Abstract
We have studied the uptake of maltose in the thermoacidophilic gram-positive bacterium Alicyclobacillus acidocaldarius, which grows best at 57°C and pH 3.5. Under these conditions, accumulation of [14C]maltose was observed in cells grown with maltose but not in those grown with glucose. At lower temperatures or higher pH values, the transport rates substantially decreased. Uptake of radiolabeled maltose was inhibited by maltotetraose, acarbose, and cyclodextrins but not by lactose, sucrose, or trehalose. The kinetic parameters (Km of 0.91 ± 0.06 μM and Vmax ranging from 0.6 to 3.7 nmol/min/mg of protein) are consistent with a binding protein-dependent ATP binding cassette (ABC) transporter. A corresponding binding protein (MalE) that interacts with maltose with high affinity (Kd of 1.5 μM) was purified from the culture supernatant of maltose-grown cells. Immunoelectron microscopy revealed distribution of the protein throughout the cell wall. The malE gene was cloned and sequenced. Five additional open reading frames, encoding components of a maltose transport system (MalF and MalG), a putative transcriptional regulator (MalR), a cyclodextrinase (CdaA), and an α-glucosidase (GlcA), were identified downstream of malE. The malE gene lacking the DNA sequence that encodes the signal sequence was expressed in Escherichia coli. The purified wild-type and recombinant proteins bind maltose with high affinity over a wide pH range (2.5 to 7) and up to 80°C. Recombinant MalE cross-reacted with an antiserum raised against the wild-type protein, thereby indicating that the latter is the product of the malE gene. The MalE protein might be well suited as a model to study tolerance of proteins to low pH.
The thermoacidophilic gram-positive bacterium Alicyclobacillus acidocaldarius was first isolated by Darland and Brock from an acidic creek in Yellowstone National Park (7). The organism grows best at pH 3.6 and 57°C and is further characterized by the presence of ω-alicyclic fatty acids in the cytoplasmic membrane (60). A. acidocaldarius can utilize a variety of organic compounds as sole sources of carbon and energy, including sugars and polysaccharides, such as starch and xylan (32, 49; U. Eckert, S. Wilken, E. Bakker, and E. Schneider, unpublished data). Since polysaccharides cannot penetrate the cell membrane, the bacteria excrete specific hydrolases that degrade the macromolecules into soluble oligomers and monomers that serve as substrates for the transport proteins. Thus, exoenzymes and other extracellular proteins of A. acidocaldarius that are exposed to the acidic environment are ideally suited as model systems to study the mechanism of tolerance of proteins to low pH (“acidostability”) on the molecular level. In particular, the comparative analysis of functionally homologous proteins from acidophilic and neutrophilic organisms on the levels of primary and, most desirably, tertiary structures, would provide hints on how acidostability is achieved. Such a study was recently performed with an amylopullulanase from A. acidocaldarius, the product of the amyA gene, and a few other proteins (35, 49). From their data, Bakker and coworkers concluded that in acidostable proteins the number of charged residues, especially in surface-exposed regions, is markedly reduced compared to that in their neutrophilic relatives (49). Whether this notion holds for acidostable proteins in general needs to be established. However, such analyses are hampered by the rather limited number of candidate proteins, which is mainly due to the fact that even acidophiles maintain a pH value in their cytoplasm close to neutrality (2). Thus, unlike in studies that are concerned with other extremophilic properties, such as thermophilicity or halophilicity, cytoplasmic enzymes are not suited for analysis of acidostability.
In an attempt to identify other extracellularly exposed proteins from A. acidocaldarius, we recently purified a maltose binding protein from the surface of maltose-grown cells that, by metabolic labeling with [14C]palmitic acid, was identified as a lipoprotein (24). The sequence of the N-terminal 20 amino acids of the purified protein was found to be almost identical to that of a peptide fragment derived from an incomplete open reading frame (ORF2) downstream of the amyA gene (32). The ORF2 product displays homology to the maltose binding protein (MalE) of Escherichia coli (32, 54). Interestingly, when compared to the translated nucleotide sequence, the purified protein lacked 23 amino acids from the amino terminus (24), most likely due to the action of an extracellular protease (49). Together, these data supported a role of the protein isolated from A. acidocaldarius as a solute binding protein component of an ATP binding cassette (ABC) transport system for maltose and maltodextrins (3). The family of ABC transporters comprises a diverse class of transport proteins that couple the energy of ATP hydrolysis to the translocation of solutes across biological membranes (27). Typically, an ABC transporter is composed of two membrane integral protein domains and two ATP-hydrolyzing domains (47). Those ABC transport systems that mediate the uptake of nutrients in bacteria and archaea are equipped with an additional component, an extracellular solute binding protein, that, in its substrate-loaded (closed) conformation initiates the transport process (3). In gram-negative bacteria, binding proteins are located in the periplasm, while in gram-positives bacteria, which lack an outer membrane, they are anchored to the cytoplasmic membrane via fatty acids that are covalently bound to the N-terminal cysteine residue (53). In the prototype maltose transporter, as is found in E. coli and Salmonella, MalE represents the maltose binding protein, while the membrane-associated transport complex is composed of one copy each of MalF and MalG and of two copies of the ATP-hydrolyzing protein, MalK (4).
Here we report on the properties of the maltose transport system of A. acidocaldarius in vivo and on the complete cloning and sequencing of six genes downstream of amyA, which encode transport components, including maltose binding protein, a transcriptional regulator, and two starch-degrading enzymes. Furthermore, the native and recombinant forms of the maltose binding protein were biochemically characterized with respect to acidostability.
MATERIALS AND METHODS
Chemicals.
[14C]maltose (13.3 GBq/mmol) was purchased from ICN (Eschwege, Germany). α-Cyclodextrin (cyclohexaamylose), β-cyclodextrin (cycloheptaamylose), and γ-cyclodextrin (cyclo-octaamylose) were purchased from Sigma (Deisenhofen, Germany). Acarbose was a generous gift of Bayer AG (Wuppertal, Germany).
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| A. acidocaldarius ATCC 27009 | Wild type | DSMZa |
| E. coli | ||
| ED169 | F− ΔlacU169 araD139 rpsL relA thi flbB ΔmalB107 | 37 |
| PD28 | pop3325 ΔmalE444 Δ(srlR-recA)306::Tn10 | 9 |
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(araA-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Plasmids | ||
| pBAD24 | Arabinose promoter, Apr | 17 |
| pBAD/HisA | Arabinose promoter, histidine fusion vector, Apr | Invitrogen |
| pQE9 | T5 promoter, histidine fusion vector, Apr | Quiagen |
| pSE380 | trc promoter, Apr | Stratagene |
| pSU19 | General cloning vector, P15A ori, Cmr | 33 |
| pUC18 | General cloning vector, Apr | Roche |
| pAH8-1 | ′malE as SstI-BamHI fragment on pUC18 | This study |
| pAH9 | ′malE malF malG malR′ as SstI fragment on pUC18 | This study |
| pAH10 | ′malF malG malR cdaA′ as KpnI fragment on pUC18 | This study |
| pAH19 | ′glcA pleD′ as EcoRV fragment on pUC18 | This study |
| pAH30 | ′amyA malE′ as BamHI-SstI fragment on pUC18 | This study |
| pJM30 | ′malR cdaA glcA′ as HindIII fragment on pUC18 | 34 |
| pAH18 | malEAa (Cys-1→Met) on pBAD24 | This study |
| pAH26-2 | malGAa on pBAD/HisA | This work |
| pAH27-3 | malF malGAa on pBAD/HisA | This work |
| pFSA12 | malF malGAa on pQE9 | This work |
| pFSA16 | malEEc (codons 1–26)′-′malEAa (codons 26–427) on pSE380 | This work |
| pFSA24 | pT5malF malGAa on pSU19 | This work |
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen.
Media and growth conditions.
A. acidocaldarius strain ATCC 27009 was grown at 57°C in minimal medium (pH 3.6) under vigorous agitation, with maltose or glucose (each at 10 mM) as the sole source of carbon and energy (49). E. coli cells were usually grown in Luria-Bertani (LB) medium (36). If required, ampicillin or chloramphenicol was added at 75 and 20 μg/ml, respectively. For overproduction of A. acidocaldarius MalE (MalEAa) strain ED169(pAH18) was grown in LB-ampicillin to an optical density at 650 nm (OD650) of 1 and supplemented with 0.08% arabinose to induce malE expression, and growth was continued for 6 h.
Gene cloning.
The DNA fragments cloned in this study were obtained by primer walking. From the nucleotide sequence downstream of the amyA gene (ORF2) that was known in the initial phase of this study (32), an oligonucleotide probe was designed and labeled with digoxigenin (MWG, Ebersberg, Germany). Genomic DNA of A. acidocaldarius was digested with BamHI and SstI, and a fragment (1.15 kb) that hybridized with the probe was identified by Southern blotting. Subsequently, genomic DNA was cleaved with BamHI and SstI on a preparative scale and subjected to agarose gel electrophoresis. Fragments of 1.0 to 1.5 kb were eluted by using the JETsorb gel extraction kit (Genomed, Bad Oeyenhausen, Germany) and ligated to BamHI- and SstI-digested and dephosphorylated pUC18. Cells of E. coli strain XL1blue were transformed with the ligation mixture, and ampicillin-resistant transformants were screened for the presence of the malE gene by colony hybridization with the probe. From one positive clone, designated pAH8-1, the nucleotide sequence of the inserted DNA fragment was determined and used to design a new oligonucleotide probe for further cloning. By this approach, four additional subgenomic DNA libraries consisting of SstI, KpnI, EcoRV, or BamHI/SstI fragments were generated that eventually yielded plasmids pAH9 (3.97-kb fragment), pAH10 (3.5-kb fragment), pAH19 (ca. 4-kb fragment), and pAH30 (1.23-kb fragment), respectively. Plasmid pJM30, derived from a HindIII digestion of chromosomal DNA, was constructed by J. Matzke (34).
Construction of plasmids.
For heterologous expression in E. coli, the malEAa gene lacking the signal-peptide-encoding fragment was amplified from genomic DNA by PCR, using VENT polymerase (NEB, Schwalbach, Germany). The 3′ oligonucleotide primer contained the recognition sequence for XbaI. The amplified DNA fragment (1,209 bp) was digested with XbaI and ligated with plasmid pBAD24 that was previously linearized with NcoI, filled in with Klenow, and subsequently digested with XbaI. As a consequence, codon 1 was changed from TGT to ATG, thereby replacing the N-terminal cysteine residue of the translated mature protein with methionine. The resulting plasmid was designated pAH18.
For complementation experiments, the signal-peptide-encoding fragment of the malEAa gene was replaced by the corresponding DNA fragment of the E. coli malE gene by a three-step PCR as described previously (26). The resulting amplification product was ligated into plasmid vector pSE380, yielding plasmid pFSA16. The primers used to amplify malEAa were designed in such a way that in the translated mature polypeptide alanine substituted for the N-terminal cysteine residue.
To clone the malF and malG genes, a fragment containing either malG or malF malG was amplified from genomic DNA by PCR using primers that introduced XhoI sites at the 5′ and 3′ ends. Amplified DNA fragments were purified, digested with XhoI, and ligated with pBAD/HisA that had been previously linearized with the same enzyme and dephosphorylated. The correct orientations of the inserted fragments in the resulting plasmids pAH26-2 (malG) and pAH27-3 (malF malG) were verified by restriction analyses using appropriate endonucleases.
For complementation analysis, a restriction fragment carrying the malF malG genes under the control of the T5 promoter was obtained by digestion of the pQE9-based plasmid pFSA12 with XhoI and SalI and ligated into pSU19. The resulting plasmid was designated pFSA24.
Standard DNA methods.
Genomic DNA from A. acidocaldarius was isolated as described previously (21). Plasmids were prepared from E. coli using a mini- or midiplasmid kit (Qiagen GmbH, Hilden, Germany). Digestion by endonucleases, ligation reactions, and PCR were performed by standard procedures (44).
Computer-aided sequence analyses.
Nucleotide sequences were analyzed by ConSequence (GATC, Konstanz, Germany) and by DNASIS (Amersham-Pharmacia, Freiburg, Germany). Protein homology searches were done with BLAST (15). Multiple alignments were performed using CLUSTAL X or W (25). SIGNAL P and TMHMM (both available at http://www.cbs.dtu.dk/services/) were used for the prediction of signal sequences and membrane-spanning peptide fragments, respectively.
Preparation of antibodies and Western blot analysis.
Antiserum against MalEAa was obtained by immunization of a rabbit with purified protein (Biogenes, Berlin, Germany). Western blot analysis was performed as described previously (57), using a 1:30,000 antiserum dilution. For immunoelectron microscopy, immunoglobulin Gs were purified from antiserum by affinity chromatography with protein A–Sepharose CL-4B (Pharmacia, Braunschweig, Germany), according to a published procedure (20).
The recombinant proteins His6-MalF and His6-MalG were detected in Western blot analyses by primary α-PentaHis antibody (Qiagen, Hilden, Germany).
Immunoelectron microscopy.
A. acidocaldarius was grown in minimal salt medium containing maltose to an OD650 1. Bacteria were pelleted and fixed by resuspension in 0.8% formaldehyde and 0.2% glutaraldehyde in phosphate-buffered saline (pH 7.4) for 1 h on ice. Fixed cells were washed three times in phosphate-buffered saline (pH 7.4) and embedded in soft agar (2%). Subsequently, cells were dehydrated in a graded series of alcohol solutions (10 to 100% [vol/vol] in H2O). During incubation in 50% ethanol, the cells were stained with uranyl acetate (2%). Bacterial pellets were embedded in LR gold resin (Plano) by light polymerization. Thin sections were prepared with a Reichert Ultracut E ultramicrotome, transferred to copper grids, and washed once with 20 mM Tris-HCl (pH 7.5) containing 0.8% NaCl. Nonspecific binding sites were blocked by incubation with 0.6% bovine serum albumin. The grids were then incubated for 2 h at room temperature with either polyclonal rabbit antibodies to MalEAa or antibodies unspecific to A. acidocaldarius protein, at a concentration of 0.76 mg/ml. Subsequently, the grids were quickly washed four times in buffer containing bovine serum albumin, followed by an incubation with 10-nm-diameter-gold-conjugated protein A/G (British-Bio Cell) (1/10 dilution) for 2 h at room temperature. After an additional washing step, the grids were poststained in 2% (vol/vol) uranyl acetate in H2O and examined in a Philips CM transmission electron microscope at 100 kV.
Transport assays.
A. acidocaldarius cells were grown in minimal salt medium (pH 3.6) with 10 mM maltose or glucose and harvested in the exponential phase (OD650 = 0.6 to 1). Cells were washed twice in minimal salt medium, resuspended in the same medium supplemented with 100 μM chloramphenicol to an OD650 of 1.2, and stored on ice until use. Aliquots (100 μl) were diluted with 890 μl of minimal salts and preheated for 1 min to 57°C (or other temperatures, as indicated) under vigorous shaking. The assay was started by addition of 10 μl of [14C]maltose (3 μM final concentration). Aliquots of 180 μl were taken at the indicated times, rapidly filtered through nitrocellulose filters (OE 67, 0.45-μm-pore-size, Schleicher & Schüll), and washed with 5 ml of minimal salt medium. Subsequently, the retained radioactivity was determined in a liquid scintillation counter. In competition experiments, unlabeled carbohydrates (1 mM final concentration) were incubated for 2 min with the cells prior to the addition of labeled maltose. To determine maltose uptake at different pH values, the minimal salt medium was adjusted with 0.5 N NaOH to the desired pH.
Binding assays.
Binding of [14C]maltose to purified MalEAa was performed by the method of Richarme and Kepes (41). Standard assay mixtures (100 μl) containing 10 mM sodium acetate buffer (pH 3.6), 9.8 mM (NH4)2SO4, and purified binding protein (5 μg) were preheated at 57°C for 1 min prior to the addition of [14C]maltose (5 μM final concentration). After 1 min, the reaction was terminated by adding 2 ml of an ice-cold saturated (NH4)2SO4 solution, and the mixture was immediately filtered through a nitrocellulose filter (0.45-μm pore size). The filter was washed once with the same solution, followed by distilled water, and retained radioactivity was determined in a liquid scintillation counter.
In competition experiments, unlabeled sugars (1 mM) were added 1 min prior to the addition of [14C]maltose. Binding experiments at various pH values were performed in citrate-phosphate buffer.
To determine the binding constant, MalE was first subjected to a denaturation-renaturation procedure using guanidinium hydrochloride (6 M) to remove excess bound unlabeled maltose (18). In all other experiments, the protein was extensively dialyzed against assay buffer prior to use.
Miscellaneous methods.
Wild-type and recombinant MalEAa proteins were purified by affinity chromatography with agarose-coupled amylose as described previously (12). MalE from S. enterica serovar Typhimurium was isolated as described previously (29). Sodium dodecyl sulfate (SDS) gel electrophoresis and protein determination were performed as described previously (57).
Nucleotide sequence accession number.
The sequences reported in this paper have been deposited in the EMBL database and assigned accession number AJ252161.
RESULTS
Maltose uptake in A. acidocaldarius is mediated by a high-affinity transport system.
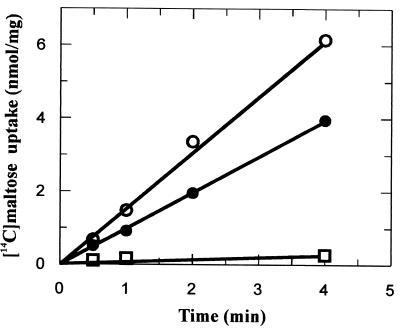
When grown with maltose as the sole source of carbon and energy, but not with glucose, cells of A. acidocaldarius produce an extracellular lipoprotein, designated MalEAa, that, when released from the cell wall by Triton X-100, binds to immobilized amylose (24). This observation was taken as evidence for a maltose and maltodextrin binding activity of the protein and thus indicated the existence of a binding-protein-dependent ABC type of transport system for these sugars in A. acidocaldarius. To obtain further proof for this notion, we studied the uptake of [14C]maltose in intact cells under growth conditions, that is, at 57°C and pH 3.6. In a representative experiment, maltose-grown cells accumulated the radiolabeled substrate at a linear rate of 1.6 nmol/min/mg of protein. In contrast, only little transport activity was observed with cells that were grown in minimal medium containing glucose (0.07 nmol/min/mg). However, when the cells were cultivated in a combination of maltose and glucose (10 mM each), a substantial transport rate (1 nmol/min/mg, corresponding to 62.5% of that of maltose-grown cells) was observed (Fig. 1). These results suggest that the mechanism of glucose repression in A. acidocaldarius is different from those operating in E. coli and Bacillus subtilis (52).
FIG. 1.
Maltose uptake by A. acidocaldarius cells. Cells were grown in minimal medium supplemented with maltose (10 mM) (○), glucose (10 mM) (□), or maltose and glucose (10 mM each) (●) as a carbon source. Uptake assays were performed at 57°C, pH 3.6, and 3 μM [14C]maltose.
Next, we investigated the temperature and pH dependencies of the observed transport activity. At 57°C and pH 5.5, the transport rate dropped to 44% relative to that of control cells measured at pH 3, while at pH 6, the transport rate corresponded to only 11%. A similar low rate of maltose uptake (7.7%) was observed at pH 3.6 and 37°C. Thus, both high temperature and low pH are required for optimal maltose uptake.
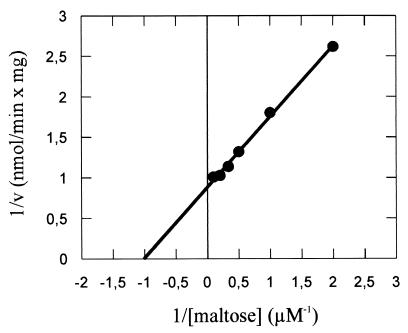
The apparent Kms for maltose uptake were found to be 0.86, 0.89, and 0.99 μM in three independent experiments at pH 3.6 and 57°C, with Vmax values ranging from 0.6 to 3.7 nmol/min/mg of protein (Fig. 2). The reason for less reproducible Vmax values is not known, but varying oxygen supply at the assay temperature, and thus changes in the energy status of the cells, seems to be a likely explanation.
FIG. 2.
Determination of apparent Km and Vmax values of maltose transport in A. acidocaldarius cells (Lineweaver-Burk plot). Cells were grown in minimal medium in the presence of 10 mM maltose and assayed at 57°C and pH 3.6 as described in Materials and Methods. Data are from a single experiment. The Km is 1 μM, and the Vmax is 1.25 nmol/min/mg of protein.
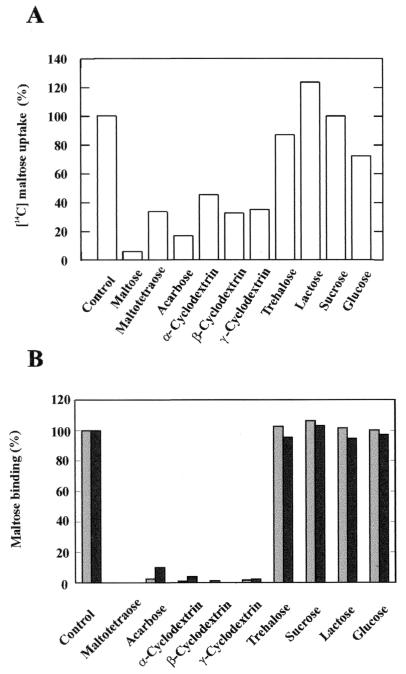
To study the specificity of the transport system, we monitored the linear rate of uptake of radiolabeled maltose (3 μM) in the presence of various sugars. As shown in Fig. 3A, transport was substantially inhibited by linear maltodextrins, such as maltotetraose and acarbose, and by α-, β-, and γ-cyclodextrins, while glucose and trehalose caused only minor inhibition (27 and 13%, respectively). In contrast, sucrose and lactose were ineffective. The strongest inhibition was achieved by acarbose, a pseudomaltotetraose consisting of an unsaturated aminocyclitol moiety, a deoxyhexose, and a maltose, which is also a substrate of the E. coli maltose transporter (5).
FIG. 3.
Maltose uptake by A. acidocaldarius cells (A) and maltose binding activity of MalEAa (B) in the presence of competing substrates. (A) Uptake was performed at 57°C, pH 3.6, and 3 μM [14C]maltose in the presence of the indicated sugars at a concentration of 1 mM each. Cells were preincubated for 2 min with nonlabeled sugars in minimal medium without a carbon source, and uptake was stopped after 1 min. (B) Binding assays were performed at 57°C, pH 3.6, and 5 μM [14C]maltose in the presence of the indicated sugars at a concentration of 1 mM each. MalEAa (5 μg) was preincubated for 1 min with nonlabeled sugars in buffer (pH 3.6), and the binding reaction was stopped after 1 min. Shaded bars, wild type MalEAa; black bars, recombinant MalEAa.
Together, these results are consistent with a binding-protein-dependent ABC type of transport system that accepts maltose and maltodextrins as substrates. Furthermore, the observed absence of maltose transport activity in glucose-grown cells suggests that the previously identified maltose binding lipoprotein might be a component of the transporter, as glucose-grown cells lack this protein (24).
Purified maltose binding protein displays high binding affinity and specificity.
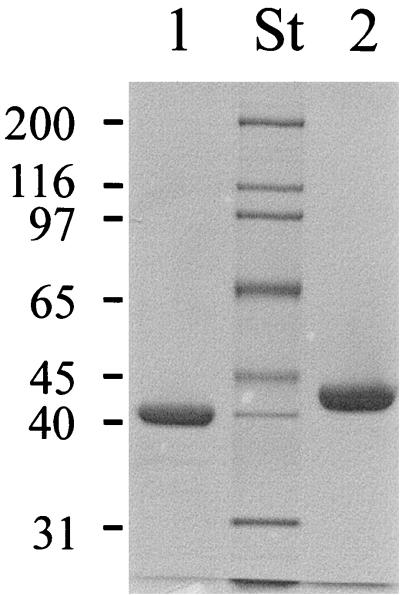
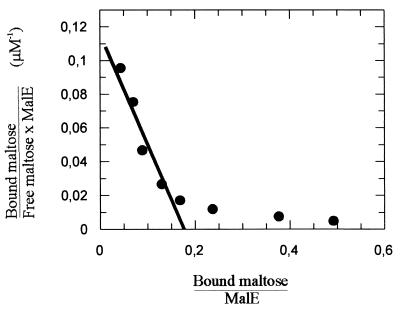
Next, we directly examined the maltose binding activity of MalEAa. While in the previous study the protein was purified from a Triton X-100 extract of intact maltose-grown cells (24), we now took advantage of the observation that after overnight growth, the majority of the protein could be recovered with the culture supernatant. After a one-step purification by affinity chromatography through an amylose column (Fig. 4, lane 1), the protein was subjected to a denaturation-renaturation procedure using 6 M guanidinium hydrochloride to remove bound unlabeled maltose (18) and subsequently was assayed for maltose binding activity. To this end, samples were incubated at pH 3.6 and 57°C in the presence of increasing concentrations of [14C]maltose, followed by ammonium sulfate precipitation, filtration, and liquid scintillation counting. As a control, maltose binding protein of S. enterica serovar Typhimurium (MalESt) was included in the study. Scatchard plot analysis of the data yielded curves with two slopes. For the high-affinity binding sites, apparent Kd values of 1.5 μM (MalEAa) (Fig. 5) and 1.3 μM (MalESt) were calculated. These data are in good agreement with dissociation constants reported for the E. coli and serovar Typhimurium proteins that were obtained by different binding assays (3, 4, 14). The nonlinear binding curve (Fig. 5) may be explained by the presence of unlabeled ligand still bound to the protein (3) or, more likely, by the correct refolding of only a subpopulation (about 20% when one binding site per polypeptide chain is assumed [Fig. 5]) of protein molecules after denaturation in 6 M guanidinium hydrochloride.
FIG. 4.
SDS gel of purified MalEAa. Wild-type and recombinant MalEAa proteins were purified from the culture supernatant of A. acidocaldarius cells and from the cytosol of E. coli strain ED169(pAH18), respectively, by affinity chromatography through agarose-coupled amylose. Aliquots were subjected to SDS-polyacrylamide gel electrophoresis, and the gel was subsequently stained with Coomassie brilliant blue. Lane 1, wild-type MalEAa (2.5 μg); lane 2, recombinant MalEAa (5 μg); lane St, molecular weight standards (in thousands). The observed difference in molecular weight is due to exposure of the wild-type protein to an extracellular protease that clips 23 amino acids from the N terminus (24).
FIG. 5.
Scatchard plot of maltose binding by MalEAa. The affinity of maltose binding was determined by a precipitation assay as described in Materials and Methods. The line fitted to a Kd of 1.5 μM is indicated.
When binding assays were performed in the presence of various unlabeled sugars, only those that were found to inhibit maltose uptake in intact cells (Fig. 3A) prevented binding of [14C]maltose to the protein (Fig. 3B). Thus, these experiments strengthen the notion that MalEAa is a component of an ABC transporter involved in maltose and maltodextrin uptake.
The majority of maltose binding protein (MalEAa) is distributed throughout the cell wall.
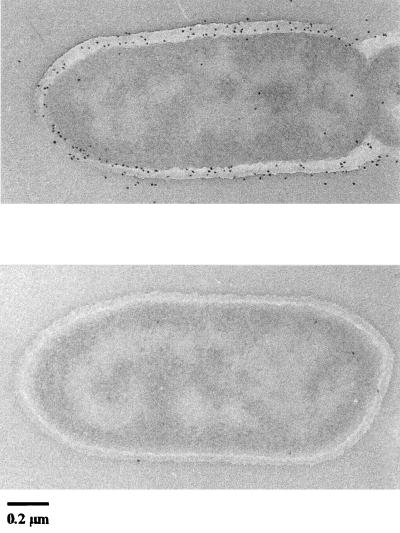
In order to determine the exact cellular location of MalEAa thin sections of maltose-grown cells from the log phase were immunogold labeled with a rabbit monospecific antibody (see Materials and Methods for details). Figure 6A shows that the majority of the protein is distributed throughout the cell wall, while only smaller quantities were located in the cytoplasmic membrane. Sections incubated with an irrelevant antiserum showed minimal labeling with protein A-gold (Fig. 6B).
FIG. 6.
Electron micrographs of thin sections of A. acidocaldarius cells reacted with monospecific antiserum to purified wild-type MalEAa. (A) Sections reacted with monospecific rabbit anti-MalEAa and protein A-gold conjugate; (B) sections reacted with irrelevant rabbit serum and protein A-gold conjugate. Representative pictures are shown.
Synthesis of MalEAa is induced by maltose and related sugars but repressed by glucose.
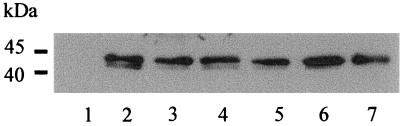
The available polyclonal antiserum raised against the purified protein allowed us to study the synthesis of MalEAa in the presence of different carbon sources. When cells of A. acidocaldarius were grown in minimal medium supplemented with maltose, linear maltodextrins, α- and β-cyclodextrins, and starch, respectively, comparable amounts of MalE protein could be detected by immunoblotting (Fig. 7, lanes 2 to 6). In contrast, but in agreement with earlier findings (24), MalE was absent in glucose-grown cells (Fig. 7, lane 1). However, when cultivated in the presence of both glucose and maltose (10 mM each), synthesis of MalE clearly occurred (Fig. 7, lane 7). Together, these data are consistent with the results from transport assays presented in Fig. 1.
FIG. 7.
Synthesis of MalEAa in the presence of different sugars. Wild-type A. acidocaldarius was grown in minimal salt medium supplemented with the following sugars (10 mM each): glucose (lane 1), maltose (lane 2), starch (0.2%, wt/vol) (lane 3), maltotetraose (lane 4), α-cyclodextrin (lane 5), β-cyclodextrin (lane 6), and maltose plus glucose (lane 7). Proteins from whole-cell extracts were separated by SDS-polyacrylamide gel electrophoresis and subsequently subjected to immunoblot analysis.
The genes encoding MalE and other ABC transport components are clustered.
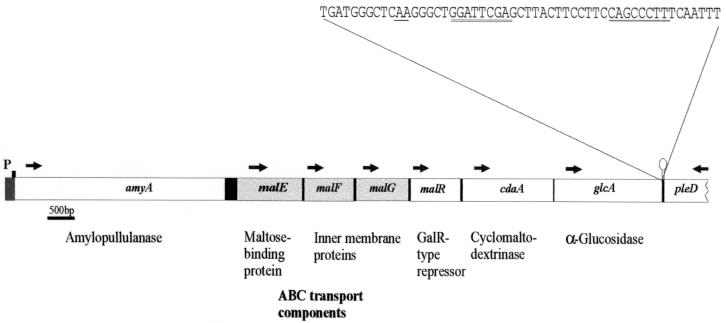
The observed sequence identity of the N terminus of MalEAa with a translated peptide fragment from ORF2 downstream of the amyA gene suggested that ORF2 was part of the gene encoding MalEAa. Consequently, an oligonucleotide probe was designed in order to clone the missing part of the putative malE gene as an SstI-BamHI fragment from genomic DNA of A. acidocaldarius into pUC18, resulting in plasmid pAH8-1. After determination of the complete nucleotide sequence of the 1.2-kb insert, chromosomal DNA was probed with an oligonucleotide that was now derived from the 3′ end of the malE gene. By the same approach, plasmids pAH9, pAH10, pAH30, pJM30 (34), and pAH19 were obtained. Together with a DNA fragment encompassing the 5′ end of the malE gene (pAH30), a total of 10.5 kb of chromosomal DNA was cloned (Table 1). Analysis of the nucleotide sequence downstream of amyA revealed, in addition to malE, five ORFs whose products (MalF, MalG, MalR, CdaA, and GlcA) resemble proteins involved in maltose and maltodextrin transport and utilization in other bacteria (Fig. 8). All of the genes are likely to be unidirectionally transcribed from the putative promoter that was identified upstream of amyA (32). Other promoter-like sequences could not be detected. At the 3′ end of the glcA gene, an imperfect palindromic sequence was predicted by DNASIS (ΔG = −28.3 kcal/mol), which might function as a transcription terminator. Moreover, adjacent to glcA, an incomplete ORF that is predicted to be divergently transcribed was identified. The putative gene product displays similarity to bacterial response regulators (22), and the gene was designated accordingly (pleD). Thus, from sequence analysis, it is tempting to speculate that amyA and the six succeeding genes may constitute an operon. However, experimental evidence from transcript analyses will be required to strengthen this view.
FIG. 8.
Organization of the genomic region around malEAa. ORFs were named according their homologs identified by database searches using BLAST. The sequence at the 3′ end of the glcA gene, including the predicted transcriptional termination structure (underlined), is also shown. Arrows indicate the predicted direction of transcription. Genes encoding maltose transport components are shaded. p, putative promoter region upstream of amyA (32).
The complete malE gene is composed of 1,284 bp that translate into a 427-amino-acid propeptide with a likely 25-amino-acid signal sequence at its N terminus (predicted using SIGNALP V1.1). Compared to the data reported by Koivula et al. (32), our sequence analysis revealed an additional nucleotide resulting in a frameshift upstream of codon −2 (relative to the codon that translates into the N-terminal cysteine residue of the mature protein). As a consequence, a GTG rather than an ATG is used as translation initiation codon. Moreover, the newly predicted signal sequence (VSVRRWGIVSTGVAALVLAGGAIAGC+1) better matches the characteristic features of signal peptides from gram-positive bacteria (11). The putative N-terminal cysteine residue of the mature protein, typical for solute binding proteins of gram-positive bacteria, is preceded by a recognition sequence for signal peptidase II (38) (underlined above). A database search revealed significant homology of the mature protein to a maltose binding protein of Thermotoga maritima (35% identical amino acids) and to a putative maltose binding protein of Deinococcus radiodurans (37% identical amino acids), but also to MalE of E. coli (33% sequence identity).
The malF and malG genes, located 69 bp downstream of malE, are composed of 966 and 906 nucleotides that translate into proteins of 321 and 301 amino acids, respectively. The genes are most likely translationally coupled, as their coding regions overlap by one nucleotide. The encoded proteins are hydrophobic membrane proteins that each may span the membrane six times (predicted by TMHMM). They share highest sequence identity with putative membrane proteins of T. maritima (MalF, 41% with TM1203; MalG, 39% with TM1202). Compared to the E. coli MalF protein and consistent with membrane components of maltose transport systems from other gram-positive bacteria, MalFAa is lacking an extended extracellularly exposed peptide loop (3). Both proteins are further characterized by the existence of the conserved EAA motif, which is common to bacterial ABC transporters involved in the uptake of solutes (3) and is thought to interact with the ATP-hydrolyzing component (30, 37).
The product of the malR gene, located 25 bp downstream of malG, is composed of 342 amino acids and displays sequence homology with bacterial transcriptional regulator proteins of the LacI-GalR family (59). MalRAa shares 41 and 40% identical amino acids with MalR of Streptococcus pneumoniae (40) and CebR of Streptomyces reticuli (45), respectively. Conserved sequence motifs, including a helix-turn-helix motif at the N terminus (ATVSRV), as well as effector binding and dimerization domains, can be identified (59).
Two genes, cdaA and glcA, occur downstream of malR (Fig. 8). The cda gene, separated from malR by 26 bp, encodes a protein of 578 amino acids that was identified as a cytoplasmic cyclomaltodextrinase with low pullulanase activity (34). This finding is consistent with the apparent lack of a signal sequence.
The product of the glcA gene, located 118 bp downstream of cdaA, is composed of 728 amino acids and resembles glycosylhydrolases of family 31 (23). Strikingly, a database search revealed most significant sequence homologies with α-glucosidases of the archaeon Sulfolobus solfataricus (42) and of mammals.
The transport components are heterologously expressed in E. coli.
At present, A. acidocaldarius cannot be stably transformed by plasmids, nor do protocols exist that would allow the isolation of mutant strains, e.g., those with defects in the chromosomally carried mal genes. Thus, in order to prove that the ORFs translate into the predicted transport proteins, we studied their heterologous expression in E. coli. To this end, we cloned malE (lacking the signal sequence-encoding fragment), malF, malG, and malG into plasmid vectors under the control of the pBAD promoter (see Materials and Methods for details). The constructed plasmids (Table 1) were introduced into E. coli strains ED169 and Top10. Subsequently, the resulting transformants were grown in LB medium, induced with arabinose, and analyzed for expression of plasmid-borne genes by immunoblotting. Using a polyclonal antiserum raised against the purified MalE protein from A. acidocaldarius as a probe, a protein of the expected size was identified in cells of ED169(pAH18). After cell fractionation, about half of the total protein was recovered with the soluble (cytoplasmic) fraction (data not shown). The observation that recombinant MalEAa protein cross-reacted with the antiserum provides further proof for its identity with the maltose binding protein isolated from wild type cells (24).
Expression of the malF and malG genes in cells of Top10(pAH27-3) and Top10(pAH26-2), respectively, was verified by using an antiserum raised against a pentahistidine peptide. In both cases, proteins with the predicted molecular masses were recovered with the cytoplasmic membrane (not shown).
The malEFG genes fail to restore the transport defect of E. coli mal mutants.
Next, we investigated the capability of the heterologously expressed A. acidocaldarius proteins to restore maltose transport in appropriate E. coli mal mutants. For this purpose, plasmid pFSA16, carrying malEAa with the fragment encoding the signal peptide replaced by the corresponding sequence of the E. coli malE gene was used to transform E. coli strain PD28 (ΔmalE444). The transformants did not grow on minimal maltose plates supplemented with IPTG (isopropyl-β-d-thiogalactopyranoside) (at 37°C), nor did transport assays reveal any maltose uptake activity. Similarly, the malF malG genes (on plasmid pFSA24) failed to substitute for the corresponding E. coli genes (not shown).
Purified native and recombinant MalEAa proteins are acidostable and bind maltose over a wide pH range.
With both, the (N-terminally truncated) wild-type and the (mature) recombinant forms of the MalE protein at hand, we characterized their binding activities for [14C]maltose with respect to pH and temperature. First, we determined the dissociation constant for maltose and the specificity of binding of the recombinant protein. To this end, the protein was purified from the cytosolic fraction of E. coli strain ED169(pAH18) by amylose affinity chromatography. From the binding data obtained under standard conditions (57°C, pH 3.6), a Kd value of 2.4 μM was calculated, which is close to that determined for the wild-type protein. Moreover, the same substrate specificity was observed (Fig. 3B).
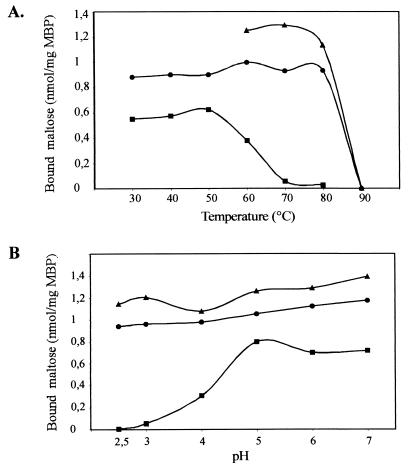
Next, binding of [14C]maltose was studied at different temperatures, ranging from 30 to 90°C (pH 3.6), and at pH values varying from 2.5 to 7 (with the temperature kept at 57°C). The results are shown in Fig. 9. The two proteins displayed similar binding activities at temperatures of up to 80°C but failed to bind the substrate at 90°C. In contrast, the purified MalE protein from S. enterica serovar Typhimurium that was used as a control (at pH 7.4!) exhibited only half of its binding activity at 60°C, while at 70°C, binding of [14C]maltose could no longer be detected (Fig. 9A). Furthermore, wild-type and recombinant MalEAa exhibited constant binding activities between pH 2.5 and 7, whereas MalESt (assayed at 37°C) showed some activity at pH 4 and full activity only at pH values of >5 (Fig. 9B).
FIG. 9.
Temperature (A) and pH (B) dependence of maltose binding to purified wild-type (▴) and recombinant (●) MalEAa. Binding assays were performed at the indicated temperatures in Na-acetate buffer (pH 3.6) and in the presence of 5 μM [14C]maltose. (B) Binding assays were performed at 57°C in citrate-phosphate buffer of the indicated pH. MalE of S. enterica serovar Typhimurium (■) was assayed at pH 7.4 (A) and 37°C (B). MBP, maltose binding protein.
The proteins were also analyzed for their functional stabilities at different temperatures and pH values. To this end, wild-type and recombinant MalEAa proteins were incubated for 3 h at pH 3.6 and 80°C or at pH 7 and 60°C. Subsequently, the assay mixtures were adjusted to 60°C and pH 3.6, respectively, and again analyzed for binding of [14C]maltose. While both proteins had lost almost all binding activity when incubated at 80°C (2 and 5% of the control value for wild-type and recombinant MalE, respectively), 96% (wild type) and 88% (recombinant protein), respectively, of their binding activities were recovered after exposure to pH 7. Together, these data suggest that MalEAa is acidostable and pH tolerant but only moderately thermostable.
DISCUSSION
In this study we have investigated the mechanism of maltose transport in the gram-positive thermoacidophilic bacterium A. acidocaldarius. Uptake of maltose is mediated by a high-affinity binding-protein-dependent ABC transport system that is specific for maltose and maltodextrins. Both, the kinetic parameters of transport and the chemical nature of inhibiting sugars clearly suggest that the transporter is more closely related to that of enterobacteria than to the maltose-trehalose transport system found in the archaeon Thermococcus litoralis (61) or to a transporter in the thermophilic anaerobic bacterium Thermoanaerobacter ethanolicus that accepts maltose, maltotriose, and trehalose (31). This notion is further supported by the properties of the purified binding protein, such as a dissociation constant for maltose in the low micromolar range and its sensitivity to the same competing sugars. Here, the protein also differs from maltose binding proteins of other thermophilic and hyperthermophilic microorganisms, which often display Kd values in the nanomolar range (1, 28, 31, 58).
The uptake of maltose was found to be optimal under experimental conditions that resemble those of the natural habitat of A. acidocaldarius. At pH values above 3.6, the transport rates substantially decreased. Similarly, strong inhibition of glucose transport was reported at higher pH values in the acidophilic archaeon Sulfolobus solfataricus (1). Acidophilic bacteria and archaea, including A. acidocaldarius, maintain their cytoplasmic pH at a value close to neutrality, resulting in a large ΔpH across the cell membrane. The latter is maintained by an active process that exchanges protons against inwardly moving potassium ions (2). Consequently, the proton motive force of acidophiles consists mainly of a ΔpH component, while the membrane potential is low. Accordingly, Albers et al. (1) explained their data by assuming that at higher pH values, the observed decrease of the internal ATP pool causes a transient solute uptake or no transport at all. In A. acidocaldarius cells, the internal ATP concentration was demonstrated to drop significantly at external pH values of above 5, even when energized by glycerol (35a). Thus, the above-described scenario is also likely to hold true for the results presented here, especially when taking into account that the transport assays were performed in the absence of an external carbon source.
The observed sensitivity to lower temperatures is most likely due to specific properties of the transport components. While the binding protein exhibited virtually the same maltose binding activity at between 30 and 80°C, the enzymatic activity of the yet-to-be identified ATP-hydrolyzing subunit of the transporter (see also below) that is required to energize the transport process might be temperature dependent. This notion is in line with the finding that the ATPase activity of MalK of the hyperthermophilic archaeon T. litoralis, which grows best at 85°C, was optimal at 80°C, while only little activity was observed at 37°C (16).
The genes encoding maltose binding protein and two putative membrane-integral components of the maltose transporter are organized in a cluster, together with genes that encode starch-degrading enzymes. Unlike the genetic organization in enterobacteria (3) but rather common to gram-positive ABC sugar transporters (39, 55), a gene encoding a cognate ATP-hydrolyzing subunit is lacking. This finding could imply that the ATPase subunit not only is engaged in maltose transport but also might power other transport systems. Experimental evidence in favor of such a view exists for the MsiK protein of Streptomycesz, which was demonstrated to assist two distinct transport systems for maltose and cellobiose (46).
The malEFG genes, when expressed in E. coli mutants lacking the homologous genes, failed to restore growth of the transformants on maltose. Possible explanations for these results may include insufficient sequence identity between the A. acidocaldarius and E. coli proteins to allow functional interactions and, in the case of MalEAa, poor translocation of the protein to the periplasm. In fact, only small amounts of MalEAa could be determined in the periplasmic fraction after treatment of cells by osmotic shock (not shown). A similar result was also reported for the maltose-trehalose binding protein of T. litoralis (28). Thus, since gene expression experiments with A. acidocaldarius are not yet feasible, the transport functions of the proteins might alternatively be analyzed in proteoliposomes. However, such experiments must await cloning of a gene encoding the cognate ATPase component.
A. acidocaldarius can utilize α- or β-cyclodextrins as sole sources of carbon and energy (Fig. 7). Together with the existence of a cyclodextrinase, the product of the cdaA gene, in the cytoplasm (34), this finding indicates that cyclodextrins are transported across the cell membrane. Whether uptake is mediated by a distinct transport system as in Klebsiella oxytoca (13) or by the maltose transporter described here remains to be established. Although cyclodextrins were demonstrated to inhibit maltose uptake and to compete with maltose for binding to purified MalEAa, these data nonetheless do not provide direct prove for uptake of cyclodextrins. The E. coli maltose binding protein binds β-cyclodextrin, which, however, is not transported due to its failure to induce closure of the binding cleft (19, 50), which is essential to initiate the transport process (3, 8).
An unusual aspect of the identified gene cluster is the localization of the malR gene, encoding a putative transcriptional regulator within the predicted operon structure. Genes encoding homologous proteins from other gram-positive bacteria are usually transcribed from their own promoter (10, 43, 45, 56). One exception appears to be the malR gene of S. pneumoniae, which probably belongs to the same transcriptional unit as malA, necessary for growth on maltotetraose (40). However, the malA-malR operon is clearly separated from the malXCD and the malMP operons, carrying genes for maltose transport and degradation, respectively (39). Interestingly enough, MalRAa shares the highest number of identical amino acids with MalR of S. pneumoniae. Clearly, further work will be required to elucidate the role of MalR in maltose utilization by A. acidocaldarius.
In this respect, it is also of interest that glucose repression of mal genes appears to be different from that in E. coli (4) and in gram-positive bacteria with low GC contents, such as B. subtilis (48). In these organisms, components of the phosphotransferase system for glucose are central to carbon catabolite repression, albeit by different mechanisms (recently reviewed in reference 52). Through complex signaling cascades, glucose largely prevents the expression of target genes, even in the presence of their respective inducers. In E. coli and other enterobacteria, the activity of transport systems for other carbon sources, including maltose, is additionally inhibited by the EIIAGlc component of the phosphotransferase system to prevent the uptake of inducer molecules. In contrast, in A. acidocaldarius, substantial rates of maltose uptake were measured in cells that were cultured in a medium containing maltose and glucose (Fig. 1), suggesting that inducer exclusion is absent. Moreover, our results also differ from mal gene regulation in B. subtilis. In this organism, no activity of the maltose-inducible α-glucosidase was detected in cells grown in glucose or in a combination of maltose and glucose (48). Rather, our findings are similar to results obtained with the ABC transporter for cellobiose of S. reticuli (45). In that study, the cellobiose binding protein CebE was demonstrated to be absent in cells grown in glucose but occurred in the presence of cellobiose or a combination of both. In Streptomyces, glucose repression is poorly understood but was proposed to be mediated by glucose kinase, which is thought to negatively influence the specific regulators of other catabolic genes (5). Whether a similar mechanism operates in A. acidocaldarius remains to be established.
Our data clearly indicate that the previously identified maltose binding lipoprotein (24) is encoded by the malE gene and thus is a component of the maltose transport system of A. acidocaldarius. Moreover, the N-terminally truncated natural gene product that can be purified from the supernatant of an A. acidocaldarius culture and the recombinant mature MalE protein are virtually indistinguishable with respect to their maltose binding activities and biochemical properties. Thus, the N-terminal 23 amino acids, including the fatty acid modification of Cys-1, are apparently dispensable for function in vitro. However, whether the truncated binding protein is capable of initiating transport remains to be established. Under the laboratory conditions used here, several forms of the protein are simultaneously detectable during cell growth. Besides the membrane-associated species that can be released by detergent (24), immunochemical analysis of the culture supernatant revealed, in addition to the truncated protein, a slower-migrating band that likely is identical to the mature maltose binding protein (data not shown). Thus, it is tempting to speculate that a certain portion of the protein might constantly be released into the cell wall, where it could assist in scavenging substrate molecules that penetrate the S-layer by which A. acidocaldarius is surrounded (unpublished observation) (E. Bakker, personal communication). The distribution of binding protein molecules throughout the cell wall as observed by immunoelectron microscopy is consistent with this notion. Whether the observed release into the medium is due to slow penetration through the pores of the S-layer is not known. Similar results have been reported for a lipoprotein involved in iron transport in Staphylococcus epidermis (6).
An unusual and most interesting aspect of the MalEAa protein is the relative insensitivity of its maltose binding activity to pH. Unlike the (neutrophilic) Salmonella MalE protein, which displayed significant maltose binding only at pH values of ≥5, MalEAa exhibited about the same maltose binding activity at pH values ranging from 2.5 to 7. This is also in contrast to the glucose binding protein of the extremely acidophilic archaeon S. solfataricus, which failed to bind glucose at above pH 3 (1). Moreover, MalEAa also displayed extreme functional stability at pH 7. Thus, MalEAa appears to be a neutrophilic protein that tolerates acidic pH rather than an acidophilic solute binding protein.
The molecular basis of acidostability and acidophilicity of proteins is still poorly understood. Sequence analysis, however, revealed a reduced density of charged residues at the surface of proteins from acidophiles compared to their functionally equivalent homologs from neutrophilic organisms. Schwermann et al. (49) interpreted this finding to mean that electrostatic repulsion at low pH is thereby prevented. Those authors also included in their study a comparative analysis of the protein sequence encoded by ORF2 of A. acidocaldarius with the corresponding E. coli MalE sequence. In accordance with their overall conclusion, the number of charged residues was decreased in favor of polar but uncharged residues. This analysis is now fully confirmed by the examination of the complete amino acid sequence of MalEAa. Compared to the E. coli protein, the total number of charged residues (K, R, H, E, and D) is reduced (MalEAa, 15%; MalEEc, 27%), while the total number of polar uncharged amino acids (N, Q, T, S, and C) is increased (MalEAa, 26%; MalEEc, 18%). Others, especially hydrophobic residues, are unchanged. Although it is attractive, ultimate proof of the above hypothesis will require a comparative analysis of the tertiary structure of MalEAa with that of E. coli MalE (51). Thus, attempts to solve the crystal structure of MalEAa are under way.
Taking our results together, we have shown that A. acidocaldarius is equipped with a maltose and maltodextrin transport system of the ABC family that allows the organism to feed on products that are released from starch by the action of its secreted amylopullulanase. In the cytoplasm, an α-glucosidase liberates glucose for further degradation. As a characteristic aspect, the transporter contains an acidostable and moderately thermostable substrate binding protein that will be a useful tool in the elucidation of the molecular mechanism of acidostability of proteins.
ACKNOWLEDGMENTS
We thank B. Sattler and H. Landmesser for excellent technical assistence, J. Matzke (Osnabrück) for providing plasmid pJM30, J.-M. Betton (Paris) for providing strain PD28, G. Lüder (Berlin) for introducing A.H. to electron microscopy technology, and E. Bakker (Osnabrück) for helpful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (SCHN 274/6-1/6-2/6-3) and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Albers S-V, Elferink M G-L, Charlebois R L, Sensen C W, Driessen A J M, Konings W N. Glucose transport in the extremely thermophilic Sulfolobus solfataricus involves a high-affinity membrane-integrated binding protein. J Bacteriol. 1999;181:4285–4291. doi: 10.1128/jb.181.14.4285-4291.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakker E P. The role of alkali-cation transport in energy coupling of neutrophilic and acidophilic bacteria: an assessment of methods and concepts. FEMS Microbiol Rev. 1990;75:319–334. [Google Scholar]
- 3.Boos W, Lucht J M. Periplasmic binding-protein-dependent ABC-transporters. In: Neidthard F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1175–1209. [Google Scholar]
- 4.Boos W, Shuman H. Maltose/maltodextrin system in Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunkhorst C, Andersen C, Schneider E. Acarbose, a pseudooligosaccharide, is transported but not metabolized by the maltose-maltodextrin system of Escherichia coli. J Bacteriol. 1999;181:2612–2619. doi: 10.1128/jb.181.8.2612-2619.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cockayne A, Hill P J, Powell N B L, Bishop K, Sims C, Williams P. Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermis ABC transporter. Infect Immun. 1998;66:3767–3774. doi: 10.1128/iai.66.8.3767-3774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darland G, Brock T D. Bacillus acidocaldarius sp. nov., an acidophilic thermophilic spore-forming bacterium. J Gen Microbiol. 1971;67:9–15. [Google Scholar]
- 8.Davidson A L, Shuman H A, Nikaido H. Mechanism of maltose transport in Escherichia coli: transmembrane signaling by periplasmic binding proteins. Proc Natl Acad Sci USA. 1992;89:2360–2364. doi: 10.1073/pnas.89.6.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duplay P, Szmelcman S, Bedouelle H, Hofnung M. Silent and functional changes in the periplasmic maltose-binding protein of Escherichia coli K12. I. Transport of maltose. J Mol Biol. 1987;194:663–673. doi: 10.1016/0022-2836(87)90243-9. [DOI] [PubMed] [Google Scholar]
- 10.Egeter E, Brückner R. Characterization of a genetic locus essential for maltose-maltotriose utilization in Staphylococcus xylosus. J Bacteriol. 1995;177:2408–2415. doi: 10.1128/jb.177.9.2408-2415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fekkes P, Driessen A J M. Protein targeting to the bacterial cytoplasmic membrane. Microbiol Mol Biol Rev. 1999;63:161–173. doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferenci T, Klotz U. Affinity chromatography of the periplasmic maltose binding protein of Escherichia coli. FEBS Lett. 1978;94:213–217. doi: 10.1016/0014-5793(78)80940-5. [DOI] [PubMed] [Google Scholar]
- 13.Fiedler G, Pajatsch M, Böck A. Genetics of a novel starch utilization pathway present in Klebsiella oxytoca. J Mol Biol. 1996;256:279–291. doi: 10.1006/jmbi.1996.0085. [DOI] [PubMed] [Google Scholar]
- 14.Ganesh C, Shah A N, Swaminathan C P, Srolia A, Varadarajan R. Thermodynamic characterization of the reversible, two-state unfolding of maltose binding protein, a large two-domain protein. Biochemistry. 1997;36:5020–5028. doi: 10.1021/bi961967b. [DOI] [PubMed] [Google Scholar]
- 15.Gish W, States D J. Identification of protein coding regions by database similarity search. Nature. 1993;385:556–559. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 16.Greller G, Horlacher R, DiRuggiero J, Boos W. Molecular and biochemical analysis of MalK, the ATP-hydrolyzing subunit of the trehalose/maltose transport system of the hyperthermophilic archeon Thermococcus litoralis. J Biol Chem. 1999;274:20259–20264. doi: 10.1074/jbc.274.29.20259. [DOI] [PubMed] [Google Scholar]
- 17.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall J A, Davidson A L, Nikaido H. Preparation and reconstitution of the membrane-associated maltose transporter complex of Escherichia coli. Methods Enzymol. 1998;292:20–29. doi: 10.1016/s0076-6879(98)92004-3. [DOI] [PubMed] [Google Scholar]
- 19.Hall J A, Gehring K, Nikaido H. Two modes of the ligand binding in maltose-binding protein of Escherichia coli. J Biol Chem. 1997;272:17605–17609. doi: 10.1074/jbc.272.28.17605. [DOI] [PubMed] [Google Scholar]
- 20.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 21.Harwood C R, Cutting S M. Molecular biological methods for Bacillus. New York, N.Y: John Wiley & Sons, Inc.; 1990. [Google Scholar]
- 22.Hecht G B, Newton A. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J Bacteriol. 1995;177:6223–6229. doi: 10.1128/jb.177.21.6223-6229.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrmann A, Schlösser A, Schmid R, Schneider E. Biochemical identification of a lipoprotein with maltose-binding activity in the thermoacidophilic grampositive bacterium Alicyclobacillus acidocaldarius. Res Microbiol. 1996;147:733–737. doi: 10.1016/s0923-2508(97)85120-0. [DOI] [PubMed] [Google Scholar]
- 25.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 26.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 27.Holland I B, Blight M A. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J Mol Biol. 1999;293:381–399. doi: 10.1006/jmbi.1999.2993. [DOI] [PubMed] [Google Scholar]
- 28.Horlacher R, Xavier K B, Santos H, DiRuggiero J, Kossmann M, Boos W. Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archeon Thermococcus litoralis. J Bacteriol. 1998;180:680–689. doi: 10.1128/jb.180.3.680-689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunke S, Dröse S, Schneider E. Vanadate and bafilomycin A1 are potent inhibitors of the ATPase activity of the reconstituted bacterial ATP-binding cassette transporter for maltose (MalFGK2) Biochem Biophys Res Commun. 1995;216:589–594. doi: 10.1006/bbrc.1995.2663. [DOI] [PubMed] [Google Scholar]
- 30.Hunke S, Mourez M, Jéhanno M, Dassa E, Schneider E. ATP modulates subunit-subunit interactions in an ATP-binding-cassette transporter (MalFGK2) determined by site-directed chemical cross-linking. J Biol Chem. 2000;275:15526–15534. doi: 10.1074/jbc.275.20.15526. [DOI] [PubMed] [Google Scholar]
- 31.Jones C R, Ray M, Dawson K A, Strobel H J. High-affinity maltose binding and transport by the thermophilic anaerobe Thermoanaerobacter ethanolicus 39E. Appl Environ Microbiol. 2000;66:995–1000. doi: 10.1128/aem.66.3.995-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koivula T T, Hemilä H, Pakkanen R, Sibakov M, Palva I. Cloning and sequencing of a gene encoding acidophilic amylase from Bacillus acidocaldarius. J Gen Microbiol. 1993;139:2399–2407. doi: 10.1099/00221287-139-10-2399. [DOI] [PubMed] [Google Scholar]
- 33.Martinez E, Bartolome B, de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZα reporter gene of pUC8/9 and pUC18/19. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 34.Matzke J, Herrmann A, Schneider E, Bakker E P. Gene cloning, nucleotide sequence and biochemical properties of a cyclomaltodextrinase (neopullulanase) from Alicyclobacillus acidocaldarius; reclassification of a group of enzymes. FEMS Microbiol Lett. 2000;183:55–61. doi: 10.1111/j.1574-6968.2000.tb08933.x. [DOI] [PubMed] [Google Scholar]
- 35.Matzke J, Schwermann B, Bakker E P. Acidostable and acidophilic proteins: the example of the α-amylase from Alicyclobacillus acidocaldarius. Comp Biochem Physiol. 1997;118A:475–479. doi: 10.1016/s0300-9629(97)00008-x. [DOI] [PubMed] [Google Scholar]
- 35a.Michels M. Energiekopplung und Kationentransport bei den acidophilen Bakterien Bacillus acidocaldarius und Thermoplasma acidophilum. Ph.D. thesis. Osnabrück, Germany: Universität Osnabrück; 1985. [Google Scholar]
- 36.Miller J H. A short course in bacterial genetics. A laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 37.Mourez M, Hofnung M, Dassa E. Subunit interaction in ABC transporters. A conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines sites of interaction with the helical domain of ABC subunits. EMBO J. 1997;16:3066–3077. doi: 10.1093/emboj/16.11.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nielsen H, Engelbrecht J, Brunak S, von Heinje G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Puyet A, Espinosa M. Structure of the maltodextrin-uptake locus of Streptococcus pneumoniae. Correlation to Escherichia coli maltose regulon. J Mol Biol. 1993;230:800–811. doi: 10.1006/jmbi.1993.1202. [DOI] [PubMed] [Google Scholar]
- 40.Puyet A, Ibañez M, Espinosa M. Characterization of the Streptococcus pneumoniae maltosaccharide regulator MalR, a member of the LacI-GalR family of repressors displaying distinctive genetic features. J Biol Chem. 1993;268:25402–25408. [PubMed] [Google Scholar]
- 41.Richarme G, Kepes A. Study of binding protein-ligand interaction by ammonium sulphate assisted adsorption on cellulose ester filters. Biochim Biophys Acta. 1983;742:16–24. doi: 10.1016/0167-4838(83)90353-9. [DOI] [PubMed] [Google Scholar]
- 42.Rolfsmeier M, Haseltine C, Bini E, Clark A, Blum P. Molecular characterization of the α-glucosidase gene (malA) from the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol. 1998;180:1287–1295. doi: 10.1128/jb.180.5.1287-1295.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell R R B, Aduse-Opoku J, Sutcliffe J C, Tao L, Ferretti J J. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J Biol Chem. 1992;267:4631–4637. [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Schlösser A, Jantos J, Hackmann K, Schrempf H. Characterization of the binding protein-dependent cellobiose and cellotriose transport system of the cellulose degrader Streptomyces reticuli. Appl Environ Microbiol. 1999;65:2636–2643. doi: 10.1128/aem.65.6.2636-2643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlösser A, Kampers T, Schrempf H. The Streptomyces ATP-binding component MsiK assists in cellobiose and maltose transport. J Bacteriol. 1997;179:2092–2095. doi: 10.1128/jb.179.6.2092-2095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 48.Schönert S, Buder T, Dahl M K. Identification and enzymatic characterization of the maltose-inducible α-glucosidase MalL (sucrase-isomaltase-maltase) of Bacillus subtilis. J Bacteriol. 1998;180:2574–2578. doi: 10.1128/jb.180.9.2574-2578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwermann B, Pfau C, Liliensiek B, Schleyer M, Fischer T, Bakker E P. Purification, properties and structural aspects of a thermoacidophilic α-amylase from Alicyclobacillus acidocaldarius ATCC 27009, insight into acidostability of proteins. Eur J Biochem. 1994;226:981–991. doi: 10.1111/j.1432-1033.1994.00981.x. [DOI] [PubMed] [Google Scholar]
- 50.Sharff A J, Rodseth L E, Quiocho F A. Refined 1,8-Å structure reveals the mode of binding of β-cyclodextrins to the maltodextrin binding protein. Biochemistry. 1993;32:10553–10559. doi: 10.1021/bi00091a004. [DOI] [PubMed] [Google Scholar]
- 51.Spurlino J C, Lu G Y, Quiocho F A. The 2,3 Å resolution structure of maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J Biol Chem. 1991;266:5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- 52.Stülke J, Hillen W. Carbon catabolite repression in bacteria. Curr Opin Microbiol. 1999;2:195–201. doi: 10.1016/S1369-5274(99)80034-4. [DOI] [PubMed] [Google Scholar]
- 53.Sutcliffe I C, Russel R B. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tam R, Saier M H. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Wezel G P, White J, Bibb M J, Postma P W. The malEFG gene cluster of Streptomyces coelicolor A3(2): characterization, disruption and transcriptional analysis. Mol Gen Genet. 1997;254:604–608. doi: 10.1007/s004380050458. [DOI] [PubMed] [Google Scholar]
- 56.van Wezel G P, White J, Young P, Postma P W, Bibb M J. Substrate induction and glucose repression of maltose utilization by Streptomyces coelicolor A3 2 is controlled by malR, a member of the lacI-galR family of regulator genes. Mol Microbiol. 1997;23:537–549. doi: 10.1046/j.1365-2958.1997.d01-1878.x. [DOI] [PubMed] [Google Scholar]
- 57.Walter C, Höner zu Bentrup K, Schneider E. Large scale purification, nucleotide binding properties and ATPase activity of the MalK subunit of Salmonella typhimurium maltose transport complex. J Biol Chem. 1992;267:8863–8869. [PubMed] [Google Scholar]
- 58.Wassenberg D, Liebl W, Jaenicke R. Maltose-binding protein from the hyperthermophilic bacterium Thermotoga maritima: stability and binding properties. J Mol Biol. 2000;295:279–288. doi: 10.1006/jmbi.1999.3367. [DOI] [PubMed] [Google Scholar]
- 59.Weickert M J, Adhya S. Family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 60.Wisotzkey J D, Jurtshuk P, Fox G E, Deinhard G, Poralla K. Comparative sequence analysis on the 16S rRNA (rDNA) of Bacillus acidocaldarius, Bacillus acidoterrestris, and Bacillus cycloheptanicus and proposal for creation of a new genus, Alicyclobacillus gen. Int J Syst Bacteriol. 1992;42:263–269. doi: 10.1099/00207713-42-2-263. [DOI] [PubMed] [Google Scholar]
- 61.Xavier K B, Martins L O, Peist R, Kossmann M, Boos W, Santos H. High-affinity maltose/trehalose transport system in the hyperthermophilic archaeon Thermococcus litoralis. J Bacteriol. 1996;178:699–704. doi: 10.1128/jb.178.16.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]