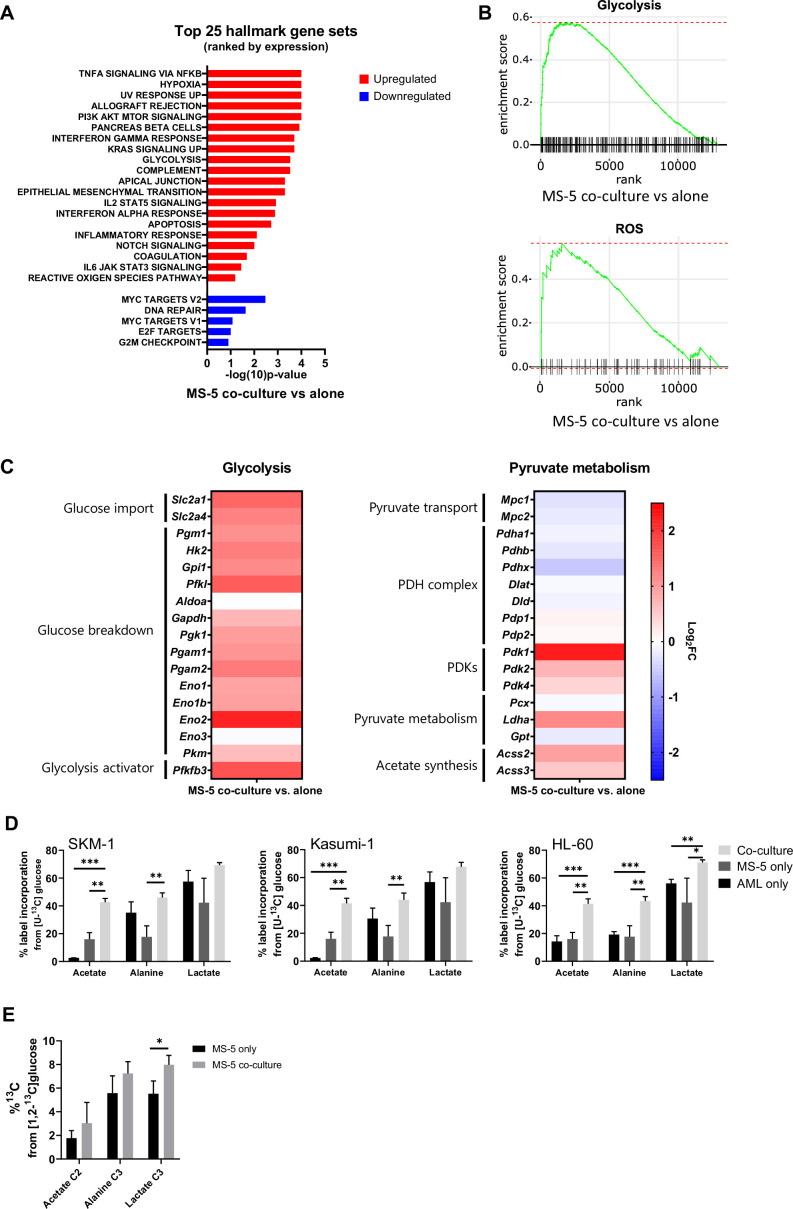
Figure 3. Transcriptomic data and [U-13C]glucose labelling reveal that stromal cell metabolism is shifted towards higher glycolysis and ROS upon co-culture with AML cells.
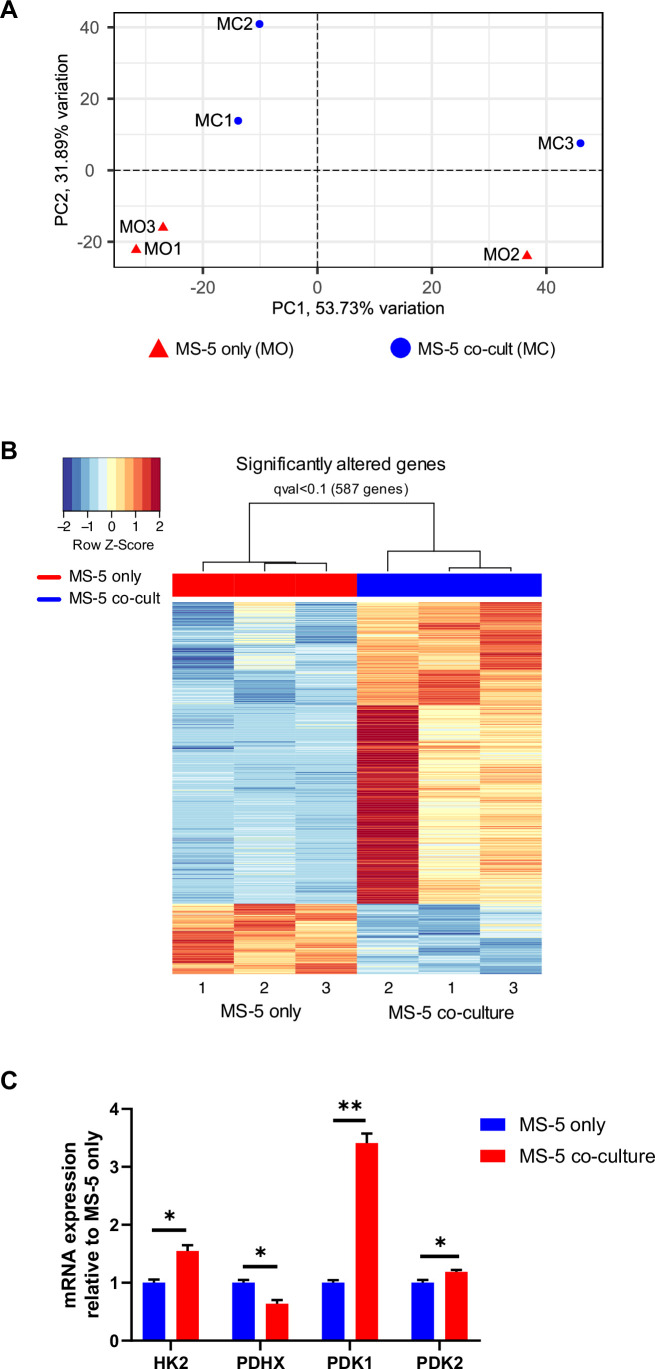
(A) Top 25 GSEA hallmark gene sets ranked by expression in MS-5 cells only vs co-culture with SKM-1 cells, analysed using the collection of hallmark gene sets from Molecular Signature Database with a false discovery rate threshold at 5%. p-Values for each pathway are presented as –log(10)p-value. (B) GSEA enrichment score plots of glycolysis and ROS generated using Sleuth 0.30.0 R statistical package. (C) Fold change values of detected gene transcripts (TPMs) related to glycolysis and pyruvate metabolism. FC values are represented as log2FC, red values indicate upregulation and blue values indicate downregulation in MS-5 cells in co-culture. (D) Label incorporation from [U-13C]glucose into extracellular metabolites in AML and MS-5 cells cultured alone or in co-culture after 24 hr. Bars represent the mean of n=3 independent experiments and error bars represent standard deviations. (E), Label incorporation from [U-13C]glucose into intracellular acetate, alanine and lactate in MS-5 cells cultured alone or in co-culture after 24 hr. Bars represent the mean of n=3 independent experiments and error bars represent standard deviations. p-Values are represented by n.s. for not significant * for p-value <0.05, ** for p-value <0.01 and *** for p-value <0.001.