Significance
Biomolecular condensates have emerged as a powerful new paradigm in cell biology with broad implications, from subcellular organization to human health and disease. Defining the spectrum of material states embodied by diverse condensates and their tunability is now an essential question in cell biology. Here, we investigate the material transitions of elastin—a major structural protein in the body. Using a model minielastin protein, we resolve the transition between liquid and solid elastin states. In addition to addressing a long-standing mechanistic gap in natural elastin assembly, this work has the potential to impact the design of elastin-inspired materials while further contributing mechanistic insight into the diverse repertoire of protein condensate maturation pathways with emerging importance in biology.
Keywords: maturation, biomolecular condensates, microrheology
Abstract
Liquid–liquid phase separation of tropoelastin has long been considered to be an important early step in the complex process of elastin fiber assembly in the body and has inspired the development of elastin-like peptides with a wide range of industrial and biomedical applications. Despite decades of study, the material state of the condensed liquid phase of elastin and its subsequent maturation remain poorly understood. Here, using a model minielastin that mimics the alternating domain structure of full-length tropoelastin, we examine the elastin liquid phase. We combine differential interference contrast (DIC), fluorescence, and scanning electron microscopy with particle-tracking microrheology to resolve the material transition occurring within elastin liquids over time in the absence of exogenous cross-linking. We find that this transition is accompanied by an intermediate stage marked by the coexistence of insoluble solid and dynamic liquid phases giving rise to significant spatial heterogeneities in material properties. We further demonstrate that varying the length of the terminal hydrophobic domains of minielastins can tune the maturation process. This work not only resolves an important step in the hierarchical assembly process of elastogenesis but further contributes mechanistic insight into the diverse repertoire of protein condensate maturation pathways with emerging importance across biology.
Elastic fibers play essential structural roles in the lung, blood vessels, skin, and bladder, where they provide tissue with the resilience and elasticity necessary for a lifetime of stretching and relaxation (1). Variations in elastic fiber structure and/or distribution result in a range of pathological conditions, including supravalvular aortic stenosis and Williams–Beuren syndrome (2). The formation of mature elastin fibers, termed elastogenesis, consists of a complex multistep process, which includes the self-assembly of the monomeric protein tropoelastin via a coacervation step, enzymatic protein cross-linking, and higher-order assembly into mature elastin fibers (1). The precise sequence of events and underlying mechanistic details associated with stages in the assembly process remain poorly understood.
Coacervation or liquid–liquid phase separation (LLPS) of tropoelastin is considered to be an early step in the elastogenesis process (3, 4). Tropoelastin is a 60- to 70-kDa protein consisting of alternating repeats of disordered hydrophobic domains and cross-linking domains (5) containing lysine residues, two to four of which are enzymatically cross-linked in mature fibers (6). Elastin coacervation is a temperature-dependent process driven by the entropic interactions between uncharged hydrophobic domains preceded by a disruption of water molecules surrounding the polymer (7, 8). These hydrophobic sequences have inspired the design of model elastin-like peptides (ELPs) with a range of biomedical and industrial applications (9, 10). Model “minielastin” constructs that additionally include cross-linking domains have also been designed to better mimic the alternating domain structure of native tropoelastin (10–12). These minielastins can undergo reversible LLPS and have similar mechanical properties to native elastin after cross-linking (10, 13–15).
While the structural properties of cross-linked mature elastin fibers have been carefully studied (4, 16–19), there is much less known about the material properties of the condensed liquid state of elastin, despite the phenomenon of elastin phase separation being well established (20, 21). Biomolecular liquids and condensates more generally are increasingly found to contribute to diverse biological contexts from the assembly of membraneless organelles to the organization of cellular contents, including structural proteins and even the genomic architecture (22, 23). As their name implies, biomolecular condensates embody a diverse spectrum of material properties from viscous fluids to elastic solids (24, 25). Transitions between material states are further implicated in condensate function, with their misregulation implicated in diseases such as amyotrophic lateral sclerosis and Huntington's disease (26, 27). A quantitative material description of the liquid phase–separated state of elastin and a mechanism that bridges its liquid and solid states are currently lacking.
Here, we study the properties and dynamics of elastin condensates using a model minielastin protein that mimics the alternating domain structure of full-length tropoelastin. We directly quantify the viscosity of the liquid state within the droplet using particle-tracking microrheology. We further demonstrate that elastin droplets mature and collapse to a solid state even in the absence of exogenous cross-linking. We quantitively resolve the transition between liquid and solid states using a combination of DIC/scanning electron microscopy, fluorescence measurements, and microrheology. We find that the homogenous, dynamic liquid state spontaneously gives rise to a heterogeneous intermediate stage marked by the coexistence of randomly distributed insoluble material within a liquid phase that remains dynamic. This intermediate stage is followed by a system collapse to a solid state. Finally, we demonstrate that varying the length of the terminal hydrophobic domains significantly tunes the maturation process. The results presented here lend mechanistic insight into the complex process of elastogenesis as well as the emerging diverse repertoire of protein condensate maturation pathways.
Results
Liquid–Liquid Phase Separation of a Minielastin.
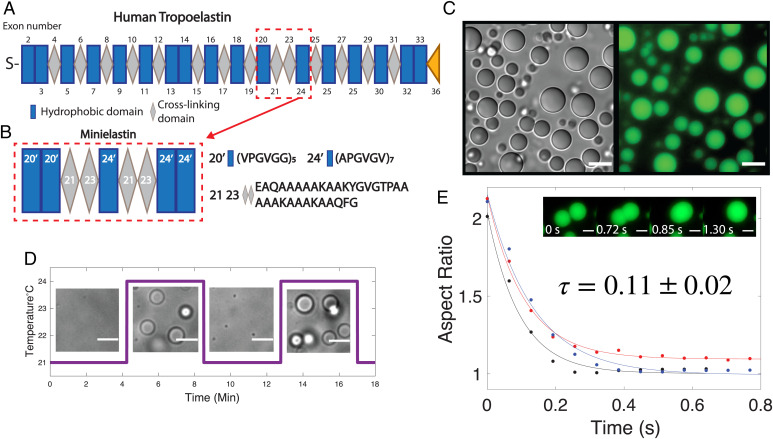
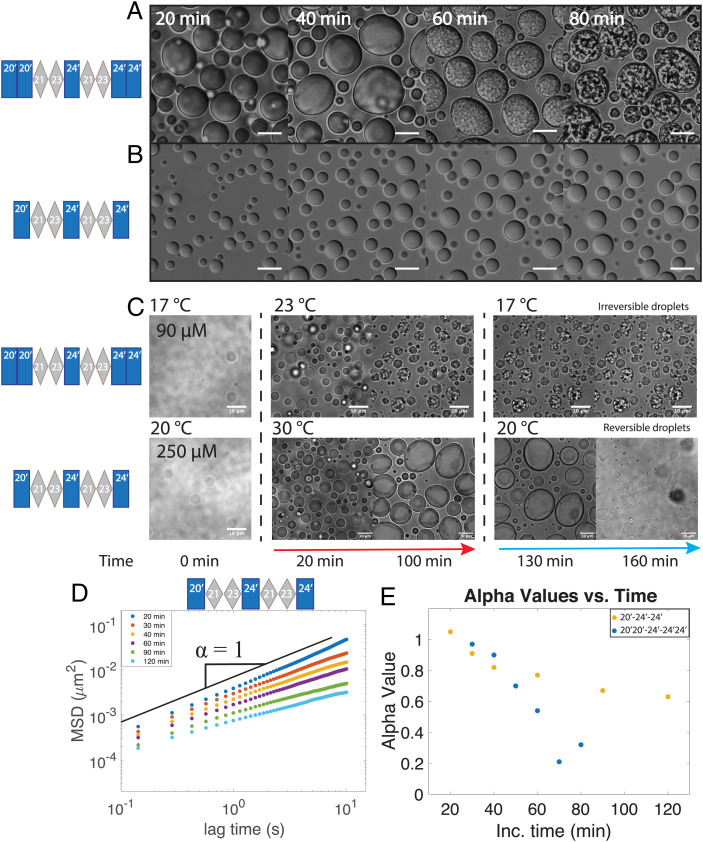
In order to gain insight into the nature of the condensed phase of elastin coacervates, we use here a model minielastin protein that contains alternating hydrophobic and cross-linking domains that mimic the domain structure of full-length tropoelastin, construct 20′20′-24′-24′24′, herein referred to simply as minielastin (Fig. 1 A and B) (28, 29). In this model protein, the quasirepeats of tropoelastin exons 20 and 24 are replaced by absolute repeats (20′ and 24′) for simplification, the cross-linking domains are composed of fused exons 21 and 23 as in previous works (12), and the termini are capped with a second respective hydrophobic domain in order to better model the full-length protein, which is flanked by expanded hydrophobic domains (28) (Fig. 1B). Consistent with synthetic hydrophobic ELP sequences and full-length tropoelastin, this minielastin can undergo a temperature-induced phase transition into liquid droplets that are reversible in response to temperature cycling at short incubation times (Fig. 1 C and D). These droplets coalesce upon contact, fusing with an approximate fusion timescale of 0.1 s (Fig. 1E). This rapid fusion is consistent with liquid behavior and results in droplets on the order of several to tens of micrometers settled rapidly to the bottom of the sample chamber. The coacervation temperature (Tc) of elastins is highly dependent on the polymer length, concentration, amino acid composition, and buffer ionic concentration (30). For experimental convenience, we tuned the polypeptide concentration and sodium chloride content to bring the Tc down to room temperature (23 °C) (Materials and Methods has details).
Fig. 1.
Minielastin behaves as a liquid. (A) Diagram of human tropoelastin domains. Hydrophobic domains are represented by blue rectangles, cross-linking domains are represented by gray diamonds, S represents domain 1 (signaling peptide), and exon 36 (the yellow triangle) represents the C-terminal domain. Exon 22 is not found in common isoforms of mature tropoelastin. Exons 34 and 35 are lost throughout human evolution. The tropoelastin structure is adapted from Muiznieks et al. (34). (B) Minielastin construct, hydrophobic domain 20′ and 24′ (blue), and lysine-rich cross-linking domain 2123 (gray). (C) DIC (Left) and fluorescent (Right) images of minielastin coacervates at 10 min of incubation time. (Scale bars: 10 μm.) (D) Temperature-induced assembly and disassembly of minielastin liquid droplets. Temperature ramps between 21 °C and 24 °C. (Scale bars: 5 μm.) (E) All three fusion events were measured at 10 min of incubation time and at room temperature without temperature cycling. Fitted exponential decay of fusion with a calculated fusion timescale (τ). Inset depicts minielastin coacervates fusing at 10 min of incubation. Buffer conditions in all images are 1.5 M NaCl, 1 mM CaCl2, and 50 mM Tris, pH 7.4. Minielastin protein was fluorescently labeled with Alexa Fluor 488. Protein concentration is 130 μM in DIC and fusion images and 115 μM in reversibility experiment. (Scale bars: 3 μm.)
Morphological Changes Accompanying Minielastin Condensate Maturation.
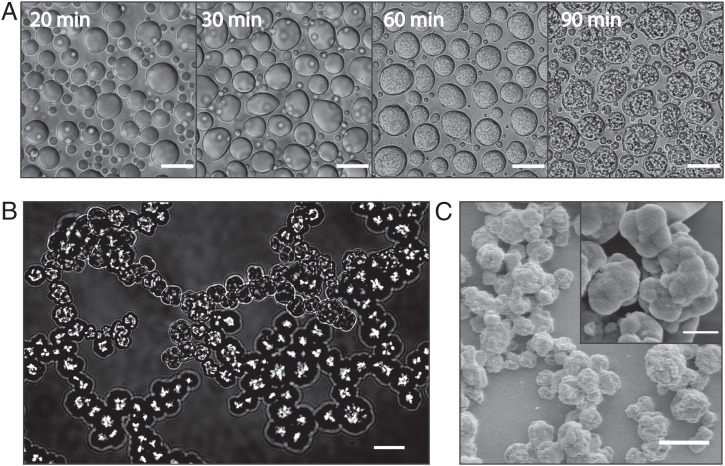
Continued incubation above the coacervation temperature results in morphological changes in the droplets over time (Fig. 2A). DIC imaging of droplets settled on the surface illustrates the appearance of submicrometer granular internal structures within minielastin droplets at about 60 to 70 min of incubation time (Fig. 2A). A similar process occurs when droplets are prevented from settling to the surface by imaging them freely diffusing in a flow chamber over time (Movies S1 and S2). In this case, the resulting granular structures are able to cluster, resulting in an interconnected network (Fig. 2B). Scanning electron microscopy of minielastin samples incubated overnight in a test tube reveals a similar interconnected network of granular particles with higher resolution (Fig. 2C). These interconnected networks, which morphologically resemble structures reported for cross-linked elastomers, occur here in the absence of any cross-linker (4, 31). Notably, these structures are not reversible even after overnight incubation in denaturant conditions (SI Appendix, Fig. S1) in contrast to the previously reported reversibility of aggregates formed without cross-linking (10).
Fig. 2.
Minielastin maturation from liquid coacervates into solid form. (A) Maturation of minielastin condensates over the course of 90 min at 23 °C. (Scale bars: 10 μm.) (B) A matured minielastin sample formed in the capillary flow chamber (Movies S1 and S2). (Scale bar: 20 μm.) (C) Scanning electron microscopy images of mature minielastin obtained after overnight incubation at 37 °C. (Inset) Enlarged image of mature minielastin droplets. Protein concentrations in DIC and scanning electron microscopy images are 130 and 230 μM, respectively. (Scale bars: C, 7 μm; C, Inset, 2 μm.)
Heterogeneous Dynamics within Droplets.
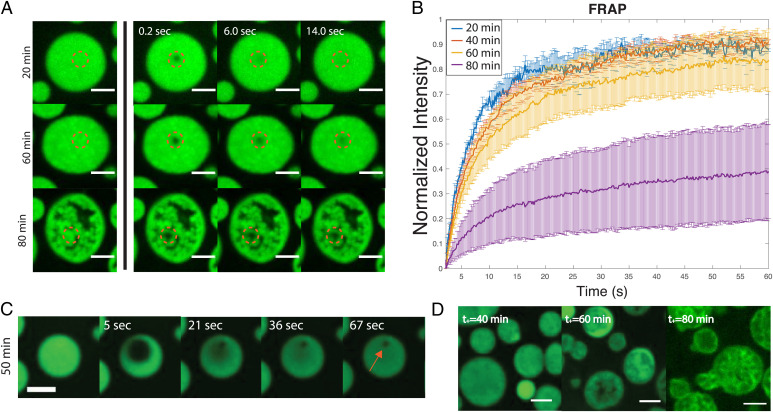
To probe the internal dynamics within the droplets, we performed fluorescence recovery after photobleaching (FRAP) experiments with ∼1% Alexa-488–tagged minielastin protein, where a submicrometer-sized region within the center of the droplet is bleached. We observe that within 20 to 40 min, the mobile fraction of the recovery curves remains relatively constant, with a 90 to 94% recovery after 60 s (Fig. 3A), suggesting the maintenance of a fluid internal environment within the droplet at early time points. However, the mobile fraction decreases consistently after 50 min, decreasing to 10 to 30% at 80 min of incubation (Fig. 3B). The decrease in recovery suggests an increased amount of immobile protein present within the droplet. Consistent with a decrease in droplet dynamics, we observe a decrease in droplet fusion rates leading to arrested coalescence at later incubation times (SI Appendix, Fig. S2). Concurrent with the changes we report in droplet morphology over time via DIC microscopy (Fig. 2A), we find that fluorescent protein becomes heterogeneously distributed within the droplet over time, giving rise to apparent regions devoid of protein at 80 min. When we increase the bleaching diameter in our FRAP experiments to ∼3 μm (Fig. 3C), we observe subregions failing to recover at the same rate as the greater bleached area even at earlier times, demonstrating that spatial heterogeneities in the droplet properties exist even before they are observable via traditional confocal imaging.
Fig. 3.
FRAP in the minielastin droplet system. (A) Image sequence of fluorescence recovery over time of minielastin coacervates at 20, 60, and 80 min of incubation time. (B) Normalized fluorescence intensity curves at 20 min (blue), 40 min (red), 60 min (yellow), and 80 min (purple) of incubation. SD was calculated for each point across three experiments. (C) Image sequence depicting fluorescence recovery over time of the minielastin droplet with a larger bleached area. The red arrow indicates the area in which fluorescent protein fails to diffuse. (D) Confocal images of minielastin after the addition of 1% labeled protein at increasing incubation times (t+). All images are single–confocal microscopy slices. All experiments were conducted with 130 μM protein concentration in buffer containing 1.5 M NaCl, 1 mM CaCl2, and 50 mM Tris, pH 7.4. Temperature was stabilized to 23 °C. (Scale bars: 5 μm.)
To further probe the apparent spatial differences in mobility within the droplet, we next investigated the diffusion pattern of fluorescent protein subsequently introduced to preformed unlabeled droplets at increasing times along the maturation pathway (Fig. 3D). We find that the addition of labeled protein at early times (∼30 to 40 min) results in a homogenous distribution of fluorescent protein across the droplet, consistent with free rapid diffusion through the droplet. However, at later incubation times, newly added fluorescent protein cannot fully distribute throughout the droplet, revealing spatial heterogeneities consistent with the presence of insoluble material as early as 60 min (Fig. 3D). In the later stages of maturation (80 min), the labeled protein takes on a more distinctive pattern, where it appears to outline the heterogeneous structures present in the collapsed droplet state. These results, along with our FRAP analysis, suggest that there is an intermediate stage (at ∼60 min) along the maturation pathway where liquid and solid-like phases coexist. Higher-resolution confocal time-lapse imaging of fluorescent droplets further captures this transition between states (Movies S3–S5).
Rheological Changes during Maturation.
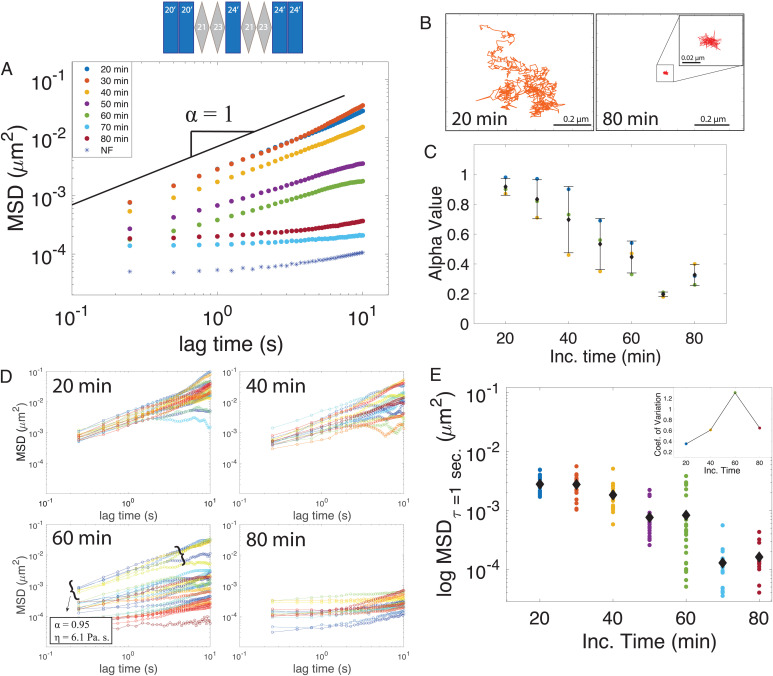
To gain further insight into the material properties of condensates during the maturation process, we next performed particle-tracking microrheology experiments (Fig. 4 and SI Appendix, Figs. S3–S5). We track the movement of fluorescent particles inside our droplet to measure their mean square displacement (MSD) (32, 33). In a simple viscous fluid, we expect the MSD to vary linearly with time, resulting in a diffusive exponent equal to one (32), whereby a viscosity can be directly extracted from the diffusion coefficient (D) using the Stokes–Einstein equation [D = KbT/6πηR], where T is temperature, η is viscosity, and R is the radius of the probe being used. At the earliest time accessible for measurement, 20 min, we find the rheological signature of minielastin droplets to resemble a viscous fluid state across the accessible lag times of our measurements with a diffusive exponent average of 0.92 ± 0.06. Applying Stokes–Einstein to these data at 20 min extracts a viscosity of 8.27 ± 1.9 Pa·s. Over time, the bead movement becomes significantly reduced (Fig. 4A and SI Appendix, Fig. S3) with representative tracks transitioning from apparent Brownian motion at 20 min, where beads can freely diffuse throughout the droplet, to fully restricted at 80 min (Fig. 4B), after which the beads end up trapped in the collapsed solid structure (SI Appendix, Fig. S5). The viscoelastic transition can be monitored by the decrease in the diffusive exponent α, which declines with time (Fig. 4C), with the slope approaching a flat line at 70 min. Note that these experiments are able to resolve deviations from a fluid state at even earlier times than other methods, as early as 30 min depending on the sample.
Fig. 4.
Rheological signatures accompanying elastin maturation. (A) Average MSD of bead particle vs. lag time within liquid droplets at increasing incubation times. The noise floor (NF) of the instrument is plotted as blue asterisks. The α-value represents the diffusive exponent, and the black solid line represents a slope of one. (B) Single-bead trajectories inside the minielastin condensate at 20 and 80 min of incubation time. Inset, zoomed in trajectory at 80 min. (C) Decreasing diffusive exponent (α) values among three individual experiments as a function of time. (D) Individual bead tracks vs. lag times at increasing incubation times of 20, 40, 60, and 80 min of incubation. (E) Black diamonds represent the average of individual MSD trajectories at increasing incubation times, with the spread of trajectories represented as scatter points at a lag time (τ) of 1 s. (Inset) Coefficient of variation among tracks at a lag time (τ) of 1 s.
Looking beyond the particle ensemble averages, the individual bead trajectories reveal more details about the sample heterogeneity. Plotting the MSDs from individual trajectories over time reveals an increase in the spread of individual bead trajectories within the droplet at intermediate incubation times (Fig. 4D and SI Appendix, Fig. S4). While the average MSD for a particular lag time continuously decreases as a function of incubation time (Fig. 4E), the SD, captured by the coefficient of variance for MSD, increases at intermediate incubation times (from 20 to 60 min) (Fig. 4E) and then, decreases as the condensate fully solidifies at 80 min (Fig. 4 E, Inset). These results indicate that individual particles experience unique diffusive environments owing to increased spatial heterogeneities. When we analyze the diffusion of only the fastest diffusing particles at an intermediate time (60 min), we extract a viscosity close to that of the ensemble at 20 min (Fig. 4D), further supporting the coexistence of liquid and solid phases.
Terminal Hydrophobic Domain Length Influences Minielastin Maturation.
The ubiquitous utility of elastin-inspired materials lies in their plasticity—where properties can be tuned by variations in sequence and/or domain architecture. We, therefore, next sought to identify domain features that influence the maturation process we report here. Interestingly, we found that previous work using a similar minielastin construct ("EP20-24-24") does not report apparent maturation of droplets without cross-linking, at least up to 60 min of incubation time (34). While composed of a similar alternating domain structure and domain types (i.e., hydrophobic domain exons 20 and 24 and cross-linking domains 21 to 23), these constructs differ 1) in the length of the hydrophobic domains at the termini, where our construct has a second hydrophobic domain capping each terminus, and 2) in that the hydrophobic domains of our minielastin are composed of perfect repeats (20′ and 24′ as defined above) instead of pseudorepeats (Fig. 1B). To examine whether terminal hydrophobic length alone can influence droplet maturation, we next compared our current minielastin (20′20′-24′-24′24′) with otherwise identical constructs with varying terminal hydrophobic lengths (SI Appendix, Fig. S6). We found that decreasing or increasing the length of the hydrophobic domain at one terminus had no significant effect on maturation based on DIC imaging (SI Appendix, Fig S6 A–C). However, decreasing the hydrophobic domain length at both termini, by removing the second hydrophobic domain at each end (i.e., construct 20′-24′-24′), had a significant effect on droplet properties (Fig. 5). DIC images of droplets composed of construct 20′-24′-24′ do not appear to change in morphology over time (Fig. 5 A and B). Additionally, in contrast to our control construct, the droplets composed of this shorter minielastin are almost completely reversible after being kept above their transition temperature for at least 100 min (Fig. 5C and SI Appendix, Fig. S7). It is worth noting that while the concentration and temperature needed to be increased for this shorter construct, which has a higher Tc, increasing either of those variables actually enhances the maturation of the control (SI Appendix, Fig. S8). Interestingly, NMR experiments indicate no significant change in secondary structure, at least on the monomeric level across constructs (SI Appendix, Fig. S9). Finally, despite no apparent morphological changes to the 20′-24′-24′ droplets, microrheology measurements do report a slow but steady decrease in the MSD and diffusive exponent over time (Fig. 5 D and E). Together, these data indicate that the length of terminal hydrophobic domains of minielastins significantly influences the maturation process of elastin condensates.
Fig. 5.
Terminal hydrophobic length modulates maturation. (A and B) DIC images of the original minielastin (20′20′-24′-24′24′) (A) and shortened minielastin (20′-24′-24′) (B) as a function of incubation time. (C) Temperature-induced reversibility of minielastin droplets after 130 min of incubation. The red arrow indicates temperature above the coacervation temperature (Tc) of construct. The blue arrow indicates temperature below Tc of the construct. (Scale bars: 10 μm.) (D) Average MSD particle trajectory against lag time of minielastin (20′-24′-24′). (E) The α-value comparison between minielastin constructs.
Discussion
In this work, we examine the condensed state of a model elastin protein that mimics the alternating domain structure of native tropoelastin. Our combined imaging, FRAP, and rheological measurements together demonstrate that the initially homogeneous viscous liquid elastin droplets spontaneously mature into a solid structure via the heterogeneous nucleation of internal insoluble material. We further demonstrate the importance of terminal hydrophobic domain length in contributing to this maturation process.
Beginning with characterization of the initial coacervates, we measure viscosities of ∼2 to 8 Pa⋅s within elastin droplets across constructs in the liquid state at early incubation times (Figs. 2 and 5 and SI Appendix, Fig. S6). While previous work has measured the global viscoelastic signatures of bulk turbid solutions of elastin composed of droplets diffusing within a dilute solution (15), this is a direct quantification of the rheological signature within an elastin liquid droplet. Our elastin droplets, which are ∼10,000× more viscous than water, are comparable with other condensates, such as LAF1, PGL-3, Whil3, and NPM1, with reported viscosities ranging from ∼1 to 30 Pa⋅s (35, 36–38).
We next probe the maturation of individual elastin droplets in real time. It should be noted that the liquid to solid transition we describe here, which we refer to as a “maturation” process, is distinct from other definitions of maturation previously described in the literature. For example, earlier works have described the ripening or increasing size of elastin droplets over time to be considered a maturation process (39, 40). Other works consider the formation of fully formed elastin fibers more broadly as a maturation process (41, 42). The maturation process we observe is also distinct from recently described aging of other biomolecular condensates. For example, PGL-3 and FUS droplets have recently been described as aging Maxwell glasses, where notably, despite drastic changes in viscoelastic properties and diffusive dynamics over time, no structural changes were detected inside the droplets by cryo-electron microscopy (36). The dramatic collapse of the droplet we observe here is not typical of other condensate aging systems (43, 44). Indeed, the drastic structural transition of this collapse (Movies S3–S5 and SI Appendix, Fig. S5), which is accompanied by the generation of a multitude of interfacial surfaces, precludes more traditional analysis of viscoelastic transitions. In fact, we propose that the resulting noise in the ensemble data specifically reflects the nucleation and growth of insoluble material and resulting interfacial surfaces (Fig. 4).
Our FRAP, fluorescence, and microrheology measurements demonstrate the development of heterogeneous insoluble assemblies within droplets preceding full collapse of the liquid state. We further show that this process can be modulated by decreasing the overall length of the terminal hydrophobic domains (Fig. 5). Previous work using a similar minielastin construct (EP20-24-24) does not report apparent maturation of droplets without cross-linking (35), despite observing apparent hardening of full-length tropoelastin. Additionally, a recent work using NMR to examine the condensed state of a minielastin reports a decrease in diffusion within the condensed phase on a timescale of days (45), in contrast with our minielastin construct that undergoes maturation within hours. The minielastin in these prior works differed from our construct in that they lacked either one (35) or both (45) terminal hydrophobic domains. Our work here (Fig. 5), which underscores the significance of the length of the hydrophobic domains at the termini to this maturation process, could explain these differences. Indeed, previous NMR studies of our construct demonstrated the stabilization of α-helical structure in the cross-linking domains when flanked between hydrophobic domains (29). This characteristic secondary structure is lacking in the absence of flanking hydrophobic domains, which can act as helical end caps that promote structure (46).
The presence or increase in secondary structure character of ELPs has generally been linked to distinct morphological characteristics in the resulting condensates (35, 47, 48), modulating their size and apparent maturation in solution. For example, proline periodicity and content in the hydrophobic domains (47) and cross-linking domains (48), respectively, have been shown to modulate elastin assembly morphology. Furthermore, increasing helix–helix interactions can enhance formation of fractal networks of mature coacervates (49). While the presence of hydrophobic termini has been shown to stabilize the helical structure of the cross-linking domains (29), our NMR experiments show no additional changes in secondary structure resulting from modulating the lengths of these domains (SI Appendix, Fig. S9), suggesting an alternate mechanism underlying the observed maturation.
Phase separation of elastins is generally thought to be driven by hydrophobic interactions between the hydrophobic domains concomitant with the release of bound water molecules. Interestingly, recent molecular dynamics simulations and NMR studies report an estimated water content of ∼40 to 60% remaining within the coacervates (45, 50), that the polypeptide backbone remains significantly hydrated within the condensed liquid phase (50), and that naturally occurring elastin fibers remain partially hydrated to retain structural integrity (51). The initial water loss supports the relative high viscosity of the coacervate in comparison with the surrounding buffer. The droplet maturation we observe finalizing with a collapse to a solid-like state likely coincides with further water loss within the coacervate as captured by high-resolution time-lapse imaging of fluorescent droplets (Movies S3–S5). We find that the initial insoluble clusters, which are revealed by our FRAP experiments (Fig. 3C) and are resolved in higher-resolution confocal images (Movie S3), persist during maturation as the surrounding liquid phase effectively dissolves (Movie S4), leaving behind a stable network of clusters (Movie S5).
Taken together, this work lends mechanistic insight into a state transition of elastin condensates with potential significance to both natural elastogenesis as well as the design of elastin-based materials with increasing complexity. Future studies that include additional structural proteins and cross-linking agents can further bridge this minimalist system with the native process of elastin fiber formation. Moreover, this work contributes to the diverse repertoire of protein condensate maturation pathways with emerging importance across biology.
Materials and Methods
Protein Expression.
Genes containing an N-terminal tobacco etch virus (TEV) protease cut site and His affinity tag were synthesized by Integrated DNA Technologies in expression vector pET32a(+) (Novagen, Inc.) as described earlier (52). Genes were transformed into NiCo21(DE3) competent Escherichia coli cells. Cells were initially grown in 5 mL of Luria Broth (LB) (per liter: 10 g of bactotyptone, 5 g of yeast extract, 10 g of NaCl, pH 7.5) with 0.01% ampicillin for 8 h at 37 °C and 225 rpm and then, transferred into 100 mL of LB with 0.01% ampicillin for 16 h at 37 °C; 50 mL of LB from the 100-mL growths was added to 1 L of TPP (per liter: 20 g of bactotryptone, 15 g of yeast extract, 8 g of NaCl, 4 g of Na2HPO4, 2 g of KH2PO4, pH 7.5; after autoclaving, 25 mL of 40% sterile glucose) with 0.01% ampicillin. The 1-L growths were incubated at 37 °C until reaching optical density (OD) 600 of 0.6 arbitrary units (AU) and then, induced with 200 mg of isopropyl-β-d-thiogalactopyranoside for 16 h at 18 °C. The cells were harvested by centrifugation at 9,000 rpm for 15 min.
For 15N-labeled proteins used in the NMR studies, cells were grown for 16 h in 5 mL of M9 minimal media (53) (per liter: 12 g of Na2HPO4, 6 g of KH2PO4, 0.5 g of NaCl, 1 g of 15N NH4Cl, pH 7.4; after autoclaving, 25 mL of 40% glucose, 2 mL of trace metal solution [per 1 L: 500 mL of EtOH, 0.8 mL of concentrated HCl, 0.5 g of FeCl2·4H2O, 18.4 mg of CaCl2·2H2O, 6.4 mg of H3BO3, 4 mg of MnCl2·4H2O, 1.8 mg of CoCl2·6H2O, 0.4 mg of CuCl2·2H2O, 34 mg of ZnCl2, 60.5 mg of Na2MoO4·2H2O, 26.8 g of MgCl2·6H2O], 1 mL of vitamin solution [per 500 mL: 1.1 mg of biotin, 1.1 mg of folic acid, 110 mg of p-aminobenzoic acid, 110 mg of riboflavin, 220 mg of pyridoxine HCl, 220 mg of thiamine HCl, 220 mg of niacinamide]) with 0.01% carbenicillin. Cells were then transferred to 1 L of M9 minimal media with carbenicillin. One-liter growths were incubated in a shaker at 37 °C until reaching an OD600 of between 0.7 and 1; then, they were induced and harvested as above.
Protein Purification.
Purification was performed as previously described (54). Cell pellets were resuspended in 20 mL of wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 3.1 mM NaN3, pH 8) with an added 1 mg of Pefabloc and 125 μL of DNase (deoxyribonuclease) solution (20 mg of DNase I in 10 mL of 20% glycerol, 75 mM NaCl) and lysed with a French pressure cell press (Glen Mills Inc.) at 1,000 psi. Cell debris was removed by centrifugation at 17,000 rpm for 30 min, and the lysate was poured through an nickel-nitrilotriacetic acid (Ni-NTA) affinity column (QIAGEN Sciences). The column was rinsed with 50 mL of wash buffer and eluted with 25 mL of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 3.1 mM NaN3, pH 8.0). The affinity tag was cut using TEV protease and removed via a second Ni-NTA column. Final purification was carried out by HPLC (high performance liquid chromotagraphy) using a 250- × 20-mm Higgins Analytical PROTO 300 C18 10 μM column with a gradient of 20 to 80% acetonitrile with 0.1% TFA over 60 min.
Protein Labeling.
The minielastin construct was fluorescently labeled using the Invitrogen Alexa Fluor 488 Microscale Protein Labeling Kit. One hundred microliters of protein in sodium phosphate buffer, pH 7.5, was mixed with 10 μL of sodium bicarbonate, pH 8.3. The solution was then mixed with 11.3 nmol/μL Alexa Fluor 488 TFP (tetrafluorophenyl) ester and incubated for 15 min at room temperature. Excess dye was then removed using a Bio-Gel P-6 fine resin column. The degree of labeling was determined by measuring the sample’s 494-nm absorbance.
Protein Resuspension and Coacervation.
All protein preparation was done at 4 °C unless otherwise specified. Lyophilized protein was resuspended the day of the experiment in buffer composed of 1.5 M NaCl, 1 mM CaCl2, and 50 mM Tris, pH 7.4. The resuspended protein solution was filtered (0.22-μm filter) to remove any potential aggregates and concentrated using a 10-kDa Amicon Ultracentrifuge Filter Unit until the desired concentration was obtained. Care was taken to avoid exposure of concentrated protein solution to room temperature to prevent phase separation and loss of protein from occurring. The microscope cover glass for imaging was washed using a 70% ethanol solution and dried thoroughly, followed by treatment with 1% Pluronics F127 for 25 to 30 min and washing five to six times with deionized water prior to protein incubation. Coacervation was induced by diluting protein in buffer to a final concentration of 130 μM at room temperature and immediately placing it under the microfluidic temperature stage (CherryTemp; CherryBiotech) at 23 °C. A final volume of 1% labeled protein was mixed with unlabeled protein for fluorescent imaging.
DIC and Confocal Microscopy Imaging.
DIC images were acquired on a wide-field Axio Observer 7 Inverted Microscope (Zeiss) with a ×63/1.4–numerical aperture (NA) Plan-Apochromat (oil immersion) objective. Single-slice confocal images were acquired using a Marianas Spinning Disk confocal microscope (Intelligent Imaging Innovations) consisting of a spinning disk confocal head (CSU-X1; Yokagawa) on a Zeiss Axio Observer inverted microscope equipped with ×63/1.4-NA Plan-Apochromat (oil immersion). Images were acquired with a Prime sCMOS camera (Photometrics) controlled by SlideBook 6 (Intelligent Imaging Innovations). Temperature was stabilized to 23 °C using the microfluidic temperature stage (CherryTemp; CherryBiotech). For fluorescent samples, protein labeled with Alexa-488 was excited with a 488-nm line from a solid-state laser (LaserStack) and collected with a 440-/521-/607-/700-nm quad emission dichroic and 525-/30-nm emission filter.
Laser Scanning Microscopy.
A Zeiss LSM800 microscope (Zeiss) was used for time lapse and z stack. Confocal z-stack images (eight bit, 512 × 512 pixels) were obtained using ×63 (NA = 1.4; oil immersion). Multichannel confocal z stacks were set to detect the following fluorophores with distinct excitation/emission properties: Alexa Fluor 488 (500/520 nm) and red fluorescent microspheres (580/605). Confocal z stacks were imported into Imaris (Oxford instruments) for visualization, three-dimensional rendering, and analysis. z stacks were captured with a 0.5-μm step size. Time-lapse videos were captured with an interval of 10 and 30 s.
NMR Spectroscopy.
NMR experiments were done as previously described (29). Briefly, protein was lyophilized and diluted to 200 μM in buffer containing 45 mM phosphate buffer, pH 6.0. NMR spectra were obtained using a 700-MHz Varian or a 800-MHz Bruker NMR spectrometer. Spectra were processed and visualized with NMRPipe (55), NMRFAM-Sparky (56), and TopSpin software. Sample integrity was monitored using 15N HSQC (heteronuclear single-quantum correlation spectroscopy).
Coacervates Fusion Experiments.
Fusion events of coacervates were collected at a rate of 65 ms per frame using a wide-field Axio Observer 7 Inverted Microscope (Zeiss) with a ×63/1.4-NA Plan-Apochromat (oil immersion) objective. Cover glass bottoms were treated with 1% Pluronics F127 for 25 to 30 min and washed with deionized water prior to protein incubation. Images were acquired with an Axiocam 506 mono camera (Zeiss) controlled by the Zen software (Zeiss). ImageJ was used for further format and images processing, and MATLAB was used to analyze fusion events as previously described (57).
FRAP.
FRAP images were taken using a spinning disk confocal microscope as with confocal imaging. Images were collected every 200 ms using 50-ms exposure time. Protein labeled with Alexa-488 was excited with the 488-nm line from a solid-state laser (LaserStack) and collected with a 440-/521-/607-/700-nm quad emission dichroic and 525-/30-nm emission filter. Circles with diameters of 1 and 3 µm were bleached with a laser of 488 nm. Fluorescence recovery was monitored for 800 time points in all experiments. The final concentration of minielastin was 130 µM with 1% protein fluorescently labeled with Invitrogen Alexa Fluor 488. Recovery curves were corrected for photobleaching, normalized, and fitted to an exponential function of the form
ImageJ was used for further processing. Data were normalized and fitted to a double-fit exponential using MATLAB to obtain the mobile fraction (I0) and Thalf [ln(2)/β] of recovery at increasing incubation times (58).
Microrheology.
Microrheology was performed using 100 to 200-nm fluorescent carboxylate-modified microspheres (Invitrogen). Bead diffusion was tracked on a Marianas Spinning Disk confocal microscope with a ×63/1.4-NA Plan-Apochromat oil immersion objective for 1,000 frames with 250-ms time intervals and exposure of 50 ms. Temperature was kept at 23 °C using a microfluidic temperature stage (CherryTemp; CherryBiotech). Analysis was done in MATLAB (equation) to obtain MSD of ELP droplets.
Particle-tracking code to locate and track bead trajectories in two dimensions (xy) was adapted from MATLAB Multiple Particle Tracking Code from The MATLAB Particle Tracking Code Repository (https://doi.org/10.1038/s41467-020-18224-y and https://doi.org/10.1073/pnas.1504822112). Custom MATLAB software was then used to analyze bead dynamics. MSD was calculated from time and ensemble averages for all trajectories:
The dependence of the MSD on lag time (τ) follows a power law; the exponent was determined as the slope of a log–log plot and diffusion coefficient as the y intercept:
where d is the number of dimensions, D is the diffusion coefficient, and is the exponent. Viscosity can be determined from the Stokes–Einstein relation, assuming a system at equilibrium and a freely diffusing Brownian particle within a solution of viscosity η. The final viscosity is the average of three values collected from individual measurements performed on 3 different days at 23 °C ± 2 °C.
Scanning Electron Microscopy Preparation and Imaging.
Protein was concentrated to ∼230 μM and incubated overnight at 37 °C. The sample was washed with 50 μL of deionized water two to three times by spinning in a tabletop microcentrifuge at 9,000 rpm and removing excess water in between washes. After a final wash, 1 µL of the sample was spotted and left to dry in a silicon wafer. The wafer containing the sample was mounted on pin stubs with double-sided copper tape and sputter coated using a Leica EM ACE600 Coater with a 5-nm-thick gold layer before imaging. Images were obtained with a Thermo Scientific (FEI) Helios NanoLab 660 FIB-SEM with HT of 5 kV, current of 25 pA, and an ETD detector.
Coefficient of Variation.
MSDs of individual bead trajectories at increasing incubation times were compared using the coefficient of variation (Cv):
where σ is the SD of the tracks and μ is the average MSD of tracks.
Supplementary Material
Acknowledgments
We acknowledge the Live Imaging and Bioenergetics Facility of the City University of New York Advanced Science Research Center for instrument use and scientific and technical assistance.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202240119/-/DCSupplemental.
Data Availability
MATLAB code files have been deposited in Zenodo (https://zenodo.org/record/6818910#.YsxnrnbMI2x) (59).
References
- 1.Yeo G. C., Keeley F. W., Weiss A. S., Coacervation of tropoelastin. Adv. Colloid Interface Sci. 167, 94–103 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Schmelzer C. E. H., Duca L., Elastic fibers: Formation, function, and fate during aging and disease. FEBS J. 289, 3704–3730 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Czirok A., et al. , Elastic fiber macro-assembly is a hierarchical, cell motion-mediated process. J. Cell. Physiol. 207, 97–106 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Tu Y., Wise S. G., Weiss A. S., Stages in tropoelastin coalescence during synthetic elastin hydrogel formation. Micron 41, 268–272 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Mithieux S. M., Weiss A. S., Elastin. Adv. Protein Chem. 70, 437–461 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Schmelzer C. E. H., Hedtke T., Heinz A., Unique molecular networks: Formation and role of elastin cross-links. IUBMB Life 72, 842–854 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Urry D. W., Entropic elastic processes in protein mechanisms. I. Elastic structure due to an inverse temperature transition and elasticity due to internal chain dynamics. J. Protein Chem. 7, 1–34 (1988). [DOI] [PubMed] [Google Scholar]
- 8.Urry D. W., Long M. M., On the conformation, coacervation and function of polymeric models of elastin. Adv. Exp. Med. Biol. 79, 685–714 (1977). [DOI] [PubMed] [Google Scholar]
- 9.Roberts S., Dzuricky M., Chilkoti A., Elastin-like polypeptides as models of intrinsically disordered proteins. FEBS Lett. 589 (19 pt A), 2477–2486 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keeley F. W., Bellingham C. M., Woodhouse K. A., Elastin as a self-organizing biomaterial: Use of recombinantly expressed human elastin polypeptides as a model for investigations of structure and self-assembly of elastin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 185–189 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miao M., et al. , Sequence and structure determinants for the self-aggregation of recombinant polypeptides modeled after human elastin. J. Biol. Chem. 278, 48553–48562 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Bellingham C. M., Woodhouse K. A., Robson P., Rothstein S. J., Keeley F. W., Self-aggregation characteristics of recombinantly expressed human elastin polypeptides. Biochim. Biophys. Acta 1550, 6–19 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Bellingham C. M., Keeley F. W., Self-ordered polymerization of elastin-based biomaterials. Curr. Opin. Solid State Mater. Sci. 8, 135–139 (2004). [Google Scholar]
- 14.Bellingham C. M., et al. , Recombinant human elastin polypeptides self-assemble into biomaterials with elastin-like properties. Biopolymers 70, 445–455 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Cirulis J. T., Keeley F. W., James D. F., Viscoelastic properties and gelation of an elastin-like polypeptide. J. Rheol. (N.Y.N.Y.) 53, 1215–1228 (2009). [Google Scholar]
- 16.Bressan G. M., et al. , Relevance of aggregation properties of tropoelastin to the assembly and structure of elastic fibers. J. Ultrastruct. Mol. Struct. Res. 94, 209–216 (1986). [DOI] [PubMed] [Google Scholar]
- 17.Gotte L., Giro M. G., Volpin D., Horne R. W., The ultrastructural organization of elastin. J. Ultrastruct. Res. 46, 23–33 (1974). [DOI] [PubMed] [Google Scholar]
- 18.Gosline J. M., The physical properties of elastic tissue. Int. Rev. Connect. Tissue Res. 7, 211–249 (1976). [DOI] [PubMed] [Google Scholar]
- 19.Fung Y.-C., Biomechanics. Mechanical Properties of Living Tissues (Springer-Verlag, New York, NY, 1993). [Google Scholar]
- 20.Tu Y., Weiss A. S., Glycosaminoglycan-mediated coacervation of tropoelastin abolishes the critical concentration, accelerates coacervate formation, and facilitates spherule fusion: Implications for tropoelastin microassembly. Biomacromolecules 9, 1739–1744 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Clarke A. W., et al. , Tropoelastin massively associates during coacervation to form quantized protein spheres. Biochemistry 45, 9989–9996 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Banani S. F., Lee H. O., Hyman A. A., Rosen M. K., Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin Y., Brangwynne C. P., Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Elbaum-Garfinkle S., Matter over mind: Liquid phase separation and neurodegeneration. J. Biol. Chem. 294, 7160–7168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alberti S., Gladfelter A., Mittag T., Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel A., et al. , A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Peskett T. R., Rau F., O'Driscoll J., Patani R., Lowe A. R., Saibil H. R., A Liquid to Solid Phase Transition Underlying Pathological Huntingtin Exon1 Aggregation. Mol. Cell. 70, 588–601.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenland K. N., et al. , Order, disorder, and temperature-driven compaction in a designed elastin protein. J. Phys. Chem. B 122, 2725–2736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carvajal M. F. C. A., et al. , Dynamics in natural and designed elastins and their relation to elastic fiber structure and recoil. Biophys. J. 120, 4623–4634 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nuhn H., Klok H. A., Secondary structure formation and LCST behavior of short elastin-like peptides. Biomacromolecules 9, 2755–2763 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Muiznieks L. D., Sharpe S., Pomès R., Keeley F. W., Role of liquid-liquid phase separation in assembly of elastin and other extracellular matrix proteins. J. Mol. Biol. 430, 4741–4753 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Selvaggi L., et al. , Multiple-particle-tracking to investigate viscoelastic properties in living cells. Methods 51, 20–26 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Schultz K. M., Furst E. M., Microrheology of biomaterial hydrogelators. Soft Matter 8, 6198–6205 (2012). [Google Scholar]
- 34.Muiznieks L. D., et al. , Modulated growth, stability and interactions of liquid-like coacervate assemblies of elastin. Matrix Biol. 36, 39–50 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Elbaum-Garfinkle S., et al. , The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U.S.A. 112, 7189–7194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jawerth L., et al. , Protein condensates as aging Maxwell fluids. Science 370, 1317–1323 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Zhang H., et al. , RNA controls PolyQ protein phase transitions. Mol. Cell 60, 220–230 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feric M., et al. , Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cirulis J. T., Keeley F. W., Kinetics and morphology of self-assembly of an elastin-like polypeptide based on the alternating domain arrangement of human tropoelastin. Biochemistry 49, 5726–5733 (2010). [DOI] [PubMed] [Google Scholar]
- 40.Cirulis J. T., et al. , Fibrillins, fibulins, and matrix-associated glycoprotein modulate the kinetics and morphology of in vitro self-assembly of a recombinant elastin-like polypeptide. Biochemistry 47, 12601–12613 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato F., et al. , Distinct steps of cross-linking, self-association, and maturation of tropoelastin are necessary for elastic fiber formation. J. Mol. Biol. 369, 841–851 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Le D. H., et al. , Self-assembly of elastin-mimetic double hydrophobic polypeptides. Biomacromolecules 14, 1028–1034 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Molliex A., et al. , Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin Y., et al. , Liquid-liquid phase separation of tau driven by hydrophobic interaction facilitates fibrillization of tau. J. Mol. Biol. 433, 166731 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reichheld S. E., Muiznieks L. D., Keeley F. W., Sharpe S., Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc. Natl. Acad. Sci. U.S.A. 114, E4408–E4415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aurora R., Rose G. D., Helix capping. Protein Sci. 7, 21–38 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muiznieks L. D., Keeley F. W., Proline periodicity modulates the self-assembly properties of elastin-like polypeptides. J. Biol. Chem. 285, 39779–39789 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumashiro K. K., Ho J. P., Niemczura W. P., Keeley F. W., Cooperativity between the hydrophobic and cross-linking domains of elastin. J. Biol. Chem. 281, 23757–23765 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Roberts S., et al. , Injectable tissue integrating networks from recombinant polypeptides with tunable order. Nat. Mater. 17, 1154–1163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rauscher S., Pomès R., The liquid structure of elastin. eLife 6, e26526 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samouillan V., André C., Dandurand J., Lacabanne C., Effect of water on the molecular mobility of elastin. Biomacromolecules 5, 958–964 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Zhang L., Andersen E. M., Khajo A., Magliozzo R. S., Koder R. L., Dynamic factors affecting gaseous ligand binding in an artificial oxygen transport protein. Biochemistry 52, 447–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neidhardt F. C., Bloch P. L., Smith D. F., Culture medium for enterobacteria. J. Bacteriol. 119, 736–747 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L., et al. , Manipulating cofactor binding thermodynamics in an artificial oxygen transport protein. Biochemistry 50, 10254–10261 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Delaglio F., et al. , NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 56.Lee W., Tonelli M., Markley J. L., NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ceballos A. V., McDonald C. J., Elbaum-Garfinkle S., Methods and strategies to quantify phase separation of disordered proteins. Methods Enzymol. 611, 31–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rapsomaniki M. A., et al. , easyFRAP: An interactive, easy-to-use tool for qualitative and quantitative analysis of FRAP data. Bioinformatics 28, 1800–1801 (2012). [DOI] [PubMed] [Google Scholar]
- 59.A. V. Ceballos, S. Elbaum-Garfinkle, MatLab code used in Vidal Ceballos et al. PNAS 2022. GitHub. https://zenodo.org/record/6818910#.YsxnrnbMI2x. Deposited 11 July 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MATLAB code files have been deposited in Zenodo (https://zenodo.org/record/6818910#.YsxnrnbMI2x) (59).