Natural Science Foundation of China (Grant No. 31972737).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.chmed.2020.06.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Cai H., Xu Q., Chang C., Song C., Yong X., Yi X.…Jiang H. The rapid discovery and identification of physalins in the calyx of Physalis alkekengi L. var. Franchetii (Mast.) Makino using ultra-high performance liquid chromatography–quadrupole time of flight tandem mass spectrometry together with a novel three-step. Journal of Chromatography A. 2014;1361:139–152. doi: 10.1016/j.chroma.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Chen J., Shi Z., Song Y., Guo X., Zhao M., Tu P., Jiang Y. Source attribution and structure classification-assisted strategy for comprehensively profiling Chinese herbal formula: Ganmaoling granule as a case. Journal of Chromatography A. 2016;1464:102–114. doi: 10.1016/j.chroma.2016.08.028. [DOI] [PubMed] [Google Scholar]
- Chingin K., Makarov A., Denisov E., Rebrov O., Zubarev R.A. Fragmentation of Positively-Charged Biological Ions Activated with a Beam of High-Energy Cations. Analytical Chemistry. 2014;86(1):372–379. doi: 10.1021/ac403193k. [DOI] [PubMed] [Google Scholar]
- Hao H., Cui N., Wang G., Xiang B., Liang Y., Xu X., Zhang H., Yang J., Zheng C., Wu L., Gong P., Wang W. Global Detection and Identification of Nontarget Components from Herbal Preparations by Liquid Chromatography Hybrid Ion Trap Time-of-Flight Mass Spectrometry and a Strategy. Analytical Chemistry. 2008;80(21):8187–8194. doi: 10.1021/ac801356s. [DOI] [PubMed] [Google Scholar]
- Hu Y., Chen M., Wang Z., Lan Y., Tang L., Liu M., Zhao J., Hu M., Zhang L., Ye L. Development of a validated UPLC-MS/MS method for determination of humantenmine in rat plasma and its application in pharmacokinetics and bioavailability studies. Biomedical Chromatography. 2017;31(12):e4017. doi: 10.1002/bmc.v31.1210.1002/bmc.4017. [DOI] [PubMed] [Google Scholar]
- Huo J.-H., Du X.-W., Sun G.-D., Dong W.-T., Wang W.-M. Identification and characterization of major constituents in Juglans mandshurica using ultra performance liquid chromatography coupled with time-of-flight mass spectrometry (UPLC-ESI-Q-TOF/MS) Chinese Journal of Natural Medicines. 2018;16(7):525–545. doi: 10.1016/S1875-5364(18)30089-X. [DOI] [PubMed] [Google Scholar]
- Jin G.-L., Su Y.-P., Liu M., Xu Y., Yang J., Liao K.-J., Yu C.-X. Medicinal plants of the genus Gelsemium (Gelsemiaceae, Gentianales)—A review of their phytochemistry, pharmacology, toxicology and traditional use. Journal of Ethnopharmacology. 2014;152(1):33–52. doi: 10.1016/j.jep.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Kobayashi H., Kogure N., Takayama H. New oxindole and indole alkaloids from Gelsemium rankinii. Tetrahedron. 2010;66(32):5987–5992. [Google Scholar]
- Lei X.-Q., Li G., Cheng L., Wang X.-L., Meng F.-Y. Identification of Ligustici Rhizoma et Radix and its adulterants based on their chemical constituents by UHPLC-Q/TOF-MS combined with data mining. Journal of Pharmaceutical and Biomedical Analysis. 2018;154:123–137. doi: 10.1016/j.jpba.2018.02.053. [DOI] [PubMed] [Google Scholar]
- Li W.L., Sun X.M., Song H., Ding J.X., Bai J., Chen Q. Hplc/q-tof-ms-based identification of absorbed constituents and their metabolites in rat serum and urine after oral administration of cistanche deserticola extract. Journal of Food Science. 2015;80(9):H2079–2087. doi: 10.1111/1750-3841.12975. [DOI] [PubMed] [Google Scholar]
- Li Z., Xiao S., Ai N.i., Luo K., Fan X., Cheng Y. Derivative multiple reaction monitoring and single herb calibration approach for multiple components quantification of traditional Chinese medicine analogous formulae. Journal of Chromatography A. 2015;1376:126–142. doi: 10.1016/j.chroma.2014.12.024. [DOI] [PubMed] [Google Scholar]
- Liang Y., Hao H., Kang A.n., Xie L., Xie T., Zheng X., Dai C., Wan L., Sheng L., Wang G. Qualitative and quantitative determination of complicated herbal components by liquid chromatography hybrid ion trap time-of-flight mass spectrometry and a relative exposure approach to herbal pharmacokinetics independent of standards. Journal of Chromatography A. 2010;1217(30):4971–4979. doi: 10.1016/j.chroma.2010.05.056. [DOI] [PubMed] [Google Scholar]
- Ling Q., Liu M., Wu M.-X., Xu Y., Yang J., Huang H.-H., Yu C.-X. Anti-allodynic and Neuroprotective Effects of Koumine, a Benth Alkaloid, in a Rat Model of Diabetic Neuropathy. Biological|Pharmaceutical Bulletin. 2014;37(5):858–864. doi: 10.1248/bpb.b13-00843. [DOI] [PubMed] [Google Scholar]
- Liu M., Huang H.-H., Yang J., Su Y.-P., Lin H.-W., Lin L.-Q., Liao W.-J., Yu C.-X. The active alkaloids of Gelsemium elegans Benth. are potent anxiolytics. Psychopharmacology (Berl) 2013;225(4):839–851. doi: 10.1007/s00213-012-2867-x. [DOI] [PubMed] [Google Scholar]
- Liu Y.-C., Lin L.i., Cheng P.i., Sun Z.-L., Wu Y., Liu Z.-Y. Fingerprint analysis of Gelsemium elegans by HPLC followed by the targeted identification of chemical constituents using HPLC coupled with quadrupole-time-of-flight mass spectrometry. Fitoterapia. 2017;121:94–105. doi: 10.1016/j.fitote.2017.07.002. [DOI] [PubMed] [Google Scholar]
- Liu Y.-C., Xiao S.a., Yang K., Ling L.i., Sun Z.-L., Liu Z.-Y. Comprehensive identification and structural characterization of target components from Gelsemium elegans by high-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry based on accurate mass databases combined with MS/M: Structural characterization of Gelsemium elegans components by LC-QqTOF MS. J. Mass Spectrom. 2017;52(6):378–396. doi: 10.1002/jms.3937. [DOI] [PubMed] [Google Scholar]
- Ma X.-D., Fan Y.-X., Jin C.-C., Wang F., Xin G.-Z., Li P., Li H.-J. Specific targeted quantification combined with non-targeted metabolite profiling for quality evaluation of Gastrodia elata tubers from different geographical origins and cultivars. Journal of Chromatography A. 2016;1450:53–63. doi: 10.1016/j.chroma.2016.04.077. [DOI] [PubMed] [Google Scholar]
- Meyer L., Boujedaini N., Patte-Mensah C., Mensah-Nyagan A.G. Pharmacological effect of gelsemine on anxiety-like behavior in rat. Behavioural Brain Research. 2013;253:90–94. doi: 10.1016/j.bbr.2013.07.010. [DOI] [PubMed] [Google Scholar]
- Mustafa A.M., Eldahmy S.I., Caprioli G., Bramucci M., Quassinti L., Lupidi G., Beghelli D., Vittori S., Maggi F. Chemical composition and biological activities of the essential oil from Pulicaria undulata (L.) C. A. Mey. Growing wild in Egypt. Natural Product Research. 2018:1–5. doi: 10.1080/14786419.2018.1534107. [DOI] [PubMed] [Google Scholar]
- Ornduff R. The systemetics and breeding system of Gelsemium (Loganiaceae) Journal of the Arnold Arboretum. 1970;51(1):1–17. [Google Scholar]
- Qiao X., Li R.u., Song W., Miao W.-J., Liu J., Chen H.-B., Guo D.-a., Ye M. A targeted strategy to analyze untargeted mass spectral data: Rapid chemical profiling of Scutellaria baicalensis using ultra-high performance liquid chromatography coupled with hybrid quadrupole orbitrap mass spectrometry and key ion filtering. Journal of Chromatography A. 2016;1441:83–95. doi: 10.1016/j.chroma.2016.02.079. [DOI] [PubMed] [Google Scholar]
- Samanipour S., Reid M.J., Bæk K., Thomas K.V. Combining a Deconvolution and a Universal Library Search Algorithm for the Nontarget Analysis of Data-Independent Acquisition Mode Liquid Chromatography−High-Resolution Mass Spectrometry Results. Environmental Science and Technology. 2018;52(8):4694–4701. doi: 10.1021/acs.est.8b00259. [DOI] [PubMed] [Google Scholar]
- Shi X.J., Yang W.Z., Qiu S., Yao C.L., Shen Y., Pan H.Q.…Guo D.A. An in-source multiple collision-neutral loss filtering based nontargeted metabolomics approach for the comprehensive analysis of malonyl-ginsenosides from Panax ginseng, p.Quinquefolius, and p.Notoginseng. Analytica Chimica Acta. 2017;952:59–70. doi: 10.1016/j.aca.2016.11.032. [DOI] [PubMed] [Google Scholar]
- Shi W., Zhang C., Zhao D., Wang L., Li P., Li H. Discovery of Hepatotoxic Equivalent Combinatorial Markers from Dioscorea bulbifera tuber by Fingerprint-Toxicity Relationship Modeling. Scientific Reports. 2018;8(1) doi: 10.1038/s41598-017-18929-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Zheng M., Duan S., Zeng Y., Zhang Z., Cui Q., Zhang J., Hong T., Bai J., Du S. Chemical profiling and screening of the marker components in the fruit of Cassia fistula by hplc and uhplc/ltq-orbitrap ms(n) with chemometrics. Molecules. 2018;23(7):1501–1507. doi: 10.3390/molecules23071501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Shi S., Wang S. Can highly cited herbs in ancient Traditional Chinese medicine formulas and modern publications predict therapeutic targets for diabetes mellitus? Journal of Ethnopharmacology. 2018;213:101–110. doi: 10.1016/j.jep.2017.10.032. [DOI] [PubMed] [Google Scholar]
- Wang L., Wen Y., Meng F. Simultaneous determination of gelsemine and koumine in rat plasma by UPLC-MS/MS and application to pharmacokinetic study after oral administration of Gelsemium elegans Benth extract. Biomedical Chromatography. 2018;32(6):e4201. doi: 10.1002/bmc.v32.610.1002/bmc.4201. [DOI] [PubMed] [Google Scholar]
- Xiao S.a., Huang Y.-J., Sun Z.-L., Liu Z.-Y. Structural elucidation of koumine metabolites by accurate mass measurements using high-performance liquid chromatography/quadrupole-time-of-flight mass spectrometry: Structural elucidation of koumine metabolites by HPLC/QqTOF-MS. Rapid Communications in Mass Spectrometry. 2017;31(3):309–314. doi: 10.1002/rcm.7797. [DOI] [PubMed] [Google Scholar]
- Xu Y., Qiu H.Q., Liu H., Liu M., Huang Z.Y., Yang J., Yu C.X. Effects of koumine, an alkaloid of gelsemium elegans benth., on inflammatory and neuropathic pain models and possible mechanism with allopregnanolone. Pharmacology Biochemistry and Behavior. 2012;101(3):504–514. doi: 10.1016/j.pbb.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Xu Y.-K., Liao S.-G., Na Z., Hu H.-B., Li Y., Luo H.-R. Gelsemium alkaloids, immunosuppressive agents from Gelsemium elegans. Fitoterapia. 2012;83(6):1120–1124. doi: 10.1016/j.fitote.2012.04.023. [DOI] [PubMed] [Google Scholar]
- Yamada Y., Kitajima M., Kogure N., Wongseripipatana S., Takayama H. Seven new monoterpenoid indole alkaloids from Gelsemium elegans. Chemistry - An Asian Journal. 2011;6(1):166–173. doi: 10.1002/asia.201000538. [DOI] [PubMed] [Google Scholar]
- Yan Y., Chai C.-Z., Wang D.-W., Yue X.-Y., Zhu D.-N., Yu B.-Y. HPLC-DAD-Q-TOF-MS/MS analysis and HPLC quantitation of chemical constituents in traditional Chinese medicinal formula Ge-Gen Decoction. Journal of Pharmaceutical and Biomedical Analysis. 2013;80:192–202. doi: 10.1016/j.jpba.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Yang K., Huang Y.-J., Xiao S.a., Liu Y.-C., Sun Z.-L., Liu Y.-S., Liu Z.-Y. Identification of gelsemine metabolites in rat liver S9 by high-performance liquid chromatography/quadrupole-time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry. 2018;32(1):19–22. doi: 10.1002/rcm.8012. [DOI] [PubMed] [Google Scholar]
- Yang K., Long X.-M., Liu Y.-C., Chen F.-H., Liu X.-F., Sun Z.-L., Liu Z.-Y. Development and in-house validation of a sensitive LC–MS/MS method for simultaneous quantification of gelsemine, koumine and humantenmine in porcine plasma. Journal of Chromatography B. 2018;1076:54–60. doi: 10.1016/j.jchromb.2018.01.019. [DOI] [PubMed] [Google Scholar]
- Yang Y., Wang H.-J., Yang J., Brantner A.H., Lower-Nedza A.D., Si N., Song J.-F., Bai B., Zhao H.-Y., Bian B.-L. Chemical profiling and quantification of Chinese medicinal formula Huang-Lian-Jie-Du decoction, a systematic quality control strategy using ultra high performance liquid chromatography combined with hybrid quadrupole-orbitrap and triple quadrupole mass spectrometers. Journal of Chromatography A. 2013;1321:88–99. doi: 10.1016/j.chroma.2013.10.072. [DOI] [PubMed] [Google Scholar]
- Zhang X., Chen Y.i., Gao B.o., Luo D., Wen Y., Ma X. Apoptotic Effect of Koumine on Human Breast Cancer Cells and the Mechanism Involved. Cell Biochemistry and Biophysics. 2015;72(2):411–416. doi: 10.1007/s12013-014-0479-2. [DOI] [PubMed] [Google Scholar]
- Zhang J.Y., Wang Z.J., Li Y., Liu Y., Cai W., Li C.…Qiao Y.J. A strategy for comprehensive identification of sequential constituents using ultra-high-performance liquid chromatography coupled with linear ion trap-orbitrap mass spectrometer, application study on chlorogenic acids in flos Lonicerae japonica. Talanta. 2016;147(62):16–27. doi: 10.1016/j.talanta.2015.09.039. [DOI] [PubMed] [Google Scholar]
- Zhang S., Hu S., Yang X., Shen J., Zheng X., Huang K., Xiang Z. Development of a liquid chromatography with mass spectrometry method for the determination of gelsemine in rat plasma and tissue: Application to a pharmacokinetic and tissue distribution study: Liquid Chromatography. Journal of Separation Science. 2015;38(6):936–942. doi: 10.1002/jssc.201401168. [DOI] [PubMed] [Google Scholar]
- Zhang W., Zhang S.-Y., Wang G.-Y., Li N.-P., Chen M.-F., Gu J.-H., Ye W.-C. Five new koumine-type alkaloids from the roots of Gelsemium elegans. Fitoterapia. 2017;118:112–117. doi: 10.1016/j.fitote.2017.03.004. [DOI] [PubMed] [Google Scholar]
- Zhang Y.u., Cheng Y., Liu Z., Ding L., Qiu T., Chai L., Chen X. Systematic screening and characterization of multiple constituents in Guizhi Fuling capsule and metabolic profiling of bioactive components in rats using ultra-high-performance liquid chromatography/quadrupole-time-of-flight mass spectrometry. Journal of Chromatography B. 2017;1061-1062:474–486. doi: 10.1016/j.jchromb.2017.07.021. [DOI] [PubMed] [Google Scholar]
- Zuo M.-T., Liu S.-S., Lin L., Wang Z.-Y., Bai X., Sun Z.-L., Liu Z.-Y. Characterization of N -methylcanadine and N -methylstylopine metabolites in rat liver S9 by high-performance liquid chromatography/quadrupole time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry. 2018;32(23):2047–2054. doi: 10.1002/rcm.8286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.