Abstract
Objective
To find a suitable ecological cultivation measure to solve the problem of root-knot nematode disease of Panax quinquefolium (Panacis Quinquefolii Radix) and the heavy metals accumulating in its roots.
Methods
Three-year-old P. quinquefolium was treated with four different combinations of microbial inoculant (MI) and garbage fermentation liquid (GFL) [the joint application of ‘TuXiu’ MI and Fifty potassium MI (TF), the combination use of ‘No. 1′ MI and Fifty potassium MI (NF), ‘Gulefeng’ poly-γ-glutamic acid MI (PGA), GFL], and the untreated control (CK). Here, high-throughput sequencing, ICP-MS and UPLC were employed to systematically characterize changes of microbial diversity and structure composition, heavy metals (As, Cd and Pb) content and ginsenoside content among different treatments.
Results
The results revealed that different MIs and GFL could increase the root dry weight of P. quinquefolium, PGA enhanced it by 83.24%, followed by GFL (49.93%), meanwhile, PGA and GFL were able to lessen root-knot nematode disease incidence by 57.25% and 64.35%. The treatment of PGA and GFL can also effectively reduce heavy metals in roots. The As content in GFL and PGA was decreased by 52.17% and 43.48% respectively, while the Cd and Pb contents of GFL and PGA was decreased somewhat. Additionally, the content of total ginsenosides was increased by 42.14% and 42.07%, in response to TF and NF, respectively. Our metagenomic analysis showed that the relative abundance of particular soil microbial community members related to the biocontrol of root-knot nematode disease and plant pathogen (i.e., Chaetomium in NF, Xylari in GFL, and Microascus in PGA), heavy metal bioremediation (Hyphomacrobium in PGA and Xylaria in GFL), and nitrogen fixation (Nordella and Nitrospira in TF) was significantly increased; notably, potential harmful microflora, such as Plectosaphaerella and Rhizobacter, were more abundant in the control group.
Conclusion
MI and GFL could improve the quality of P. quinquefolium by modifying its rhizosphere microbial community structure and composition, both of them are beneficial to the development of ecological cultivation of P. quinquefolium.
Keywords: garbage fermentation liquid, heavy metal, microbial inoculants, Panax quinquefolium L., nematodes disease
1. Introduction
Panax quinquefolium L., a perennial medicinal plant also known as American ginseng, is famous for its pharmacological activities (Li et al., 2019, Szczuka et al., 2019). However, it frequently suffers from severe root diseases. The P. quinquefolium nematode disease is one such root diseases, capable of reducing its yield by more than 60% (Zhang et al., 2009). This disease also aggravates the severity of other co-occurring root diseases and affects the plant quality adversely (Al-Hazmi & Al-Nadary, 2015). To date, the main plant parasitic nematodes are Meloidogyne, Heterodera, and Globodera spp. (East et al., 2019, Vieira and Gleason, 2019, Wilkes and Kirkpatrick, 2020), whose prevention and control depends primarily on organochlorine pesticides (Zhang et al., 2009). Yet this kind of pesticide features stable chemical properties, so it easily accumulates in the roots of P. quinquefolium, thus threatening its consumption and therapeutic use. For example, 15 different organochlorine pesticides were detected in 60 samples of P. quinquefolium, namely benzene hexachloride (BHC, including a-BHC, b-BHC, c-BHC, d-BHC), hexachlorobenzene, heptachlor, heptachlor epoxide, aldrin, p,p’-DDE, p,p’-DDD, o,p’-DDT, p,p’-DDT, mirex, endrin, and dieldrin (Wu, Liu, Zhao, & Xu, 2011). Recently, Zhao et al. (2018) reported 16 pesticides in P. quinquefolium, of which 15 were organochlorine pesticides. Furthermore, the steady input of pesticides will also worsen soil ecology, as well as diminishing the disease-controlling effect of pesticides. For instance, Liu, Li, Li, and Chen (2008) reported that pentachlorophenol could significantly reduce the diversity of bacterial community and the number of ammonia oxidizing bacteria in soil. Hill and Hausbeck (2008) found that 82% of 210 strains of Phytophthora cactorum isolated from P. quinquefolium soil in Wisconsin, USA, were resistant to metalaxyl. To sum up, there are sufficient and mounting evidences to show that it affects medicinal plant quality adversely to apply pesticides to treat the nematode diseases of P. quinquefolium. Therefore, it is imperative to find a safer way, to inhibit the population of pathogenic nematodes and reduce the production risk in the cultivation process.
Biological control is an environment-friendly and low-cost plant disease control way that is increasingly gaining attention. Using a microbial inoculant (MI) which contains plant growth-promoting microorganisms (PGPM), is becoming extremely popular in agriculture. When the PGPM is applied to plants or soils, it can assist plants’ growth by enhancing their nutrient absorption or alleviating diseases, such as root rot and nematode diseases (Jiang et al., 2018, Shen et al., 2018). Bacillus is the most commonly used PGPM. When B. subtilis EA-CB0575 was incubated to tomato rhizosphere, it can produce indole and iron-carrier compounds, dissolve phosphate, and fix nitrogen, was shown to improve the dry weight of tomato by 34.60% (Franco-Sierra et al., 2020). The B. amyloliquefaciens AK-0 can release antibiotic compounds to slow down the root rot of P. ginseng caused by Cylindrocarpon destructans (Kim, Balaraju, & Jeon, 2017). Another PGPM, B. cereus BCM2, is able to adhere itself to the surface of nematodes, and its metabolites can quickly identify and kill the second-stage juveniles of Meloidogyne incognita (Hu et al., 2020).
Garbage fermentation liquid (GFL) is an organic fermentation product, which is fermented by mixing plant tissues or other biological garbage with water and brown sugar (Arun & Sivashanmugam, 2017). Some PGPM, namely Lactobacillus and Candida, are the dominant microorganisms produced in this fermentation process (Du et al., 2017). Lactobacillus spp. are generally regarded as beneficial microorganisms, which can be used to improve the stress resistance and the control of plant parasitic nematodes (Hussein and Joo, 2018, Seo et al., 2019). Candida members can promote the growth of Vigna radiata under drought stress, and biodegrade 4-nitroaniline in contaminated land (Silambarasan & Vangnai, 2017). In recent years, more researches are showing that GFL can play a pivotal role in removing impurities and pathogenic bacteria from dairy waste-activated sludge, and improving the effective components of medicinal plants (Arun and Sivashanmugam, 2015, Ouyang and Gu, 2017, Wei et al., 2020). Under conditions of Cd stress, GFL was capable of reducing the Cd content of Salvia miltiorrhiza by 25.31% and increasing it total tanshinone content by 40.08% (Wei, Cao, et al., 2020). Therefore, GFL has a broad application prospect in the cultivation of medicinal plant.
Soil microbial community composition and structure is closely associated with plants’ growth and health. The diversity of Panax notoginseng soil fungi is decreased significantly after planted three successive years in the same field (Dong, Xu, Feng, Li, & Chen, 2016). Additionally, significant microbial structural differences could be found between the healthy and rusty P. ginseng rhizosphere soil. The genera Fusarium, Cylindrocarpn, Acrophialophora, Altenaria and Doratomices were mainly concentrated in the rhizosphere of diseased plants, while Xenopolyscytalum, Arthrobotrys, Chalara, Cryptococus, and Scutellinia were mainly enriched in healthy ones (Wei, Wang et al., 2020). In the present study, we carried out a metagenomics analysis to study the quality-improvement effect upon P. quinquefolium as driven by MIs and GFL. Our aim was (i) to compare the differences of rhizosphere soil microflora of P. quinquefolium treated by different MIs and GFL, and (ii) to explore the potential mechanism(s) of biological control of root-knot nematode, biological removal of heavy metals, and promotion of ginsenoside content by various MIs and GFL from the perspective of microorganisms. It is anticipated this study will prove helpful for better understanding the effects of MI and GFL on the quality improvement of medicinal plants.
2. Materials and methods
2.1. Description of microbial inoculants and garbage fermentation liquid
Four kinds of commercially available MIs and GFL were used. The ‘Tuxiu’ MI (Beijing Chenao Runze Technology Co., Ltd., China) is a microbial mixture containing B. licheniformis, B. amylolyticus, B. subtilis, actinomycetes and yeast over 500 million propagules per milliter. The ‘Fifty potassium’ MI (Beijing Chenao Runze Technology Co., Ltd.) is mainly composed of nitrogen, phosphorus, and potassium (16:3:12) nutrients, whose viable bacteria number more than 500 million propagules per gram. The ‘No. 1′ MI (Beijing Chenao Runze Technology Co., Ltd.) contains a liquid suspension, comprising of B. subtilis, B. licheniformis, and actinomycetes, at a concentration exceeding 2 billion propagules per milliter. The ‘Gulefeng’ poly-γ-glutamic acid MI (Nanjing Xuankai Biotechnology Co., Ltd., China) is composed of B. subtilis, B. amylolytica, and B. gelatinosus over 5 billion propagules per milliliter, and rich in a great quantity of poly-γ-glutamic acid. Finally, garbage fermentation liquid (GFL), is a hand-made brown liquid fermented by Citrus aurantium fruits, together with brown sugar and water, as described by in Ouyang and Gu (2017), from January to April. After completing the fermentation period, the liquid was separated from the solids for further investigation, which pH is 3.67, and the dissolved oxygen is 1.69 mg/L. The microbial community composition of GFL was tested prior to implementing the experiment via high-throughput sequencing technology: the main microbes in it were Lactobacillus, Pichia, and Candida spp., among others.
2.2. Field experiment and soil collection
P. quinquefolium had been cultivated for 3 years in Rongcheng City, Shandong Province (37.2027 N, 122.4298E, mean elevation: 52 m), in eastern China, the region which is well known as the country’s main cultivation area of this medicinal plant. The As, Cd, and Pb soil concentrations at this site were 2.73 μg/g, 0.03 μg/g and 11.42 μg/g, respectively. Five treatments were applied in a field divided into 15 plots randomly (each 8 m × 1.5 m): TuXiu MI + Fifty potassium MI (Tuxiu irrigation + Fifty potassium fertilizer application), No. 1 MI + Fifty potassium (No. 1 irrigation + Fifty potassium fertilizer application), Gulefeng poly-γ-glutamic acid MI (Gulefeng poly-γ-glutamic acid irrigation), GFL (GFL irrigation), and the untreated control (water irrigation), hereon TF, NF, PGA, GFL, and CK respectively, with three plot replicates per treatment. A 1.5 × 1.5 isolation zone was set between adjacent replicate. These soil treatments were applied according to the manufacturer’s instructions and the principles described previously (Ouyang & Gu, 2017). At harvest time, 15 rhizosphere soil (0–20 cm depth layer) of P. quinquefolium roots were collected by the five-spot-sampling method and labeled accordingly (i.e., TF, NF, PGA, GFL, and CK respectively) (Wang, Li, Li, Pan, & Zhang, 2018). All soil samples were homogenized by 2-mm sieve and then stored at − 80 °C for further study.
2.3. DNA extraction, PCR amplification and sequencing
Total soil DNA was extracted by using the soil genomic DNA kit (Tiangen Biotech Beijing Co., China) and following the manufacturer’s instructions. The V4-V5 region of the bacterial 16S rRNA gene was amplified with the primer pair 515F/907R (5′- GTGCCAGCMGCCGCGG-3′, 5′- CCGTCAATTCMTTTRAGTTT-3′) (Fu et al., 2017), while the internal transcribed spacer (ITS) region of the rRNA gene was amplified by the fungal-specific primer pair ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA) and ITS2R (5′-GCTGCGTTCTTCATCGATGC) (Li, Ma, Mark Ibekwe, Wang, & Yang, 2018). Both PCR amplification and purification were carried out following previously (Rodrigues et al., 2013), with the ensuing amplicons pooled in equimolar ratios for sequencing. The purified PCR products were sequenced on an Illumina HiSeq platform (Shanghai Biozeron Co., Ltd., China).
2.4. Herb yield and health status
In October, the 4.5 × 1.5 area of P. quinquefolium roots in the middle of each replicate was harvested. The roots were removed from soil to assess its incidence of disease (i.e., of the root-knot nematode disease). For a given individual replicate, the number of infected plants and total plants of harvested was recorded to calculate the incidence of disease. The roots were rinsed and oven-dried at 50 °C to a constant weight for counting the average dry weight of each root in different duplicons.
2.5. Detection of ginsenosides (Rb1, Re, Rg1) and heavy metals
Next, the concentrations of three ginsenosides (Rb1, Re and Rg1) were determined by ultra-performance liquid chromatography (UPLC). The samples and standards were prepared, according to the guidelines established by the Pharmacopoeia of the People’s Republic of China (Pharmacopoeia Committee of P. R. China, 2015). The standards of ginsenoside Rb1, Re, and Rg1 were purchased from the National Institute for Food and Drug Control (China). Acetonitrile and methanol were obtained from Fisher Scientific, with an ACQUITY UPLC BEH C18 chromatographic column (2.1 mm × 50 mm, 1.7 µm) used in the UPLC (WATERS, USA). The mobile phase consisted of 100% acetonitrile (solvent A) and 100% water (solvent B) (gradient elution: 0–2 min, 18% acetonitrile; 2–5 min, 20% acetonitrile; 5–11 min, 40% acetonitrile; 11–11.1 min, 18% acetonitrile; 11.1–12 min, 18% acetonitrile; the flow rate was set to 0.45 mL/min; the column temperature was 30 °C with an injection volume of 2 µL. All samples were detected at 203 nm (Yang, Zheng, Jiang, Zhou, & Xiao, 2008).
Arsenic (As) cadmium (Cd), and lead (Pb) in P. quinquefolium roots were then detected by inductively coupled plasma-mass spectrometry (ICP-MS). The fine powder of dried roots of P. quinquefolium (20–30 mg) was digested in 5 mL of concentrated HNO3 for 24 h. After adding 3 mL H2O2 to each tube, all samples were heated at 180 °C for 4–5 h until the tissues were completely digested. Then the digested samples were diluted with ultrapure water and analyzed by ICP-MS (Thermo X7, Waltham, MA, USA) (Xu, Xin, Lin, & Rui, 2010).
2.6. Statistical analyses
After the FASTQ data is converted into the fasta format, the original metagenome file is demultiplexed and quality filtered by QIIME according to standard pipeline (Caporaso et al., 2010). Sequences sharing 97% identity were assigned to the same operational taxonomic unit (OTU), by using the UPARSE tool (Edgar, 2013). The online resources Silva (http://www.arb-silva.de) and Unite (http://unite.ut.ee/index.php) were used to determining the taxonomic identities of the bacteria and fungi, respectively (Koljalg et al., 2013, Quast et al., 2013). Alpha diversity of the bacterial and fungal communities was analyzed using the Shannon (H’) and Chao I indices (Schloss et al., 2009), both calculated in Mothur software (http://www.mothur.org/wiki/Schloss_SOP#Alpha_diversity). A principal coordinate analysis (PCoA) was used to illustrate the β-diversity based on Bray-Curtis distances metric (Oh et al., 2020). The linear discriminant analysis (LDA) effect size (LEfSe) was used to further confirm whether differences in the abundances of individual taxa existed among different groups, which were considered significant if they had an LDA score greater than 2 and P-value < 0.05 (Segata et al., 2011). The metagenome functional of all bacterial communities was predicted by phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) software package using 16S rRNA gene sequencing based on the kyoto encyclopedia of genes and genomes (KEGG) database (Liu et al., 2020).
The five treatments’ data were expressed here as means ± standard errors (n = 3 replicates). Statistical significance was determined using a one-way analysis of variance (ANOVA) when P < 0.05 between the treatment groups and the control, implemented in SPSS v22 (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Effects of MIs and GFL on dry weight and nematode disease
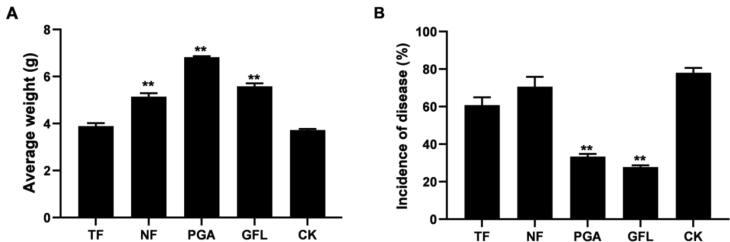
At harvest time of P. quinquefolium, its average dry weight (i.e., root weight) and the incidence of disease (i.e., root-knot nematode) were determined. Compared with the control group, the average dry weights of treatment groups (TF, NF, PGA, and GFL) was greater than that of CK; NF, PGA, and GFL groups respectively increased it by 38.16%, 83.24%, and 49.93% (5.14, 6.82, and 5.58 g/plant) (P < 0.01), while TF increased only slightly, by 4.59% (Fig. 1A). The incidences of nematode disease in P. quinquefolium root of all treatments (TF, NF, PGA, and GFL) were lower than that of CK, which ranked as follows: GFL < PGA < TF < NF < CK. Relative to the control group, PGA and GFL significantly reduced the incidence by 57.25% and 64.35% respectively (P < 0.01), while the TF and NF treatments decreased it less so, by 22.08% and 9.55%, respectively (Fig. 1B).
Fig. 1.
Average weight (A) and incidence of nematode disease (B) of the roots of Panax quinquefolium (mean ± SE, n = 3). TF, NF, PGA, and GFL represent the rhizosphere soil of P. quinquefolium treated by TuXiu microbial inoculum + Fifty potassium, No. 1 microbial inoculum + Fifty potassium, Gulefeng poly-γ-glutamic acid microbial inoculum, and garbage fermentation liquid, respectively, with CK as the control group. *P < 0.05; **P < 0.01 vs CK group.
3.2. MIs and GFL affect content of As, Cd and Pb
The contents of As, Cd, and Pb in P. quinquefolium roots were determined by ICP-MS. The effects of MIs and GFL on heavy metal content differed in some degree. The contents of As were decreased by treatment groups (TF, NF, PGA and GFL) than that of CK, which ranked as follows: CK > NF > TF > PGA > GFL. In the GFL and PGA groups, the content of As were significantly reduced by 52.17% and 43.48% (0.0867 and 0.0733 μg/g; P < 0.05), while TF and NF treatments decreased it by 23.91% and 19.57% (0.1167 and 0.1233 μg/g), respectively, when compared with the control (0.1533 μg/g) (Fig. 2A). The Cd content of PGA and GFL groups was lower than that of CK, for which it was diminished by 8.33% and 6.25% (0.1467 and 0.1500 μg/g) versus the control group (0.1600 μg/g) (Fig. 2B). For Pb content, the treatments had a ranking of NF > TF > CK > PGA > GFL. Similar to As and Cd, the Pb content was lowered by most in GFL and PGA groups, by 45.66% and 35.26% (0.3733 and 0.3133 μg/g) compared with the control group (0.5767 μg/g) (Fig. 2C). In short, the least amount of As, Cd, and Pb in the medicinal root materials occurred under GFL, followed by PGA.
Fig. 2.
Contents of As (A), Cd (B), and Pb (C) in roots of Panax quinquefolium (mean ± SE, n = 3). TF, NF, PGA, and GFL represent the rhizosphere soil of P. quinquefolium treated by TuXiu microbial inoculum + Fifty potassium, No. 1 microbial inoculum + Fifty potassium, Gulefeng poly-γ-glutamic acid microbial inoculum, and garbage fermentation liquid, respectively, with CK as the control group. *P < 0.05; **P < 0.01 vs CK group.
3.3. MIs and GFL affect ginsenosides content
Next, UPLC was used to accurately measure the ginsenoside content in the experimental medicinal root materials. The Re content of treatment groups (TF, NF, PGA, and GFL) was greater than that of CK, which was increased by 65.05% and 67.27% (0.0149 and 0.0151 μg/g) in the TF and NF groups (P < 0.05), while PGA and GFL increased by 35.30% and 12.29% (0.0122 and 0.0101 μg/g), respectively, relative to the control (0.0090 μg/g) (Fig. 3A). The content of Rb1 of treatment groups (TF, NF, PGA, and GFL) was greater than that of CK, which was increased by 31.03%, 27.64%, 9.19%, and 9.19% (0.0138, 0.0135, 0.0119, and 0.0119 μg/g) over the control (0.0111 μg/g) (Fig. 3B). Generally, it was found that ginsenoside Re and Rb1 contents were significantly higher in the roots treated with TF and NF than those of other treatments (PGA, GFL or CK); moreover, Rg1 was only detectable in the TF group.
Fig. 3.
Ginsenoisdes Re (A) and Rb1 (B) contents in roots of P. quinquefolium (mean ± SE, n = 3). TF, NF, PGA, and GFL represent the rhizosphere soil of P. quinquefolium treated by TuXiu microbial inoculum + Fifty potassium, No. 1 microbial inoculum + Fifty potassium, Gulefeng poly-γ-glutamic acid microbial inoculum, and garbage fermentation liquid, respectively, with CK as the control group. *P < 0.05; **P < 0.01 vs CK group
3.4. Overall bacterial and fungal community structure in rhizosphere of P. quinquefolium
For bacteria, a total of 880 048 classified reads were obtained from the 15 soil samples, with an average of 58 670 reads per sample. H’ and Chao I indices were used to estimate the alpha diversity of microbial communities. The results showed that MIs and GFL led to a changed soil bacterial diversity (Fig. 4A and B). As the Venn diagram shows, the shared number of bacterial OTUs in the five groups was 3403, while the number of OTUs unique to the control, TF, NF, PGA, and GFL groups was 356, 679, 425, 351, and 268, respectively (Fig. 5A). The PCoA revealed some notable differences among bacterial communities in different treatments (Fig. 6A). Along the first principal component (35.18% contribution), the bacterial communities of TF were separated from those of NF, PGA, GFL, and CK. The bacterial communities of the NF, PGA, and GFL groups differed significantly from the control group. According to the distribution of bacterial taxa (Fig. 7A), the relative abundances of Proteobacteria and Gemmatimonadetes were lower in the treatment groups (TF, NF, PGA, and GFL) than the control, but those of Actinobacteria, Chloroflexi, Planctomyces and Firmicutes had the opposite pattern. Compared with the control group, the relative abundance of Acidobacteria was higher in both TF and GFL groups, while that of Nirospirae was greater in the TF, NF, and PGA groups, yet lower in the GFL. These results provided crucial evidence for studying how bacterial community structure affected by different MIs and GFL.
Fig. 4.
Chao I (A) and H′ values (B) for bacterial and Chao I (C) and H′ values (D) for fungal diversity in TF, NF, PGA, GFL, and CK soils (mean ± SE, n = 3). TF, NF, PGA, and GFL represent the rhizosphere soil of P. quinquefolium treated by TuXiu microbial inoculum + Fifty potassium, No. 1 microbial inoculum + Fifty potassium, Gulefeng poly-γ-glutamic acid microbial inoculum, and garbage fermentation liquid, respectively, with CK as the control group. *P < 0.05; **P < 0.01 vs CK group.
Fig. 5.
Venn diagrams showing the core rhizosphere microbiome (OTUs) of bacterial (A) and fungal (B) communities among the TF, NF, PGA, GFL and CK groups. TF, NF, PGA, and GFL represent the rhizosphere soil of P. quinquefolium treated by TuXiu microbial inoculum + Fifty potassium, No. 1 microbial inoculum + Fifty potassium, Gulefeng poly-γ-glutamic acid microbial inoculum, and garbage fermentation liquid, respectively, with CK as the control group.
Fig. 6.
Principal coordinate analysis (PCoA) plot based on bacterial (A) and fungal (B) community scores from the TF, NF, PGA, GFL, and CK soil samples. The scatterplot is of principal coordinate 1 (PC1) vs. principal coordinate 2 (PC2). TF, NF, PGA, and GFL represent the rhizosphere soil of P. quinquefolium treated by TuXiu microbial inoculum + Fifty potassium, No. 1 microbial inoculum + Fifty potassium, Gulefeng poly-γ-glutamic acid microbial inoculum, and garbage fermentation liquid, respectively, with CK as the control group.
Fig. 7.
Taxonomic classification of bacterial (A) and fungal (B) reads retrieved from the TF, NF, PGA, GFL and CK soil samples (pooled) at the phylum level. TF, NF, PGA, and GFL represent the rhizosphere soil of P. quinquefolium treated by TuXiu microbial inoculum + Fifty potassium, No. 1 microbial inoculum + Fifty potassium, Gulefeng poly-γ-glutamic acid microbial inoculum, and garbage fermentation liquid, respectively, with CK as the control group.
For fungi, a total of 932 347 classifiable reads were obtained from the 15 soil samples, averaging 62 156 reads per sample. Applying the MIs and GFL contributed to changed Chao I and H’ indices (Fig. 4C and D). The Venn diagram revealed that the shared number of fungi OTUs in the five groups totaled 538, while their unique number of fungi OTUs was 83, 222, 138, 133, and 92 (Fig. 5B). Their corresponding PCoA analysis indicated that fungal communities in the control group and NF group were unalike (separated), while GFL and PGA groups were most similar (closer in ordination space). Along the first principal component (22.1% contribution), the TF’s fungal communities were positioned apart from the other treatment groups, suggesting their dissimilarity (Fig. 6B). The distribution of fungal taxa is shown in Fig. 7B, Ascomycota, Mucoromycota and Basidiomycota are the dominant phyla of all rhizosphere soil samples, accounting for 73.60%, 14.67%, and 5.62% of all OTUs, on average. Compared with the control group, the abundance of Ascomycota in the treatment groups (TF, NF, PGA, and GFL) increased by 14.62%, 5.45%, 6.92% and 1.83%, respectively. The relative abundances of Chytridiomycota and Mucoromycota in all four treatment groups (TF, NF, PGA, and GFL) were lower than in the control group.
3.5. Special bacterial and fungal communities
The LEfSe analysis was used to identify those bacteria with significant differences between different treatments (Fig. 8A). At the genus level, four characteristic species were significantly more abundant in the NF samples, where Nitrospira showed the highest abundance, followed by Ktedonobacter, Streptacidiphilus, and Nordella. In particular, the relative abundances of Nordella and Nitrospira were significantly increased, by 75.68% and 158.10%, compared with the control; in the PGA group, the abundance of Hyphomicrobium was significantly increased (by 23.61%). Chujaibacter was most abundant in the GFL treatment, while Rhizobacter, Rhodanobacter, Dokdonella, Aeromicrobium, Rhodopseudomonas, and Shimazuella were richer in the control than any treatments. Importantly, the relative abundance of Bacillus under three MI treatments (TF, NE, PGA) exceeded that under the control. Lactobacillus microorganisms were detected in the GFL treatment, but not in the control group.
Fig. 8.
Linear discriminant analysis effect size analysis of bacterial (A) and fungal (B) taxa for TF, NF, PGA, GFL and CK. Significance was defined as a P-value < 0.05 and an LDA score greater than 2.0. TF, NF, PGA, and GFL represent the rhizosphere soil of P. quinquefolium treated by TuXiu microbial inoculum + Fifty potassium, No. 1 microbial inoculum + Fifty potassium, Gulefeng poly-γ-glutamic acid microbial inoculum, and garbage fermentation liquid, respectively, with CK as the control group.
Likewise, the LEfSe analysis was used to detect differential fungi occurring in the 15 soil samples (Fig. 8B). The Conlarium, Corynascus, Endophragmiella, Helicoma and Sarocladium genera were significantly more abundant in the TF group, while Chaetomium, Gymnascella, and Pseudaleuria dominated the NF soil samples. In this NF group, the relative abundance of Chaetomium increased substantially, by 153.82%, over the control. GFL sustained a greater abundance of Arachnomyces, Crassicarpon, and Arcopilus than the control group, and this treatment also significantly augmented the relative abundance of Xylaria, by 219.39%. Additionally, the control group harbored a community richer in Neurospora, Plectosphaerella, and Phialemonium. Notably, the Helicoma, Halenospora, Pithoascu, and Microascus genera were all detected in every treatment group (TF, NF, PGA, GFL), yet not in the control group. The respective relative abundances of Helicoma under TF, of Halenospora, Pithoascus under NF, and of Microascus under PGA group surpassed those in the other groups. Finally, the relative abundance of Candida increased by 44.96% in the GFL compared to the control group.
3.6. Predictive functions of bacteria community
To verify the direct effect of MIs and GFL on soil microbial function, PICRUSt was used to analyze the sequence of 16S rRNA marker genes. The results showed that the gene copy number related to mineral absorption, plant-pathogen interaction, photosynthesis, oxidative phosphorylation and nitrogen metabolism in the rhizosphere soil of P. quinquefolium increased significantly after applying MIs and GFL. As shown in Fig. 9, the numbers of gene copies involved to mineral absorption from TF (8187), NF (8238), PGA (7023) and GFL (8199) were higher than that of CK (6468); Importantly, compared with control (79140), the gene copy number of involved to plant-pathogen interaction in response to TF (101717), NF (104137), PGA (98165) and GFL (109319) increased significantly, which was 28.53%, 31.59% and 24.04% respectively; TF, NF, PGA and GFL treatments also increased the photosynthetic gene copy number by 28.55%, 4.93%, 22.74% and 33.90%, respectively; For oxidative phosphorylation, the gene copy numbers of TF (1224097), NF (1244360), PGA (1187971) and GFL (1309243) were higher than those of CK (946360); the gene copy number of nitrogen metabolism from TF (574229), NF (589035), PGA (55168) and GFL (61600) were all higher than CK (450829).
Fig. 9.
Relative abundance of predictive metagenome functions from samples. TF, NF, PGA, and GFL represent the rhizosphere soil of P. quinquefolium treated by TuXiu microbial inoculum + Fifty potassium, No. 1 microbial inoculum + Fifty potassium, Gulefeng poly-γ-glutamic acid microbial inoculum, and garbage fermentation liquid, respectively, with CK as the control group.
4. Discussion
4.1. MIs and GFL could modify microbial community structure and composition in rhizosphere of P. quinquefolium
PGPM can play a direct or indirect role in plant growth. Some genera of PGPM are extensively studied, such as Bacillus, Lactobacillus, Chaetomium, and Trichoderma. On the one hand, PGPM can improve soil fertility by fixing nitrogen and dissolving potassium and phosphorus, thus contributing to improve the plant growth (Bononi et al., 2020, Ghadam Khani et al., 2019, Liu et al., 2019). On the other hand, they can release IAA, ACC deaminase, and other bioactive substances into the environment to stimulate plant growth (Barnawal et al., 2017, Khan et al., 2020). Moreover, some strains show strong and stable activity against plant diseases or pathogenic organisms (Dong et al., 2018, Luo et al., 2018). Furthermore, some microorganisms have shown good application prospects for the bioremediation of heavy metal-contaminated land (Banerjee et al., 2020, Pandey et al., 2019). However, one thing cannot be ignored: the influence of some beneficial microorganisms upon plants is not necessarily directly caused by its inoculation, but may be related to inducement or inhibition facilitated by other resident microbial populations (Trabelsi & Mhamdi, 2013). Because of these special traits, many microorganisms are sought after and used as active strains of a microbial inoculant, and now widely utilized in agricultural production and environmental management.
In this study, MIs and GFL modified the microbial structure and composition in the rhizosphere of P. quinquefolium. Firstly, they changed the microbial diversity index and the number of specific OTUs (Fig. 4, Fig. 5). Importantly, MIs and GFL clearly contributed to the structural changes in rhizosphere microbial communities of P. quinquefolium. At the phylum level, the relative abundance of Proteobacteria was reduced by 2.93%−11.76%, while that of Ascomycota was increased by 1.83%−14.62%. Additionally, treating the plant cultivation with TF, NF, and PGA promoted the relative abundance of Nitrospirae in rhizosphere soil (Fig. 6). Our LEfSe analysis uncovered significant changes in the genera forming under different treatments. Among these, some microorganisms can participate in the biological nitrogen fixation process, control of plant diseases, and degradation of contaminated soil. The relative abundance of Nordella and Nitrospira in the TF group was significantly increased by 75.68% and 158.10%. Recently, Yu et al. (2019) reported Nordella and Rhizobium were increased significantly after using legumes for bioremediation of soil, and Nitrospira spp. are known to play a key role in the global nitrogen cycle via their nitrification activity (Koch, van Kessel, & Lucker, 2019). Moreover, NF increased the relative abundance of Chaetomium by 153.81%; this genus and their metabolites can generally be used for the biological control of plant pathogenic organisms (Khan et al., 2019, Zhou et al., 2019). Furthermore, we found that PGA significantly increased the relative abundance of Hyphomicrobium by 23.61%. It was reported that Hyphomicrobium spp. harbored bioremediation potential for degrading organic reagents (methamidophos and dimethylsulfoxide) in the environment, and they can be applied to remediate polluted environments (Fukushima et al., 2013, Wang et al., 2010). In our experiment, Microascus was also detected in four treatment groups (TF, NF, PGA, and GFL), but not in the control group. Species of Microascus are usually used as plant biocontrol strains to control pathogens (Fotso et al., 2018). Additionally, GFL increased the relative abundance of Xylaria by a staggering 219.39%; in this respect, a number of previous studies have reported that Xylaria spp. may be used to control plant root-knot nematodes (Kim et al., 2018). Furthermore, the results of our LEfSe analysis revealed a significantly greater abundance of Plectosphaerella and Rhizobacter in the control group than in the treatment groups (i.e., TF, NF, PGA, and GFL). Interestingly, these two genera are often linked to plant diseases of crops (Hu et al., 2019, Song et al., 2017). In summary, MIs and GFL promoted the accumulation of potential beneficial microorganisms and the reduction of potential harmful microorganisms, thus modifying the microbial structure and composition in the rhizosphere of P. quinquefolium.
4.2. MIs and GFL could increase yield of P. quinquefolium
In recent years, MIs have been widely applied in crop cultivation management, for example, to bananas, tea, and rice (Jha et al., 2020, Shen et al., 2018, Zhou et al., 2020). This formulation not only can reduce the application of chemical fertilizers, but it also plays a certain role in the prevention and control of plant diseases. Besides the use in environmental management, GFL also has potential advantages for improving the quality of medicinal herbs, such as Salvia miltiorrhiza (Wei, Cao, et al., 2020). Our research results suggested that MIs and GFL are beneficial for increasing the root dry weight of P. quinquefolius, in that these applications promoted a 4.59%−83.24% greater root biomass accumulation. We also found that some special microbial taxa were changed by the introduction of MIs and GFL, such as Xylaria in GFL (an increase of 219.39%) and Chaetomium in NF (an increase of 153.82%), which may contribute to the growth promotion of plants. According to previous reports, inoculation with X. regalis can significantly increase the stem length, root length, and stem root dry matter yield of pepper seedlings (Adnan et al., 2018). The inoculation of C. cuprum in the rhizosphere of Eucalyptus globulus promoted this herb to express auxin biosynthesis gene (Ortiz et al., 2019). Moreover, Bacillus sp. and Lactobacillus sp. existed in the MIs and GFL, that are beneficial to plant biomass accumulation. B. megaterium, which can be used as a zinc-dissolving agent to enhance the yield of pepper (Bhatt & Maheshwari, 2020). Under 100 and 150 mmol/L NaCl stress, Lactobacilli sp. can promote the germination of lettuce seeds by 25% and 15%, respectively (Hussein & Joo, 2018). Collectively, these studies underscore the indispensable role of beneficial microorganisms in plant growth, and also point to why MIs and GFL can offer environmentally friendly and novel strategies for improving the yield of P. quinquefolius.
Our study also proved that MIs and GFL lessen the severity of nematode disease in P. quinquefolius, especially those grown in GFL-treated plots, where the disease was inhibited by 64.35%. According to our LEfSe analysis results, GFL could increase the relative abundance of Xylaria by 219.39%. It has been reported that lectin produced by X. hypoxylon can inhibit the plant parasitic nematode Ditylenchus dipsaci (Zhao, Guo, Liu, Wang, & Ng, 2009). So, we speculate this certain genus may work as potential bio-control agent against plant nematode diseases. Moreover, Bacillus and Lactobacillus are already in use as biocontrol agents of plant parasitic nematodes. The lethal concentration (LC50) of dichloromethane extract of B. subtilis and B. cereus was 204 g/ml, being far superior to that of furantan, a professional nematicide (Oliveira et al., 2014). The L. brevis WiKim0069 isolated from Kimchi has strong nematicidal activity against the second-stage juveniles of M. incognita, M. arenaria, and M. hapla, as well as M. incognita eggs. Moreover, the fermentation of WiKim0069 can also reduce the gall formation of melons under field conditions, and its control effect (62.8%) exceeds that of fosthiazate (32.8%) (Seo et al., 2019). These studies implied that MIs and GFL are effective at alleviating the nematode disease of P. quinquefolium. To our best knowledge, this study is the first to report on the successful biological control of plant root-knot nematode disease by GFL.
4.3. MIs and GFL could increase content of Rb1 and Re in P. quinquefolium roots
In our study, ginsenoside (Rb1 and Re) content increased by 9.05%−42.14% in the P. quinquefolium roots treated with different MIs and GFL. This result is consistent with previous findings that microbial inoculum and garbage fermentation liquid can improve the quality of Chinese medicinal herbs (Bao et al., 2019, Dong et al., 2019). Additionally, many studies have reported on endophytic fungi promoting the accumulation of metabolites in medicinal plants (Park et al., 2019, Song et al., 2017). Chaetomium spp. are among those taxa beneficial to the biosynthesis of active ingredients in medicinal plants. It has been reported that C. globosum can significantly stimulate the secondary metabolism of Salvia miltiorrhiza (Zhai et al., 2017), Anoectochilus roxburghii (Ye et al., 2020), and Chrysanthemum morifolium (Song, Dai, Liu, & Cai, 2010). In our study, the abundance of Chaetomium was increased by 66.30%−153.82% in the four treatment groups. Although no study has yet to directly report that Chaetomium is associated with ginsenoside synthesis, the above studies do imply that Chaetomium can promote the accumulation of secondary metabolites in medicinal plants. Therefore, it could be possible to regulate the microflora related to metabolite accumulation of medicinal plants via the appropriate use of MIs and GFL, to stimulate metabolites’ accumulation. In this study, under the TF and NF treatments, the relative abundances of Nitrospira, Ktedonobacter, Conlarium, Corynascus, and Pithoascus were significantly increased. It is estimated that only about 0.1%−10% of soil microorganisms have been studied well (Berg & Smalla, 2009), so we boldly speculate that some of these microorganisms we detected may be related to the accumulation of metabolites of P. quinquefolius, but more experimental studies are needed to test this hypothesis.
4.4. MIs and GFL could reduce content of as in P. quinquefolium roots
In this study, MIs and GFL contributed to a decrease in the content of the heavy metal As in P. quinquefolium, especially under GFL (52.17%) and the PGA (43.48%) microbial inoculant. Furthermore, lower Cd and Pb contents were also found in the PGA and GFL groups. Xylaria is a genus that responds positively to the introduction of GFL, which involves the biodegradation of heavy metals (Wong et al., 2018). Hyphomicrobium is a genus whose members were significantly increased by the PGA microbial inoculant in our field experiment. Hyphomicrobium spp. may be used as Mn-oxidizing bacteria in biofilters, and they can participate in the removal of both As and Mn metallic elements (Yang, Li, Chu, Ren, & Zhang, 2014). These studies jointly suggest that these taxa of beneficial microorganisms may contribute to the reduction of heavy metal content in plant roots. Often Bacillus and Lactobacillus are used to bioremediate heavy metal-contaminated soils. B. nealsonii ARP2 can adsorb 93% of As (V) from an arsenic-containing medium, while B. tequilensis ART2 can adsorb 77% of As (III), and this bacterial strain’s cell membrane can interact with As and accumulate in its cells (Pandey et al., 2019). L. casei DSM 20,011 is capable of removing As(V) (Halttunen, Finell, & Salminen, 2007). Overall, beneficial microorganisms related to the introduction of MIs and GFL, figured prominently in enabling the biological removal of heavy metals in this study. Therefore, both MIs and GFL should be considered as effective microbial remediation means for soil contaminated by heavy metals.
PICRUSt analysis based on 16S rRNA showed that the introduction of MIs and GFL provided a suitable rhizosphere environment for the bacteria which were related to nitrogen fixation, phosphorus solubilization, plant photosynthesis, mineral element absorption and plant pathogen interaction. Taken together, we speculated that application of microbial inoculants and garbage fermentation liquid, promoted P. quinquefolium yield and reduced the As content by improving the beneficial microorganisms and inhibiting pathogenic microbes in soil, as shown in Fig. 10, thus the introducing microbial inoculants and garbage fermentation liquid into P. quinquefolium cultivation has a broad application prospect.
Fig. 10.
Potential mechanism of microbial inoculants and garbage fermentation liquid regulating the soil microecology to enhance yield and reduced As content of P. quinquefolium. The green and red circles represent the beneficial and pathogenic microorganisms, respectively.
5. Conclusion
In this study, field applications of microbial inoculants and garbage fermentation liquid were able to reduce the occurrence of nematode diseases, promote the yield and effective components of P. quinquefolium, and reduce the As content in the medicinal materials. Metagenomics analysis confirmed the application of microbial inoculants and garbage fermentation liquid resulted in more potentially beneficial soil microorganisms—namely Nordella, Nitrospira, Chaetomium, Hyphomicrobium, Microascus, and Xylaria—and fewer potential harmful microorganisms, thus changing the structure and composition of the rhizosphere community. These results suggest microbial inoculants and garbage fermentation liquid could improve the structure and composition of the rhizosphere microbiota, and have positive effects on the quality of P. quinquefolium, both of them are beneficial to the development of ecological cultivation of P. quinquefolium.
Authors’ Contributions
XW and JH designed the research. PC and XW performed the research. PC and XW analyzed the data. PC wrote the manuscript. All authors have read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (No. 2019YFC1604701).
Contributor Information
Jianping Han, Email: jphan@implad.ac.cn.
Yuan Li, Email: yli@ippcaas.cn.
References
- Adnan M., Alshammari E., Ashraf S.A., Patel K., Lad K., Patel M. Physiological and molecular characterization of biosurfactant producing endophytic fungi Xylaria regalis from the cones of Thuja plicata as a potent plant growth promoter with Its potential application. BioMed Research International. 2018;2018:7362148. doi: 10.1155/2018/7362148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hazmi A.S., Al-Nadary S.N. Interaction between Meloidogyne incognita and Rhizoctonia solani on green beans. Saudi Journal of Biological Sciences. 2015;22(5):570–574. doi: 10.1016/j.sjbs.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun C., Sivashanmugam P. Investigation of biocatalytic potential of garbage enzyme and its influence on stabilization of industrial waste activated sludge. Process Safety and Environmental Protection. 2015;94:471–478. [Google Scholar]
- Arun C., Sivashanmugam P. Study on optimization of process parameters for enhancing the multi-hydrolytic enzyme activity in garbage enzyme produced from preconsumer organic waste. Bioresource Technology. 2017;226:200–210. doi: 10.1016/j.biortech.2016.12.029. [DOI] [PubMed] [Google Scholar]
- Banerjee A., Hazra A., Das S., Sengupta C. Groundwater inhabited Bacillus and Paenibacillus strains alleviate arsenic-induced phytotoxicity of rice plant. International Journal of Phytoremediation. 2020;22(10):1048–1058. doi: 10.1080/15226514.2020.1725871. [DOI] [PubMed] [Google Scholar]
- Bao W., Bu L., Liu Z., Ha D. Effects of garbage enzyme on the growth and stress resistance of Perilla frutescens. Journal of Southern Agriculture. 2019;13:145–146. [Google Scholar]
- Barnawal D., Pandey S.S., Bharti N., Pandey A., Ray T., Singh S., et al. ACC deaminase-containing plant growth-promoting rhizobacteria protect Papaver somniferum from downy mildew. Journal of Applied Microbiology. 2017;122(5):1286–1298. doi: 10.1111/jam.13417. [DOI] [PubMed] [Google Scholar]
- Berg G., Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiology Ecology. 2009;68(1):1–13. doi: 10.1111/j.1574-6941.2009.00654.x. [DOI] [PubMed] [Google Scholar]
- Bhatt K., Maheshwari D.K. Zinc solubilizing bacteria (Bacillus megaterium) with multifarious plant growth promoting activities alleviates growth in Capsicum annuum L. 3. Biotech. 2020;10(2):36. doi: 10.1007/s13205-019-2033-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bononi L., Chiaramonte J.B., Pansa C.C., Moitinho M.A., Melo I.S. Phosphorus-solubilizing Trichoderma spp. from Amazon soils improve soybean plant growth. Scientific Reports. 2020;10(1):2858. doi: 10.1038/s41598-020-59793-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Li Y., Xu J., Yang J., Wei G., Shen L., et al. Biofertilizers regulate the soil microbial community and enhance Panax ginseng yields. Chinese Medcine. 2019;14:20. doi: 10.1186/s13020-019-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Xu J., Feng G., Li X., Chen S. Soil bacterial and fungal community dynamics in relation to Panax notoginseng death rate in a continuous cropping system. Scientific Reports. 2016;6:31802. doi: 10.1038/srep31802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Xu J., Zhang L., Cheng R., Wei G., Su H., et al. Rhizospheric microbial communities are driven by Panax ginseng at different growth stages and biocontrol bacteria alleviates replanting mortality. Acta Pharmaceutica Sinica B. 2018;8(2):272–282. doi: 10.1016/j.apsb.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Liu Y., Jiao Y., Ma L., Xiao D., Guan Y. Polymerase chain reactiondenaturing gradient gel electrophoresis analysis of microbial diversity of papaya enzyme preparation during natural fermentation. Modern Food Science & Technology. 2017;33(08):80–87. [Google Scholar]
- East K.E., Zasada I.A., Schreiner R.P., Moyer M.M. Developmental dynamics of Meloidogyne hapla in Washington wine grapes. Plant Disease. 2019;103(5):966–971. doi: 10.1094/PDIS-07-18-1195-RE. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nature Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Fotso S., Graupner P., Xiong Q., Gilbert J.R., Hahn D., Avila-Adame C., et al. Alveolarides: Antifungal peptides from Microascus alveolaris active against phytopathogenic fungi. Journal of Natural Products. 2018;81(1):10–15. doi: 10.1021/acs.jnatprod.7b00337. [DOI] [PubMed] [Google Scholar]
- Franco-Sierra N.D., Posada L.F., Santa-Maria G., Romero-Tabarez M., Villegas-Escobar V., Alvarez J.C. Bacillus subtilis EA-CB0575 genome reveals clues for plant growth promotion and potential for sustainable agriculture. Functional and Integrative Genomics. 2020;20(4):575–589. doi: 10.1007/s10142-020-00736-x. [DOI] [PubMed] [Google Scholar]
- Fu L., Penton C.R., Ruan Y., Shen Z., Xue C., Li R., et al. Inducing the rhizosphere microbiome by biofertilizer application to suppress banana Fusarium wilt disease. Soil Biology and Biochemistry. 2017;104:39–48. [Google Scholar]
- Fukushima T., Whang L.M., Chen P.C., Putri D.W., Chang M.Y., Wu Y.J., et al. Linking TFT-LCD wastewater treatment performance to microbial population abundance of Hyphomicrobium and Thiobacillus spp. Bioresource Technology. 2013;141:131–137. doi: 10.1016/j.biortech.2013.03.122. [DOI] [PubMed] [Google Scholar]
- Ghadam Khani A., Enayatizamir N., Norouzi Masir M. Impact of plant growth promoting rhizobacteria on different forms of soil potassium under wheat cultivation. Letters in Applied Microbiology. 2019;68(6):514–521. doi: 10.1111/lam.13132. [DOI] [PubMed] [Google Scholar]
- Halttunen T., Finell M., Salminen S. Arsenic removal by native and chemically modified lactic acid bacteria. International Journal of Food Microbiology. 2007;120(1–2):173–178. doi: 10.1016/j.ijfoodmicro.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Hill S.N., Hausbeck M.K. Virulence and fungicide sensitivity of Phytophthora cactorum isolated from American ginseng gardens in Wisconsin and Michigan. Plant Disease. 2008;92(8):1183–1189. doi: 10.1094/PDIS-92-8-1183. [DOI] [PubMed] [Google Scholar]
- Hu H., Gao Y., Li X., Chen S., Yan S., Tian X. Identification and nematicidal characterization of proteases secreted by endophytic bacteria Bacillus cereus BCM2. Phytopathology. 2020;110(2):336–344. doi: 10.1094/PHYTO-05-19-0164-R. [DOI] [PubMed] [Google Scholar]
- Hu W., Strom N.B., Haarith D., Chen S., Bushley K.E. Seasonal variation and crop sequences shape the structure of bacterial communities in cysts of soybean cyst nematode. Frontiers in Microbiology. 2019;10:2671. doi: 10.3389/fmicb.2019.02671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein K.A., Joo J.H. Plant growth-promoting rhizobacteria improved salinity tolerance of Lactuca sativa and Raphanus sativus. Journal of Microbiology and Biotechnology. 2018;28(6):938–945. doi: 10.4014/jmb.1712.12027. [DOI] [PubMed] [Google Scholar]
- Jha P.N., Gomaa A.B., Yanni Y.G., El-Saadany A.Y., Stedtfeld T.M., Stedtfeld R.D., et al. Alterations in the endophyte-enriched root-associated microbiome of rice receiving growth-promoting treatments of urea fertilizer and Rhizobium Biofertilizer. Microbial Ecology. 2020;79(2):367–382. doi: 10.1007/s00248-019-01406-7. [DOI] [PubMed] [Google Scholar]
- Jiang C.H., Xie P., Li K., Xie Y.S., Chen L.J., Wang J.S., et al. Evaluation of root-knot nematode disease control and plant growth promotion potential of biofertilizer Ning shield on Trichosanthes kirilowii in the field. Brazilian Journal of Microbiology. 2018;49(2):232–239. doi: 10.1016/j.bjm.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan B., Yan W., Wei S., Wang Z., Zhao S., Cao L., et al. Nematicidal metabolites from endophytic fungus Chaetomium globosum YSC5. FEMS Microbiology Letters. 2019;366(14):fnz169. doi: 10.1093/femsle/fnz169. [DOI] [PubMed] [Google Scholar]
- Khan M.S., Gao J., Chen X., Zhang M., Yang F., Du Y., et al. The endophytic bacteria Bacillus velezensis Lle-9, isolated from Lilium leucanthum, harbor antifungal activity and plant growth-promoting effects. Journal of Microbiology and Biotechnology. 2020;30(5):668–680. doi: 10.4014/jmb.1910.10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.Y., Jang J.Y., Yu N.H., Chi W.J., Bae C.H., Yeo J.H., et al. Nematicidal activity of grammicin produced by Xylaria grammica KCTC 13121BP against Meloidogyne incognita. Pest Management Science. 2018;74(2):384–391. doi: 10.1002/ps.4717. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Balaraju K., Jeon Y.H. Biological characteristics of Bacillus amyloliquefaciens AK-0 and suppression of ginseng root rot caused by Cylindrocarpon destructans. Journal of Applied Microbiology. 2017;122(1):166–179. doi: 10.1111/jam.13325. [DOI] [PubMed] [Google Scholar]
- Koch H., van Kessel M., Lucker S. Complete nitrification: Insights into the ecophysiology of comammox Nitrospira. Applied Microbiology and Biotechnology. 2019;103(1):177–189. doi: 10.1007/s00253-018-9486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koljalg U., Nilsson R.H., Abarenkov K., Tedersoo L., Taylor A.F., Bahram M., et al. Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology. 2013;22(21):5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- Li L., Ma J., Mark Ibekwe A., Wang Q., Yang C.H. Influence of Bacillus subtilis B068150 on cucumber rhizosphere microbial composition as a plant protective agent. Plant and Soil. 2018;429(1):519–531. [Google Scholar]
- Li Y., Hu H., Li Z., Li R., Xu F., Zhao C., et al. Pharmacokinetic characterizations of ginsenoside ocotillol, RT5 and F11, the promising agents for Alzheimer's disease from American ginseng, in rats and beagle dogs. Pharmacology. 2019;104(1–2):7–20. doi: 10.1159/000499595. [DOI] [PubMed] [Google Scholar]
- Liu H., Li G., Li X., Chen J. Molecular characterization of bacterial community in aerobic granular sludge stressed by pentachlorophenol. Journal of Environmental Sciences (China) 2008;20(10):1243–1249. doi: 10.1016/s1001-0742(08)62216-0. [DOI] [PubMed] [Google Scholar]
- Liu W., Wang F., Sun Y., Yang L., Chen H., Liu W., et al. Influence of dragon bamboo with different planting patterns on microbial community and physicochemical property of soil on sunny and shady slopes. Journal of Microbiology. 2020;58(11):906–914. doi: 10.1007/s12275-020-0082-8. [DOI] [PubMed] [Google Scholar]
- Liu X., Li Q., Li Y., Guan G., Chen S. Paenibacillus strains with nitrogen fixation and multiple beneficial properties for promoting plant growth. PeerJ. 2019;7 doi: 10.7717/peerj.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T., Hou S., Yang L., Qi G., Zhao X. Nematodes avoid and are killed by Bacillus mycoides-produced styrene. Journal of Invertebrate Pathology. 2018;159:129–136. doi: 10.1016/j.jip.2018.09.006. [DOI] [PubMed] [Google Scholar]
- Oh J.K., Amoranto M.B.C., Oh N.S., Kim S., Lee J.Y., Oh Y.N., et al. Synergistic effect of Lactobacillus gasseri and Cudrania tricuspidata on the modulation of body weight and gut microbiota structure in diet-induced obese mice. Applied Microbiology and Biotechnology. 2020;104(14):6273–6285. doi: 10.1007/s00253-020-10634-8. [DOI] [PubMed] [Google Scholar]
- Oliveira D.F., Santos Junior H.M., Nunes A.S., Campos V.P., Pinho R.S., Gajo G.C. Purification and identification of metabolites produced by Bacillus cereus and B. subtilis active against Meloidogyne exigua, and their in silico interaction with a putative phosphoribosyltransferase from M. incognita. Anais Da Academia Brasileira De Ciencias. 2014;86(2):525–538. doi: 10.1590/0001-3765201402412. [DOI] [PubMed] [Google Scholar]
- Ortiz J., Soto J., Fuentes A., Herrera H., Meneses C., Arriagada C. The endophytic fungus Chaetomium cupreum regulates expression of genes involved in the tolerance to metals and plant growth promotion in Eucalyptus globulus roots. Microorganisms. 2019;7(11):490. doi: 10.3390/microorganisms7110490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang C., Gu L. Experiment on the control effect of garlic enzyme on two common diseases of strawberry in greenhouse. Journal of Anhui Agricultural Sciences. 2017;23:73–74. [Google Scholar]
- Pandey N., Manjunath K., Sahu K. Screening of plant growth promoting attributes and arsenic remediation efficacy of bacteria isolated from agricultural soils of Chhattisgarh. Archives of Microbiology. 2019;202(3):567–678. doi: 10.1007/s00203-019-01773-2. [DOI] [PubMed] [Google Scholar]
- Park Y.H., Chandra Mishra R., Yoon S., Kim H., Park C., Seo S.T., et al. Endophytic Trichoderma citrinoviride isolated from mountain-cultivated ginseng (Panax ginseng) has great potential as a biocontrol agent against ginseng pathogens. Journal of Ginseng Research. 2019;43(3):408–420. doi: 10.1016/j.jgr.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharmacopoeia Committee of P. R. China . (Vol. 2020). China Medical Science Press; Beijing, China: 2015. (Pharmacopoeia of the People’s Republic of China). [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Research. 2013;41(Database issue):590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues J.L., Pellizari V.H., Mueller R., Baek K., Jesus Eda C., Paula F.S., et al. Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(3):988–993. doi: 10.1073/pnas.1220608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., et al. Metagenomic biomarker discovery and explanation. Genome Biology. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.J., Park A.R., Kim S., Yeon J., Yu N.H., Ha S., et al. Biological control of root-knot nematodes by organic acid-producing Lactobacillus brevis WiKim0069 isolated from Kimchi. The Plant Pathology Journal. 2019;35(6):662–673. doi: 10.5423/PPJ.OA.08.2019.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Xue C., Taylor P.W.J., Ou Y., Wang B., Zhao Y., et al. Soil pre-fumigation could effectively improve the disease suppressiveness of biofertilizer to banana Fusarium wilt disease by reshaping the soil microbiome. Biology and Fertility of Soils. 2018;54(7):793–806. [Google Scholar]
- Silambarasan S., Vangnai A.S. Plant-growth promoting Candidasp. AVGB4 with capability of 4-nitroaniline biodegradation under drought stress. Ecotoxicology and Environmental Safety. 2017;139:472–480. doi: 10.1016/j.ecoenv.2017.02.018. [DOI] [PubMed] [Google Scholar]
- Song W.L., Dai C.C., Liu X.Z., Cai X.Z. The effect of different endophytic fungi on Chrysanthemum morifolium output and quality. Journal of Chinese Medicinal Materials. 2010;33(1):4–7. [PubMed] [Google Scholar]
- Song X., Wu H., Yin Z., Lian M., Yin C. Endophytic bacteria isolated from Panax ginseng improves ginsenoside accumulation in adventitious ginseng root culture. Molecules. 2017;22(6):837. doi: 10.3390/molecules22060837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczuka D., Nowak A., Zaklos-Szyda M., Kochan E., Szymanska G., Motyl I., et al. American ginseng (Panax quinquefolium L.) as a source of bioactive phytochemicals with pro-health properties. Nutrients. 2019;11(5):1041. doi: 10.3390/nu11051041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabelsi D., Mhamdi R. Microbial inoculants and their impact on soil microbial communities: A review. BioMed Research International. 2013;2013 doi: 10.1155/2013/863240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira P., Gleason C. Plant-parasitic nematode effectors — insights into their diversity and new tools for their identification. Current Opinion in Plant Biology. 2019;50:37–43. doi: 10.1016/j.pbi.2019.02.007. [DOI] [PubMed] [Google Scholar]
- Wang L., Wen Y., Guo X., Wang G., Li S., Jiang J. Degradation of methamidophos by Hyphomicrobium species MAP-1 and the biochemical degradation pathway. Biodegradation. 2010;21(4):513–523. doi: 10.1007/s10532-009-9320-9. [DOI] [PubMed] [Google Scholar]
- Wang W., Li J., Li Z., Pan J., Zhang Y. Eliminating redundant spatial variation to better understand the variance of interest of soil potentially toxic elements at different sampling scales in different soil types south of Nanjing, China. Environmental Science and Pollution Research. 2018;25(29):29038–29053. doi: 10.1007/s11356-018-2872-7. [DOI] [PubMed] [Google Scholar]
- Wei X., Cao P., Wang G., Han J. Microbial inoculant and garbage enzyme reduced cadmium (Cd) uptake in Salvia miltiorrhiza (Bge.) under Cd stress. Ecotoxicology and Environmental Safety. 2020;192 doi: 10.1016/j.ecoenv.2020.110311. [DOI] [PubMed] [Google Scholar]
- Wei X., Wang X., Cao P., Gao Z., Chen A.J., Han J. Microbial community changes in the rhizosphere soil of healthy and rusty Panax ginseng and discovery of pivotal fungal genera associated with rusty roots. BioMed Research International. 2020;2020:8018525. doi: 10.1155/2020/8018525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes J.E., Kirkpatrick T.L. The effects of Meloidogyne incognita and Heterodera glycines on the yield and quality of edamame (Glycine max L.) in Arkansas. J Nematol. 2020;52:1–15. doi: 10.21307/jofnem-2020-012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C., Tan L.T., Mujahid A., Lihan S., Wee J.L.S., Ting L.F., et al. Biosorption of copper by endophytic fungi isolated from Nepenthes ampullaria. Letters in Applied Microbiology. 2018;67(4):384–391. doi: 10.1111/lam.13049. [DOI] [PubMed] [Google Scholar]
- Wu J., Liu Y., Zhao R., Xu R. Fast pesticide multiresidue analysis in American ginseng (Panax quinquefolium L.) by gas chromatography with electron capture detection. Journal of Natural Medicines. 2011;65(2):406–409. doi: 10.1007/s11418-010-0500-z. [DOI] [PubMed] [Google Scholar]
- Xu F., Xin S., Lin Q., Rui Y. Determination of trace elements, heavy metals and rare earths in Pteris emipinnata L. by ICP—MS. Lishizhen Medicine and Materia Medica Research. 2010;21(1):48–49. [Google Scholar]
- Yang L., Li X., Chu Z., Ren Y., Zhang J. Distribution and genetic diversity of the microorganisms in the biofilter for the simultaneous removal of arsenic, iron and manganese from simulated groundwater. Bioresource Technology. 2014;156:384–388. doi: 10.1016/j.biortech.2014.01.067. [DOI] [PubMed] [Google Scholar]
- Yang L., Zheng C., Jiang Z., Zhou H., Xiao S. Determination of ginsenoside Rg1, Re and Rb1 in Panax quinquefolium by ultra high performance liquid chromatography. Journal of Chinese Medicinal Materials. 2008;31(1):55–57. [Google Scholar]
- Ye B., Wu Y., Zhai X., Zhang R., Wu J., Zhang C., et al. Beneficial effects of endophytic fungi from the Anoectochilus and Ludisia species on the growth and secondary metabolism of Anoectochilus roxburghii. ACS Omega. 2020;5(7):3487–3497. doi: 10.1021/acsomega.9b03789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Kang X., Li Y., Cui Y., Tu W., Shen T., et al. Rhizobia population was favoured during in situ phytoremediation of vanadium-titanium magnetite mine tailings dam using Pongamia pinnata. Environmental Pollution. 2019;255(Pt 1) doi: 10.1016/j.envpol.2019.113167. [DOI] [PubMed] [Google Scholar]
- Zhai X., Luo D., Li X., Han T., Jia M., Kong Z., et al. Endophyte Chaetomium globosum D38 promotes bioactive constituents accumulation and root production in Salvia miltiorrhiza. Frontiers in Microbiology. 2017;8:2694. doi: 10.3389/fmicb.2017.02694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang G., Zhang H., Peng D., Du Z., Tang C. Effects of soil treatments on the quantitative dynamics of rhizospheric nematodes and their control efficacy to root rot of American ginseng caused by Ditylenchus destructor. Acta Phytopathologica Sinica. 2009;39(5):555–560. [Google Scholar]
- Zhao S., Guo Y.X., Liu Q.H., Wang H.X., Ng T.B. Lectins but not antifungal proteins exhibit anti-nematode activity. Environmental Toxicology and Pharmacology. 2009;28(2):265–268. doi: 10.1016/j.etap.2009.05.003. [DOI] [PubMed] [Google Scholar]
- Zhao W., Wang Y., Jia Z., Sun L., Jin H., Ma S. Rapid screening and verification of 71 pesticide residues in Chinese medicinal materials by APGC-OT. Journal of Pharmaceutical Analysis. 2018;38(12):2152–2159. [Google Scholar]
- Zhou C.Y., Piao X.M., Yan M.X., Wang Y.P. Isolation, screening and identification of endophytic fungi and detection of its antifungal effects against Alternaria panax. China Journal of Chinese Materia Medica. 2019;44(2):274–277. doi: 10.19540/j.cnki.cjcmm.20181106.003. [DOI] [PubMed] [Google Scholar]
- Zhou S., Zeng X., Xu Z., Bai Z., Xu S., Jiang C., et al. Paenibacillus polymyxa biofertilizer application in a tea plantation reduces soil N2O by changing denitrifier communities. Canadian Journal of Microbiology. 2020;66(3):214–227. doi: 10.1139/cjm-2019-0511. [DOI] [PubMed] [Google Scholar]