ABSTRACT
Micrococcus yunnanensis TT9 was isolated from the forehead of human skin. This strain can grow on Triton X-100. We report the complete whole-genome sequence of this strain, which has one chromosome of 2,470,932 bp (73.0% G+C content) with 2,151 coding sequences.
ANNOUNCEMENT
Micrococcus yunnanensis is a high-GC-content, Gram-positive coccus phylogenetically belonging to the phylum Actinobacteria. Its involvement in the degradation of pollutants such as polycyclic aromatic hydrocarbons on the human skin has been reported (1). This article reports the isolation and whole-genome sequence of M. yunnanensis TT9 from human skin, which can grow on Triton X-100.
A swab sample from the skin of a male college student’s forehead was directly streaked on minimal salts basal medium (MSB) agar (2) containing 0.02% yeast extract and 0.5% Triton X-100 (Sigma-Aldrich). The agar medium was aerobically incubated for 1 week at 28°C for growth. One strain, named TT9, was further purified by streaking on tryptic soy broth (TSB) agar (Kisan) and incubation at 28°C for 2 days. This strain forming light-yellow colored colonies was deposited in the Korean Collection for Type Cultures as KCTC 49797. Ethical approval for subject sampling was granted by the institutional review board of Changwon National University. A similarity search for sequences in the type material of the NCBI database produced the best hits with the 16S rRNA gene sequences of Micrococcus luteus NCTC 2665 (accession number NR_075062.2), M. aloeverae AE-6 (NR_134088.1), and M. yunnanensis YIM 65004 (NR_116578.1), with more than 99% identities.
For DNA extraction, cells were cultured in a flask in TSB for 48 h at 28°C with shaking at 140 rpm. Total genomic DNA was purified using the phenol extraction method (3). Genomic DNA was sequenced with Illumina and Oxford Nanopore Technologies. Illumina sequencing was performed at DNALink Co. (Seoul, Korea). The whole-genome sequencing was performed using a TruSeq DNA PCR-free 550 bp library kit (Illumina) and demultiplexing by bcl2fastaq2 (ver. 2.20) on the Illumina NovaSeq6000 sequencer with default settings. The quality of the raw sequencing data was checked using FastQC with ASCII Qscore offset 33 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) (4). The read length was 2 × 151 of approximately 550 bp insert size. A total of 40,082,478 reads with 6,052 Mbp with a mean Phred quality score of 35.52 were produced. According to the instructions, libraries for Nanopore sequencing were prepared with the SQK-LSK109 kit and multiplexed using the EXP-NBD104 barcoding kit. Sequencing was performed on a MinION sequencer (v20.10.3) using an R9.4.1 flow cell with default settings. Reads were base called and demultiplexed using Guppy v3.4.1 in high accuracy mode with the minimum Qscore of 8. The final data set (reads, 8,756; total base number, 140.36 Mbp; mean length, 16,029 bp; minimum, 112 bp; maximum, 161,826 bp; N50, 30,795 bp) was yielded and was assembled de novo following a hybrid Nanopore–Illumina program using Unicycler v0.4.9 (https://github.com/rrwick/Unicycler) (5) with default settings. The quality of the assembled genome sequences was evaluated using CheckM v1.1.3 (https://github.com/Ecogenomics/CheckM/releases/tag/v1.1.3) (6). Gene predictions and annotations were provided by NCBI using the best-placed reference protein set and GeneMarkS-2+ of the NCBI Prokaryotic Genome Annotation Pipeline 6.1 (7). In addition, the subsystem features in the genome were analyzed by Rapid Annotations using Subsystems Technology (RAST) (https://rast.nmpdr.org/) (8). The relationship of the BLAST average nucleotide identity (ANIb) values of strain TT9 and type species in the database was searched in the JSpeciesWS server (https://jspecies.ribohost.com/jspeciesws/) (9). Digital DNA–DNA hybridization (dDDH) values were calculated by applying the Genome-to-Genome Distance calculator (GGDC 3.0) using formula 2 (https://ggdc.dsmz.de/ggdc.php) (10).
The final genome assembly of TT9 was found to be closed, circular, and rotated to place dnaA at the origin of replication and consists of one chromosome of 2,470,932 bp (73.0% G+C content), with 2,507 × mean coverage. The plasmid was not detected. Analysis with CheckM showed 98.70% completeness and 0% strain heterogeneity. TT9 exhibited the highest similarity with M. yunnanensis DSM 21948T with an ANI value of 98.17% (84.0% coverage) and dDDH of 85.5% (formula 2).
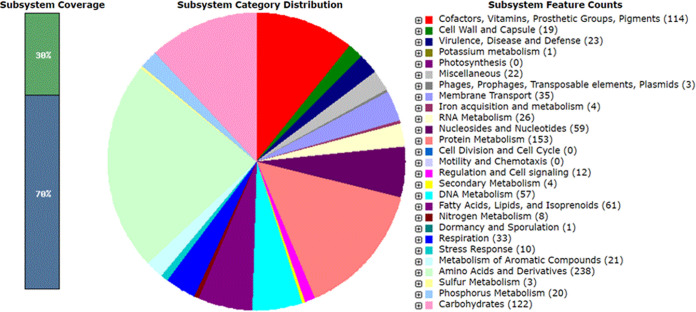
The chromosome contains 2,151 protein-coding sequences, two copies of rRNA genes (5S, 16S, and 23S), 48 coding regions of tRNAs, and three ncRNA genes. An overview of the subsystem features (functional category distribution) assigned to the genome of M. yunnanensis TT9 is shown in Fig. 1. A total of 237 subsystems (30% of coding genes) were identified. The M. yunnanensis TT9 genome contained genes coding for the glyoxylate cycle required for Triton X-100 degradation by Pseudomonas nitroreducens TX1 (11).
FIG 1.
Functional category distribution assigned to the genome of M. yunnanensis TT9 according to RAST analysis.
Data availability.
The M. yunnanensis TT9 genome sequence has been deposited in GenBank under accession number CP097650.1. Raw sequence data used for assembly were deposited in GenBank under SRA accession numbers SRX15389513 (MinIon seq) and SRX15389512 (Illumina Nova seq).
ACKNOWLEDGMENTS
This research was funded by the Financial Program for Self-Directed Research Capacity in 2022 of Changwon National University and by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (No. 2016R1D1A1B01007775).
Contributor Information
Kyoung Lee, Email: kyounglee@changwon.ac.kr.
Steven R. Gill, University of Rochester School of Medicine and Dentistry
REFERENCES
- 1.Sowada J, Schmalenberger A, Ebner I, Luch A, Tralau T. 2014. Degradation of benzo [a] pyrene by bacterial isolates from human skin. FEMS Microbiol Ecol 88:129–139. doi: 10.1111/1574-6941.12276. [DOI] [PubMed] [Google Scholar]
- 2.Stanier RY, Palleroni NJ, Doudoroff M. 1966. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol 43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel FM, B R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1990. Current protocols in molecular biology. John Wiley and Sons, New York, NY. [Google Scholar]
- 4.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom. [Google Scholar]
- 5.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meier-Kolthoff JP, Carbasse JS, Peinado-Olarte RL, Göker M. 2022. TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res 50:D801–D807. doi: 10.1093/nar/gkab902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen TN, Yeh CW, Tsai PC, Lee K, Huang SL. 2016. Transposon mutagenesis identifies genes critical for growth of Pseudomonas nitroreducens TX1 on octylphenol polyethoxylates. Appl Environ Microbiol 82:6584–6592. doi: 10.1128/AEM.01907-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The M. yunnanensis TT9 genome sequence has been deposited in GenBank under accession number CP097650.1. Raw sequence data used for assembly were deposited in GenBank under SRA accession numbers SRX15389513 (MinIon seq) and SRX15389512 (Illumina Nova seq).