Abstract
Objective
Why are different medicinal parts including heads, bodies and tails of Angelicae Sinensis Radix (ASR) distinct in pharmaceutical activities? Here we explored their discrepancy in chemical constituents and transcriptome.
Methods
ASR were separated into three medicinal parts: heads (rootstocks with petiole traces of ASR), bodies (taproots of ASR) and tails (lateral roots of ASR), and chemical and transcriptomic analyses were conducted simultaneously.
Results
High performance liquid chromatography (HPLC) fingerprint results showed that five widely used active ingredients (ferulic acid, senkyunolide H, senkyunolide A, n-butylphathlide, and ligustilide) were distributed unevenly in the three ASR medicinal parts. Partial least squares-discriminant analysis (PLS-DA) demonstrated that the heads can be differentiated from the two other root parts due to different amounts of the main components. However, the content of ferulic acid (a main quality marker) was significantly higher in tails than in the heads and bodies. The transcriptome analysis found that 25,062, 10,148 and 29,504 unigenes were specifically expressed in the heads, bodies and tails, respectively. WGCNA analysis identified 17 co-expression modules, which were constructed from the 19,198 genes in the nine samples of ASR. Additionally, we identified 28 unigenes involved in two phenylpropanoid biosynthesis (PB) pathways about ferulic acid metabolism pathways, of which 17 unigenes (60.7%) in the PB pathway were highly expressed in the tails. The expression levels of PAL, C3H, and CQT transcripts were significantly higher in the tails than in other root parts. RT-qPCR analysis confirmed that PAL, C3H, and CQT genes were predominantly expressed in the tail parts, especially PAL, whose expression was more than doubled as compared with that in other root parts.
Conclusion
Chemical and transcriptomic analyses revealed the distribution contents and pivotal transcripts of the ferulic acid biosynthesis-related pathways. The spatial gene expression pattern partially explained the discrepancy of integral medicinal activities of three medicinal root parts.
Keywords: Angelicae Sinensis Radix, biosynthesis, comparative transcriptomics, ferulic acid, three medicinal root parts
1. Introduction
Angelicae Sinensis Radix (ASR; Danggui in Chinese) is a representative traditional Chinese medicine (TCM) with a history of two thousand years in Shennong's Classic of Materia Medica (Liu et al., 2014; Wei et al., 2015 & Wei, Zeng, Gu, Qu & Huang, 2016), a paragon classic in China. ASR is also known as ‘female ginseng’, as it is mainly used in the therapy of gynecological disorders (Wang et al., 2007; Xu et al., 2013; Zhan et al., 2014 & Wang et al., 2017). Moreover, ASR has been used as a tonic hematopoietic agent for thousands of years in TCM prescriptions (Lin, He, Lian, King & Elliott, 1998; Yang, Zhao, Li, Wang, & Lv, 2008 & Tan et al., 2015). Interestingly, the pharmaceutical effects of the heads, bodies and tails of ASR are distinct. In the Ming Dynasty of China, Shi-zhen Li indicated that the “heads of ASR were mainly utilized for stopping bleeding, bodies of ASR were used for nourishing blood, while the tails were used for disintegrating blood stasis and eliminating congestion”, which is supported by clinical cases (Nino, Yang, Franz, Anita & Caroline, 2017 & Tong et al., 2017; Wen, Niu, & Wang, 2014; Wu et al., 2014; Xue, Hua, & Guo, 2012; Yan, Duan, Shang, Hua, & Qian, 2014; Zhang et al., 2015). Yang, Deng and Yang (2018) reported that ASR tails are stronger in promoting blood in the acute blood stasis rat than ASR bodies and heads. The tendency of clinical effects, revealed by statistical data (Wu et, al.,2016; Yang et al., 2018), largely agreed with the ancient conclusion. Despite that the pharmaceutical discrepancy among heads, bodies and tails of ASR is of important clinical value, why different parts of ASR show the pharmaceutical discrepancy remains largely unknown. The diverse pharmacological activities could be related with the variations of main effective constituents in different root parts of ASR, and the enrichment site of active substances of ASR could be associated with vital genes of secondary metabolism pathways. In order to optimize the therapeutic efficacy, it is essential to elucidate the localization pattern of active ingredients in ASR and the biosynthesis mechanism in three medicinal root parts.
Ferulic acid is an essential bioactive compound in ASR, which is responsible for suppressing aggregation of platelet, reducing consumption of oxygen of myocardia and increasing supply of O2 and blood to heart cardiac muscles, and promoting thromboprophylaxis (Fang, Xiao, Liu & He, 2012 & Lin et al., 2017). It is also a commonly used marker for quality assessment of ASR (Chen, Li, Xiao, Zhang, & Liu, 2013 & Bai et al., 2014). Experimental and clinical data indicated the uneven accumulation pattern of ferulic acid in various medicinal parts, possibly reflecting the multiple differential expressions of key genes in the biosynthesis pathway of plant secondary metabolites. Ferulic acid skeleton is synthesized via the catechol-O-methyltransferase (COMT) as well as caffeoyl-CoA-3-O-methyltransferase (CCoAOMT) pathways, and multiple genes are involved in ferulic acid biosynthesis. We hypothesize that the differential expressions of the phenylpropanoid biosynthesis (PB) pathway genes could cause the variation of ferulic acid levels in the ASR heads, bodies and tails, which could partially explain the distinct action mechanisms of three root parts.
In this study, A. sinensis (AS) roots were separated into three medicinal parts, i.e., heads, bodies and tails, and chemical and transcriptomic analyses were conducted synchronously. First, the chemical composition of three root parts of ASR was analyzed by HPLC fingerprint method. Second, the gene expression differences, with respect to therapeutic effects of each part, were explored based on RNA-Seq data. In particular, differentially expressed genes (DEGs) in the PB pathway were identified to elucidate the regulatory networks of the biosynthesis of ferulic acid, a quality marker of ASR. The comprehensive analyses of the HPLC fingerprints and the transcriptome contribute novel understandings regarding the overall characteristics of the main active compounds generated in ASR biosynthesis.
2. Materials and methods
2.1. Plant materials
The harvested ASR of two years old was collected from Min County of Gansu Province, China, in August 2017. Every main body of the root was carefully peeled into three portions: heads, bodies and tails (Fig. 1A); 27 samples from nine roots were subjected to liquid nitrogen freezing and −80 °C storage. Alternatively, the freeze drying was done for RNA isolation and/or HPLC analysis. A mortar was used to grind root samples into powder, and RNeasy Plant Mini kit (Biteke, Beijing, China) was used to isolate total RNA.
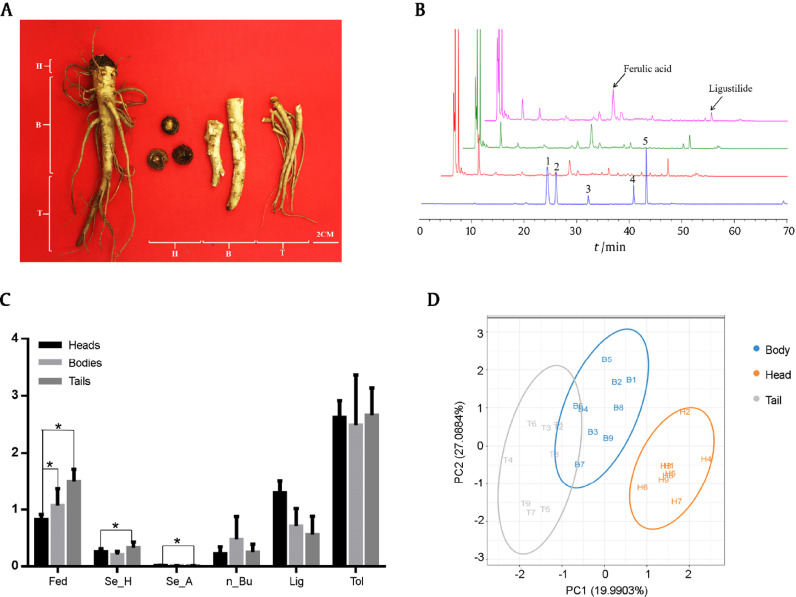
Fig. 1.
Content of main active components in three medicinal root parts of ASR.
(A) Roots of A. sinensis with heads (H), bodies (B) and tails (T).
(B) HPLC fingerprint profile of three root parts of ASR. 1. Ferulic acid; 2. Senkyunolide H; 3. Senkyunolide A; 4. n-Butylphathlide; 5. Ligustilide.
(C) Content of five main components. Fed: Ferulic acid; Se-H: Senkyunolide H; Se-A: Senkyunolide A; Bu:n-butylphathlide, Lig: Ligustilide, Tol: Total amount of five main components of ASR. (*P < 0.05 represents significant differences between the three medicinal parts).
(D) Differences of chemical profiles of three root parts based on PLS-DA.
2.2. HPLC
A protocol described earlier was used with small modification of the ultrasonic extraction in extracting effective constituents (Bao, 2014). Nearly 200 mg crushed sample was subjected to overnight soaking in 2 mL of 70% methanol. After centrifugation, samples were subjected to ultrasound lysis at room temperature for 30 min (500 W, 40 kHz), the extracts were centrifuged for 5 min (14,000 r/min) and the supernatant was transferred to another test tube. The above process was repeated for the supernatant followed by mixing of the two supernatants and dried 70% methanol at 100 °C in water bath. And then, two millilitres of 70% methanol was used to dissolve the residue followed by filtration using an organic micropore membrane of 0.45 µm before HPLC. The Agilent 1260 series HPLC system with 35 °C column temperature was utilized for chemical analysis, and the optimal separation was performed by BDS Hypersil C18 (250 mm × 4.6 mm, 5 µm). The elution was done with solvent system A: acetonitrile and solvent system B: 0.5% methanoic acid in ultra-pure water. A gradient system involved: 0 − 20 min: 90%−80% B; 20−35 min: 80%−50% B; 35−40 min: 50%−30% B; 40−60 min: 30%−0% B; 60−70 min: 0% B; and 70−80 min: 0%−10% B. The external standards were used for quantification at wavelength of 281 nm and with 10 µL injection volume.
2.3. PLS-DA
Multivariate statistical approaches were applied to process the data of the five major components. The partial least squares discriminant analysis (PLS-DA) is an effective classification, clustering, and regression method; Chemometric analyses were implemented in the R language version 3.0.1, using the packages ChemoSpec, Chemometrics, mixOmics, e1071, and pls. The R package PLS-DA was employed for all studies, and the data from nine repetitions (n = 9) were expressed as mean ± standard deviation. In multiple comparisons, one-way ANOVA and Tukey HSD tests were performed.
2.4. Transcriptome sequencing and analysis
Skewer and FastQC were utilized to process raw sequencing data to yield clean reads. The reads from all nine samples were subjected to de novo assembly with Trinity using default parameters. The transcriptome sequencing reads were deposited in NCBI SRA with accession number SUB 4972114. The transcript coding for the protein of maximum length was used to define the representative sequence for gene annotation. The sequence similarity and homology were studied using BLASTX (E-value < 10−5) in NCBI (ftp://ftp.ncbi.nih.gov/blast/db) and Swiss-Prot (http://www.uniprot.org/). The functional classification of predicted proteins was performed using GO (http://www.geneontology.org/) and KEGG databases (http://www.genome.jp/tools/kaas/).
The transcript abundance was estimated by Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/) (Lao et al., 2004), and DEG identification (fold change (FC) > 2 and false discovery rate (FDR) < 0.05) and quantification of gene expression were performed by calculating fragments per kilobase of exon model per million mapped reads (FPKM). Goatools (https://github.com/tanghaibao/Goatools) was utilized to perform the directed acyclic graph of the GO enrichment studies. Co-expression modules were identified with R package weighted gene co-expression network analysis (WGCNA) hat was visualized on Cytoscape; R programming and software were used to construct heat maps of gene expression. The Illumina sequencing data were used to identify PB pathway genes followed by GenBank database searching using candidate gene name in a functional annotation file.
2.5. Real-time qPCR of PAL, C3H, and CQT
The total RNA from ASR root parts was converted into a single-stranded cDNA by using a FastQuant RT kit with gDNase (TIANGEN, Beijing, China). Quantitative reactions were performed using a real-time qPCR system (Biorad 7500 System). The reaction mixture contained 10 µL of SYBR Green qPCR Master Mix (Toyobo, Osaka, Japan), 1 µL of 10 µmoL/L each of the forward and reverse primers, and 2 µL of the cDNA template. Sterile water was added to obtain a final volume of 20 µL. PCR amplification was conducted under the following conditions: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 60 °C for 34 s, and with a dissociation stage of 95 °C for 15 s, 55 °C for 60 s and 95 °C for 15 s.The forward and reverse primer sequences are as follows: Actin: 5′-TGGTATTGTGCTGGATTCTGGT-3′ and 5′-TGAGATCACCACCAGCAAGG-3′; PAL: 5′-GGAACCATCACAGCCTCAG-3′ and 5′-CAACTCAAAGAACCCACCA-3′; C3H: 5′-CAATCCAAGTTGACGACGAA-3′ and 5′-CGAAGGCGAAACATAGGC-3′; CQT: 5′-GTTTGGTTCACGCACTC-3′ and 5′-TTCCTCAAGACAATCCC. The relative expression levels of the key genes were calculated using the 2−△△Ct method.
3. Results
3.1. Chemical composition of three medicinal root parts of ASR
The five major components varied among three medicinal root parts (Fig. 1B and Table 1). Quantities of therapeutic compounds ferulic acid and ligustilide significantly differed among three ASR root parts. The ferulic acid was the highest in tails (1.131 mg/g), followed by the bodies (1.085 mg/g) and the heads (0.806 mg/g). However, the ligustilide was the highest in heads (1.299 mg/g). The amount of ligustilide was comparable between the bodies (0.713 mg/g) and tails (0.858 mg/g). Other active ingredients such as senkyunolide H, senkyunolide A and n-butylphathlide were found in varying amounts in three medicinal parts (Fig. 1C and Table 1). Hence, the active compounds were unevenly distributed in different root parts of ASR.
Table 1.
Content, calibration curves, linearity, precision, repeatability, stability and recovery rate content of five component in ASR different medical parts (Heads,Bodies,Tails).
| Saponins | Calibration curves | R2 | Linearity (μg) | RSD% (n = 6) |
Recovery rate/% (RSD%) | Content of each component (mg/g; n = 9) |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| Precision | Repeatability | Stability | Heads | Bodies | Tails | |||||
| Ferulic acid | Y = 4.15X−41.3320 | 0.9996 | 0.45−3.20 | 0.21 | 0.96 | 1.67 | 101.78 (1.74) | 0.806 ± 0.068 | 1.085 ± 0.289 | 1.131 ± 0.356 |
| Senkyunolide H | Y = 2.48X+18.6377 | 0.9994 | 0.10−2.25 | 0.32 | 2.16 | 3.11 | 98.75 (1.85) | 0.268 ± 0.027 | 0.207 ± 0.059 | 0.269 ± 0.083 |
| Senkyunolide A | Y = 98.59X+6.1494 | 0.9992 | 0.01−1.21 | 0.09 | 1.60 | 2.23 | 98.34 (2.21) | 0.018 ± 0.004 | 0.015 ± 0.002 | 0.015 ± 0.004 |
| n-butylphathlide | Y = 0.999X−5.5651 | 0.9998 | 0.15−2.55 | 0.29 | 1.18 | 1.96 | 100.36 (1.01) | 0.201 ± 0.105 | 0.457 ± 0.409 | 0.304 ± 0.271 |
| Ligustilide | Y = 2.881X+21.5355 | 0.9998 | 0.65−3.54 | 0.77 | 2.62 | 2.72 | 96.73 (1.77) | 1.299 ± 0.221 | 0.713 ± 0.308 | 0.858 ± 0.425 |
| Total amount | 2.592 ± 0.085 | 2.477 ± 0.213 | 2.577 ± 0.228 | |||||||
Note: The linearity of all calibration curves were R2 > 0.999. The precision variations, repeatability variations and stability variations were 0.09%–0.77%, 0.96%–2.62%, and 1.01%–2.21%, respectively. The recoveries ranged from 96.73% – 101.78%, with variations of 1.01%–2.21%. These results showed that the established HPLC method was accurate and sensitive, which was appropriate for the quantitative analyses.
3.2. PLS-DA of major components
R language (version 3.0.1) was used to establish a PLS-DA model for preprocessed HPLC fingerprint data, in order to classify and identify three medicinal root parts of ASR. The scattered dots representing different medicinal parts were disparate (Fig. 1D). The heads can be differentiated from other two root parts due to different amounts of organic acids and phthalide according to HPLC fingerprint data. Within each group, the active compounds displayed instability relatively along with dissimilarity with respect to the five major components. PLS-DA results corroborated that the amount of organic acids and their esters, ligustilide and phthalide dimers differed greatly among AS three root parts, thereby explaining their variations in the therapeutic effects (Table 1).
3.3. De novo assembly of ASR transcriptome
The transcriptome analysis of the AS three root parts was performed based on sequencing platform (Illumina, San Diego, CA, USA). A total of 68.43 Gb of data were obtained, and the average output of each sample was 7.60 Gb of data. The numbers of raw reads from the heads, bodies and tails were 162,175,492, 171,048,182 and 139,994,798 respectively. After filtering was performed, 456.24 M clean reads with an average size of 150 bp were used for the de novo assembly. The read numbers of all replicates are more than 40,000,000, with the maximal value of 53 million reads. And then, 157,447,128, 165,945,584 and 132,849,116 high quality (Q30 > 93.09%) reads were generated respectively. These reads were adequate for further quantification of gene expression. Overall, totally 219,500 unigenes were assembled, and the percentage of GC was 43.93% with an average contig size of 531 bp and an N50 contig size of 801 bp (Table 2).
Table 2.
Summary of transcriptome assembly of ASR root parts.
| Items | No. of sequences |
|---|---|
| Clean reads / M | 456.24 |
| Clean bases / G | 68.43 |
| No. of contig>500 bp | 137,595 |
| Total unigenes | 219,500 |
| Total length / bp | 116,508,544 |
| Average contig size / bp | 531 |
| N50 contig size / bp | 801 |
| GC /% | 43.93 |
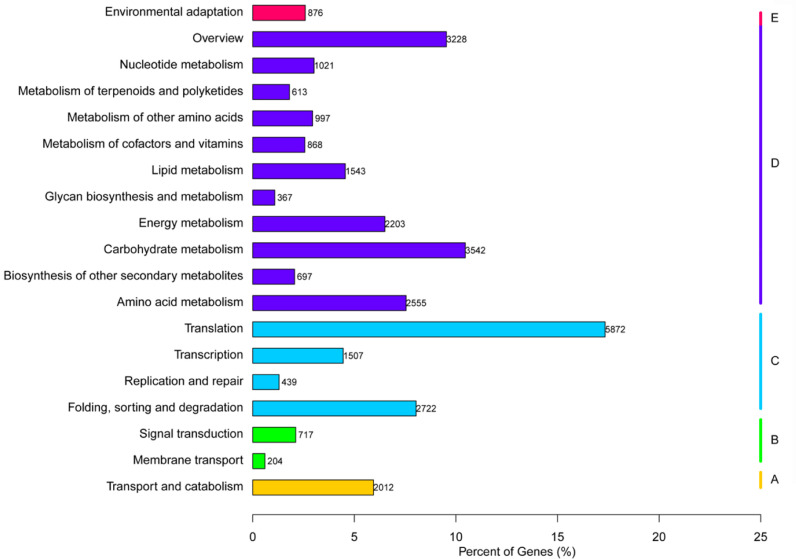
The homology search of assembled unigenes was conducted against NCBI non-redundant (Nr) protein database, Swiss-Prot, Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG), with 10−5 E-value as a cutoff. Totally 61.89% of all unigenes (135,855) were annotated with a minimum of one significant match in the database search; 73,249 (33.37%) unigenes had the match in the Nr database. Similarly, 79,024, 72,724, and 31,983 unigenes had significant homologs in Swiss-Prot, GO, and KEGG databases, respectively (Fig. 2 and Table 3). Neither generation nor deposit into databases of approximately 50% of the unigenes was done that was suggestive of genes specific to certain tissues in ASR (Yang, Wang, Ding, Li, & Liu, 2015). This new and substantial set of unigenes serve as a significant tool to facilitate additional studies of ASR and interpreting the heterogeneous distribution of active ASR ingredients.
Fig. 2.
Functional classification of ASR unigenes based on KEGG pathway.
Table 3.
Homology and protein sequence similarity search of ASR unigene set.
| Homology search | No. of unigenes | Percentage/% |
|---|---|---|
| Nr | 73,249 | 33.37 |
| Nt | 91,664 | 41.76 |
| Pfam | 71,234 | 32.45 |
| Swiss-Prot | 79,024 | 36.00 |
| GO | 72,724 | 33.13 |
| KEGG | 31,983 | 14.57 |
| Annotated in all databases | 14,294 | 6.51 |
| At least one significant match | 135,855 | 61.89 |
3.4. Functional analysis of unigenes
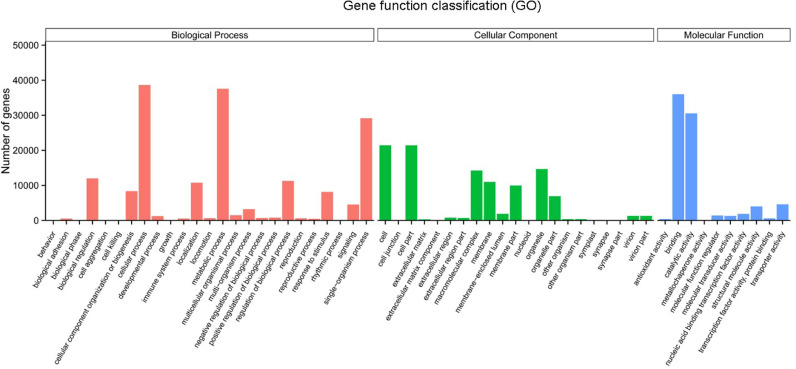
GO assignments were used to classify the functions of the predicted ASR genes. A total of 72,724 unigenes were subjected to the assignment of three GO categories: biological process, cellular component and molecular function. Three categories were classified into 56 subcategories. The majority of assigned GO terms were biological processes, followed by cellular components and molecular function categories. Binding and catalytic activity were the most abundant GO terms within the molecular function category. In the biological process category, the dominant subcategories were in response to cellular and metabolic processes. “Cell” and “Cell part” were the most highly represented terms in the cellular component category (Fig. 3). In addition, 39,365 sequences were annotated as a metabolic process category, suggesting that these genes were involved in the metabolite synthesis pathways.
Fig. 3.
GO analysis of ASR unigenes.
3.5. Individual transcriptome of three root parts of ASR
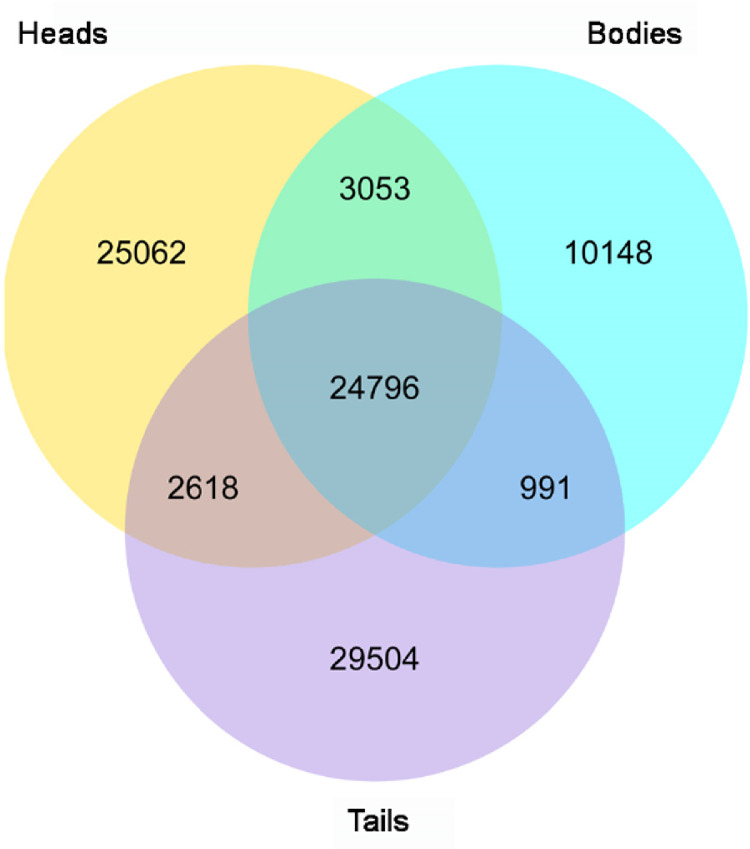
We compared the gene expression profiles of three root parts and characterized their relationship. FPKM ≥ 2 was used to assess the gene expression level and select the high expression genes. A total of 96,172 genes were chosen from the transcriptome data; 55,529, 38,998, and 57,909 genes were from heads, bodies and tails, respectively, 24,796 of which (25.8% of total) were shared by three root parts (Fig. 4). A total of 10,148, 25,062, and 29,504 unigenes were solely expressed in bodies, heads, and tails, respectively.
Fig. 4.
Venn diagram showing shared and unique unigenes among three ASR root parts.
Using FDR ≤ 0.05 and FC ≥ 2 as the criteria, Differential expression genes (DEGs) were identified. There were 2112, 1856, and 864 DEGs between tails and bodies, tails and heads, and heads and bodies, respectively (Table 4). Overall, the transcription profiles of three root parts were distinct, possibly corresponding to their distinct therapeutic functions. Most DEGs between tails and heads and between tails and bodies were associated with single-organism and carbohydrate metabolic processes in the GO enrichment analysis (Fig. 3 and Table 4). Unigenes with comparable and highly variable expression levels in three root parts of ASR reflect the evolutionary conservation as well as environmental responses and physiological functions (Breschi et al., 2016). After filtering, 19,198 DEGs had the expression level of FPKM ≥ 2 in all root parts. WGCNA was utilized to construct a hierarchical cluster tree. Weighted factors were the contents of ferulic acid and ligustilide. Genes were grouped into 17 expression modules.
Table 4.
DEGs between ASR root parts (FDR ≤ 0.05 & FC≥ 2).
| Pair of root parts | No. of unigenes with significant expression changes | No. of up-regulated unigenes | No. of down-regulated unigenes |
|---|---|---|---|
| Head-VS-Body | 864 | 333a | 531 |
| Body-VS-Tail | 2112 | 424a | 1688 |
| Tail-VS-Head | 1856 | 1529a | 327 |
Note:a The expressions of these genes were up-regulated in the former part as compared with the latter one.
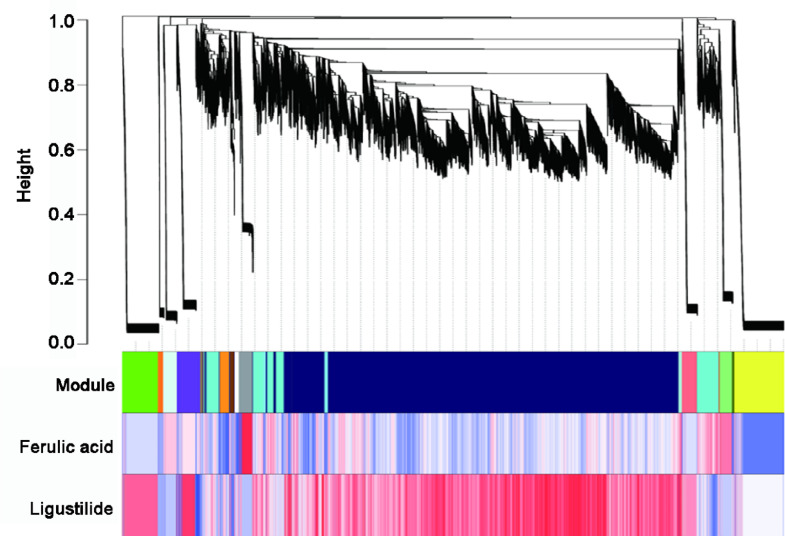
The modules corresponding to the branches are represented by colors in the first color band underneath the tree, and the remaining color bands reveal the correlation between transcripts and ferulic acid and ligustilide. The correlation for modules is as follows: Red for highly positive, white for weak and blue for highly negative. Hence, this conclusion showed the complex synthesis and accumulation mechanisms of ferulic acid and ligustilide (Fig. 5).
Fig. 5.
Co-expression analysis of 19,198 unigenes through WGCNA.
Note: The modules corresponding to the branches are represented by colors in the first color band underneath the tree, and the remaining color bands reveal the correlation between transcripts and therapeutic compounds (ferulic acid and ligustilide). The red module indicates a significant positive correlation with the corresponding gene, the white module denotes a weak correlation, and the blue module represents a significant negative correlation.
3.6. Pivotal genes involved in ferulic acid biosynthesis in ASR
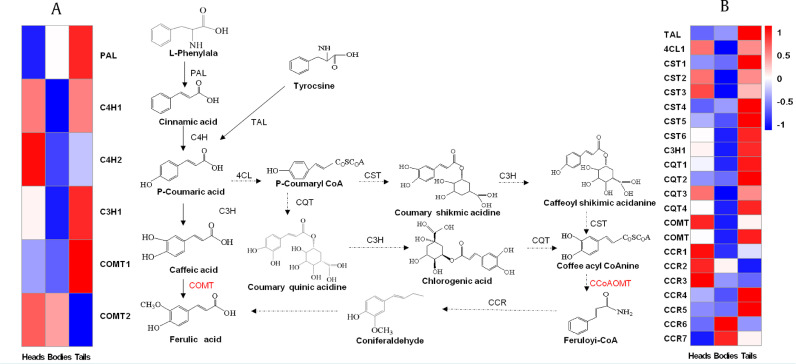
Ferulic acid, one of the most important pharmaceutical compounds of ASR, is produced in the PB pathway. In order to explore the regulatory networks of ferulic acid biosynthesis, accumulation, and transportation in three AS root parts, the enzyme genes involved in ferulic acid biosynthesis and modification were investigated. Twelve genes (28 transcripts) of PB pathway were identified in the transcriptome datasets. The COMT and CCoAOMT pathway genes responsible for the biosynthesis of ferulic acid skeleton were identified. These genes were seen as isoforms and numerous copies (ranging from one to seven copies) associated with a large spectrum of regulatory pathways to control AS plants secondary metabolism. Furthermore, the expression data of the 28 transcripts were extracted at FPKM ≥ 2 followed by heat map analysis to assess their relationships (Fig. 6).
Fig. 6.
Heat map depicting expression profile of ferulic acid biosynthesis genes in heads, bodies and tails of ASR.
Note: (A) Transcript abundance profiles of enzyme genes of COMT pathway; (B) Transcript abundance profiles of enzyme genes of CCoAOMT pathway.
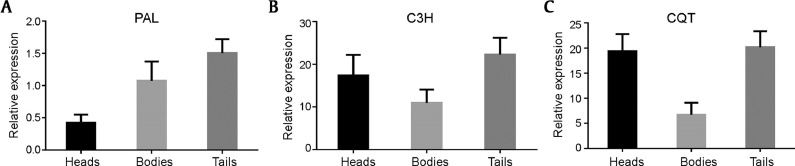
The expression patterns of ferulic acid biosynthesis genes in the COMT and CCoAOMT pathway were analyzed. Levels of main transcripts of the COMT pathway varied among three root parts. The enzyme gene of the first step of PB pathway, phenylalanine ammonia lyase (PAL), showed maximal expression in tails followed by the bodies. The expression patterns of ASR coumarate 3-hydroxylase (C3H) were similar as those of PAL, i.e., its expression in tails was higher than that in bodies and heads. Ferulic acid skeleton biosynthesis genes of ASR, AS cinnamate 4-hydroxylase (C4H) and one transcript encoding COMT showed high expression in heads, while these genes had lower expression levels in bodies and tails. In another pathway of ferulic acid biosynthesis, i.e., the CCoAOMT pathway, the expression patterns were similar when blocking COMT. Tyrosine ammonia lyase (TAL), quinate hydroxycinnamoyl transferase (CQT) and C3H transcripts were highly expressed in the tails. One transcript encoding CCoAOMT and two transcripts encoding cinnamoyl-CoA reductase (CCR) were highly expressed in the heads. The irregular distribution of ferulic acid in three root parts of ASR may be due to the differential expression patterns of relevant biosynthetic genes. The RT-qPCR results showed that PAL, C3H, and CQT were most highly expressed in the tails, which were consistent with the FPKM values (Figs. 6 and 7).
Fig. 7.
Relative expression levels of PAL (A), C3H (B) and CQT (C) genes quantified by RT-qPCR.
4. Discussion
4.1. Herbgenomics provides a new platform for researching traditional herbal medicines
Herbgenomics provides an unparalleled platform to revolutionize our understandings and the utilities of traditional herbal medicines (Chen et al., 2015 & Chen & Song, 2016). It is becoming widely used in the various fields of herbal research. For example, Xu et al. (2017) reported the next-generation sequencing of the whole genome sequence of Panax ginseng. Zhang et al., 2017 reported that the accumulation and distribution of ginsenosides were closely correlated with genes involved in their biosynthesis and regulation. Wei et al. (2018, 2019) identified pivotal genes regulating saponin distribution and their complex mechanisms in Panax quinquefolius and Panax notoginseng. These works help comprehend the biosynthesis of active compounds in key herbs and provide valuable resources to improve breeding. An integration of transcriptome sequencing and chemical analysis for an overall characterization of the main active components was conducted in this study, and the results are useful in explaining the differences of bioactive compounds in heads, bodies and tails of ASR.
4.2. Chemical analysis revealed metabolic characteristics of three root parts of A. sinensis
The HPLC fingerprints comparison of ASR revealed that the distribution of its main active ingredients vary in its three medicinal parts (Breschi et al., 2016 & Tian, Hao, Xu, Yang & Sun, 2017; Cao et al., 2014). The heads can be differentiated from other two parts due to different amounts of main components. These results are basically consistent with previous findings (Wen, Niu, & Wang, 2014; Xue, Hua, & Guo, 2012; Yan, Duan, Shang, Hua, & Qian, 2014; Zhang et al., 2015 & Nino et al., 2017).
Among more than 80 isolated and identified compounds of ASR (Wang et al., 2007; Xu et al., 2013; Zhan et al., 2014 & Wang et al., 2017), ferulic acid and ligustilide are the most associated with pharmacological activities (Zhao et al., 2003, 2013). They, along with polysaccharides compounds, are receiving the spotlight from both the industry and academics because of their medicinal features (Cao et al., 2014 & Tian et al., 2017). The three main active ingredients exert distinct pharmacological effects. Firstly, the pharmacological activities of ferulic acid could be related to decreased blood viscosity, improved blood circulation and anti-thrombotic functions (Lü et al., 2009; Zeng et al., 2014 & Wang et al., 2015). Secondly, ligustilide, a major bioactive constituent of ASR (Zhong et al., 2014), exhibits vasodilation activities, the effects of antiplatelet aggregation, analgesia and smooth muscle relaxant, as well as antifungal, antibacterial, antiasthmatic, anti-inflammatory and antioxidant activities (Li et al., 2015, Li et al., 2015b). Butylidenephthalide, senkyunolide H and senkyunolide A also showed similar bioactivities (Gu, Chen, Wang, Wen & Sun, 2014; Li et al., 2014 & Li et al., 2015, Li et al., 2015b). Thirdly, ASR polysaccharides can effectively enhance haematopoiesis via stimulation of muscles and hematopoietic cells by augmenting the level of haematopoietic growth factors, e.g., erythropoietin (Liu et al., 2010). Polysaccharides are effective medicinal components and display heterogeneous distribution in ASR roots (Li et al., 2014). Our HPLC results showed that ferulic acid was more abundant in the tail samples when compared with the body and head samples.
The distinct pharmacological activity of three root parts of ASR might be caused by the differential composition of the effective pharmaceutical components, such as organic acids, their esters, and essential oil, etc. Polysaccharides and other molecules of three root parts should be further investigated. According to Chinese medicine theories, the clinical effects of each of three root parts were revealed by the statistical data analysis, and were mostly consistent with the TCM documentations (Dare et al., 2017 & Lin et al., 2017). Hence, ASR has multiple pharmacological activities. In this regard, additional analytical and biological studies are necessary to elucidate substances responsible for the effects of different parts of ASR.
4.3. Transcriptome analysis revealed biosynthesis characteristics of three root parts of A. sinensis
Analysis of transcriptomes revealed variations and similarities of the unigene expressions in three root parts. A total of 25,062, 10,148 and 29,504 unigenes were expressed in the heads, bodies and tails, respectively. Different expression patterns of three parts are similar to previous findings (Lao et al., 2004), and could partially explain the biological effects of three parts of ASR. Furthermore, more than 50% of unigenes were novel. The transcriptome data size reported in this study exceeds that of the biggest ASR transcriptome reported recently (Yang, Wang, Ding, Li, & Liu, 2015). Our dataset can complement previous transcriptome data and facilitate deeper analysis.
Ferulic acid, an essential intermediate metabolite in the PB pathway, is often associated with the pharmacological activities of ASR. Genes involved in the PB pathway were identified from transcriptome dataset; They could be responsible for the metabolism of ferulic acid. Candidate genes of COMT and CCoAOMT associated with the PB pathway were also identified. Overall, the transcript expressions varied greatly in three root parts. Twenty eight unigenes with FPKM ≥ 2, which could be involved in the PB pathway, were identified, 17 of which (60.7%) had the highest expressions in tails. The result may be partly revealed that contents of ferulic acid were higher in the tails medical parts of AS.
In PB and relevant metabolic pathways, cinnamate-4-hydroxylase (C4H), coumarate-3-hydroxylase (C3H), caffeic acid O-methyl transferase (COMT), 4-coumarate-CoA ligase (4CL) and cinnamoyl-CoA reductase (CCR) had significant impacts on the biosynthesis of therapeutic compounds (Dare et al., 2017). These PB pathway transcripts with high expression and the differential expression profiles amongst different medical parts may provide useful information for investigating the functions and expression patterns of unigenes in ASR. In higher plants, PAL is the rate-limiting enzyme at the initial step of ferulic acid biosynthesis. Luo et al. (2014) found that PAL was expressed at the highest level in AS leaf, followed by stem and root, and the relative expression levels in leaves and stems were 7.5 and 2.7 times that of roots, respectively. The intermediate metabolites of this pathway are probably synthesized actively in the aboveground part of AS, then they reach the roots. AS C3H participates in the lignin biosynthesis through the multi-step catalysis of PB pathway. The PAL, C3H and CQT transcripts were identified in the transcriptome datasets of three root parts of AS, and all were more abundant in tails than in other parts.
COMT is the crucial enzyme in catalyzing methylation and lignin biosynthesis. Li et al., 2015, Li et al., 2015b suggest ed that LcCOMT might be used as a target to improve the content of ferulic acid of Ligusticum chuanxiong. The COMT gene expression in the stem of Lolium perenne had a sharp rise during early stages of elongation, while it dipped at the mid and later stages, implying its role in growth regulation (Giordano et al., 2014). In this study, two COMT transcripts were identified in three root parts; One transcript was more abundant in tails, and the other was a little more expressed in heads. The phenolic acid could be more actively synthesized and stored in tails, which helps understand the discrepancy of integral medicinal activities and provides promising candidates for functional genomics studies in ASR.
In this paper, ligustilide is another marker compound of ASR and is most often associated with the pharmacological activities. The interrelated studies on the ligustilide biosynthetic pathway have not yet been identified previously, so ligustilide and the other active component scientific metabolism of ligustilide should be further investigated.
5. Conclusions
This study, for the first time, systematically reports transcriptome variations of three medicinal root parts (heads, bodies and tails) of AS. Twenty eight unigenes involved in the PB pathway were identified from transcriptome datasets. The biosynthesis and regulation of ferulic acid are associated with the expression of PAL, C3H and CQT. This genetic resource will help improve the understanding of complex mechanisms of ferulic acid biosynthesis, accumulation, and transportation. Our results could contribute to elucidating the molecular mechanisms for discrepant pharmaceutical activities of three medicinal parts of AS.
Declaration of Competing Interest
All authors have no conflict interest.
Acknowledgments
Acknowledgments
This work was supported by the grants from National Science Foundation of China (grant number 81903782), Hubei Provincial Department of Education Plan for Science & Technology Project (Q20171705) and Scientific Research Project of Wuhan Polytechnic University (2018Y10).
Author Contributions
R.X. wrote the original manuscript. S.Z. and Z.L. carried out the data curation. L.Y. and G.W. performed the experimental data analysis. J.X. and Y.D. revied & edited the manuscript. S.C. and L.D designed the research and organized the manuscript. All authors approved the final manuscript.
Contributor Information
Lin-lin Dong, Email: lldong@icmm.ac.cn.
Shi-lin Chen, Email: slchen@icmm.ac.cn.
References
- Bai Y.J., Kong M., Xu J.D., Zhang X.L., Zhou S.S., Wang X.N., et al. Effect of different drying methods on the quality of Angelicae Sinensis Radix evaluated through simultaneously determining four types of major bioactive components by high-performance liquid chromatography photodiode array detector and ultra-high performance liquid chromatography quadrupole time-of-flight mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis. 2014;94:77–83. doi: 10.1016/j.jpba.2014.01.036. [DOI] [PubMed] [Google Scholar]
- Bao X.P. Determination and processing of ferulic acid in different parts of Angelica sinensis. Strait Pharmaceutical Journal. 2014;26:73–74. (in Chinese) [Google Scholar]
- Breschi A., Djebali S., Gillis J., Pervouchine D.D., Dobin A., Davis C.A., et al. Gene-specific patterns of expression variation across organs and species. Genome Biology. 2016;17:151. doi: 10.1186/s13059-016-1008-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G., Cai H., Lou Y.J., Tu S.C., Liu X., Qin K.M., et al. Analysis of the influence of sulfur fumigation on the volatile components of Angelicae Sinensis Radix by comprehensive two dimensional gas chromatograph time of flight mass spectrometry. Pharmacognosy Magazine. 2014;39:304–313. doi: 10.4103/0973-1296.137371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.L., Song J.Y. Herbgenomics. China Journal of Chinese Materia Medica. 2016;41(21):3881–3889. doi: 10.4268/cjcmm20162101. (in Chinese) [DOI] [PubMed] [Google Scholar]
- Chen S.L., Song J.Y., Sun C., Xu J., Zhu Y., Verpoorte R., et al. Herbal genomics: Examining the biology of traditional medicines. Science. 2015;347:S27–S29. [Google Scholar]
- Chen X.P., Li W., Xiao X.F., Zhang L.L., Liu C.X. Phytochemical and pharmacological studies on Radix Angelicae Sinensis. Chinese Journal of Natural Medicines. 2013;11:577–587. doi: 10.1016/S1875-5364(13)60067-9. [DOI] [PubMed] [Google Scholar]
- Dare A.P., Yark K.Y., Tomes S., McGhie T.K., Rebstock R.S., Cooney J.M., et al. Silencing a phloretin-specific glycosyltransferase perturbs both general phenylpropanoid biosynthesis and plant development. The Plant Journal. 2017;91:237–250. doi: 10.1111/tpj.13559. [DOI] [PubMed] [Google Scholar]
- Fang L., Xiao X.F., Liu C.X., He X. Recent advance in studies on Angelica sinensis. Chinese Herbal Medicines. 2012;4:12–25. [Google Scholar]
- Giordano A., Liu Z.Q., Panter S.N., Dimech A.M., Shang Y.J., Wijesinghe H., et al. Reduced lignin content and altered lignin composition in the warm season forage grass Paspalum dilatatum by down-regulation of a cinnamoyl coa reductase gene. Transgenic Research. 2014;23:503–517. doi: 10.1007/s11248-014-9784-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z.R., Chen H., Wang Y.L., Wen X.Y., Sun Y.J. Study on identification of smoked Angelica sinensis by TLC. Journal of Gansu College of Traditional Chinese Medicine. 2014;31:27–29. (in Chinese) [Google Scholar]
- Lao S.C., Li S.P., Kan K.K.W., Li P., Wan J.B., Wang Y.T., et al. Identification and quantification of 13 components in Angelica sinensis (Danggui) by gas chromatography-mass spectrometry coupled with pressurized liquid extraction. Analytica Chimica Acta. 2004;526:131–137. [Google Scholar]
- Li B.X., Wang C.Q., Xi L.L., Wei Y.H., Duan H.H., Wu X.H. Qualitative and quantitative analysis of Angelica sinensis using near infrared spectroscopy and chemometrics. Analytical Methods. 2014;6:9691–9697. [Google Scholar]
- Li J.J., Zhang G., Yu J.H., Li Y.Y., Huang X.H., Wang W.J., et al. Molecular cloning and characterization of caffeic acid 3-O-methyltransferase from the rhizome of Ligusticum chuanxiong. Biotechnology Letters. 2015;37:2295–2302. doi: 10.1007/s10529-015-1917-y. [DOI] [PubMed] [Google Scholar]
- Li J.J., Zhu Q., Lu Y.P., Zhao P., Feng Z.B., Qian Z.M., et al. Ligustilide prevents cognitive impairment and attenuates neurotoxicity in d-galactose induced aging mice brain. Brain Research. 2015;1595:19–28. doi: 10.1016/j.brainres.2014.10.012. [DOI] [PubMed] [Google Scholar]
- Lin H.Q., Gong A.G.W., Wang H.Y., Duan R., Dong T.T.X., Zhao K.J., et al. Danggui Buxue Tang (Astragali Radix and Angelicae Sinensis Radix) for menopausal symptoms: A review. Journal of Ethnopharmacology. 2017 doi: 10.1016/j.jep.2017.01.044. [DOI] [PubMed] [Google Scholar]
- Lin L.Z., He X.G., Lian L.Z., King W.N., Elliott J. Liquid chromatographic–electrospray mass spectrometric study of the phthalides of Angelica sinensis and chemical changes of Z-ligustilide. Journal of Chromatography A. 1998;810:71–79. [Google Scholar]
- Liu C., Li J.Q., Meng F.Y., Liang Simon X., Deng R.X., Li C.K., et al. Polysaccharides from the root of Angelica sinensis promotes hematopoiesis and thrombopoiesis through the PI3K/AKT pathway. BMC Complementary and Alternative Medicine. 2010;10:79. doi: 10.1186/1472-6882-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wang W.X., Yang Y.J., Yan Y.N., Wan W.Y., Wu H.Z., et al. A rapid discrimination of authentic and unauthentic Radix Angelicae Sinensis growth regions by electronic nose coupled with multivariate statistical analyses. Sensors. 2014;14:20134–20148. doi: 10.3390/s141120134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü J.L., Zhao J., Duan J.A., Yan H., Tang Y.P., Zhang L.B. Quality evaluation of Angelica sinensis by simultaneous determination of ten compounds using LC-PDA. Chromatographia. 2009;70:455–465. [Google Scholar]
- Luo J., Wang Y.Q., Wen S.C., Li J., Zhang J.L., Xia Q. Cloning and tissue-specific expression analysis of phenylalanine ammonia-lyase gene fragment in Angelica sinensis. Acta Ecologica Sinica. 2014;23:130–137. [Google Scholar]
- Nino G., Yang Y.P., Franz K.H., Anita A., Caroline S.W. Angelica sinensis (Oliv.) Diels: Influence of value chain on quality criteria and marker compounds ferulic acid and Z-ligustilide. Medicines. 2017;4:14. doi: 10.3390/medicines4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H.S., Hu D.D., Song J.Z., Xu Y., Cai S.F., Chen Q.L., et al. Distinguishing Radix Angelica sinensis from different regions by HS-SFME/GC-MS. Food Chemistry. 2015;186:200–206. doi: 10.1016/j.foodchem.2014.05.152. [DOI] [PubMed] [Google Scholar]
- Tian S.Y., Hao C.C., Xu G.K., Yang J.J., Sun R.G. Optimization conditions for extracting polysaccharide from Angelica sinensis and its antioxidant activities. Journal of Food and Drug Analysis. 2017;25:766–775. doi: 10.1016/j.jfda.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J.Y., Chao J., Dai Y.T., Fan Z.Q., Wang D.D., Li Q., et al. Quality evaluation of standard decoction of Angelicae Sinensis Radix. Chinese Journal of Experimental Traditional Medical Formulae. 2017;23:18–23. (in Chinese) [Google Scholar]
- Wang L.Y., Tang Y.P., Liu X., Zhu M., Tao W.W., Li W.X., et al. Effects of ferulic acid on antioxidant activity in Angelicae sinensis Radix, Chuanxiong Rhizoma, and their combination. Chinese Journal of Natural Medicines. 2015 doi: 10.1016/S1875-5364(15)30032-7. [DOI] [PubMed] [Google Scholar]
- Wang S., Ma H.Q., Sun Y.J., Qiao C.D., Shao S.J., Jiang S.X. Fingerprint quality control of Angelica sinensis (Oliv.) Diels by high-performance liquid chromatography coupled with discriminant analysis. Talanta. 2007;72:434–436. doi: 10.1016/j.talanta.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Wang Y., Chen X.T., Zhao C.C., Miao J., Mao X.H., Li X., et al. Effects of temperature during processing with wine on chemical composition, antioxidant capacity and enzyme inhibition activities of Angelica Sinensis Radix. International Journal of Food Science and Technology. 2017;52:1324–1332. [Google Scholar]
- Wei F.G., Yuan C., Chen Z.J., Wang Y., Xu J., Zhang Y.Q., et al. Integrated chemical and transcriptomic analysis reveals the distribution of protopanaxadiol- and protopanaxatriol-type saponins in Panax notoginseng. Molecules (Basel, Switzerland) 2018;23:1773. doi: 10.3390/molecules23071773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G.F., Yang F., Wei F.G., Zhang L.J., Gao Y., Qian J., et al. Metabolomes and transcriptomes revealed the saponin distribution in root tissues of Panax quinquefolius and Panax notoginseng. Journal of Ginseng Research. 2019;16:1–13. doi: 10.1016/j.jgr.2019.05.009. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W.L., Huang L.F. Simultaneous determination of ferulic acid and phthalides of Angelica sinensis based on UPLC-Q-Tof/Ms. Molecules (Basel, Switzerland) 2015;20:4681–4694. doi: 10.3390/molecules20034681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W.L., Zeng R., Gu C.M., Qu Y., Huang L.F. Angelica sinensis in China-A review of botanical profile, ethnopharmacology, phytochemistry and chemical analysis. Journal of Ethnopharmacology. 2016;190:116–141. doi: 10.1016/j.jep.2016.05.023. [DOI] [PubMed] [Google Scholar]
- Wen X.X., Niu Y., Wang S.F. HPLC analysis and characteristic chromatograms of active components in different medicinal parts of Angelicae Sinensis Radix. Chinese Journal of Pharmaceutical Analysis. 2014;34:317–324. [Google Scholar]
- Wu G.X., Deng Y. Advances in pharmacological studies of promoting blood circulation effect of medicinal root parts of Angelica sinensis. TCM Research. 2016;29(9):73–77. (in Chinese) [Google Scholar]
- Wu Y.Y., Wang L., Liu G.X., Xu F., Shang M.Y., Cai S.Q. Characterization of principal compositions in the roots of Angelica sinensis by HPLC-ESI-MSn and chemical comparison of its different parts. Journal of Chinese Pharmaceutical Sciences. 2014;23:393–402. [Google Scholar]
- Xu J., Chen H.B., Liu J., Kwok K.Y., Yue R.Q., Yi T., et al. Why are Angelicae Sinensis Radix and Chuanxiong Rhizoma different? An explanation from a chemical perspective. Food Research International. 2013;54:439–447. [Google Scholar]
- Xu J., Chu Y., Liao B.S., Xiao S.M., Yin Q.G., Bai R., et al. Panax ginseng genome examination for ginsenoside biosynthesis. Gigascinence. 2017;6:1–15. doi: 10.1093/gigascience/gix093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W.X., Hua Y.L., Guo Y.S. Changes of compositon in different parts of Angelicae sinensis based on geoherbs. Journal of Gansu Agricultural University. 2012;1:149–154. (in Chinese) [Google Scholar]
- Yan H., Duan J.A., Shang E.X., Hua Y.Q., Qian D.W. Study on chemical materials and drug nature association of efficacy orientation of different parts from Angelicae Sinensis Radix. Chinese Traditional and Herbal Drugs. 2014;45:3208–3211. (in Chinese) [Google Scholar]
- Yang J., Wang J.Q., Ding W.J., Li W.H., Liu H.W. Study on transcriptome characteristic of Angelica Sinensis Radix by Illumina Hi Seq 2000 sequencing. Chinese Traditional and Herbal Drugs. 2015;46(8):1216–1222. [Google Scholar]
- Yang X.B., Zhao Y., Li G.L., Wang Z.Z., Lv Y. Chemical composition and immuno-stimulating properties of polysaccharide biological response modifier isolated from Radix Angelica Sinensis. Food Chemistry. 2008;106:269–276. [Google Scholar]
- Yang X.J., Deng Y., Yang Z., et al. Effect of different medicinal part in Angelica sinensis on the promoting blood in acute blood stasis rats. The Chinese Journal of Clinical Pharmacology. 2018 [Google Scholar]
- Zeng M., Zhang J., Yang Y., Jin Y., Xiao W., Wang Z., et al. An automated dual-gradient liquid chromatography–MS/MS method for the simultaneous determination of ferulic acid, ligustrazine and ligustilide in rat plasma and its application to a pharmacokinetic study. Journal of Pharmaceutical and Biomedical Analysis. 2014;88:354–363. doi: 10.1016/j.jpba.2013.08.038. [DOI] [PubMed] [Google Scholar]
- Zhan J.Y., Ya P., Cathy W.C.B., Ken Y.Z., Wendy L.Z., Jian P.C., et al. The sulfur-fumigation reduces chemical composition and biological properties of Angelicae Sinensis Radix. Phytomedicine. 2014;21:1318–1324. doi: 10.1016/j.phymed.2014.07.002. [DOI] [PubMed] [Google Scholar]
- Zhang J.J., Su H., Zhang L., Liao B.S., Xiao S.M., Dong L.L., et al. Comprehensive characterization for ginsenosides biosynthesis in ginseng root by integration analysis of chemical and transcriptome. molecules. 2017;22:889. doi: 10.3390/molecules22060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Y., Gu Z.R., Ding J.X., Wang Y.P., Sun Y.J., Wang Y.L. NIR fingerprints of different medicinal parts of Angelicae Sinensis Radix. Journal of Chinese Medicinal Materials. 2015;38:1413–1415. [PubMed] [Google Scholar]
- Zhao K.J., Dong T.T.X., Tu P.F., Song Z.H., Lo C.K., Tsim K.W.K. Molecular genetic and chemical assessment of radix Angelica sinensis in China. Journal of agricultural and food chemistry. 2003;51:2576–2583. doi: 10.1021/jf026178h. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Sun J.H., Yu L.L., Chen P. Chromatographic and mass spectrometric fingerprinting analyses of Angelica sinensis (Oliv.) Diels-derived dietary supplements. Analytical and Bioanalytical Chemistry. 2013;405:4477–4485. doi: 10.1007/s00216-012-6668-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W.Z., Shi J.S. Evaluation of the quality of Angelica sinensis by high-performance liquid chromatography. Analytical Letters. 2014;47:2857–2870. [Google Scholar]