Abstract
Previous studies of the 16S rRNA genes from Mycobacterium ulcerans and Mycobacterium marinum have suggested a very close genetic relationship between these species (99.6% identity). However, these organisms are phenotypically distinct and cause diseases with very different pathologies. To investigate this apparent paradox, we compared 3,306 nucleotides from the partial sequences of eight housekeeping and structural genes derived from 18 M. ulcerans strains and 22 M. marinum strains. This analysis confirmed the close genetic relationship inferred from the 16S rRNA data, with nucleotide sequence identity ranging from 98.1 to 99.7%. The multilocus sequence analysis also confirmed previous genotype studies of M. ulcerans that have identified distinct genotypes within a geographical region. Single isolates of both M. ulcerans and M. marinum that were shown by the sequence analysis to be the most closely related were then selected for further study. One- and two-dimensional pulsed-field gel electrophoresis was employed to compare the architecture and size of the genome from each species. Genome sizes of approximately 4.4 and 4.6 Mb were obtained for M. ulcerans and M. marinum, respectively. Significant macrorestriction fragment polymorphism was observed between the species. However, hybridization analysis of DNA cleaved with more frequently cutting enzymes identified significant preservation of the flanking sequence at seven of the eight loci sequenced. The exception was the 16S rRNA locus. Two high-copy-number insertion sequences, IS2404 and IS2606, have recently been reported in M. ulcerans, and significantly, these elements are not present in M. marinum. Hybridization of the AseI restriction fragments from M. ulcerans with IS2404 and IS2606 indicated widespread genome distribution for both of these repeated sequences. Taken together, these data strongly suggest that M. ulcerans has recently diverged from M. marinum by the acquisition and concomitant loss of DNA in a manner analogous to the emergence of M. tuberculosis, where species diversity is being driven mainly by the activity of mobile DNA elements.
Mycobacterium ulcerans is an emerging human pathogen that causes a chronic, necrotic skin lesion in humans. Its prevalence throughout West Africa appears to have increased dramatically since the late 1980s (35). The organism is unlike other mycobacterial pathogens in that it appears to maintain an extracellular location during infection (23). The disease is usually treated by surgical excision of infected and surrounding tissue, as the organism in situ is unresponsive to drug therapy (31). Possible explanations for the increased occurrence of this disease include environmental changes that have led to proliferation of the organism followed by increased human contact (22, 30) and adaptation of the organism to a changed environment and coincidental acquisition of increased virulence. Despite several extensive investigations over the past 30 years, the mode of transmission of M. ulcerans has not been determined (2, 46). Recent detection of M. ulcerans-specific DNA sequences in water from swamps in southeastern Australia and aquatic insects in Benin have confirmed that it is an environmental organism (47, 53, 60).
The etiology and epidemiology of Mycobacterium marinum are much better understood. It has long been recognized as a fish pathogen and has been isolated from swimming pools, fish aquaria, and marine environments worldwide (12, 15, 25). It is an intracellular pathogen, and in humans it usually causes a limited granulomatous skin infection at the extremities, probably via direct inoculation at the site of minor cuts and abrasions (15, 17). The infection can usually be treated with antimycobacterial drugs (19). M. marinum is relatively fast growing, has nonfastidious growth requirements, and produces a light-inducible pigment, presumably for protection against incident UV irradiation (50). The picture built up from these findings is one of a widespread and robust environmental organism which is capable of withstanding some of the extremes of aquatic environments such as sunlight exposure, varying temperatures, and nutrient limitation. Conversely, the profile of M. ulcerans includes a worldwide but highly focal environmental distribution, slow growth, UV sensitivity, optimal growth under microaerophilic conditions, and the production of an unusual cytotoxic type I polyketide (18, 40, 45; W. M. Meyers, personal communication). These characteristics suggest an organism that has adapted to a specific environmental niche.
Several studies have highlighted an apparently paradoxical relationship between these two species, where their striking phenotypic differences are contradicted by a high degree of genetic similarity. It has been known for some time that M. ulcerans and M. marinum have identical signature sequences through the two hypervariable regions of the 16S rRNA gene (6, 52) and that the only sequence differences within this locus are two nucleotides at the 3′ end of the gene (48, 64). Furthermore, the nucleotide at one of these positions varies from that in M. marinum in only some strains of M. ulcerans (48). Sequence analysis of a partial groEL fragment (51) and analyses of cell wall mycolate composition (11, 64) have also confirmed the close genetic relationship between these species. However, DNA-DNA hybridization studies have shown a relative binding ratio of approximately 37% between M. ulcerans and M. marinum strains (64). This does suggest that there is a fundamental genetic basis for the significant phenotypic differences observed. Recently, two high-copy-number insertion sequences, IS2404 and IS2606, were identified in M. ulcerans (59). Neither of these elements was present in M. marinum, but they were present in M. ulcerans isolates collected from around the world (58). Thus, the presence of these sequences appears to be a defining and important characteristic of M. ulcerans.
Our hypothesis is that M. ulcerans has recently diverged from M. marinum by the recruitment of foreign DNA from the environment. Such a scenario is in accord with the mosaic genome structure identified within other mycobacteria (43) and their ability to evolve rapidly by the transposition of insertion sequences, such as IS6110 in Mycobacterium tuberculosis (62), IS900 in Mycobacterium avium subsp. paratuberculosis (20), and IS1512 in Mycobacterium gordonae (44).
In the current study, our overall aim was to learn more about the emergence of M. ulcerans as a pathogen by comparing it at a genetic level with M. marinum. This was accomplished by employing multilocus sequence typing, two-dimensional pulsed-field gel electrophoresis (PFGE), and restriction fragment hybridization analysis to compare both structural and sequence compositions of the genomes of these species.
MATERIALS AND METHODS
Bacterial strains.
The details of the 18 M. ulcerans isolates and 22 M. marinum isolates used in this study are listed in Table 1. Culture media and conditions were as previously described (59).
TABLE 1.
Strain information
| Species | Strain | Yr isolated | Origin | Sourcea | 2426 typeb | Sequence type |
|---|---|---|---|---|---|---|
| M. ulcerans | 144727 | 1989 | Victoria, Australia | VIDRL | Victorian | Victorian |
| ATCC 19423 | 1948 | Victoria, Australia | ATCC | Victorian | Victorian | |
| 11878/70 | 1971 | Papua New Guinea | QDRL | PNG(I)c | SE Asian | |
| MD | ||||||
| 94-1331 | 1994 | Papua New Guinea | ITM | PNG(II)c | SE Asian | |
| 13822/70 | 1971 | North Queensland, Australia | QDRL | Queensland | SE Asian | |
| MD | ||||||
| 94-1328 | 1994 | Malaysia | ITM | Malaysian | SE Asian | |
| 186510 | 1992 | Malaysia | VIDRL | Malaysian | SE Asian | |
| 96-658 | 1996 | Angola | ITM | African | African | |
| 94-856 | 1994 | Benin | ITM | African | African | |
| 97-111 | 1997 | Benin | ITM | African | African | |
| 5152 | 1976 | Congo | ITM | African | African | |
| 97-610 | 1997 | Ghana | ITM | African | African | |
| 97-680 | 1997 | Togo | ITM | African | African | |
| 98-912 | 1997 | China | ITM | Asian | Asian | |
| ATCC 33728 | 1980 | Japan (also called M. shinshuense) | ITM | Asian | Asian | |
| 5114 | 1953 | Mexico | ITM | Mexico | Mexican | |
| 5143 | 1967 | Mexico | ITM | Mexican | Mexican | |
| 842 | 1986 | Surinam | ITM | Surinam | Surinam | |
| NCTC 2275 | 1926 | Saltwater fish, Philadelphia (same as ATCC 927) | NCTC | I | ||
| M. marinum | ATCC 11565 | 1958 | Human, Sweden | ATCC | I | |
| 99/84 | 1999 | Bilby, western Australia | PC | I | ||
| 99/88 | 1993 | Human, western Australia | PC | I | ||
| Mon10 | 1996 | Human, Philadelphia, Pa. | RML | I | ||
| 472 | 1993 | Water, Norway | RML | I | ||
| JKD2394 | 1998 | Human, Victoria, Australia | VIDRL | II | ||
| 991831797 | 1999 | Human, New South Wales, Australia | ICPMR | II | ||
| 471 | Human, Norway | RML | III | |||
| 99/87 | 1996 | Human, western Australia | PC | IV | ||
| 993362605 | 1999 | Human, New South Wales, Australia | ICPMR | IV | ||
| 99/86 | 1993 | Human, Tasmania, Australia | PC | V | ||
| 99/89 | 1994 | Human, Tasmania, Australia | PC | V | ||
| 99/90 | 1997 | Human, Tasmania, Australia | PC | V | ||
| JKD2395 | 1998 | Human, Victoria, Australia | VIDRL | V | ||
| JKD2396 | 1998 | Human, Victoria, Australia | VIDRL | V | ||
| JKD2397 | 1998 | Human, Victoria, Australia | VIDRL | V | ||
| 0500525 | 1999 | Human, Canberra, Australia | ICPMR | V | ||
| 0412214 | 1999 | Human, New South Wales, Australia | ICPMR | V | ||
| 1542578 | 1999 | Human, New South Wales, Australia | ICPMR | V | ||
| 992092077 | 1999 | Human, New South Wales, Australia | ICPMR | V | ||
| 991961552 | 1999 | Human, New South Wales, Australia | ICPMR | V |
VIDRL, Victorian Infectious Diseases Reference Laboratory; QDRLMD, Queensland Diagnostic and Reference Laboratory for Mycobacterial Diseases; ITM, Institute for Tropical Medicine; PC, Western Australian Centre for Pathology and Medical Research; RML, NIH/NIAID/DIR Rocky Mountain Laboratories; ICPMR, Institute of Clinical Pathology and Medical Research.
2426 type, genotype designation as determined by 2426-PCR (58).
PNG(I) and PNG(II), Papua New Guinea 2426 types (I) and (II), respectively.
Multilocus sequence analysis.
PCR was used to amplify internal fragments from eight genes in M. ulcerans and M. marinum. The oligonucleotide primers for amplification of the rrs, groEL, sod, and fbpA loci were those used previously (48, 55, 61, 69) (Table 2). Primers for adk, aroE, and ppk were designed by alignment of sequences obtained from the Mycobacterium leprae and M. tuberculosis genome databases (10; http://www.sanger.ac.uk/Projects/M_leprae/blast_server.shtml). It was reasoned that regions of sequence conservation between these two distantly related mycobacteria would permit the design of genus-level primers. The names of each of the eight genes, the putative gene products, and the positions sequenced are given in Table 2. GenBank accession numbers are also given in Table 2 for the sequences obtained from the type strains of M. ulcerans and M. marinum. The sequences obtained from the other 38 isolates have also been deposited in GenBank. The accession numbers for these additional sequences are available from the authors or by searching GenBank.
TABLE 2.
Oligonucleotides used for PCR amplification and nucleotide sequencing of the internal regions of genes from M. ulcerans and M. marinum
| Oligonucleotide | Sequence, 5′→3′ | Expected PCR product size and putative gene function | Reference | Nucleotide positions sequenceda | GenBank accession no.b |
|---|---|---|---|---|---|
| adk-P1 | G(GT)ATCCCGCAGATCTCCACC | adk-P1 + adk-P2, amplification of a 442-bp product from adk (adenylate kinase) | This study | 114–486 | AF271093 |
| adk-P2 | CAC(CT)TCGTCCATGGTGCCGA | AF271342 | |||
| aroE-P1 | CCCGGTGAACTGCTCCACCT | aroE-P1 + aroE-P2, amplification of a 467-bp product from aroE (shikimate dehydrogenase) | This study | 304–748 | AF271094 |
| aroE-P2 | TGGCGGGCCGACAACACCGA | AF271343 | |||
| crtB-P1 | CGACGACATTCTGGACTCCT | crtB-P1 + crtB-P2, amplification of a 469-bp product from crtB (phytoene synthase) | This study | 184–638 | AF271095 |
| crtB-P2 | GACACCACATCAGCACATCC | AF271344 | |||
| MT1 | TTCCTGACCAGCGAGCTGCCG | MT1 + MT2, amplification of a 508-bp product from fbpA (32-kDa surface antigen) | 55 | 476–893 | AF271092 |
| MT2 | CCCCAGTACTCCCAGCTGTGC | AF271345 | |||
| Tb11 | ACCAACGATGGTGTGTCCAT | Tb11 + Tb12, amplification of a 439-bp product from groEL (65-kDa heat shock protein) | 61 | 159–540 | AF271096 |
| Tb12 | CTTGTCGAACCGCATACCCT | AF271346 | |||
| 1004R | AGGAATTCTGGGTTTGACATGCACAGGA | 1004R + rRog, amplification of a 517-bp product from rrs (3′ region of the 16S rRNA gene) | 48 | 1038–1491 | AF27302 |
| rRog | AAGGAGGTGATCCAGCCGCA | AF271347 | |||
| ppk-P1 | AGTTGCTGCTGCGTGAGC | ppk-P1 + ppk-P2, amplification of a 421-bp product from ppk (polyphosphate kinase) | This study | 999–1395 | AF271097 |
| ppk-P2 | GATGTTGGCCTGCTCGTC | AF271348 | |||
| Z212 | TCG(GT)CCCAGTTCACGAC(GA)TTCCA | Z212 + Z261, amplification of a 434-bp product from sod (superoxide dismutase) | 69 | 144–534 | AF271098 |
| Z261 | CCAA(AG)CTCGAAGAGGCGCG(CG)GCCAA | AF271349 |
Numbering based on M. tuberculosis H37Rv except for crtB, which was based on M. marinum sequence (accession no. U92075) and rrs, which was based on E. coli 16S rRNA.
Accession numbers are provided for the type strains of each species, M. ulcerans ATCC 19423 (upper line) and M. marinum NCTC 2275 (lower line).
DNA extraction and PCR.
Mycobacterial DNA was extracted from 5 to 25 mg (wet weight) of cell pellet by glass bead cell homogenization in the presence of Triton X-100 and chloroform-isoamyl alcohol (24:1) as previously described (58). A 2-μl volume of the Triton X-100 aqueous phase was then used as a template for PCR. Reaction conditions used for the PCR amplification of all fragments were as follows: each reaction mixture (50 μl) contained 1× PCR buffer II (10× PCR buffer II contained 500 mM KCl, 100 mM Tris-HCl [pH 8.3]), 1.5 mM MgCl2, 0.5 mM deoxynucleoside triphosphates (dNTPs; 0.5 mM each dATP, dTTP, dCTP, and dGTP), 10% dimethyl sulfoxide, 0.5 μM each primer, and 1 U of Ampli-Taq DNA polymerase (Applied Biosystems, Foster City, Calif.). Thermal cycling was performed in an FTS-960 thermal sequencer (Corbett Research, Sydney, Australia) with five cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min, 30 cycles of 95°C for 20 s, 58°C for 30 s, and 72°C for 45 s, followed by a final extension step at 72°C for 5 min. The reactions were held at 4°C until analyzed by 1.5% agarose gel electrophoresis with ethidium bromide staining. QIAquick spin columns (Qiagen Inc., Valencia, Calif.) were used to purify the PCR products prior to cycle sequencing. The products were sequenced on both strands with the primers used for PCR, according to the protocols supplied with the Prism Big Dye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems). Extension products were analyzed with a PE Applied Biosystems model 373 automated sequencer, and the sequences were compiled with Sequencher 3.1.1 software (Gene Codes Corporation).
Nucleotide sequence analysis.
Strains were grouped according to their combination of alleles, and each unique allelic pattern was identified as a sequence type (genotype). A representative strain from each genotype was then selected for phylogenetic analysis. The sequences from the seven protein-encoding loci were concatenated in frame to produce a 2,853-bp semantide for each genotype, which were aligned with Clustal W (63). Phylogenetic analysis was performed with MEGA software version 1.1.2 (33) and Splits Tree version 3.1 (26). P distances were used throughout, as the overall level of sequence divergence was small. Values for synonymous (dS) and nonsynonymous (dN) mutation frequencies were calculated with Nei and Gojobori's method (38), and standard errors of the means of these values were estimated by the method of Nei and Jin (39). All calculations of dS and dN were performed using the dSdNqw program (14). The G+C% at each codon position was determined using Web-based software (Murdoch University Bioinformatics Research Institute, http://arginine.it.murdoch.edu.au/research).
PFGE.
Mycobacterial DNA plugs were prepared as previously described (54) with the following modifications. Ampicillin and d-cycloserine were added to the culture 24 h prior to harvesting at final concentrations of 0.1 and 1.0 mg/ml, respectively (8). The step requiring vortexing of the cells in the presence of 3-mm glass beads was omitted, and the Bio-Rad Genepath wash solution was replaced with TE buffer (10 mM Tris, 1 mM EDTA [pH 8.0]). Restriction endouclease digestion of the DNA in the plugs was performed as described previously (42). For DraI digestion, MgCl2 was added to a final concentration of 10 mM. First- and second-dimension PFGE were performed using the Bio-Rad CHEF DRII system (Bio-Rad, Richmond, Calif.) with 1.0% agarose in 0.5× Tris-borate-EDTA (TBE) at 200 V, with 10 to 35 s switching times for 25 h. DNA was visualized by staining with ethidium bromide (0.5 μg/ml) overnight at 4°C. Southern hybridization analysis was performed as described previously (59), and DNA restriction fragment sizes from both PFGE and Southern blots were estimated with Sigmagel software (Jandel Scientific).
RESULTS
Multilocus sequence typing.
A collection of 18 M. ulcerans isolates and 22 M. marinum isolates was used in this study (Table 1). These isolates originated from a variety of sources and represent both temporal and geographic diversity. The majority of the isolates were of human origin. However, among the M. marinum strains, one was isolated from a fish, another from a bilby (Macrotis lagotis, a small Australian native marsupial), and another from water (Table 1). For the sequence typing, a panel of seven unlinked genes were used (see the hybridization results below). The 3′ region of the 16S rRNA gene from each isolate was also sequenced, but only the data from the seven protein-encoding loci were included in the subsequent phylogenetic analyses. The allelic profiles for some isolates differed at more than three of the seven loci, so phylogeny was inferred by using a distance method rather than a pairwise comparison of the allelic profiles (56). The sequences from the seven loci were concatenated in the order crtB, adk, fbpA, aroE, groEL, ppk, and sod to produce a 951-codon semantide.
The 40 isolates were represented by 11 different genotypes, where a unique combination of the seven alleles defined a particular genotype. A summary of all the variable sites for each genotype and the division between synonymous and nonsynonymous substitutions is shown in Fig. 1. Five M. marinum genotypes were identified and named types I to V (Table 1, Fig. 1). There was no obvious correlation between strain origin and genotype, although no genotype IV or V isolates were detected among the strains obtained from the Northern Hemisphere. There were six M. ulcerans genotypes, and in accord with previous studies, these were named according to their geographic origin. There was only one variable position across all eight loci that discriminated between the species. This site was within the fbpA gene at position 1128 of the concatenated sequences (Fig. 1). As has been reported previously, no variation was detected in the 3′ region of the 16S rRNA gene for any of the M. marinum isolates, and there were five alleles of the gene among the M. ulcerans strains (48, 64).
FIG. 1.
Alignment of the 2,853-bp sequences derived from the seven concatenated protein-encoding loci for each of the 11 genotypes. Only variable nucleotides are shown, and the numbers at the top of figure indicate their positions in the sequence. A period indicates identity with the M. ulcerans Surinam strain, and nonsynonymous mutations are highlighted with gray shading.
M. ulcerans and M. marinum have been shown by 16S rRNA analysis to be most closely related to M. tuberculosis (64). The percent nucleotide identity between M. ulcerans ATCC 19423, M. marinum NCTC 2275, and M. tuberculosis H37Rv was calculated at each locus to indicate the general relatedness between each species. Identity scores ranged from 96.3 to 99.6% (average, 98.7%) between M. ulcerans and M. marinum, compared to 77.2 to 99.3% (average, 86.9%) between M. ulcerans or M. marinum and M. tuberculosis.
Split decomposition analysis was used to examine the phylogenetic relationship between the M. marinum and M. ulcerans strains. The treelike structure shown in the splits graph and the absence of networks (Fig. 2) are clear evidence of a bifurcating phylogeny. These observations, combined with a high level of statistical support for each node in the splits graph and complete congruence with a dendrogram derived by the neighbor-joining method (data not shown), provide good evidence for an evolutionary link between M. ulcerans and M. marinum via a series of de novo point mutations within each locus. M. marinum could be categorized into two distinct and divergent groups (I and II versus III, IV, and V). The discrete clustering of all M. ulcerans strains suggests that M. ulcerans is a derivative of an M. marinum type III, IV, or V ancestor. There was also significantly less sequence variation within the M. ulcerans cluster compared to M. marinum (Fig. 1), supporting the proposition that M. marinum is the ancestral species. A close genetic relationship was also evident between the southeast Asian, African, and Victorian (Australian) genotypes of M. ulcerans (Fig. 2). This observation is in accord with previous findings based on PCR amplification of inter-IS sequences (2426-PCR) (58). No sequence differences were detected among any of the African isolates. Overall, there was good correlation between multilocus sequence analysis and 2426-PCR, but the 2426-PCR offered additional resolution among isolates of the southeast Asian genotype (Table 1).
FIG. 2.
Splits graph of the phylogenetic relationship among the six M. ulcerans and five M. marinum genotypes. The vertices are labeled with each genotype. (MM, M. marinum; MU, M. ulcerans). The graph was generated from the concatenated sequences of the seven protein-encoding loci. All edges in the graph had greater than 80% bootstrap support (1,000 iterations) with the exception of the edges marked with an asterisk. These edges had greater than 60% bootstrap support.
Synonymous and nonsynonymous substitution frequencies.
A high frequency of nonsynonymous substitutions (dN) compared to synonymous substitutions (dS) within a particular gene or locus can indicate the presence of positive selection pressure (16, 65). From the data presented in Fig. 1, this difference (dS − dN) was calculated across all loci for both species. For the M. marinum genotypes, the value for dS − dN was 2.8 ± 0.5 (z = 5.57, P < 0.001, dS = 3.0 ± 0.5, dN = 0.2 ± 0.08). That is, the frequency of synonymous mutation was significantly higher than the nonsynonymous mutation frequency, suggesting that there is no obvious selection pressure. However, among the M. ulcerans genotypes, the value for dS − dN of 0.32 ± 0.18 (z = 1.76, P > 0.05, dS = 0.54 ± 0.17, dN = 0.22 ± 0.07) was much lower, and the dS and dN values were not significantly different. Expressed another way, the ratio of dN to dS was 6.8 times higher in M. ulcerans than in M. marinum, suggesting the presence of positive or purifying selection pressure acting on M. ulcerans. This observation lends support to a theory that M. ulcerans has adapted to a changed or changing environment, particularly given that the two species appear to have a common genetic backbone and therefore should exhibit similar theoretical mutation rates. The presence of five rrs alleles among the six M. ulcerans strains compared with only a single rrs allele for all the M. marinum genotypes is also consistent with an organism in a state of evolutionary flux and adaptation.
The evolutionary age of M. ulcerans was estimated by determining dS across the 951 codons of the seven loci (rrs excluded). By using previous estimates of bacterial synonymous substitution rates of 0.58 to 0.78 substitutions per 100 sites per million years (32), the time needed to accumulate the amount of synonymous mutation observed within the M. ulcerans genotypes was calculated. This analysis indicated that M. ulcerans emerged between 470,000 and 1,200,000 years ago. To check that there were no codon biases, which can indicate reduced rates of substitution (7), the GC content at the third codon position (GC3%) was compared with the overall GC content for each genotype across both species. The values obtained (average GC% = 65.5, standard deviation [sd] = 0.1; average GC3% = 85.9, sd = 0.1) were very similar to those reported for M. tuberculosis, suggesting that the rate at which M. ulcerans and M. marinum accumulate synonymous substitutions is the same as that observed in M. tuberculosis (4). This estimate assumes that there are no significant in vivo growth rate differences between species. However, fluctuations in growth rates have been suggested to be inconsequential over a geological time scale and given actual environmental generation times (37).
Comparisons of genome structure.
To further investigate the hypothesis that M. ulcerans has recently diverged from M. marinum, a southeast Asian isolate of M. ulcerans (isolate 13822/70) and a type V isolate of M. marinum (isolate 99/86) were selected for genome structure comparisons.
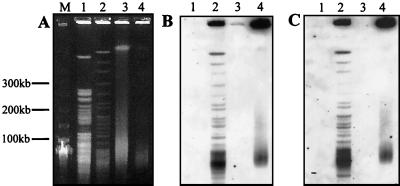
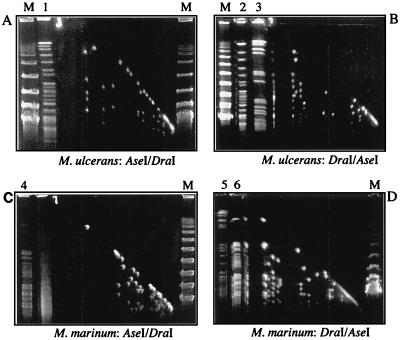
PFGE was used to compare macrorestriction fragment patterns and to obtain estimates of the genome sizes. The restriction enzymes PacI, PmeI and SwaI, which have eight-base AT-rich recognition sites, were tried first in an attempt to obtain a simple pattern of fragments that would permit straightforward genome size estimations. Unfortunately, these enzymes failed to cut the genome of either M. marinum or M. ulcerans. AseI and DraI gave the most useful array of fragments (Fig. 3). No plasmid bands were detected in either isolate (Fig. 4A). However, with these enzymes there were probable doublets and areas of significant compression that prevented accurate sizing. These regions could not be resolved satisfactorily with altered electrophoretic separation parameters. Two-dimensional PFGE was used to improve resolution. Reciprocal AseI and DraI digests were performed for each organism, and these are shown in Fig. 5. Indicative genome sizes were obtained by summing the averages of the AseI and DraI restriction fragments length estimates from both one- and two-dimensional pulsed-field arrays (Table 3). This indicated a genome size for M. ulcerans of approximately 4.4 Mb and a slightly larger genome for M. marinum of approximately 4.6 Mb. This latter figure is comparable to other genome size estimates for M. marinum (1).
FIG. 3.
PFGE analysis of genomic DNA from M. marinum 99/86 (lanes 1 and 2) and M. ulcerans 13822/70 (lanes 3 and 4) digested with AseI (lanes 1 and 3) and DraI (lanes 2 and 4). Lanes M, 50-kb lambda DNA size ladder.
FIG. 4.
PFGE (A) and Southern hybridization (B and C) analyses of M. marinum 99/86 (lanes 1 and 3) and M. ulcerans 13822/70 (lanes 2 and 4), probed with IS2606 (B) and IS2404 (C). Lanes 1 and 2, AseI digest; lanes 3 and 4, undigested DNA; lane M, 50-kb lambda DNA size ladder.
FIG. 5.
Two-dimensional PFGE analysis of genomic DNA from M. ulcerans 13822/70 (A and B) and from M. marinum 99/86 (C and D), reciprocally digested with the restriction enzymes AseI and DraI as indicated on each panel. Lanes 1, 2, 4, and 6, first-dimension separations of genomic DNA digested with the restriction enzyme AseI; lanes 3 and 5, first-dimension separations of genomic DNA digested with the restriction enzyme DraI; lane M, 50-kb lambda DNA size ladder.
TABLE 3.
Estimated sizes of restriction fragments from AseI and DraI digests of M. marinum and M. ulcerans
|
AseI
|
DraI
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
M. marinum
|
M. ulcerans
|
M. marinum
|
M. ulcerans
|
|||||
| Fragment | Size (kb) | Fragment | Size (kb) | Fragment | Size (kb) | Fragment | Size (kb) | |
| A | 441 | A1 | 510 | A | 924 | A1 | 1,044 | |
| B1 | 248 | A2 | 510 | B | 531 | B | 490 | |
| B2 | 248 | B | 370 | C1 | 420 | C1 | 412 | |
| C | 245 | C | 330 | C2 | 420 | C2 | 412 | |
| D1 | 240 | D | 278 | D | 370 | D | 360 | |
| D2 | 240 | E | 245 | E | 340 | E1 | 250 | |
| E | 220 | F | 216 | F | 235 | E2 | 240 | |
| F | 208 | G | 200 | G | 206 | F | 205 | |
| G | 200 | H1 | 170 | H | 201 | G | 140 | |
| H | 180 | H2 | 167 | I | 187 | H | 129 | |
| I | 175 | I | 160 | J | 163 | I | 122 | |
| J | 170 | J1 | 150 | K | 117 | J | 104 | |
| K | 160 | J2 | 147 | L | 106 | K | 95 | |
| L | 155 | K | 134 | M | 99 | L | 87 | |
| M | 148 | L | 120 | N | 84 | O | 78 | |
| N | 137 | M1 | 109 | O | 70 | M | 72 | |
| O | 132 | M2 | 107 | P | 47 | N | 49 | |
| P | 112 | N | 104 | Q | 40 | O | 43 | |
| Q1 | 103 | O1 | 75 | R1 | 31 | P1 | 41 | |
| Q2 | 102 | O2 | 74 | R2 | 28 | P2 | 37 | |
| R1 | 92 | P1 | 63 | S | 20 | Q | 6 | |
| R2 | 92 | P2 | 62 | T | 7 | Total | 4,416 | |
| S1 | 78 | Q | 43 | Total | 4,646 | |||
| S2 | 76 | R1 | 40 | |||||
| S3 | 75 | R2 | 29 | |||||
| T | 71 | R3 | 22 | |||||
| U | 55 | S | 5 | |||||
| V | 45 | T | 2 | |||||
| W1 | 40 | Total | 4,442 | |||||
| W2 | 36 | |||||||
| X | 28 | |||||||
| Y | 10 | |||||||
| Z | 9 | |||||||
| Total | 4,571 | |||||||
From the one-dimensional pulsed-field patterns, there appeared to be little similarity in AseI and DraI restriction patterns between strains. One explanation for observing nucleotide sequence similarity with genomic structural diversity is the presence of mobile DNA in one or both species. Insertion sequences are well known to promote genome rearrangements (34), and IS2404 and IS2606 are two elements present in M. ulcerans but absent from M. marinum that could act as substrates for such rearrangements. Hybridization of IS2404 and IS2606 probes against M. ulcerans digested with AseI indicated the widespread distribution of both elements around the genome (Fig. 4B and C). As expected, M. marinum did not hybridize to either probe. All M. marinum isolates were also screened by PCR and found not to contain either IS2404 or IS2606 (data not shown).
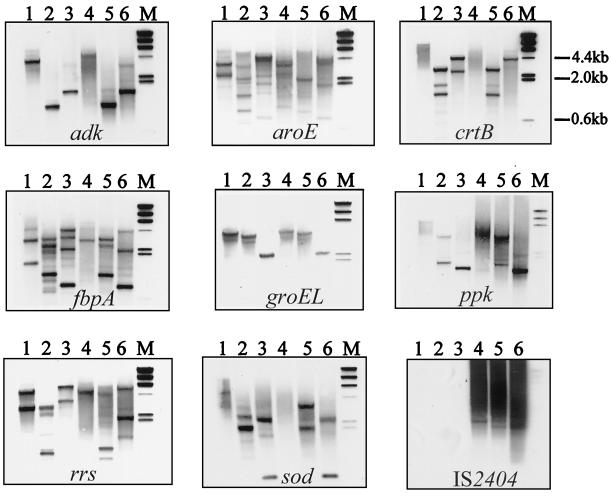
If, as suggested by the restricted sequence polymorphism, large-scale genome rearrangements have occurred recently, then some preservation of genomic subarchitecture could be expected between each species. The restriction enzymes NcoI, PvuII, and PstI were predicted to cut no more than once within the entire coding region of each gene used for multilocus sequence analysis. When full-length M. ulcerans or M. marinum gene sequences were not available, this prediction was based on the M. tuberculosis genome sequences (10). These enzymes were then used to digest genomic DNA from M. marinum and M. ulcerans. The DNA was hybridized against probes from each of the eight loci described above, and the sizes of the hybridizing fragments were estimated and compared. All loci appeared to hybridize to different-sized fragments for all three enzymes, indicating that none of the targets selected for multilocus testing were linked. A significant degree of conservation of the DNA flanking most of the loci between the two species was revealed (Fig. 6). One exception was the 16S rRNA locus, for which multiple polymorphisms were detected with all three enzymes. The presence of two hybridizing fragments with each enzyme against M. marinum DNA suggests that M. marinum may possess at least two copies of the rRNA operon. Multiple bands also hybridized to the probes derived from the fbpA and aroE genes. However, from an analysis of the M. tuberculosis genome, the presence of these bands is probably due to cross-hybridization with other genes of similar sequence, such as fbpC and other dehydrogenase genes.
FIG. 6.
Southern hybridization analysis of genomic DNA from M. marinum 99/86 (lanes 1, 2, and 3) and from M. ulcerans 13822/70 (lanes 4, 5, and 6). The DNA was digested with the restriction enzymes NcoI (lanes 1 and 4), PvuII (lanes 2 and 5), and PstI (lanes 3 and 6) and then probed with sequences derived from each locus as indicated. Lane M, lambda HindIII-digested DNA size markers.
DISCUSSION
In this study we have used multilocus sequence analysis to clearly establish for the first time the population structure of and evolutionary relationship between M. ulcerans and M. marinum. The data we have gathered suggest the recent divergence of M. ulcerans from an M. marinum progenitor. Overall, M. marinum and M. ulcerans have very high nucleotide homology. Their close genetic relationship is highlighted by the presence of only one species-discriminating variable site among the 3,306 bp from the eight loci (Fig. 1). The level of intraspecies nucleotide sequence divergence was higher between M. marinum strains than M. ulcerans strains, and this observation correlates well with previous DNA-DNA hybridization studies (64). An increased level of nucleotide sequence divergence and the absence of IS2404 and IS2606 from all M. marinum strains are the expected states for the ancestral species of M. ulcerans.
Insertion sequences and other repetitive DNA elements play an important role in mycobacterial genetics (13, 49). In M. tuberculosis, IS6110 is responsible for the rapid evolution of distinct clones (57). Similarly, IS900 and IS901/902 are defining characteristics for M. avium subsp. paratuberculosis and M. avium subsp. silvaticum, organisms with a high degree of genetic identity to the M. avium complex (20). M. ulcerans has acquired at least two IS elements, IS2404 and IS2606, and their pattern of widespread genome distribution and high copy number indicate the potential for these elements to act as substrates for ongoing genome rearrangements. The detection of variations in inter-IS distances between strains of M. ulcerans is evidence of such rearrangements (58).
Interestingly, both IS2404 and IS2606 are related to elements in the genus Streptomyces. The transposase from IS2404 has 31% amino acid identity (45% amino acid similarity) with that from IS1629, an IS associated with mobilization of the nec1 virulence determinant in plant-pathogenic strains of various Streptomyces spp. (24). Recently, a homolog of IS2606 has been identified in Streptomyces albus. The putative transposase from this IS has 47% amino acid identity (57% amino acid similarity) with that from IS2606 (C. M. Smith, personal communication). The transposition of an IS from Streptomyces coelicolor into a mycobacterial genome has been demonstrated (5).
While the IS elements may play an important role in promoting rearrangements and modifying gene expression, the presence of the unusual type 1 polyketide mycolactone (18) in M. ulcerans means that it is unlikely that IS2404 and IS2606 are the only sequences that M. ulcerans has acquired. A large amount of specific genetic material is predicted to be required for the synthesis of this molecule. From the M. tuberculosis genome sequence data, mycobacteria are known to contain several polyketide synthase operons, but none of these operons resemble the predicted modular composition of the genes required to synthesize mycolactone (10). It is possible that M. ulcerans may have appropriated an additional polyketide synthase locus, and interestingly, the streptomycetes are a rich source of these enzymes (68). We are currently performing genomic subtractions between M. ulcerans and M. marinum to identify additional M. ulcerans-specific sequences.
Environmental PCR-based surveys have shown that M. ulcerans is present in water and detrital material from swamps in M. ulcerans-endemic areas in southeastern Australia (53, 60). In West Africa, aquatic insects appear to be a source of the organism rather than water or plant material (47). These data suggest that M. ulcerans may occupy different environmental niches in different geographical regions. The multilocus sequencing data (Fig. 1 and 2) and previous molecular typing studies (29, 48, 58) have demonstrated unique genotypes within a geographic region. Variations in genotype according to locale also correlate with phenotypic differences between strains. For example, there are consistent growth rate differences between the African and Australian isolates (41). Combining the findings from the environmental surveys, the genotype data, and the phenotype data, it appears likely that M. ulcerans is adapting to the unique conditions of a particular region. The presence of multiple 16S rRNA alleles also suggests that strains may be in the process of local adaptation. Point mutations within the rRNA operon of mycobacteria that have only a single copy of this operon can confer significant biological effects, such as antibiotic resistance (66).
The PFGE data demonstrated that the M. ulcerans genome was approximately 200 kb smaller than that of M. marinum. Considering that the M. ulcerans genome contains approximately 180 kb of DNA not present in M. marinum (based on 40 copies of IS2606 and 50 to 100 copies of IS2404) (59), there is likely to be at least 380 kb of difference in genetic material between these species. Therefore, in addition to M. ulcerans's having acquired DNA, it may have also undergone a deletion event(s). Other evidence that might suggest deletion of genetic material includes the presence of only a single copy of the 16S rRNA gene in M. ulcerans compared to two copies in M. marinum. This observation may also explain the substantial growth rate differences observed between these species. It also suggests that slow growth may be of selective advantage to M. ulcerans. These advantages may include facilitation of growth as an endosymbiont (9, 28) and survival under nutrient-poor conditions (27). The presence of two copies of the rRNA operon in M. marinum also has taxonomic implications for its current classification as a slow-growing species (67).
M. ulcerans may perhaps best be thought of as an ecotype of M. marinum, that is, an M. marinum progenitor genotype that has adapted to a particular ecological niche (36). The presence of unique M. ulcerans genotypes or subecotypes based on geographic origin represents the continuing evolution and adaptation of the organism to varying environments. This would explain the general process by which isolates from temperate regions of southeastern Australia have evolved differently from strains inhabiting tropical regions.
It has been proposed that M. ulcerans is a legacy of the microbial ecology from the Jurassic Period and that its global distribution can be attributed to the breakup of the supercontinents 150 million years ago (21). However, the global history of M. ulcerans suggested by this study is one of the organism's originating less than 1.2 million years ago and then spreading throughout the world. The absence of any sequence differences or inter-IS variation (58) among African strains of M. ulcerans is evidence of even more recent distribution of the organism across this continent. The level of nucleotide sequence variation observed among isolates from Africa is the same as that reported for M. tuberculosis globally (57), and thus it appears that the African strain may have arisen in the past 18,000 years. Multilocus analysis of more strains from Africa would confirm this proposition.
Future work should now be directed towards whole-genome studies of M. ulcerans and M. marinum using microarray-based comparative techniques similar to those recently applied to strains of Mycobacterium bovis BCG (3). Whole-genome comparisons should reveal the fundamentals of pathogenesis in each of these species, particularly given their close genetic relationship and contrasting phenotypes.
ACKNOWLEDGMENTS
We are grateful to Françoise Portaels, Pam Small, William Chew, David Dawson, Aina Sievers, and Frank Haverkort for the provision of mycobacterial isolates. We also thank Carol Smith and Wayne Meyers for the provision of unpublished data.
This work was supported by a grant from the Australian Research Council.
REFERENCES
- 1.Baess I, Mansa B. Determination of genome size and base ratio on deoxyribonucleic acid from mycobacteria. Acta Microbiol Scand Sect B Microbiol. 1978;86B:309–312. doi: 10.1111/j.1699-0463.1978.tb00049.x. [DOI] [PubMed] [Google Scholar]
- 2.Barker D J. Epidemiology of Mycobacterium ulcerans infection. Trans R Soc Trop Med Hyg. 1973;67:43–50. doi: 10.1016/0035-9203(73)90317-9. [DOI] [PubMed] [Google Scholar]
- 3.Behr M A, Wilson M A, Gill W P, Salamon H, Schoolnik G K, Rane S, Small P M. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 4.Bellgard M I, Gojobori T. Significant differences between the G+C content of synonymous codons in orthologous genes and the genomic G+C content. Gene. 1999;238:33–37. doi: 10.1016/s0378-1119(99)00318-2. [DOI] [PubMed] [Google Scholar]
- 5.Bhatt A, Kieser T. Transposition of IS117 of Streptomyces coelicolor A3(2) in Mycobacterium smegmatis. Microbiology. 1999;145:1201–1207. doi: 10.1099/13500872-145-5-1201. [DOI] [PubMed] [Google Scholar]
- 6.Boddinghaus B, Rogall T, Flohr T, Blocker H, Bottger E C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britten R J. Forbidden synonymous substitutions in coding regions. Mol Biol Evol. 1993;10:205–220. doi: 10.1093/oxfordjournals.molbev.a039996. [DOI] [PubMed] [Google Scholar]
- 8.Burki D R, Bernasconi C, Bodmer T, Telenti A. Evaluation of the relatedness of strains of Mycobacterium avium using pulsed-field gel electrophoresis. Eur J Clin Microbiol Infect Dis. 1995;14:212–217. doi: 10.1007/BF02310358. [DOI] [PubMed] [Google Scholar]
- 9.Cirillo J D, Falkow S, Tompkins L S, Bermudez L E. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect Immun. 1997;65:3759–3767. doi: 10.1128/iai.65.9.3759-3767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 11.Daffe M, Laneelle M A, Lacave C. Structure and stereochemistry of mycolic acids of Mycobacterium marinum and Mycobacterium ulcerans. Res Microbiol. 1991;142:397–403. doi: 10.1016/0923-2508(91)90109-n. [DOI] [PubMed] [Google Scholar]
- 12.Dailloux M, Laurain C, Weber R, Hartemann P. Water and nontuberculous mycobacteria. Water Res. 1999;33:2219–2228. [Google Scholar]
- 13.Dale J W. Mobile genetic elements in mycobacteria. Eur Respir J. 1995;8:S633–S648. [PubMed] [Google Scholar]
- 14.da Silva J, Hughes A L. dSdNqw, version 1.0. University Park, Pa: Pennsylvania State University; 1998. [Google Scholar]
- 15.Dobos K M, Quinn F D, Ashford D A, Horsburgh C R, King C H. Emergence of a unique group of necrotizing mycobacterial diseases. Emerg Infect Dis. 1999;5:367–378. doi: 10.3201/eid0503.990307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endo T, Ikeo K, Gojobori T. Large-scale search for genes on which positive selection may operate. Mol Biol Evol. 1996;13:685–690. doi: 10.1093/oxfordjournals.molbev.a025629. [DOI] [PubMed] [Google Scholar]
- 17.Falkinham J O. Epidemiology of infection by nontuberculous mycobacteria. Clin Microbiol Rev. 1996;9:177–215. doi: 10.1128/cmr.9.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.George K M, Chatterjee D, Gunawardana G, Welty D, Hayman J, Lee R, Small P L. Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science. 1999;283:854–857. doi: 10.1126/science.283.5403.854. [DOI] [PubMed] [Google Scholar]
- 19.Gluckman S J. Mycobacterium marinum. Clin Dermatol. 1995;13:273–276. doi: 10.1016/0738-081x(95)00023-9. [DOI] [PubMed] [Google Scholar]
- 20.Green E P, Tizard M L, Moss M T, Thompson J, Winterbourne D J, McFadden J J, Hermon-Taylor J. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 1989;17:9063–9073. doi: 10.1093/nar/17.22.9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayman J. Mycobacterium ulcerans: an infection from Jurassic time? Lancet. 1984;ii:1015–1016. doi: 10.1016/s0140-6736(84)91110-3. [DOI] [PubMed] [Google Scholar]
- 22.Hayman J. Postulated epidemiology of Mycobacterium ulcerans infection. Int J Epidemiol. 1991;20:1093–1098. doi: 10.1093/ije/20.4.1093. [DOI] [PubMed] [Google Scholar]
- 23.Hayman J, McQueen A. The pathology of Mycobacterium ulcerans infection. Pathology. 1985;17:594–600. doi: 10.3109/00313028509084759. [DOI] [PubMed] [Google Scholar]
- 24.Healy F G, Bukhalid R A, Loria R. Characterization of an insertion sequence element associated with genetically diverse plant-pathogenic Streptomyces spp. J Bacteriol. 1999;181:1562–1568. doi: 10.1128/jb.181.5.1562-1568.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horsburgh C R., Jr Epidemiology of disease caused by nontuberculous mycobacteria. Semin Respir Infect. 1996;11:244–251. [PubMed] [Google Scholar]
- 26.Huson D H. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- 27.Iivanainen E, Sallantaus T, Katila M L, Martikainen P J. Mycobacteria in runoff waters from natural and drained peatlands. J Environ Qual. 1999;28:1226–1234. [Google Scholar]
- 28.Inglis T J J, Rigby P, Robertson T A, Dutton N S, Henderson M, Chang B J. Interaction between Burkholderia pseudomallei and Acanthamoeba species results in coiling phagocytosis, endamebic bacterial survival, and escape. Infect Immun. 2000;68:1681–1686. doi: 10.1128/iai.68.3.1681-1686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson K, Edwards R, Leslie D E, Hayman J. Molecular method for typing Mycobacterium ulcerans. J Clin Microbiol. 1995;33:2250–2253. doi: 10.1128/jcm.33.9.2250-2253.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson P D, Veitch M G, Leslie D E, Flood P E, Hayman J A. The emergence of Mycobacterium ulcerans infection near Melbourne. Med J Aust. 1996;164:76–78. doi: 10.5694/j.1326-5377.1996.tb101352.x. [DOI] [PubMed] [Google Scholar]
- 31.Johnson P D R, Stinear T P, Hayman J A. Mycobacterium ulcerans—a mini review. J Med Microbiol. 1999;48:511–513. doi: 10.1099/00222615-48-6-511. [DOI] [PubMed] [Google Scholar]
- 32.Kapur V, Whittam T S, Musser J M. Is Mycobacterium tuberculosis 15,000 years old? J Infect Dis. 1994;170:1348–1349. doi: 10.1093/infdis/170.5.1348. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S, Tamura K, Nei M. MEGA—Molecular Evolutionary Genetics Analysis, version 1.02. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 34.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marston B J, Diallo M O, Horsburgh C R, Jr, Diomande I, Saki M Z, Kanga J M, Patrice G, Lipman H B, Ostroff S M, Good R C. Emergence of Buruli ulcer disease in the Daloa region of Côte d'Ivoire. Am J Trop Med Hyg. 1995;52:219–224. doi: 10.4269/ajtmh.1995.52.219. [DOI] [PubMed] [Google Scholar]
- 36.Maynard-Smith J. Population genetics: an introduction. In: Neidhardt F C, Curtiss R, Ingraham V C, Lin E C C, Brookslow K, Magasanik B, Reznikoff W S, Ritey M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. II. Washington, D.C.: ASM Press; 1996. pp. 2685–2690. [Google Scholar]
- 37.Moran N A, Munson M A, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc London B Biol Sci. 1993;253:167–171. [Google Scholar]
- 38.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 39.Nei M, Jin L. Variances of the average numbers of nucleotide substitutions within and between populations. Mol Biol Evol. 1989;6:290–300. doi: 10.1093/oxfordjournals.molbev.a040547. [DOI] [PubMed] [Google Scholar]
- 40.Palomino J C, Obiang A M, Realini L, Meyers W M, Portaels F. Effect of oxygen on growth of Mycobacterium ulcerans in the Bactec system. J Clin Microbiol. 1998;36:3420–3422. doi: 10.1128/jcm.36.11.3420-3422.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palomino J C, Portaels F. Effects of decontamination methods and culture conditions on viability of Mycobacterium ulcerans in the Bactec system. J Clin Microbiol. 1998;36:402–408. doi: 10.1128/jcm.36.2.402-408.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philipp W J, Gordon S, Telenti A, Cole S T. Pulsed-field gel electrophoresis for mycobacteria. Methods Mol Biol. 1998;101:51–63. doi: 10.1385/0-89603-471-2:51. [DOI] [PubMed] [Google Scholar]
- 43.Philipp W J, Schwartz D C, Telenti A, Cole S T. Mycobacterial genome structure. Electrophoresis. 1998;19:573–576. doi: 10.1002/elps.1150190418. [DOI] [PubMed] [Google Scholar]
- 44.Picardeau M, Bull T J, Vincent V. Identification and characterization of IS-like elements in Mycobacterium gordonae. FEMS Microbiol Lett. 1997;154:95–102. doi: 10.1111/j.1574-6968.1997.tb12629.x. [DOI] [PubMed] [Google Scholar]
- 45.Portaels F. Epidemiology of mycobacterial diseases. Clin Dermatol. 1995;13:207–222. doi: 10.1016/0738-081x(95)00004-y. [DOI] [PubMed] [Google Scholar]
- 46.Portaels F. Etude d'Actinomycetales isolées de l'homme et de son environnement en Afrique Centrale. Ph.D. thesis. Brussels, Belgium: Faculté des Sciences, Université Libre de Bruxelles; 1978. [Google Scholar]
- 47.Portaels F, Elsen P, Guimares-Peres A, Fonteyne P A, Meyers W M. Insects in the transmission of Mycobacterium ulcerans infection. Lancet. 1999;353:986. doi: 10.1016/S0140-6736(98)05177-0. [DOI] [PubMed] [Google Scholar]
- 48.Portaels F, Fonteyne P A, de Beenhouwer H, de Rijk P, Guedenon A, Hayman J, Meyers M W. Variability in 3′ end of 16S rRNA sequence of Mycobacterium ulcerans is related to geographic origin of isolates. J Clin Microbiol. 1996;34:962–965. doi: 10.1128/jcm.34.4.962-965.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poulet S, Cole S T. Repeated DNA sequences in mycobacteria. Arch Microbiol. 1995;163:79–86. doi: 10.1007/BF00381780. [DOI] [PubMed] [Google Scholar]
- 50.Ramakrishnan L, Valdivia R H, McKerrow J H, Falkow S. Mycobacterium marinum causes both long-term subclinical infection and acute disease in the leopard frog (Rana pipiens) Infect Immun. 1997;65:767–773. doi: 10.1128/iai.65.2.767-773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts B, Hirst R. Immunomagnetic separation and PCR for detection of Mycobacterium ulcerans. J Clin Microbiol. 1997;35:2709–2711. doi: 10.1128/jcm.35.10.2709-2711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogall T, Flohr T, Bottger E C. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J Gen Microbiol. 1990;136:1915–1920. doi: 10.1099/00221287-136-9-1915. [DOI] [PubMed] [Google Scholar]
- 53.Ross B C, Johnson P D, Oppedisano F, Marino L, Sievers A, Stinear T, Hayman J A, Veitch M G, Robins-Browne R M. Detection of Mycobacterium ulcerans in environmental samples during an outbreak of ulcerative disease. Appl Environ Microbiol. 1997;63:4135–4138. doi: 10.1128/aem.63.10.4135-4138.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh S P, Salamon H, Lahti C J, Farid-Moyer M, Small P M. Use of pulsed-field gel electrophoresis for molecular epidemiologic and population genetic studies of Mycobacterium tuberculosis. J Clin Microbiol. 1999;37:1927–1931. doi: 10.1128/jcm.37.6.1927-1931.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soini H, Viljanen M K. Diversity of the 32-kilodalton protein gene may form a basis for species determination of potentially pathogenic mycobacterial species. J Clin Microbiol. 1997;35:769–773. doi: 10.1128/jcm.35.3.769-773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spratt B G, Maiden M C. Bacterial population genetics, evolution and epidemiology. Proc R Soc London B Biol Sci. 1999;354:701–710. doi: 10.1098/rstb.1999.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stinear T, Davies J K, Jenkin G A, Portaels F, Ross B C, Oppedisano F, Purcell M, Hayman J A, Johnson P D R. A simple PCR method for rapid genotype analysis of Mycobacterium ulcerans. J Clin Microbiol. 2000;38:1482–1487. doi: 10.1128/jcm.38.4.1482-1487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stinear T, Ross B C, Davies J K, Marino L, Robins-Browne R M, Oppedisano F, Sievers A, Johnson P D R. Identification and characterization of IS2404 and IS2606: two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J Clin Microbiol. 1999;37:1018–1023. doi: 10.1128/jcm.37.4.1018-1023.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stinear T P, Davies J K, Jenkin G A, Hayman J A, Oppedisano F, Johnson P D R J. The identification of Mycobacterium ulcerans in the environment from regions in which it is endemic in southeastern Australia with sequence capture-PCR. Appl Environ Microbiol. 2000;66:3206–3213. doi: 10.1128/aem.66.8.3206-3213.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Telenti A, Marchesi F, Balz M, Bally F, Bottger E C, Bodmer T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thierry D, Cave M D, Eisenach K D, Crawford J T, Bates J H, Gicquel B, Guesdon J L. IS6110, an IS-like element of Mycobacterium tuberculosis complex. Nucleic Acids Res. 1990;18:188. doi: 10.1093/nar/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tonjum T, Welty D B, Jantzen E, Small P L. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J Clin Microbiol. 1998;36:918–925. doi: 10.1128/jcm.36.4.918-925.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wagner R R, Riley M A. Low synonymous site variation at the lacY locus in Escherichia coli suggests the action of positive selection. J Mol Evol. 1996;42:79–84. doi: 10.1007/BF02198831. [DOI] [PubMed] [Google Scholar]
- 66.Wallace R J, Jr, Meier A, Brown B A, Zhang Y, Sander P, Onyi G O, Bottger E C. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob Agents Chemother. 1996;40:1676–1681. doi: 10.1128/aac.40.7.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wayne L G, Good R C, Bottger E C, Butler R, Dorsch M, Ezaki T, Gross W, Jonas V, Kilburn J, Kirschner P, Krichevsky M I, Ridell M, Shinnick T M, Springer B, Stackebrandt E, Tarnok I, Tarnok Z, Tasaka H, Vincent V, Warren N G, Knott C A, Johnson R. Semantide- and chemotaxonomy-based analyses of some problematic phenotypic clusters of slowly growing mycobacteria, a cooperative study of the International Working Group on Mycobacterial Taxonomy. Int J Syst Bacteriol. 1996;46:280–297. doi: 10.1099/00207713-46-1-280. [DOI] [PubMed] [Google Scholar]
- 68.Xue Y Q, Sherman D H. Alternative modular polyketide synthase expression controls macrolactone structure. Nature. 2000;403:571–575. doi: 10.1038/35000624. [DOI] [PubMed] [Google Scholar]
- 69.Zolg J W, Philippischulz S. The superoxide dismutase gene, a target for detection and identification of mycobacteria by PCR. J Clin Microbiol. 1994;32:2801–2812. doi: 10.1128/jcm.32.11.2801-2812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]