Abstract
The UGA codon, which usually acts as a stop codon, can also direct the incorporation into a protein of the amino acid selenocysteine. This UGA decoding process requires a cis-acting mRNA element called the selenocysteine insertion sequence (SECIS), which can form a stem-loop structure. In Escherichia coli, selenocysteine incorporation requires only the 17-nucleotide-long upper stem-loop structure of the fdhF SECIS. This structure carries a bulged nucleotide U at position 17. Here we asked whether the single bulged nucleotide located in the upper stem-loop structure of the E. coli fdhF SECIS is involved in the in vivo interaction with SelB. We used a genetic approach, generating and characterizing selB mutations that suppress mutations of the bulged nucleotide in the SECIS. All the selB suppressor mutations isolated were clustered in a region corresponding to 28 amino acids in the SelB C-terminal subdomain 4b. These selB suppressor mutations were also found to suppress mutations in either the loop or the upper stem of the E. coli SECIS. Thus, the E. coli SECIS upper stem-loop structure can be considered a “single suppressible unit,” suggesting that there is some flexibility to the nature of the interaction between this element and SelB.
The UGA codon, which usually acts as a stop codon, can also direct incorporation of the amino acid selenocysteine (for reviews, see references 4, 5, 7, and 25). This UGA decoding process requires a cis-acting mRNA element called the selenocysteine insertion sequence (SECIS), which can form a stem-loop structure (4, 9, 15; for reviews, see references 2 and 20). In Escherichia coli, a number of genes have been identified in which the UGA directs the incorporation of selenocysteine. These include genes fdhF (27) and fdnG (3), encoding the selenocysteine-containing enzymes formate dehydrogenase H and N, respectively. Immediately downstream from the selenocysteine-specifying UGA in the mRNA of each of these polypeptides is found a SECIS that has been described as consisting of at least 40 nucleotides capable of forming a stem-loop RNA structure (2, 9). In later work, it was suggested that an extended fdhF SECIS was required, consisting of an additional helix of 7 bp in which the U and G residues of the UGA codon are included and the A residue is bulged out (10). After carrying out an extensive mutational analysis of the fdhF SECIS DNA, we found that for in vivo UGA-directed selenocysteine incorporation, there is no requirement for the whole stem-loop RNA structure of the E. coli fdhF SECIS (including the extended form) (18), as thought previously. Instead, the 17-bp upper stem-loop structure is sufficient to permit selenocysteine incorporation on the condition that it is located 11 nucleotides downstream from the UGA codon (Fig. 1). This mini upper stem-loop structure contains a bulged nucleotide, a U residue, located 17 nucleotides downstream from the UGA (Fig. 1). Selenocysteine incorporation into an fdhF-lacZ′ fusion polypeptide depends on both (i) the specificity of nucleotide 17 as a U residue and (ii) its presence as a bulged nucleotide (18). The importance of the bulged U17 has also been shown using the SELEX procedure (12).
FIG. 1.
Minimal E. coli fdhF SECIS required for SelB binding and selenocysteine incorporation. The upper stem-loop structure (boxed) is the minimal region required for SelB binding (13). UGA-directed selenocysteine incorporation requires that this structure be located 11 nucleotides (nt) from the UGA codon (bold) (18). The U residue at position 17 is bulged (12, 18). The pairing of boxes C20 and G27 is questionable (18) and is therefore designated by a dot instead of by a dash.
The UGA-directed selenocysteine incorporation into a polypeptide in E. coli also requires a number of trans elements. These include the selC-specified tRNASec (16), which is a specialized tRNA that contains a UCA anticodon, and also a special protein elongation factor (EF) called SelB (reviewed in reference 2). SelB binds both GTP and selenocysteyl-tRNASec and also binds the mRNA stem-loop structure formed by fdhF SECIS mRNA (1, 8, 9, 11, 23). In vitro experiments (13) have shown that selenocysteyl-tRNASec binds the N-terminal part of SelB (homologous to EF-Tu) and that the C-terminal subdomain of SelB binds the fdhF SECIS. Furthermore, the efficiency of SelB binding is not reduced when the mRNA motif is reduced to a 17-nucleotide-long minihelix. That minihelix is the same 17-bp upper stem-loop structure that we have shown to be the minimal requirement for in vivo selenocysteine incorporation into a polypeptide (Fig. 1) (18). Recently, this interaction between SelB and the loop of the fdhF SECIS upper minihelix has also been demonstrated by genetic analysis (14).
Here we asked whether the single bulged nucleotide in the upper minihelix of the E. coli fdhF SECIS is involved in the in vivo interaction with SelB. We used a genetic approach in which we generated and characterized selB mutations that suppress mutations in the bulged nucleotide. All the selB mutations that we isolated were clustered in a region corresponding to 28 amino acids in the C-terminal 4b subdomain of SelB (31 amino acids before the end of the protein). Our results further support the importance of the bulged nucleotide (U17) of the upper stem-loop of E. coli SECIS in the interaction of the SECIS with the SelB elongation factor.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli strains and plasmids used in this study are described in Table 1.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description or relevant genotype | Source or reference |
|---|---|---|
| E. coli strain | ||
| MC4100 | araD139 Δ(argF-lac)205 flbB3501 ptsF25 rpsL150 deoC1 relA1 | 6 |
| WL81300 | MC4100 Δ(selB300::Kmr), Δ(srl-recA)306::Tn10 | 26; A. Böck |
| RM1 | ΔselC derivative of MC4100 | 22 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE relA1 lac [F′ proAB lacIqZΔM15 Tn10(Tetr)] | Stratagene |
| Plasmids | ||
| pRM4 | pBR322 derivative (Ampr) carrying the fused genes λcI′-lac′I"Z into which the TGA region of the SECIS DNA (from −9 to +47) of E. coli fdhF gene has been inserted at the junction of λcI′-lac′I"Z | 22 |
| pZL38 | pRM4 derivative in which bulged nucleotide U18 was changed to A18 | 18 |
| pZL42 | pRM4 derivative in which bulged nucleotide U17 was changed to A17 | 18 |
| pZL44 | pRM4 derivative in which bulged nucleotide U17 was changed to A17 and A29 to U29 | 18 |
| pZL70 | pRM4 derivative in which bulged nucleotide U17 was changed to C17 | 18 |
| pL24A | pRM4 derivative in which nucleotide U24 in loop was changed to A24 | This work |
| pLC1 | pACYC184 derivative (Cmr) in which selB was cloned into the HindIII and BamHI sites of the tetracycline resistance gene | This work |
| pLC2* | Pool of pLC1 derivatives carrying PCR-generated random mutations in the C-terminal part of selB | This work |
Media.
Bacteria were grown in liquid or solid Luria-Bertani (LB) medium, M9 minimal medium supplemented with a mixture of amino acids, each at a final concentration of 20 μg/ml (21), or solid LB medium with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, 40 μg/ml). Ampicillin (100 μg/ml) or chloramphenicol (35 μg/ml) was added to the media in which the plasmid-carrying strains were grown. Sodium selenite was obtained from Sigma Chemical Co. (St. Louis, Mo.).
Molecular cloning.
All recombinant DNA manipulations were carried out by standard procedures (24). Site-directed mutagenesis was carried out as we have described previously (18, 19). Restriction enzymes and other enzymes used in the recombinant DNA experiments were obtained from New England Biolabs (Beverly, Mass.). DNA sequencing was done using an automated fluorescence DNA 373 sequencer.
Bacterial growth, transformations, and measurements of β-galactosidase activity.
E. coli cells were transformed (24) by the plasmid of choice. Single colonies of freshly transformed cells were grown on LB plates at 37°C overnight and then in M9 liquid medium at 37°C in a rolling drum for 8 to 10 h until the cultures reached an optical density at 600 nm of 0.7 to 1.0. β-Galactosidase activity was determined as we have described previously (18).
Generation of mutations in E. coli selB.
Plasmid pLC1(Cmr) was constructed by cloning the 1,867-bp PCR fragment of the whole selB gene from E. coli strain MC4100 into the HindIII and BamHI sites in the tetracycline resistance gene of pACYC184 (Table 1). We used the PCR-based random mutagenesis technique in a reaction mixture including 3.5 mM MgCl2 to introduce random point mutations in the C-terminal part of selB (17). The C-terminal part includes the last 872 bp of selB from the EcoRV site until immediately after the termination codon, corresponding to the end of SelB subdomain 3 and the whole of SelB subdomains 4a and 4b (13). We replaced the last 872 bp of the selB in pLC1 with the PCR library-generated mutations in selB. We used these ligation mixtures to transform the XL1-Blue strain and picked 4,000 colonies, which we then divided into 40 groups. Each group was grown in 2 ml of LB medium containing chloramphenicol (35 μg/ml) for 6 h. The DNA was extracted from plasmids pLC2*1 to pLC2*40 bearing the pool of mutated selB alleles (Table 1) and used as described below.
Selecting mutations in selB that can suppress mutations in the bulged nucleotide U17 in E. coli fdhF SECIS.
In previous studies, we generated mutations in the bulged nucleotide U17 in the fdhF-lacZ fusions of the Ampr plasmid pRM4 (18). They are on plasmids pZL42 and pZL70 (Table 1 and Fig. 2). The incorporation of selenocysteine was prevented in MC4100 harboring each one of these fusions (18), and the colonies appeared as Lac− on X-Gal plates. Here, we first used each of these plasmids separately to transform strain WL81300, a ΔselB derivative of MC4100 (Table 1). Then we transformed the Ampr transformants using plasmid DNA prepared from each group (1 to 40) of the pLC2* (Cmr) plasmid that carried random mutations in the C-terminal part of selB. Treated cells were plated on LB agar containing ampicillin (100 μg/ml), chloramphenicol (35 μg/ml), X-Gal (40 μg/ml), and 10−6 M sodium selenite. Of the Ampr Cmr Lac+ transformants, we selected one colony from each plate. We further confirmed the Lac+ phenotype by measuring β-galactosidase activity. The Lac+ colonies contain two types of plasmids, one carrying the fdhF-lacZ fusion mutations in bulged U17 (Ampr) and the second carrying suppresser mutations in selB (Cmr). To isolate the plasmid that was Cmr Amps, we used the plasmid DNA that was extracted from the doubly transformed cells to transform MC4100 cells and selected Cmr Amps Lac− colonies. The final characterization of suppressor mutations in selB was done by DNA sequencing.
FIG. 2.
Location of the mutations in the upper stem and loop of the E. coli fdhF SECIS on the plasmids used in this study. The mutated nucleotides are boxed. In pZL44, U18 is bulged (circle). The numbers represent the distance of the nucleotide from the UGA codon of E. coli fdhF SECIS.
RESULTS
Characterizing selB mutations that can suppress mutations in the bulged nucleotide at position 17 of the E. coli fdhF SECIS.
For this work we used our plasmid pRM4 (22), which we constructed previously to carry the TGA codon context of the E. coli fdhF gene fused to lac′Z (lacZ lacking the first eight codons). Selenocysteine incorporation into the gene product of the lac′Z fusion was studied in two ways: (i) qualitatively by the appearance of Lac+ colonies on X-Gal plates, and (ii) quantitatively by measuring the UGA-directed (selC-dependent) β-galactosidase activity.
To select mutations in selB that can suppress mutations in the bulged nucleotide at position 17, two constructs were used. In plasmid pZL70, the nucleotide U17 was changed to C, and in pZL42 U17 was changed to A (Table 1 and Fig. 2). We prepared plasmid DNA from each of the 40 groups of pCL2* (Cmr) plasmids that carried random mutations corresponding to the 4a and 4b subdomains of SelB (see Materials and Methods). We used the DNA of each plasmid separately to doubly transform cells that already contained either pZL70 or pZL42. The transformation procedure using pZL70 resulted in 10 Lac+ transformants that were called p70bm[1] to p70bm[10]. Similarly for the other plasmid, pZL42, seven transformants were found from which the plasmids were isolated that were designated p42bm[1] to p42bm[7]. In each case bm stands for bulged mutated. We examined the suppression efficiency of these selB mutations quantitatively by measuring the level of UGA-directed (selC-dependent) β-galactosidase activity in cells cotransformed by each one of the plasmid groups p70bm[1–10] and p42bm[1–7]. The level of enzymatic activity driven by the unmutated (wild-type) selB plasmid pLC1 was only 1%. The presence of the selB suppressor mutations that we isolated led to an increase in β-galactosidase activity in the range of 7 to 29% for series p70bm[1–10] and 12 to 40% for series p42bm[1–7] (Table 2).
TABLE 2.
selB mutationsa
| Plasmid used for screening | Plasmid carrying wt or mutated selBb | Nucleotide(s) changed in selBc | Amino acid(s) changed in SelBd | Relative β-galactosidase activitye (% of control) |
|---|---|---|---|---|
| pZL70 | pLC1 | None (wt) | None (wt) | 1 |
| p70bm[1] | GCA→aCA(1747) | Ala→Thr(583) | 7 | |
| p70bm[2] | ATG→ATa(1668) | Met→Ile(556) | 21 | |
| p70bm[3] | ATC→tTC(1669) | Ile→Phe(557) | 17 | |
| p70bm[4] | GAG→GgG(1559) | Glu→Gly(520) | 10 | |
| CTC→CcC(1679) | Leu→Pro(560) | |||
| p70bm[5] | GTA→GaA(1733) | Val→Glu(578) | 18 | |
| CTG→CcG(1574) | Leu→Pro(525) | |||
| p70bm[6] | TGC→cGC(1702) | Cys→Arg(568) | 25 | |
| p70bm[7] | GTA→GaA(1733) | Val→Glu(578) | 29 | |
| p70bm[8] | GAC→GtC(1820) | Aap→Val(607) | 11 | |
| ATG→gTG(1666) | Met→Val(556) | |||
| p70bm[9] | TTC→TaC(1715) | Phe→Tyr(572) | 16 | |
| p70bm[10] | TGC→TtC(1703) | Cys→Phe(568) | 16 | |
| GCC→tCC(1249) | Ala→Ser(417) | |||
| pZL42 | pLC1 | None (wt) | None (wt) | 1 |
| p42bm[1] | ATG→ATa(1668) | Met→Ile(556) | 22 | |
| p42bm[2] | GTA→GaA(1733) | Val→Glu(578) | 15 | |
| CTG→CcG(1574) | Leu→Pro(525) | |||
| p42bm[3] | GAT→GAa(1683) | Asp→Glu(561) | 17 | |
| GAC→GtC(1532) | Asp→Val(511) | |||
| p42bm[4] | GTA→GcA(1733) | Val→Ala(578) | 40 | |
| p42bm[5] | ATG→tTC(1669) | Ile→Phe(557) | 14 | |
| p42bm[6] | TGC→cGC(1702) | Cys→Arg(568) | 23 | |
| p42bm[7] | TGC→TtC(1703) | Cys→Phe(568) | 12 | |
| GCC→tCC(1249) | Ala→Ser(417) |
Strain E. coli WL81300 (a ΔselB derivative of MC4100) harboring either pZL70 or pZL42 was freshly transformed by one of each group of plasmids pLC*1–40. Lac+ Ampr Cmr transformants were selected, and β-galactosidase activity was determined as described in Materials and Methods. The level of β-galactosidase activity was derived after subtracting the β-galactosidase values of RM1 cells (a ΔselC derivative of MC4100) transformed by pRM4, pZL70, or pZL42. The level of β-galactosidase activity in WL81300 cells cotransformed by pRM4 and pLC1 (a pACYC184 derivative carrying the selB gene) was designated as 100% activity. wt, wild type.
E. coli WL81300 carrying either pZL70 or pZL42 was freshly transformed by plasmids pLC2*1–40. Lac+ Ampr Cmr cells carrying plasmids p70bm[1–10] or p42bm[1–7] were selected. Plasmids carrying a single nucleotide change in selB are marked in bold letters.
The numbers in brackets represent the position of the changed nucleotide determined by DNA sequencing. Lowercase letters indicate the changed nucleotide.
The numbers in brackets represent the position of the changed amino acid determined according to the DNA sequence.
The β-galactosidase activity directed by the gene fusions in plasmids pZL70 and pZL42 is given relative to that of pRM4. The numbers represent the average of the results of at least three experiments and were derived after subtracting the level of β-galactosidase activity in the ΔselC derivative RM1 from that in MC4100.
We separated suppressor plasmids bearing the putative selB mutations from the reporter plasmids, p70bm[1–10] from pZL70 and p42bm[1–7] from pZL42 (Materials and Methods). Subsequently, the nature of the mutations in selB of each plasmid was characterized by DNA sequencing. We found that p70bm[1], p70bm[2], p70bm[3], p70bm[6], and p70bm[7] each carry a single mutation in selB, while p70bm[4], p70bm[5], p70bm[8], and p70bm[10] each carry a double mutation. However, one of the two mutations in the selB of p70bm[5] is identical to the single mutation in selB of p70bm[7]. In addition, one of the two mutations in p70bm[8] and p70bm[10] causes a change in the amino acid located in the same place as the single mutations in p70bm[2] and p70bm[6], respectively. In all, we isolated six single mutations in selB that suppressed bulged nucleotide 17 when it was mutated from U to C. We found that each of these single selB mutations was located in a separate specific site of the gene, in the region of nucleotides 1668 to 1747 (Table 2). The corresponding changes in the selB amino acids spanned amino acids 556 to 583 (Fig. 3). Thus, we found that changes in a 28-amino-acid-long region of SelB can suppress the effects of the change from U to C in bulged nucleotide 17. At least one change within this region was found even in the double mutations in selB that was obtained in p70bm[5], p70bm[7], and p70bm[10] (Table 2).
FIG. 3.
Schematic representation of the region in SelB in which the mutations are clustered that suppress the mutated U17 bulged nucleotide in E. coli fdhF SECIS. selB DNA corresponding to SelB domains 4a (amino acids [aa] 343 to 474) and 4b (amino acids 472 to 614) (13) (dotted region) was subjected to PCR random mutagenesis. Selection was made for mutations in selB that could suppress mutations in the SECIS U17 bulged nucleotide; these mutations were subsequently sequenced (Table 2 and Materials and Methods). The selected single mutations are located in 28 amino acids of the 4b region (between 556 and 583 of SelB) (shaded rectangle). Mutated amino acids are underlined.
When we studied plasmids p42bm[1–7] that carried selB mutations that suppressed mutations in the bulged nucleotide from U17 to A17, four single mutations in selB were isolated (Table 2). It is particularly interesting that these mutations are each identical to one of the single selB mutations obtained by selection with pZL70, in which the bulged nucleotide was mutated from U17 to C17: p42bm[1] is identical to p70bm[2], p42bm[4] is identical to p70bm[7], p42bm[5] is identical to p70bm[3], and p42bm[6] is identical to p70bm[6]. Thus, selecting with either pZL70 or pZL42 produced mutations located in a region of 28 amino acids of selB that can suppress bulged nucleotide U17 (Fig. 3).
selB mutations selected by the suppression of mutations in bulged nucleotide U17 of E. coli SECIS also suppress other mutations in the SECIS upper stem-and-loop structure.
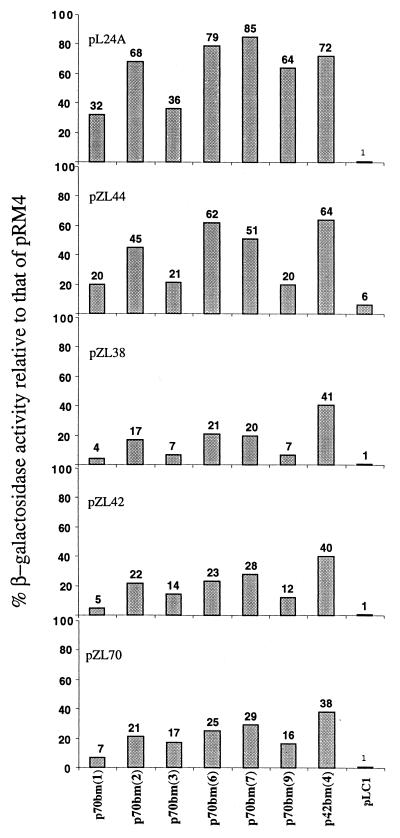
Here we asked whether the selB mutations that were selected by their ability to suppress the bulged nucleotide U17 of SECIS can also suppress other mutations in this cis element. To answer this question, we used the following plasmids (Fig. 2): (i) in pL24A the U24 in the loop was changed to A24; (ii) in pZL44 the wild-type U17 bulged nucleotide was mutated to A17 so that it paired with a U29 that was changed from A29, and the bulged U17 nucleotide was replaced with a bulged U18 of the wild type; and (iii) in pZL38 we changed U18 to A18 so that A18 became the bulged nucleotide. The level of suppression of the selB mutations (selected with U17 mutated bulged nucleotide) of these three mutations in the upper stem-loop structure is shown in Fig. 4. It is noteworthy that mutations in selB that were selected to suppress a mutation in U17 could also suppress mutations in other locations of the SECIS. The mutation in the SECIS loop (pL24A) caused the highest level of suppression. The mutation in the SECIS that generated the bulged U18 (pZL44) caused an intermediate level of suppression, and the mutation in which U18 was changed to generate a bulged A18 (pZL38) caused the lowest level of suppression. The last level of suppression was similar to the level of suppression caused by the two original mutations, U17 to C17 (pZL70) and U17 to A17 (pZL42), which we used to select the original selB mutations. The latter case is similar to the level of suppression of a mutation in bulged U17 to which the selB mutations were originally selected.
FIG. 4.
Selected mutations in selB suppress mutations in different positions of the upper stem-loop of E. coli SECIS. E. coli WL83100 cells were double transformed, first by each of plasmids pL24A, pZL44, pZL38, pZL42, and pZL70 and then by each of plasmids p70bm[1], p70bm[2], p70bm[3], p70bm[6], p70bm[7], p70bm[9], p42bm[4], or the wild-type pLC1. The level of suppression (represented by the number above the columns) was determined by the level of β-galactosidase as described in Table 2, footnote e.
DISCUSSION
Among the functions of the special elongation factor SelB is that it binds to the E. coli mRNA at the SECIS (9, 23). The results of in vitro experiments have shown that subdomain 4b of the SelB C terminus binds to the upper stem-loop structure of the SECIS (13). Previously, we showed that this minihelix is the same 17-bp upper stem-loop structure that we showed is the minimal requirement for in vivo selenocysteine incorporation into a polypeptide (18). This minimal SECIS has a bulged nucleotide that has been shown to be crucial for selenocysteine incorporation (12, 18).
Here, we used a genetic analysis to examine whether this single bulged U17 nucleotide is also involved in the in vivo interaction with SelB. We used PCR-based random mutagenesis to generate point mutations in selB. These mutations were characterized for their ability to suppress mutations in the bulged nucleotide U17 when it was borne on plasmid pZL42 or pZL70 (Table 1 and Fig. 2). DNA sequencing was used to determine the exact position of the nucleotide of the suppressing mutation in selB. We found that the changed amino acids in the selB suppressor mutants were clustered in a region corresponding to 28 amino acids in the C-terminal subdomain 4b of the SelB protein, the last of which is located 31 amino acids before the end of the protein (Fig. 3).
Kromayer and colleagues (14) obtained similar results using a different genetic approach. Rather than seeking suppressors of mutations in the bulged nucleotide as we did, they isolated selB mutations that suppress defined mutations in the loop of E. coli SECIS. They found that most of the selB mutations correspond to a region of 23 amino acids included in the region that we have described above. Thus, our results reported here combined with those of Kromayer and colleagues (14) suggest that C-terminal subdomain 4b is involved in the interaction with both the SECIS U17 bulged nucleotide and the SECIS upper stem-and-loop structure. We found further confirmation for this suggestion in the results of our experiments that showed that the selB mutations that were selected by their ability to suppress mutations in the bulged U17 were also able to suppress mutations in the loop (pL24A) and even in the stem (pZL38 and pZL44) of the upper minihelix of E. coli SECIS (Fig. 4). The mutations with the highest efficiencies of suppression were found in the loop and the intermediate efficiencies in the stem region. The mutation with the lowest level of suppression efficiency was found in the bulged nucleotide either in position 18 or, as in the wild type, in position 17 (Fig. 4). Thus, the E. coli SECIS upper stem-loop structure can be regarded as a single suppressible unit, suggesting that there is a flexible interaction between this cis mRNA element and SelB.
Our selection procedure allowed us to increase the number of known changes in the amino acids in SelB that can suppress mutations in SECIS (Fig. 3 and Table 2). Both Kromayer and colleagues (14) and our group selected for mutations in the SelB protein in amino acids 556, 568, and 578 (Fig. 3). Furthermore, both groups found similar amino acid changes: Met556 to Ile556, Cys568 to Arg568, and Val578 to Ala578. However, we also found a mutation in which Val578 was changed to Glu578. Moreover, in addition to the amino acid changes reported by Kromayer and colleagues (14), we found changes in the 28-amino-acid region at the C terminus of SelB (between amino acids 556 and 583), including Phe572 to Tyr572, Ile557 to Phe557, and Ala583 to Thr583 (Table 2). Recall that all of the mutations in selB that we have reported here that were able to suppress mutations in E. coli SECIS were clustered in the 28-amino-acid region of the 4b subdomain SelB (Fig. 3). Based on in vitro studies (13), the 4b subdomain of SelB has been thought to be the functional domain in the interaction of SelB with the E. coli SECIS. However, Kromayer and colleagues (14) have found an additional mutation (Glu437 to Lys437) that lies outside 4b, in the 4a subdomain of the SelB protein. We found that single mutations in selB suppress mutations in the SECIS loop at higher efficiency than do mutations in the bulged nucleotide position 17 (Fig. 4). It seems possible that the screening test that we used here for selB mutations, using the bulged nucleotide in the upper stem-loop structure of E. coli SECIS, was more rigid than that used by Kromayer and colleagues (14) to select suppressor mutations in the SECIS loop.
In summary, using a genetic approach, we have increased the repertoire of the amino acids in SelB that are important for a direct interaction between SelB and the 17-nucleotide-long upper stem-loop-structure of the minimal E. coli SECIS mRNA. All of these amino acids were clustered in a region of 28 amino acids, the last one of which was 31 amino acids before the C-terminal end of SelB. The direct functional role of each of these amino acids will have to be determined in further experiments by physical means such as X-ray diffraction.
ACKNOWLEDGMENTS
We are deeply grateful to F. R. Warshaw-Dadon (Jerusalem, Israel) for her critical reading of the manuscript.
This research was supported by grants from the United States-Israel Binational Science Foundation (BSF), from the German-Israel Foundation for Scientific Research and Development (GIF), and the Ministry of Science and Culture of the State of Niedersachsen, Germany.
REFERENCES
- 1.Baron C, Heider J, Böck A. Interactions of translation factor SelB with the formate dehydrogenase H selenopolypeptide mRNA. Proc Natl Acad Sci USA. 1993;90:4181–4185. doi: 10.1073/pnas.90.9.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron C, Böck A. In: tRNA: structure, biosynthesis and function. Söll D, RhajBhandary U, editors. Washington, D.C.: ASM Press; 1995. pp. 529–544. [Google Scholar]
- 3.Berg B L, Li J, Heider J, Stewart V. Nitrate-inducible formate dehydrogenase in Escherichia coli K-12. I. Nucleotide sequence of the fdnGHI operon and evidence that opal (UGA) encodes selenocysteine. J Biol Chem. 1991;266:22380–22385. [PubMed] [Google Scholar]
- 4.Berry M J, Larsen P R. Recognition of UGA as a selenocysteine codon in eukaryotes: a review of recent progress. Biochem Soc Trans. 1993;21:827–832. doi: 10.1042/bst0210827. [DOI] [PubMed] [Google Scholar]
- 5.Böck A, Fochhammer K, Heider J, Leinfelder W, Sawers G, Veprek B, Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- 6.Casadaban M J, Cohen S N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers I, Harrison P R. A new puzzle in selenoprotein biosynthesis: selenocysteine seems to be encoded by the 'stop' codon, UGA. Trends Biochem Sci. 1987;12:255–256. [Google Scholar]
- 8.Forchhammer K, Leinfelder W, Böck A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature. 1989;342:453–456. doi: 10.1038/342453a0. [DOI] [PubMed] [Google Scholar]
- 9.Heider J, Baron C, Böck A. Coding from a distance: dissertion of the mRNA determinants required for the incorporation of selenocysteine into protein. EMBO J. 1992;11:3759–3766. doi: 10.1002/j.1460-2075.1992.tb05461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hüttenhofer A, Westhof E, Böck A. Solution structure of mRNA hairpins promoting selenocysteine incorporation in Escherichia coli and their base-specific interactions with special elongation factor SelB. RNA. 1996;2:354–366. [PMC free article] [PubMed] [Google Scholar]
- 11.Hüttenhofer A, Heider J, Böck A. Interaction of the Escherichia coli fdhF mRNA hairpin promoting selenocysteine incorporation with the ribosome. Nucleic Acids Res. 1996;24:3903–3910. doi: 10.1093/nar/24.20.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klug S J, Huttenhofer A, Kromayer M, Famulok M. In vitro and in vivo characterization of novel mRNA motifs that bind special elongation factor SelB. Proc Natl Acad Sci USA. 1997;94:6676–6681. doi: 10.1073/pnas.94.13.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kromayer M, Wilting R, Tormay P, Böck A. Domain structure of the prokaryotic selenocysteine-specific elongation factor SelB. J Mol Biol. 1996;262:413–420. doi: 10.1006/jmbi.1996.0525. [DOI] [PubMed] [Google Scholar]
- 14.Kromayer M, Neuhierl B, Friebel A, Böck A. Genetic probing of the interaction between the translation factor SelB and its mRNA binding element in Escherichia coli. Mol Gen Genet. 1999;262:800–806. doi: 10.1007/s004380051143. [DOI] [PubMed] [Google Scholar]
- 15.Lee B J, Worland P J, Davis J N, Stadtman T C, Hatfield D L. Identification of a selenocysteyl-tRNA (Ser) in mammalian cells that recognizes the nonsense codon, UGA. J Biol Chem. 1989;264:9724–9727. [PubMed] [Google Scholar]
- 16.Leinfelder W, Zehelein E, Mandrand-Berthelot M A, Böck A. Gene for a novel tRNA species that accepts L-serine and cotranslationally inserts selenocysteine. Nature. 1988;331:723–725. doi: 10.1038/331723a0. [DOI] [PubMed] [Google Scholar]
- 17.Lin-Goerke J L, Robbins D J, Burczak J D. PCR-based random mutagenesis using manganese and reduced dNTP concentration. Biotechniques. 1997;23:409–412. doi: 10.2144/97233bm12. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, Reches M, Groisman I, Engelberg-Kulka H. The nature of the minimal “selenocysteine insertion sequence” (SECIS) in Escherichia coli. Nucleic Acids Res. 1998;26:896–902. doi: 10.1093/nar/26.4.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Reches M, Engelberg-Kulka H. A sequence in the Escherichia coli fdhF “selenocysteine insertion sequence” (SECIS) operates in the absence of selenium. J Mol Biol. 1999;294:1073–1086. doi: 10.1006/jmbi.1999.3307. [DOI] [PubMed] [Google Scholar]
- 20.Low S C, Berry M J. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 22.Reches M, Zhao C, Engelberg-Kulka H. A bio-assay based on recombinant DNA technology for determining selenium concentration. J Appl Environ Microbiol. 1994;60:45–50. doi: 10.1128/aem.60.1.45-50.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ringquist S, Schneider D, Gibson T, Baron C, Böck A, Gold L. Recognition of the mRNA selenocysteine insertion sequence by the specialized translational elongation factor SELB. Genes Dev. 1994;8:376–85. doi: 10.1101/gad.8.3.376. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Stadtman T C. Selenocysteine. Annu Rev Biochem. 1996;65:83–100. doi: 10.1146/annurev.bi.65.070196.000503. [DOI] [PubMed] [Google Scholar]
- 26.Tormay P, Sawers A, Böck A. Role of stoichiometry between mRNA, translation factor SelB and selenocysteyl-tRNA in selenoprotein synthesis. Mol Microbiol. 1996;21:1253–9. doi: 10.1046/j.1365-2958.1996.881450.x. [DOI] [PubMed] [Google Scholar]
- 27.Zinoni F, Heider J, Böck A. Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc Natl Acad Sci USA. 1990;87:4660–4664. doi: 10.1073/pnas.87.12.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]