Abstract
Objective
Pulmonary infectious diseases (PID) include viral pneumonia (VP) and pulmonary tuberculosis (PT). Mongolian medicine (MM) is an effective treatment option in China, however, the core group medicines (CGMs) in the treatment of PID and their underlying therapeutic mechanisms remain unclear. In this study, through the method of data mining, the CGMs of MM for the treatment of PID were excavated, and the possible mechanism of action of the CGMs in the treatment of PID was explored by using network pharmacology.
Methods
First, 89 MM formulae for the treatment of pulmonary infectious diseases collected from Gan Lu Si Bu, Meng Yi Jin Kui, People's Republic of China Ministry of Health Drug Standards (Mongolian Medicine Volume), Standard of Mongolian Medicine Preparations in Inner Mongolia (2007 Edition), and Standard of Mongolian Medicine Preparations in Inner Mongolia (2014 Edition). The CGMs of MM for PID were excavated through association rule analysis and cluster analysis. Then, the active ingredients and potential targets of the CGMs were obtained from TCMSP, TCMIP, BATMAN-TCM databases. PID targets information was collected from OMIM, GeneCards, and DrugBank databases. The possible targets of CGMs treatment for PID were obtained by intersection. The PPI network was constructed through the STRING database, and the topology analysis of the network was performed. Through the enrichment analysis of the intersection targets by R language, the main action pathways and related target proteins of CGMs in the treatment of PID were screened out. The results were verified by molecular docking.
Results
A total of 89 formulae were included, involving 164 MM herbs. The efficacy of the drugs was mainly cough-suppressing and panting-calming herbs, and heat-clearing herbs. The nature and flavor were mainly bitter and cold. The CGMs of MM to treatment of PID was excavated as the classic famous formula Sanzi Decoction (Toosendan Fructus-Chebulae Fructus-Gardeniae Fructus). A total of 28 candidate components and 237 predicted targets of CGMs were collected, and 61 common targets with PID were obtained, including key compounds such as quercetin, kaempferol, β-sitosterol and stigmastero and key targets such as VEGFA, IL6, TP53, AKT1. KEGG enrichment analysis yielded AGE-RAGE signaling pathways, IL-17 signaling pathways, and TNF signaling pathways. Molecular docking results showed that the key targets were well matched with the potential active ingredients of CGMs.
Conclusion
This study found that MM commonly used cough-suppressing and panting-calming herbs in combination with heat-clearing herbs to treat PID, and the CGMs for the treatment of PID is “Toosendan Fructus-Chebulae Fructus-Gardeniae Fructus”. CGMs mainly play a role in the treatment of PID by acting on VEGFA, IL6, TP53, AKT1 and other targets, regulating AGE-RAGE signaling pathways, IL-17 signaling pathways, and TNF signaling pathways.
Keywords: data mining, Mongolian medicine, network pharmacology, pulmonary tuberculosis, viral pneumonia
1. Introduction
Pulmonary infectious diseases (PID) include viral pneumonia (VP) and pulmonary tuberculosis (PT). According to estimates by the World Health Organization (WHO), approximately 450 million patients are diagnosed with pneumonia worldwide each year, of whom approximately 4 million (7% of pneumonia deaths) die from VP (Mathers et al., 2003). Viral infections account for 25%–50% of non-bacterial pneumonia and are its main cause of death (Kao et al., 2017, Burk et al., 2016). Primary symptoms of VP include fever, cough with or without phlegm, shortness of breath, and pharyngalgia. Despite its significance, there is a lack of relevant research data on the pathogenesis of VP, which limits its clinical treatment. The U.S. Food and Drug Administration (FDA) has approved the use of antiviral neuraminidase inhibitors such as oseltamivir and peramivir (Dandachi & Rodriguez-Barradas, 2018) in the treatment of VP. However, antiviral drug efficacy is negatively affected by the high degree of respiratory virus variability, emergence of drug-resistant strains, and adverse drug reactions. According to the “Global Pulmonary Tuberculosis Report 2019” released by the WHO, tuberculosis is one of the top 10 causes of death globally and is responsible for the greatest number of deaths caused by a single infectious pathogen. Currently, antibiotics remain the primary treatment for patients with PT. However, with the widespread application of anti-tuberculosis drugs, the global incidence of drug-resistant PT is gradually increasing. As a result, the multidrug resistance rate is as high as 82%, which has made the treatment of PT significantly more difficult (Liu et al., 2021). Thus, the search for highly effective antiviral drugs and anti-tubercle bacilli drugs are gaining an increasing level of research interest.
The curtain rose on the application of traditional medicine millennia ago, and a >2800-year history of traditional Mongolian medicine (MM) has continued thereafter (Robesan, 1922). Throughout history, the use of MM has developed distinctive ethnic, regional, and clinical characteristics, and it is characterized by the incorporation of theories and advanced clinical experiences of various ethnic groups in Han, Tibet, and other regions in China (Zhang et al., 2015). MM has a long history of treating PID. MM believes that “VP” belongs to the category of “Pulmonary fever”, which is due to the fact that “Badagan” (Badagan is feminine, belongs to water, and its attribute is cold) is prevalent, and “Xila” (Xila is masculine, is a fire source, and its attribute is heat) fever is flourishing in the pulmonary. Treatment should focus on clearing heat; PT belongs to the category of “pulmonary carbuncle” is due to the fact that “Badagan” is too prosperous, and “Xi la” fever is blazing in the pulmonary, so this disease occurs. Therefore, “VP” and “PT” have a common cause. MM has distinctly affected the treatment of PID, and has had a therapeutic impact on the disease treatment via multiple targets and pathways dysregulated in PID. However, the underlying mechanisms by which MM therapies affect PID treatment remain unclear, which restricts the internationalization and standardization of MM use in the treatment of PID.
Data mining technology has the potential to improve the assessment of the medication mode of the clinical upper agent, identify potential compatibility rules from existing medication methods, and provide references for subsequent clinical applications (Tang, Shen, & Yang, 2019, Wu et al., 2021, Wu et al., 2021). However, these studies do have limitations. Effects of MM occur via multicomponent, multitarget, and multipathway mechanisms, it makes MM uniquely effective in the treatment of complex diseases. However, it also brings out difficulty with regard to clarifying the therapeutic mechanisms of MM. The emergence of network pharmacology has helped clarify the therapeutic effects of MM (Cao et al., 2020). Network pharmacology is a new method for studying the biologically active components, potential drug targets, and molecular mechanisms of MM (Wan et al., 2021). In addition, it can be used to elucidate the pharmacological mechanisms of candidate drugs from an overall perspective, which is consistent with the overall concept of MM theory (Hopkins, 2011). Research has shown that network pharmacology is a new and feasible method for identifying drug candidates in the early stages of drug screening (Deb, 2019). In addition, molecular docking is based on the docking of small molecules to target proteins and is used in basic research studies (Lohning, Levonis, Williams, & Schweiker, 2017).
In this study, representative classes of MM therapies from the Qing Dynasty to the modern period (CE 1751–2014) were selected, which included therapies described in Gan Lu Si Bu (Qing dynasty, CE 1751–1785, One of the three classic works of Mongolian medicine), Meng Yi Jin Kui (Qing dynasty, CE 1829. Mongolian medical prescription monograph, one of the three classic works of mongolian medicine), People's Republic of China Ministry of Health Drug Standards (Mongolian Medicine Volume), and The Standard of Mongolian Medicine Preparations in Inner Mongolia (Technical standards for the preparation, use, inspection and supervision and management of Mongolian medicinal preparations in medical institutions in Inner Mongolia). Gan Lu Si Bu is the one of the three classic works of Mongolian medicine, which laid a solid theoretical foundation for the development of MM. It was written in 1751–1785. Meng Yi Jin Kui also known as Fang Hai, is particularly important since it was the first systematic and complete book of prescriptions in the history of MM. The People's Republic of China Ministry of Health Drug Standards (Mongolian Medicine Volume) was the first, highly scientific Mongolian medicine standard, which was issued by the Ministry of Health in 1998. Moreover, the Standard of Mongolian Medicine Preparations in Inner Mongolia was the first book to include prescription specifications that was issued by Inner Mongolia, and included prescriptions that basically reflect the clinical use of MM therapeutics at the time. In our study, core group medicines (CGMs) in PID treatment were determined by mining PID prescription data from classic MM texts. Then, network pharmacology and molecular docking were used to explore CGMs therapeutic mechanisms in the treatment of PID to provide data needed for further research (Sun, Sun, Yan, Li, & Xin, 2020, Sun, Jiang, Wang, & Liu, 2020, Yu et al., 2021).
2. Materials and methods
2.1. Data source and normalization
In this study, 89 MM formulae for the treatment of PID and 164 MMs were collected from Gan Lu Si Bu (Qing dynasty, CE 1751–1785), Meng Yi Jin Kui (Qing dynasty, CE 1829), People's Republic of China Ministry of Health Drug Standards (Mongolian Medicine Volume) (1998), Standard of Mongolian Medicine Preparations In Inner Mongolia (2007 Edition) (2007), and Standard of Mongolian Medicine Preparations In Inner Mongolia (2014 Edition) (2014) texts. Data were used to establish a database of MMs and perform subsequent analyses. Moreover, the name of each prescription used in MM was standardized based on conventions detailed in the Pharmacopeia of the People's Republic of China (2020 Edition) (Chinese Pharmacopoeia Commission, 2020) and Nei Meng Gu Meng Yao Cai Biao Zhun (1987 Edition) (Mongolian herbal medicine standards promulgated by Inner Mongolia Autonomous Region) (Inner Mongolia Department of Health, 1987).
2.2. Data processing and analysis
A database of MM formulae for the treatment of PID was established using Excel 2013 and was converted into a format compatible with the data mining software. Based on MM medication rules, categories, properties, and tastes associated with single drugs included in MM prescriptions, Studio software was used for implementing a hierarchical clustering algorithm to categorize high-frequency drugs. Further, the Apriori algorithm was used to assess associations between high-frequency drugs.
2.3. CGMs active compound collection
Compounds comprising all herb were identified using the following three traditional Chinese medicine databases: Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) (Ru et al., 2014), Integrative Pharmacology-based Research Platform of TCM (TCMIP) (Xu et al., 2017), and Bioinformatics Analysis Tool for Molecular Mechanisms of Traditional Chinese Medicine Database (BATMAN-TCM) (Liu et al., 2016). Screening criteria for bioactive compounds were set based on the TCMSP database and included oral bioavailability (OB) ≥ 30% and drug-likeness (DL) ≥ 0.18. Putative targets of bioactive compounds were collected from the three previously mentioned TCM databases, and the gene name of each target protein was searched using the UniProt database. Conditions associated with data were reviewed; duplicate, non-human targets were removed; the corresponding gene abbreviation of targets were identified to facilitate follow-up research. A simultaneous herb-compound-target network was constructed to explore active compounds and their potential targets. Core compounds were identified using an herb-compound-target network.
2.4. Therapeutic targets of CGMs in treatment of PID
PID-related therapeutic targets were obtained from OMIM (Amberger & Hamosh, 2017), Genecards (Safran et al., 2010), and DrugBank (Wishart et al., 2018) databases; the search terms used were “Viral Pneumonia” (including Corona Virus Disease 2019), and “Pulmonary Tuberculosis”, which was defined as the key word. Results were restricted to human genes and proteins. Targets with a relevance score above the median value were selected from the GeneCards database. CGMs and PID targets that overlapped were identified using a Venny 2.1. Overlapping CGMs and PID targets identified were considered the best potential targets of CGMs in the treatment of PID.
2.5. Network construction and analysis
First, using the Cytoscape software, an open-source software platform for visualizing complex networks, a “disease chemical composition target” network of CGMs in the treatment of VP and PT was constructed. Second, using the same software, a common mechanism network of CGMs in the treatment of VP and PT was constructed. Third, to obtain protein–protein interaction (PPI) data, candidate targets (common targets) were used as inputs in STRING with a minimum interaction score of >0.7, and the search was limited to “Homo sapiens” visual network graphs were created using Cytoscape. Target interaction network parameters were calculated using NetworkAnalyzer, and core genes of the network were predicted.
2.6. Protein functional enrichment analysis
To elucidate the therapeutic mechanisms of the CGMs on PID, we used the R language Bioconductor biology data package to perform gene GO function and KEGG pathway enrichment analysis on common targets in the PPI network. The three functional categories cell component, molecular function, and biological process were separately assessed to identify target protein functions. According to the statistical method, set P to 0.05 and q to 0.05 to perform GO (gene ontology) biological enrichment analysis and KEGG pathway enrichment analysis, which were visualized using the “ggplot2” package in R.
2.7. Molecular docking
Molecular docking was conducted to validate whether CGMs were capable of binding targets. Two-dimensional structures of the top 10 core compounds (top 10 degrees in the herb-compound-target network) were downloaded from TCMSP and PubChem databases. Structures were charged and displayed rotatable keys using AutoDock Tools. The protein crystal structures corresponding to the core target genes (top 10 degrees in the PPI network) were downloaded from the Protein Data Bank (PDB) (Burley et al., 2017). The screening conditions were set as follows: 1) the protein structure was obtained using X-ray diffraction; 2) the protein crystal resolution was <3 Å; 3) the species was Homo sapiens. Water and other hetero molecules of proteins were removed using AutoDock Tools, and hydrogen atoms and charge operations of proteins were added. The 3D Grid box for molecular docking simulation was also obtained using AutoDock tools and displayed using AutoDock Vina (Trott & Olson, 2010). Pymol software was used to visualize docking results.
3. Results
3.1. Herb frequency and analysis
The 89 formulae in the analysis included 164 MM herbs, of which 43 were used more than five times, and that with the highest frequency was Glehniae Radix, which occurred 45 times at a frequency of 50.56%. Next, Bambusae Concretio Silicea was identified 44 times (49.43%), and Aucklandiae Radix was identified 41 times (46.06%). There were 43 MM that were used >5 times. Details regarding MM herb identification are included in Table 1.
Table 1.
Frequency analysis of main Mongolian medicines formulae for treatment of pulmonary infectious diseases.
| Mongolian medicines | Frequency | Rate (%) | Mongolian medicines | Frequency | Rate (%) |
|---|---|---|---|---|---|
| Glehniae Radix | 45 | 50.56 | Amomi Fructus Rotundus | 9 | 10.11 |
| Bambusae Concretio Silicea | 44 | 49.43 | Cinnamomi Cortex | 9 | 10.11 |
| Aucklandiae Radix | 41 | 46.06 | Coptidis Rhizoma | 8 | 8.98 |
| Glycyrrhizae Radix et Rhizom | 39 | 43.82 | Pterocephali Herba | 8 | 8.98 |
| Chebulae Fructus | 36 | 40.44 | Arnebiae Radix | 8 | 8.98 |
| Carthami Flos | 32 | 35.95 | Tsaoko Fructus | 7 | 7.86 |
| Hippophae Fructus | 31 | 34.83 | Dianthi Herba | 7 | 7.86 |
| Santali Albi Lignum | 29 | 32.58 | Cornu Rhinoceri | 7 | 7.86 |
| Gardeniae Fructus | 28 | 31.46 | Gentianae Macrophyllae Radix | 7 | 7.86 |
| Bistortae Rhizoma | 23 | 25.84 | Mel | 7 | 7.86 |
| Artemisiae Scopariae Herba | 20 | 22.47 | Inulae Radix | 6 | 6.74 |
| Lanceae Herba | 20 | 22.47 | Entire Meconopsis | 6 | 6.74 |
| Caryophylli Flos | 19 | 21.34 | Fructus Seu Semen Granati | 6 | 6.74 |
| Piperis Longi Fructus | 19 | 21.34 | Picrorhizae Rhizoma | 5 | 5.61 |
| Toosendan Fructus | 17 | 19.10 | Gypsum Fibrosum | 5 | 5.61 |
| Bovis Calculus | 15 | 16.85 | Akebiae Caulis | 5 | 5.61 |
| Lignum Pterocarpi | 13 | 14.60 | Saigae Tataricae Cornu | 5 | 5.61 |
| Alpine Gentian | 13 | 14.60 | Corydalis Bungeanae Herba | 5 | 5.61 |
| Aconiti Kusnezoffii Folium | 11 | 12.35 | Aconitum Naviculare | 5 | 5.61 |
| Lacca | 11 | 12.35 | Moschus | 5 | 5.61 |
| Rubiae Radix et Rhizoma | 9 | 12.35 | Folium Rhododendri | 5 | 5.61 |
| Myris Ticae Semen | 9 | 12.35 |
3.2. Frequency and analysis of Mongolian medicinal categories
According to the classification standard of Nei Meng Gu Meng Yao Cai Biao Zhun (Mongolian herbal medicine standards promulgated by Inner Mongolia Autonomous Region), we analyzed the efficacy of high-frequency drugs (use frequency ≥5), and sorted 43 MMs with eight functions used 659 times. Among them, the top three most frequently used herbs had cough-suppressing and anti-hyperventilation (245 times, 37.17%), heat-clearing (199 times, 30.19%), and Qu “Badagan” (78 times, 11.83%) effects. The results of the analyses are shown in Table 2.
Table 2.
Frequency of Mongolian medicinal categories.
| Herb category | Frequency | Rate (%) |
|---|---|---|
| Cough-suppressing and panting-calming herbs | 245 | 37.17 |
| Heat-clearing herbs | 199 | 28.37 |
| Qu “Badagan” herbs | 78 | 11.83 |
| Zhen “Heyi” herbs | 53 | 8.04 |
| Qi ta herbs (Chebulae Fructus) | 36 | 5.46 |
| Zao “Xie ri wu su” herbs | 24 | 3.64 |
| “Sha nian” herbs | 24 | 3.64 |
Qu “Badagan” herbs: Anti-cold herbs; Zhen “He yi” herbs: Regulating Qi movement herbs; Zao “Xie ri wu su” herbs: Dryness and dampness herbs; “Sha nian” herbs: Insecticidal and antifungal herbs.
3.3. Analysis of MM properties
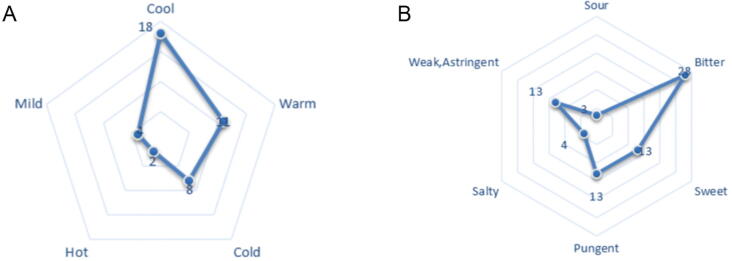
According to the classification system of Neimenggu Mengyaocai Biaozhun, properties and tastes associated with high-frequency drugs (use frequency ≥5) were statistically analyzed. If multiple properties and tastes of MM were identified, they were then counted. Among the 43 MMs, 43 different “nature” and 74 “flavor” were identified; the principal properties of MMs were either cool (observed 18 times, 41.86%) or warm (observed 11 times, 25.58%). The taste most frequently associated with MMs was bitter (28 times, 37.83%). The properties and tastes associated with MMs are shown in Fig. 1.
Fig. 1.
Medicinal natures (A) and flavors (B) of 43 MMs.
3.4. Results of association rules analysis
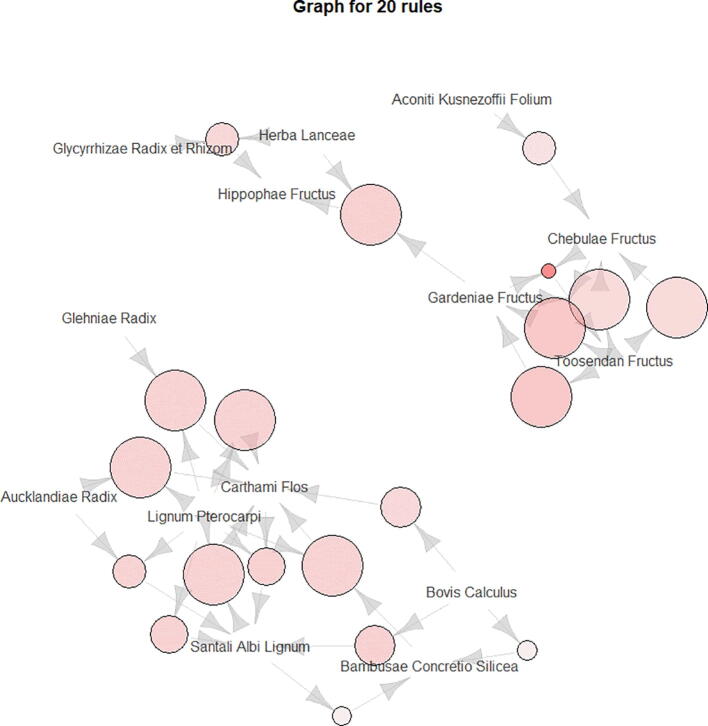
In this study, the R data mining software was used for modeling, while the Apriori algorithm was used to analyze association rules of core herbs. The following parameters were applied: support degree ≥10% and confidence level ≥85% (Xia et al., 2020). A total of 61 herb pairs and herb suit association rules were obtained (Table 3); correlation rules are shown in Fig. 2. The herb pair identified with the highest degree of confidence and strong support was Chebulae Fructus -Toosendan Fructus -Gardeniae Fructus, with a confidence of 100% and a support of 19.31%. According to the Apriori algorithm in association rule, it is considered that the two drug pairs with the highest support are the two drug pairs with the highest probability of simultaneous occurrence in prescription. Therefore, we believe that Chebulae Fructus -Toosendan Fructus -Gardeniae Fructus are the core group medicines of Mongolian medicine for the treatment of PID.
Table 3.
Top 15 association rules of herbs in Mongolian medicine formulae.
| No. | Herbal suits | Support (%) | Confidence (%) | Lift |
|---|---|---|---|---|
| 1 | Toosendan Fructus=>Gardeniae Fructus | 19.31 | 100 | 3.14 |
| 2 | Toosendan Fructus=>Chebulae Fructus | 19.31 | 100 | 2.44 |
| 3 | Gardeniae Fructus, Toosendan Fructus=>Chebulae Fructus | 19.31 | 100 | 2.44 |
| 4 | Chebulae Fructus, Toosendan Fructus=>Gardeniae Fructus | 19.31 | 100 | 3.14 |
| 5 | Lignum Pterocarpi=>Carthami Flos | 14.77 | 100 | 2.75 |
| 6 | Lignum Pterocarpi, Santali Albi Lignum=>Carthami Flos | 13.63 | 100 | 2.75 |
| 7 | Aucklandiae Radix, Lignum Pterocarpi=>Carthami Flos | 12.50 | 100 | 2.75 |
| 8 | Bistortae Rhizoma, Bovis Calculus=>Santali Albi Lignum | 11.36 | 100 | 3.03 |
| 9 | Bistortae Rhizoma, Bovis Calculus=>Carthami Flos | 11.36 | 100 | 2.75 |
| 10 | Glehniae Radix, Lignum Pterocarpi=>Carthami Flos | 10.22 | 100 | 2.75 |
| 11 | Bambusae Concretio Silicea, Lignum Pterocarpi=>Carthami Flos | 10.22 | 100 | 2.75 |
| 12 | Gardeniae Fructus, Lanceae Herba=>Hippophae Fructus | 10.22 | 100 | 2.83 |
| 13 | Lanceae Herba, Santali Albi Lignum=>Bambusae Concretio Silicea | 10.22 | 100 | 2.00 |
| 14 | Bovis Calculus, Chebulae Fructus=>Santali Albi Lignum | 10.22 | 100 | 3.03 |
| 15 | Bovis Calculus, Glycyrrhizae Radix et Rhizom=>Santali Albi Lignum | 10.22 | 100 | 3.03 |
Fig. 2.
Association rule diagram. The circle size represents Confidence and the color represents Lift. The larger the circle, the darker the color, and the denser the arrows, indicating the stronger the connection between the drugs and the higher the credibility.
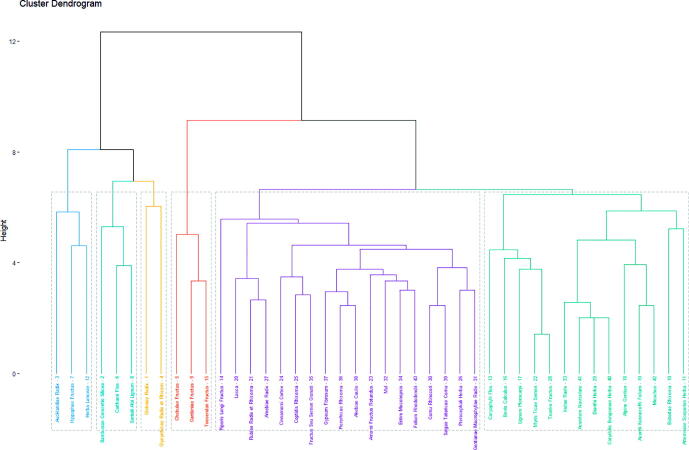
3.5. Cluster analysis results
The R hierarchical clustering algorithm was used to cluster 43 high-frequency drugs (use frequency ≥5), using the R Studio software. The drugs were clustered into six categories, and a cluster dendrogram was generated. The cluster analysis is shown in Fig. 3. We found that the core group medicines screened by association rule analysis were clustered into one group (the fourth group), further illustrating the importance of core group medicine and the accuracy of association rule analysis. Therefore, we took Chebulae Fructus -Toosendan Fructus -Gardeniae Fructus as the core group medicine to further explore its mechanism of action in the treatment of PID.
Fig. 3.
Cluster analysis tree diagram. The first group: Aucklandiae Radix, Hippophae Fructus, Lanceae Herba. The second group: Bambusae Concretio Silicea, Carthami Flos, Santali Albi Lignum. The third group: Glehniae Radix, Glycyrrhizae Radix et Rhizom. The fourth group: Chebulae Fructus, Toosendan Fructus, Gardeniae Fructus. The fifth group: Piperis Longi Fructus, Lacca, Rubiae Radix et Rhizoma, Arnebiae Radix, Cinnamomi Cortex, Coptidis Rhizoma, Fructus Seu Semen Granati, Gypsum Fibrosum, Picrorhizae Rhizoma, Akebiae Caulis, Amomi Fructus Rotundus, Mel, Entire Meconopsis, Folium Rhododendri, Cornu Rhinoceri, Saigae Tataricae Cornu, Pterocephali Herba, Gentianae Macrophyllae Radix. The sixth group: Caryophylli Flos, Bovis Calculus, Lignum Pterocarpi, Myris Ticae Semen, Tsaoko Fructus, Inulae Radix, Aconitum Naviculare, Dianthi Herba, Corydalis Bungeanae Herba, Alpine Gentian, Aconiti Kusnezoffii Folium, Moschus, Bistortae Rhizoma, Artemisiae Scopariae Herba.
3.6. Screening active ingredients of CGMs
Active ingredients of each herb included in the three TCM databases (TCMSP, TCMIP, and BATMAN-TCM databases) were combined. Toosendan Fructus (Chuanlianai in Chinese, number CLZ) and Chebulae Fructus (Hezi in Chinese, number HZ) have seven chemical components, respectively, while Gardeniae Fructus (Zhizi in Chinese, number ZZ) has 14. In addition, the assessment revealed that quercetin (number Q) is found in both Toosendan Fructus and Gardeniae Fructus. Results of CGMs ingredient screening are shown in Table 4.
Table 4.
CGMs active ingredient list.
| No. | Compounds | OB | DL | Chemical structures |
|---|---|---|---|---|
| CLZ1 | Melianone | 40.73 | 0.81 | 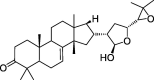 |
| CLZ2 | Stigmasterol | 43.41 | 0.76 | 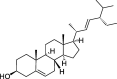 |
| CLZ3 | Balanophonin | 54.74 | 0.40 | 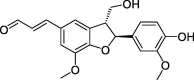 |
| CLZ4 | Ethyl linolenate | 46.10 | 0.20 | |
| CLZ5 | Medioresil | 57.20 | 0.62 | 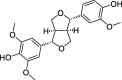 |
| CLZ6 | Nimbolidin B | 30.22 | 0.61 | 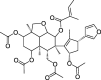 |
| HZ1 | Ellagic acid | 43.06 | 0.43 | 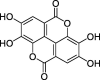 |
| HZ2 | 7-Dehydrosigmasterol | 37.42 | 0.75 | 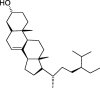 |
| HZ3 | Chebulic acid | 72.00 | 0.32 | 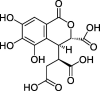 |
| HZ4 | Ellipticine | 30.82 | 0.28 |  |
| HZ5 | Peraksine | 82.58 | 0.78 | 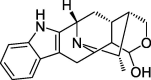 |
| HZ6 | (R)-(6-methoxy-4-quinolyl)-[(2R,4R,5S)-5-vinylquinuclidin-2-yl]methanol | 55.88 | 0.40 | 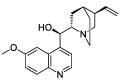 |
| HZ7 | Cheilanthifoline | 46.51 | 0.72 | 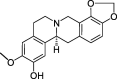 |
| ZZ1 | Crocetin | 35.30 | 0.26 | |
| ZZ2 | Ammidin | 34.55 | 0.22 | 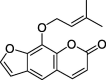 |
| ZZ3 | Sudan III | 84.07 | 0.59 | 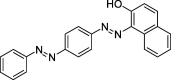 |
| ZZ4 | Beta-sitosterol | 36.91 | 0.75 | 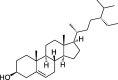 |
| ZZ5 | Kaempferol | 41.88 | 0.24 | 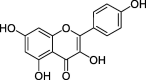 |
| ZZ6 | Stigmasterol | 43.83 | 0.76 | 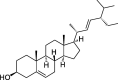 |
| ZZ7 | Mandenol | 42.00 | 0.19 | |
| ZZ8 | Isoimperatorin | 45.46 | 0.23 | 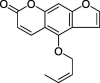 |
| ZZ9 | Ethyl oleate (NF) | 32.40 | 0.19 | |
| ZZ10 | 5-hydroxy-7-methoxy-2-(3,4,5-trimethoxyphenyl)chromone | 51.96 | 0.41 | 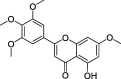 |
| ZZ11 | 3-Methylkempferol | 60.16 | 0.26 | 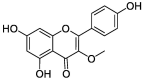 |
| ZZ12 | Artemisetin | 49.55 | 0.48 | 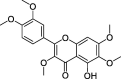 |
| ZZ13 | Sitosterol | 36.91 | 0.75 | 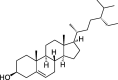 |
| Q | Quercetin | 46.43 | 0.28 |  |
Note: CLZs (1–6) represent active compounds in Toosendan Fructus, HZs (1–7) represent the active compounds of Chebulae Fructus, and ZZs (1–13) represent the active compounds of Gardeniae Fructus. Q is Toosendan Fructus and Gardeniae Fructus common active compounds.
3.7. Prediction of the potential targets of active ingredients
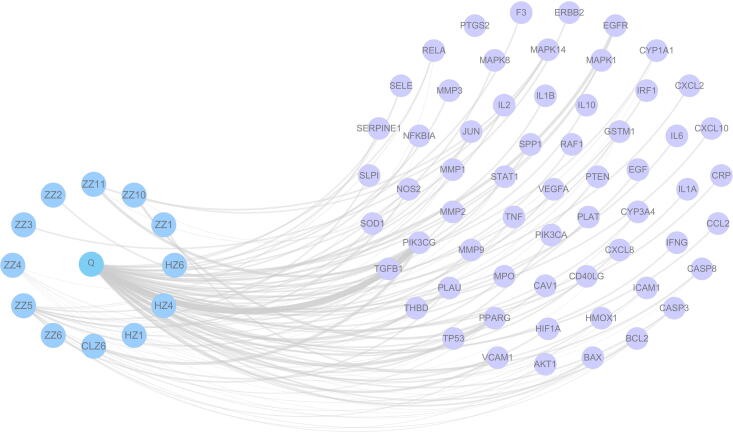
Twenty-nine active ingredients and 702 predicted targets of CGMs were identified; after eliminating overlapping targets, 237 remained. To further clarify interactions between compound and targets, a compound-target network was constructed. As shown in Fig. 4, the MM-compound-target network included a total of 267 nodes (three drugs, 28 compounds, and 237 target nodes) and 686 edges.
Fig. 4.
Network diagram of the compound-targets of the CGMs. Compound-target network. Three herbs are shown as pink triangles, 28 active compounds are shown as circles with different colors, blue quadrilateral is a compound shared by ZZ and ClZ, and 237 target genes are shown as blue hexagon.
3.8. Disease prediction
Using OMIM, GeneCards, and DrugBank databases, 653 and 638 disease targets related to VP and PT, were respectively identified. Targets of both diseases were paired with CGMs component targets, and 79 and 72 CGMs for treating VP and PT, respectively, were identified. Further, 61 targets of CGMs, VP, and PT were shared (Fig. 5).
Fig. 5.
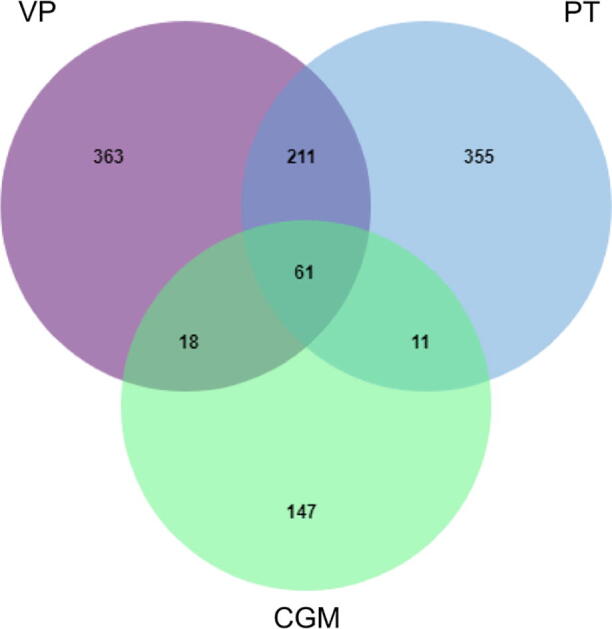
Venn diagram of the targets of the CGMs in the treatment of VP and PT.Venn diagram of drug targets of CGMs and VP and PT disease targets, screened out common targets.
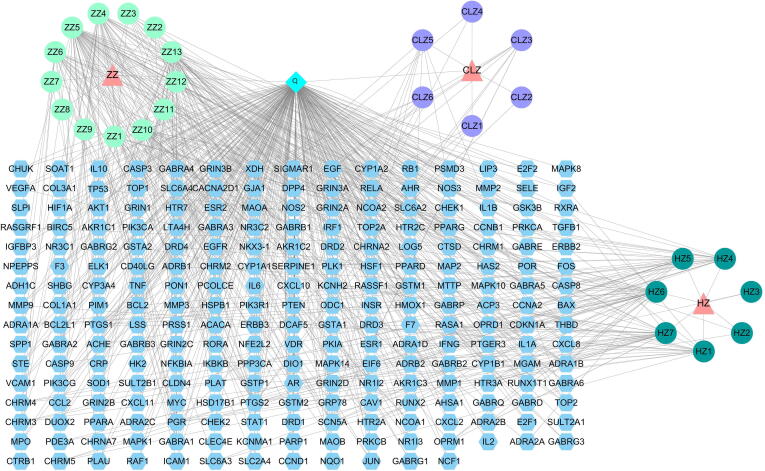
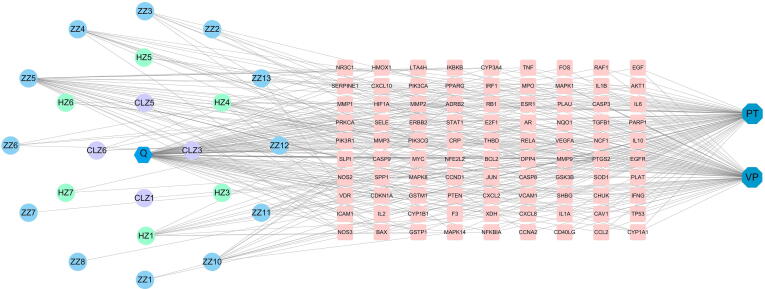
3.9. Common target multi-level network construction
Cytoscape software was used to construct a compound-target network of active ingredients of CGMs associated with VP, PT, and common targets (Fig. 6, Fig. 7). Key targets are arranged according to degree value in Table 5. By including common targets of CGMs in the treatment of VP and PT in the network, it was possible to obtain a network diagram of the common mechanisms of VP and PT treatment (Fig. 7). The network diagram revealed that CGMs co-treatment effects on VP and PD mainly involved 13 chemical components, including quercetin, kaempferol, and ellipticine, and 61 targets, including PTGS2, PPARG, and EGFR.
Fig. 6.
CGMs treatment of VP and PT “disease-compound-target” network. Disease-compound-target network; 23 active compounds are shown as hexagon or circles with different colors, two diseases are shown as a dark blue octagonal, and 90 target genes are shown as pink quadrilateral.
Fig. 7.
CGMs treatment of PID “disease-compound-target” network. Common target mechanism network; 13 active compounds are shown as blue circles and 61 common target genes are shown as purple circles.
Table 5.
CGMs's targets for treatment of PT, VP and PID for three diseases (Top ten degree).
| Diseases | Number of targets | Target name (Top 10 degree) |
|---|---|---|
| PT | 79 | PTGS2, ADRB2, PIK3CG, BAX, BCL2, CASP3, NOS2, GSTM1, CYP1A1, MAPK14 |
| VP | 72 | PTGS2, DPP4, PIK3CG, ESR1, AR, BAX, BCL2, CASP3, NOS3, NOS2 |
| PID | 61 | VEGFA, IL6, TP53, AKT1, TNF, CXCL8, IL1B, PTGS2, MMP9, JUN |
Note: TNF, tumor necrosis factor; CXCL8, interleukin-8; IL1B, interleukin-1 beta; PTGS2, prostaglandin G/H synthase 2; MMP9, matrix metalloproteinase-9; JUN, transcription factor AP-1.
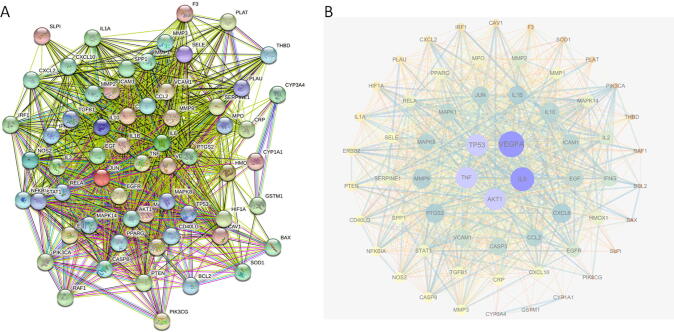
Fig. 8 showed a PPI network of 61 targets. We initially entered common targets into the STRING database and imported nodes 1 and 2, and the binding score information from result files into Cytoscape 3.6.1. The resultant network contained 61 nodes and 1,091 edges, which denoted proteins and the interrelations among proteins, respectively. The larger and darker the node, the greater the value indicated. Similarly, the thicker the edge, the greater the combined score of the connected targets. Using a Network Analyzer topology attribute analysis, we found that nodes with higher betweenness and closeness values tended to have a greater degree of connection than those with lower values. Core targets with betweenness of >0.00432403, closeness of >0.73170732, and degree values of >38 were screened. For example, those with values greater than their respective median values were selected. As a result of this analysis, 35 key proteins were identified, and proteins with the top five scores were vascular endothelial growth factor A (VEGFA), interleukin-6 (IL-6), cellular tumor antigen p53 (TP53), and serine/threonine-protein kinase akt-1 (AKT).
Fig. 8.
PPI network of common targets by STRING database (A) and Cytoscape software (B).
3.10. Module and functional enrichment analysis
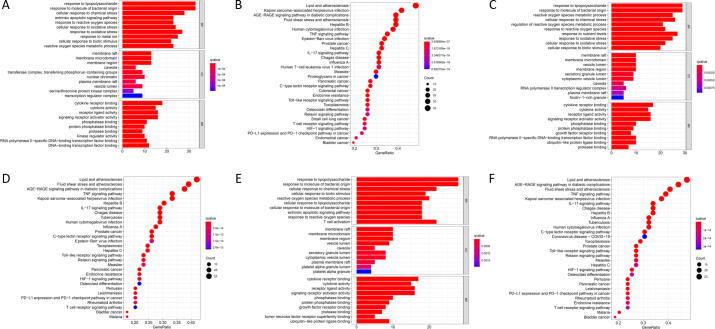
GO and KEGG enrichment analyses of common targets were performed (P < 0.05). The mechanism of drug pair action in the treatment of the disease was also explored. GO function analysis of CGMs in the treatment of VP produced a total of 2,540 GO entries (P < 0.05), and the top 10 biological processes (BP), cellular components (CC), and molecular functions (MF) were selected for visualization (Fig. 9A). In the histogram shown, the redder the color, the higher the degree of enrichment and the greater the potential for the component to be used as a drug target. The results of the BP assessment showed that the active components of modified drug pairs in the human body mainly include the response to lipopolysaccharide (GO:0032496), response to the molecule of bacterial origin (GO:0002237), cellular response to chemical stress (GO:0062197), extrinsic apoptotic signaling pathway (GO:0097191), and response to oxidative stress (GO:0006979). Enriched CC mainly included membrane rafts (GO:0045121), membrane microdomains (GO:0098857), membrane regions (GO:0098589), transferase complexes, and the transfer of phosphorus-containing groups (GO:0061695). Enriched MF mainly included cytokine receptor binding (GO:0005126), cytokine activity (GO:0005125), receptor ligand activity (GO:0048018), signaling receptor activator activity (GO:0030546), and protein phosphatase binding (GO:0019903).
Fig. 9.
GO enrichment analysis (A, C, E) and KEGG analysis (B, D, F) of therapeutic targets of CGMs for VP (A, B), PT (C, D) and common targets (E, F).
Through a KEGG pathway enrichment analysis, a total of 172 related signaling pathways affected by CGMs in the treatment of VP were identified (P < 0.05), and the top 30 items were listed for visual analysis (Fig. 9B). In the bubble diagram shown, the abscissa indicates the gene ratio, the color corresponds to the degree of enrichment, and the bubble size indicates the number of included genes. Enriched pathways identified mainly included the AGE-RAGE signaling in diabetic complications, lipid and atherosclerosis, fluid shear stress and atherosclerosis, Kaposi sarcoma-associated herpesvirus infection TNF signaling, and IL-17 signaling.
A GO function analysis of drug pairs used to treat PT produced a total of 2,435 GO entries (P < 0.05); the top 10 BP, CC, and MF are visualized in Fig. 9C. BP affected by drug pairs included the response to lipopolysaccharide (GO:0032496), response to molecules of bacterial origin (GO:0002237), reactive oxygen species metabolic process (GO:0072593), cellular response to chemical stress (GO:0062197), and regulation of reactive oxygen species metabolic process (GO:2000377). CC identified mainly included membrane rafts (GO:0045121), membrane microdomains (GO:0098857), the vesicle lumen (GO:0031983), and the membrane region (GO:0098589). Enriched MF mainly included cytokine receptor binding (GO:0005126), cytokine activity (GO:0005125), receptor ligand activity (GO:0048018), and signaling receptor activator activity (GO:0030546).
Through a KEGG pathway enrichment analysis, a total of 162 related signaling pathways of drug pairs in the treatment of PT were obtained (P < 0.05); the top 30 items were listed for visual analysis (Fig. 9D). Pathways determined to be enriched mainly included AGE-RAGE signaling in diabetic complications, fluid shear stress and atherosclerosis, lipid and atherosclerosis, TNF signaling, and IL-17 signaling.
A GO function analysis of common targets of drug pairs produced 2,391 GO entries (P < 0.05), and the top 10 BP, CC, and MF were visualized (Fig. 9E). The BP analysis revealed that active components of modified drug pairs in the human body mainly affect the response to lipopolysaccharides (GO:0032496), response to molecules of bacterial origin (GO:0002237), response to chemical stress (GO:0062197), cellular response to biotic stimulus (GO:0071216), and metabolism of reactive oxygen species (GO:0072593). These results suggest that CGMs may affect the treatment of PID by regulating the biological processes identified. CC identified mainly included membrane rafts (GO:0045121), membrane microdomains (GO:0098857), membrane regions (GO:0098589), and the vesicle lumen (GO:0031983). Further, MF identified mainly included cytokine receptor binding (GO:0005126), cytokine activity (GO:0005125), receptor ligand activity (GO:0048018), and signaling receptor activator activity (GO:0030546). These MF may play important roles in the occurrence and development of PID.
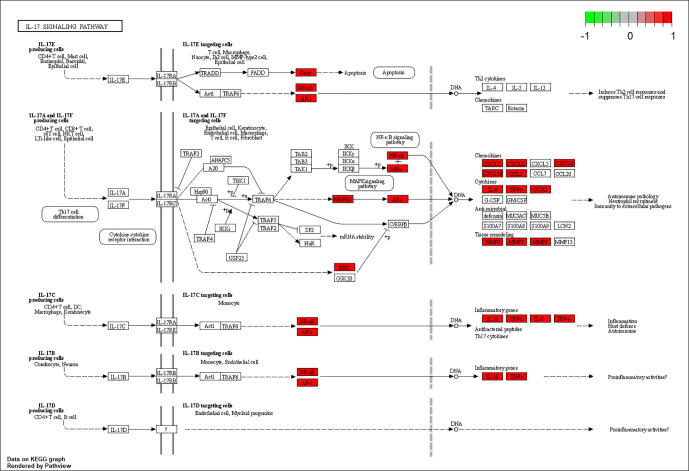
Through KEGG pathway enrichment analysis, a total of 159 related signaling pathways of drug pairs of common targets were obtained (P < 0.05), and the top 30 items were listed for visual analysis (Fig. 9F). Enriched pathways mainly included the AGE-RAGE signaling pathway in diabetic complications, lipid and atherosclerosis, TNF signaling, fluid shear stress, atherosclerosis, and IL-17 signaling. The IL-17 signaling pathway, which plays crucial roles in both acute and chronic inflammatory responses, was selected as an example (Fig. 10). Red labeled nodes indicate the targets of CGMs, which shows that CGMs play a key role in modulating the IL-17 signaling pathway by regulating marker targets.
Fig. 10.
IL-17 signaling pathway. CGMs may further regulate IL-17 signaling pathway by affecting IL6, PTGS2, TNF, JUN, CXCL8, MMP9 and other targets to achieve anti-inflammatory purposes.
3.11. Molecular docking
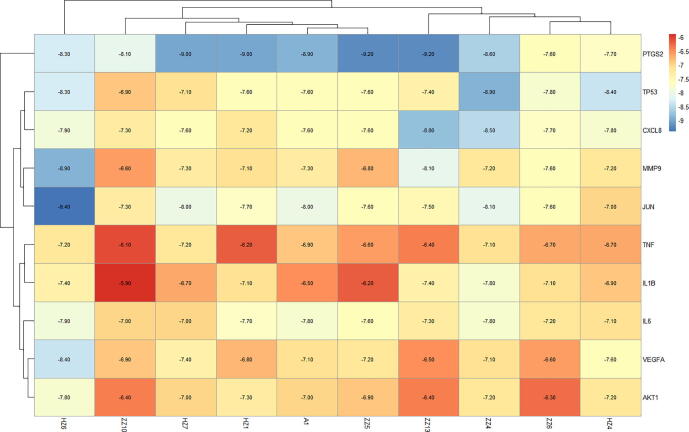
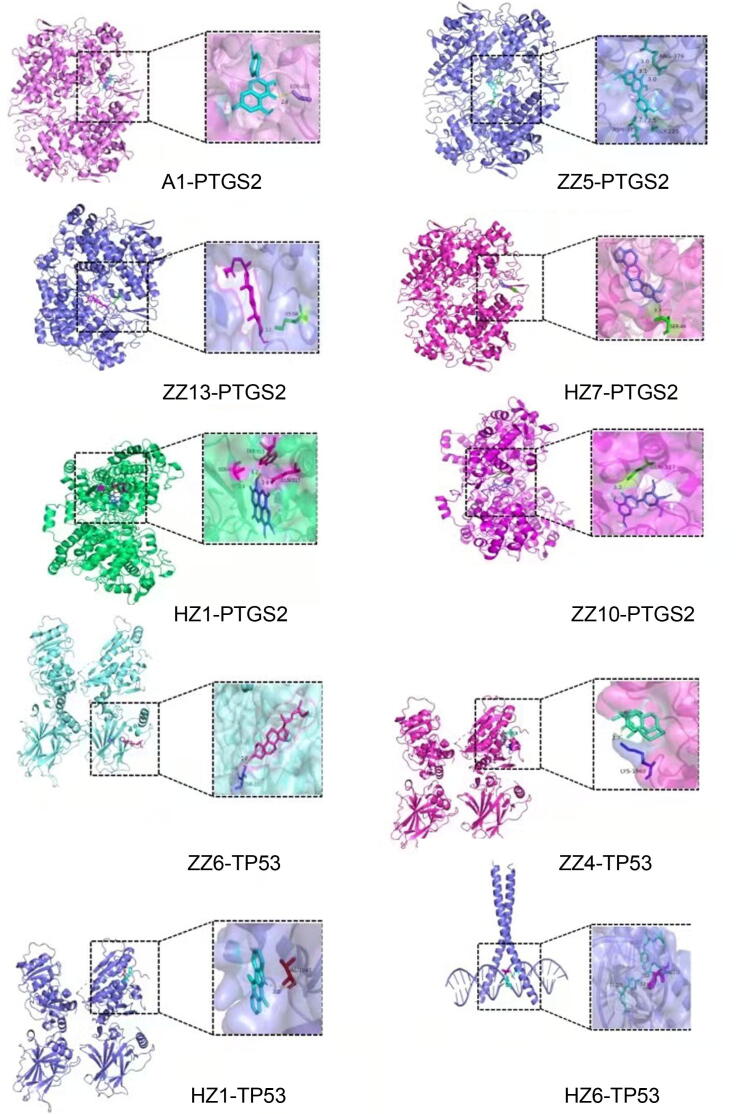
Among the core targets of the PPI network (Fig. 8), we selected those with the top 10 scores for subsequent molecular docking analyses. To analyze the docking results, findings were depicted using a heat map, as shown in Fig. 11. The minimum binding energy needed for binding of each compound to a key protein was determined (Fig. 12). As a result, a semiflexible docking model was adopted. For example, a rigid configuration of the protein versus a flexible configuration of small-molecules was considered. Grid energy was calculated using AutoGrid, and the docking operation was based on a Lamarck genetic algorithm. Docking results showed that the binding energy required for the binding of a compound and a protein was <−5 kcal·mol−1, indicating that each compound and protein was capable of binding tightly.
Fig. 11.
Heat map of binding energy of active ingredient and core target molecule docking. The horizontal is the active ingredient, and the vertical is the core target. Each number represents the binding energy between the active ingredient and the core target. The darker the color, the lower the binding energy and the more stable the binding.
Fig. 12.
Docking modl diagram of compound and key target molecule. The combination with the lowest binding energy to the core target for each active ingredient.
4. Discussion
4.1. Three core medicines we mined are Sanzi Decoction, a famous classical formula in Monglian medicine
In this study, we found that Toosendan Fructus-Chebulae Fructus-Gardeniae Fructus was the core group medicine of Mongolian medicine for the treatment of PID. “Toosendan Fructus-Chebulae Fructus-Gardeniae Fructus” is the composition of the famous classical formulas of Mongolian medicine Sanzi Decoction (it is composed of three herbs of Toosendan Fructus, Chebulae Fructus, Gardeniae Fructu) (Sa & Siqin, 2019), which was first published in the Four Medical Tantras (Wu, Du, & Jin, 2015).
Gardeniside is one of the active and toxic components of Chebulae Fructus-Gardeniae Fructus in Sanzi Decoction. The combination of the three medicines can increase the curative effect and reduce the toxicity of gardeniaside. (Wang, Bao, & Wang, 2015). Sanzi Decoction has the functions of clearing away heat and detoxification and cooling blood, and can be used to treat plague. The Mongolian Medicine Jingui also records that the plague can be treated with Sanzi Decoction for 4–6 d, with heat-clearing and detoxifying (Li & Tu, 2020). Pharmacological studies have found that Sanzi Decoction has the effects of improving immunity, anti-inflammatory and antibacterial (Luo, Liu, Zhou, & Yun, 2001). It has significant antibacterial effect on Staphylococcus aureus, Pseudomonas aeruginosa and Streptococcus (Zhang, Wang, & Bao, 2009). Therefore, it is the traditional basis for analysing the thareputic mechanism of “Toosendan Fructus-Chebulae Fructus-Gardeniae Fructus” in the treatment of pulmonary infectious diseases by network pharmacology.
4.2. CGMS treat PID mainly through anti-inflammatory and antibacterial effects
4.2.1. Quercetin, kaempferol, β-sitosterol and stigmastero might be responsible compounds
In this study, the potential mechanism of “Toosendan Fructus-Chebulae Fructus-Gardeniae Fructus” in treating PID was explored by using network pharmacology method. Among the active ingredients screened, quercetin has a wide range of biological activities, including those that are antiviral, anti-inflammatory, immunomodulatory (Li et al., 2016), and antioxidant (Xu, Hu, Wang, & Cui, 2019). It can reduce inflammatory damage in lipopolysaccharide-caused WI-38 lung fibroblasts by inhibiting NF-κB and JNK signaling pathways, and also inhibit early-stage influenza A virus infection and viral invasion (Wu et al., 2015). Kaempferol has anti-inflammatory, antibacterial, antioxidative stress activity, and antiviral effects. (Zhang et al., 2017). It can inhibit the expression of inflammatory cytokines TNF-α, IL-6, IL-1β and PGE2, improve pulmonary edema, and also inhibit the phosphorylation of NFκB and MAP kinase, exerting anti-inflammatory effects in vitro and in vivo (Sun et al., 2018). Studies showed that β-sitosterol has immunomodulatory, anti-inflammatory, antioxidant and anti-tumor effects (Liu, Ji, & Huang, 2019, Zhi et al., 2020). It can play an anti-inflammatory effect by inhibiting the synthesis of NO and the activity of interleukin-6 (IL-6), and reducing the secretion of inflammatory factors such as IL-1 and TNF-α (Choi et al., 2012). Stigmasterol can exert anti-inflammatory, immune regulation, and increase blood oxygen saturation by acting on CASP3, PTGS2, NOS2, NOS3 and other targets (Lin et al., 2006). Stigmasterol and β-sitosterol also have certain anti-mycobacterial activity (Dai, 2009). At the same time, we found that quercetin, kaempferol, β-sitosterol and stigmasterol are also key compounds of Qingfei Paidu Decoction (Ephedrae Herba, Glycyrrhizae Radix et Rhizoma Praeparata Cum Melle, Armeniacae Semen Amarum, Gypsum Fibrosum, Cinnamomi Ramulus, Alismatis Rhizoma, Polyporus, Atractylodis Macrocephalae Rhizoma, Poria, Bupleuri Radix, Scutellariae Radix, Pinelliae Rhizoma Praeparatum cum Zingibere et Alumine, Zingiberis Rhizoma Recens, Asteris Radix et Rhizoma, Farfarae Flos, Belamcandae Rhizoma, Asari Radix et Rhizoma, Dioscoreae Rhizoma, Aurantii Fructus Immaturus, Citri Reticulatae Pericarpium, Pogostemonis Herba) in the treatment of COVID-19 (Ren, Yang, Zhang, & Gao, 2021). It shows that these compounds may have the effect of inhibiting virus replication and inhibiting the binding of virus spike protein to ACE2, which can prevent SARS-CoV-2 from infecting cells.
4.2.2. VEGFA, IL6, TP53, AKT1, and TNF might be key targets for treatment of PID
The core targets of VEGFA, IL6, TP53, AKT1and TNF were obtained by protein–protein interaction network analysis, suggesting that these targets play a key role in the treatment of PID with “Toosendan Fructus-Chebulae Fructus-Gardeniae Fructus”. VEGFA is one of the main factors that stimulate angiogenesis, and plays an important role in activating signaling pathways and inducing endothelial cell proliferation and migration (Claesson & Wslsh, 2013). Studies showed that elevated VEGFA levels are likely to be associated with resolution of lung inflammation (Strouvalis et al., 2018). As a pro-inflammatory cytokine, IL-6 can stimulate the inflammatory process in various diseases (Tanaka, Narazaki, & Kishimto, 2014). Cytokine storm is an important part of the severe response of COVID-19 patients, and IL6 plays a key role in cytokine release syndrome, which has been used as a clinically important indicator of patient deterioration (Zhang, Wu, Li, Zhao, & Wang, 2020). At the same time, WHO studies have confirmed that IL-6 inhibitors can reduce the risk of death in hospitalized patients with COVID-19 (Tharaux, 2021). TP53 can regulate cell cycle arrest and apoptosis. The experimental study of mycobacterium tuberculosis infection of alveolar epithelial cells showed that, p53 can resist the invasion of Mycobacterium tuberculosis by inhibiting the activation of TLR-4 and NF-κB and reducing the secretion of inflammatory factors (Wang et al., 2019). AKT1 can regulate the proliferation and growth of cells. By acting on JNK, it further promotes cell apoptosis and reduces the viability of mycobacterium tuberculosis (Zhang, et al., 2021). TNF can mediate cell apoptosis, inflammatory response and other processes, and can activate other downstream inflammatory factors, attracting inflammatory cells to the respiratory tract (Tang et al., 2002). At the same time, it was found that TNF is closely related to lung injury and pulmonary fibrosis (Liang, Yuan, Wang, Lei, & Ouyang, 2020).
4.2.3. AGE-RAGE signaling pathways, IL-17 signaling pathways, and TNF signaling pathways might be regulated by CGMs
The results of the KEGG pathway enrichment analysis of “Toosendan Fructus-Chebulae Fructus-Gardeniae Fructus” in the treatment of PID showed that it was mainly enriched in AGE-RAGE signaling pathways, IL-17 signaling pathways, and TNF signaling pathways. The AGE-RAGE signaling pathway can enhance the expression of VGEF, TGF-β, and MCP-1, and promote the release of inflammatory factors, thereby accelerating the inflammatory response, cell apoptosis, and tissue damage (Wendt et al., 2003). Relevant studies showed that the overexpression of IL-17 can cause a large number of neutrophils to aggregate and secrete inflammatory factors, activate the inflammatory response, and cause alveolar endothelial damage and increased tissue permeability (Liu & Wen, 2021). In addition, IL-17 in the peripheral blood of patients with severe pneumonia was significantly increased, and the level of IL-17 was closely related to the development and severity of the patient's disease, suggesting that IL-17 may be a cytokine that plays an important role in the inflammatory response system (Cai et al., 2017). TNF signaling pathway plays an important role in regulating inflammation and immune response in vivo. Relevant studies have shown that the level of TNF-α and the incidence of ARDS in severe patients with COVID-19 are higher than those in non-severe patients (Li et al., 2020). At the same time, the levels of TNF-α and TNF-β in the serum of pulmonary tuberculosis patients were significantly increased, which made Mycobacterium tuberculosis in a relatively static and stable state by promoting tissue damage and further activation of macrophages and phagocytosis of bacteria (Li et al., 2016).
4.3. Limitations of this study
First, due to the small number of included prescriptions, the reliability of the results was affected to a certain extent. Therefore, it is expected that more prescriptions can be collected in the future to improve the accuracy of the results. Second, network pharmacology is a research technology based on network data and computer simulation analysis, and the database used is not updated timely enough. Therefore, the obtained results still need further experimental verification.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (No. 81874336); National Key Research and Development Program of China (No. 2021YFE0190100); Inner Mongolia Autonomous Region Science and Technology Plan Project (No. 2020GG0128); Inner Mongolia natural science (No. 2020MS08123).
Contributor Information
Yuan Gao, Email: gaoyuan_0524@sina.com.
Minhui Li, Email: prof_liminhui@yeah.net.
References
- Amberger J.S., Hamosh A. Searching online mendelian inheritance in man (omim): A knowledgebase of human genes and genetic phenotypes. Current Protocols in Bioinformatics. 2017;58(1):1–12. doi: 10.1002/cpbi.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk M., El-Kersh K., Saad M., Wiemken T., Ranirez J., Cavallazzi R. Viral infection in community-acquired pneumonia: A systematic review and meta-analysis. The European Respiratory Review. 2016;25(140):178–188. doi: 10.1183/16000617.0076-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S.K., Berman H.M., Kleywegt G.J., Markley J.L., Nakamura H., Velankar S. Protein data bank (PDB): The single global macromolecular structure archive. Methods in Molecular Biology. 2017;1607:627–641. doi: 10.1007/978-1-4939-7000-1_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S.S., Han Y.Y., Li Q.F., Chen Y.J., Zhu D., Su Z.H., Guo H.W. Mapping pharmacological network of multi-targeting litchi ingredients in cancer therapeutics. Frontiers in Pharmacology. 2020;11:451. doi: 10.3389/fphar.2020.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B.L., Lin G.H., Xie H.W., Chen J., Chen M.D., Ceng W.C. Study of clinical expression significance of IL-17 in peripheral blood of patients with severe pneumonia. China Modern Medicine. 2017;24(13):17–19. [Google Scholar]
- Choi J.N., Choi Y.H., Lee J.M., Noh I.C., Park J.W., Choi W.S., Choi J.H. Anti-inflammatory effects of β-sitosterol-β-D-glucoside from Trachelospermum jasminoides (apocynaceae) in lipopolysaccharide-stimulated RAW 264.7 murine macrophages. Natural Product Research. 2012;26(24):2340–2343. doi: 10.1080/14786419.2012.654608. [DOI] [PubMed] [Google Scholar]
- Claesson W.L., Wslsh M. VEGFA and tumour angiogenesis. Journal of Internal Medicine. 2013;273(2):114–127. doi: 10.1111/joim.12019. [DOI] [PubMed] [Google Scholar]
- Dai L. Antituberculous triterpenoids and phytosterols in Pandanus tectorius Soland. var. laevis. Drugs & Clinic. 2009;24(2):113. [Google Scholar]
- Dandachi D., Rodriguez-Barradas M.C. Viral pneumonia: Etiologies and treatment. Journal of Investigative Medicine. 2018;66(6):957–965. doi: 10.1136/jim-2018-000712. [DOI] [PubMed] [Google Scholar]
- Deb P.K. Recent updates in the computer aided drug design strategies for the discovery of agonists and antagonists of adenosine receptors. Current Pharmaceutical Design. 2019;25(7):747–749. doi: 10.2174/1381612825999190515120510. [DOI] [PubMed] [Google Scholar]
- Hopkins A.L. Network pharmacology: The next paradigm in drug discovery. Nature Biotechnology. 2011;29(1):14–22. [Google Scholar]
- Kao K.C., Chiu L.C., Hung C.Y., Chang C.H., Yang C.T., Huang C.C., Hu H.C. Coinfection and mortality in pneumonia-related acute respiratory distress syndrome patients with bronchoalveolar lavage: A prospective observational study. Shock Injury, Inflammation and Sepsis. 2017;47(5):615–620. doi: 10.1097/SHK.0000000000000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.C., Xu S.Y., Yu M.Q., Wang K., Tao Y., Zhou Y., Zhao J.P. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. Journal of Allergy and Clinical Immunology. 2020;146(1):110–118. doi: 10.1016/j.jaci.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.Q., Tu Y. Prevention and treatment of Mongolian medicine and Mongolian medicine against new crown pneumonia and epidemic diseases. Modernization of Traditional Chinese Medicine and Materia Medica-World Science and Technology. 2020;22(03):642–644. [Google Scholar]
- Li T., Xiang Y.G., Fan R.H., Ma X.H., Shi G.M., Yu R., Peng X.F. Research progress on cytokines in pulmonary tuberculosis patients. Practical Preventive Medicine. 2016;23(7):894–897. [Google Scholar]
- Li Y., Yao J.Y., Han C.Y., Yang J.X., Chaudhry M.T., Wang S.N., Yin Y.L. Quercetin, inflammation and immunity. Nutrients. 2016;8(3):167. doi: 10.3390/nu8030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Yuan S., Wang X., Lei Y., Ouyang H. Attenuation of pristimerin on TNF-α-induced endothelial inflammation. International Immunopharmacology. 2020;82(5):106326. doi: 10.1016/j.intimp.2020.106326. [DOI] [PubMed] [Google Scholar]
- Lin L.G., Zhong Q.X., Cheng T.Y., Tang C.P., Ke C.Q., Lin G., Ye Y. Stemoninines from the roots of Stemona tuberosa. Journal of Natural Products. 2006;69(7):1051–1054. doi: 10.1021/np0505317. [DOI] [PubMed] [Google Scholar]
- Liu L., Li J., Ma Y., Wu T., Dong N., Cui B., Zhang N. Investigation of drug resistance in 348 cases of pulmonary tuberculosis and its influencing factors. Shaanxi Medical Journal. 2021;50(05):626–629. [Google Scholar]
- Liu L., Wen X.H. Preterm neonatal respiratory distress syndrome serum CD62E, IL-17 expression levels and its relationship with the risk and the degree of disease. Journal of Clinical Pulmonary Medicine. 2021;26(5):713–718. [Google Scholar]
- Liu W.L., Ji Y., Huang A.X. Research and development progress of β-sitosterol. Farm Products Processing. 2019;1 [Google Scholar]
- Liu Z.Y., Guo F.F., Wang Y., Li C., Zhang X.L., Li H.L., He F.C. BATMAN-TCM: A bioinformatics analysis tool for molecular mechANism of Traditional Chinese medicine. Scientific Reports. 2016;6(1):21146. doi: 10.1038/srep21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohning A.E., Levonis S.M., Williams N.B., Schweiker S.S. A practical guide to molecular docking and homology modelling for medicinal chemists. Current Topics in Medicinal Chemistry. 2017;17(18):2023–2040. doi: 10.2174/1568026617666170130110827. [DOI] [PubMed] [Google Scholar]
- Luo S.Q., Liu L.L., Zhou Y.X., Yun X.Y. Study on the content and antiphlogosis of trace elements in Mongolian medicine Sanzi Tang. Guangdong Trace Elements Science. 2001;03:39–41. [Google Scholar]
- Mathers C.D., Bernard C., Iburg K.M., Inoue M., Fat D.M., Shibuya K., Xu H. Global burden of disease in 2002: Data sources, methods and results. Moesgaard Ik, Inoue M, Fat Dm, Shibuya K, Stein C, Tomijima N & Xu H. 2003;54:1–116. [Google Scholar]
- Ren X., Yang J., Zhang W.T., Gao R. Discussion on the mechanisms of Qinfei Paidu decoction in the treatment of early stage of COVID-19 based on network pharmacology. World Chinese Medicine. 2021;16(19):2845–2850. [Google Scholar]
- Robesan . Shanghai Science and Technology Press; 1922. Mongolian medicine; p. 247. [Google Scholar]
- Ru J.L., Li P., Wang J.N., Zhou W., Li B.H., Huang C., Yang L. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. Journal of Cheminformatics. 2014;6(1):1–6. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sa C.L., Siqin T.Y. Research progress of Mongolian medicine compound Sanzi decoction. Journal of Medicine & Pharmacy of Chinese Minorities. 2019;25(03):58–60. [Google Scholar]
- Safran M., Dalah I., Alexander J., Rosen N., Iny S.T., Shmoish M., Lancet D. GeneCards version 3: The human gene integrator. Database. 2010;2010:020. doi: 10.1093/database/baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strouvalis I., Routsi C., Adamopoulou M., Raftogiannis M., Renieris G., Orfanos S.E., Giamarellos B.E.J. Early increase of VEGF-A is associated with resolution of ventilator-associated pneumonia: Clinical and experimental evidence. Respirology. 2018;23(10):942–949. doi: 10.1111/resp.13320. [DOI] [PubMed] [Google Scholar]
- Sun J.H., Sun F., Yan B., Li J.Y., Xin D.L. Data mining and systematic pharmacology to reveal the mechanisms of Traditional Chinese medicine in mycoplasma pneumoniae pneumonia treatment. Biomedicine & Pharmacotherapy. 2020;125(c):109900. doi: 10.1016/j.biopha.2020.109900. [DOI] [PubMed] [Google Scholar]
- Sun X.L., Jiang J.H., Wang Y., Liu S.Y. Exploring the potential therapeutic effect of traditional Chinese medicine on coronavirus disease 2019 (COVID-19) through a combination of data mining and network pharmacology analysis. European Journal of Integrative Medicine. 2020;40:101242. doi: 10.1016/j.eujim.2020.101242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z.J., Li Q., Hou R.R., Sun H.X., Tang Q.H., Wang H.X., Wu S. Kaempferol-3-O-glucorhamnoside inhibits inflammatory responses via MAPK and NF-κB pathways in vitro and in vivo. Toxicology and Applied Pharmacology. 2018;364:22–28. doi: 10.1016/j.taap.2018.12.008. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Narazaki M., Kishimto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harbor Perspectives in Biology. 2014;6(10) doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S.H., Shen D., Yang H.J. Analysis on composition rules of Chinese patent drugs treating pain-related diseases based on data mining method. Chinese Journal of Integrative Medicine. 2019;25(11):861–866. doi: 10.1007/s11655-017-2957-z. [DOI] [PubMed] [Google Scholar]
- Tang S.J., Xiao H.P., Fan Y.H., Wu F.R., Zhang Z.S., Li H., Yang Y. Changes of proinflammatory cytokines and their receptors in serum from patients with pulmonary tuberculosis. Chinese Journal of Tuberculosis and Respiratory Diseases. 2002;06:8–12. [PubMed] [Google Scholar]
- Tharaux P.L. Association between administration of IL-6 antagonists and mortality among patients hospitalized for COVID-19: A meta-analysis. The Journal of the American Medical Association. 2021;326(12):499–518. doi: 10.1001/jama.2021.11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.Y., Bao Y.M., Wang Y.Y. HPLC determination of the content of geniposide in Mongolia medicine complex soup of Gardenia, Toosendan Fruit and Terminalia Fruit. Lishizhen Medicine and Materia Medica Research. 2015;26(06):1321–1322. [Google Scholar]
- Wang Y., Bai G.B., Wang J., Zeng J., Liu X.M., Wang Y. Effect of p53 signaling pathway on AEC Ⅱ line a549 infected with mycobacterium tuberculosis (MTB) Journal of Northwest A & F University (Natural Science Edition) 2019;47(8):002. [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q., Shan D., Hu P.F., Liang C.G.L., Zhao M., Zhang Q.S. Based on network pharmacology and molecular docking explore the mechanism of Mongolian medicine Sanwei Tanxiang decoction in the treatment of ischemic heart disease. Modernization of Traditional Chinese Medicine and Materia Medica-World Science and Technology. 2021;23(11):3971–3985. [Google Scholar]
- Wendt T., Tanji N., Guo J.C., Hudson B.I., Bierhaus A., Ramasamy R., Schmidt A.M. Glucose, glycation, and RAGE: Implications for amplification of cellular dysfunction in diabetic nephropathy. Journal of the American Society of Nephrology. 2003;14(5):1383–1395. doi: 10.1097/01.asn.0000065100.17349.ca. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Wilson M. DrugBank 5.0: A major update to the drugbank database for 2018. Nucleic Acids Research. 2018;6(D1):D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.D., Du G.E., Jin H. Research progress and prospect of Mongolian medicine Sanzi decoction. Journal of Medicine & Pharmacy of Chinese Minorities. 2015;21(09):49–51. [Google Scholar]
- Wu W.J., Li R.C., Li X.L., He J., Jiang S.B., Liu S.W., Yang J. Quercetin as an antiviral agent inhibits influenza a virus (IAV) entry. Viruses. 2015;8(1):6. doi: 10.3390/v8010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.Q., Wang X., Pan R.L., Huang X.F., Li Y.H., Jiang L. Study of the relationship between ICU patient recovery and TCM treatment in acute phase: A retrospective study based on python data mining technology. Evidence-Based Complementary and Alternative Medicine. 2021;2021(1):1–6. doi: 10.1155/2021/5548157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D.Y., Hou X.T., Xia Z.S., Hao E.W., Xie J.J., Liang J.Y., Deng J.G. Analysis on oral medication rules of traditional Chinese medicine prescriptions for prevention of COVID-19. Chinese Herbal Medicines. 2021;13(4):502–517. doi: 10.1016/j.chmed.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Gao K., Xie J.D., Sun W., Shi M., Li W., He W.M. Data Mining-Based Analysis of Chinese Medicinal Herb Formulae in Chronic Kidney Disease Treatment. 2020;1(24):9719872. doi: 10.1155/2020/9719872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Hu M.J., Wang Y.Q., Cui Y.L. Antioxidant activities of quercetin and its complexes for medicinal application. Molecules. 2019;24(6):1123. doi: 10.3390/molecules24061123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H.Y., Liu Z.M., Fu Y., Zhang Y.Q., Yu J.J., Guo F.F., Yang H.J. Exploiture and application of an internet-based computation platform for integrative pharmacology of traditional Chinese medicine. China Journal of Chinese Materia Medica. 2017;42(18):3633–3638. doi: 10.19540/j.cnki.cjcmm.2017.0141. [DOI] [PubMed] [Google Scholar]
- Yu M.X., Song X., Ma X.Q., Hao C.X., Huang J.J., Yang W.H. Investigation into molecular mechanisms and high-frequency core TCM for pulmonary fibrosis secondary to COVID-19 based on network pharmacology and data mining. Annals of Palliative Medicine. 2021;10(4):3960–3975. doi: 10.21037/apm-20-1384. [DOI] [PubMed] [Google Scholar]
- Zhang C., Wu Z., Li J.W., Zhao H., Wang G.Q. Cytokine release syndrome in severe COVID-19: Interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. International Journal of Antimicrobial Agents. 2020;55(5):105954. doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.H., Zhao Z.Y., HaSi B.T.E., Li Z.H., Wu M.M., Zou D.Z., Li M.H. Mongolian folk medicine—from traditional practice to scientific development. China Journal of Chinese Materia Medica. 2015;40(13):2492–2495. [PubMed] [Google Scholar]
- Zhang Y.W., Shao D.Y., Shi J.L., Zhu J., Huang Q.S., Yang H. A review on biological activities of kaempferol. Chinese Bulletin of Life Sciences. 2017;29(4):400–405. [Google Scholar]
- Zhang D.L., Wang T., Bao C.H. Experimental study on antibacterial activity of Mongolian medicine Sanzi decoction in vivo. Journal of Medicine & Pharmacy of Chinese Minorities. 2009;15(5):31–32. [Google Scholar]
- Zhang G.W., Ou M., Zhou X.F., Tao X.Y., Fu J.P., Fu X.D. MiR-20a-5p negatively regulates jnk2 gene expression in human macrophages infected with mycobacterium tuberculosis. Electronic Journal of Emerging Infectious Diseases. 2021;6(3):198–201. [Google Scholar]
- Zhi H.J., Jin X., Zhu H.Y., Li H., Zhang Y.Y., Lu Y., Chen D.F. Exploring the effective materials of flavonoids-enriched extract from Scutellaria baicalensis roots based on the metabolic activation in influenza a virus induced acute lung injury. Journal of Pharmaceutical and Biomedical Analysis. 2020;177:112876. doi: 10.1016/j.jpba.2019.112876. [DOI] [PubMed] [Google Scholar]