Abstract
Objective
Plant hormones act as chemical messengers in the regulation of plant development and metabolism. The production of ginsenosides in Panax hybrid is promoted by auxins that are transported and accumulated by PIN-FORMED (PIN) and PIN-LIKES (PILS) auxin transporters. However, genome-wide studies of PIN/PILS of ginseng are still scarce. In current study, identification and transcriptional profiling of PIN/PILS gene families, as well as their potential relationship with ginsenoside biosynthesis in Panax ginseng were investigated.
Methods
PIN/PILS genes in P. ginseng was identified via in silico genome-wide analysis, followed by phylogenetic relationships, gene structure, and protein profiles investigation. Moreover, previously reported RNA-sequence data from various tissues and roots after infection were utilized for PIN/PILS genes expression pattern analysis. The Pearson’s correlation analysis of specific PIN/PILS genes expression level and main ginsenoside contents were taken to reveal the potential relationship between auxin transports and ginsenoside biosynthesis in P. ginseng.
Results
A genome-wide search of P. ginseng genome for homologous auxin transporter genes identified a total of 17 PIN and 11 PILS genes. Sequence alignment, putative motif organization, and sub-cellular localization indicated redundant and complementary biological functions of these PIN/PILS genes. Most PIN/PILS genes were differentially expressed in a tissue-specific manner, and showed significant correlations with ginsenoside content correspondingly. Eight auxin transporter genes, including both PIN and PILS subfamily members, were positively correlated with ginsenoside content (cor > 0.60; P-value <0.05). The expression levels of eleven auxin transporter genes were increased dramatically in the early stage (0–0.5 DPI) after Cylindrocarpon destructans infection, accompanied with various overall expression patterns, implying the dynamic auxin transport in response to biotic stress.
Conclusion
Based on the results, we speculate that the accumulation or depletion in temporal or spatial manner of auxin by PIN/PILS transporters involved in the regulation of HMGR activity and subsequent ginsenoside biosynthesis.
Keywords: auxin, genome-wide, ginsenoside, PILS, PIN, transcriptome
1. Introduction
Ginseng (Panax ginseng C.A. Meyer) has been used clinically as tonic or adaptogenic agent for millennia, and nowadays incorporated in functional foods, cosmetics, and beverages (Mancuso and Santangelo, 2017, Yan et al., 2014). Ginsenosides, almost exclusively be found in Panax plants, exert the major pharmacological activities (Kim et al., 2015, Leung and Wong, 2010). Tissue culture, bioconversion, and other synthetic biology techniques have been applied to produce ginsenosides to overcome the resource scarcity and meet the market demand (Bulgakov et al., 1998, Palazón et al., 2003). The biosynthetic pathway of ginsenosides and expression pattern of related regulatory genes have been investigated by comparative transcriptome (Cao et al., 2015). Xu et al. (2017) performed whole-genome sequencing of P. ginseng, that facilitates genome-wide investigation and reference genome-based transcriptomic profiling of ginsenoside biosynthesis and regulation.
The phytohormone auxin participates in plant growth, development, and responses to environmental stress through its spatiotemporally regulated local accumulation or depletion (Benková et al., 2003). As defense molecules in plant stress and pathogen interactions, ginseng saponins production in the hairy roots of a Panax hybrid increased approximately 1.7-fold with the addition of auxins (0.5 μmol/L 3-indole butyric acid and 1.0 μmol/L 1-naphthaleneacetic acid) (Washida, Shimomura, Takido, & Kitanaka, 2004). The activity of 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR), the first rate-limiting enzyme in triterpene ginsenoside biosynthesis through the mevalonic acid (MVA) pathway, is modified at the transcriptional and post-translational level, including Pleiotropic Regulatory Locus1 (PRL1) (Tholl & Lee, 2011) and negative regulation by protein phosphatase 2A (PP2A) (Leivar et al., 2011). Since PRL1 functions as a regulator of stress and hormone response and PP2A is indeed involved in abscisic acid and auxin signaling, it is assumed that phytohormone and stress response contribute to regulate the HMGR activity (Úrsula et al., 2010; Kim et al., 2015). Furthermore, both MVA and methylerythritol phosphate (MEP) pathways have been shown to be coordinated by hormone molecules via PRL1 (Flores-Pérez et al., 2010). However, the regulatory mechanism controlling genes/enzymes relative to plant triterpene biosynthesis is still little known.
The multiple functions exerted by auxin are largely dependent on its uneven distribution, which is primarily achieved via the vascular system and polarized auxin transport system (Friml et al., 2003, Tanaka et al., 2006). As the prominent auxin transporters, the PIN-FORMED (PIN) protein family consists of eight members in Arabidopsis thaliana (Paponov, Teale, Trebar, Blilou, & Palme, 2005), among which PIN1–4 and PIN7 are localized to the plasma membrane and govern directional, cell-to-cell auxin transport (Adamowski & Friml, 2015). In contrast, the endoplasmic reticulum-localized PIN5, 6, and 8, together with members of the PIN-LIKE (PILS) family of auxin efflux transporters, modulate intracellular auxin homeostasis (Barbez et al., 2012, Cazzonelli et al., 2013, Ding et al., 2012, Mravec et al., 2009). PIN genes from rice (Wang et al., 2009), sorghum (Shen et al., 2010), maize (Yue et al., 2015), potato (Efstathios, Bjorn, Marian, Visser, & Bachem, 2013), and soybean (Wang et al., 2015) and their functions have been characterized. Liu et al. (2017) found the tissue specific expression profile of the PIN genes (PIN2, PIN3, and PIN6) in P. ginseng, and speculated that these three genes probably involved in the tropism growth of ginseng roots and the growth and development of the aerial part. To the contrary, the role of PILS genes remains quite unclear (Barbez et al., 2012, Mohanta et al., 2015).
Regulating ginseng yield and ginsenoside content are essential to meet the demands in clinical treatment and health care. Considering the pivotal roles of auxin and its transporters in growth regulation and ginsenoside biosynthesis, we performed a genome-wide analysis and characterized the PIN/PILS auxin transporter gene families, as well as tissue expression patterns, their potential correlation with ginsenoside synthesis, and the gene expression profile after infection with Cylindrocarpon destructans. The current results provide a new perspective to understanding the molecular basis and regulatory mechanisms of PIN/PILS in P. ginseng, including their roles in ginsenoside production and response to environmental stress.
2. Materials and methods
2.1. Sequence retrieval and identification
After generating the draft assembly of the P. ginseng genome and predicting a total of 42,006 protein-coding genes in our previous work (Xu et al., 2017), all annotated genes were initially screened by BLAST using the genomic sequences of PIN and PILS genes from A. thaliana as queries to identify 28 auxin transport candidates. The amino acid sequences of these candidates were then screened for the presence of a transmembrane domain with the local PEAM profile hidden Markov model (Pfam: PF03547.16), which identified 11 PIN and 17 PILS candidates considered to be authentic targets (alignment length ≥55 amino acids [aa], E-value ≤0.0001). The PIN/PILS dataset of A. thaliana was obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org/index.jsp), while Oryza sativa, Medicago truncatula, and Populus trichocarpa datasets were retrieved from Phytozome 12.1.4 (https://phytozome.jgi.doe.gov/pz/portal.html). Accession numbers of PIN/PILS proteins were reported in Table S1.
2.2. Multiple alignment and phylogenetic analyses
Multiple protein sequence alignments of PIN or PILS were performed using ClustalW in BioEdit Sequence Alignment Editor and were visualized with the WEBLOGO program (http://weblogo.berkeley.edu) to identify conserved amino acid residues. Full-length protein amino acid sequences were used to construct a phylogenetic tree, by employing Jones, Taylor, and Thornton substitution models with parameters of estimated proportion of invariable sites (I) and estimated g-distribution (G). An unrooted neighbor-joining phylogenetic tree of 111 PIN/PILS proteins was constructed using MEGA 7.0 with the following parameters: pairwise deletion option, 1000 bootstrap replicates, and Poisson correction distance. The display, manipulation, and annotation of phylogenetic trees were performed using the Interactive Tree of Life (iTOL, http://itol.embl.de/).
2.3. Sequence features and gene structure
The exon–intron structure of each PIN/PILS gene from P. ginseng (PgPIN/PgPILS) was determined by aligning the full-length cDNA sequence or predicted coding sequence (CDS) with the genomic sequence. Gene structure was displayed by the cross-platform program TBtools 0.53 (http://cj-chen.github.io/tbtools). Subsequently, the online ExPASy server (https://www.expasy.org/) was used to predict the theoretical isoelectric point (pI) and molecular weight of PgPIN/PgPILS proteins. Conserved motifs in 17 PgPINs and 11 PgPILSs were identified using MEME (Suite version 4.12.0, http://meme-suite.org/index.html) with previously reported criteria (Chu et al., 2018). Finally, the MAST program was used to search for detected motifs (Bailey & Gribskov, 1998).
The subcellular localization of PgPIN/PgPILS proteins was predicted by WoLFPSORT (Horton et al., 2007) based on the 14 nearest neighbors’ features (https://wolfpsort.hgc.jp/). HMMTOP 2.0 (Tusnady and Simon, 2001) (http://www.enzim.hu/hmmtop/html/submit.html) and TMHMM Server 2.0 (Krogh, Larsson, von Heijne, & Sonnhammer, 2001) (http://www.cbs.dtu.dk/services/TMHMM/) were used to predict the structures of PgPIN/PgPILS proteins, which were visualized by TMRPres2D (Spyropoulos, Liakopoulos, Bagos, & Hamodrakas, 2004).
2.4. Digital gene expression analysis
Gene expression patterns across different organs of P. ginseng were analyzed using 11 RNA-Seq datasets from NCBI (SRP066368) (Wang et al., 2015), while nine additional RNA-Seq datasets (PRJNA369187 and PRJNA381509) were utilized to compare different tissues (Zhang et al., 2017). Clean reads were separately aligned to assemble the P. ginseng genome in the orientation mode using TopHat (http://tophat.cbcb.umd.edu/) (Langmead & Salzberg, 2012). Cuffdiff (http://cufflinks.cbcb.umd.edu/) was used for differential expression analysis (Trapnell et al., 2013), using the fragments per kilobase of exon per million mapped reads (FPKM) method to calculate the expression level for each transcript and identify differentially expressed genes (DEGs) among different samples (Wang, Feng, Wang, Wang, & Zhang, 2010). DEGs were identified based on log2 fold change > 2 and false discovery rate (FDR) < 0.05. C. destructans infection was analyzed in seven datasets from P. ginseng fibrous roots, where firstly showed the corresponding symptoms (such as reddish-brown spots) at 0.25, 0.5, 1, 4, 7, and 12 days post incubation (DPI) (SRR1639601) (Gao et al., 2016). The fibrous roots harvested from the uninfected P. ginseng plants served as controls (0 DPI). At each time point, the fibrous roots from 3 to 5 P. ginseng plants were rinsed with distilled water and then pooled to address the problems of insufficient sample biomass and lack of biological repetition.
Pearson’s correlation test was performed using the R package “Psych” between ginsenoside content and the FPKM of PgPIN/PgPILS genes in the nine tissues of the main roots of P. ginseng (Zhang et al., 2017). Co-expression analyses between PgPIN/PgPILS genes and triterpene saponin biosynthetic enzyme-encoding genes were performed in all 27 RNA-Seq datasets.
2.5. Availability of data and materials
The necessary information of public data used in this study are present within the article. Genome sequencing data of P. ginseng are available via NCBI under the project number PRJNA385956. The latest versions of the genome assemblies and annotation are available through our website at http://ginseng.vicp.io:23488/. The GenBank accession numbers for the PgPIN/PgPILS genes sequences reported in this paper is summarized in Table S2.
3. Results
3.1. Sequence features and phylogenetic analysis
Seventeen PgPINs containing open reading frames (ORFs) were identified, with the ORFs being more similar to those of P. trichocarpa than of A. thaliana, M. truncatula, and O. sativa (Fig. 1 and Table 1). PgPINs gene length varied from 1,373 bp (PgPIN5a) to 5,693 bp (PgPIN6a), while the length of cDNAs ranged from 864 bp (PgPIN5a) to 3,043 bp (PgPIN3a) (Table 2). The molecular weights of the predicted proteins ranged from 32,003.30 Da (PgPIN5c) to 71,248.39 Da (PgPIN3b), and their isoelectric points were predicted to range from 6.42 (PgPIN5a) to 9.73 (PgPIN8b) (Table 2). Similarly, 11 PgPILSs containing ORFs were also characterized (Fig. 1 and Table 2). The number of PILS genes in P. ginseng was similar to that of P. trichocarpa and Medicago truncatula but greater than that of A. thaliana and O. sativa (Fig. 1 and Table 1). PgPILSs gene length varied from 1380 bp (PgPILS2a and PgPILS2b) to 11,577 bp (PgPILS6b), while the length of cDNAs ranged from 747 bp (PgPILS6a) to 1812 bp (PgPILS1b) (Table 2). The molecular weights of the predicted proteins ranged from 27,130.08 Da (PgPILS6a) to 50,871.47 Da (PgPILS2a), and the isoelectric points were predicted to range from 4.94 (PgPILS1e) to 9.75 (PgPILS1b) (Table 2).
Fig. 1.

Phylogenetic analysis of PILS and PIN proteins. Phylogenetic relationships of the PINs and PILSs auxin transporters from Oryza sativa (Os), Arabidopsis thaliana (At), Medicago truncatula (Mt), Populus trichocarpa (Pt), and Panax ginseng (Pg). The bootstrap value is represented by the radius of blackspot on branch. Accession numbers of PIN/PILS proteins are reported in Supplementary material Table S1. The ginseng genes are shown in bold font.
Table 1.
Size of PIN/PILS gene families in different plant species.
| Species | No. of PINs | No. of PILSs |
|---|---|---|
| Oryza sativa | 12 | 6 |
| Arabidopsis thaliana | 8 | 7 |
| Medicago truncatula | 12 | 9 |
| Panax ginseng | 17 | 11 |
| Populus trichocarpa | 16 | 13 |
Table 2.
Gene and protein features of 28 PgPINs/PgPILSs.
| No. | Gene length/bp | mRNA length/bp | Protein length/aa | Molecular weight | pI | Exon No. |
|---|---|---|---|---|---|---|
| PgPILS1a | 8261 | 1657 | 370 | 40 502.34 | 9.41 | 11 |
| PgPILS1b | 5660 | 1812 | 312 | 34 258.37 | 9.75 | 11 |
| PgPILS1c | 3357 | 1146 | 382 | 41 071.45 | 9.14 | 10 |
| PgPILS1d | 1633 | 828 | 276 | 30 465.82 | 5.89 | 6 |
| PgPILS1e | 4397 | 1059 | 353 | 38 608.46 | 4.94 | 9 |
| PgPILS2a | 1380 | 1380 | 460 | 50 841.29 | 6.01 | 1 |
| PgPILS2b | 1380 | 1380 | 460 | 50 871.47 | 6.27 | 1 |
| PgPILS5a | 2529 | 1340 | 363 | 39 681.61 | 5.33 | 10 |
| PgPILS5b | 2241 | 1479 | 379 | 41 661.24 | 6.89 | 10 |
| PgPILS6a | 2734 | 747 | 249 | 27 130.08 | 8.99 | 5 |
| PgPILS6b | 11,577 | 1563 | 353 | 38 232.51 | 8.15 | 9 |
| PgPIN1a | 3580 | 1949 | 543 | 59 505.59 | 9.13 | 4 |
| PgPIN1b | 5472 | 2547 | 608 | 66 563.21 | 9.17 | 6 |
| PgPIN1c | 3785 | 2349 | 601 | 64 672.86 | 9.16 | 7 |
| PgPIN1d | 3379 | 2092 | 595 | 63 947.06 | 9.10 | 7 |
| PgPIN1e | 2957 | 2283 | 546 | 58 912.69 | 9.18 | 4 |
| PgPIN2a | 2455 | 1923 | 641 | 70 664.68 | 8.71 | 6 |
| PgPIN2b | 2599 | 1848 | 616 | 66 969.12 | 8.89 | 5 |
| PgPIN3a | 3882 | 3043 | 657 | 71 216.34 | 7.28 | 7 |
| PgPIN3b | 3826 | 2995 | 657 | 71 248.39 | 7.28 | 6 |
| PgPIN3c | 2899 | 2231 | 650 | 70 706.57 | 6.90 | 6 |
| PgPIN5a | 1373 | 864 | 288 | 32 435.75 | 6.42 | 2 |
| PgPIN5b | 3161 | 1033 | 344 | 38 009.73 | 6.43 | 6 |
| PgPIN5c | 2953 | 873 | 291 | 32 003.30 | 8.76 | 3 |
| PgPIN6a | 5693 | 1996 | 544 | 59 605.75 | 8.71 | 7 |
| PgPIN6b | 1927 | 1053 | 351 | 38 656.57 | 6.44 | 2 |
| PgPIN8a | 2499 | 930 | 310 | 34 360.91 | 9.39 | 4 |
| PgPIN8b | 2670 | 1023 | 341 | 37 189.51 | 9.73 | 5 |
An unrooted phylogenetic tree was constructed using the PIN and PILS proteins in P. ginseng, M. truncatula, O. sativa, A. thaliana, and P. trichocarpa for subfamily design (Fig. 1). All P. ginseng proteins were named based on their relationship with known A. thaliana PINs and PILSs, namely, the cluster of PIN and PILS families from A. thaliana. The 111 proteins from the above five plant species can be divided into two separate subtrees, suggesting that PILS and PIN proteins are highly conserved in subfamilies but evolutionarily distinct from each other among angiosperms. It was also found that PINs could be divided into eight groups, PIN1, PIN2, PIN3, PIN5, PIN6, PIN8, PIN9, and PIN10, while PILSs could be divided into four groups, PILS1, PILS2, PILS5, and PILS6. No P. ginseng members were found in the PIN9 or PIN10 groups, which may have evolved independently in monocots (Adamowski and Friml, 2015, Balzan et al., 2014). PIN3 and PIN6 exhibited dicot-specificity. Within other groups, PINs from each species clustered separately. PIN1 and PILS1 proteins exhibited more distant evolutionary relationships, suggesting that additional clusters could be obtained. This neighbor-joining tree contained 28 PIN/PILS members identified in P. ginseng, which indicated a total of 10 distinct subfamilies (Fig. 1). In addition, PIN1 subfamily members from 1 to 5 were dramatically extended in P. ginseng compared to that in A. thaliana.
3.2. Gene structure and protein profile analysis
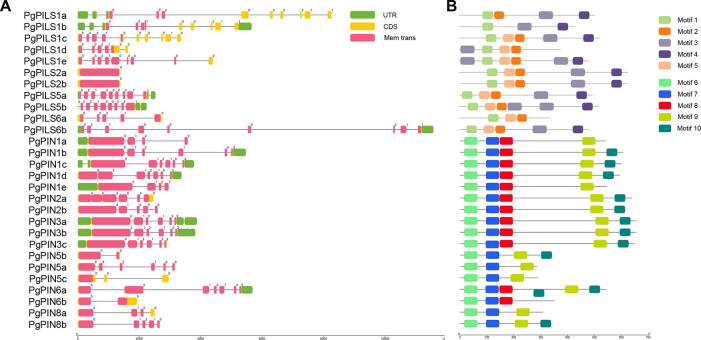
The PIN/PILS genes of P. ginseng contained a conserved intron–exon organization in several groups (Fig. 2A). The average exon numbers for PgPIN2, PgPIN5, PgPILS2, PgPILS3, and PgPILS8 were 1, 10, 5.5, 6.3, and 4.5, respectively, with the standard deviation being <1. However, gene structures in the PgPIN1 and PgPILS1 groups were more variable. The PgPIN1 group contained five members with the number of exons varying from 6 to 11, with a standard deviation of up to 2.07. Similarly, the PgPILS1 group contained five members with the number of exons varying from 4 to 7, with a standard deviation of 1.52. As a major factor affecting gene structure, the variations in total intron length (2925 bp vs. 674 bp) were primarily responsible for the striking differences in gene structure between the largest gene (PgPILS1b, 5472 bp) and the smallest gene (PgPILS1e, 2957 bp) in the PgPILS1 group.
Fig. 2.
Structural features and conserved motifs of PgPINs and PgPILSs. (A) Structural features of PgPINs and PgPILSs. The UTR is represented by green round-cornered rectangles with black lines connecting as introns. The exons were emphasized by a yellow color with transmembrane domains in pink. The numbers above the rectangles corresponded to the exon phase; (B) the conserved motifs of PgPINs and PgPILSs. A representation of each protein is shown with conserved motifs drawn as color boxes. The sequences of each motif in individual proteins are given in the supplementary materials.
Similar to other plants, the P. ginseng PIN/PILS proteins exhibited a highly conserved hydrophobicity profile, with two hydrophobic segments located at the N- and C-termini being linked with a central hydrophilic loop (Figs. 2B and 3). All PgPIN and PgPILS proteins possessed 8–10 transmembrane segments (Fig. 2A and Table S3). P. ginseng PINs and PILSs could be classified as long and short based on the length of the predicted protein and the central hydrophilic loop (Feraru et al., 2012, Viaene et al., 2013). The long PINs consisted of 10 members (543–657 aa), including all genes from the PIN1, PIN2, and PIN3 groups; the short PINs and PILSs contained 14 members from the PIN5, PIN8, PILS1, PILS5, and PILS6 (249–379 aa) groups (Fig. 3). Four members of the PIN6 and PILS2 groups had proteins with lengths (351–544 aa) between the typical long and short types. The length of the central hydrophilic loop was approximately 300 aa for members of the long PINs and 50–100 aa for members of the short PINs/PILSs (Fig. 3C). Multiple sequence alignment and conserved motifs revealed that the sequences of the N- and C-terminal transmembrane segments were highly conserved in PgPIN and PgPILS proteins, but with high heterogeneity in the central hydrophilic loop (Figs. S1 and S2). Ten conserved PIN and PILS protein motifs were discovered using MEME and were characterized with high conservation in both their combination and relative position (Fig. 2B and Table S41). Approximately 64% of PgPILS members contained five motifs in a fixed order (motif 1-5-2-3-4) at the C- and N-termini. Motif combinations such as motif 6-7-8 were found in all long-type PgPINs at the C-terminal, while motif 9-10 was found at the N-terminal in 80% of the long-type PgPINs. Motif 8 was absent in two typical short PIN subfamilies (PgPIN5 and PgPIN8). To understand the molecular function of these proteins in P. ginseng, we analyzed their predicted subcellular localization. Most long PgPIN proteins were predicted to be localized in the plasma membrane, while the majority of the short proteins were predicted to be localized in the vacuoles, plasma membrane, and endoplasmic reticulum (Fig. 3B and Table S5).
Fig. 3.
Structure of typical PIN/PILS proteins in P. ginseng. (A) Predicted topology of three typical PIN/PILS proteins. These predictions were made using TMHMM 2.0 and visualized by TMRPres2D; (B) Predicted location of PgPIN/PgPILS proteins depending on the 14 nearest neighbor features by WoLFPSORT; (C) Predicted topology of three typical PIN/PILS proteins. These predictions were made by HMMTOP 2.0.
3.3. Expression profile of PgPIN/PgPILS genes in various organs and tissues
Upon analyzing the expression levels of the 28 PgPIN/PgPILS genes in samples from 11 different organs of P. ginseng, all genes were detected in at least one organ, and seven transport genes were highly expressed in one organ (Fig. 4C and Table S6). Among these seven genes, the seeds expressed three specific PgPINs (PgPIN1d, PgPIN5a, and PgPIN5b), while the roots expressed two specific PgPINs and one PgPILS that were associated with the growth period (PgPIN2b in 12-year root and PgPIN6b in 25-year root; PgPILS1c in 12-year root). PgPIN5c was highly expressed in the fruit pedicel. Five of the seven specific transport genes were not of the typical long type. The remaining 21 PgPIN/PgPILS genes also exhibited altered expression patterns in different organs. As seen in the heatmap (Fig. 4), all identified PgPIN/PILS genes clustered into four distinct groups, while eleven samples clustered into three individual groups with specific expression patterns. A total of 13 PgPIN/PgPILS genes belonging to typical short-type subfamilies (PILS1, PILS5, PILS6, PIN5, and PIN8) were differentially expressed, with high levels in the leaf blade and pedicel. Ten PgPIN/PgPILS genes, including six long and two short genes, were more highly expressed in the roots (Fig. 4), while PgPILS1d and PgPILS6b were more highly expressed in the stem.
Fig. 4.
Heatmap representing partial expression profiles of PgPIN/PgPILS genes. (A) Heatmap showing the FPKM values of 28 PgPIN/PgPILS genes upon Cylindrocarpon destructans infection; (B) Clustering and differential expression analysis of 28 PgPIN/PgPILS genes in cortex, periderm, and stele; (C) Clustering and differential expression analysis of 28 PgPIN/PgPILS genes in various organs. The FPKM values were transformed to log2 (value + 1). The color scale is shown at the right, and higher expression levels are shown in red. The genes with FPKM values of 0 in all samples were not represented.
To further analyze PgPIN/PgPILS expression in P. ginseng roots, the expression of 28 PgPIN/PgPILS genes was examined in three tissues of the main root: the cortex, periderm, and stele. Based on the expression patterns of the 28 PgPIN/PgPILS genes, nine samples were clustered into three distinct groups (Fig. 4B). Four PgPIN/PgPILS genes, including three seed-specific genes, were not detected in any tissue, and 24 PgPIN/PgPILS genes were detected in at least one tissue. Ten PgPINs and seven PgPILSs across eight subfamilies were more highly expressed in periderm tissue; 10 of the 17 PgPIN/PgPILS genes were of the short type. In the stele, three PgPIN1 and all PgPILS2 members were more highly expressed, none of which were of the short type. Comparing to stele, both PgPILS5 members were more highly expressed in the cortex and periderm (Table S6).
3.4. Expression profile of PgPIN/PgPILS genes after C. destructans infection
Given that the phytohormone signaling network coordinates the production of defense-related proteins and secondary metabolites during the plant stress response (Gao et al., 2016, Park et al., 2017, Rahman and Punja, 2005), we performed a dynamic transcriptome analysis following infection with C. destructans. Based on their expression patterns at 0, 0.25, 0.5, 1, 4, 7, and 12 DPI, 28 genes were clustered into four distinct groups. At T0 (0 DPI), PgPIN1a, PgPIN5a, PgPIN8a, PgPILS1a, and PgPILS6a showed significantly higher expression, but their expression decreased quickly after infection with C. destructans. PgPIN2a, PgPIN2b, PgPIN8b, PgPILS2a, and PgPILS5a expression increased dramatically at T1 (0.25 DPI), with PgPIN2b and PgPIN8b only being expressed temporarily. At T2 (0.5 DPI), the expression levels of six PIN/PILS genes, namely, PgPIN1c, PgPIN1d, PgPIN3a, PgPIN3b, PgPIN5b, and PgPILS1e, increased, while those of PgPIN1e, PgPIN3c, and PgPIN5c decreased. During the later stages of infection (T3–T6), the expression of PgPIN6a and five PgPILSs, namely, PgPILS1b, PgPILS1d, PgPILS2b, PgPILS5b, and PgPILS6b, increased tardily. Notably, at T5 (4 DPI), the expression levels of PgPIN1b, PgPIN6b, and PgPILS1c increased temporarily (Fig. 4 and Table S6).
3.5. Co-expression analyses of PgPIN/PgPILS genes and their relationship with ginsenoside biosynthesis
To explore the potential correlation between these PgPIN/PgPILS proteins and ginsenoside accumulation, Pearson’s correlation test was performed between individual or total ginsenoside levels and expression of the 28 PgPIN/PgPILS genes (Table S7). Eight auxin transporter genes, including four PgPIN and four PgPILS subfamily members, with all four PgPILSs belonging to the PILS1 group, were positively correlated with ginsenoside content (cor > 0.60; P-value <0.05). Furthermore, PgPILS1a, PgPILS1b, and PgPILS1c were more strongly correlated than PgPILS1e, while PgPIN2a, PgPIN3c, PgPIN5c, and PgPIN1e were more strongly correlated with protopanaxadiol (PPD)-type ginsenosides than with protopanaxatriol (PPT)-type ginsenosides (Fig. 5A). Among these eight positively correlated genes, five encoded typical short-type auxin transporters. Additionally, two PgPILS2 members, three PgPIN1 members, and PgPIN3b, four of which encoded typical long-type auxin transporters, were negatively correlated with ginsenoside accumulation (cor < −0.60; P-value <0.05). Typically, a gene regulated by a specific metabolite is often co-expressed with the genes encoding the enzymes involved in its biosynthetic pathway (De Geyter, Gholami, Goormachtig, & Goossens, 2012). Therefore, the co-expression of eight candidates that positively regulated PgPIN/PgPILSs and target genes in the MEP and MVA pathways of ginsenoside biosynthesis was analyzed (Fig. 6 and Table S7). As previously mentioned, PILS proteins adjust intracellular auxin accumulation, whereas canonical PIN proteins, except PIN5/PIN8s, are involved in the rate-limiting efflux of cellular auxin. Pearson’s correlation test was used to confirm the correlation between PgPINs and PgPILSs (Fig. 5B and Table S8). Two PgPILS6 genes showed moderate negative correlations with most PgPINs (cor < −0.40; P-value <0.05). In contrast, PgPILS1c was positively correlated with PgPINs. Two PgPIN8s exhibited a strong positive correlation with PgPILS1d and PgPILS2b and a moderate negative correlation with PgPILS1a, PgPILS1b, PgPILS5a, and PgPILS1b (Table S8).
Fig. 5.
Co-expression analyses of PgPIN/PgPILS genes for ginsenoside biosynthesis. (A) Pearson’s correlation coefficients of selected PgPIN/PgPILS expression levels and main ginsenoside contents; (B) Pearson’s correlation coefficients of expression pattern between PgPINs and PgPILSs in 27 RNA-Seq datasets. The size of the circles represents the value of Pearson’s correlation coefficients. The negative correlation showed in blue, while positive correlation in red.
Fig. 6.
Biosynthesis pathway for ginsenosides with the Pearson’s correlation coefficients of 8 PgPIN/PgPILS genes. AACT, acetyl-CoA C-acetyltransferase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; MVK, mevalonate kinase; MVP, mevalonate phosphate; PMK, phosphomevalonate kinase; MVPP, diphosphomevalonate; MVD, mevalonate diphosphate decarboxylase; DXP, 1-deoxy-D-xylulose-5-phosphate; DXS, 1-deoxy-D-xylulose-5-phosphate synthase; MEP, 2-C-methyl-D-erythritol 4-phosphat; DXR, DXP reducto-isomerase; CDP-ME, methylerythritol cytidyl diphosphate; IspD, CDP-ME synthetase; CDP-MEP, 4-diphosphocytidyl-2 -C-methyl-D- erythritol-2-phosphat; IspE, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; IspF, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; MEcPP, 2-C-methyl-D-erythritol-2,4-cyclodiphosphate; IspG, (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase; HMBPP, 4-hydroxy-3-methyl-butenyl-1-diphosphate; IspH, 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; IDI, isopentenyl-diphosphate delta-isomerase; FPS, farnesyl diphosphate synthase; FPP, farnesyl diphosphate; SS, squalene synthase; SE, squalene epoxidase; β-AS, β-amyrin synthase; DDS, dammarenediol synthase; LAS, lanosterol synthase; CAS, cycloartenol synthase; OAS, oleanolic acid synthase; PPDS, protopanaxadiol synthase; PPTS, protopanaxatriol synthase.
The PgPILS1 members PgPILS1a and PgPILS1b exhibited similar co-expression patterns to triterpene saponin biosynthesis enzyme-encoding genes. In particular, PgPILS1a exhibited a strong positive correlation with both genes in the MEP pathway and squalene synthase (SS) but not with genes in the MVA pathway (cor > 0.8). Unlike PgPILS1a and PgPILS1b, PgPILS1d and PgPILS1e were moderately positively correlated with 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase (IspH) and strongly positively correlated with PPD synthase (PPDS), PPT synthase (PPTS), cycloartenol synthase (CAS), and β-amyrin synthase (β-AS). PgPIN1e showed moderate or strong correlations with 1-deoxy-d-xylulose-5-phosphate synthase (DXS) and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), while PgPIN2a showed a similar but weaker co-expression pattern than PgPIN1e. Unexpectedly, PgPIN5c was positively correlated with both the MEP and MVA pathways. PgPIN3c was negatively correlated with HMGR and strongly positively correlated with the MEP pathway. PgPILS1a and PgPILS1b also showed strong negative correlations with 3-hydroxy-3-methylglutaryl-CoA synthase (HMGS), phosphomevalonate kinase (PMK), SS, oleanolic acid synthase (OAS), and CAS. Based on the expression profile, we hypothesized that PgPILS1 may be involved in regulating ginsenoside biosynthesis (Fig. 6 and Table S9).
4. Discussion
4.1. Herbgenomics facilitates to uncover the genetic information of herbs
Herbgenomics, which aims to explore the genetics and biology of herbs at the genomic level, is proposed as a global platform to identify the synthetic pathways of bioactive compounds (Chen and Song, 2016, Chen et al., 2012, Chen et al., 2015, Huang et al., 2016). Ginseng is one of the most widely used herbal medicines with increasing market demand, on account of its extraordinary ability to maintain physical vitality and increase resistance to aging. Nowadays ginseng has even been developed into healthy nutrition, dietary supplements, and natural cosmetics (Yun, 2001). Furthermore, methods such as BLAST, sequence alignment, phylogenetic analysis, and domain analysis can facilitate the genome-wide identification of genes related to ginsenoside biosynthesis. In this study, we present the genome-wide identification and transcriptional profiling analysis of PIN/PILS auxin transporter gene families in P. ginseng.
4.2. Structural features of PgPIN/PgPILS contribute to auxin regulation
To coordinate plant growth, development, and secondary metabolism, multiple auxin transporter proteins, including PIN and PILS, function to maintain the directional flow of auxin within different organs through the formation of a spatial and temporal gradient (Adamowski and Friml, 2015, Barbez et al., 2012). In current study, all PgPIN and PgPILS proteins were predicted to localize to membranes and contain putative auxin efflux transport domains (Muday & Murphy, 2002). In silico analysis of protein topology indicated that PILS proteins contain a central hydrophilic loop flanked on each side by five transmembrane domains, similar to PIN proteins (Fig. 3). The high similarity in predicted protein topology of PIN and PILS proteins implies similar biological functions in auxin transport (Barbez et al., 2012). Furthermore, PIN proteins can be grouped into long (PIN1-type) and short (PIN5-type) types based on the length of the central hydrophilic loop (Zazimalova et al., 2010). Canonical PIN1-type auxin efflux transporters localize to the plasma membrane and are rate-limiting transporters mediating cellular auxin efflux (Petrášek et al., 2006), while PIN5-type transporters are localized to the endoplasmic reticulum and manage intracellular auxin distribution and homeostasis (Bosco et al., 2012, Ding et al., 2012, Mravec et al., 2009). In our study, PgPIN1, PgPIN2, and PgPIN3 proteins were found to possess a long hydrophilic loop and were, therefore, assumed to localize to the plasma membrane. PgPIN5, PgPIN8, PgPILS1, PgPILS5, and PgPILS6 were predicted to have a short hydrophilic loop and localize to the endomembranes (primarily the endoplasmic reticulum and vacuoles) (Fig. 3). Barbez et al. (2012) reported that all PILS proteins in A. thaliana localize to the endoplasmic reticulum, similar to that in O. sativa (Mohanta et al., 2015). The majority of PgPILS localized to the endoplasmic reticulum or vacuoles, which may facilitate auxin accumulation inside the cells (Mohanta et al., 2015).
Liu et al. (2017) analyzed the expression patterns of auxin efflux transporter PIN genes in 4-year-old Jilin ginseng and hypothesized that PgPIN2 and PgPIN6 are likely involved in the development and tropism growth in ginseng roots, while PIN3 may be related to the growth and development of the aerial parts of plants. The organ-specific expression profile of PIN/PILS was also confirmed in current study (Fig. 4).
4.3. PgPIN/PgPILS probably involved in ginsenosides biosynthesis pathway
In addition to its role in development, auxin is likely involved in secondary metabolism (Sauerwein et al., 1991, Sauerwein et al., 1991). Ginsenosides show a tissue-specific accumulation and distribution and are even heterogeneously distributed in ginseng roots (Liang et al., 2015, Xu et al., 2017, Zhang et al., 2014). At gene level, multiple genes involved in ginsenoside synthesis are expressed in the phloem and xylem, while ginsenosides are primarily distributed and stored in the periderm (Kim et al., 2015, Zhang et al., 2017). More specifically, we measured the ginsenoside content and gene expression in the periderm, cortex, and stele, which were separated from the ginseng main root (Fig. 4). Ten PgPINs and seven PgPILSs were distributed across eight subfamilies with higher expression levels in periderm tissue, with 10 of the 17 PgPIN/PgPILS being of the short type. This implies the potential role of auxin and its transporters in the distribution and accumulation of ginsenosides (Fig. 4).
The production of ginsenosides and root growth in hairy roots in Panax hybrids is promoted by the addition of auxins (Washida et al., 2004); however, little is known about the potential mechanism. As stated earlier, it is possible that HMGR activity is regulated by phytohormones and stress (Fig. 7). Furthermore, both the MVA and MEP pathways are coordinated by hormone molecules via PRL1 (Úrsula et al., 2010). In the current study, we found a significant correlation between PIN/PILS expression and expression of the genes encoding enzymes involved in ginsenoside biosynthesis (Fig. 6), especially DXS and HMGR. Additionally, triterpenes function in plant growth and development, both of which are regulated by auxin (Kim et al., 2015); for example, the allelopathic effect of ginsenosides on growth (Zhang, Lei, Fang, Jia, & Zhang, 2011). Total ginsenosides, panaxadiol ginsenosides, and ginsenosides-Rb group inhibited the seedling growth of P. quinquefolius at high concentrations but had stimulatory effects at low concentrations. In contrast, panaxatriol ginsenosides stimulate seedling growth at various concentrations. However, the spatiotemporal distribution of auxin depends on a complex interplay between auxin metabolism and intercellular auxin transport. We just found the correlation between PIN/PILS and the ginsenoside biosynthetic pathways in the current study, the underlying mechanism and more details involved in auxin transport and ginsenoside biosynthesis remains to be elucidated (Fig. 7).
Fig. 7.
Speculated possible regulatory pathway of ginsenosides biosynthesis by PIN/PILS auxin transporters. Auxin transporter PIN/PILS family proteins, mainly locating on plasma membrane and endoplasmic reticulum membrane, maintain the distribution and homeostasis of auxin. PP2A, a heterotrimeric enzyme that binds the N-terminal region of HMGR, is involved in auxin, abscisic acid and ethylene signaling and emerges as a positive and negative multilevel regulator of plant HMGR. Meanwhile, PRL1 functions as a regulator of stress, and hormone response. From the above, it is assumed that phytohormone and stress response contribute to regulate the HMGR activity and subsequent ginsenoside production.
5. Conclusion
In summary, a genome-wide search of the P. ginseng genome for homologous auxin transporter genes identified 17 PIN and 11 PILS genes. Sequence analysis, putative motif organization, and sub-cellular localization indicated potential redundant and complementary biological functions of these PIN/PILS genes. Furthermore, organ/tissue expression patterns, together with the significant correlation between PIN/PILS expression and ginsenoside content, suggested that auxin transporters are potentially involved in the regulation of ginsenoside biosynthesis and its local accumulation. Genome-wide comparative analysis of PIN/PILS genes and their expression profile provide a new perspective to understand the molecular basis and regulatory mechanisms of PIN/PILS in P. ginseng, including their roles in ginsenoside production and stress response.
Editor Note
Shilin Chen is Editorial Board Members of Chinese Herbal Medicines. He was blinded from reviewing or making decisions on the manuscript. The article was subject to the journal's standard procedures, with peer review handled independently of this Editorial Board Member and their research groups.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grants from the National Science and Technology Major Project (No. 2018ZX09201-011, 2017ZX09301060-012), the Fundamental Research Funds for the Central Public Welfare Research Institutes (ZXKT17027, ZXKT19027), the National Nature Science Foundation of China (81803672) and the National Key Research and Development Program of China (2017YFC1702100). We sincerely thank He Su, Rui Bai and Xiao-yan Zhang for providing abundance of helpful on data generation and analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.chmed.2021.08.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Adamowski M., Friml J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell. 2015;27(1):20–32. doi: 10.1105/tpc.114.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Gribskov M. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics. 1998;14(1):48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Balzan S., Johal G.S., Carraro N. The role of auxin transporters in monocots development. Frontiers in Plant Science. 2014;5:393. doi: 10.3389/fpls.2014.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbez E., Kubes M., Rolcik J., Beziat C., Pencik A., Wang B., Rosquete M.R., Zhu J., Dobrev P.I., Lee Y., Zazimalova E., Petrasek J., Geisler M., Friml J., Kleine-Vehn J. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature. 2012;485(7396):119–122. doi: 10.1038/nature11001. [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115(5):591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Bosco, C. D., Dovzhenko, A., Liu, X., Woerner, N., Rensch, T., Eismann, M., Eimer S., Hegermann J., Paponov I.A., Ruperti B. (2012). The endoplasmic reticulum localized PIN8 is a pollen‐specific auxin carrier involved in intracellular auxin homeostasis. The Plant Journal, 71(5), 860-870. [DOI] [PubMed]
- Bulgakov V.P., Khodakovskaya M.V., Labetskaya N.V., Chernoded G.K., Zhuravlev Y.N. The impact of plant rolC oncogene on ginsenoside production by ginseng hairy root cultures. Phytochemistry. 1998;49(7):1929–1934. [Google Scholar]
- Cao H., Nuruzzaman M., Xiu H., Huang J., Wu K., Chen X., Li J., Wang L., Jeong J.H., Park S.J., Yang F., Luo J., Luo Z. Transcriptome analysis of methyl jasmonate-elicited Panax ginseng adventitious roots to discover putative ginsenoside biosynthesis and transport genes. International Journal of Molecular Sciences. 2015;16(2):3035–3057. doi: 10.3390/ijms16023035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli C.I., Vanstraelen M., Simon S., Yin K., Carron-Arthur A., Nisar N., Tarle G., Cuttriss A.J., Searle I.R., Benkova E., Mathesius U., Masle J., Friml J., Pogson B.J., Muday G. Role of the Arabidopsis PIN6 auxin transporter in auxin homeostasis and auxin-mediated development. PLoS One. 2013;8(7):e70069. doi: 10.1371/journal.pone.0070069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.L., Song J.Y. Herbgenomics. China Journal of Chinese Materia Medica. 2016;41(21):3881–3889. doi: 10.4268/cjcmm20162101. [DOI] [PubMed] [Google Scholar]
- Chen S., Song J., Sun C., Xu J., Zhu Y., Verpoorte R., Fan T.P. Herbal genomics: Examining the biology of traditional medicines. Science. 2015;347(6219):S27–S29. [Google Scholar]
- Chen S., Xu J., Liu C., Zhu Y., Nelson D.R., Zhou S.…Sun C. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nature Communications. 2012;3(1):913. doi: 10.1038/ncomms1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y., Xiao S., Su H.e., Liao B., Zhang J., Xu J., Chen S. Genome-wide characterization and analysis of bHLH transcription factors in Panax ginseng. Acta Pharmaceutica Sinica B. 2018;8(4):666–677. doi: 10.1016/j.apsb.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Geyter N., Gholami A., Goormachtig S., Goossens A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends in Plant Science. 2012;17(6):349–359. doi: 10.1016/j.tplants.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Ding Z., Wang B., Moreno I., Dupláková N., Simon S., Carraro N.…Friml J. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nature Communications. 2012;3(1):941. doi: 10.1038/ncomms1941. [DOI] [PubMed] [Google Scholar]
- Efstathios R., Bjorn K., Marian O., Visser R.G.F., Bachem C.W.B. The PIN family of proteins in potato and their putative role in tuberization. Frontiers in Plant Science. 2013;4:524. doi: 10.3389/fpls.2013.00524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E., Vosolsobě S., Feraru M.I., Petrášek J., Kleine-Vehn J. Evolution and structural diversification of PILS putative auxin carriers in plants. Frontiers in Plant Science. 2012;3:227. doi: 10.3389/fpls.2012.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Pérez Ú., Pérez-Gil J., Closa M., Wright L.P., Botella-Pavía P., Phillips M.A.…Rodríguez-Concepción M. Pleiotropic regulatory locus 1 (PRL1) integrates the regulation of sugar responses with isoprenoid metabolism in Arabidopsis. Molecular Plant. 2010;3(1):101–112. doi: 10.1093/mp/ssp100. [DOI] [PubMed] [Google Scholar]
- Friml J., Vieten A., Sauer M., Weijers D., Schwarz H., Hamann T.…Jürgens G. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature. 2003;426(6963):147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- Gao Y., He X., Wu B., Long Q., Shao T., Wang Z.i.…Chen S. Time-course transcriptome analysis reveals resistance genes of Panax ginseng induced by Cylindrocarpon destructans infection using RNA-seq. PLoS One. 2016;11(2):e0149408. doi: 10.1371/journal.pone.0149408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. WoLF PSORT: Protein localization predictor. Nucleic Acids Research. 2007;35(suppl_2):W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z.H., Xu J., Xiao S.M., Liao B.S., Gao Y., Zhai C.C.…Chen S.L. Comparative optical genome analysis of two pangolin species: Manis pentadactyla and Manis javanica. GigaScience. 2016;5(1):1–5. doi: 10.1093/gigascience/giw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-J., Zhang D., Yang D.-C. Biosynthesis and biotechnological production of ginsenosides. Biotechnology Advances. 2015;33(6):717–735. doi: 10.1016/j.biotechadv.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. Journal of Molecular Biology. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar, P., Antolínllovera, M., Ferrero, S., Closa, M., Arró, M., Ferrer, A., ... Campos N. (2011). Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A. Plant Cell, 23(4), 1494-1511. [DOI] [PMC free article] [PubMed]
- Leung K., Wong A. Pharmacology of ginsenosides: A literature review. Chinese Medicine. 2010;5(1):20. doi: 10.1186/1749-8546-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z., Chen Y., Xu L., Qin M., Yi T., Chen H., Zhao Z. Localization of ginsenosides in the rhizome and root of Panax ginseng by laser microdissection and liquid chromatography–quadrupole/time of flight-mass spectrometry. Journal of Pharmaceutical and Biomedical Analysis. 2015;105:121–133. doi: 10.1016/j.jpba.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Liu J., Chen T., Yuan Y., Ji R.F., Guo J., Wang Y.P.…Huang L.Q. Expression analysis of polar auxin transport mediated gene PIN in Panax ginseng. Acta Pharmaceutica Sinica. 2017;52(4):641–646. [Google Scholar]
- Mancuso C., Santangelo R. Panax ginseng and Panax quinquefolius: From pharmacology to toxicology. Food and Chemical Toxicology. 2017;107(Pt A):362–372. doi: 10.1016/j.fct.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanta T.K., Mohanta N., Bae H. Identification and expression analysis of PIN-Like (PILS) gene family of rice treated with auxin and cytokinin. Genes. 2015;6(3):622–640. doi: 10.3390/genes6030622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J., Skůpa P., Bailly A., Hoyerová K., Křeček P., Bielach A.…Stierhof Y.D. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459(7250):1136–1140. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- Muday G.K., Murphy A.S. An emerging model of auxin transport regulation. Plant Cell. 2002;14(2):293–299. doi: 10.1105/tpc.140230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazón J., Cusidó R.M., Bonfill M., Mallol A., Moyano E., Morales C., Piñol M.T. Elicitation of different Panax ginseng transformed root phenotypes for an improved ginsenoside production. Plant Physiology and Biochemistry. 2003;41(11-12):1019–1025. [Google Scholar]
- Paponov I., Teale W., Trebar M., Blilou I., Palme K. The PIN auxin efflux facilitators: Evolutionary and functional perspectives. Trends in Plant Science. 2005;10(4):170–177. doi: 10.1016/j.tplants.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Park Y.H., Kim Y., Mishra R.C., Bae H. Fungal endophytes inhabiting mountain-cultivated ginseng (Panax ginseng Meyer): Diversity and biocontrol activity against ginseng pathogens. Scientific Reports. 2017;7(1):16221. doi: 10.1038/s41598-017-16181-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášek J., Mravec J., Bouchard R., Blakeslee J.J., Abas M., Seifertová D.…Čovanová M. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;312(5775):914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- Rahman M., Punja Z.K. Factors influencing development of root rot on ginseng caused by Cylindrocarpon destructans. Phytopathology. 2005;95(12):1381–1390. doi: 10.1094/PHYTO-95-1381. [DOI] [PubMed] [Google Scholar]
- Sauerwein M., Ishimaru K., Shimomura K. Indole alkaloids in hairy roots of Amsonia elliptica. Phytochemistry. 1991;30(4):1153–1155. [Google Scholar]
- Sauerwein M., Yamazaki T., Shimomura K. Hernandulcin in hairy root cultures of Lippia dulcis. Plant Cell Reports. 1991;9(10):579–581. doi: 10.1007/BF00232336. [DOI] [PubMed] [Google Scholar]
- Shen C.J., Bai Y.H., Wang S.K., Zhang S.N., Wu Y.R., Chen M.…Qi Y.H. Expression profile of PIN, AUX/LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS Journal. 2010;277(14):2954–2969. doi: 10.1111/j.1742-4658.2010.07706.x. [DOI] [PubMed] [Google Scholar]
- Spyropoulos I.C., Liakopoulos T.D., Bagos P.G., Hamodrakas S.J. TMRPres2D: High quality visual representation of transmembrane protein models. Bioinformatics. 2004;20(17):3258–3260. doi: 10.1093/bioinformatics/bth358. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Dhonukshe P., Brewer P.B., Friml J. Spatiotemporal asymmetric auxin distribution: A means to coordinate plant development. Cell and Molecular Life Sciences. 2006;63(23):2738–2754. doi: 10.1007/s00018-006-6116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D., Lee S. Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book. 2011;9:e0143. doi: 10.1199/tab.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature Biotechnology. 2013;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady G.E., Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17(9):849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Viaene T., Delwiche C.F., Rensing S.A., Friml J. Origin and evolution of PIN auxin transporters in the green lineage. Trends in Plant Science. 2013;18(1):5–10. doi: 10.1016/j.tplants.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Wang Y.Q., Chai C.L., Valliyodan B., Maupin C., Annen B., Nguyen H.T. Genome-wide analysis and expression profiling of the PIN auxin transporter gene family in soybean (Glycine max) BMC Genomics. 2015;16(1):951. doi: 10.1186/s12864-015-2149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.K., Feng Z.X., Wang X., Wang X.W., Zhang X.G. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Wang J.R., Hu H., Wang G.H., Li J., Chen J.Y., Wu P. Expression of PIN genes in rice (Oryza sativa L.): Tissue specificity and regulation by hormones. Molecular Plant. 2009;2(4):823–831. doi: 10.1093/mp/ssp023. [DOI] [PubMed] [Google Scholar]
- Wang K.Y., Jiang S.C., Sun C.Y., Lin Y.P., Yin R., Wang Y., Zhang M.P. The spatial and temporal transcriptomic landscapes of ginseng, Panax ginseng C A. Meyer. Scientific Reports. 2015;11(5):18283. doi: 10.1038/srep18283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washida D., Shimomura K., Takido M., Kitanaka S. Auxins affected ginsenoside production and growth of hairy roots in Panax hybrid. Biological and Pharmaceutical Bulletin. 2004;27(5):657–660. doi: 10.1248/bpb.27.657. [DOI] [PubMed] [Google Scholar]
- Xu, J., Chu, Y., Liao, B., Xiao, S., Yin, Q., Bai, R., ...Chen S. (2017). Panax ginseng genome examination for ginsenoside biosynthesis. GigaScience, 6(11), 1-15. [DOI] [PMC free article] [PubMed]
- Yan X., Fan Y., Wei W., Wang P., Liu Q., Wei Y.…Zhou Z. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Research. 2014;24(6):770–773. doi: 10.1038/cr.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue R.Q., Tie S.G., Sun T., Zhang L., Yang Y.J., Qi J.S.…Shen C.J. Genome-wide identification and expression profiling analysis of ZmPIN, ZmPILS, ZmLAX and ZmABCB auxin transporter gene families in maize (Zea mays L.) under various abiotic stresses. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0118751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun T.-K. Panax ginseng—a non-organ-specific cancer preventive? Lancet Oncology. 2001;2(1):49–55. doi: 10.1016/S1470-2045(00)00196-0. [DOI] [PubMed] [Google Scholar]
- Zazímalová, E., Murphy, A. S., Yang, H., Hoyerová, K., & Hosek, P. (2010). Auxin transporters-why so many? Cold Spring Harbor Perspectives in Biology, 2(3), a001552. [DOI] [PMC free article] [PubMed]
- Zhang A.H., Lei F.J., Fang S.W., Jia M.H., Zhang L.X. Effects of ginsenosides on the growth and activity of antioxidant enzymes in American ginseng seedlings. Journal of Medicinal Plant Research. 2011;5(14):3217–3223. [Google Scholar]
- Zhang Y.C., Li G., Jiang C., Yang B., Yang H.J., Xu H.Y., Huang L.Q. Tissue-specific distribution of ginsenosides in different aged ginseng and antioxidant activity of ginseng leaf. Molecules. 2014;19(11):17381–17399. doi: 10.3390/molecules191117381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.J., Su H., Zhang L., Liao B.S., Xiao S.M., Dong L.L., Chen S.L. Comprehensive characterization for ginsenosides biosynthesis in ginseng root by integration analysis of chemical and transcriptome. Molecules. 2017;22(6):889. doi: 10.3390/molecules22060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The necessary information of public data used in this study are present within the article. Genome sequencing data of P. ginseng are available via NCBI under the project number PRJNA385956. The latest versions of the genome assemblies and annotation are available through our website at http://ginseng.vicp.io:23488/. The GenBank accession numbers for the PgPIN/PgPILS genes sequences reported in this paper is summarized in Table S2.