Abstract
The histidine kinase (HK) component of many two-component regulatory systems exhibits regulated ability to phosphorylate itself and to participate in transfer of phosphate to and from its cognate response regulator. The signaling system that controls expression of the UhpT sugar phosphate transporter in Escherichia coli in response to external glucose 6-phosphate includes the HK protein UhpB and the polytopic membrane protein UhpC, a UhpT homolog which is required for responsiveness to an inducer and activation of UhpB. The existence of a UhpBC signaling complex is suggested by the requirement for UhpC for the activity of certain constitutively active variants of UhpB, the dominance and epistasis relationships of uhp alleles, and the finding that expression of UhpB in excess of UhpC has a strong dominant-negative effect. Expression of a hybrid protein containing the cytoplasmic C-terminal half of UhpB fused to glutathione S-transferase (GST) also interfered with Uhp signaling. This interference phenotype could not result solely from the phosphatase activity of UhpB, because interference affected both overexpressed UhpA and UhpA variants which are active in the absence of phosphorylation. Variant forms of UhpB which were active in the absence of UhpC carried amino acid substitutions near motifs conserved in HK proteins. The GST fusion protein inhibited the ability of UhpA to bind and activate transcription at the uhpT promoter. Unlike the wild-type situation, a GST fusion variant carrying one of the UhpB-activating substitutions, R324C, displayed autokinase activity and phosphate transfer to UhpA but retained the ability to sequester UhpA when it was altered in the conserved residues important for phosphate transfer. Thus, the default state of UhpB is kinase off, and activation of its phosphate transfer activity requires either the action of UhpC or the occurrence of certain mutations in UhpB. The interference phenotype shown by UhpB in excess of UhpC appears to include the binding and sequestration of UhpA.
The control of gene expression by two-component regulatory systems occurs through changes in the phosphorylation of specific response regulator (RR) proteins (7, 20). Their phosphorylation is mediated by a multifunctional cognate sensor kinase or histidine kinase (HK) protein, which has autokinase activity to provide the phosphate moiety for transfer to the response regulator and which also can accelerate the rate of its dephosphorylation. Many HKs are controlled by external signals detected by their periplasmic domains. Separation of the kinase domain from the transmembrane and external portions can often be achieved without loss of protein kinase activity, and in many cases the liberated soluble cytoplasmic domain confers high and unregulated protein kinase activity (4, 9, 21, 23).
Induction by extracellular glucose-6-phosphate (Glu6P) of the sugar phosphate transporter UhpT requires transcription activation by the response regulator UhpA (3, 18, 29). UhpA has high sequence similarity to the nitrate-responsive RR NarL (1), whose structure reveals a two-domain protein with an N-terminal CheY-like phosphorylation domain and a C-terminal DNA-binding helix-turn-helix domain. UhpA can be phosphorylated on aspartate-54 by acetyl-phosphate, and phosphorylation stimulates its binding to the uhpT promoter (2, 3). Transcription of the uhpT promoter in vitro requires the presence of UhpA, but its phosphorylation is not required when it is overexpressed or carries certain substitutions (18). The uhpB and uhpC genes are also required for UhpT expression in vivo (11). The bipartite UhpB protein has a hydrophobic amino-terminal half (residues 1 to 273) expected to span the membrane eight times and a carboxyl-terminal half (residues 274 to 500) which is exposed to the cytoplasm and contains the conserved sequence elements common to HK proteins, i.e., the H box around the phosphorylated histidine, the N box, and the G box comprising the ATP-binding and phosphate transfer region (20). The UhpC protein is related in sequence and topology to UhpT and other organophosphate antiporters but plays a role only in regulation and is required for responsiveness to Glu6P. Some mutations that insert tetrapeptide sequences into UhpB and UhpC result in altered regulation, including constitutive behavior. Since the constitutive phenotype of some of these UhpB mutations was expressed only in the presence of functional UhpC, Island and Kadner (10) proposed that UhpB and UhpC act jointly in a membrane-embedded signaling complex. If UhpC is the signal receptor, then some mechanism must operate to prevent activation of UhpA by any UhpB molecules except those in complex with UhpC molecules with bound external Glu6P, i.e., the default state of UhpB must be kinase off.
This model is tested here by examination of the dominance and epistasis properties of some UhpB and UhpC variants. In particular, it is shown that overexpression of UhpB, either the full-length protein or the liberated C-terminal cytoplasmic domain, results in a strong dominant-negative phenotype, indicating interference with signal transduction by UhpB in excess relative to UhpC. Variant forms of UhpB that are active in the absence of UhpC were isolated, and some have lost the dominant-negative phenotype when overexpressed. The restoration of the interference phenotype to these constitutively active UhpB variants following mutagenesis of conserved residues needed for autokinase activity, and the interference of UhpB with phosphorylation-independent variants of UhpA, indicate that the interference by UhpB involves both cophosphatase and sequestration activity on UhpA.
MATERIALS AND METHODS
Plasmids and strains.
The Escherichia coli strains and the plasmids used in this study are listed in Table 1. The uhp deletions used in this study remove most of the coding sequence but leave the reading frame intact to eliminate polar effects on expression of distal genes (11). The isolation of uhp variants containing 12-bp PstI oligonucleotide linker insertions and encoding variant Uhp proteins with a tetrapeptide insertion was described previously (10). These mutant alleles are designated by the position of the amino acid residue preceding the site of insertion and the number of amino acids inserted; thus, the genotype uhpA15:4 indicates that the encoded UhpA protein contains a four-amino-acid insertion between amino acids 15 and 16. Plasmids carrying these uhp mutations were integrated by homologous recombination into the chromosomal uhpT gene in the polA hosts RK1294 and RK1300, which are unable to support plasmid replication.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| JM109 | e14(mcrA) recA gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lac-proAB) (F′ traD36 proAB+ lacIqΔZM15) | Stratagene |
| MC4100 | Δ(argF-lac)U169 araD139 rpsL 150 relA1 flbB5301 deoC1 ptsF25 rbsR22 | |
| CW235 | MC4100 λRZ5[Km]uhpTp-lacZ recA56 srl300::Tn10 uhpAH170Y | 25 |
| RK1294 | MC4100 λRZ5[Km]uhpTp-lacZ polA1 Δuhp(A14-C437) | This work |
| RK1300 | MC4100 λRZ5[Km]uhpTp-lacZ polA1 uhp+ | |
| RK1310 | MC4100 λRZ5[Km]uhpTp-lacZ recA56 srl300::Tn10 uhp+ | |
| RK1302 | MC4100 λRZ5[Km]uhpTp-lacZ recA56 srl300::Tn10 ΔuhpB(60-489) | |
| RK1305 | MC4100 λRZ5[Km]uhpTp-lacZ recA56 srl300::Tn10 ΔuhpC(41-437) | |
| RK1306 | MC4100 λRZ5[Km]uhpTp-lacZ recA56 srl300::Tn10 Δuhp(A15-B489) | |
| RK1320 | ||
| XL-1 Blue | recA endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F′ proAB+ lacIq ΔZM15 Tn10) | Stratagene |
| Plasmids | ||
| pCAW8 | pACYC184 carrying uhpA coding sequence; Cm | 25 |
| pA-D54N | pCAW8 carrying uhpA D54N | This work |
| pGEX3x | lacIqlacP-gst; Ap | Pharmacia |
| pGEX3X-TEV | pGEX3X with TEV protease site | This work |
| pJSW141 | pGEX3x-TEV uhpB(273-500) and derivatives | This work |
| pRJK10 | pBR322 carrying uhpABCT; Ap | 28 |
| pSu19 | Cloning vector ori p15A from pACYC184 lac promoter; Cm | 13 |
| pSuB | pSu19 carrying lacP-(uhpA189-uhpB+-uhpC91); Cm | |
| pSuBC | pSu19 carrying lacP-(uhpA189-uhpB+-uhpC+); Cm | |
| pUT1 | pUC18 carrying uhpT promoter from −135 to +90 | 18 |
Construction of GST-Bc, UhpB, and UhpA plasmids.
The pGEX3x plasmid vector (Amersham-Pharmacia Biotech Inc.) was modified to introduce the tobacco etch virus (TEV) protease recognition site (ENLYFQS) by ligating the complementary oligonucleotide pair 5′-GATCGAAAACCTGTACTTCCAGTCAGGGATCCATATG and 5′-AATTCATATGGATCCCTGACTGGAAGTACAGGTTTTC into BamHI- and EcoRI-digested pGEX3x to create pGEX3x-TEV. This linker insertion destroyed the upstream BamHI site, restored it downstream in the correct reading frame, added an NdeI site between the BamHI and EcoRI sites, and caused loss of the unique SmaI site. Residues 273 to 500 of uhpB were amplified by PCR using VENT polymerase (New England Biolabs), the primers 5′-GGCGCTGGGATCCAGCGGTT-3′ containing a BamHI site and 5′-CGGCAGGCGAATTCAGAAACGGCAA-3′ containing an EcoRI site, and pRJK10 (uhpABCT) as a template. The PCR product was digested and ligated into pGEX3x-TEV to create pJSW141. The correct nucleotide sequence was confirmed by DNA sequencing at the University of Virginia Biomolecular Research Facility.
Plasmids pGEX3x and pGEX3x-TEV were used interchangeably as controls and are designated pGST. The glutathione S-transferase (GST)-UhpB fusion proteins expressed from pJSW141 and its derivatives are designated GST-Bc.
The uhpA gene under control of its own promoter had been cloned into pACYC184 to generate pCAW8 (25). Restriction fragment exchange was used to transfer the uhpA(D54N) mutation from pALTER-UhpAD54N (26) into pCAW8 to generate pA-D54N.
Plasmids pSuB and pSuBC carry the 3′ end of uhpA, the entirety of uhpB, and either the 5′ segment or the entirety of uhpC, respectively. These were constructed by ligation into the PstI-digested plasmid pSU19 of PstI fragments extending from the coding region for residue 189 of UhpA to the coding region for residue 91 of UhpC or to the uhpT promoter region.
Site-directed mutagenesis.
Site-directed mutagenesis of pJSW141 was performed using the ExSite kit (Stratagene) and Pfu DNA polymerase. Mutagenized plasmids were transformed into E. coli XL-1 Blue and screened for the presence of a silent restriction site that was incorporated by the mutagenic primers. The sequences of primers used for plasmid construction are available upon request. All mutations were confirmed by DNA sequencing (University of Virginia Biomolecular Research Facility).
Purification of GST-Bc.
Plasmids pGEX3x-TEV, encoding GST, and pJSW141, encoding GST-Bc and its variants, were expressed in JM109 (Amersham-Pharmacia Biotech Inc.) and purified using the manufacturer's recommendations with minor modifications. An overnight culture in Luria-Bertani broth was inoculated into 1 liter of Luria-Bertani broth. The cells were grown at 30°C to an optical density at 595 nm of approximately 0.8, and protein expression was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 to 3 h. The cells were harvested by centrifugation and stored at −70°C. Cells suspended in phosphate-buffered saline were incubated with 1 mg of lysozyme/ml for 30 min on ice and disrupted by sonication or by three passes through a French pressure cell. DNase at 10 μg/ml and RNase at 5 μg/ml were added to the lysed cells without Triton X-100. Cleared lysates were incubated with glutathione-conjugated Sepharose 4B (Amersham-Pharmacia Biotech Inc.) for 30 min at 25°C. Beads were washed three times with 10 volumes of phosphate-buffered saline each time and loaded onto a 10-ml disposable column. Reduced glutathione was added and incubated with the beads for 10 min before fractions were collected. The fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), pooled, and dialyzed overnight in 50 mM HEPES (pH 8.0) and 10% glycerol at 4°C. The protein concentration was determined using the Bradford dye-binding protein assay (Bio-Rad) with bovine serum albumin as a standard. The proteins were aliquoted and stored at −70°C.
β-Galactosidase assays.
Regulation of the uhpT promoter was determined from the level of β-galactosidase expressed from a uhpTp-lacZ transcriptional fusion carried as a single lysogen of phage λRZ5 (15). β-Galactosidase assays were performed as described previously (11). Overnight cultures were diluted 1:100 in minimal medium A supplemented with 1% (vol/vol) glycerol, 0.5% Casamino Acids, and 1.5 mM MgSO4. IPTG was added at the time of subculture when indicated. Cells in logarithmic growth were induced with 0.25 mM Glu6P in 96-well microtiter plates containing 200 μl of culture per well. After induction for 40 min at 37°C, the cells were lysed with CHCl3-SDS and mixed with Z buffer (16) and 2 mM o-nitrophenyl-β-d-galactopyranoside. The rate of hydrolysis was measured at 415 nm over 5 min at 37°C in a microplate reader (Molecular Devices) and was normalized for cell density. All values are the average of at least three experiments and agree within 10%.
In vitro transcription and DNase I footprinting assay.
Transcription assays were performed by preincubation of 1 nM plasmid pUT1 DNA with 220 nM UhpA and various concentrations (50 to 400 nM) of GST-Bc or GST for 10 min in TXN buffer (40 mM Tris-HCl, pH 8.0, 50 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol) before the addition of 30 nM RNA polymerase (Amersham-Pharmacia Biotech Inc.) as described previously (18). After 15 min, transcription was initiated by the addition of 40 μM [α-32P]UTP (2.5 Ci/nmol), 50 μg of heparin/ml, and 200 μM (each) ATP, CTP, and GTP. After 10 min, the reaction was terminated by the addition of stop solution (7 M urea, 0.1 M EDTA, 0.4% [wt/vol] SDS, 40 mM Tris-HCl [pH 8.0], 0.5% [wt/vol] bromophenol blue, and 0.5% xylene cyanol), separated by electrophoresis in 5% polyacrylamide–7 M urea gels in 1× Tris-borate-EDTA buffer, and analyzed by PhosphorImager (Molecular Dynamics).
DNase I footprinting reactions of the uhpT promoter were performed as described previously (18), using 800 nM UhpA and 1,600 nM GST or GST-Bc, which were incubated in TXN buffer with 1 nM P32-end-labeled uhpT promoter DNA for 30 min at 37°C prior to DNase I digestion. DNA fragments were separated on sequencing gels and analyzed by PhosphorImager.
Phosphate transfer reactions.
To assay autokinase activity, various GST-Bc variant proteins were used at 2.8 μM concentration in a reaction volume of 50 μl containing 50 mM HEPES, pH 8.0, 50 mM KCl, 5 mM MgCl2, and 1 mM dithiothreitol. The reaction mixture was incubated at 25°C for 5 min before the addition of 2 μl of [γ-32P]ATP (330 μCi; ca. 1 nM; ICN Pharmaceuticals). The reaction mixtures were incubated at 37°C, and 8-μl portions were removed at intervals, mixed with 2 μl of 6× sample buffer (350 mM Tris-HCl [pH 6.8], 10% SDS, 30% [vol/vol] glycerol, 0.6 M dithiothreitol, 50 mM EDTA), and placed on ice. Samples were resolved by SDS-PAGE, and the radioactivity on dried gels was localized with a PhosphorImager. To assay phosphotransfer activity, the autophosphorylation reaction was allowed to proceed for 20 min for maximal phosphorylation of UhpB. UhpA protein was added to 2.8 μM. Portions were removed and processed as described above. When indicated, UhpA-D54N was used at 5 μM concentration instead of UhpA. Unlabeled 1 mM ATP or 10 mM EDTA was incubated with the UhpA prior to its addition to P-UhpB.
RESULTS
Dominance and epistasis properties of Uhp regulatory mutants.
Mutations that result in tetrapeptide insertions in UhpB or UhpC proteins exhibited a range of regulatory phenotypes, including normal inducible behavior, loss of expression, elevated basal levels, or fully constitutive behavior (10). The dominance and epistasis relationships of some of these mutations were investigated. Each mutation in uhpA or uhpB was combined with one of three uhpC alleles: uhpC+, the uhpC91:4 allele expressing a tetrapeptide insertion after amino acid 91 and conferring high-level constitutive expression, or the uhpC91:8 allele expressing an 8-residue insertion after amino acid 91 and unable to express uhpT. These sets of plasmids were integrated into the chromosomal uhpT locus in polA strains in such a way that the wild-type uhpABC locus was either present in tandem with the integrated plasmid or deleted.
The regulatory behavior elicited by the presence of these uhp alleles was determined from expression of a uhpT-lacZ transcriptional reporter (Table 2). The Uhpc phenotype of the uhpC91:4 allele was dominant to uhpC+, and the Uhp− phenotype of uhp91:8 was recessive, indicating that uhpC91:4 encodes a gain-of-function variant and uhpC91:8 is a null variant. The loss of Uhp expression resulting from tetrapeptide insertions in uhpA, typified by uhpA15:4, was recessive to uhpA+ and epistatic to uhpC, as expected for the loss of function of the required uhpT transcription activator.
TABLE 2.
Dominance and epistasis relationships among uhp regulatory gene alleles
|
uhp genotype of integrated plasmidb
|
β-Galactosidase activity from uhpT-lacZ reportera
|
|||||
|---|---|---|---|---|---|---|
| RK1294 (ΔuhpABC)
|
RK1300 (uhp+)
|
|||||
| uhpA | uhpB | uhpC | − | + | − | + |
| + | + | + | 2.5 | 660 | 3.5 | 450 |
| + | + | 91:4 | 2,610 | 2,070 | 970 | 960 |
| + | + | 91:8 | 6 | 6 | 2 | 170 |
| 15:4c | + | + | 1.5 | 1 | 2 | 265 |
| 15:4c | + | 91:4 | 1.5 | 1 | 835 | 710 |
| 15:4c | + | 91:8 | 1.5 | 1 | 25 | 240 |
| + | 151:4 | + | 2,250 | 1,700 | 530 | 570 |
| + | 151:4 | 91:4 | 1,490 | 1,115 | 975 | 890 |
| + | 151:4 | 91:8 | 1,900 | 1,690 | 970 | 960 |
| + | 250:4d | + | 750 | 1,060 | 610 | 800 |
| + | 250:4d | 91:4 | 1,630 | 1,240 | 1,470 | 1,580 |
| + | 250:4d | 91:8 | 25 | 25 | 215 | 375 |
| + | 87:4e | + | 20 | 15 | 5 | 445 |
| + | 87:4e | 91:4 | 40 | 30 | 470 | 565 |
| + | 87:4e | 91:8 | 60 | 55 | 40 | 285 |
β-Galactosidase activity was measured in the indicated uhp+ polA or ΔuhpABC polA strains into which the indicated plasmids bearing PstI linker insertions in uhp genes were introduced by transformation. Stable integrants, probably resulting from homologous recombination into the uhp locus, were selected by ampicillin resistance. Cells were grown in the absence (−) or presence (+) of Glu6P, as indicated, and β-galactosidase activity was measured as an average from three independent transformants, with variation of <10%.
Plasmids are derivatives of pMI29 carrying PstI linker insertions (10). The location of the insertion is denoted by the amino acid residue preceding the site of insertion and the number of amino acids inserted. +, wild type.
Similar results were obtained with the uhpA169:4 and uhpA189:4 alleles.
Similar regulatory behavior was seen with the uhpB60:4 and uhpB387:4 alleles, which confer basal levels that are lower than those with uhpB250:4 but higher than with the wild type.
Similar results were obtained with the uhpB411:4 allele.
Insertions in uhpB resulted in three general phenotypes. The uhpB151:4 allele conferred high constitutive expression regardless of the status of uhpC, consistent with activation of UhpB kinase activity that was independent of the presence of UhpC. Some insertions in uhpB, typified by uhpB87:4, conferred very low Uhp expression which was somewhat higher than that of the uninduced wild type, presumably owing to loss of P-UhpA phosphatase activity, but was unaffected by the presence of Glu6P, as is seen for uhpB null mutations (11). This phenotype was recessive to uhpB+ and was indifferent to uhpC. Thirdly, several insertions in the transmembrane portion of UhpB, typified by uhpB250:4, exhibited basal expression which was elevated to various degrees and which was further inducible by Glu6P. This phenotype was dominant to uhpB+ but required the presence of an active form of uhpC, suggesting the existence of a UhpB-UhpC signaling complex.
UhpC-independent mutants.
All of the tetrapeptide insertions in uhpB that conferred elevated or constitutive behavior affected the membrane-spanning N-terminal half of UhpB (10). Mutants active in the absence of UhpC were obtained by selection for spontaneous or mutagen-induced Uhp+ variants of the uhpC91::Km or uhpC409::Km parents. All mutants chosen for study retained the kanamycin resistance of the parental strains, exhibited constitutive uhpT-lacZ expression, and carried second-site mutations which were responsible for the constitutive behavior and were closely linked to the kanamycin resistance determinant by P1-mediated transduction. The uhpAB regions from 12 independently derived mutants were cloned and sequenced.
The 12 UhpC-independent variants included 11 with single-amino-acid substitutions and one with double-amino-acid substitutions in the C-terminal half of UhpB. These UhpC-independent uhpB mutants were represented by five isolates with the E299K substitution, three with R366C, two with R324C, one with G479D, and one isolate with the double substitution E295G plus E302K. All isolates with the same amino acid substitution had the same nucleotide substitution. The constitutive uhpT-lacZ expression level in the presence of the E299K variants was several times higher than that of the other mutants. These results show that the requirement for UhpC to activate UhpB can be bypassed by certain substitutions, resulting in a change of charge near the conserved motifs in the C-terminal portion of UhpB.
Effect of overexpression of UhpB.
To examine the effect on Uhp regulation of coexpression of uhpB and uhpC, a series of plasmids was made that encode intact or N-terminally truncated forms of UhpB, with or without intact UhpC. Construction of these variants made use of the set of PstI linker insertions in the uhp region (11). Expression of uhpB may be coupled to translation of uhpA, because their ends overlap in the sequence TGATG. Hence, the transcription of uhpB was driven by the lac promoter in the pSU19 vector, and translation initiated at the lacZ′ gene of the vector was engineered to continue in frame through the distal end of the uhpA coding sequence. In plasmid pSuB, the uhpB gene is followed by the proximal portion of uhpC encoding the first 91 amino acids. In plasmid pSuBC, the uhpB gene is followed by the intact uhpC gene. The plasmids were introduced into host strains carrying a uhpT-lacZ reporter and the intact uhp+ chromosomal locus or the ΔuhpB allele, respectively. A ΔuhpBC host showed regulatory properties similar to those of the ΔuhpB strain (data not shown). Expression of β-galactosidase after growth in the absence and presence of Glu6P was measured (Table 3).
TABLE 3.
Effect of uhpB truncation derivatives on uhp expression
| Plasmid genotype
|
β-Galactosidase activity from uhpT-lacZ reportera
|
||
|---|---|---|---|
| uhpB | uhpC | RK1310 (uhp+) (−/+) | RK1302 (ΔuhpB) (−/+) |
| Vector | 1/350 | 40/34 | |
| B+:1-500 | Δ | 1/2 | 2/3 |
| + | 1/279 | 15/262 | |
| B′:182-500 | Δ | 2/7 | 1/1 |
| + | 1/1 | 1/1 | |
| B′:288-500 | Δ | 32/108 | 115/102 |
| + | 1/213 | 1/1 | |
| B′:345-500 | Δ | 4/189 | 5/4 |
| + | 1/241 | 4/2 | |
β-Galactosidase units are presented for cells grown in the absence (−) or presence (+) of Glu6P.
The presence of plasmid pSuB encoding wild-type UhpB (residues 1 to 500) completely blocked Uhp expression in the uhp+ strain RK1310, whereas the empty vector plasmid or plasmid pSuBC expressing both uhpBC genes allowed Glu6P-inducible expression. The pSuBC plasmid conferred inducible Uhp expression in host strains with uhpB or uhpBC deleted, whereas the uhpB+ plasmid pSuB did not. The inability of the chromosome-encoded UhpC protein to allow the function of plasmid-encoded UhpB is consistent with the interference seen in the uhp+ strain, indicating that the excess UhpB blocks the activating effect of smaller amounts of the UhpBC complex.
To test whether the entire UhpB protein was required for this interference phenomenon, plasmids encoding N-terminally truncated forms of UhpB, designated B′:182-500, B′:288-500, and B′:345-500, were analyzed (Table 3). In these plasmids, the distal portion of uhpA was fused in frame at several sites to the lacZ′ gene using the PstI linker insertions. As in the cases of the pSuB and pSuBC plasmids, the uhpB coding region was followed by either the proximal portion of the uhpC gene or the intact gene. The UhpB′:182-500 variant lacks about two-thirds of the transmembrane portion of UhpB and conferred a strong dominant-negative interference effect. When UhpC was coexpressed, this UhpB variant was unable to activate Uhp expression, showing that it retained the sequences required for interference but was unable to recognize the activating signal transmitted by UhpC. The UhpB′:288-500 variant has the entire transmembrane region and 16 residues of the cytoplasmic domain deleted. This variant conferred an unusual response of partially constitutive uhpT expression when it was expressed alone but strong interference when it was coexpressed with UhpC. The UhpB′:345-500 variant lacks the transmembrane portion and 73 residues of the cytoplasmic segment, including the site of phosphorylation. This variant had no effect on Uhp expression determined by the chromosomal locus and did not activate by itself. Thus, Uhp expression is blocked by the presence of intact UhpB in excess of UhpC or by the C-terminal half of UhpB beyond residue 182 but including residues between positions 182 and 345.
Interference by the cytoplasmic portion of UhpB.
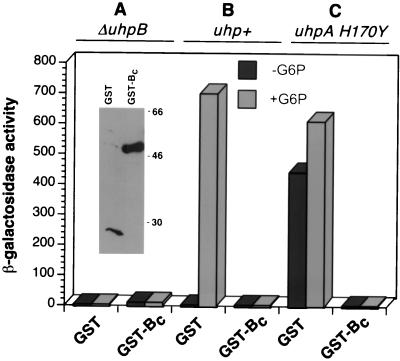
To facilitate the enzymatic analysis of UhpB function, the C-terminal cytoplasmic portion of UhpB from residues 273 to 500 was expressed as a C-terminal fusion to GST, designated GST-Bc. Unlike many other HKs, GST-Bc was unable to activate Uhp expression in a ΔuhpB host, even though appreciable levels of the fusion protein were produced (Fig. 1, inset). As was seen with intact UhpB, the presence of GST-Bc completely blocked uhpT activation by the chromosomal uhp+ locus, whether assayed by growth on Glu6P (data not shown) or by uhpT-lacZ reporter activity (Fig. 1B).
FIG. 1.
Effect of GST-Bc expression on uhpT-lacZ activation. Plasmids expressing GST or GST-Bc were introduced into uhpT-lacZ strains RK1305 (ΔuhpB) (A), RK1310 (uhp+) (B), and CW235 (uhpA H170Y) (C). As indicated, the cells were grown in the absence (solid bars) and presence (shaded bars) of Glu6P. The inset shows a Western blot of equal numbers of cells of RK1305 expressing GST or GST-Bc probed with anti-GST antibody.
This interference phenotype could be explained by the ability of the GST-Bc protein to accelerate dephosphorylation of P-UhpA that was phosphorylated by the chromosome-encoded UhpBC complex. To test this model, GST-Bc was expressed in strain CW235 carrying the chromosomal phosphorylation-independent uhpA(H170Y) allele. This variant was obtained as a UhpB-independent mutant and allows constitutive Uhp expression in the absence or presence of the uhpBC genes (26). Its constitutive Uhp expression was completely abrogated when GST-Bc was expressed (Fig. 1C).
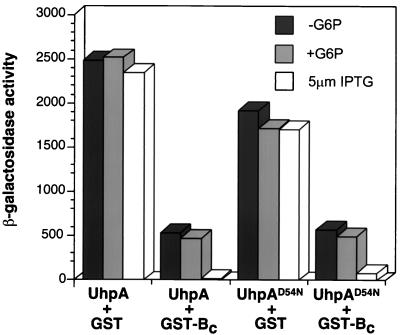
To show further that the negative action of GST-Bc operated by a different mechanism than dephosphorylation of UhpA, the effect of GST-Bc was tested in a strain in which UhpA was overexpressed from a compatible plasmid carrying uhpA in the ΔuhpAB host RK1306 (Fig. 2). Multicopy expression of uhpA resulted in phosphorylation-independent and constitutive activity of the uhpT-lacZ reporter. Coexpression of GST-Bc resulted in an 80% decrease in β-galactosidase activity, which remained independent of Glu6P induction. Addition of 5 μM IPTG to increase transcription of the GST-Bc gene from its tac promoter resulted in almost complete loss of Uhp expression. Similar results were obtained with the UhpA-D54N variant lacking the site of phosphorylation. Taken together, these results show that the interference phenotype conferred by GST-Bc is not dependent on the state of phosphorylation of UhpA but is affected by the relative amounts of the Uhp regulatory proteins. We propose that, in addition to having cophosphatase activity for P-UhpA, UhpB and GST-Bc might also affect UhpA action by binding and sequestering it.
FIG. 2.
Effect of GST-Bc expression when UhpA is also overexpressed. Plasmids pGEX3x-TEV and pJSW141 expressing GST or GST-Bc and plasmids pCAW8 and pA-D54N expressing UhpA or UhpA-D54N, respectively, were introduced into strain RK1306 (ΔuhpAB λuhpT-lacZ). The level of β-galactosidase was measured following growth in the absence (−) or presence (+) of Glu6P or 5 μM IPTG.
GST-Bc inhibits DNA binding and transcription activation by UhpA.
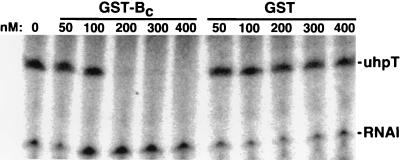
We have previously shown that UhpA can bind to sites in the uhpT promoter and activate its transcription in an in vitro system with purified components. These activities do not require phosphorylation of UhpA (18). DNase I footprinting of UhpA at the uhpT promoter and in vitro transcription were assayed in the presence of GST and GST-Bc. In the footprinting assay (data not shown), the protection or enhancement of DNase cleavage of sites in the −80 to −32 region by the presence of 800 nM UhpA was completely reversed by the presence of 1,600 nM GST-Bc but was not affected by 1,600 nM GST. The uhpT-specific in vitro transcription activated by 220 nM UhpA was completely blocked by the presence of 200 nM GST-Bc but was unaffected by GST (Fig. 3). These results show that GST-Bc has a direct inhibitory effect on UhpA that is independent of its state of phosphorylation. The stoichiometry of the inhibition of transcription suggests the formation of an inactive 1:1 complex of UhpA and UhpB.
FIG. 3.
Effect of GST-Bc on UhpA-dependent transcription of the uhpT promoter. The conditions for in vitro transcription were 30 nM RNA polymerase, 220 nM UhpA, and 1 nM plasmid pUT1 DNA, along with the indicated final concentrations of GST or GST-Bc. The synthesis of RNA species was determined by PhosphorImager analysis of electropherograms. The locations of the uhpT-specific transcript and the vector-encoded RNAI are indicated on the right.
Effect of activating mutations in GST-Bc.
Each of the mutations described above that conferred UhpC-independent constitutive expression was transferred into the GST-Bc coding region by oligonucleotide-directed mutagenesis. The presence of the R324C and the double E295G-plus-E302K (both of which were necessary) substitutions resulted in loss of the dominant-negative interference phenotype and in constitutive Uhp expression in the absence of chromosome-encoded UhpB, as measured by growth on Glu6P or expression of the uhpT-lacZ reporter.
To test whether these activating mutations resulted in loss of the sequestration effect, these mutations were combined with changes in the motifs that are conserved among HK proteins, namely, H313E and H313Q at the site of phosphorylation, N424D and H428Q in the N box, and four substitutions in the ATP-binding G box, D451A, D452A, G453A, and G455A. The presence of these mutations in otherwise-wild-type GST-Bc protein did not affect its strong interference phenotype (Table 4), suggesting that interference was unrelated to the phosphate-transfer activity of UhpB (see below). When these mutations in the conserved motifs were combined with either of the two activating substitutions, the resulting GST-Bc variants were unable to activate uhpT expression and displayed a strong dominant interference phenotype. These results show that each of the conserved motifs is necessary for Uhp activation and that the activating mutations do not block sequestration but instead enhance UhpB autokinase activity.
TABLE 4.
Effect on uhpT-lacZ expression of amino acid substitutions in conserved residues in GST-Bc with activating mutations
| Substitution in conserved residues | β-Galactosidase activity in uhpT-lacZ host straina
|
|||||
|---|---|---|---|---|---|---|
|
uhp+
|
ΔuhpBCb
|
|||||
| WT | E295G + E302K | R324C | WT | E295G + E302K | R324C | |
| Vector | 3/497 | 7/454 | 7/450 | 7/3 | 7/8 | 7/8 |
| WT GST-Bc | 1/2 | 490/482 | 285/281 | 1/2 | 481/457 | 381/343 |
| H313E | 3/2 | 3/2 | 2/22 | ND | 4/4 | 5/3 |
| H313Q | 4/2 | 3/4 | 2/3 | ND | 3/3 | 3/4 |
| N424D | 4/2 | 2/17 | 2/2 | ND | 3/3 | 2/3 |
| H428Q | 1/2 | 1/3 | 2/2 | ND | 3/2 | 3/3 |
| G boxc | 2/2 | 2/51 | 1/12 | ND | 3/3 | 3/2 |
β-Galactosidase units are presented for cells grown in the absence/presence of Glu6P in host strains with the indicated activating mutation or wild type (WT). ND, not determined.
Identical results were obtained with host strains carrying ΔuhpB or ΔuhpC alleles.
G box substitutions comprise the following four substitutions: D451A, D452A, G453A, and G455A.
Phosphate transfer activity of GST-Bc.
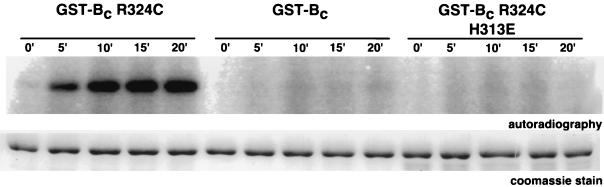
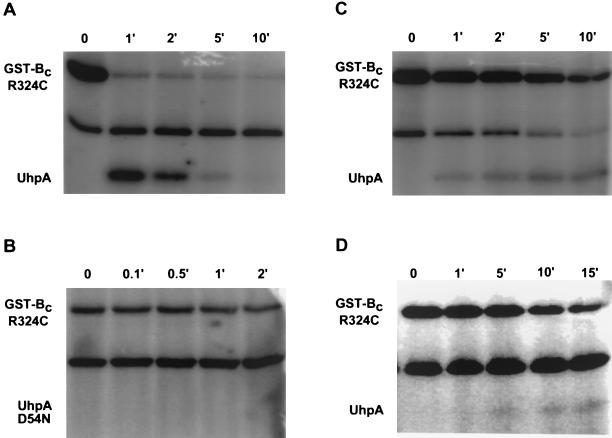
Many HK proteins exhibit autokinase activity and phosphate transfer to their cognate response regulator. To test for autokinase activity, purified GST-Bc protein and several of its variants were incubated with [γ-32P]ATP, followed by electrophoretic resolution and detection of radioactive proteins. The GST-Bc protein exhibited very low levels of autophosphorylation (Fig. 4). However, the active variant, GST-Bc R324C, exhibited substantial autokinase activity, which was completely lost when the site of phosphorylation was altered by the H313E substitution.
FIG. 4.
Autophosphorylation of GST-Bc variants. Purified samples of GST-Bc R324C, GST-Bc, and GST-Bc R324C H313E, each at 2.8 μM, were incubated for the indicated periods (in minutes) with [γ-32P]ATP. The samples were resolved by SDS-PAGE, the distribution of radioactivity was determined by PhosphorImager analysis (top panel), and the amount of GST-Bc protein was estimated by Coomassie blue staining of the electropherogram (bottom panel).
The ability of phosphorylated GST-Bc R324C to mediate phosphate transfer to UhpA was shown by allowing formation of P-GST-Bc by autophosphorylation with [γ-32P]ATP for 20 min followed by the addition of UhpA. At intervals, samples were removed and resolved by SDS-PAGE (Fig. 5). Under these conditions, P-GST-Bc R324C exhibited almost complete transfer of phosphate to UhpA within 1 min, but the P-UhpA thus formed underwent dephosphorylation, presumably by release of inorganic phosphate, at a much higher rate than the rate of dephosphorylation of P-UhpA alone, i.e., <5 min and >60 min, respectively (3). When P-GST-Bc R324C was incubated with UhpA D54N, lacking the site of phosphorylation (Fig. 5B), there was no transfer of labeled phosphate to UhpA, and the amount of label on GST-Bc was retained.
FIG. 5.
Phosphate transfer from P-UhpB to UhpA. Purified GST-Bc R324C was incubated for 20 min with [γ-32P]ATP under the same conditions for autophosphorylation specified in the legend to Fig. 4. After the period for labeling of Uhp, the following were added: 2.8 μm UhpA (A), 5 μM UhpA D54N (B), 2.8 μM UhpA plus 1 mM ATP (C), or 2.8 μM UhpA plus 10 mM EDTA. Samples were removed at the indicated times (in minutes), mixed with 6× SDS sample buffer on ice, and resolved by SDS-PAGE, and the distribution of radioactivity was determined by PhosphorImager analysis. The labeled band in the middle of each panel is not a Uhp protein but a very minor contaminant that copurifies with GST-Bc.
In the presence of 1 mM unlabeled ATP added together with UhpA, the transfer of labeled phosphate from P-GST-Bc to wild-type UhpA was greatly slowed, but the retention of label on both Uhp proteins was much more prolonged than in the absence of added ATP (Fig. 5C). Similar behavior was seen with the addition of 10 mM EDTA (Fig. 5D).
DISCUSSION
The Uhp system provides a simple but atypical model for study of two-component signaling, involving a single, specific, and easily controlled input signal that leads to an extensive change in expression of a single tightly regulated promoter. HK proteins typically possess competing kinase and phosphatase activities, whose balance is controlled by the specific input signal. The default state of most HK proteins, revealed by liberation of the cytoplasmic phosphotransfer domain, exhibits high unregulated kinase activity.
UhpB represents an unusual situation, owing to its requirement for the participation of another transmembrane protein, UhpC, probably as a signal receptor. Since a uhpC null mutant is phenotypically Uhp−, it appears that UhpB must exist in a form that is inactive as an autokinase in the absence of UhpC. To test the dominance and epistasis properties of peptide insertion variants on uhp regulation, diploid strains carrying combinations of mutant and wild-type uhp regulatory genes were constructed by homologous recombination into the uhpT locus of polA strains. The Uhpc phenotype of certain insertions in uhpB and uhpC was dominant, but the induced level of expression was reduced under conditions where the copy number of uhpB exceeded that of uhpC, consistent with a negative action of UhpB that is not in complex with UhpC. Consistent with this view was the finding that a plasmid carrying the uhpB coding region cannot complement a ΔuhpB or ΔuhpBC strain. Complementation for restoration of inducible behavior occurred only when the plasmid carried both uhpB and uhpC. This result agreed with the properties of the constitutive but UhpC-dependent uhpB mutants (10), which suggested that both proteins function as a complex and that the molecules of UhpB present in excess of UhpC have a dominant-negative interference effect. The relative amounts of UhpB and UhpC proteins have not been measured, but they are expressed as an operon and could be translationally coupled.
The interference phenotype was localized to the C-terminal half of UhpB, since liberation of this HK domain from the transmembrane N-terminal half did not result in activation of UhpA but retained the strong dominant-negative interference. Although we initially suspected that the interference was a reflection of the phosphatase activity of the UhpB HK domain, several lines of evidence indicated that this effect is independent of the state of phosphorylation of UhpA and probably involves the binding and sequestration of UhpA. There was strong interference by GST-Bc on activation by the UhpB-independent UhpA H170Y variant or by overexpressed UhpA-D54N lacking the site of phosphorylation. GST-Bc inhibited the ability of UhpA to bind and to activate the in vitro transcription at the uhpT promoter; both processes were measured in the absence of phosphorylation. Inhibition of transcription occurred with roughly equimolar amounts of UhpA and GST-Bc, and complex formation between GST-Bc and UhpA has been seen by a GST pulldown assay (30).
HK proteins function as homodimers, as indicated by their capability for intersubunit phosphorylation (17), by the occurrence of examples of dominant-negative interactions, and by direct structure determinations. Truncations of VanS that inhibit PhoB activation by functional VanS appeared to form inactive heterodimers with VanS rather than to inhibit PhoB directly (4). The EnvZ HK domain forms a dimer through subdomain A (19, 24). In contrast, the dominant-negative effect of GST-Bc does not require the presence of intact UhpB and appears to occur through direct action on UhpA.
Complex formation between other HK proteins and their cognate response regulators has been described. Surface plasmon resonance studies showed that binding of CheA to CheY occurs with affinity around 30 nM (22) and that the CheA P2 domain forms a stable complex with CheY (14, 27). The VanS-VanR protein pair forms a complex with a dissociation constant around 30 nM (5), and VanS was an efficient inhibitor of the binding of P-VanR to DNA (8). Subdomain A of EnvZ was shown to interact with OmpR (19).
Some of the UhpC-independent forms of UhpB showed relief of the dominant-negative effect when expressed as a GST-Bc fusion. These activating substitutions occurred near the H box, in the region related to subdomain A of EnvZ, which forms a four-helix bundle during EnvZ dimerization (24). The UhpB R324C variant is active in the absence of UhpC and results in the appearance of autokinase activity in the GST-Bc context. This activating change does not appear to alter the interaction of UhpB with UhpA, since GST-Bc-R324C still showed a strong dominant-negative effect when its autokinase function was abrogated by the substitutions in conserved regions important for phosphotransfer activity. As expected from studies of other HK proteins, conserved residues in the H box, the N box, and the G box were required for autokinase activity. However, they were not required for the interference phenotype. The active variant of GST-Bc could participate in phosphate transfer to UhpA, but it also seems to mediate the dephosphorylation of P-UhpA, as shown by the relatively rapid loss of labeled phosphate from UhpA in the presence of UhpB. As expected, phosphate transfer to UhpA was blocked by the presence of EDTA, which removes the essential Mg ion from the active site of RR proteins (12).
Taken together, these results show that the default state of UhpB is kinase off and that activation of autokinase activity requires either the presence of UhpC or certain mutations near the H box of UhpB. The requirement for UhpC to activate kinase activity allowed the demonstration that the unactivated form of UhpB confers a very strong interference with UhpA action. This interference cannot be explained solely by UhpB phosphatase activity and must involve the binding and sequestration of UhpA by inactive UhpB. Whether this sequestration activity is a general feature of HK proteins remains to be demonstrated, but tethering could provide a general mechanism for a quick response to environmental signals. The RR protein could be docked with the HK protein, ready for immediate phosphorylation upon receipt of the appropriate signal. Studies with the Bacillus subtilis KinA HK and its cognate RR, Spo0F, demonstrated that KinA autophosphorylation is enhanced by the presence of Spo0F (6), an indication that HK-RR tethering may have physiological and kinetic value.
ACKNOWLEDGMENTS
The important contributions of Beiyang Wei, Michael Island, and Carol Webber to initial portions of this work are gratefully acknowledged.
This work was supported by research grant GM38681 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Baikalov I, Schröder I, Kaczor-Grzeskowiak M, Grzeskowiak K, Gunsalus R P, Dickerson R E. Structure of the Escherichia coli response regulator NarL. Biochemistry. 1996;35:11053–11061. doi: 10.1021/bi960919o. [DOI] [PubMed] [Google Scholar]
- 2.Chen Q, Kadner R J. Effect of altered spacing between uhpT promoter elements on transcription activation. J Bacteriol. 2000;182:4430–4436. doi: 10.1128/jb.182.16.4430-4436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahl J L, Wei B Y, Kadner R J. Protein phosphorylation affects binding of the Escherichia coli transcription activator UhpA to the uhpT promoter. J Biol Chem. 1997;272:1910–1919. doi: 10.1074/jbc.272.3.1910. [DOI] [PubMed] [Google Scholar]
- 4.Fisher S L, Jiang W, Wanner B L, Walsh C T. Cross-talk between the histidine protein kinase VanS and the response regulator PhoB. J Biol Chem. 1995;270:23142–23149. doi: 10.1074/jbc.270.39.23143. [DOI] [PubMed] [Google Scholar]
- 5.Fisher S L, Kim S-K, Wanner B L, Walsh C T. Kinetic comparison of the specificity of the vancomycin resistance kinase VanS for two response regulators, VanR and PhoB. Biochemistry. 1996;35:4732–4740. doi: 10.1021/bi9525435. [DOI] [PubMed] [Google Scholar]
- 6.Grimshaw C E, Huang S, Hanstein C G, Strauch M A, Burbulys D, Wang L, Hoch J A, Whiteley J M. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry. 1998;37:1365–1375. doi: 10.1021/bi971917m. [DOI] [PubMed] [Google Scholar]
- 7.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 8.Holman T R, Wu Z, Wanner B L, Walsh C T. Identification of the DNA-binding site for the phosphorylated VanR protein required for vancomycin resistance in Enterococcus faecium. Biochemistry. 1994;33:4625–4631. doi: 10.1021/bi00181a024. [DOI] [PubMed] [Google Scholar]
- 9.Igo M M, Silhavy T J. EnvZ, a transmembrane environmental sensor of Escherichia coli K-12, is phosphorylated in vitro. J Bacteriol. 1988;170:5971–5973. doi: 10.1128/jb.170.12.5971-5973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Island M D, Kadner R J. Interplay between the membrane-associated UhpB and UhpC regulatory proteins. J Bacteriol. 1993;175:5028–5034. doi: 10.1128/jb.175.16.5028-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Island M D, Wei B-Y, Kadner R J. Structure and function of the uhp genes for the sugar phosphate transport system in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1992;174:2754–2762. doi: 10.1128/jb.174.9.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lukat G S, Stock A M, Stock J B. Divalent metal ion binding to the CheY protein and its significance to the phosphotransfer in bacterial chemotaxis. Biochemistry. 1990;29:5436–5442. doi: 10.1021/bi00475a004. [DOI] [PubMed] [Google Scholar]
- 13.Martinez E, Bartolome B, de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 14.McEvoy M M, Hausrath A C, Randolph G B, Remington S J, Dahlquist F W. Two binding modes reveal flexibility in kinase/response regulator interactions in the bacterial chemotaxis pathway. Proc Natl Acad Sci USA. 1998;95:7333–7338. doi: 10.1073/pnas.95.13.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merkel T J, Nelson D M, Brauer C L, Kadner R J. Promoter elements required for positive control of transcription of the Escherichia coli uhpT gene. J Bacteriol. 1992;174:2763–2770. doi: 10.1128/jb.174.9.2763-2770.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 17.Ninfa E G, Atkinson M R, Kamberov E S, Ninfa A J. Mechanism of autophosphorylation of Escherichia coli nitrogen regulator II (NRII or NtrB): trans-phosphorylation between subunits. J Bacteriol. 1993;175:7024–7032. doi: 10.1128/jb.175.21.7024-7032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olekhnovich I N, Dahl J L, Kadner R J. Separate contributions of UhpA and CAP to activation of transcription of the uhpT promoter of Escherichia coli. J Mol Biol. 1999;292:973–986. doi: 10.1006/jmbi.1999.3127. [DOI] [PubMed] [Google Scholar]
- 19.Park H, Soumitra K S, Inouye M. Two-domain reconstitution of a functional protein histidine kinase. Proc Natl Acad Sci USA. 1998;95:6728–6732. doi: 10.1073/pnas.95.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 21.Schröder I, Wolin C D, Cavicchioli R, Gunsalus R P. Phosphorylation and dephosphorylation of the NarQ, NarX, and NarL proteins of the nitrate-dependent two-component regulatory system of Escherichia coli. J Bacteriol. 1994;176:4985–4992. doi: 10.1128/jb.176.16.4985-4992.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuster S C, Swanson R V, Alex L A, Bourret R B, Simon M I. Assembly and function of a quaternary signal transduction complex monitored by surface plasmon resonance. Nature. 1993;365:343–347. doi: 10.1038/365343a0. [DOI] [PubMed] [Google Scholar]
- 23.Shi L, Hulett F M. The cytoplasmic kinase domain of PhoR is sufficient for the low phosphate-inducible expression of Pho regulon genes in Bacillus subtilis. Mol Microbiol. 1999;31:211–222. doi: 10.1046/j.1365-2958.1999.01163.x. [DOI] [PubMed] [Google Scholar]
- 24.Tomomori C, Tanaka T, Dutta R, Park H, Saha S K, Zhu Y, Ishima R, Liu D, Tong K I, Kurokawa H, Qian H, Inouye M, Ikura M. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat Struct Biol. 1999;6:729–734. doi: 10.1038/11495. [DOI] [PubMed] [Google Scholar]
- 25.Webber C A, Kadner R J. Action of receiver and activator modules of UhpA in transcriptional control of the Escherichia coli sugar phosphate transport system. Mol Microbiol. 1995;15:883–893. doi: 10.1111/j.1365-2958.1995.tb02358.x. [DOI] [PubMed] [Google Scholar]
- 26.Webber C A, Kadner R J. Involvement of the amino-terminal phosphorylation module of UhpA in activation of uhpT transcription in Escherichia coli. Mol Microbiol. 1997;24:1039–1048. doi: 10.1046/j.1365-2958.1997.4021765.x. [DOI] [PubMed] [Google Scholar]
- 27.Welch M, Chinardet N, Mourey L, Birck C, Samama J-P. Structure of the CheY-binding domain of histidine kinase CheA in complex with CheY. Nat Struct Biol. 1997;5:25–29. doi: 10.1038/nsb0198-25. [DOI] [PubMed] [Google Scholar]
- 28.Weston L A, Kadner R J. Identification of Uhp polypeptides and evidence for their role in exogenous induction of the sugar phosphate transport system of Escherichia coli K-12. J Bacteriol. 1987;169:3546–3555. doi: 10.1128/jb.169.8.3546-3555.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weston L A, Kadner R J. Role of uhp genes in expression of the Escherichia coli sugar-phosphate transport system. J Bacteriol. 1988;170:3375–3383. doi: 10.1128/jb.170.8.3375-3383.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright J S. Ph.D. thesis. Charlottesville: University of Virginia; 2000. [Google Scholar]