Significance
Alzheimer’s disease (AD) is a neurodegenerative disease characterized by the presence of amyloid-β plaques and Tau neurofibrillary tangles in affected brains. Detection of different phosphorylated species and isoforms of Tau has shown promise as biomarkers for AD. Here, we identified a new C-terminal truncated Tau-11i species caused by premature stop codon in the retained intron 11. Tau11i protein is more abundant in AD brain tissues tested and exhibits biochemical properties that resemble pathological Tau. Recent studies showed that dysregulation of RNA splicing and intron retention is a hallmark of AD pathogenesis, suggesting that Tau11i might be downstream of these events. Further studies with larger cohorts could help assess if Tau11i may be potential biomarker for AD.
Keywords: Tau, intron retention, Alzheimer disease
Abstract
Alzheimer’s disease (AD) is characterized by the accumulation of amyloid-β plaques and Tau tangles in brain tissues. Recent studies indicate that aberrant splicing and increased level of intron retention is linked to AD pathogenesis. Bioinformatic analysis revealed increased retention of intron 11 at the Tau gene in AD female dorsal lateral prefrontal cortex as compared to healthy controls, an observation validated by quantitative polymerase chain reaction using different brain tissues. Retention of intron 11 introduces a premature stop codon, resulting in the production of truncated Tau11i protein. Probing with customized antibodies designed against amino acids encoded by intron 11 showed that Tau11i protein is more enriched in AD hippocampus, amygdala, parietal, temporal, and frontal lobe than in healthy controls. This indicates that Tau messenger RNA with the retained intron is translated in vivo instead of being subjected to nonsense-mediated decay. Compared to full-length Tau441 isoform, ectopically expressed Tau11i forms higher molecular weight species, is enriched in Sarkosyl-insoluble fraction, and exhibits greater protein stability in cycloheximide assay. Stably expressed Tau11i also shows weaker colocalization with α-tubulin of microtubule network in human mature cortical neurons as compared to Tau441. Endogenous Tau11i is enriched in Sarkosyl-insoluble fraction in AD hippocampus and forms aggregates that colocalize weakly with Tau4R fibril-like structure in AD temporal lobe. The elevated level of Tau11i protein in AD brain tissues tested, coupled with biochemical properties resembling pathological Tau species suggest that retention of intron 11 of Tau gene might be an early biomarker of AD pathology.
Alzheimer’s disease (AD) is a familial and sporadic neurodegenerative disease that encompasses many risk factors such as genetic makeup and aging (1). Synaptic and neuronal loss in AD is caused by interplay between the pathology of amyloid-β plaques and Tau neurofibrillary tangles with impairment of innate clearance pathways (2). Accumulated evidence indicates that epigenetic alterations (3) and aberrant RNA splicing events at several AD susceptibility loci during aging might predispose healthy brains to AD pathogenesis (4–6), with reports showing differential intron retention (IR) events at the Tau gene (Fig. 1A) (7–9). Given that Tau pathology may initiate and regulate sporadic AD (10–12), as well as the successful use of different phosphorylated Tau species as diagnostic AD biomarker (13–15), we examined how IR at the Tau gene might affect its biochemical properties and expression level in AD tissues.
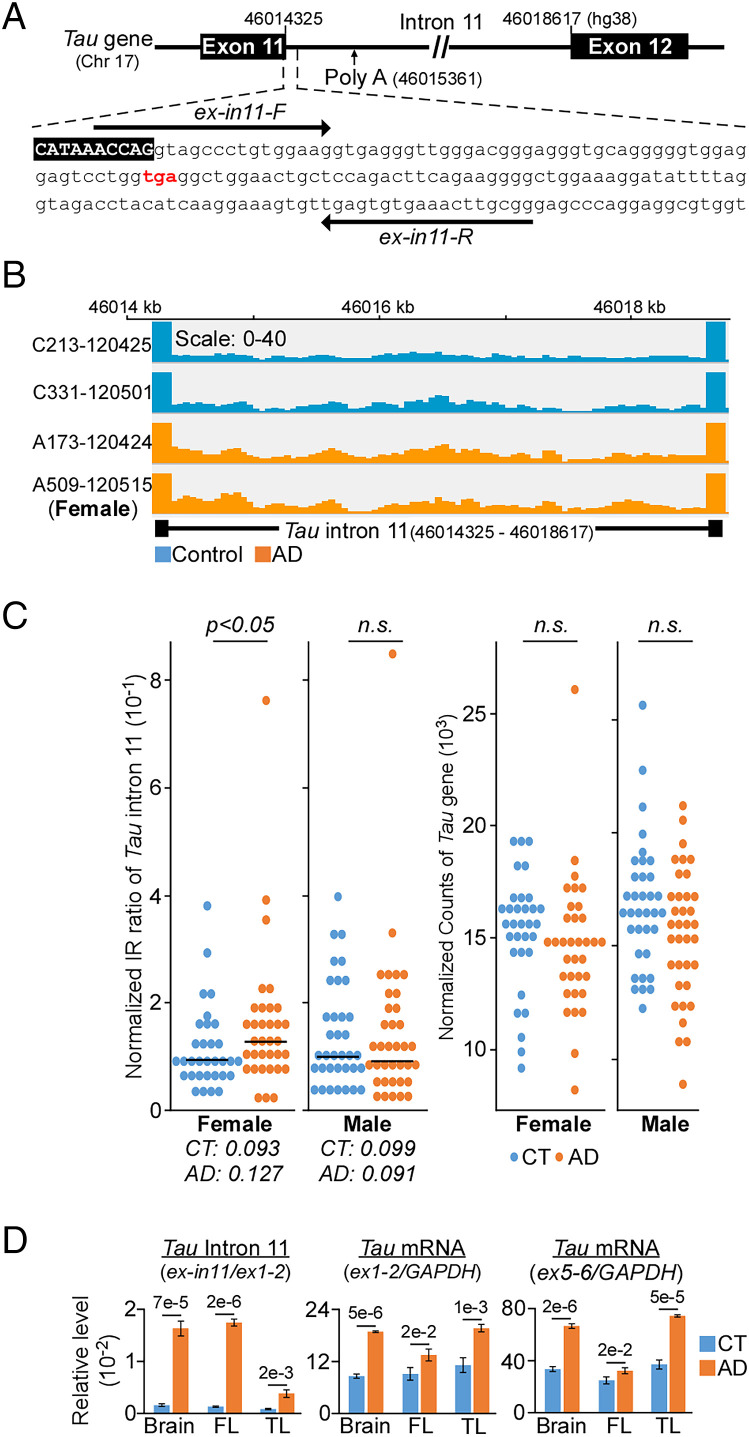
Fig. 1.
Increased retention of intron 11 of Tau gene in AD female. (A) Intron 11 contains a premature stop codon (red) with a canonical polyadenylation site that is located 1 kb downstream. Arrows indicate qPCR primers. (B) Integrative genomic viewer of intron 11 from control (blue) and AD (orange) patients. (C, Left) Statistically significant higher intron retention (IR) is observed only in female AD dorsal lateral prefrontal cortex (n = 34) as compared to control (CT, n = 32). Each dot represents individual normalized IR ratio. Right: No differential expression in the Tau gene between CT and AD cohorts. (D) qPCR validation of intron 11 (relative to Tau exon1/exon2) and Tau expression (exon1/exon 2 and exon5/exon6 relative to GAPDH) from three distinct pairs of brain tissues. FL, frontal lobe; TL, temporal lobe. Data presented as mean of triplicates ± SD, P values calculated by two-tailed t test.
Results
Increased Retention of Tau Intron 11 in Female AD Cohorts.
Analysis of more than 30 pairs of healthy controls and AD dorsolateral prefrontal cortex (DLPFC) (16) using IRFinder (17) revealed increased retention of Tau intron 11 (Fig. 1A) in female, but not male AD samples when compared to healthy controls (Fig. 1 B and C). These changes appeared to be unique to AD since no significant difference in intron retention (IR) ratio was observed in Parkinson’s disease (18) and progressive supranuclear palsy (19) datasets (SI Appendix, Fig. S1A and Dataset S01). Increased IR in AD samples was next validated by quantitative polymerase chain reaction (qPCR) of brain, frontal, and temporal lobe from three distinct pairs of control and AD patients (Fig. 1D). The marked differences observed in qPCR could be explained by targeted amplification of intron 11 as compared to RNA-sequencing of the entire transcriptome where low abundance intronic read counts may be underrepresented. Although qPCR detected an elevated level of Tau messenger RNA (mRNA) in the three AD brain tissues samples as compared to control (Fig. 1D), we observed no differential expression of Tau gene in the DLPFC (Fig. 1C).
Truncated Tau11i Oligomerizes and Is More Enriched in Sarkosyl-Insoluble Fraction.
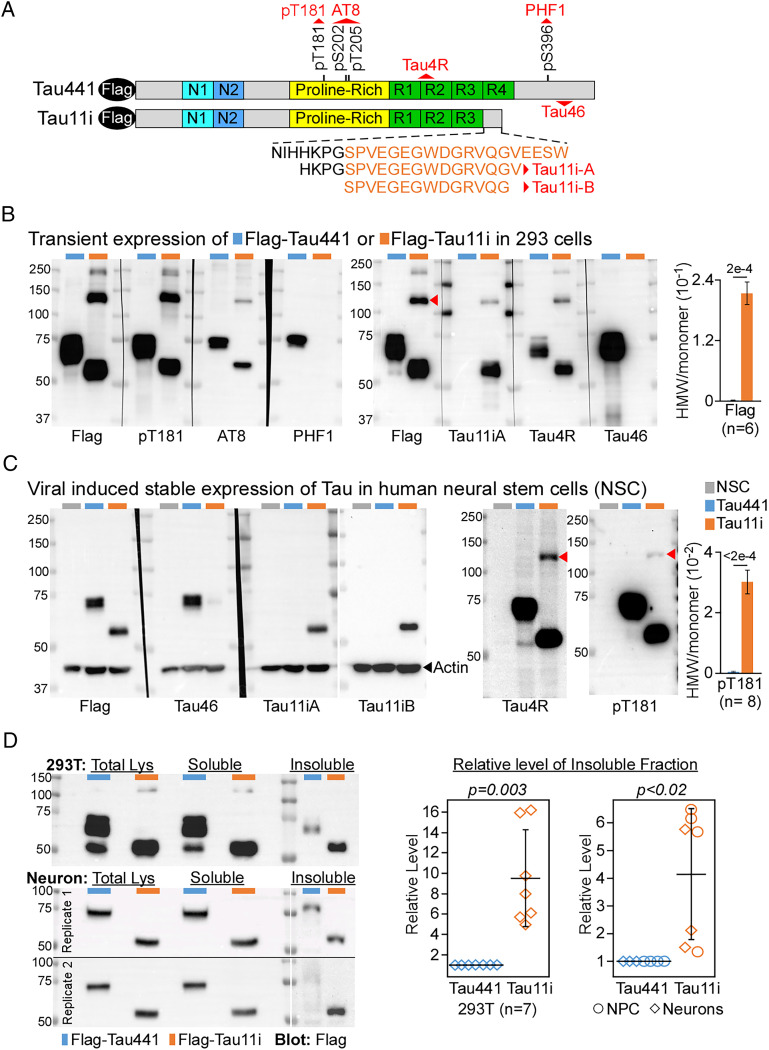
In the brain, full-length Tau encodes for 441 amino acids (herein Tau441) (20) whereas the premature stop codon in intron 11 (Fig. 1A) may lead to protein truncation (herein Tau11i, amino acids encoded by retained intron is colored orange) (Fig. 2A). The presence of canonical polyadenylation site 1 kb downstream of stop codon within intron 11 further suggests that the Tau mRNA transcript with IR might escape nonsense-mediated decay (NMD) (8, 21, 22). To study the effect caused by IR, we expressed either Flag-tag full-length Tau441 or truncated Tau11i in 293T cells. As predicted, truncated Tau11i runs below Tau441 at ∼55 kDa size and cannot be detected by PHF1 or Tau46 antibodies which recognize Tau C-terminal domain. Importantly, it can be detected specifically by Tau11i-A antibody raised against the amino acid sequences encoded by the retained intron 11 (Fig. 2B). Interestingly, Tau11i protein also migrates as higher molecular weight (HMW) species on denaturing gels and was readily detected by pT181 antibody, suggesting it undergoes oligomerization in 293T cells (Fig. 2B and SI Appendix, Fig. S1B). This observation is also consistent with the C-terminal domain inhibiting polymerization of Tau monomer (23–25) and HMW-Tau undergoes phosphorylation at Thr181 (26). To negate the effect of transient expression, we used lentiviral infection to stably express Flag-Tau441 or Flag-Tau11i in human neural stem cells (NSC). Similar to 293T cells, Flag-Tau11i protein was specifically recognized by two different Tau11i antibodies but not Tau46 (Fig. 2C). Tau11i also consistently oligomerizes to form HMW species in NSC, albeit at a lower level, as detected by Tau4R and pT181 antibodies (Fig. 2C and SI Appendix, Fig. S1B). However, the two customized Tau11i antibodies are unable to detect the HMW species in the neuronal context. As formation of HMW species is one of the characteristics of pathological Tau in AD (26), we further examined other biochemical properties of Tau11i in these cells. Extraction by ionic detergent N-lauryl-sarcosine (Sarkosyl), which solubilizes natively folded protein effectively, has been widely used to isolate detergent-insoluble protein aggregates from brain tissues with neurodegenerative diseases such as AD (27). Compared to Flag-Tau441, there was more ectopically expressed Flag-Tau11i in the Sarkosyl-insoluble fraction from 293T cells, neuronal progenitor cells (NPC) and neurons (Fig. 2D and SI Appendix, Fig. S1 C and D). Taken together, our results indicate that ectopically expressed Tau11i undergoes multiple phosphorylation, oligomerizes to form HMW species and is enriched in Sarkosyl-insoluble fractions.
Fig. 2.
Truncated Tau11i oligomerizes and is found in Sarkosyl-insoluble fraction. (A) Schematic of Flag-tag full-length and truncated Tau proteins with specific antibody (red) recognition sites. Amino acids encoded by intron 11 are colored orange. pT181, phosphor-Tau (T181); Tau4R, 275–291; Tau46, 404–421; AT8, phosphor-Tau (S202/T205). (B and C) Immunoblot and quantification of Tau11i HMW species in (B) 293T transfected with Tau441 or Tau11i, and (C) NSC expressing Tau441 or Tau11i proteins. Data presented as mean ± SEM. (D) Immunoblot and quantification showed elevated Sarkosyl-insoluble fraction of Tau11i in 293T cells (n = 7), NPC and day 44 neurons (n = 7) expressing Flag-Tau441 or Flag-Tau11i. The level of insoluble Tau was first normalized to total lysate. The relative level of insoluble fraction from each biological replicate was combined and presented as mean ± SD. All P values were calculated by two-tailed t test.
Detection of a Higher Level of Tau11i Species in AD Brain Regions.
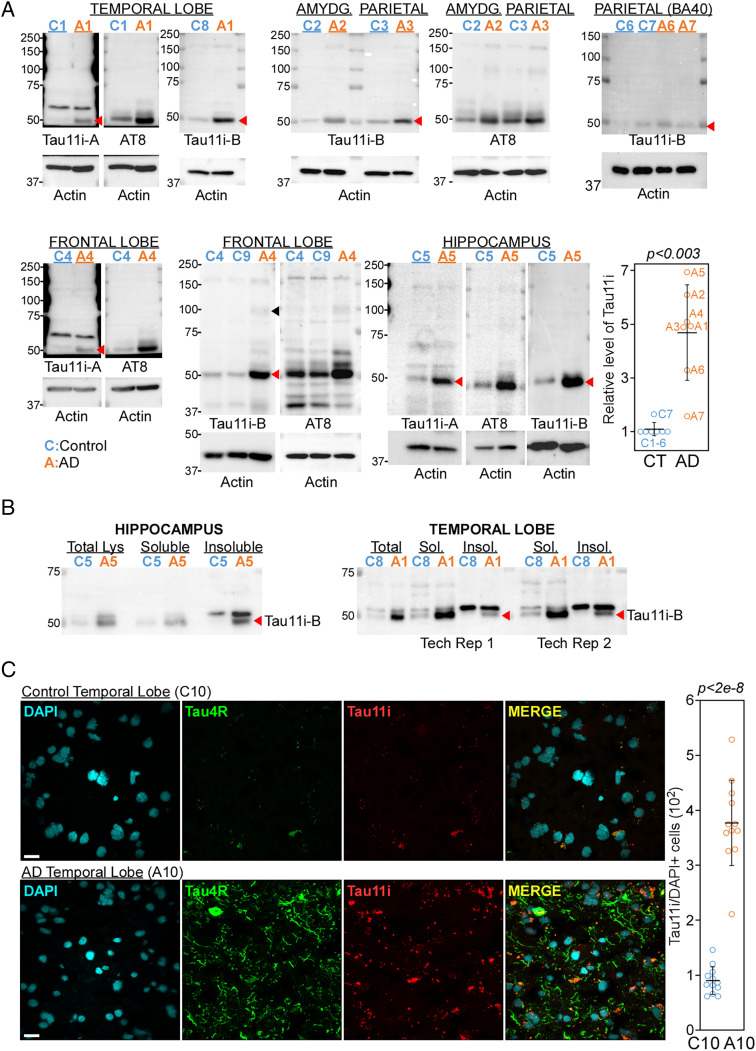
To test whether Tau mRNA transcript with IR is degraded by NMD or translated in vivo, we probed hippocampus, amygdala, parietal, frontal, and temporal lobes from nine healthy controls and seven AD patients with Tau11i and AT8 antibodies. Consistent with the increased retention of intron 11 in AD samples (Fig. 1), two different Tau11i antibodies detected significantly higher level of ∼50 kDa band, which corresponds to AT8 band (phosphor-Tau at S202/T205), in the AD samples when compared to controls (Fig. 3A, red arrowhead. P < 0.003, paired 2-tailed t test, n = 7). Although Tau11i antibodies failed to detect HMW form in NSC (Fig. 2C), we observed weak higher migrating bands of different sizes that correspond to AT8 immunoblot in AD frontal lobe lysates (Fig. 3A, black arrowhead). These results indicate that Tau mRNA with retained intron 11 escapes NMD and is translated into protein. Despite qualitative analysis with a limited sample size, the higher level of Tau11i protein in AD tissues examined corroborates with increased IR observed with transcriptomic analysis and qPCR validation. The similar migrating pattern on denaturing gel further suggests that Tau11i protein may undergo phosphorylation at serine 202 and threonine 205 (AT8) and oligomerize in AD tissues.
Fig. 3.
Elevated level of Tau11i in different AD brain tissues. (A) Immunoblot of control (blue) and AD (orange) temporal lobe, amygdala (AMYDG.), parietal lobe, frontal lobe, and hippocampus showed enrichment of Tau11i (red arrowhead) in AD samples. Samples were also immunoblotted with AT8 and actin antibodies. Each number depicts unique human subject. Quantification of seven pairs of control and AD samples with data presented as mean ± SD. The level of Tau11i in control for each brain region was set as 1. Sample quantified was underlined. The P value was calculated by paired two-tailed t test. (B) Immunoblots showed that Tau11i is found in Sarkosyl-insoluble fraction (red arrow) in AD hippocampus and temporal lobe. 10% of total lysate (Lys), 15% of soluble fraction (Sol.) and 50% of Sarkosyl-insoluble fraction (Insol.) were loaded. Tech Rep, technical replicate. (C) Immunofluorescence showed weak colocalization of Tau11i aggregates with Tau4R fibrils in AD temporal lobe. (Scale bar: 20 μm.) Each circle represents one field. Data presented as mean ± SD with P values calculated by two-tailed t test.
We next investigate the solubility of endogenous Tau11i protein by Sarkosyl extraction. Compared to AD temporal lobe, a higher portion of Tau11i protein is found to be enriched in Sarkosyl-insoluble fraction in AD hippocampus (Fig. 3B). This suggests that IR-induced C-terminal truncation may reduce the solubility of monomeric Tau11i protein in AD brain tissues. To better understand the cellular localization patterns of Tau11i in brain tissues, we stained control and AD temporal lobe sections with Tau11i and Tau4R (recognizes amino acid 275–291) (Fig. 3C) or α-tubulin (SI Appendix, Fig. S2). Consistent with immunoblot result, there was higher level of Tau11i staining in AD as compared to control temporal lobe (Fig. 3C and SI Appendix, Fig. S2). Interestingly, Tau11i appears to form granular aggregates that colocalize weakly with Tau4R fibril-like structures in AD temporal lobe, suggesting that Tau11i might contribute to pretangle assembly (28) rather than pathological neurofibrillary tangle burden (Fig. 3C).
Tau11i Has Reduced Association with Microtubule and Increased Protein Stability.
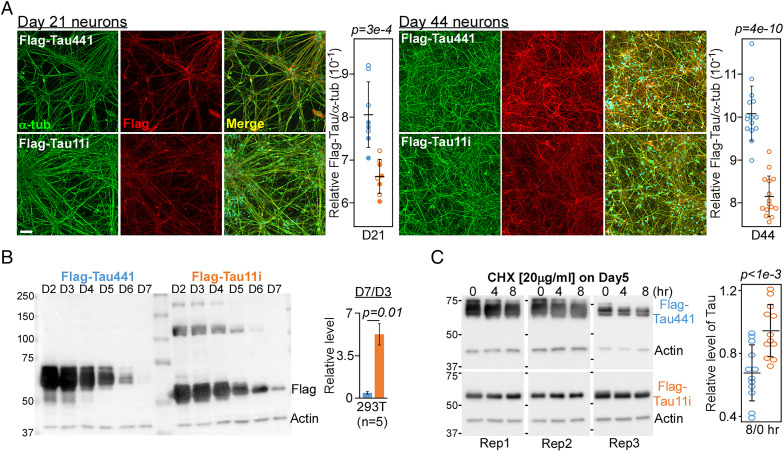
The association of Tau to microtubule can be disrupted by extensive phosphorylation and mutations (29–32). Therefore, we sought to investigate whether IR-induced protein truncation may impair its association with microtubules in neurons as well as alter its protein stability in cells. NSC expressing Flag-Tau441 and Tau11i were differentiated into cortical neurons (day 14) and cultured for additional 1 week (day 21, n = 2) or 1 month (day 44, n = 5). Immunostaining with Flag and α-tubulin antibodies showed significant reduction in the colocalization signals of Tau11i with microtubules networks in neurons as compared to Tau441 (Fig. 4A and SI Appendix, Figs. S3 and S4A), suggesting that Tau11i has lower binding efficiency to microtubules. The stability of Tau441 and Tau11i proteins was next investigated by time course monitoring and cycloheximide assay of transiently transfected Tau protein. The level of Tau11i protein on day 7 was ∼5% of that at day 3 whereas Tau441 level dropped to ∼0.4% (Fig. 4B and SI Appendix, Fig. S4B). Consistent with time course experiment, Tau11i was also more stable and exhibited less protein degradation than Tau441 following 8 h of cycloheximide treatment (Fig. 4C).
Fig. 4.
Tau11i protein associates poorly with microtubule network and is more stable. (A) Images of Day 21 and 44 neurons expressing Tau441 or Tau11i stained with α-tubulin and Flag antibodies. (Scale bar: 50 μm.) Quantification showed lower colocalization of Tau11i with α-tubulin as compared to Tau441. Each circle represents one field with filled circle from second biological replicate. Data presented as mean ± SD (B) Immunoblot using Flag and actin antibodies in 293T cells harvested on different day “D” after transfection. Quantification of Tau level (D7 relative to D3) is presented as mean ± SEM (n = 5). (C) Immunoblot of Tau level after cycloheximide (CHX) treatment of 293T cells on day 5 after transfection. Rep, replicate. Quantification of Tau level (8 h relative to 0 h) is presented as mean ± SD where each circle represents one replicate (n = 12). All P values were calculated by two-tailed t test.
Discussion
Aberrant splicing and intron retention have been linked to AD pathogenesis (4–7, 33). In this study, significant differential retention of Tau intron 11 was observed only in female AD patients as compared to healthy control in the DLPFC cohort (n > 30 for each group). Such sex-specific effect could possibly be explained by several mechanisms. Splicing of nascent RNA occurs during transcription and is intricately regulated by the interaction between transcriptional machineries and the spliceosome (34–36). This provides the framework for epigenetic features such as DNA methylation to impact RNA splicing through regulating the rate of transcription or spliceosome assembly. Consistent with this notion, binding of MeCP2 to methylated DNA has been reported to facilitate the recruitment of splicing factors which in turn inhibits aberrant IR during granulocyte differentiation (37). Interestingly, methylome profiling of different human brain tissues (38–40) and blood (41) revealed widespread sex-specific differences in DNA methylation patterns across both sex and autosomal chromosomes. Therefore, it is conceivable that sex-specific DNA methylation patterns might account for the difference in the IR patterns between female and male cohorts. In addition, sexual dimorphic gene expression observed across many different human tissues (42), which reflects differences in the rate of transcription, could also affect RNA splicing and hence IR patterns in a sex-specific manner. Further integrative analysis that incorporates DNA methylome data, differential gene expression and IR patterns from the same brain regions may provide definitive answers to this hypothesis and help address the prevalence of AD in women (43–45). Secondly, protein aggregation has been demonstrated to sequester different components of spliceosome and impact splicing efficiency in AD tissues (5, 46). As increased amyloid plaque and neurofibrillary burden was reported in female AD patients (47), it is plausible to speculate that the elevated protein aggregation might lead to more aberrant splicing or IR events in female AD brains.
The presence of premature stop codon and downstream canonical polyadenylation signal in intron 11 is likely to facilitate NMD evasion and the subsequent translation of novel truncated Tau11i protein in vivo. Characterization of Tau11i in different cell types revealed several biochemical properties that resemble pathological Tau (11, 12). Compared to full-length Tau441, truncated Tau11i oligomerizes to form HMW species, is enriched in Sarkosyl-insoluble fraction, has reduced association with microtubule networks in neurons and exhibits lower rate of protein turnover.
Analysis of different brain regions from limited number of distinct donors by qPCR (n = 3), immunoblot (five brain regions, control = 9, AD = 7) and immunofluorescence (four temporal lobe sections, n = 1) showed that Tau11i protein was significantly elevated in AD tissues as compared to healthy control. In AD hippocampus, endogenous Tau11i is also observed in Sarkosyl-insoluble fraction, consistent with its association with protein aggregates. The formation of Tau11i aggregates in AD temporal lobe that colocalize infrequently with Tau4R fibril-like structures suggests that Tau11i might not be deposited into neurofibrillary tangles. Recent studies showed that dysregulation of RNA splicing and intron retention is a hallmark of AD pathogenesis, suggesting that Tau11i might function downstream of these aberrant events and potentially serves as an early biomarker during AD progression. Follow-up studies using different brain regions from larger AD cohorts will provide correlation of Tau11i level with the different Braak stages of AD progression and insight to its tissue-specific effects.
Materials and Methods
High-Throughput Analysis of Intron Retention.
DLPFC datasets of healthy control subjects and AD patients (syn21088596) (16) in the Religious Orders Study/Memory and Aging Project (ROSMAP) were downloaded from AD Knowledge Portal. RNA-sequencing reads were first mapped against human reference genome (GRCh38.100) and IR was quantified by IRFinder pipeline using default setting (17). The IR ratio was calculated by dividing intronic abundance by the sum of intronic and exonic abundance. Intron 11 of the Tau (MAPT) gene (MAPT/ENSG00000186868/clean/17:46014324-46018617: +) investigated in this study has a minimal splicing sequencing depth of greater than five reads. Generalized linear model from DESeq2 (48) was applied to measure the differential IR ratio between control subjects and AD patients, where P value <0.05 was considered significant. Normalized IR ratio from individual human subject determined from DESeq2 was used to generate dot plot with “ggplot” function in “ggplot2” package (https://ggplot2.tidyverse.org). Counts for Tau gene were generated using featureCounts (v.2.0.1.) and differential expression was calculated by DESeq2 (v.1.30.1).
Cell Lines and Human Brain Tissues.
Human NSC (XCL-1, SC-001–1V) was purchased from XCell Science Inc. and differentiated according to manufacturer’s protocol. Brain total RNA, protein lysates and frozen tissue sections were purchased from MYBIOSOURCE and BIOCHAIN. Parietal lobe (BA40) samples C6, C7, A6, and A7 were previously described (49). Donors’ information is provided in SI Appendix.
Customized Tau11i Antibodies.
Two Tau11i antibodies were generated by GENSCRIPT using different amino acids peptides as antigens where C is the conjugated cysteine.
Tau11i-A: HKPGSPVEGEGWDGRVQGV-C.
Tau11i-B: C-SPVEGEGWDGRVQG.
Sarkosyl Extraction.
Sarkosyl-soluble and insoluble fractions were prepared as described (27).
Immunofluorescence.
Immunostaining of neurons and image quantification were performed as previously described (50). Images were captured with confocal microscope (FV3000) and quantified by Imaris software (Bitplane). Detailed procedures are described in the SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by Temasek Core Funding (3160). Post-mortem parietal lobe (BA40) of control and neuropathologically diagnosed AD subjects were obtained from the Newcastle Brain Tissue Resource, part of the Brains for Dementia Research network.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204179119/-/DCSupplemental.
Data, Materials, and Software Availability
Individual normalized IR ratio is available in SI Appendix, Dataset 01. All study data are included in the article and/or SI Appendix.
References
- 1.Sims R., Hill M., Williams J., The multiplex model of the genetics of Alzheimer’s disease. Nat. Neurosci. 23, 311–322 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Knopman D. S., et al. , Alzheimer disease. Nat. Rev. Dis. Primers 7, 33 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith A. R., Wheildon G., Lunnon K., Invited review—A 5-year update on epigenome-wide association studies of DNA modifications in Alzheimer’s disease: Progress, practicalities and promise. Neuropathol. Appl. Neurobiol. 46, 641–653 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Raj T., et al. , Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat. Genet. 50, 1584–1592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai B., et al. , U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 110, 16562–16567 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tollervey J. R., et al. , Analysis of alternative splicing associated with aging and neurodegeneration in the human brain. Genome Res. 21, 1572–1582 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adusumalli S., Ngian Z. K., Lin W. Q., Benoukraf T., Ong C. T., Increased intron retention is a post-transcriptional signature associated with progressive aging and Alzheimer’s disease. Aging Cell 18, e12928 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.García-Escudero V., et al. , A new non-aggregative splicing isoform of human Tau is decreased in Alzheimer’s disease. Acta Neuropathol. 142, 159–177 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trabzuni D., et al. , MAPT expression and splicing is differentially regulated by brain region: Relation to genotype and implication for tauopathies. Hum. Mol. Genet. 21, 4094–4103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnsten A. F. T., Datta D., Del Tredici K., Braak H., Hypothesis: Tau pathology is an initiating factor in sporadic Alzheimer’s disease. Alzheimers Dement. 17, 115–124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Götz J., Halliday G., Nisbet R. M., Molecular pathogenesis of the tauopathies. Annu. Rev. Pathol. 14, 239–261 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Iqbal K., Liu F., Gong C. X., Tau and neurodegenerative disease: The story so far. Nat. Rev. Neurol. 12, 15–27 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Palmqvist S., et al. ; Alzheimer’s Disease Neuroimaging Initiative, Prediction of future Alzheimer’s disease dementia using plasma phospho-tau combined with other accessible measures. Nat. Med. 27, 1034–1042 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Barthélemy N. R., et al. ; Dominantly Inherited Alzheimer Network, A soluble phosphorylated tau signature links tau, amyloid and the evolution of stages of dominantly inherited Alzheimer’s disease. Nat. Med. 26, 398–407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chong J. R., et al. , Plasma P-tau181 to Aβ42 ratio is associated with brain amyloid burden and hippocampal atrophy in an Asian cohort of Alzheimer’s disease patients with concomitant cerebrovascular disease. Alzheimers Dement. 17, 1649–1662 (2021). [DOI] [PubMed] [Google Scholar]
- 16.De Jager P. L., et al. , A multi-omic atlas of the human frontal cortex for aging and Alzheimer’s disease research. Sci. Data 5, 180142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Middleton R., et al. , IRFinder: Assessing the impact of intron retention on mammalian gene expression. Genome Biol. 18, 51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumitriu A., et al. , Integrative analyses of proteomics and RNA transcriptomics implicate mitochondrial processes, protein folding pathways and GWAS loci in Parkinson disease. BMC Med. Genomics 9, 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen M., et al. , Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci. Data 3, 160089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goedert M., Spillantini M. G., Jakes R., Rutherford D., Crowther R. A., Multiple isoforms of human microtubule-associated protein tau: Sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron 3, 519–526 (1989). [DOI] [PubMed] [Google Scholar]
- 21.Silva A. L., Ribeiro P., Inácio A., Liebhaber S. A., Romão L., Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA 14, 563–576 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martins R., et al. , Alternative polyadenylation and nonsense-mediated decay coordinately regulate the human HFE mRNA levels. PLoS One 7, e35461 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abraha A., et al. , C-terminal inhibition of tau assembly in vitro and in Alzheimer’s disease. J. Cell Sci. 113, 3737–3745 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Gu J., et al. , Truncation of Tau selectively facilitates its pathological activities. J. Biol. Chem. 295, 13812–13828 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y. P., Biernat J., Pickhardt M., Mandelkow E., Mandelkow E. M., Stepwise proteolysis liberates tau fragments that nucleate the Alzheimer-like aggregation of full-length tau in a neuronal cell model. Proc. Natl. Acad. Sci. U.S.A. 104, 10252–10257 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y., et al. , Relevance of phosphorylation and truncation of tau to the etiopathogenesis of Alzheimer’s Disease. Front. Aging Neurosci. 10, 27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diner I., Nguyen T., Seyfried N. T., Enrichment of detergent-insoluble protein aggregates from human postmortem brain. J. Vis. Exp. (128): (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moloney C. M., Lowe V. J., Murray M. E., Visualization of neurofibrillary tangle maturity in Alzheimer’s disease: A clinicopathologic perspective for biomarker research. Alzheimers Dement. 17, 1554–1574 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alonso A. D., Grundke-Iqbal I., Barra H. S., Iqbal K., Abnormal phosphorylation of tau and the mechanism of Alzheimer neurofibrillary degeneration: Sequestration of microtubule-associated proteins 1 and 2 and the disassembly of microtubules by the abnormal tau. Proc. Natl. Acad. Sci. U.S.A. 94, 298–303 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alonso A. D., et al. , Interaction of tau isoforms with Alzheimer’s disease abnormally hyperphosphorylated tau and in vitro phosphorylation into the disease-like protein. J. Biol. Chem. 276, 37967–37973 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Alonso A. C., Grundke-Iqbal I., Iqbal K., Alzheimer’s disease hyperphosphorylated tau sequesters normal tau into tangles of filaments and disassembles microtubules. Nat. Med. 2, 783–787 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Hasegawa M., Smith M. J., Goedert M., Tau proteins with FTDP-17 mutations have a reduced ability to promote microtubule assembly. FEBS Lett. 437, 207–210 (1998). [DOI] [PubMed] [Google Scholar]
- 33.Li H. D., et al. , Integrative functional genomic analysis of intron retention in human and mouse brain with Alzheimer’s disease. Alzheimers Dement. 17, 984–1004 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herzel L., Ottoz D. S. M., Alpert T., Neugebauer K. M., Splicing and transcription touch base: Co-transcriptional spliceosome assembly and function. Nat. Rev. Mol. Cell Biol. 18, 637–650 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gehring N. H., Roignant J. Y., Anything but ordinary - emerging splicing mechanisms in eukaryotic gene regulation. Trends Genet. 37, 355–372 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Ong C. T., Adusumalli S., Increased intron retention is linked to Alzheimer’s disease. Neural Regen. Res. 15, 259–260 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong J. J., et al. , Intron retention is regulated by altered MeCP2-mediated splicing factor recruitment. Nat. Commun. 8, 15134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H., et al. , Sex-biased methylome and transcriptome in human prefrontal cortex. Hum. Mol. Genet. 23, 1260–1270 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gross J. A., et al. , Characterizing 5-hydroxymethylcytosine in human prefrontal cortex at single base resolution. BMC Genomics 16, 672 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lister R., et al. , Global epigenomic reconfiguration during mammalian brain development. Science 341, 1237905 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solomon O., et al. , Meta-analysis of epigenome-wide association studies in newborns and children show widespread sex differences in blood DNA methylation. Mutat. Res. Rev. Mutat. Res. 789, 108415 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliva M., et al. ; GTEx Consortium, The impact of sex on gene expression across human tissues. Science 369, eaba3066 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisher D. W., Bennett D. A., Dong H., Sexual dimorphism in predisposition to Alzheimer’s disease. Neurobiol. Aging 70, 308–324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medeiros A. M., Silva R. H., Sex differences in Alzheimer’s disease: Where do we stand? J. Alzheimers Dis. 67, 35–60 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Toro C. A., Zhang L., Cao J., Cai D., Sex differences in Alzheimer’s disease: Understanding the molecular impact. Brain Res. 1719, 194–207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsieh Y. C., et al. , Tau-mediated disruption of the spliceosome triggers cryptic RNA splicing and neurodegeneration in Alzheimer’s disease. Cell Rep. 29, 301–316.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barnes L. L., et al. , Sex differences in the clinical manifestations of Alzheimer disease pathology. Arch. Gen. Psychiatry 62, 685–691 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chua X. Y., et al. , Elevation of inactive cleaved annexin A1 in the neocortex is associated with amyloid, inflammatory and apoptotic markers in neurodegenerative dementias. Neurochem. Int. 152, 105251 (2022). [DOI] [PubMed] [Google Scholar]
- 50.Rao V. K., et al. , Phosphorylation of Tet3 by cdk5 is critical for robust activation of BRN2 during neuronal differentiation. Nucleic Acids Res. 48, 1225–1238 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Individual normalized IR ratio is available in SI Appendix, Dataset 01. All study data are included in the article and/or SI Appendix.