Abstract
The radC102 mutation causes mild UV and X-ray sensitivity and was mapped previously to near pyrE and recG at 82 min on the Escherichia coli chromosome (I. Felzenszwalb, N. J. Sargentini, and K. C. Smith, Radiat. Res. 97:615–625, 1984). We report that radC102 has two striking phenotypes characteristic of recG mutations. First, it causes dramatically increased RecA-dependent mutation in a stationary-phase mutation assay. Second, it causes extreme UV sensitivity in combination with ruv mutations affecting the RuvABC Holliday junction resolution system. DNA sequencing of the radC and recG genes in radC102 strains revealed that the radC102 mutation creates a stop codon in recG that is predicted to truncate the RecG protein at 410 of 603 amino acids. A low-copy-number plasmid carrying the radC+ gene did not affect the UV sensitivity of a wild-type strain, a radC102 strain, or a recG258::Tn10mini-kan strain. We conclude that radC102 is an allele of recG and that the function of the RadC protein remains to be determined.
The radC102 mutation causes a mild UV and X-ray sensitivity and was mapped by transduction previously to the pyrE recG region at 82 min on the Escherichia coli chromosome (11). Further work identified a novel open reading frame (ORF), designated radC, as the site of the radC102 mutation (9, 10). We present here several lines of evidence that radC102 is an allele of recG. The recG gene was identified originally by mutations that cause mild UV sensitivity and slight defects in transductional and conjugational recombination (20, 22, 35). recG encodes a helicase capable of binding and unwinding strand exchange recombination intermediates (such as Holliday junctions) in vitro (39, 40) and probably carries out branch migration of recombination substrates in vivo (reviewed in reference 21). The importance of the RecG protein in recombination became clear with the discovery that cells lacking both RecG and RuvA, -B, or -C are extremely UV sensitive and recombination defective (18). Because the absence of either RecG or RuvABC has only slight effects on transductional and conjugational recombination (in the RecBCD pathway of recombination [21]), they were thought to play functionally redundant roles in recombination of linear substrates. However, they do not play identical roles, because their substrate specificities and directions of branch migration differ in vitro (1, 38, 39) and their effects on stationary-phase mutation (13, 17) and on recombination in some assays (19, 20, 25; M. Motamedi and S. M. Rosenberg, unpublished results) differ in vivo.
We began this work to ask whether radC102 might affect lac frameshift mutation in stationary-phase E. coli cells, a mutational process dependent upon the recombination proteins RecA, RecBC, and RuvABC (13, 16, 17). In this stationary-phase (or adaptive) Lac+ mutation process, recombination intermediates are proposed to promote DNA replication and mutation (16, 23, 30). In the course of these experiments we found that the phenotypes of radC102 strains mimicked those of recG mutations in two assays, stationary-phase mutation and UV sensitivity. Subsequently, sequencing revealed that radC102 strains carry a mutation in the recG gene and that the radC gene is wild type in radC102 strains. We conclude that radC102 is an allele of recG. The function of the E. coli radC gene (and its many bacterial homologs) remains to be determined.
MATERIALS AND METHODS
Bacterial strains.
Strains used in this work are shown in Table 1. All strains were constructed by standard transformation or P1 transduction techniques (28). Antibiotics were used as necessary at the following concentrations: tetracycline, 15 μg/ml; ampicillin, 100 μg/ml. The radC102 allele was transduced from SR1187 (11) with selection for nearby pyrE+ into SMR5426. Eleven of 12 Pyr+ transductants tested were mildly UV sensitive on YENB medium (11), consistent with the linkage and UV sensitivity described previously (11). One of these transductants (SMR5441) was used for strain constructions and experiments. The radC102 ruvC53 strain was constructed and grown at 30°C. Initial constructions at 37°C gave widely varying colony sizes, UV sensitivities, and stationary-phase mutation phenotypes. Instability of such phenotypes in ruv recG strains was observed previously (13, 17, 24).
TABLE 1.
E. coli K-12 strains
| Strain | Genotype | Source and/or reference |
|---|---|---|
| BW229 | pyrE70 rfa-209::Tn10 lac rpsL gltS metB thi | E. coli Genetic Stock Center (Yale University) |
| CS85 | ruvC53 eda-51::Tn10 his-4 argE3 leuB6 proA2 thr-1 thi-1 rpsL31 galK2 lacY1 ara-14 xyl-5 mtl-1 kdgK51 supE44 tsx-33 | 34 |
| FC29 | Δ(lac-proB)XIIIara thi/F′ ΔlacIZ proAB+ | 4 |
| GY8322 | Δ(srlR-recA)306::Tn10 sfiA11 (sulA) thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 qsr′ supE44 galK2 hisG4 rfbD1 mgl-51 rpsL31 kdgK51 xylA5 mtl-1 argE3 thi-1/mini-F K5353 (recA+) | S. Sommer; ENZ280 (6) carrying the K5353 mini-F plasmid (7) |
| RDK2641 | ruvA59::Tn10 recB21 recC22 sbcB15 sbcC201 hsdR(rK− mK+)a | R. Kolodner |
| RSH154 | SMR4562 ruvA59::Tn10 | 17 |
| RSH316 | SMR4562 recG258::Tn10mini-kan | 17 |
| SMR624 | SMR4562 Δ(srlR-recA)306::Tn10 | 16 |
| SMR2041 | SMR4562 recG258::Tn10mini-kan ruvC53 eda-51::Tn10 | 15 |
| SMR4562 | Δ(lac-proB)XIIIara thi Rifr/F′ lacI33ΩlacZ proAB+ | Identical in genotype to FC40 (4) |
| SMR5426 | SMR4562 pyrE70 rfa-209::Tn10 | Tetr Pyr− transductant of P1(BW229) × SMR4562 |
| SMR5441 | SMR4562 radC102 | Pyr+ Tets UVs transductant of P1(SR1187) × SMR5426 |
| SMR5447 | SMR4562 radC102 Δ(srlR-recA)306::Tn10 | Tetr UVs transductant of P1(GY8322) × SMR5441 |
| SMR5509 | SMR4562 radC102 ruvA59::Tn10 | Tetr UVs transductant of P1(RDK2641) × SMR5441 |
| SMR5517 | SMR4562 ruvC53 eda-51::Tn10 | Tetr UVs transductant of P1(CS85) × SMR4562 |
| SMR5518 | SMR4562 radC102 ruvC53 eda-51::Tn10 | Tetr UVs transductant of P1(CS85) × SMR5441 |
| SMR5677 | SMR4562/pLG338-30 (Ampr) | Ampr transformant of pLG338-30 into SDMR4562 |
| SMR5678 | SMR4562/pMJ10 (radC+ Ampr) | Ampr transformant of pMJ10 into SMR4562 |
| SMR5681 | SMR5441/pLG338-30 | Ampr transformant of pLG338-30 into SMR5441 |
| SMR5682 | SMR5441/pMJ10 | Ampr transformant of pMJ10 into SMR5441 |
| SMR5685 | RSH316/pLG338-30 | Ampr transformant of pLG338-30 into RSH316 |
| SMR5686 | RSH316/pMJ10 | Ampr transformant of pMJ10 into RSH316 |
| SR1187 | radC102 thr-1 leu-6 proA2 his-4 thi-1 lacY1 galK2 ara-14 xyl-5 gpt mtl-1 txs-33 strA31 supE44 | N. Sargentini (11) |
This genotype may not be complete.
Mutation and UV sensitivity assays.
Stationary-phase mutation assays were as described previously (17). Briefly, multiple independent cultures of each strain were grown to saturation in minimal glycerol medium, washed twice in minimal medium with no carbon source, and plated on minimal lactose medium. Lac− scavenger cells, incapable of reverting to Lac+ (FC29 [4]), were plated along with each strain at approximately a 20-fold excess cell number to prevent growth on any contaminating nonlactose carbon sources (4). Plates were incubated at 37°C, and Lac+ colonies were counted each day for 5 days. recA strains were concentrated 10-fold prior to plating to obtain enough Lac+ colonies. Viability of the Lac− frameshift-bearing cells on the selection plates was monitored each day as described previously (16, 17). There was no net change in the total number of Lac− frameshift-bearing cells on the plates during the course of these experiments (data not shown).
UV sensitivity was determined using saturated LBH (36) cultures. When plasmid-bearing strains were tested, ampicillin was included in the broth and in plates. Cultures were diluted and plated on LBH or LBH-ampicillin, irradiated or not, incubated at 37°C for approximately 24 h, and then counted. The fraction surviving was calculated as cells surviving/cells plated. All cultures were grown at 37°C, except when radC102 ruv and recG ruv strains were involved (see Fig. 1), in which case all cultures were incubated at 32°C for all steps of the experiment to prevent faster-growing suppressor mutants from accumulating (17).
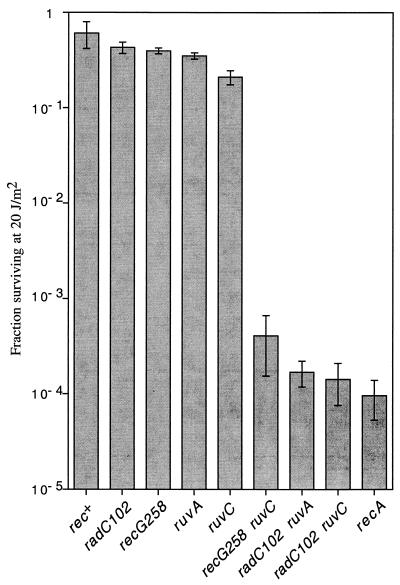
FIG. 1.
radC102 confers extreme UV sensitivity in combination with ruv mutations. Values are averages from one experiment with three independent cultures of each strain. The error bars represent one standard deviation. Similar results were obtained in another experiment (data not shown). All strains were grown at 32°C to prevent accumulation of suppressor mutants or revertants (detected as heterogeneity of colony size and UV resistance) in the double ruv recG mutants (17). The eda-51::Tn10 transposon linked with ruvC53 has no phenotype on its own (data not shown). The strains are (from left to right): SMR4562, SMR5441, RSH316, RSH154, SMR5517, SMR2041, SMR5509, SMR5518, and SMR624.
Construction of a radC+ plasmid and DNA sequence analysis.
The entire radC+ gene (as annotated in the E. coli genome sequence [Swiss-Prot no. P25531,3]) and its promoter region (14) were amplified by PCR from E. coli SMR4562 cells using primers 5′CGTAGTGGTATAGAAGTGACCAGTA3′ and 5′ACCAGAAACCGCCTGCAAGCTAAGT3′. This product was cut at an AatII site flanking the radC gene and ligated as a 1,489-bp AatII blunt fragment into AatII-SmaI digested pLG338-30, a pSC101-derived low-copy-number plasmid (5). The resulting radC+ plasmid was designated pMJ10. DNA sequencing confirmed that the entire fragment cloned is identical to the sequence in the published E. coli genome (3). Cloning of the same PCR product into AatII-ScaI-digested pBR322 (a higher-copy-number plasmid) gave small sickly transformants, suggesting that radC+ is toxic in high copy number. Similar toxicity in high copy was reported for a plasmid carrying the complete radC+ gene and flanking sequences (10). One radC plasmid previously reported to complement the UV sensitivity of the radC102 mutation does not contain the entire ORF (a site within the radC ORF was used as the cloning site) (10). A predicted RadC protein of 99 amino acids was proposed to be expressed from that plasmid (8, 10).)
The chromosomal radC and recG genes of several strains (see Results and Discussion) were sequenced using PCR-generated templates (Lone Star Labs, Inc., Houston, Tex.). Primers for radC amplification were the same as those used for cloning (see above). Primers for recG amplification were 5′AGCAACAACGCCTGTTGTTTGAAG3′ and 5′GTGATGAATCGCATCCGGCAGGAA3′. Additional primers were designed for sequencing. The absence of the reported frameshift mutation in the radC102 strain SR1187 (9) was confirmed by sequencing across the mutation site in five independent PCR products.
RESULTS AND DISCUSSION
radC102 greatly elevates stationary-phase mutation of a lac frameshift allele.
Stationary-phase (or adaptive) mutation in the lac frameshift assay system requires the recombination proteins RecA, RecBC, and RuvABC (13, 16, 17). In this mutation assay (4), cells carrying a lac frameshift allele on an F′ plasmid are placed on lactose minimal medium and Lac+ mutant colonies are scored each day for several days. New colonies appear each day due to mutations that occur on the selective plate and not prior to plating (27). The recombination gene dependence (13, 16, 17), sequence spectrum (12, 31), and other features of Lac+ stationary-phase mutation support the hypothesis that the mutations are formed by DNA polymerase errors during synthesis primed from recombination intermediates (16, 30). We wondered whether radC102 might affect stationary-phase mutation, given its effects on UV sensitivity (11) and recombination between tandem repeats (33). We find that radC102 dramatically increases the frequency of Lac+ mutations in this assay (Table 2). The increase is completely dependent on the RecA protein (Table 2). These phenotypes of radC102 are very similar to those of recG mutations, which also stimulate RecA-dependent Lac+ stationary-phase mutation strongly (13, 17).
TABLE 2.
radC102 elevates RecA-dependent stationary-phase mutation of a lac frameshift allele
| Relevant genotypeb | Lac+ colonies per 108 cells plateda
|
|||||
|---|---|---|---|---|---|---|
| Expt 1
|
Expt 2
|
|||||
| Day 2 | Day 2–5 | Fold difference | Day 2 | Day 2–5 | Fold difference | |
| rec+ | 9.4 ± 2.4 | 200 ± 35 | 1 | 13 ± 2.5 | 220 ± 30 | 1 |
| radC102 | 10 ± 7 | 5,000 ± 850 | 25 | 64 ± 19 | 6,400 ± 610 | 29 |
| recA | 0.95 ± 0.30 | 5.2 ± 0.52 | 0.026 | 3.0 ± 0.77 | 14 ± 1.8 | 0.064 |
| radC102 recA | 1.2 ± 0.37 | 4.3 ± 0.61 | 0.022 | 4.3 ± 1.9 | 27 ± 7 | 0.12 |
See Materials and Methods. The values given are the average ± one standard error of the mean from experiments with six independent cultures of each strain. The effects of radC102 correspond closely to those obtained by Harris et al. (17) and Foster et al. (13) with the recG258::Tn10mini-kan allele. Values for days 2 to 5 are the total number of Lac+ colonies on day 5, and the fold differences of the values for days 2 to 5 relative to rec+ are also given.
From top to bottom, these strains are SMR4562, SMR5441, SMR624, and SMR5447.
radC102 ruv mutants are extremely UV sensitive.
We constructed strains carrying radC102 and mutations in ruvA or ruvC. Both of these strains are extremely UV sensitive, as shown in Fig. 1. radC102 strains carrying ruvA or ruvC mutations are as sensitive as the recA deletion strain and as a recG258 ruvC53 strain at this dose. In contrast, radC102 or the single ruv mutations alone cause little or no UV sensitivity at this dose. Extreme UV sensitivity is a phenotype of recG258 ruv strains (18). Thus, radC102 also behaves like a recG mutation in combination with ruv mutations. In addition, the radC102 ruv strains show an absence of stationary-phase mutation (data not shown), as do the recG258 ruv combinations (13, 17).
The radC102 mutation is in the recG gene.
The striking similarity between radC102 and recG in stationary-phase mutation and in UV sensitivity led us to consider that, contrary to published work (9, 10), radC102 might be an allele of the nearby recG gene. We sequenced the entire radC coding region (Swiss-Prot accession no. P25531) in two isolates of the radC102 strain SR1187 (obtained on separate occasions from N. Sargentini [9]) in SMR5441 (radC102) and in SMR4562 (radC+). SR1187 is the source of the radC102 allele for the experiments reported here and those of Saveson and Lovett (33). All of these radC sequences were identical to that in the genome sequence. None displayed the frameshift mutation in the radC ORF that was reported to be the radC102 mutation in SR1187 (9). The recG genes of the two radC102 strains (SR1187 and SMR5441) and the radC+ strain SMR4562 were also sequenced completely. The radC102 strains both contain a substitution mutation in recG (TGG to TGA) that changes Trp411 to a stop codon, predicting translation of a truncated RecG of 410 amino acids rather than 603 amino acids. Based on these data, we conclude that radC102 is an allele of recG.
The radC102 allele and recG258 alleles of recG have similar UV sensitivity phenotypes.
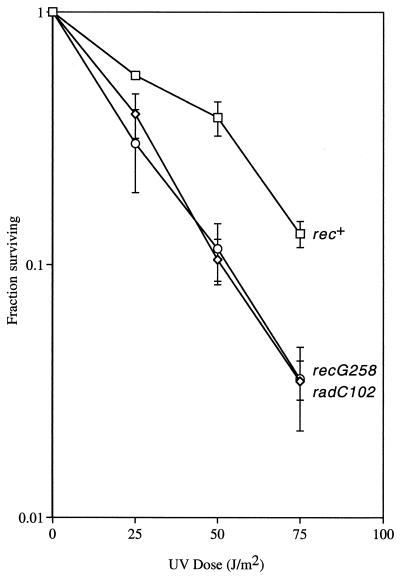
Because radC102 had not been compared directly with recG alleles previously, we assayed the UV sensitivities of a set of isogenic radC102, recG258::Tn10mini-kan, and recG+ strains (Fig. 2). The radC102 strain is as sensitive to UV as the strain carrying recG258::Tn10mini-kan, a commonly used null allele of recG (20).
FIG. 2.
radC102 and recG258 confer similar UV sensitivities. Values are the averages from one experiment with three independent cultures. The error bars represent one standard deviation. A similar result was obtained in a second experiment (data not shown). The strains are as follows: recG+ (rec+), SMR4562; radC102, SMR5441; recG258::Tn10mini-kan (recG258), RSH316.
Effect of radC overexpression on UV sensitivity.
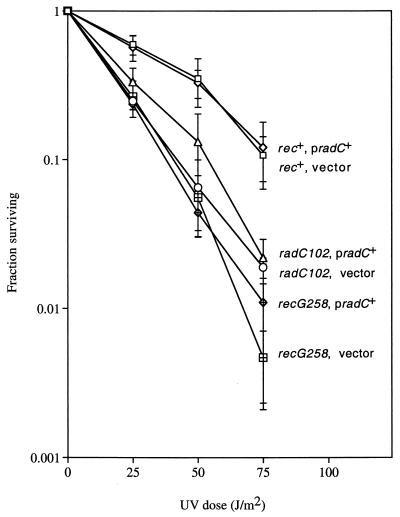
With the above findings, the evidence that E. coli radC encodes a DNA repair gene becomes limited to the report that expression of the complete radC+ gene (and of a fragment of radC) from a low-copy-number vector complements the UV sensitivity of the radC102 (recG) mutation (10). The plasmid carrying the complete radC+ gene was also reported to confer UV sensitivity to a wild-type strain (10). We cloned the complete radC+ gene on a related low-copy-number vector and found that this radC+ plasmid does not alter the UV sensitivity of either a radC102 strain, a recG258::Tn10mini-kan strain, or a recG+ strain (Fig. 3). This result is consistent with our finding that the radC102 mutation affects the recG gene. The apparent conflict between these overexpression results and those reported previously (10) might be due to differences between the plasmid constructs. An observed high-copy toxicity of the radC+ gene (see Materials and Methods), similar to that seen previously (10), suggests that radC+ is expressed.
FIG. 3.
Effect of pradC+ on UV sensitivity. Values are averages from one experiment with three independent cultures of each strain. The error bars represent one standard deviation. Within each set of strains (rec+, radC, and recG258), the error bars are overlapping, with the exception of the radC102 set at the 25 J/m2 dose. Similar results were obtained in a second experiment (data not shown). The strains used are (from top to bottom) SMR5678, SMR5677, SMR5682, SMR5681, SMR5686, and SMR5685.
Implications for RecG and RadC function.
The radC102 mutation causes a large decrease in tandem repeat recombination stimulated by a dnaB107 mutation (33). Our results demonstrating that radC102 is an allele of recG indicate that RecG is required for that RecA-dependent process. The dnaB107-stimulated recombination assay (33) provides an example in which the RuvABC and RecG systems appear to play dramatically different roles in vivo, in contrast to their overlapping functions in recombination of linear DNA substrates (18). The ruv dnaB107 combination is inviable, suggesting that RecG cannot process some lethal recombination substrate(s) created in the dnaB107 strains or that RecG processes them to lethal intermediates (33). In contrast, the radC102 dnaB107 combination appears to be viable (33), indicating that RuvABC deals efficiently with the potentially lethal recombination substrates produced in a dnaB107 strain lacking RecG.
There are several other recombinational assay systems in which RecG and RuvABC appear to play different roles, including recombination in the RecF pathway (19, 20), recombination-dependent stationary-phase Lac+ mutation (13, 17), homeologous recombination (25), λ Red-mediated recombination (29), and double-strand break repair (M. Motamedi and S. M. Rosenberg, unpublished results). In stationary-phase Lac+ mutation, RuvA, -B, and -C are required for mutation and RecG is inhibitory (13, 17). In that system, these opposing roles are proposed to reflect different effects of RuvABC and RecG on initiation of DNA synthesis from 3′ strand invasion intermediates, with RuvABC facilitating and RecG inhibiting due to opposite polarities of branch migration (17). This proposal is supported by work with the λ Red system, a known 3′ end invasion system inhibited by RecG and requiring RuvC (29).
The distinct functions of the Ruv and RecG systems in vivo are likely to be dictated by two classes of factors. First, their intrinsic properties, such as different polarities of branch migration (37, 40) or different affinities for particular recombination substrates such as D-loops and three-stranded junctions (26, 37), will govern which substrates can be bound and whether branch migration will facilitate or abort the reaction. Second, competition with other proteins for recombination intermediates will help define which substrates will be accessible. For example, genetic and biochemical evidence indicates that RecG and PriA, a primosome assembly protein with important roles in replication restart and double-strand break repair (reviewed in reference 32), probably compete for D-loop recombination intermediates in vivo (1, 26). Further investigation of assay systems in which RuvABC and RecG have differential effects will help to reveal their functions and substrates in vivo.
With the finding that the radC102 mutation affects recG, the evidence that E. coli RadC is involved in DNA repair becomes very limited, as discussed above. Construction and characterization of radC mutations will be necessary to reveal its function and to provide clues to the function of the multiple bacterial radC homologs. These include three E. coli homologs (ykfG, yfiY, and yeeS), which lack the putative helix-hairpin-helix DNA binding motif in the N terminus of RadC (2).
ACKNOWLEDGMENTS
We thank N. Sargentini for radC102 strains, R. Montelaro for pLG338-30, the E. coli Genetic Stock Center for bacterial strains, and M. Price for medium preparation.
This work was supported by National Institutes of Health grants F32-GM19909 (to M.-J. L.), R01-GM53158, and R01-AI43917.
REFERENCES
- 1.Al-Deib A A, Mahdi A A, Lloyd R G. Modulation of recombination and DNA repair by the RecG and PriA helicases of Escherichia coli K-12. J Bacteriol. 1996;178:6782–6789. doi: 10.1128/jb.178.23.6782-6789.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind L, Walker D R, Koonin E V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glassner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Cairns J, Foster P L. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cunningham T P, Montelaro R C, Rushlow K E. Lentivirus envelope sequences and proviral genomes are stabilized in Escherichia coli when cloned in low-copy-number plasmid vectors. Gene. 1993;124:93–98. doi: 10.1016/0378-1119(93)90766-v. [DOI] [PubMed] [Google Scholar]
- 6.Dri A M, Rouviere-Yaniv J, Moreau P L. Inhibition of cell division in hupA hupB mutant bacteria lacking HU protein. J Bacteriol. 1991;173:2852–2863. doi: 10.1128/jb.173.9.2852-2863.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutreix M, Moreau P L, Bailone A, Galibert F, Battista J R, Walker G C, Devoret R. New recA mutations that dissociate the various RecA protein activities in Escherichia coli provide evidence for an additional role for RecA protein in UV mutagenesis. J Bacteriol. 1989;171:2415–2423. doi: 10.1128/jb.171.5.2415-2423.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felzenszwalb I, Boiteux S, Laval J. Cloning of the Escherichia coli radC gene: identification of the RadC protein. Braz J Med Biol Res. 1993;26:1261–1268. [PubMed] [Google Scholar]
- 9.Felzenszwalb I, Boiteux S, Laval J. Identification of the radC102 mutation. Order of the genes in the 81.5-82.0 min region of the Escherichia coli chromosome. Nucleic Acids Res. 1992;20:366. doi: 10.1093/nar/20.2.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felzenszwalb I, Boiteux S, Laval J. Molecular cloning and DNA sequencing of the radC gene of Escherichia coli K-12. Mutat Res. 1992;273:263–269. doi: 10.1016/0921-8777(92)90088-k. [DOI] [PubMed] [Google Scholar]
- 11.Felzenszwalb I, Sargentini N J, Smith K C. Characterization of a new radiation-sensitive mutant, Escherichia coli K-12 radC102. Radiat Res. 1984;97:615–625. [PubMed] [Google Scholar]
- 12.Foster P L, Trimarchi J M. Adaptive reversion of a frameshift mutation in Escherichia coli by simple base deletions in homopolymeric runs. Science. 1994;265:407–409. doi: 10.1126/science.8023164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster P L, Trimarchi J M, Maurer R A. Two enzymes, both of which process recombination intermediates, have opposite effects on adaptive mutation in Escherichia coli. Genetics. 1996;142:25–37. doi: 10.1093/genetics/142.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gifford C M, Wallace S S. The genes encoding formamidopyrimidine and MutY DNA glycosylases in Escherichia coli are transcribed as part of complex operons. J Bacteriol. 1999;181:4223–4236. doi: 10.1128/jb.181.14.4223-4236.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris R S. Ph.D. thesis. Edmonton, Canada: University of Alberta; 1997. [Google Scholar]
- 16.Harris R S, Longerich S, Rosenberg S M. Recombination in adaptive mutation. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- 17.Harris R S, Ross K J, Rosenberg S M. Opposing roles of the Holliday junction processing systems of Escherichia coli in recombination-dependent adaptive mutation. Genetics. 1996;142:681–691. doi: 10.1093/genetics/142.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lloyd R G. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli depends on recG. J Bacteriol. 1991;173:5414–5418. doi: 10.1128/jb.173.17.5414-5418.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lloyd R G, Benson F E, Shurvinton C E. Effect of ruv mutations on recombination and DNA repair in Escherichia coli K12. Mol Gen Genet. 1984;194:303–309. doi: 10.1007/BF00383532. [DOI] [PubMed] [Google Scholar]
- 20.Lloyd R G, Buckman C. Genetic analysis of the recG locus of Escherichia coli K-12 and of its role in recombination and DNA repair. J Bacteriol. 1991;173:1004–1011. doi: 10.1128/jb.173.3.1004-1011.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd R G, Low K B. Homologous recombination. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2236–2255. [Google Scholar]
- 22.Lloyd R G, Sharples G J. Molecular organization and nucleotide sequence of the recG locus of Escherichia coli K-12. J Bacteriol. 1991;173:6837–6843. doi: 10.1128/jb.173.21.6837-6843.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lombardo M-J, Rosenberg S M. Hypermutation in stationary-phase E. coli: tales from the lac operon. J Genet. 1999;78:13–21. [Google Scholar]
- 24.Mandal T N, Mahdi A A, Sharples G J, Lloyd R G. Resolution of Holliday intermediates in recombination and DNA repair: indirect suppression of ruvA, ruvB, and ruvC mutations. J Bacteriol. 1993;175:4325–4334. doi: 10.1128/jb.175.14.4325-4334.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matic I, Rayssiguier C, Radman M. Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell. 1995;80:507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]
- 26.McGlynn P, Al-Deib A A, Liu J, Marians K J, Lloyd R G. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J Mol Biol. 1997;270:212–221. doi: 10.1006/jmbi.1997.1120. [DOI] [PubMed] [Google Scholar]
- 27.McKenzie G J, Lombardo M-J, Rosenberg S M. Recombination-dependent mutation in Escherichia coli occurs in stationary phase. Genetics. 1998;149:1163–1165. doi: 10.1093/genetics/149.2.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 29.Poteete A R, Fenton A C, Murphy K C. Roles of RuvC and RecG in phage λ Red-mediated recombination. J Bacteriol. 1999;181:5402–5408. doi: 10.1128/jb.181.17.5402-5408.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenberg S M. Mutation for survival. Curr Opin Genet Dev. 1997;7:829–834. doi: 10.1016/s0959-437x(97)80047-0. [DOI] [PubMed] [Google Scholar]
- 31.Rosenberg S M, Longerich S, Gee P, Harris R S. Adaptive mutation by deletions in small mononucleotide repeats. Science. 1994;265:405–407. doi: 10.1126/science.8023163. [DOI] [PubMed] [Google Scholar]
- 32.Sandier S J, Marians K J. Role of PriA in replication fork reactivation in Escherichia coli. J Bacteriol. 2000;182:9–13. doi: 10.1128/jb.182.1.9-13.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saveson C J, Lovett S T. Tandem repeat recombination induced by replication fork defects in Escherichia coli requires a novel factor, RadC. Genetics. 1999;152:5–13. doi: 10.1093/genetics/152.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shurvinton C E, Lloyd R G, Benson F E, Attfield P V. Genetic analysis and molecular cloning of the Escherichia coli ruv gene. Mol Gen Genet. 1984;194:322–329. doi: 10.1007/BF00383535. [DOI] [PubMed] [Google Scholar]
- 35.Storm P K, Hoekstra W P, de Haan P G, Verhoef C. Genetic recombination in Escherichia coli. IV. Isolation and characterization of recombination-deficiency mutants of Escherichia coli K12. Mutat Res. 1971;13:9–17. doi: 10.1016/0027-5107(71)90121-7. [DOI] [PubMed] [Google Scholar]
- 36.Torkelson J, Harris R S, Lombardo M-J, Nagendran J, Thulin C, Rosenberg S M. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitby M C, Lloyd R G. Branch migration of three-strand recombination intermediates by RecG, a possible pathway for securing exchanges initiated by 3′-tailed duplex DNA. EMBO J. 1995;14:3302–3310. doi: 10.1002/j.1460-2075.1995.tb07337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitby M C, Lloyd R G. Targeting Holliday junctions by the RecG branch migration protein of Escherichia coli. J Biol Chem. 1998;273:19729–19739. doi: 10.1074/jbc.273.31.19729. [DOI] [PubMed] [Google Scholar]
- 39.Whitby M C, Ryder L, Lloyd R G. Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell. 1993;75:341–350. doi: 10.1016/0092-8674(93)80075-p. [DOI] [PubMed] [Google Scholar]
- 40.Whitby M C, Vincent S D, Lloyd R G. Branch migration of Holliday junctions: identification of RecG protein as a junction specific DNA helicase. EMBO J. 1994;13:5220–5228. doi: 10.1002/j.1460-2075.1994.tb06853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]