Abstract
CD47 regulates the trafficking of specific coding and noncoding RNAs into extracellular vesicles (EVs), and the RNA contents of CD47+ EVs differ from that of CD63+ EVs released by the same cells. Single particle interferometric reflectance imaging sensing combined with immunofluorescent imaging was used to analyse the colocalization of tetraspanins, integrins, and CD47 on EVs produced by wild type and CD47‐deficient Jurkat T lymphoblast and PC3 prostate carcinoma cell lines. On Jurkat cell‐derived EVs, β1 and α4 integrin subunits colocalized predominantly with CD47 and CD81 but not with CD63 and CD9, conserving the known lateral interactions between these proteins in the plasma membrane. Although PC3 cell‐derived EVs lacked detectable α4 integrin, specific association of CD81 with β1 and CD47 was preserved. Loss of CD47 expression in Jurkat cells significantly reduced β1 and α4 levels on EVs produced by these cells while elevating CD9+, CD63+, and CD81+ EVs. In contrast, loss of CD47 in PC3 cells decreased the abundance of CD63+ and CD81+ EVs. These data establish that CD47+ EVs are mostly distinct from EVs bearing the tetraspanins CD63 and CD9, but CD47 also indirectly regulates the abundance of EVs bearing these non‐interacting tetraspanins via mechanisms that remain to be determined.
Keywords: CD47, EV heterogeneity, integrins, single particle analysis, surface markers, tetraspanins
1. BACKGROUND
CD47 is a cell surface receptor for thrombospondin‐1 (Gao & Frazier, 1994; Isenberg et al., 2009; Soto‐Pantoja et al., 2015) and the counter‐receptor for signal regulatory protein‐α (SIRPα) on macrophages (Oldenborg et al., 2000; Vernon‐Wilson et al., 2000). Based on its specific lateral association with several integrins, CD47 was originally named integrin associated protein (IAP) (Brown et al., 1990). Thrombospondin‐1 interaction with CD47 induces activation of the associated integrins, thereby stimulating cell adhesion and migration (Gao et al., 1996; Wang & Frazier, 1998). Recently, CD47 and several integrins have been identified as ubiquitous and abundant membrane components of EVs from a variety of cell types including platelets, myeloid‐derived suppressor cells, mesenchymal stem cells, and blood plasma (Fenselau & Ostrand‐Rosenberg, 2021; Kibria et al., 2016; Kim et al., 2012; Kugeratski et al., 2021; Sadallah et al., 2011). Functional roles of CD47 in EVs have been identified in Jurkat T cells and endothelial cells (Kaur et al., 2014), breast cancer cells (Kaur et al., 2018), ovarian cancers (Shimizu et al., 2021), and pancreatic cancer (Kamerkar et al., 2017).
EVs are ubiquitously produced by all types of cells but are heterogenous in their size, surface molecules, and internal composition (Willms et al., 2018; Witwer & Thery, 2019). Various techniques have been used to identify subsets of EVs based on biophysical properties or surface marker expression (Chiang & Chen, 2019; Panagopoulou et al., 2020; Zhang et al., 2018), and standardization protocols have been proposed (Thery et al., 2018). However, challenges remain for EV characterization using currently available techniques (Li et al., 2017; Lobb et al., 2015; Patel et al., 2019; Ramirez et al., 2018). In addition to expressing shared cell surface markers, EVs also express surface markers specific to their cells of origin (Edgar, 2016). Tetraspanins have been widely used as EV markers, and some commercial purification methods selectively purify EVs bearing the tetraspanin CD63. CD63 selectively associates with exosomes and regulates their secretion via multivesicular bodies, whereas CD9 is primarily associated with ectosomes released from the plasma membrane (Hurwitz et al., 2018; Mathieu et al., 2021). Effects of CD9 gene deletion also suggested a role for this tetraspanin in EV biogenesis (Suarez et al., 2021). Tetraspanins associate with specific integrins and regulate their functions including in development, immunity, and cancer metastasis (Smith & Holick, 1987; Vences‐Catalan and Levy, 2018; Yeung et al., 2018). Tetraspanins play multiple roles in regulating immune cell functions (Rocha‐Perugini et al., 2015; Yeung et al., 2018).
Our previous analysis of CD47+, MHC1+and CD63+ subsets of EVs produced by Jurkat cells revealed that each subset contains distinct small noncoding RNAs (Kaur et al., 2018). We also found that loss of CD47 alters the abundance of some tetraspanins on EVs, which is consistent with our findings that CD47 regulates the RNA contents of CD63+ EVs produced by Jurkat T cells (Kaur et al., 2021). However, CD47 was detected only on a subset of vesicles in multivesicular bodies. To further examine the distribution and colocalization of CD47 with tetraspanins and integrins on EVs, we have analysed EVs released by Jurkat T lymphoblast and PC3 prostate carcinoma cell lines and their respective CD47‐deficient (JinB8) and CD47‐null (CRISPR knockout) mutants. Antibody arrays were employed to capture single vesicles bearing tetraspanins (CD63, CD81 and CD9), integrins CD29(β1), CD49d (α4), CD61(β3), and CD47 in conditioned media produced by wild type (WT) and CD47 mutant Jurkat T lymphoblast and PC3 prostate carcinoma cells. Co‐expression of tetraspanins with CD47 and integrins was evaluated by staining captured 50–200 nm EVs with fluorescent antibodies specific for CD47, CD29, CD63, CD81 and CD9 along with the respective isotype control antibodies. Our results show that the integrin subunits β1 and α4 preferentially colocalize with CD47+ and CD81+ as compared to CD63+ and CD9+ subsets of EVs and that CD47 regulates the abundance of EVs bearing these tetraspanins and integrins in a cell‐type dependent manner.
2. MATERIALS AND METHODS
2.1. Cell culture and reagents
The human T lymphoblast Jurkat clone E6.1 (American Type Culture Collection), a CD47‐negative Jurkat mutant (JinB8) (Reinhold et al., 1999), PC3 human prostate carcinoma cells (ATCC) and a Cas9/CRISPR‐derived CD47‐knockout (Kaur et al., 2019) were routinely cultured using complete RPMI 1640 medium (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum, glutamine, penicillin, and streptomycin with a 5% CO2 atmosphere at 37°C. Jurkat and JinB8 cells were maintained for a maximum of ∼4 weeks for experiments. Both JinB8 and PC3‐CD47 cell lines were tested for lack of CD47 expression and cell viability along with their parental cells prior to use.
Custom chips were made using purified anti‐human CD47 Antibody, clone CC2C6(Biolegend), purified anti‐human CD61 Antibody, clone VI‐PL2(Biolegend), purified anti‐human CD29 Antibody, clone TS2/16(Biolegend), purified anti‐human CD49d Antibody, clone 9F10(Biolegend) and Tetraspanins antibodies for CD63, CD81, CD9, clones H5C6, JS81 and HI9a, respectively (Unchained Labs). Fluorescent antibodies AF488 anti‐CD29, CF647 anti‐CD47, clone B6H12(BD Biosciences), CF555 anti‐CD81, CF647 anti‐CD63, AF488 anti‐CD9, and CF647 mouse IgG1, kappa isotype, were used to phenotype EVs in the ExoView® R100(Unchained Labs).
2.2. Collection of serum free conditioned media for EVs
Jurkat and JinB8 cells (∼1*106/ml) were cultured using serum free HITES medium (Kaur et al., 2011) for 24 h. The cells were centrifuged at 300 × g (Thermo Sorvall Legend XTR centrifuge) to remove cells, and supernatants were transferred to new 50 ml Falcon Tubes. The conditioned media were further centrifuged for 10 min at 2000 × g (Thermo Sorvall Legend XTR centrifuge), and further concentrated using Amicon Ultra‐15 Centrifugal Filter Units (Millipore Sigma) to ∼1 ml volume and transferred to safe seal microcentrifuge tubes (Eppendorf, 1.7 ml) for storage at −80°C until use. Similarly, non‐concentrated conditioned media was processed and re‐centrifuged at 2500 × g using a Thermo Sorvall Legend XTR centrifuge for 10 min twice before freezing at −80°C. Conditioned media were collected into 50 ml tubes from PC3 cells plated on 75 cm2 flasks, centrifuged, and processed as described above for Jurkat cells. Both concentrated (∼1 ml) and non‐concentrated (∼15 ml aliquots) supernatants from Jurkat, JinB8, PC3, PC3‐CD47 cell lines were kept at −80°C until samples were shipped on dry ice to analysing facility (Unchained Labs).
2.3. Single vesicle analysis using conditioned media from T lymphoblast and prostate cancer cell lines
Label free capture, imaging, and size determination of EVs were performed by utilizing the ExoView® imaging platform (Unchained Labs), which detects single vesicles down to 50 nm in diameter using single particle interferometric reflectance imaging sensing (SP‐IRIS) (Daaboul et al., 2016). Customized Antibody arrays (panel of tetraspanins [anti‐CD63, anti‐CD81 and anti‐CD9], negative control [anti‐MigG], integrins β1, α4 and β3[anti‐CD29, anti‐CD49d and anti‐CD61] along with anti‐CD47) were used to isolate EVs on the chip based on surface membrane bound markers/proteins. Antibody arrays were incubated with EV‐containing samples (samples were diluted 1:5 in incubation solution I except for the samples stained for CD9. These samples were diluted 2:5 and had their particle counts normalized for comparison) to allow immobilization of EVs on the chips, and all capture spots were used in triplicates (n = 3) throughout the experimentations. After EV capture, chips were incubated with a fluorescent antibody cocktail to enable three‐colour sequential imaging with single binding event sensitivity, revealing all small EVs captured on the chips, and SP‐IRIS measured their size. The data reported per antibody array spot quantifies the number, size (50–200 nm), and phenotype of EVs bound. After detecting particles, abundance of the indicated proteins was determined based on fluorescence intensity at emission wavelengths of the respective conjugated antibodies. As positive controls, validation of CD61 and tetraspanins were performed using HEK293 human embryonic kidney, PANC‐1 pancreatic epithelioid carcinoma, and plasma EVs on customized antibody arrays.
Two CD47 antibody clones were screened for their ability to capture and detect CD47‐positive EVs. Following overnight capture of EVs from conditioned media, the CC2C6 clone captured markedly more IM (label free) particles compared to the B6H12 clone (S1A). Next, fluorescent CD47 counts on the CC2C6 capture probe were assessed(S1B). The CF647‐conjugated B6H12 clone detected more particles than the CF647‐conjugated CC2C6 clone. In addition, relatively fewer fluorescent CD47 particles were observed on the B6H12 capture probe regardless of the CD47 clone used for detection (S1B), consistent with the IM results above. Few SP‐IRIS or fluorescent particles were detected on the MIgG isotype probe, indicating specificity of CD47 capture (S1A & S1B), and few fluorescent particles were detected across all capture probes when probed with a CF647‐conjugated‐isotype antibody, mIgG1,κ, which indicates specificity of the CD47 detection antibody (S1C). Taken together, utilizing the CC2C6 clone for capture and the B6H12 clone for fluorescent detection is effective at identifying CD47‐positive EVs.
Colocalization analysis of captured EVs: Colocalization tables, which bin particles based on being positive or negative for each fluorescent channel and size, were generated in ExoView Analyzer 3.0 for each sample on each capture probe. For these analyses, only particles with a corresponding size (i.e. having an SP‐IRIS signal) were analysed, and each particle was assumed to be positive for the capture probe that it bound to (e.g. a CD47 single‐positive particle bound to the CD81 probe is considered a CD47/CD81 dual positive particle). In addition, SP‐IRIS particles were quantified before sample incubation for background subtraction, and this total was subtracted from the SP‐IRIS‐only group in the colocalization table. For each probe ligand (Lx) and capture ligand (Lc), the % colocalization
2.4. Flow cytometry analysis
Jurkat, JinB8, PC3, PC3‐CD47 cell lines were cultured as indicated above. The cells were stained with antibodies to the tetraspanins CD9, CD81 and CD63(Unchained Labs), CD47‐APC (clone CC2C6), integrin β1(CD29‐AF488, clone TS2/16), integrin α2(CD49b‐FITC, clone P1E6‐C5), integrin α5(CD49e‐PE, clone NKI‐SAM‐1), integrin α6(CD49f‐APC Cyanine7, clone GoH3), integrin α4(CD49d‐647, clone 9F10), and integrin β3(CD61‐PerCP‐Cy™5.5, clone VI‐PL2) according to the manufacturer's instructions (BioLegend, USA). The flow cytometry analyses were performed on a FACSCanto II (BD Bioscience, San Jose, CA) instrument, and data analysis was done with FlowJo v10.2(BD Bioscience, San Jose, CA) as described (Kaur et al., 2021)
2.5. Statistical analysis
The total number of particles (IM) for each indicated capture probes (n = 3 technical replicates per chip) were calculated using Student t‐test: Two‐Sample Assuming Equal Variances used. For illustration of p values, asterisks symbols are used with these criteria 0.05(*), 0.0001(**) and 0.000001(***) on the bar graphs. Similarly, Fluorescent labelling of integrin β1, CD47, CD63, CD81 and CD9 on capture spots were analysed for significance values for Figure 1, 2 panel E and F, Figure 3, Panel A–D. Standard errors for percent colocalization based on Figures 2 and 4 data were calculated using propagation of error methods.
FIGURE 1.
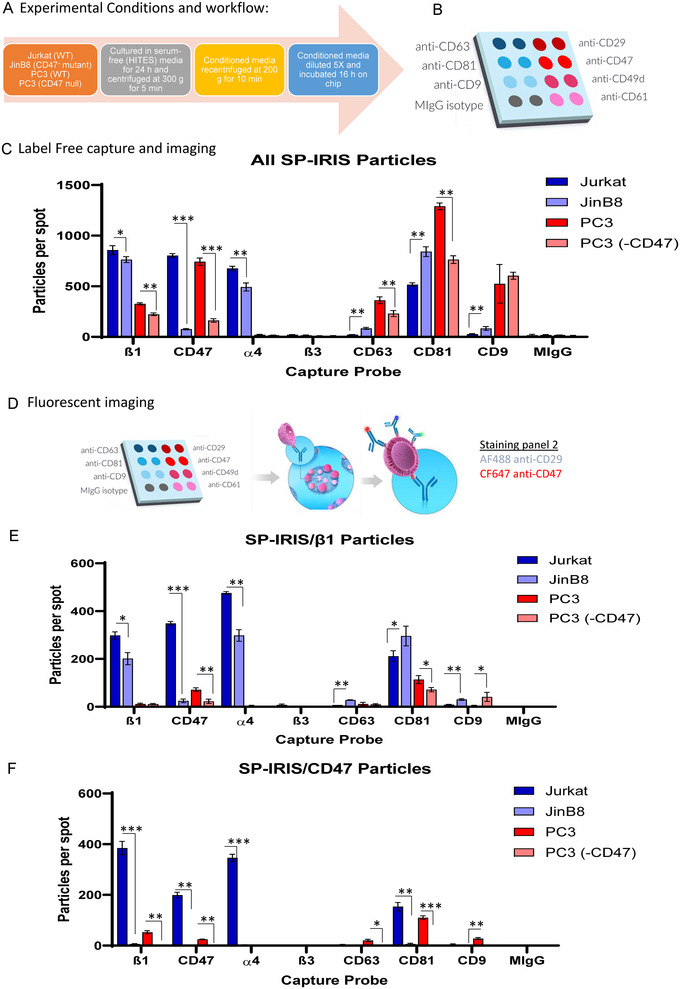
(A) Experimental conditions for collection of supernatants from WT and CD47 mutant Jurkat T Lymphoblast and PC3 prostate carcinoma cells. (B) Custom antibody array on ExoView Chip. (C) Total label‐free, SP‐IRIS particle analysis of 50–200 nm EVs released from Jurkat and PC3 cells captured on spots with antibodies specific for β1 integrins (CD29), CD49d (α4), CD61 (β3), CD47, and the tetraspanins CD63, CD81 and CD9 EVs. Error bars indicate SD, N = 3. (D) Overview of workflow labelling captured EVs using CF647‐anti‐CD47 and AF488‐anti‐CD29. (E) Fluorescent imaging using CF647‐anti‐CD47 to detect CD47+ EVs captured on customized antibody array chip (Panel C). (F) Fluorescent imaging using AF488‐anti‐CD29 to detect β1+ EVs captured on the customized antibody array chip (Panel C). Significant values were calculated using t‐test: two‐sample assuming equal variances for each capture spot (n = 3). Asterisks indicate p‐values: * = P > 0.05, ** = P > 0.0002, and *** = P > 0. 00003
FIGURE 2.
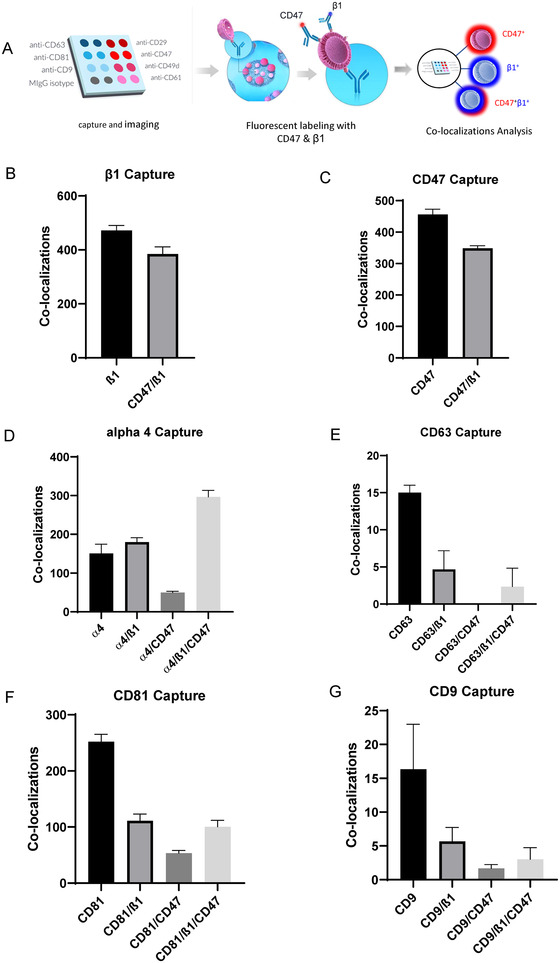
(A–G) Single particle fluorescent imaging to detect colocalization of β1 and CD47 using fluorescent antibodies CF647‐anti‐CD47 and AF488‐anti‐CD29 on 50–200 nm Jurkat EVs captured on β1, CD47, α4, β3, CD63, CD81 and CD9 antibodies. Three replicates (n = 3) per capture spot were used for each panel, Error bars indicate SD, N = 3
FIGURE 3.
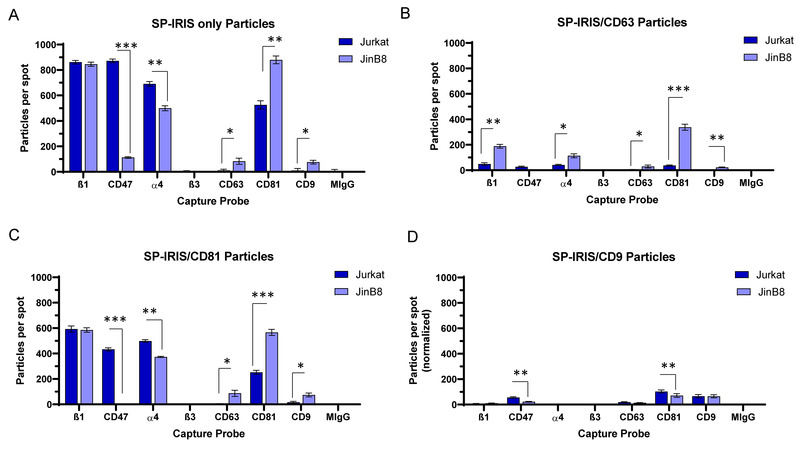
(A) Total label‐free SP‐IRIS particle analysis of 50–200 nm EVs captured on spots with antibodies specific for β1, CD47, α4, β3, CD63, CD81 and CD9 using Jurkat and JinB8 supernatants. (B) Fluorescent imaging of CD63+ EVs using CF647‐anti‐CD63, (C) CF555‐anti‐CD81 for CD81+ EVs and, (D) CF488‐anti‐CD9 for CD9+ EVs captured on β1, CD47, α4, β3, CD63, CD81 and CD9 antibody spots as shown in Panel A. Significant values were calculated using a two‐sample t‐test assuming equal variances for each capture spots (n = 3), and Asterisks value indicate p‐values: * = P > 0.05, ** = P > 0.0002, and *** = P > 0.00003
FIGURE 4.
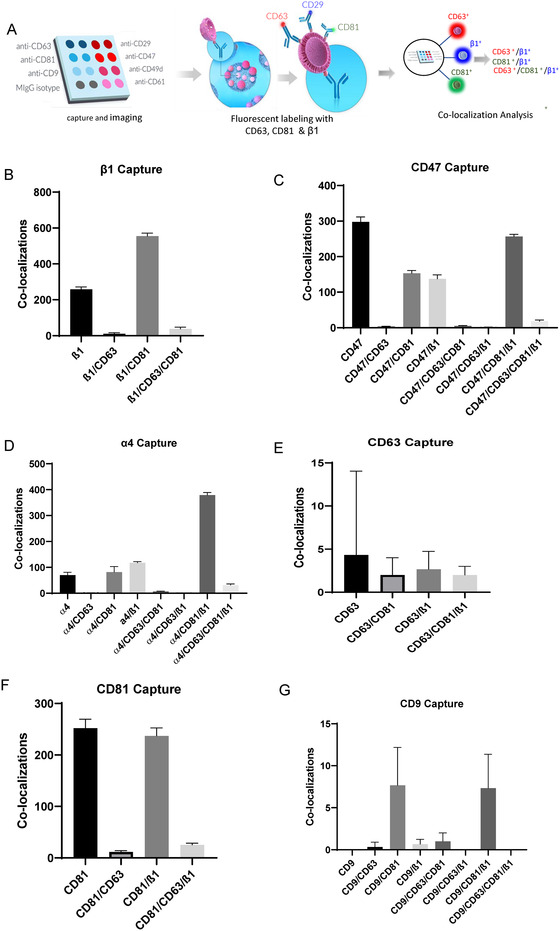
(A–G) Colocalization between β1+, CD63+ and CD81+ on 50–200 nm Jurkat EVs using AF488‐anti‐ CD29, CF647‐anti‐CD63 and CF555‐anti‐CD81 antibodies on EVs captured on anti‐ β1, CD47, α4, β3, CD63, CD81 and CD9. Three replicate spots were used per capture for each panel. Error bars indicate SD, N = 3
3. RESULTS AND DISCUSSION
3.1. Detection and quantification of CD47, integrins and tetraspanins on EVs via single vesicle analysis
Flow cytometry using APC‐conjugated CD47 antibody confirmed complete loss of plasma membrane CD47 expression on the Jurkat JinB8 and PC3(CD47−) CRISPR mutant (Figure S2A–1F). Cell supernatants containing EVs released from WT Jurkat and PC3 cells and the respective CD47− mutants were collected as shown in (Figure 1A).
The supernatants were incubated on a customized antibody array to capture EVs expressing tetraspanins (anti‐CD63, anti‐CD81 and anti‐CD9), β1, α4 and β3 integrins (anti‐CD29, anti‐CD49d and anti‐CD61) along with anti‐CD47 and respective negative control (anti‐MIgG), for 16 h (Figure 1B). Label‐free counting and sizing of the captured EVs from Jurkat and PC3 via single particle interferometric reflectance imaging sensing (SP‐IRIS) identified the size distribution of particles in the range of 50–200 nm captured on each immobilized antibody spot (Figure S2G–L, n = 3). Overall, EVs captured on integrins were larger than those captured on CD47 or tetraspanin antibodies. β1+ EVs had the highest mean size (Figure S2G), followed by α4+ (Figure S2H), CD47+ (Figure S1I), CD81+ (Figure S1J), CD9+ (Figure S2K), and CD63+ EVs (Figure S2L). Captured Jurkat EVs were larger than PC3 EVs, though this difference was modest on the tetraspanins spots. On the other hand, captured EVs produced by JinB8 cells were smaller than those from WT Jurkat cells except on the immobilized CD9 antibody (Figure S2K). EVs captured from CD47‐ PC3 cells were larger than those from WT PC3 cells except on CD47 and α4 capture spots (Figure S2H, I).
3.2. SP‐IRIS of total EVs
α4β1 is the major integrin expressed by Jurkat T cells that associates with CD47, and this integrin is activated by CD47 signalling in Jurkat cells (Azcutia et al., 2013; Barazi et al., 2002). Immobilized β1 and α4 integrin antibodies efficiently captured EVs produced by WT Jurkat cells, whereas minimal capture was observed on anti‐β3, which is poorly expressed by these cells (Figure 1C). In contrast, fewer PC3 EVs expressed β1 integrin, and anti‐α4 capture was minimal. On the other hand, greater capture on the pooled tetraspanin antibodies was observed in PC3‐derived EVs. SP‐IRIS of EVs captured on β1 spots (n = 3) showed that Jurkat cells express more β1 integrin+ EVs than PC3. Although the JinB8 and PC3(‐CD47) cells lack CD47 expression, some EV capture above mIgG background was detected on the CD47 antibody spots. Loss of CD47 slightly increased β1 integrin levels on the surface of Jurkat cells as detected by flow cytometry (Figure S3A), but the number of β1+ EVs was significantly reduced in supernatants from JinB8 cells (Figure 1C). The β1 subunit pairs with multiple α subunits to form functional dimers. We examined the expression of four α subunits that pair with β1 and have been reported to interact physically or functionally with CD47(Figure S3E–H). Only α4 showed a decrease in JinB8 cells, which is consistent with the decreased number of α4+ EVs released by JinB8 cells (Figure 1C). This suggests that lack of CD47 significantly reduced the trafficking of α4β1 integrin into EVs. This is consistent with the strong physical interaction between α4β1 and CD47 and the defect in integrin activation in CD47‐deficient Jurkat cells (Azcutia et al., 2013; Barazi et al., 2002).
PC3 cells also showed a selective decrease in α4 integrin expression on the CD47− mutant (Figure S3I–L), but α4‐positive EVs were not detected for these cells (Figure 1C). Consistent with the low expression of β3 integrin on Jurkat cells (Figure S3B), Jurkat supernatants contained minimal numbers of β3+ EVs. Although β3 integrin is expressed on prostate cancer cells and their EVs (Krishn et al., 2019; Quaglia et al., 2020), the level on PC3 cells was similar to that on Jurkat cells (Figure S3D), and β3+ EVs were not detected (Figure 1C). Function of the β3 capture probe was confirmed using HEK‐derived EVs, which are negative for β3, and plasma derived EVs, which have abundant β3+ EVs (Figure S3M).
Flow cytometry analysis indicated that Jurkat cells express high levels of CD63 and CD81 and low levels of CD9 (Figure S1E–J). In contrast, PC3 cells strongly express all three tetraspanins. Consistent with its interaction with α4β1 integrin, CD81 was moderately lower in JinB8 cells, whereas CD63 and CD9 were moderately higher. Overall, PC3 cells secreted more tetraspanin‐positive EVs than Jurkat cells (Figure 1C). Lack of CD47 on JinB8 cells led to a significant increase of CD63+ EVs, whereas PC3(CD47‐null) supernatants contained less CD63+ EVs than WT supernatants. Similarly, CD81+ and CD9+ EVs were more abundant in JinB8 relative to Jurkat supernatants, whereas CD81+ EVs were significantly reduced in CD47‐deficient PC3 cell conditioned media compared to supernatants from parental PC3 cells. PC3 supernatants contained more CD9+ EVs than Jurkat cell supernatants, but the loss of CD47 did not significantly alter their numbers. Thus, CD63+ and CD81+ EVs were impacted due to lack of CD47, which is consistent with analysis of EVs from WT Jurkat and JinB8 cells isolated using size exclusion chromatography (Figure S1 in Kaur et al., 2021).
3.3. Colocalization of CD47, tetraspanins, and β1 integrin on EVs using single vesicle analysis
Probing captured EVs on each antibody with a fluorescent‐tagged CD47 antibody enabled analysis of CD47 colocalization on EVs with integrins and tetraspanins (Figure 1D–F). CF647‐conjugated anti‐CD47 bound to Jurkat and PC3 EVs captured on anti‐CD47 but not to EVs from the respective CD47 mutants (Figure 1E). Similarly, CF647‐conjugated anti‐CD47 bound significantly to Jurkat and PC3 EVs captured on anti‐β1, α4, CD47 and CD81 spots (n = 3); however, CD47 was not detected on EVs captured on anti‐CD63 or CD9 spots (Figure 1E). This suggested that CD47+ EVs are distinct from CD63+ and CD9+ subsets produced by the same cells, which is consistent with our previous finding that CD47+ EVs contain distinct small RNA contents compared to CD63+ and MHCI+ EVs released from Jurkat cells (Kaur et al., 2018).
CD47 interacts specifically with a subset of β1 integrins in various types of cells (Barazi et al., 2002; Brown & Frazier, 2001; Ticchioni et al., 2001), but their interaction within EVs has not been examined. CD47 interacts with α4β1 in Jurkat T cells, and CD47 ligation regulates activation of this integrin in these cells (Barazi et al., 2002). As shown in Figure 1C, significantly fewer β1+ EVs were detected in JinB8 and PC3(‐CD47) supernatants than in Jurkat and PC3 supernatants, which suggested a role for CD47 in trafficking of these associated integrins to EVs. EVs from JinB8 cells were captured on immobilized anti‐β1 less efficiently than EVs from WT Jurkat cells (Figure 1C). Correspondingly, binding of AF488‐conjugated anti‐CD29 to JinB8‐derived EVs on immobilized anti‐β1 spots was reduced compared to WT Jurkat‐derived EVs (Figure 1F). Therefore, CD47 is not necessary for α4β1 integrin to be present on EVs, but its abundance is decreased in the absence of CD47. The low abundance of β1 integrin in PC3 cells limited the ability to evaluate colocalization, but AF488‐conjugated anti‐CD29 labelling was strongest on CD47+ and CD81+ EVs. Consistent with the total capture in Figure 1C, the highest number of β1 integrin+ Jurkat EVs were captured on anti‐CD81 spots relative to EVs captured on anti‐CD63 or anti‐CD9, and higher numbers of JinB8 EVs were captured on each of the tetraspanin antibodies (Figure 1F). This suggests that lack of CD47 on cells and/or EVs released from JinB8 cells leads to significantly increased production of β1+ EVs that bear these tetraspanins. A similar significant CD47‐dependence was observed for PC3 cell EVs (Figure 1F). On the other hand, loss of CD47 in PC3(‐CD47) cells resulted in fewer β1+ EVs detected on anti‐CD81 capture spots compared to EVs from WT PC3 cells (compare CD81 bar graphs in Figure 1C and F).
3.4. Colocalization of CD47 and CD29 in vesicles on capture spots
Colocalization of CD47 with β1 integrin on single captured EVs derived from Jurkat cells was further analyzed by counting β1+ positive EVs that were positive or negative for CD47 detected using a fluorescent antibody (Figure 2A). Consistent with its known association with α4β1 integrin on Jurkat cells (Barazi et al., 2002) CD47 was detected on 44.9 ± 2.1% of the β1+ EVs captured on anti‐β1 (Figure 2B), and 43.3 ± 1.3% of CD47+ Jurkat EVs were labelled with the anti‐β1 antibody (Figure 2C). As expected, 70.3 ± 1.7% of β1 integrin subunit was colocalized with its known partner α4 on EVs, and 50.8 ± 1.4% of the α4+ EVs were labelled with the CD47 antibody (Figure 2D). Parallel colocalization analysis of EVs from JinB8 cells showed the expected absence of CD47+ EVs captured on anti‐β1 (Figure S4A) and minimal capture of these EVs on anti‐CD47 spots (Figure S4B).
In contrast, the EVs captured on each tetraspanin showed less colocalization with the integrins and CD47. Consistent with its preferential physical and functional association with α4β1 integrin (Feigelson et al., 2003; Mannion et al., 1996; Serru et al., 1999; Spring et al., 2013), 41 ± 1.8% of CD81+ EVs were β1+ and 29,8 ± 1.3% were CD47+ (Figure 2F). Jurkat EVs were poorly captured on either anti‐CD63 or CD9 spots. On CD63+ EVs, β1 was colocalized on 30 ± 9% and CD47 only on 10 ± 6.6% (Figure 2E). On CD9+ EVs, β1 was colocalized on 32.5 ± 5.8% and CD47 only on 17.5 ± 3.9% (Figure 2G). The minimal colocalization of CD47 with CD63 further suggests that CD63+ EVs are distinct from CD47+ EVs. However, the low levels of CD63 and CD9 limits further interpretation of their colocalizations.
PC3 EVs also expressed β1 integrin and CD47, but less colocalization of CD47 with β1 integrin was observed compared to Jurkat EVs (Figure S4C,E). This is consistent with the minimal α4 subunit expression in these cells and the weaker lateral association of CD47 with other β1 integrins. As expected, no CD47+ EVs were captured on anti‐β1 in the PC3 CD47 null supernatant, (Figure S4B), and capture of these EVs on anti‐CD47 was much lower than for the WT PC3 cell‐derived EVs (Figure S4F).
3.5. CD47‐dependence of tetraspanin colocalization with integrins and other tetraspanins
While tetraspanins were more abundant on PC3 EVs as compared to Jurkat EVs (Figure 1A), the abundance of β1 and α4 integrin‐positive EVs released from PC3 cells was below the limit of detection (Figure 1F). Therefore, we selected Jurkat EVs to further examine tetraspanin colocalization with α4β1 integrin and other tetraspanins (Figure 3). Total Jurkat EV capture was higher on anti‐β1, α4, and CD81 compared to anti‐CD63 or CD9 spots (Figure 3A). JinB8 EV capture on anti‐CD47 was significantly reduced and close to the background threshold (Figure 3A). Further analysis of JinB8 EV total particle capture showed that loss of CD47 did not alter total capture on anti‐β1 but significantly reduced capture efficacy on anti‐α4 integrin spots, consistent with Figure 1C and indicating that loss of its interaction partner CD47 impairs the biogenesis of EVs containing α4β1 integrin. In contrast, JinB8 EV capture was significantly increased on anti‐CD63, CD81, and CD9 (Figure 3A). Comparing Figures 3A to 1A showed chip to chip reproducibility for total particle capture.
Tetraspanins are known to form lateral intermolecular complexes, including those involving CD9 with CD81 (Rubinstein et al., 1996). Fluorescent labelling of EVs with anti‐CD63 (Figure S5A) showed modest capture of CD63+ Jurkat EVs on anti‐CD81 spots, less capture by anti‐β1 and α4, and minimal signal on anti‐CD63 or CD9 spots (Figure 3B). Remarkably, all colocalization signals were significantly increased using CD47− JinB8 EVs, with the expected lack of signal for anti‐CD47 capture. This suggested that the presence of CD47 inhibits the colocalization of CD63 on EVs with α4β1 integrin and the tetraspanins CD9 and CD81.
Significant increases in positivity for the three tetraspanins were observed for CD63+ JinB8 EVs (Figure 3B) and CD81+ JinB8 EVs (Figure 3C). However, comparable increases were not found for CD9+ JinB8‐deried EVs relative to the respective Jurkat‐derived EVs (Figure 3D). Thus, in contrast to the β1 and CD47 labeling data in Figure 1E,F, the absence of CD47 increases the number of CD63+, CD81+ and CD9+ EVs that are captured on the three tetraspanin antibodies.
3.6. Colocalization of β1 with tetraspanins on EVs from lymphoma cells
Colocalization of tetraspanins with integrins and CD47 on single captured EVs derived from Jurkat cells was further analysed by counting EVs positive for various combinations of fluorescent labelled CD63, CD81, CD9, and β1 antibodies (Figure 4A). Consistent with the data in Figure 2E and F, 68.7 ± 1.2% of Jurkat EVs captured on anti‐β1 were positive for CD81, whereas only 5.6 ± 0.7% were positive for CD63 (Figure 4B). In Jurkat cells, β1 exists in functional dimers primarily with α4 and α5 and minor amounts of other α subunits. Analysis of EVs captured on anti‐α4 supported published evidence that CD81 selectively associates with α4β1 (Figure 4D). The 72.2 ± 1.9% colocalization of CD81 and 6 ± 0.4% colocalization of CD63 with α4 closely match the percentages for the respective colocalizations with β1 in panel B. Similarly, EVs captured with anti‐CD47, which specifically interacts with α4β1 integrin in Jurkat cells (Barazi et al., 2002), showed similar colocalizations of CD47 with CD81(49.6 ± 0.6%) as with β1(47.4 ± 0.8%), Figure 4B). Consistent with Figure 2E, CD63 was almost completely absent from CD47+ EVs (3.1 ± 0.2%). Although CD63+ EVs can be readily purified from Jurkat cells and their RNA contents analysed (Kaur et al., 2018), and anti‐CD63 spots captured more than 70 particles, relatively few had sufficient CD63 to also bind labelled anti‐CD63, and minimal labelling with anti‐CD81 or anti‐β1 prohibited further analysis.
CD81 contributes to CD3‐dependent T cell activation and is required for regulating CD3ζ, ZAP‐70, LAT, CD69 surface expression, extracellular signal‐regulated kinase (ERK) phosphorylation, and interleukin‐2 (IL‐2) secretion (Rocha‐Perugini et al., 2013), which overlap with functional roles of the CD47‐integrin complex on T cells (Azcutia et al., 2013; Barazi et al., 2002). Consistent with Figure 1C and E, CD81 was the most abundant tetraspanin on EVs detected by simultaneous capture and labelling (Figure 4F), and of these 49.8% ± 1.7% showed colocalization with β1 integrin, but only 7 ± 0.4% colocalized with CD63.
The association of CD81 with α4β1 integrin does not require CD47 because similar colocalization as seen in WT Jurkat cells was seen in JinB8 cells (Figure S5B,D,F). In contrast, JinB8 cells showed much higher colocalizations of CD81 with CD63 (Figure S5E,F) and CD9 (Figure S5G). Combined with the data in Figure 1C showing a higher overall abundance of CD9+, CD63+ and CD81+ EVs produced by JinB8 cells, the increased colocalization of CD81 with CD9 and CD63 suggests that loss of CD47 signalling results in selective accumulation of CD81 in an EV subset that contains the other tetraspanins but not in the α4β1 integrin+ subset of EVs,
Consistent with Figure 1C and E, few Jurkat EVs were captured on anti‐CD9 spots (Figure 4G). and 44 ± 13% of the CD9+ EVs colocalized with β1 but minimally with CD63 (1.3 ± 3.7%). However, 89 ± 20% of CD9+ EVs were colocalized with CD81 and 44 ± 13% of these were β1+ (Figure 4G). The low numbers of Jurkat EVs captured by anti‐CD9 prohibited further analysis.
Although PC3 cells produced more abundant CD63+ and CD9+ EVs than Jurkat cells (Figure 1C), the analysis in Figure 1E indicated that relatively few of these EVs contain β1 integrins. However, label‐free counting via SP‐IRIS analysis further indicated that the selective association of β1 integrins with CD81 is conserved in PC3 cell EVs. WT PC3 EVs captured on CD81 showed greater colocalization in vesicles with β1 integrins (8.8 ± 1.5%) than the respective EVs captured on anti‐CD9 (0.8 ± 0.1%) or CD63 (3.3 ± 2.0%) (Figure S4G–I). EVs from the CD47− mutant PC3 cells showed similar colocalization of β1 integrins with the tetraspanins as observed for the WT PC3 cell EVs, with 9.4 ± 1.6%, 6.7 ± 2.8%, and 4.1 ± 0.8% for CD81, CD9 and CD63, respectively. Conversely, CD47 showed limited selectivity for colocalization with CD81 in EVs from PC3 cells: 8.5 ± 0.5%, 5.6 ± 1.5%, and 5.6 ± 0.9% for CD81, CD9 and CD63, respectively.
Label‐free counting via SP‐IRIS analysis was also performed using EVs derived from HEK 293 and PANC‐1 human pancreatic cancer cells, which produce EVs with high levels of the tetraspanins CD63 and CD9 (Figure S6A). The preferential association of β1 integrins with CD81+ EVs extended to EVs released by HEK‐293 cells (Figure S6). Vesicles captured on anti‐CD81 spots were enriched in β1+ EVs relative to those captured on anti‐CD63 or anti‐CD9 (Figure S6B). SP‐IRIS analysis of HEK‐293 cell EVs identified 16 ± 5.4% of CD9, 8 ± 0.7% of CD63, and 28 ± 0.5% of CD81 EVs colocalized to vesicles with β1 integrin (Figure S6E–H). In contrast, PANC‐1 cell EVs showed a higher relative abundance of β1 integrin+ vesicles captured on anti‐CD63 (Figure S6B), Correspondingly, SP‐IRIS analysis showed 73 ± 5% of CD9, 53 ± 0.5% of CD63, and 68 ± 0.5% of CD81 EVs colocalized with β1 integrin (Figure S6E–G). Therefore, the preferential colocalization of β1 integrins with CD81+ EVs is not unique to T lymphoblast cells and is conserved in PC3 and HEK‐293 cells, but the divergence of PANC‐1 EVs demonstrates that β1 integrins can colocalize with CD63 or CD9 on EVs from other cell types. This is consistent with reports that specific integrins form complexes with these tetraspanins in several cell types (Israels et al., 2001; Mitsuzuka et al., 2005; Okochi et al., 1997; Radford et al., 1996).
4. CONCLUSIONS
Taken together, the present data demonstrate that β1 and α4 integrins colocalize predominantly with CD47 and CD81 on Jurkat T cell‐derived EVs but not with CD63 and CD9 (Figure 5A). Therefore, CD47+ EVs are mostly distinct from EVs bearing the tetraspanins CD63 and CD9. This divergence between CD47+ and CD63+ EVs released by Jurkat cells is consistent with the divergent small RNA contents of the CD47+ and CD63+ EVs released by these cells (Kaur et al., 2018). We also demonstrate that loss of CD47 significantly reduces the abundance of β1+ and α4 integrin+ EVs but increases the abundance of CD63+, CD9+ and CD81+ EVs (Figure 5A). This shift in the abundance of tetraspanin‐rich versus integrin‐rich subsets of EVs is consistent with our finding that EVs released by WT and CD47‐deficient Jurkat cells have divergent effects on the activation of recipient T cells and angiogenic responses in recipient endothelial cells (Kaur et al., 2014) and that CD63+ and MHC1+ subsets of EVs from JinB8 are enriched in small non‐coding RNAs relative to the respective EVs from Jurkat cells and differentially altered gene expression in target human umbilical vein endothelial cells in a CD47‐dependent manner to regulate VEGF and inflammatory signalling, cell cycle, and lipid and cholesterol metabolism pathways (Kaur et al., 2021).
FIGURE 5.
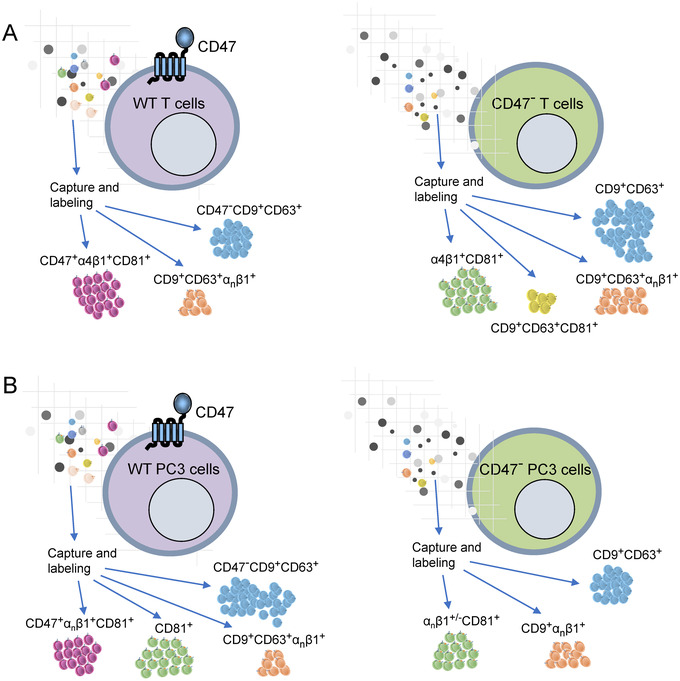
(A) Single vesicle imaging indicates that WT Jurkat T cells produce several distinct subsets of EVs. The CD47+ subset is highly enriched in α4β1 and the tetraspanin CD81. The tetraspanins CD9 and CD63 are in a distinct subset of EVs that lacks CD47, some of which contain other β1 integrins. Loss of CD47 by mutation in these cells results in increased release of EVs bearing one or more of the three tetraspanins. CD81 remains preferentially associated with α4β1 integrin, but more CD81+ EVs are present that lack α4β1 integrin. (B) PC3 prostate carcinoma cells do not express α4β1 integrin. Therefore, although CD81+ EVs are more abundant than in Jurkat cells, fewer colocalize with CD47 and β1 integrins. PC3 cells also produce more abundant CD9+ and CD63+ EVs, which do not colocalize with CD47. In contrast to Jurkat cells, CRISPR/Cas9 deletion of CD47 in PC3 cells decreases the abundance of CD81+ and CD63+ EVs, and more of the CD9+ EVs colocalize with β1 integrins
Supramolecular protein complexes can self‐assemble in the plasma membrane of cells based on direct lateral interactions between membrane proteins or can be mediated by specific interactions of transmembrane proteins with cytoskeletal elements that stabilize such complexes (Serge, 2016). CD47 is known to interact directly with specific integrins including α4β1, but it also exists in supramolecular complexes in cells that lack integrins, such as the Rh antigen complex on red blood cells (Soto‐Pantoja et al., 2015). Because cytoskeletal elements required for these interactions in cells may not transfer to EVs, supramolecular complexes in EVs may differ from those in their cells of origin. However, our data demonstrates that the CD47 interaction with α4β1 integrin is maintained in EVs and that the preferential association of α4β1 integrin with CD81 also enriches this tetraspanin in CD47+ EVs. Therefore, the abilities of CD47 and CD81 to modulate integrin activation may also be conserved in EVs. In addition to the T cell line studied most extensively, enrichment of β1 integrins in EVs containing CD81 was also demonstrated for prostate carcinoma and embryonic kidney cells, but not for a pancreatic carcinoma cell line.
In addition to colocalization based on direct interactions, analysis of EVs produced by CD47‐defiicient mutants revealed its indirect regulation of the tetraspanin contents of EVs (Figure 5). The absence of CD47 in Jurkat cells resulted in increased numbers of EVs bearing CD9, CD63, and CD81 released by Jurkat cells, whereas the absence of CD47 in PC3 cells decreased the numbers of EVs bearing CD63 and CD81 without affecting CD9+ EVs. One potential mechanism for the increased abundance of CD9+ EVs released by JinB8 cells could involve CD47 dependent regulation of CD9+ EVs produced by the ectosome pathway (Mathieu et al., 2021). Investigation of additional markers of budding from the plasma membrane in Jurkat versus JinB8 cells and EVs could be used to further evaluate effects of CD47 signalling on this pathway. Based on the colocalization data, the divergent effects of CD47 loss on integrin‐rich versus tetraspanin‐rich EV subsets in the prostate carcinoma cells may result from the absence of α4β1 integrin (Figure 5B), which appears to retain CD81 in the CD47/α4β1 integrin+ EVs produced by Jurkat cells (Figure 5A).
AUTHOR CONTRIBUTIONS
Sukhbir Kaur: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Writing – original draft; Writing – review & editing. George Daaboul: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Visualization; Writing – review & editing. Leif Anderson: Conceptualization; Formal analysis; Methodology; Visualization; Writing – review & editing. David D. Roberts: Conceptualization; Funding acquisition; Project administration; Supervision; Writing – review & editing.
CONFLICT OF INTEREST
S.K., F.L., and D.R. do not have any conflict of interest.
Supporting information
Figure S1: (A,B) Standardization of CD47 antibodies. (C) Evaluation of non‐specific interaction of CF647 anti‐mIgG1κ with Jurkat, JinB8, PC3, and PC3 (‐CD47) EVs on captured spots β1, CD47, α4, β3, CD63, CD81 and CD9. (F) CD29 (β1), co‐stained with CD47 antibodies. (E–J) Flow cytometry analysis of CD9, CD63 and CD81 surface expression on Jurkat, JinB8, PC3 and CRISPR targeted PC3(‐CD47) cells relative to unstained controls (grey). Figure S2: (A–F) Surface expression of CD47 using Jurkat, JinB8, PC3 and CRISPR targeted PC3(‐CD47) cells along with unstained control via flow cytometry. (G–L) Sizing of the Label Free captured Jurkat, JinB8, PC3, and PC3 (‐CD47) EVs on spots β1, CD47, α4, β3, CD63, CD81 and CD9 via SP‐IRIS. Figure S3: (A–L) Surface expression of CD29/β1 integrin, CD61/β3 integrin, CD49b/α2 integrin, CD49d/α4 integrin, CD49e/α5 integrin, and CD49f/α6 integrin using Jurkat, JinB8, PC3 and PC3(‐CD47) cells along with unstained control via flow cytometry. (M) Validation of CD61 antibody using human embryonic kidney (HEK) cell conditioned media and plasma incubated on anti‐β1, CD47, α4, β3, CD63, CD81 and CD9 capture spots. Figure S4: (A–F) Fluorescent imaging was used to determine colocalization of β1 and CD47 on vesicles using fluorescent antibodies CF647‐anti‐CD47 and AF488‐anti‐CD29 on 50–200 nm EVs from JinB8, PC3 and PC3 (‐CD47) cells captured on anti‐β1 and CD47 antibody spots. (G–I) Fluorescent imaging was used to determine colocalization of β1 and CD47 with tetraspanins on vesicles captured on CD81, CD9 and CD63 antibody spots. Figure S5: (A) Visualization of labelled tetraspanins extracted from Figure 3B–D graphs for Jurkat Panel only. (B–G) Colocalization between β1+, CD63+ and CD81+ EVs using anti‐ AF488‐CD29, anti‐CF647‐CD63 and CF555‐CD81 antibodies to stain Jurkat and JinB8 EVs captured on spots coated with β1, CD47, α4, β3, CD63, CD81 and CD9 antibodies. Error bars indicate SD, N = 3. Figure S6: (A) Total label‐free, SP‐IRIS particle analysis of 50–200 nm EVs released from HEK293, and PANC‐1 cells captured on spots with antibodies specific for CD63, CD81 and CD9. (B–D) Fluorescent detection of EVs containing β1 integrins (CD29) or CD47 captured on the indicated tetraspanins. Error bars indicate SD, N = 3. (E–G) Vesicle colocalization of β1+ integrins and CD47+ with CD63, CD81 and CD9 on captured spots. (H) Percentage of colocalization of tetraspanins with β1+/CD47+.
ACKNOWLEDGEMENTS
L.A. and G.D. are employees Unchained Labs. G.D. is a shareholder of Unchained Labs. Research funding was provided by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (DDR, project ZIA SC009172). Flow cytometry analyses were performed at the CCR/LGI Flow Cytometry Core which supported by the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Kaur, S. , Livak, F. , Daaboul, G. , Anderson, L. , & Roberts, D. D. (2022). Single vesicle analysis of CD47 association with integrins and tetraspanins on extracellular vesicles released by T lymphoblast and prostate carcinoma cells. Journal of Extracellular Vesicles, 11, e12265. 10.1002/jev2.12265
Contributor Information
Sukhbir Kaur, Email: kaurs@mail.nih.gov.
David D. Roberts, Email: droberts@mail.nih.gov.
DATA AVAILABILITY STATEMENT
All data is included in the manuscript or supplemental figures. Primary images are available upon request.
REFERENCES
- Azcutia, V. , Routledge, M. , Williams, M. R. , Newton, G. , Frazier, W. A. , Manica, A. , Croce, K. J. , Parkos, C. A. , Schmider, A. B. , Turman, M. V. , Soberman, R. J. , & Luscinskas, F. W. (2013). CD47 plays a critical role in T‐cell recruitment by regulation of LFA‐1 and VLA‐4 integrin adhesive functions, Molecular Biology of the Cell, 24, 3358–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazi, H. O. , Li, Z. , Cashel, J. A. , Krutzsch, H. C. , Annis, D. S. , Mosher, D. F. , & Roberts, D. D. (2002). Regulation of integrin function by CD47 ligands. Differential effects on alpha vbeta 3 and alpha 4beta1 integrin‐mediated adhesion, Journal of Biological Chemistry, 277, 42859–42866. [DOI] [PubMed] [Google Scholar]
- Brown, E. , Hooper, L. , Ho, T. , & Gresham, H. (1990). Integrin‐associated protein: A 50‐kD plasma membrane antigen physically and functionally associated with integrins, Journal of Cell Biology, 111, 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, E. J. , & Frazier, W. A. (2001). Integrin‐associated protein (CD47) and its ligands, Trends in Cell Biology, 11, 130–135. [DOI] [PubMed] [Google Scholar]
- Chiang, C. Y. , & Chen, C. (2019). Toward characterizing extracellular vesicles at a single‐particle level, Journal of Biomedical Science, 26, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daaboul, G. G. , Gagni, P. , Benussi, L. , Bettotti, P. , Ciani, M. , Cretich, M. , Freedman, D. S. , Ghidoni, R. , Ozkumur, A. Y. , Piotto, C. , Prosperi, D. , Santini, B. , Unlu, M. S. , & Chiari, M. (2016). Digital detection of exosomes by interferometric imaging, Science Reports, 6, 37246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, J. R. (2016). Q&A, what are exosomes, exactly?, Bmc Biology, 14, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feigelson, S. W. , Grabovsky, V. , Shamri, R. , Levy, S. , & Alon, R. (2003). The CD81 tetraspanin facilitates instantaneous leukocyte VLA‐4 adhesion strengthening to vascular cell adhesion molecule 1 (VCAM‐1) under shear flow, Journal of Biological Chemistry, 278, 51203–51212. [DOI] [PubMed] [Google Scholar]
- Fenselau, C. , & Ostrand‐Rosenberg, S. (2021). Molecular cargo in myeloid‐derived suppressor cells and their exosomes, Cellular Immunology, 359, 104258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, A. G. , & Frazier, W. A. (1994). Identification of a receptor candidate for the carboxyl‐terminal cell binding domain of thrombospondins, Journal of Biological Chemistry, 269, 29650–29657. [PubMed] [Google Scholar]
- Gao, A. G. , Lindberg, F. P. , Dimitry, J. M. , Brown, E. J. , & Frazier, W. A. (1996). Thrombospondin modulates alpha v beta 3 function through integrin‐associated protein, Journal of Cell Biology, 135, 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz, S. N. , Cheerathodi, M. R. , Nkosi, D. , York, S. B. , & Meckes, D. G. Jr. (2018). Tetraspanin CD63 bridges autophagic and endosomal processes to regulate exosomal secretion and intracellular signaling of epstein‐barr virus LMP1, Journal of Virology, 92(5), e01969–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg, J. S. , Annis, D. S. , Pendrak, M. L. , Ptaszynska, M. , Frazier, W. A. , Mosher, D. F. , & Roberts, D. D. (2009). Differential interactions of thrombospondin‐1, ‐2, and ‐4 with CD47 and effects on cGMP signaling and ischemic injury responses, Journal of Biological Chemistry, 284, 1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israels, S. J. , McMillan‐Ward, E. M. , Easton, J. , Robertson, C. , & McNicol, A. (2001). CD63 associates with the alphaIIb beta3 integrin‐CD9 complex on the surface of activated platelets, Thrombosis Haemostasis, 85, 134–141. [PubMed] [Google Scholar]
- Kamerkar, S. , LeBleu, V. S. , Sugimoto, H. , Yang, S. , Ruivo, C. F. , Melo, S. A. , Lee, J. J. , & Kalluri, R. (2017). Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer, Nature, 546, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, S. , Elkahloun, A. G. , Arakelyan, A. , Young, L. , Myers, T. G. , Otaizo‐Carrasquero, F. , Wu, W. , Margolis, L. , & Roberts, D. D. (2018). CD63, MHC class 1, and CD47 identify subsets of extracellular vesicles containing distinct populations of noncoding RNAs, Science Reports, 8, 2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, S. , Elkahloun, A. G. , Petersen, J. D. , Arakelyan, A. , Livak, F. , Singh, S. P. , Margolis, L. , Zimmerberg, J. , & Roberts, D. D. (2021). CD63(+) and MHC class I(+) subsets of extracellular vesicles produced by wild‐type and CD47‐deficient jurkat T cells have divergent functional effects on endothelial cell gene expression, Biomedicines, 9(11), 1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, S. , Elkahloun, A. G. , Singh, S. P. , Arakelyan, A. , & Roberts, D. D. (2018). A function‐blocking CD47 antibody modulates extracellular vesicle‐mediated intercellular signaling between breast carcinoma cells and endothelial cells, Journal of Cell Communication and Signaling, 12, 157–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, S. , Kuznetsova, S. A. , Pendrak, M. L. , Sipes, J. M. , Romeo, M. J. , Li, Z. , Zhang, L. , & Roberts, D. D. (2011). Heparan sulfate modification of the transmembrane receptor CD47 is necessary for inhibition of T cell receptor signaling by thrombospondin‐1, Journal of Biological Chemistry, 286, 14991–15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, S. , Saldana, A. C. , Elkahloun, A. G. , Petersen, J. D. , Arakelyan, A. , Singh, S. P. , Jenkins, L. M. , Kuo, B. , Reginauld, B. , Jordan, D. G. , Tran, A. D. , Wu, W. , Zimmerberg, J. , Margolis, L. , & Roberts, D. D. (2021). CD47 interactions with exportin‐1 limit the targeting of m(7)G‐modified RNAs to extracellular vesicles, Journal of Cell Communication and Signaling, 16, 397–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, S. , Schwartz, A. L. , Jordan, D. G. , Soto‐Pantoja, D. R. , Kuo, B. , Elkahloun, A. G. , Griner, L. M. , Thomas, C. J. , Ferrer, M. , Thomas, A. , Tang, S. W. , Rajapakse, V. N. , Pommier, Y. , & Roberts, D. D. (2019). Identification of schlafen‐11 as a target of CD47 signaling that regulates sensitivity to ionizing radiation and topoisomerase inhibitors, Frontiers in Oncology, 9, 994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, S. , Singh, S. P. , Elkahloun, A. G. , Wu, W. , Abu‐Asab, M. S. , & Roberts, D. D. (2014). CD47‐dependent immunomodulatory and angiogenic activities of extracellular vesicles produced by T cells, Matrix Biology, 37, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibria, G. , Ramos, E. K. , Lee, K. E. , Bedoyan, S. , Huang, S. , Samaeekia, R. , Athman, J. J. , Harding, C. V. , Lotvall, J. , Harris, L. , Thompson, C. L. , & Liu, H. (2016). A rapid, automated surface protein profiling of single circulating exosomes in human blood, Science Reports, 6, 36502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H. S. , Choi, D. Y. , Yun, S. J. , Choi, S. M. , Kang, J. W. , Jung, J. W. , Hwang, D. , Kim, K. P. , & Kim, D. W. (2012). Proteomic analysis of microvesicles derived from human mesenchymal stem cells, Journal of Proteome Research, 11, 839–849. [DOI] [PubMed] [Google Scholar]
- Krishn, S. R. , Singh, A. , Bowler, N. , Duffy, A. N. , Friedman, A. , Fedele, C. , Kurtoglu, S. , Tripathi, S. K. , Wang, K. , Hawkins, A. , Sayeed, A. , Goswami, C. P. , Thakur, M. L. , Iozzo, R. V. , Peiper, S. C. , Kelly, W. K. , & Languino, L. R. (2019). Prostate cancer sheds the alphavbeta3 integrin in vivo through exosomes, Matrix Biology, 77, 41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugeratski, F. G. , Hodge, K. , Lilla, S. , McAndrews, K. M. , Zhou, X. , Hwang, R. F. , Zanivan, S. , & Kalluri, R. (2021). Quantitative proteomics identifies the core proteome of exosomes with syntenin‐1 as the highest abundant protein and a putative universal biomarker, Nature Cell Biology, 23, 631–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Kaslan, M. , Lee, S. H. , Yao, J. , & Gao, Z. (2017). Progress in exosome isolation techniques, Theranostics, 7, 789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobb, R. J. , Becker, M. , Wen, S. W. , Wong, C. S. , Wiegmans, A. P. , Leimgruber, A. , & Moller, A. (2015). Optimized exosome isolation protocol for cell culture supernatant and human plasma, Journal of Extracellular Vesicles, 4, 27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion, B. A. , Berditchevski, F. , Kraeft, S. K. , Chen, L. B. , & Hemler, M. E. (1996). Transmembrane‐4 superfamily proteins CD81 (TAPA‐1), CD82, CD63, and CD53 specifically associated with integrin alpha 4 beta 1 (CD49d/CD29), Journal of Immunology, 157, 2039–2047. [PubMed] [Google Scholar]
- Mathieu, M. , Nevo, N. , Jouve, M. , Valenzuela, J. I. , Maurin, M. , Verweij, F. J. , Palmulli, R. , Lankar, D. , Dingli, F. , Loew, D. , Rubinstein, E. , Boncompain, G. , Perez, F. , & Thery, C. (2021). Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9, Nature Communication, 12, 4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuzuka, K. , Handa, K. , Satoh, M. , Arai, Y. , & Hakomori, S. (2005). A specific microdomain ("glycosynapse 3") controls phenotypic conversion and reversion of bladder cancer cells through GM3‐mediated interaction of alpha3beta1 integrin with CD9, Journal of Biological Chemistry, 280, 35545–35553. [DOI] [PubMed] [Google Scholar]
- Okochi, H. , Kato, M. , Nashiro, K. , Yoshie, O. , Miyazono, K. , & Furue, M. (1997). Expression of tetra‐spans transmembrane family (CD9, CD37, CD53, CD63, CD81 and CD82) in normal and neoplastic human keratinocytes: An association of CD9 with alpha 3 beta 1 integrin, British Journal of Dermatology, 137, 856–863. [PubMed] [Google Scholar]
- Oldenborg, P. A. , Zheleznyak, A. , Fang, Y. F. , Lagenaur, C. F. , Gresham, H. D. , & Lindberg, F. P. (2000). Role of CD47 as a marker of self on red blood cells, Science, 288, 2051–2054. [DOI] [PubMed] [Google Scholar]
- Panagopoulou, M. S. , Wark, A. W. , Birch, D. J. S. , & Gregory, C. D. (2020). Phenotypic analysis of extracellular vesicles: A review on the applications of fluorescence, Journal of Extracellular Vesicles, 9, 1710020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, G. K. , Khan, M. A. , Zubair, H. , Srivastava, S. K. , Khushman, M. , Singh, S. , & Singh, A. P. (2019). Comparative analysis of exosome isolation methods using culture supernatant for optimum yield, purity and downstream applications, Science Reports, 9, 5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaglia, F. , Krishn, S. R. , Daaboul, G. G. , Sarker, S. , Pippa, R. , Domingo‐Domenech, J. , Kumar, G. , Fortina, P. , McCue, P. , Kelly, W. K. , Beltran, H. , Liu, Q. , & Languino, L. R. (2020). Small extracellular vesicles modulated by alphaVbeta3 integrin induce neuroendocrine differentiation in recipient cancer cells, Journal of Extracellular Vesicles, 9, 1761072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford, K. J. , Thorne, R. F. , & Hersey, P. (1996). CD63 associates with transmembrane 4 superfamily members, CD9 and CD81, and with beta 1 integrins in human melanoma, Biochemical and Biophysical Research Communications, 222, 13–18. [DOI] [PubMed] [Google Scholar]
- Ramirez, M. I. , Amorim, M. G. , Gadelha, C. , Milic, I. , Welsh, J. A. , Freitas, V. M. , Nawaz, M. , Akbar, N. , Couch, Y. , Makin, L. , Cooke, F. , Vettore, A. L. , Batista, P. X. , Freezor, R. , Pezuk, J. A. , Rosa‐Fernandes, L. , Carreira, A. C. O. , Devitt, A. , Jacobs, L. , … Dias‐Neto, E. (2018). Technical challenges of working with extracellular vesicles, Nanoscale, 10, 881–906. [DOI] [PubMed] [Google Scholar]
- Reinhold, M. I. , Green, J. M. , Lindberg, F. P. , Ticchioni, M. , & Brown, E. J. (1999). Cell spreading distinguishes the mechanism of augmentation of T cell activation by integrin‐associated protein/CD47 and CD28, International Immunology, 11, 707–718. [DOI] [PubMed] [Google Scholar]
- Rocha‐Perugini, V. , Sanchez‐Madrid, F. , & Martinez Del Hoyo, G. (2015). Function and dynamics of tetraspanins during antigen recognition and immunological synapse formation, Frontiers in Immunology, 6, 653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha‐Perugini, V. , Zamai, M. , Gonzalez‐Granado, J. M. , Barreiro, O. , Tejera, E. , Yanez‐Mo, M. , Caiolfa, V. R. , & Sanchez‐Madrid, F. (2013). CD81 controls sustained T cell activation signaling and defines the maturation stages of cognate immunological synapses, Molecular and Cellular Biology, 33, 3644–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein, E. , Naour, F. L. , Lagaudriere‐Gesbert, C. , Billard, M. , Conjeaud, H. , & Boucheix, C. (1996). CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA‐DR and VLA integrins, European Journal of Immunology, 26, 2657–2665. [DOI] [PubMed] [Google Scholar]
- Sadallah, S. , Eken, C. , Martin, P. J. , & Schifferli, J. A. (2011). Microparticles (ectosomes) shed by stored human platelets downregulate macrophages and modify the development of dendritic cells, Journal of Immunology, 186, 6543–6552. [DOI] [PubMed] [Google Scholar]
- Serge, A. (2016). The molecular architecture of cell adhesion: dynamic remodeling revealed by videonanoscopy, Frontiers in Cell and Developmental Biology, 4, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serru, V. , Naour, F. L. , Billard, M. , Azorsa, D. O. , Lanza, F. , Boucheix, C. , & Rubinstein, E. (1999). Selective tetraspan‐integrin complexes (CD81/alpha4beta1, CD151/alpha3beta1, CD151/alpha6beta1) under conditions disrupting tetraspan interactions, Biochemical Journal, 340(Pt 1), 103–111. [PMC free article] [PubMed] [Google Scholar]
- Shimizu, A. , Sawada, K. , Kobayashi, M. , Yamamoto, M. , Yagi, T. , Kinose, Y. , Kodama, M. , Hashimoto, K. , & Kimura, T. (2021). Exosomal CD47 plays an essential role in immune evasion in ovarian cancer, Molecular Cancer Research, 19, 1583–1595. [DOI] [PubMed] [Google Scholar]
- Smith, E. L. , & Holick, M. F. (1987). The skin: The site of vitamin D3 synthesis and a target tissue for its metabolite 1,25‐dihydroxyvitamin D3, Steroids, 49, 103–131. [DOI] [PubMed] [Google Scholar]
- Soto‐Pantoja, D. R. , Kaur, S. , & Roberts, D. D. (2015). CD47 signaling pathways controlling cellular differentiation and responses to stress, Critical Reviews in Biochemistry and Molecular Biology, 50, 212–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring, F. A. , Griffiths, R. E. , Mankelow, T. J. , Agnew, C. , Parsons, S. F. , Chasis, J. A. , & Anstee, D. J. (2013). Tetraspanins CD81 and CD82 facilitate alpha4beta1‐mediated adhesion of human erythroblasts to vascular cell adhesion molecule‐1, Plos One, 8, E62654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez, H. , Andreu, Z. , Mazzeo, C. , Toribio, V. , Perez‐Rivera, A. E. , Lopez‐Martin, S. , Garcia‐Silva, S. , Hurtado, B. , Morato, E. , Pelaez, L. , Arribas, E. A. , Tolentino‐Cortez, T. , Barreda‐Gomez, G. , Marina, A. I. , Peinado, H. , & Yanez‐Mo, M. (2021). CD9 inhibition reveals a functional connection of extracellular vesicle secretion with mitophagy in melanoma cells, Journal of Extracellular Vesicles, 10, E12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J. M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines, Journal of Extracellular Vesicles, 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticchioni, M. , Raimondi, V. , Lamy, L. , Wijdenes, J. , Lindberg, F. P. , Brown, E. J. , & Bernard, A. (2001). Integrin‐associated protein (CD47/IAP) contributes to T cell arrest on inflammatory vascular endothelium under flow, FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 15, 341–350. [DOI] [PubMed] [Google Scholar]
- Vences‐Catalan, F. , & Levy, S. (2018). Immune targeting of tetraspanins involved in cell invasion and metastasis, Frontiers in Immunology, 9, 1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon‐Wilson, E. F. , Kee, W. J. , Willis, A. C. , Barclay, A. N. , Simmons, D. L. , & Brown, M. H. (2000). CD47 is a ligand for rat macrophage membrane signal regulatory protein SIRP (OX41) and human SIRPalpha 1, European Journal of Immunology, 30, 2130–2137. [DOI] [PubMed] [Google Scholar]
- Wang, X. Q. , & Frazier, W. A. (1998). The thrombospondin receptor CD47 (IAP) modulates and associates with alpha2 beta1 integrin in vascular smooth muscle cells, Molecular Biology of the Cell, 9, 865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willms, E. , Cabanas, C. , Mager, I. , Wood, M. J. A. , & Vader, P. (2018). Extracellular vesicle heterogeneity: Subpopulations, isolation techniques, and diverse functions in cancer progression, Frontiers in Immunology, 9, 738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K. W. , & Thery, C. (2019). Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature, Journal of Extracellular Vesicles, 8, 1648167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung, L. , Hickey, M. J. , & Wright, M. D. (2018). The many and varied roles of tetraspanins in immune cell recruitment and migration, Frontiers in Immunology, 9, 1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Freitas, D. , Kim, H. S. , Fabijanic, K. , Li, Z. , Chen, H. , Mark, M. T. , Molina, H. , Martin, A. B. , Bojmar, L. , Fang, J. , Rampersaud, S. , Hoshino, A. , Matei, I. , Kenific, C. M. , Nakajima, M. , Mutvei, A. P. , Sansone, P. , Buehring, W. , … Lyden, D. (2018). Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field‐flow fractionation, Nature Cell Biology, 20, 332–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: (A,B) Standardization of CD47 antibodies. (C) Evaluation of non‐specific interaction of CF647 anti‐mIgG1κ with Jurkat, JinB8, PC3, and PC3 (‐CD47) EVs on captured spots β1, CD47, α4, β3, CD63, CD81 and CD9. (F) CD29 (β1), co‐stained with CD47 antibodies. (E–J) Flow cytometry analysis of CD9, CD63 and CD81 surface expression on Jurkat, JinB8, PC3 and CRISPR targeted PC3(‐CD47) cells relative to unstained controls (grey). Figure S2: (A–F) Surface expression of CD47 using Jurkat, JinB8, PC3 and CRISPR targeted PC3(‐CD47) cells along with unstained control via flow cytometry. (G–L) Sizing of the Label Free captured Jurkat, JinB8, PC3, and PC3 (‐CD47) EVs on spots β1, CD47, α4, β3, CD63, CD81 and CD9 via SP‐IRIS. Figure S3: (A–L) Surface expression of CD29/β1 integrin, CD61/β3 integrin, CD49b/α2 integrin, CD49d/α4 integrin, CD49e/α5 integrin, and CD49f/α6 integrin using Jurkat, JinB8, PC3 and PC3(‐CD47) cells along with unstained control via flow cytometry. (M) Validation of CD61 antibody using human embryonic kidney (HEK) cell conditioned media and plasma incubated on anti‐β1, CD47, α4, β3, CD63, CD81 and CD9 capture spots. Figure S4: (A–F) Fluorescent imaging was used to determine colocalization of β1 and CD47 on vesicles using fluorescent antibodies CF647‐anti‐CD47 and AF488‐anti‐CD29 on 50–200 nm EVs from JinB8, PC3 and PC3 (‐CD47) cells captured on anti‐β1 and CD47 antibody spots. (G–I) Fluorescent imaging was used to determine colocalization of β1 and CD47 with tetraspanins on vesicles captured on CD81, CD9 and CD63 antibody spots. Figure S5: (A) Visualization of labelled tetraspanins extracted from Figure 3B–D graphs for Jurkat Panel only. (B–G) Colocalization between β1+, CD63+ and CD81+ EVs using anti‐ AF488‐CD29, anti‐CF647‐CD63 and CF555‐CD81 antibodies to stain Jurkat and JinB8 EVs captured on spots coated with β1, CD47, α4, β3, CD63, CD81 and CD9 antibodies. Error bars indicate SD, N = 3. Figure S6: (A) Total label‐free, SP‐IRIS particle analysis of 50–200 nm EVs released from HEK293, and PANC‐1 cells captured on spots with antibodies specific for CD63, CD81 and CD9. (B–D) Fluorescent detection of EVs containing β1 integrins (CD29) or CD47 captured on the indicated tetraspanins. Error bars indicate SD, N = 3. (E–G) Vesicle colocalization of β1+ integrins and CD47+ with CD63, CD81 and CD9 on captured spots. (H) Percentage of colocalization of tetraspanins with β1+/CD47+.
Data Availability Statement
All data is included in the manuscript or supplemental figures. Primary images are available upon request.


