Abstract
Objective
In order to obtain new glycosyltransferases with highly efficient catalysis, the glycosyltransferases from Carthamus tinctorius which contains diverse types of glycosides were mined. Methods: A new glycosyltransferase gene (UGT88B2) with full length was obtained by PCR and further transformed into Escherichia coli for heterologous expression. The catalytic activity of recombinant UGT88B2 was determined by HPLC-MSn. The structures of representative catalytic products were elucidated by MS and NMR.
Results
UGT88B2 exhibited catalytic promiscuity and various patterns in glycosylation of flavonoids with high efficiency.
Conclusion
A new glycosyltransferase named UGT88B2 was successfully mined and can be employed as enzymatic tools in glycosylation of flavonoids.
Keywords: Carthamus tinctorius L., enzyme promiscuity, glycosyltransferase, O-glycosides
1. Introduction
Safflower (Carthamus tinctorius L.) is widely cultivated in China as traditional herbs (Hu et al., 2013). Its dried pedals have been used in traditional Chinese medicine (TCM) clinically for the prevention and treatment of cardiovascular and cerebrovascular diseases for a long time (Guo et al., 2017). Many studies showed that the main active ingredients in safflower flowers were structurally diverse flavonoids and their glycosides (Hu et al., 2013), indicating the existence of various glycosyltransferases (GTs). GTs are recognized as powerful synthetic tools for bioactive glycosylated products, which can alleviate many disadvantages by chemical glycosylation such as poor regio- and stereoselectivities, tedious protection and deprotection of functional groups (Xie et al., 2017). In our previous works, we have obtained several GTs with good catalytic properties from C. tinctorius (Xie et al., 2014, Xie et al., 2017, Zhang et al., 2019). In the course of discovering GTs with novel catalytic properties from this plant, a new GT named UGT88B2 was obtained and employed as enzymatic tools in glycosylation of selected flavonoids.
2. Materials and methods
2.1. Chemicals and reagents
Chemical reagents were purchased from Sigma Aldrich (St. Louis, MO, USA), J & K Scientific Ltd. (Beijing, China), InnoChem Science & Technology Co., Ltd. (Beijing, China) and BioBioPha (Kunming, China). KOD-Plus DNA polymerase was purchased from Toyobo Biotech Co., Ltd. (Shanghai, China). Primer synthesizing and DNA sequencing were performed by Sangon Biotech Co., Ltd. (Shanghai, China). Restriction enzymes and DNA ligase were purchased from Takara Biotech Co., Ltd. (Dalian, China).
2.2. Analytical procedures
Enzymatic products were detected on an Agilent 1260 series HPLC system (Agilent Technologies, Germany) coupled with an LCQ Fleet ion trap mass spectrometer (Thermo Electron Corp., USA) equipped with an electrospray ionization (ESI) source. Compounds were characterized by 1H NMR at 400 MHz and 13C NMR at 100 MHz on Mercury-400 spectrometers. Chemical shifts (δ) were referenced to internal solvent resonances and given in parts per million (ppm). Coupling constants (J) were given in hertz (Hz).
2.3. Molecular cloning of UGT88B2
The total RNA of C. tinctorius fresh pedals was prepared by using E.Z.N.A.TM Plant RNA Kit (Omega Bio-Tek, USA) and reverse-transcribed (RT) to cDNA with RACE cDNA Amplification Kit (Clontech, USA). The RT product was subjected to rapid amplification of cDNA ends (RACE) according to the manufacturer’s protocol. The full-length UGT88B2 was amplified by PCR and the PCR forward primer (FP) and reverse primer (RP) were 5'-ATGGAATCAGCCACGGTGGTTATGTATCCTTCGC-3' and 5'-TCATTTCAAAATTTCATTTTCCGATCCCTTGCGCCC-3', respectively.
2.4. Expression and purification of UGT88B2
The coding region of UGT88B2 was amplified with the primer pairs UGT88B2-F-BamH I and UGT88B2-R-EcoR I harboring BamH I and EcoR I restriction sites, respectively. The PCR product was purified, digested with the corresponding enzymes and cloned into pET-28a vector. After the verification of the sequences, the recombinant plasmid was transformed into Transetta (DE3) Escherichia coli (TransGen Biotech, China) for heterologous expression. Then the recombinant E. coli strains were cultivated in 1 L shake flasks at 37 °C and 200 rpm using 400 mL LB-media containing 50 µg/mL kanamycin and 34 µg/mL chloromycetin until OD600 reached 0.6. The recombinant N-terminal His6-UGT88B2 expression was achieved by induction with 0.1 mmol/L isopropyl β-D-thiogalactoside (IPTG) for 18 h at 17 °C. The cells were harvested by centrifugation at 10 000 g for 5 min at 4 °C and resuspended in 20 mL binding buffer (20 mmol/L phosphate buffer, 0.5 mol/L NaCl, 20 mmol/L imidazole, pH 7.4) containing 1 mmol/L phenylmethanesulfonyl fluoride (PMSF). Cells were disrupted by sonication in ice bath and the cell debris was removed by centrifugation at 10 000 g and 4 °C for 60 min. The soluble fraction was passed through a 0.45 µm syringe filter unit and the cleared supernatant was immediately applied to 1 mL of Ni-NTA resin (GE, USA) loaded in a column pre-equilibrated with binding buffer. The resin column was subsequently eluted with 10 mL binding buffer. Elution was carried out with 5 mL different elution buffer (20 mmol/L phosphate buffer, 0.5 mol/L NaCl, 50−500 mmol/L imidazole, pH 7.4). The protein purification was performed under the flow rate of 1 mL/min and the temperature of 4 °C. The protein was concentrated and buffer exchanged to desalting buffer (50 mmol/L Tris-HCl buffer, 50 mmol/L NaCl, 1 mmol/L DTT, 5% glycerol, pH 7.4) using centrifugal concentrator with Amicon Ultra-30K (Millipore). Protein purity was confirmed by SDS-PAGE to be >90% and protein concentration for all studies was determined by the Protein Quantitative Kit (Bradford) (TransGen Biotech, China).
2.5. HPLC-MS based glycosyltransferase activity assay
The reaction mixture containing 0.8 mmol/L uridine diphosphate glucose (UDP-glc), 0.4 mmol/L aglycon and 216 µg purified UGT88B2 in a final volume of 100 µL were incubated at 30 °C for 12 h. The reactions were terminated by the addition of 200 µL ice-cold methanol. Subsequently, samples were centrifuged at 15 000 g for 30 min and analyzed by HPLC-MS. The supernatants were performed on Polar RP-18 column (250 mm × 4.6 mm I.D., 5 µm, Shiseido Co., Ltd., Japan) at a flow rate of 1 mL/min at 30 °C. The HPLC conditions were as follows: 15%−100% methanol linear gradient (30 min) in 0.1% formic acid aqueous solution for aglycons 2−6; or 15%−35% acetonitrile linear gradient (30 min) in 0.1% formic acid aqueous solution, then acetonitrile reached to 100% in 1 min, followed by 100% B for 5 min for aglycon 1. For quantification, three parallel assays were routinely carried out.
2.6. Effects of pH, temperature, and divalent metal ions
To assay the optimal reaction temperature, the reactions were incubated at different temperatures (20−60 °C) for 30 min. To investigate the optimal pH, the enzymatic reaction was performed in various reaction buffers with pH values in the range of 5.0−6.0 (citric acid-sodium citrate buffer), 6.0−8.0 (Na2HPO4-NaH2PO4 buffer), and 8.0−10.0 (Tris-HCl buffer) at 40 °C for 10 min. To test the necessity of divalent metal ions for UGT88B2 activity, CaCl2, FeCl2, ZnCl2, MgCl2, CuCl2, MnCl2, BaCl2, CoCl2, NiSO4, and EDTA were used individually in the final concentration of 5 mmol/L. All determinations were performed with UDP-glc (0.8 mmol/L) as a donor and kaempferol as an acceptor (0.4 mmol/L) at 40 °C for 10 min. Three parallel assays were routinely conducted. Aliquots were quenched by adding ice-cold methanol and centrifuged at 15 000 g for 30 min. Supernatants were analyzed by HPLC-MSn as described above.
2.7. Determination of kinetic parameters
For kinetic studies of UGT88B2, a typical assay contained 50 mmol/L Tris-HCl (pH 8.0), saturating UDP-glc (0.8 mmol/L), 1 µg purified UGT88B2 and varying concentration of kaempferol (0.02−0.4 mmol/L) at 40 °C in a total volume of 100 μL for reaction in 10 min. Aliquots were quenched by adding ice-cold methanol and centrifuged at 15 000 g for 30 min. Supernatants were analyzed by HPLC-MSn as described above. All experiments were performed in triplicate.
2.8. Effects of protein amount on kaempferol glycosylation pattern
The reaction mixture contained 0.4 mmol/L UDP-glc, 0.2 mmol/L kaempferol, 50 mmol/L Tris-HCl (pH 8.0) and different amount (2.4, 4.8, 12, 24, 48, 96, 192, and 232 µg, respectively) of purified UGT88B2 (1 µg/µL) in a final volume of 100 µL were incubated at 40 °C for 12 h. The reactions were terminated by the addition of 200 µL ice cold methanol. Subsequently, samples were centrifuged at 15 000 g for 30 min and analyzed by HPLC. The HPLC conditions were the same as above for aglycon 1.
2.9. Products identification by MS and NMR
Reactions were up-scaled and carried out with following alterations: 28.6 mg of kaempferol (1) and UDP-glc (120 mg) were added in 21 mL UGT88B2 (9 mg/mL protein) and 95 mL 50 mmol/L Tris-HCl (pH 7.4), incubated at 30 °C for 12 h, followed by extracting with ethyl acetate (300 mL × 5). The organic layer was evaporated under reduced pressure to afford fraction 1. The water layer was centrifuged at 10 000 g for 30 min, and the resulting supernatant was subjected to an Amberlite XAD-16 macroporous resin column (50 mL, Rohm & Haas Corp., USA) eluting with water, 50% (volume percentage) ethanol, and 100% (volume percentage) ethanol. The fraction of 50% (volume percentage) ethanol containing products tested by HPLC-MSn was concentrated to give fraction 2. Fraction 1 was further purified by reversed-phase semi-preparative HPLC (solvent A, 0.1% formic acid aqueous solution; solvent B, acetonitrile; flow rate, 3 mL/min; gradient, 22%−25% B in 20 min followed by 100% B for 5 min) to afford 1a (3.0 mg), 1b (9.4 mg), 1c (1.0 mg). Fraction 2 was further purified by reversed-phase semipreparative HPLC (solvent A, pure water; solvent B, methanol; flow rate, 3 mL/min; gradient, 15%−70% B in 20 min) to afford 1d (1.2 mg), 1e (0.5 mg), 1f (3.6 mg). The purified products were analyzed by 1H-NMR and 13C-NMR (in DMSO-d6).
3. Results and discussion
3.1. UGT88B2 expression and determination of enzymatic properties
The full-length of UGT88B2 (1431bp, GenBank accession number MH631206) was amplified by RT-PCR using the total RNA from C. tinctorius fresh pedals as template and heterologously expressed in E. coli. The recombinant His6-UGT88B2 was purified using His6-tagged affinity chromatography and analyzed by SDS-PAGE (Fig. 1).
Fig. 1.
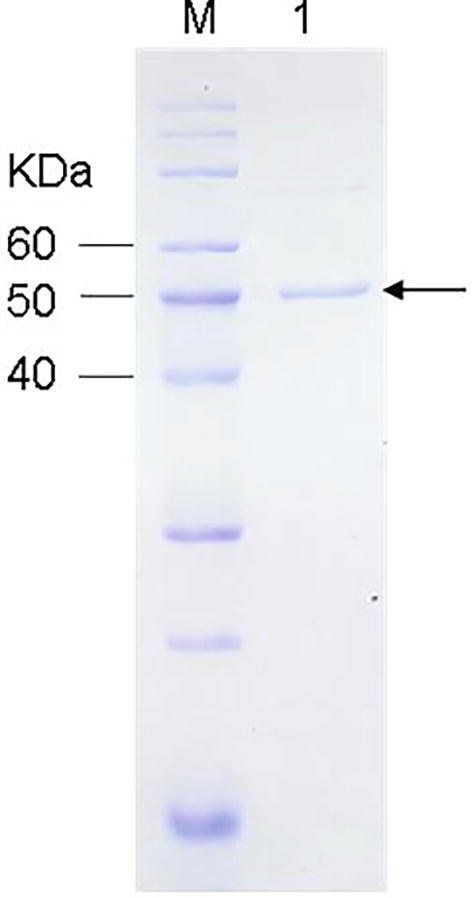
SDS-PAGE analysis of purified recombinant UGT88B2. Lane M: Protein marker; Lane 1: His-tagged UGT88B2 (indicated with an arrow; predicted M. W., 51.5 kDa).
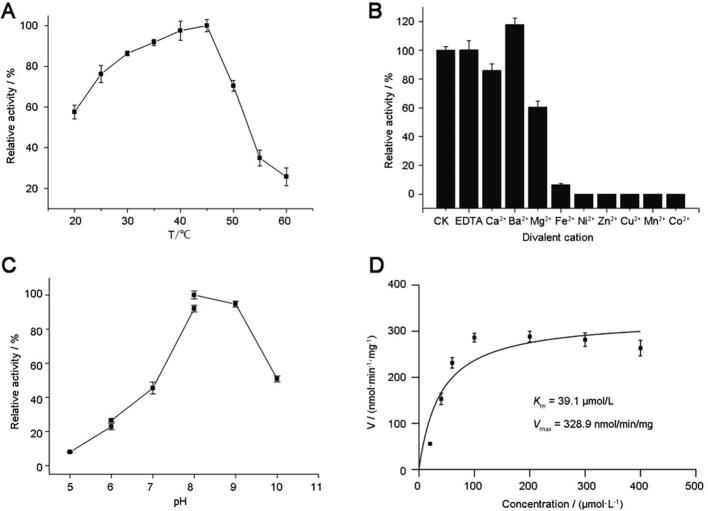
To determine the optimal reaction condition for UGT88B2, the biochemical characteristics including temperatures, pH and divalent metal ions were investigated by using kaempferol as receptor substrate. As shown in Fig. 2A, the catalytic activity of UGT88B2 increased with an increasing temperature in the 15−45 °C interval but decreased with increasing temperature in the 45−65 °C interval, and the optimum temperature was 45 °C. Analysis of the enzyme activity over the pH range of 5.0−10.0 revealed that the maximum activity was at pH 8.0 (Fig. 2B). The assay of divalent cations showed that UGT88B2 was metal ion-independent (Fig. 2C), while Ba2+ could enhance the catalytic activity. The kinetic parameter of the enzyme in the catalytic activity was determined under the optimum temperature and pH condition. The Km value of UGT88B2 toward kaempferol was determined to be 39.1 μmol/L. The Vmax value of UGT88B2 toward kaempferol was 328.9 nmol/min/mg (Fig. 2D).
Fig. 2.
Effects of temperature (A), pH (B), and divalent metal ions (C) on activity of UGT88B2. Nonlinear regression (D) of Michaelis-Menten equation for UGT88B2. Kaempferol (1) was used as the acceptor, UDP-glc was used as the sugar donor.
3.2. Probing glycosylation pattern of UGT88B2 to kaempferol
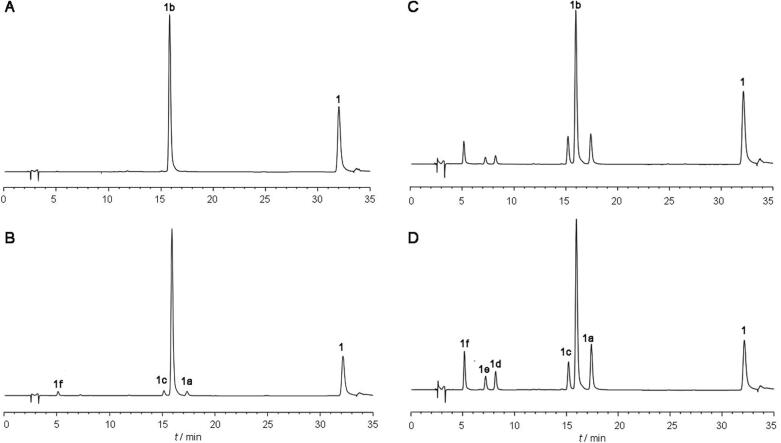
Interestingly, we observed that the amount of UGT88B2 influenced the glycosylation pattern of kaempferol (Table 1). When small amount of enzyme (for example 4.8 µg) was used for reaction, only one mono-O-glycosylated product, kaempferol 7-O-β-D-glucoside (1b) was detected at high conversion (Fig. 3A). Two other mono-O-glucosides (1a, 1c) and one di-O-glucoside (1f) were monitored when the amount of enzyme was increased to 12 µg (Table 1). However, two additional di-O-glucosides (1d, 1e) were also observed when enzyme 192 µg used (Table 1). Besides, the conversion of di-O-glucosides increased as the amount of enzyme was increased (Fig. 3B−D).
Table 1.
Conversions of kaempferol glucosides catalyzed by different amount of UGT88B2
| Enzyme amount /µg | 1a /% | 1b /% | 1c /% | 1d /% | 1e /% | 1f /% |
|---|---|---|---|---|---|---|
| 2.4 | 0 | 60.96 | 0 | 0 | 0 | 0 |
| 4.8 | 0 | 78.43 | 0 | 0 | 0 | 0 |
| 12 | 0.52 | 77.16 | 0.46 | 0 | 0 | 0.40 |
| 24 | 0.98 | 72.29 | 0.97 | 0 | 0 | 0.79 |
| 48 | 1.67 | 68.51 | 1.76 | 0 | 0 | 1.33 |
| 96 | 2.91 | 45.12 | 3.07 | 0 | 0 | 1.40 |
| 192 | 8.99 | 42.29 | 6.95 | 2.14 | 1.68 | 5.12 |
| 232 | 12.68 | 43.83 | 6.47 | 4.22 | 3.06 | 7.81 |
Fig. 3.
HPLC of reaction of different amount of UGT88B2 with kaempferol (1) and UDP-glc. (A−D). Amount of UGT88B2 was 4.8, 48, 192, and 232 µg, respectively.
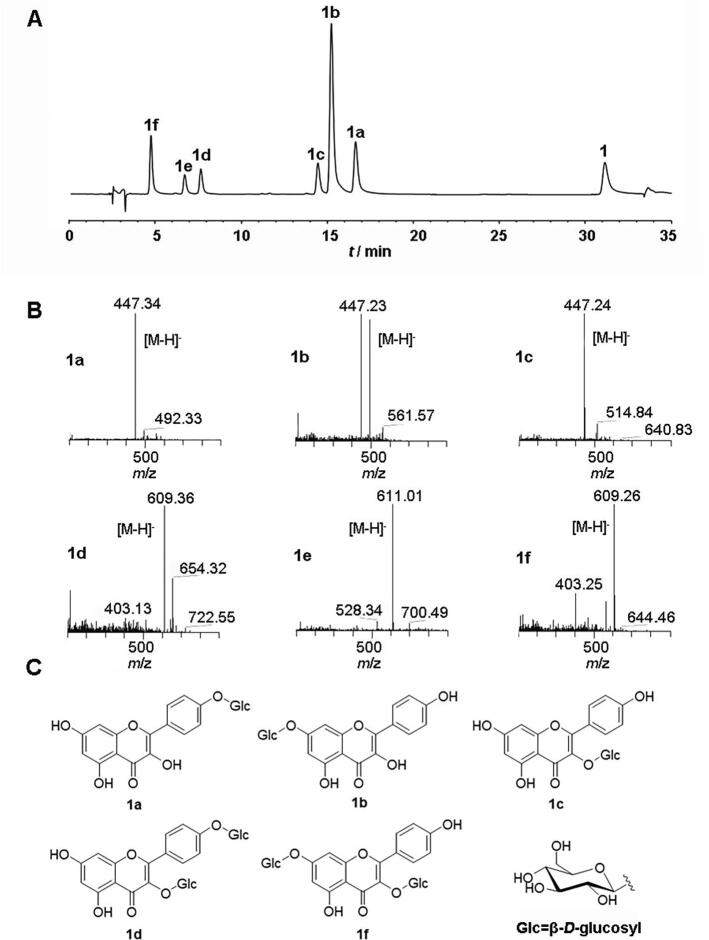
As an important aglycon from C. tinctorius, kaempferol was glycosylated by UGT88B2 to produce six products, among which, products (1a, 1b, 1c) were three mono-O-glucosides and products (1d, 1e, 1f) were three di-O-glucosides (Fig. 4). It is interesting that there is only one kaempferol mono-O-glucoside (kaempferol 3-O-β-D-glucoside) found in C. tinctorius (Hu et al., 2013), while di-O-kaempferol glucosides have not been reported in C. tinctorius.
Fig. 4.
HPLC-DAD/ESI-MS analysis of UGT88B2 enzymatic products using kaempferol (1) as an aglycon acceptor. (A) HPLC chromatogram of 1 and enzyme products 1a−1f; (B) MS spectra of 1a−1f in negative or positive mode successively; (C) Structures of enzymatic products of 1.
In order to elucidate the structures of kaempferol glucosides, products were prepared by up-scaled reaction in vitro and structurally determined. Five kaempferol glucosides, kaempferol 4’-O-β-D-glucoside (1a) (Scheer & Wichtl, 1987), kaempferol 7-O-β-D-glucoside (1b) (Matlawska & Sikorska, 2004), kaempferol 3-O-β-D-glucoside (1c) (Kazuma et al., 2000), kaempferol 3, 4’-di-O-β-D-glucoside (1d) (Guo et al., 2009), kaempferol 3, 7-di-O-β-D-glucoside (1f) (Liang et al., 2015) were elucidated on the basis of their spectroscopic data and comparison with those reported in the literatures. 1e was a di-O-glucoside by ESI-MS at m/z 611.01 [M + H]+, whose structure was not characterized due to the limited amount.
Kaempferol 4’-O-β-D-glucoside (1a): yellow amorphous power. ESI-MS (m/z): 447.34 [M−H]−. 1H-NMR (400 MHz, DMSO-d6)δ: 6.17 (1H, d, J = 2.0 Hz, H-6), 6.44 (1H, d, J = 2.0 Hz, H-8), 8.16 (2H, d, J = 9.0 Hz, H-2’/H-6’), 7.18 (2H, d, J = 9.0 Hz, H-3’/H-5’); 4’-O-Glc, 4.98 (1H, d, J = 7.2 Hz, H-1’’), 3.16−3.71 (6H, m, H-2’’−H-6’’). 13C-NMR (100 MHz, DMSO-d6) δ: 145.7 (C-2), 136.7 (C-3), 176.4 (C-4), 160.7 (C-5), 98.5 (C-6), 164.8 (C-7), 93.6 (C-8), 156.3 (C-9), 102.8 (C-10), 124.7 (C-1’), 129.0 (C-2’/C-6’), 116.1 (C-3’/C-5’), 158.3 (C-4’); 4’-O-Glc, 100.0 (C-1’’), 73.2 (C-2’’), 76.7 (C-3’’), 69.7 (C-4’’), 77.2 (C-5’’), 60.6 (C-6’’) (Scheer & Wichtl, 1987).
Kaempferol 7-O-β-D-glucoside (1b): yellow amorphous power. ESI-MS (m/z): 447.23 [M−H]−. 1H-NMR (400 MHz, DMSO-d6) δ: 6.42 (1H, d, J = 2.0 Hz, H-6), 6.80 (1H, d, J = 2.0 Hz, H-8), 8.08 (2H, d, J = 8.8 Hz, H-2’/H-6’), 6.94 (2H, d, J = 8.8 Hz, H-3’/H-5’); 7-O-Glc, 5.07 (1H, d, J = 7.2 Hz, H-1’’), 3.17−3.71 (6H, m, H-2’’−H-6’’). 13C-NMR (100 MHz, DMSO-d6) δ: 147.5 (C-2), 136.2 (C-3), 176.2 (C-4), 160.4 (C-5), 98.8 (C-6), 162.7 (C-7), 94.4 (C-8), 155.8 (C-9), 104.7 (C-10), 121.6 (C-1’), 129.7 (C-2’/C-6’), 115.5 (C-3’/C-5’), 159.4 (C-4’); 7-O-Glc, 100.0 (C-1’’), 73.1 (C-2’’), 76.5 (C-3’’), 69.6 (C-4’’), 77.2 (C-5’’), 60.6 (C-6’’) (Matlawska & Sikorska, 2004).
Kaempferol 3-O-β-D-glucoside (1c): yellow amorphous power. ESI-MS (m/z): 447.24 [M−H]−. 1H-NMR (400 MHz, DMSO-d6) δ: 6.11 (1H, brs, H-6), 6.44 (1H, brs, H-8), 8.02 (2H, d, J = 8.8 Hz, H-2’/H-6’), 6.87 (2H, d, J = 8.8 Hz, H-3’/H-5’); 3-O-Glc, 5.42 (1H, d, J = 7.2 Hz, H-1’’), 3.07−3.57 (6H, m, H-2’’−H-6’’). 13C-NMR (100 MHz, DMSO-d6) δ: 156.6 (C-2), 133.0 (C-3), 177.5 (C-4), 161.2 (C-5), 99.0 (C-6), 164.0 (C-7), 93.6 (C-8), 156.6 (C-9), 104.1 (C-10), 120.9 (C-1’), 130.7 (C-2’/C-6’), 115.1 (C-3’/C-5’), 160.0 (C-4’); 3-O-Glc, 101.1 (C-1’’), 74.2 (C-2’’), 76.5 (C-3’’), 69.9 (C-4’’), 77.5 (C-5’’), 60.8 (C-6’’) (Kazuma et al., 2000).
Kaempferol 3, 4’-di-O-β-D-glucoside (1d): yellow amorphous power. ESI-MS (m/z): 609.36 [M−H]−. 1H-NMR (400 MHz, DMSO-d6) δ: 6.04 (1H, d, J = 2.4 Hz, H-6), 6.26 (1H, d, J = 2.4 Hz, H-8), 8.09 (2H, d, J = 8.8 Hz, H-2’/H-6’), 7.14 (2H, d, J = 8.8 Hz, H-3’/H-5’); 3-O-Glc, 5.43 (1H, d, J = 7.6 Hz, H-1’’), 3.07−3.57 (6H, m, H-2’’−H-6’’); 4’-O-Glc, 5.01 (1H, d, J = 7.2 Hz, H-1’’’), 3.16−3.71 (6H, m, H-2’’’−H-6’’’). 13C-NMR (100 MHz, DMSO-d6) δ: 156.5 (C-2), 133.5 (C-3), 177.7 (C-4), 161.1 (C-5), 98.6 (C-6), 164.2 (C-7), 93.5 (C-8), 156.6 (C-9), 104.1 (C-10), 123.9 (C-1’), 130.5 (C-2’/C-6’), 115.8 (C-3’/C-5’), 159.0 (C-4’); 3-O-Glc, 101.4 (C-1’’), 74.2 (C-2’’), 76.5 (C-3’’), 69.9 (C-4’’), 77.6 (C-5’’), 60.9 (C-6’’) ; 4’-O-Glc, 100.8 (C-1’’’), 73.2 (C-2’’’), 76.4 (C-3’’’), 69.6 (C-4’’’), 77.2 (C-5’’’), 60.6 (C-6’’’) (Guo et al., 2009).
Kaempferol 3, 7-di-O-β-D-glucoside (1f): yellow amorphous power. ESI-MS (m/z): 609.26 [M−H]−. 1H-NMR (400 MHz, DMSO-d6) δ: 6.44 (1H, brs, H-6), 6.79 (1H, brs, H-8), 8.05 (2H, d, J = 8.8 Hz, H-2’/H-6’), 6.90 (2H, d, J = 8.8 Hz, H-3’/H-5’); 3-O-Glc, 5.48 (1H, d, J = 7.2 Hz, H-1’’), 3.07-3.55 (6H, m, H-2’’−H-6’’); 7-O-Glc, 5.07 (1H, d, J = 6.8 Hz, H-1’’’), 3.07−3.70 (6H, m, H-2’’’−H-6’’’). 13C-NMR (100 MHz, DMSO-d6) δ: 156.0 (C-2), 133.5 (C-3), 177.6 (C-4), 160.9 (C-5), 99.4 (C-6), 162.7 (C-7), 94.5 (C-8), 156.8 (C-9), 105.7 (C-10), 120.7 (C-1’), 130.9 (C-2’/C-6’), 115.2 (C-3’/C-5’), 160.3 (C-4’); 3-O-Glc, 100.7 (C-1’’), 74.2 (C-2’’), 76.5 (C-3’’), 69.9 (C-4’’), 77.5 (C-5’’), 60.8 (C-6’’); 7-O-Glc, 99.7 (C-1’’’), 73.1 (C-2’’’), 76.5 (C-3’’’), 69.6 (C-4’’’), 77.2 (C-5’’’), 60.6 (C-6’’’) (Liang et al., 2015).
3.3. Exploring glycosylation capability of UGT88B2
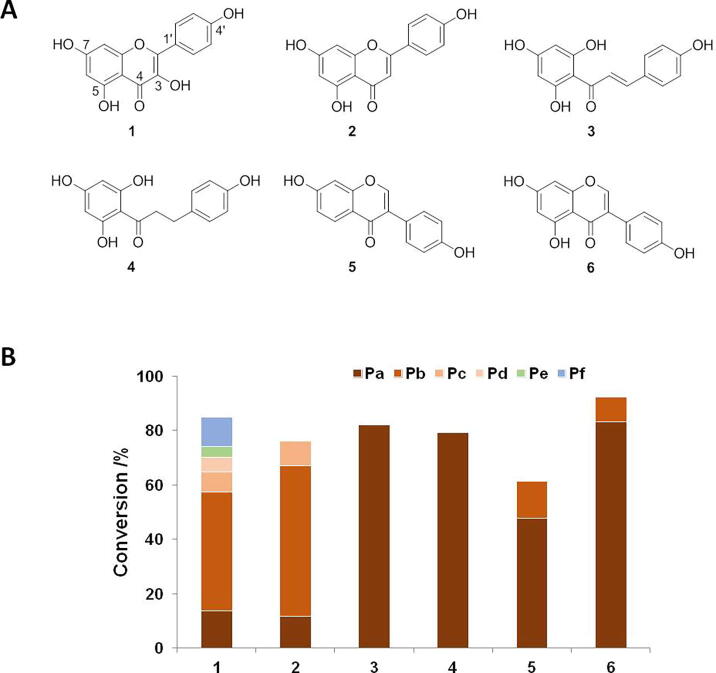
To investigate the catalytic activity of UGT88B2, flavonoids with several skeletons including flavonol (kaempferol, 1), flavone (apigenin, 2), chalcone (naringenin chalcone, 3), dihydrochalcone (phloretin, 4), and isoflavone (daidzein, 5; genistein, 6) were employed for the enzymatic assays (Fig. 5A). Surprisingly, UGT88B2 was able to recognize all selected flavonoids with high conversion rates from the first analysis with HPLC-MS (Fig. 5B). UGT88B2 not only recognized flavonoid aglycons (1−3) isolated from C. tinctorius, also accepted flavonoid aglycons (4−6) undiscovered in C. tinctorius.
Fig. 5.
Exploration of catalytic capabilities of UGT88B2. (A) Structures of aglycons; (B) Conversions of glycosylated products catalyzed by UGT88B2. Pa−Pf: means products a−f of substrates listed in part A.
4. Conclusion
In summary, we have mined a promiscuous flavonoid O-GT, UGT88B2, from plant C. tinctorius, which can catalyze various flavonoids with different structures. Especially, this enzyme exhibits multiple glycosylation patterns including mono-, di-O-glycosylation at different positions when kaempferol is used as receptor substrate. UGT88B2 with catalytic promiscuity and various glycosylation patterns might be exploited as a powerful biocatalyst for the enzymatic synthesis of various bioactive glycosides for drug discovery.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
This work is financially supported by the National Natural Science Foundation of China (No. 81573317).
Contributor Information
Lin Yang, Email: jgdai@imm.ac.cn.
Jun-gui Dai, Email: jgdai@imm.ac.cn.
References
- Guo D.D., Xue Y.R., Li D.Q., He B.X., Jia X.L., Dong X., Guo M.L. Overexpression of CtCHS1 increases accumulation of quinochalcone in safflower. Frontiers in Plant Science. 2017;8(8):1–14. doi: 10.3389/fpls.2017.01409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.L., Zhang P.C., Zhang Z.W. Studies on chemical constituents from bee-collected rape pollen. China Journal of Chinese Materia Medica. 2009;34(10):1235–1237. [PubMed] [Google Scholar]
- Hu X.J., Yin S., Yuan T.T., Huang Z.R., Lu Y. Chemical constituents and pharmacological activities of Carthamus tinctorius L. Journal of Pharmaceutical. Practice. 2013;31 (3), 161–168:197. [Google Scholar]
- Kazuma K., Takahashi T., Sato K., Takeuchi H., Matsumoto T., Okuno T. Quinochalcones and flavonoids from fresh florets in different cultivars of Cartaamus tinctorius L. Bioscience Biotechnology and Biochemistry. 2000;64(8):1588–1599. doi: 10.1271/bbb.64.1588. [DOI] [PubMed] [Google Scholar]
- Liang D., Liu Y.F., Hao Z.Y., Luo H., Wang Y., Zhang C.L.…Yu D.Q. Flavonol glycosides from Lysimachia clethroides. China Journal of Chinese Materia Medica. 2015;40(1):103–107. [PubMed] [Google Scholar]
- Matlawska I., Sikorska M. Flavonoids from flowers of Malva crispa L. (Malvaceae) Acta Poloniae Pharmaceutica. 2004;61(1):65–68. [PubMed] [Google Scholar]
- Scheer T., Wichtl M. On the occurrence of kaempfero1-4’-O-3-D- glucopy-ranoside in Filipendula ulmaria and Allium cepa. Planta Medica. 1987;53(6):573–574. doi: 10.1055/s-2006-962817. [DOI] [PubMed] [Google Scholar]
- Xie K.B., Chen R.D., Li J.H., Wang R.S., Chen D.W., Dou X.X., Dai J.G. Exploring the catalytic promiscuity of a new glycosyltransferase from Carthamus tinctorius. Organic Letters. 2014;16(18):4874–4877. doi: 10.1021/ol502380p. [DOI] [PubMed] [Google Scholar]
- Xie K.B., Chen R.D., Chen D.W., Li J.H., Wang R.S., Yang L., Dai J.G. Enzymatic N-glycosylation of diverse arylamine aglycones by a promiscuous glycosyltransferase from Carthamus tinctorius. Advanced Synthesis & Catalysis. 2017;359(4):603–608. [Google Scholar]
- Zhang Y.J., Xie K.B., Liu A.J., Chen R.D., Chen D.W., Yang L., Dai J.G. Enzymatic biosynthesis of benzylisoquinoline alkaloid glycosides via promiscuous glycosyltransferases from Carthamus tinctorius. Chinese Chemical Letters. 2019;30(2):447–450. [Google Scholar]