Allogeneic hematopoietic stem cell transplantation (HSCT) recipients have a higher risk of developing severe coronavirus disease (COVID-19) and a higher mortality rate compared with the general population (Ljungman et al.1) potentially also as a consequence of their reduced ability to respond to vaccination (Mamez et al.2; Redjoul et al.3; Einarsdottir et al.4). To evaluate the magnitude and breadth of T-cell responses against SARS-CoV-2 in allogeneic HSCT recipients, we carried out high-throughput T cell receptor (TCR) repertoire profiling on cells recovered from allogeneic HSCT recipients or healthy controls (HC) after COVID-19 natural infection or messenger RNA (mRNA)-based vaccination.
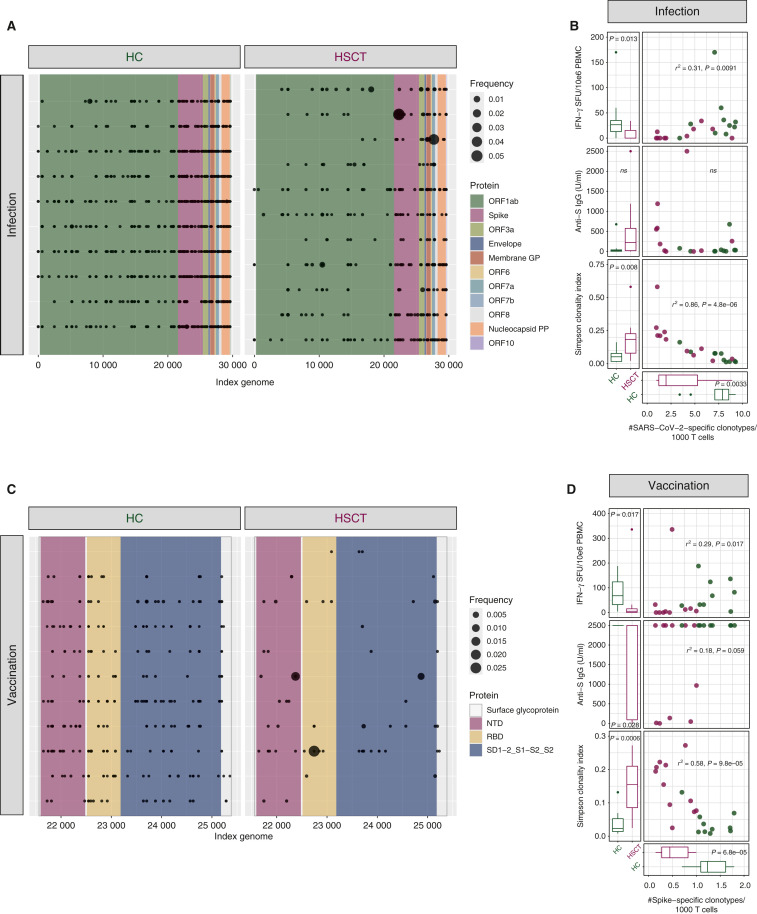
Peripheral blood samples were obtained after COVID-19 infection from allogeneic HSCT recipients (n = 11; Supplementary Tables S1 and S2, available at https://doi.org/10.1016/j.annonc.2022.09.153) or HC (n = 10; Supplementary Material, available at https://doi.org/10.1016/j.annonc.2022.09.153). A total of 6 out of 11 patients were under immunosuppression for active (n = 3) or resolved (n = 3) graft-versus-host disease. T-cell receptor (TCR) beta sequencing (Supplementary Material, available at https://doi.org/10.1016/j.annonc.2022.09.153) identified SARS-CoV-2 specific T-cell clonotypes in both HC and HSCT recipients after COVID-19 infection (Figure 1 A). No difference was observed in the proportion of T cells specific for SARS-CoV-2 between HSCT recipients and HC (data not shown). The diversity of the SARS-CoV-2-specific T-cell clonotypes, a measure previously shown to be inversely associated with severity of the disease (Elyanow et al.5), however, was significantly reduced in HSCT recipients compared with HC (Figure 1B). Enzyme-Linked ImmunoSpot (ELISpot) assay (Supplementary Material, available at https://doi.org/10.1016/j.annonc.2022.09.153) showed significantly lower numbers of interferon- γ (IFN-γ) spot forming units (SFU) after stimulation of PBMCs from HSCT recipients with peptides from both the SARS-CoV-2 spike (S) protein (Figure 1B, upper panel) and the membrane glycoprotein (M) plus the nucleocapsid phosphoprotein (N) proteins (data not shown) compared with HC. A significant positive correlation between SARS-CoV-2-specific T-cell clonotypes and IFN-γ SFU was observed (Figure 1B, upper panel). Conversely, we detected no significant difference in anti-S immunoglobulin G (IgG) titers and no correlation between antibody titers and different clonotypes (Figure 1B, middle panel). HSCT recipients displayed a less diverse TCR repertoire compared with HC as revealed by higher Simpson clonality and the Simpson clonality negatively correlated with the number of different SARS-CoV-2-specific T-cell clonotypes (Figure 1B, lower panel).
Figure 1.
Reduced SARS-CoV2-specific T-cell clonotypes after COVID-19 infection and vaccination in allogeneic HSCT recipients. (A, C) SARS-CoV-2-specific T-cell clonotypes visualized based on the putative sequence of the SARS-CoV-2 genome recognized. (B, D) Scatter plots and marginal bar plots correlating and comparing the number of different SARS-CoV-2-specific T-cell clonotypes/1000 T cells, the anti-S IFN-γ SFU, the anti-S IgG titers and the Simpson clonality index in HC and HSCT. Differences between groups were assessed using the Mann–Whitney U test. Correlations were evaluated using a Spearman rank correlation coefficient test.
GP, glycoprotein; HC, healthy controls; HSCT, hematopoietic stem cell transplantation; IFN, interferon; IgG, immunoglobulin G; NTD, N-terminal domain; PP, phosphoprotein; RBD, receptor binding domain; SFU, spot forming units.
We next carried out the same analysis on samples recovered from allogeneic HSCT recipients (n = 11; Supplementary Tables S1 and S2, available at https://doi.org/10.1016/j.annonc.2022.09.153) or from healthy controls (n = 10) after vaccination with three doses of mRNA-based SARS-CoV-2 vaccines (Supplementary Material, available at https://doi.org/10.1016/j.annonc.2022.09.153). We observed a significant reduction in different S-protein-specific T-cell clonotypes in allogeneic HSCT recipients compared with HC (Figure 1C and D). ELISpot analysis revealed significantly lower numbers of IFN-γ SFU in HSCT recipients compared with HC and a slightly significant positive correlation between the ELISpot and the TCR-seq results (Figure 1D, upper panel). We observed slightly reduced anti-S titers in HSCT recipients compared with HC and a trend toward a positive correlation between S-specific clonotypes and anti-S titers (Figure 1D, middle panels). We detected a negative correlation between the Simpson clonality and the number of different S-protein-specific T-cell clonotypes after vaccination (Figure 1D).
Our results indicate that allogeneic HSCT recipients display reduced breadth of SARS-CoV-2-specific T-cell clonotypes after COVID-19 infection and vaccination. No clear correlation was detected between TCR clonal breadth and anti-S IgG titers. The clonal breadth defect was associated with increased T-cell clonality after HSCT, pointing to the reduced diversity of the TCR repertoire as a mechanism leading to impaired cellular responses against SARS-CoV-2 in HSCT recipients.
Acknowledgments
Funding
This work was supported by the Dubois-Ferrière-Dinu-Lipatti Foundation (no grant number) to ACM, the Geneva University Hospitals’ Private Foundation (no grant number) to ACM, the Geneva University Hospitals' Clinical Research Center [grant number PRD 13-21-I] to ACM, the Choose Life Foundation (no grant number) to YC and FS, the Fondation Gustave & Simone Prévot (no grant number) to FS and the Geneva Cancer League [grant number LGC 20 11] to FS.
Disclosure
The authors have declared no conflicts of interest.
Supplementary Material
References
- 1.Ljungman P., de la Camara R., Mikulska M., et al. COVID-19 and stem cell transplantation; results from an EBMT and GETH multicenter prospective survey. Leukemia. 2021;35(10):2885–2894. doi: 10.1038/s41375-021-01302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mamez A.C., Pradier A., Giannotti F., et al. Antibody responses to SARS-CoV2 vaccination in allogeneic hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 2021;56(12):3094–3096. doi: 10.1038/s41409-021-01466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redjoul R., Le Bouter A., Parinet V., et al. Antibody response after third BNT162b2 dose in recipients of allogeneic HSCT. Lancet Haematol. 2021;8(10):e681–e683. doi: 10.1016/S2352-3026(21)00274-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Einarsdottir S., Martner A., Waldenström J., et al. Deficiency of SARS-CoV-2 T-cell responses after vaccination in long-term allo-HSCT survivors translates into abated humoral immunity. Blood Adv. 2022;6(9):2723–2730. doi: 10.1182/bloodadvances.2021006937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elyanow R., Snyder T.M., Dalai S.C., et al. T cell receptor sequencing identifies prior SARS-CoV-2 infection and correlates with neutralizing antibodies and disease severity. JCI Insight. 2022;7(10) doi: 10.1172/jci.insight.150070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.