Abstract
The gene encoding a thermoactive pullulanase from the hyperthermophilic anaerobic archaeon Desulfurococcus mucosus (apuA) was cloned in Escherichia coli and sequenced. apuA from D. mucosus showed 45.4% pairwise amino acid identity with the pullulanase from Thermococcus aggregans and contained the four regions conserved among all amylolytic enzymes. apuA encodes a protein of 686 amino acids with a 28-residue signal peptide and has a predicted mass of 74 kDa after signal cleavage. The apuA gene was then expressed in Bacillus subtilis and secreted into the culture fluid. This is one of the first reports on the successful expression and purification of an archaeal amylopullulanase in a Bacillus strain. The purified recombinant enzyme (rapuDm) is composed of two subunits, each having an estimated molecular mass of 66 kDa. Optimal activity was measured at 85°C within a broad pH range from 3.5 to 8.5, with an optimum at pH 5.0. Divalent cations have no influence on the stability or activity of the enzyme. RapuDm was stable at 80°C for 4 h and exhibited a half-life of 50 min at 85°C. By high-pressure liquid chromatography analysis it was observed that rapuDm hydrolyzed α-1,6 glycosidic linkages of pullulan, producing maltotriose, and also α-1,4 glycosidic linkages in starch, amylose, amylopectin, and cyclodextrins, with maltotriose and maltose as the main products. Since the thermoactive pullulanases known so far from Archaea are not active on cyclodextrins and are in fact inhibited by these cyclic oligosaccharides, the enzyme from D. mucosus should be considered an archaeal pullulanase type II with a wider substrate specificity.
Pullulanases (pullulan-6-glucanohydrolase [EC 3.2.1.41]) are classified as type I or type II (amylopullulanase) depending on their ability to degrade α-1,4 glycosidic linkages in starch, amylopectin, and related oligosaccharides. Unlike type II, pullulanase type I is unable to attack α-1,4 glycosidic linkages. Both pullulanase type I and type II attack α-1,6 glycosidic linkages in pullulan, producing maltotriose. All pullulanases known to date are unable to degrade cyclodextrins. In contrast, pullulan hydrolase type I (neopullulanase) and type II (isopullulanase) are able to cleave α-1,4 glycosidic linkages in pullulan, releasing panose and isopanose, respectively, and are highly active on cyclodextrins (32). Enzymes that degrade cyclodextrins faster than starch are designated cyclodextrinases (EC 3.2.1.54; cyclomaltodextrinase) (30).
In recent years, a large number of pullulanases have been isolated, particularly from thermophilic microorganisms (32). The research on pullulanases from thermophiles is interesting not only for understanding enzyme stability but also for discovering improved enzymes for application in the industrial starch hydrolysis process. The first step in the conversion of starch to glucose is liquefaction, which runs at temperatures of 95 to 105°C in the presence of a thermostable α-amylase. Due to the absence of thermoactive enzymes, saccharification follows at a temperature of about 60°C using a pullulanase type I (debranching enzyme) and glucoamylase. The discovery of a thermostable debranching enzyme that is active above 80°C will significantly improve the starch bioconversion process (11).
The pullulanases studied in detail to date are type II enzymes and are derived from extreme thermophilic archaea. Among these enzymes, only those from Pyrococcus woesei (39), Pyrococcus furiosus (7, 14), Thermococcus hydrothermalis (15, 18), and Thermococcus aggregans (33) have been studied at the genetic level, and their corresponding genes have been cloned and expressed in Escherichia coli. Among the thermoactive archaeal enzymes, only the α-amylase from P. furiosus has been expressed in Bacillus subtilis, but no data concerning the expression level or the purification procedure for the enzyme are available (21).
Pullulanase activity was detected in the crude cellular extract of the extreme thermophilic anaerobic archaeon Desulfurococcus mucosus (8). In this article, we report on the cloning and sequencing of a pullulanase gene from D. mucosus. Furthermore, to our knowledge, we present for the first time data concerning the expression of this thermophilic archaeal enzyme in B. subtilis and the purification of the recombinant amylopullulanase. Interestingly, the enzyme possesses a substrate specificity which differs from that of the archaeal pullulanases described to date.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
D. mucosus DSM2162 was grown on DSM184 medium at pH 5.8 and 85°C. Starch (0.5%, wt/vol) was added as the sole carbohydrate source, and sulfur and tryptone were omitted from the medium. The cell density achieved was >108 cells/ml. E. coli DH10B electrocompetent cells were obtained from Stratagene (La Jolla, Calif.). E. coli FD120 expressing the recombinant pullulanase from D. mucosus was grown aerobically at 37°C in Luria-Bertani (LB) medium (40) containing ampicillin (100 μg/ml) and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). The B. subtilis host strain is DN1885 (13). Plasmid pJA803 is a derivative of pDN2801 (13) in which the polylinker region has been modified. B. subtilis JA803 was grown in shake flasks containing BPX medium with chloramphenicol (6 μg/ml) at 37°C, and the supernatant was harvested after 6 days of growth. BPX medium was prepared as a suspension of 100 g of potato flour, 50 g of barley flour, 0.1 g of BAN 5000 SKB (α-amylase [Novo Nordisk A/S], which is inactivated during autoclaving of the medium), 10 g of sodium caseinate, 20 g of soy bean extract, 9 g of Na2HPO4 · 12H2O, and 0.1 g of pluronic acid per liter (final volume). The starting pH of BPX medium was 7.0 (20).
Enzyme activity around the colonies was detected using medium containing 0.07% (wt/vol) AZCL-amylose and/or 0.07% (wt/vol) AZCL-pullulan (Megazyme, Bray, Ireland). Positive clones were identified by halos around the colonies.
Cloning of pullulanase from D. mucosus DSM 2162.
Chromosomal DNA was isolated from D. mucosus DSM2162 by the method of Ramakrishnan and Adams (38). The DNA was partially digested with Sau3A and fractionated on an agarose gel. Fractions of between 1.5 and 8 kb were used for making the DNA library in ZAP express (Stratagene). Phagemids were excised as described in the instructions for users. The mass excised library (7,100 clones) was grown in microtiter plates inoculated at 0.7 CFU/well. After growth for 3 days at 37°C, 50 μl of culture was added to the assay plates, and the remaining microtiter plates containing the E. coli clones were stored at 4°C. The assay plates contained 150 μl of AZCL-amylose (0.07% wt/vol) plus AZCL-pullulan (0.07%, wt/vol) in 50 mM sodium acetate buffer (pH 5.5) and incubated at 80°C for 24 to 48 h. Positive clones were restreaked onto LB plates containing AZCL-amylose (0.07%) and AZCL-pullulan. Plasmid DNA was isolated from the four positive clones using Qiagen miniprep kits (Qiagen, Munich, Germany).
The insert from the E. coli clone AMY1011 containing the pullulanase gene was tagged using the primer island transposition kit (Perkin Elmer Applied Biosystems), and pullulanase-negative, trimethoprim- and kanamycin-resistant clones were selected. The clones were sequenced with primers specific for the transposon using an ABI sequencer. Primer walking was used to fill any gaps.
Subcloning and expression of ORF1 encoding pullulanase in E. coli.
PCR amplification was carried out using the Expand Long Template PCR system (Boehringer Mannheim) at the following temperature profile: 94°C for 2 min and 30 cycles of 94°C for 30 s, 55°C for 45 s, and 68°C for 3 min. Plasmid pSE420 containing the IPTG-inducible trc promoter (Invitrogen) was used for expression in E. coli. Pullulanase activity could be seen after overnight growth of the positive clones on LB medium containing AZCL-pullulan at 37°C and incubation at 70°C for 16 h. Sequencing confirmed the correct insertion of the gene.
E. coli FD120 with the pullulanase gene from D. mucosus (apuA) cloned into pSE420 was inoculated from an overnight culture (LB medium containing ampicillin [100 μg/ml]) into Terrific Broth (40) containing ampicillin (100 μg/ml) and incubated by shaking at 37°C. The cultures were induced with 1 mM IPTG upon reaching an optical density of 0.8 at 600 nm. The cells were harvested after 18 h. The cell pellets were resuspended in 50 mM sodium acetate buffer (pH 5.5) (2 ml/g [wet weight]) and sonicated for 15 min. Following centrifugation, the pullulanase-containing supernatant was assayed for activity and protein concentration as described below.
B. subtilis JA803 expression.
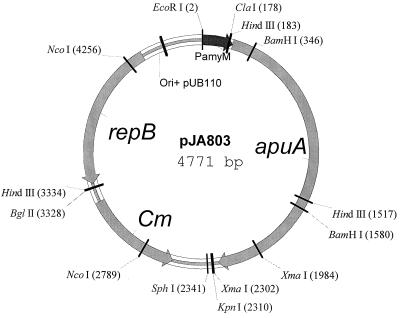
The apuA gene was amplified by PCR and cloned into plasmid pJA803, a derivative of pDN2801 (13) in which the polylinker region has been modified. The chloramphenicol acetyltransferase gene of this plasmid is derived from pC194 of Staphylococcus aureus repB, the gene encoding the replication protein, and the replication origin is from plasmid pUB110 of S. aureus. The host strain is B. subtilis DN1885 (13). The apuA gene encoding the signal and mature protein of the pullulanase was fused to the ATG codon of the promoter region of the maltogenic α-amylase (PamyM) and the restriction site, BamHI, of the cloning vector. PamyM is the promoter of the maltogenic α-amylase gene from Bacillus stearothermophilus (12) (Fig. 1). We monitored enzyme production at 1-day intervals over 6 days by measuring pullulanase activity. Samples of B. subtilis culture fluid were withdrawn and tested for enzymatic activity as described below.
FIG. 1.
Map of plasmid JA803 used for expression of pullulanase type II from D. mucosus. PamyM is the promoter of the maltogenic α-amylase gene from B. stearothermophilus. apuA represents the gene encoding the signal and mature protein of the pullulanase. Cm is the chloramphenicol acetyltransferase gene of plasmid pC194 of S. aureus; repB is the gene encoding the replication protein on plasmid pUB110; and the replication origin is from plasmid pUB110 of S. aureus.
Enzyme assay.
After cultivation of D. mucosus, the culture fluid was centrifuged at 12,000 × g for 30 min at 4°C, and the cell-free supernatant was concentrated up to 100-fold using an Amicon Ultrafiltration system. The cell pellet was resuspended in 50 mM sodium acetate buffer (pH 5.5) and sonicated three times for 3 min at 50% cycle in a Branson 450 sonifier. The cell debris was separated after centrifugation at 10,000 × g for 30 min at 4°C.
Pullulanase activity was determined by measuring the amount of reducing sugars released during incubation with pullulan. To 50 μl of 1% (wt/vol) pullulan dissolved in 50 mM sodium acetate buffer (pH 5.5), 25 or 50 μl of enzyme solution was added, and the samples were incubated at different temperatures for 10 min to 60 min. The reaction was stopped by cooling on ice, and the amount of reducing sugars released was determined by the dinitrosalicylic acid method (3). Sample blanks were used to correct for the nonenzymatic release of reducing sugars. One unit of pullulanase activity is defined as the amount of enzyme that releases 1 μmol of reducing sugars (with maltose as the standard) per min under the assay conditions specified. Pullulanase activity was routinely determined in 50 mM sodium acetate buffer (pH 5.5) at 80°C with 0.5% (wt/vol) pullulan. Protein concentration was determined according to Bradford (6). Amylase activity was determined as above with the substitution of 0.2% (wt/vol) amylose-starch for pullulan. In addition, pullulanase activity was also detected by observing halo formation on agar plates containing the dye substrates AZCL-amylose and AZCL-pullulan.
Purification of recombinant pullulanase from B. subtilis JA803.
The supernatant of 1 liter of culture broth of B. subtilis was centrifuged at 15,000 × g for 20 min and dialyzed against 100 volumes of 50 mM Tris-HCl (pH 8) (buffer A). The supernatant was then loaded onto a Q-Sepharose column (2.5 by 20 cm) equilibrated in buffer A. The column was washed with buffer A, and proteins were eluted with a linear 0 to 0.5 M NaCl gradient in buffer A. Active fractions were pooled, concentrated by ultrafiltration, and dialyzed overnight against 100 volumes of 50 mM potassium phosphate (pH 6) (buffer B) containing 1 M KCl. The enzyme preparation was subjected to hydrophobic interaction chromatography on a HiLoad phenyl-Sepharose column (2.5 by 6 cm) equilibrated in buffer B containing 1 M KCl. After washing the column with 60 ml of starting buffer, a linear reverse gradient of 1 to 0 M KCl in 150 ml of buffer A was applied to the column. The column was then washed with buffer B until no absorbance at 280 nm was detectable. The pullulanase was eluted in a linear gradient of 0 to 40% (vol/vol) dimethyl sulfoxide in buffer B. The active fractions were combined, concentrated by ultrafiltration, and dialyzed overnight against 50 mM sodium acetate buffer (pH 5) (buffer C) containing 0.1 M NaCl. The concentrated sample was applied to a Pharmacia fast protein liquid chromatography (FPLC) Superdex S-200 prep column (1.5 by 96 cm; Pharmacia Biotech Inc.) connected to a Pharmacia FPLC system equilibrated in buffer C. Elution was performed with the same buffer, and 1-ml fractions were collected at a flow rate of 1 ml/min. Fractions containing enzyme were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Gel electrophoresis.
Native polyacrylamide gels containing a gradient of 5 to 27% polyacrylamide were prepared as described by Koch et al. (27). Gels were run at 300 V for 24 h at 4°C. High-molecular-weight marker proteins (Pharmacia Biotech) were used as standards. In order to examine the subunit composition of the pullulanase, protein samples were also analyzed by SDS–12% PAGE as described by Laemmli (29) after heating the samples at 100°C for 5 min. Low-molecular-weight marker proteins (Pharmacia Biotech) were used as standards. Following native PAGE and SDS-PAGE, the proteins were stained with silver as described by Blum et al. (4). Zymogram staining for pullulytic activity was performed according to Furegon et al. (17). The amylolytic activity was detected as follows. After rinsing with water, the native gel was soaked in 50 mM sodium acetate buffer (pH 5) (buffer C) containing 1% (wt/vol) starch at 4°C for 60 min. The gel was incubated further at the optimum temperature for enzymatic activity in buffer C. After 2 min, the gel was incubated in a solution containing 0.15% (wt/vol) iodine and 1.5% (wt/vol) potassium iodide solution until clear bands became visible. The stained gels were stored in 10% (vol/vol) ethanol at room temperature.
Influence of pH and temperature.
In order to study the influence of pH and temperature, experiments were carried out with the purified recombinant enzyme (26 U/mg). The influence of pH on pullulanase activity was determined by using the protocol described above but replacing sodium acetate buffer with 0.12 M Universal buffer to obtain pHs from 3.5 to 10.0. All of the assays were performed at 80°C. To determine the influence of temperature on the enzymatic activity, samples were incubated at temperatures from 40 to 100°C for 10 min. For temperatures above 90°C, an oil bath was used. Thermostability was investigated after incubation of the samples at different temperatures at pH 6.0. In all cases, incubations were carried out in closed Hungate tubes in order to prevent boiling of the solutions. After various time intervals, samples were withdrawn and clarified by centrifugation, and the enzymatic activity was measured.
Characterization of the hydrolysis products.
Hydrolysis products arising after the action of pullulanase on various linear and branched polysaccharides were analyzed by high-performance liquid chromatography (HPLC) using an Aminex HPX-42A column (300 by 78 mm; Bio-Rad, Hercules, Calif.). Double-distilled water was used as the mobile phase at a flow rate of 0.3 ml/min (27, 42). The purified pullulanase was incubated at different temperatures with 0.2% (wt/vol) pullulan, starch, glycogen, amylopectin, maltodextrin, panose, or amylose. Samples were withdrawn at different time intervals, and the reaction was stopped by incubation on ice. In order to distinguish between maltotriose (contains only α-1,4 bonds) and panose or isopanose (contains α-1,4 and α-1,6 bonds), incubation was performed with α-glucosidase from Saccharomyces cerevisiae in 50 mM potassium phosphate buffer (pH 6.0) at 37°C. This enzyme is capable of hydrolyzing α-1,4 but not α-1,6 linkages in short-chain oligosaccharides.
Effect of metal ions and other reagents on pullulanase activity.
The effect of various substances on pullulanase activity was examined after incubation of the purified enzyme (0.2 U/ml, final concentration) with metal ions and other reagents in various concentrations at 80°C for 10 min. Aliquots were withdrawn, cooled on ice, and tested for pullulanase activity.
DNA sequencing.
Plasmid DNA was isolated using Qiagen spin columns (Qiagen). DNA sequencing was performed with an ABI automatic DNA sequencer using primer extension in both directions.
Sequence analysis.
DNA sequence analysis was carried out using the Lasergene program for Windows (DNAStar Inc.). Multiple alignments were carried out using the ClustalW algorithm (44). The BLAST and FASTA algorithms were used to search the databases (1). Signal sequence prediction was carried out using the SignalP program for the Unix (34).
Chemicals.
All chemicals were reagent grade and were obtained from Merck (Darmstadt, Germany) unless otherwise stated. Chemicals for gel electrophoresis were from Serva (Heidelberg, Germany).
RESULTS
Cloning and sequencing of the 3.5-kb insert encoding the pullulanase from D. mucosus DSM2162.
E. coli clone AMY1011, producing a thermostable pullulanase, was obtained as described in Materials and Methods. The entire 3,529-bp insert was sequenced in both directions, and three open reading frames (ORFs) of 2,061, 651, and 360 bp were identified. The large ORF encodes the pullulanase, but no function could be assigned to the smaller ORFs based on sequence homology. PCR amplification of the pullulanase from D. mucosus chromosomal DNA confirmed the origin of the insert DNA (data not shown).
The large ORF was confirmed as encoding a pullulanase by subcloning into plasmid pSE420 and observing activity of the E. coli transformants on AZCL-pullulan plates. This gene is referred to as apuA. It is 1,974 bp in length and encodes a protein of 686 amino acids with a predicted molecular mass of 77 kDa before processing. A Shine-Dalgarno-like sequence (GGAGG) is present 11 to 15 bp upstream from the predicted GTG translational start site. The G+C content of apuA is 52%. A signal sequence of 27 amino acids was predicted, with cleavage taking place between amino acids Ala and Ser, resulting in a protein with a predicted mass of 74 kDa. It was not possible to determine the N terminus of the mature pullulanase isolated from D. mucosus or of the recombinant pullulanase after expression in B. subtilis JA803.
The codon bias of the pullulanase from D. mucosus differs strongly from that of E. coli; arginine is mainly encoded by AGG, leucine by CTA/CTC, and isoleucine by ATA. In fact, nine rare codons are present in the first 22 amino acids (positions 4, 12, 13, 14, 16, 17, 19, 21, and 22).
The nucleotide and deduced amino acid sequences of apuA have been submitted to GenBank under accession number AF247191.
Expression of pullulanase in E. coli clone FD120.
The apuA gene with the signal sequence and GTG start site altered to ATG was subcloned into expression vector pSE420 under control of the trc promoter, yielding the E. coli clone FD120. Pullulanase activity could be detected after overnight growth on LB containing AZCL-pullulan and subsequent incubation at 70°C, confirming the correct reading frame of the gene. However, due to the low intracellular expression levels, the pullulanase could not be purified from the E. coli culture.
Expression and purification of pullulanase from B. subtilis clone JA803.
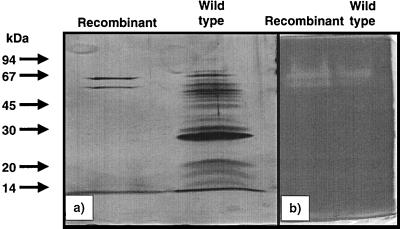
The apuA gene of D. mucosus containing its full signal sequence was cloned into B. subtilis under the control of the PamyM promoter as described in Materials and Methods. A map of the plasmid construct is shown in Fig. 1. Samples were collected at 1-day intervals for the 6-day growth period in shake flasks to determine optimal pullulanase expression. Supernatant from 3 liters of culture broth was collected after 6 days of cultivation and used for purification. The results of the purification procedure are summarized in Table 1. After anion-exchange chromatography, hydrophobic interaction, and gel filtration, a pullulanase was purified 76-fold, with a specific activity of 26 U/mg and a final yield of 8.2%. Proteins from the purification steps were separated using SDS-PAGE (12%). The sample after gel filtration revealed two protein bands with apparent masses of 66 and 50 kDa (Fig. 2a). The zymogram staining shows that both bands are active on dyed pullulan by forming yellow spots (Fig. 2b). Since the mass of the native protein calculated using native gradient gel electrophoresis is 110 kDa, the enzyme is hence a dimer, apparently composed of two subunits (data not shown). The 66-kDa size is closest to that predicted for the apuA gene (74 kDa), whereas the smaller band is probably due to proteolytic degradation of the large 66-kDa protein. The N-terminal sequence could not be determined for either polypeptide, and so the nonrecombinant enzyme was partially purified from D. mucosus to compare the molecular masses. Although the native enzyme from D. mucosus was not purified to homogeneity, the band with the highest molecular mass in the active fraction of the gel filtration comigrated with the purified recombinant enzyme from B. subtilis. According to SDS molecular weight markers, both the wild-type and recombinant enzymes are 66 kDa (Fig. 2a). In addition, wild-type and recombinant enzymes exhibited identical zymogram patterns in native (data not shown) and denaturing acrylamide gel electrophoresis (Fig. 2b).
TABLE 1.
Purification of the cloned pullulanase of D. mucosus after expression in B. subtilisa
| Purification step | Total protein (mg) | Total activityb (U) | Spec. act. (U/mg) | Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|
| Crude extract | 1,376 | 474 | 0.34 | 100 | 1 |
| Q-Sepharose | 232 | 204 | 1.14 | 43 | 3.35 |
| Phenyl-Sepharose | 4.4 | 92 | 20.9 | 19.4 | 69.7 |
| Superdex S-200 | 1.5 | 39 | 26 | 8.2 | 76 |
After the aerobic growth of B. subtilis at 37°C, an 800-ml culture was centrifuged, and the supernatant was applied to the column.
One unit of pullulanase catalyzes the formation of 1 μmol of reducing sugars per min under the defined conditions; maltose was used as a standard.
FIG. 2.
Gel electrophoretic analysis and zymogram of the pullulanase from D. mucosus. Arrows indicate positions of molecular size markers. (a) Comparison of molecular mass between the purified cloned and partially purified pullulanases from D. mucosus. Proteins were detected by silver staining (5). (b) Zymogram of the same samples, as described in Materials and Methods.
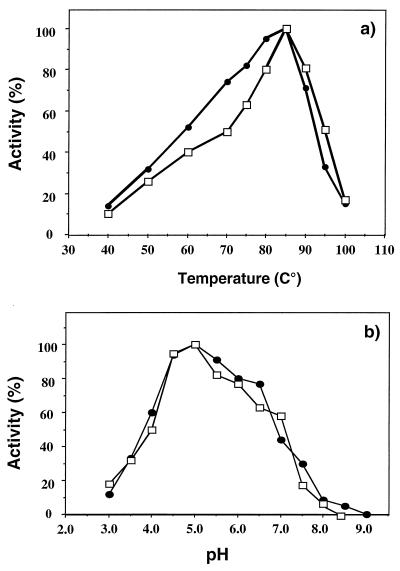
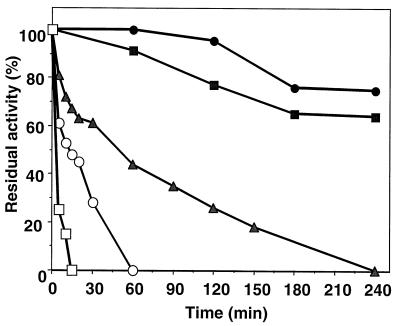
The recombinant full-length D. mucosus pullulanase (rapuDm) is active between 40 and 90°C. The temperature optimum of the purified enzyme is 85°C, and a rapid decrease in pullulanase activity was observed above this point. (Fig. 3a). RapuDm shows activity over a broad pH range, with an optimum at pH 5.0. Interestingly, the enzyme retains 60% of the initial activity at pH 4.0 (Fig. 3b). In order to evaluate the thermostability of rapuDm, we carried out the experiments after prolonged incubation at temperatures from 70 to 90°C. RapuDm was stable at 80°C for 4 h and exhibited a half-life of 50 min at 85°C (Fig. 4).
FIG. 3.
Influence of temperature (a) and pH (b) on the activity of the cloned pullulanase from D. mucosus with pullulan (●) and starch (□) as the substrate. For determination of the temperature optimum, the purified enzyme (0.01 mg/ml) was incubated in sodium acetate buffer (pH 5.0), and incubation was performed for 20 min at various temperatures from 35 to 100°C. The reported values are the percentages of the maximal activity.
FIG. 4.
Thermostability of the recombinant pullulanase from D. mucosus. The enzyme (0.2 U/ml) was incubated at 75°C (●), 80°C (■), 85°C (▴), 90°C (○), and 100°C (□). Samples were withdrawn and tested for pullulanase activity.
Substrate specificity and analysis of hydrolysis product.
A number of different α-glucans were incubated with purified rapuDm in order to ascertain the substrate specificity. As shown in Table 2, rapuDm preferentially hydrolyzed pullulan. Amylose was hydrolyzed with a high velocity, while amylopectin was cleaved at very low rate. The enzyme did not cleave glycogen and dextran, the latter containing only α-1,6 glycosidic linkages.
TABLE 2.
Hydrolysis of various substrates by the recombinant pullulanase from D. mucosusa
| Glucose polymer | Relative rate of hydrolysis (%) |
|---|---|
| Pullulan | 100 |
| Amylose | 77 |
| Starch | 62 |
| Amylopectin | 35 |
| Glycogen | 0 |
| Dextran | 0 |
| α-Cyclodextrin | 0 |
| β-Cyclodextrin | 21 |
| γ-Cyclodextrin | 71 |
Substrates were dissolved in 50 mM sodium acetate buffer (pH 5.0) at a final concentration of 0.2%. Enzyme (0.5 U) was added, and incubation was performed at 85°C for 20 min.
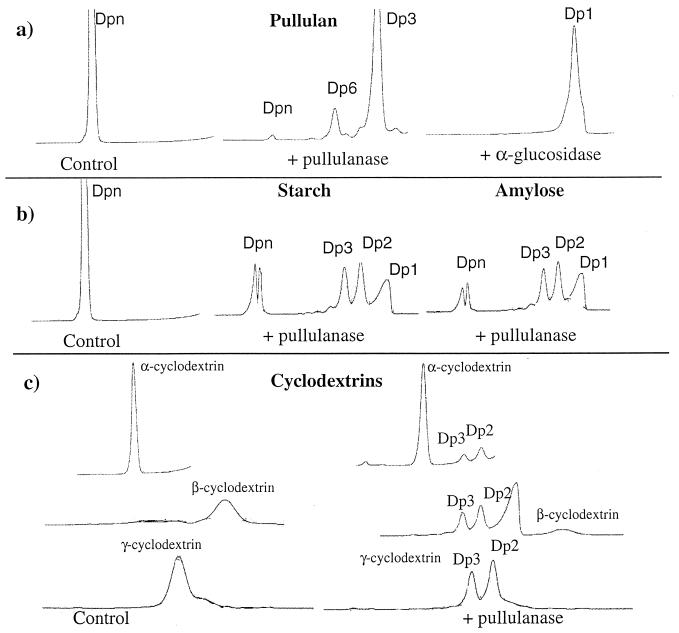
Individual polysaccharides and oligosaccharides were incubated with the pullulanase, and the products were separated and identified by HPLC analysis. The thermostable pullulanase hydrolyzed pullulan after 1 h of incubation at 85°C to generate maltotriose as the major product and maltohexaose as a minor product (Fig. 5a). In order to confirm that the hydrolysis product from pullulan was maltotriose (possessing two α-1,4 glycosidic linkages) and not panose or isopanose (possessing α-1,4 and α-1,6 glycosidic linkages), we incubated the hydrolysis products of pullulan with α-glucosidase from S. cerevisiae. Under these conditions, glucose was formed as the major product, indicating that maltotriose was completely converted by α-glucosidase to glucose. Accordingly, the pullulanase attacks pullulan and releases maltotriose. (Fig. 5a).
FIG. 5.
HPLC analysis of hydrolysis products formed after incubation of the purified recombinant pullulanase in the presence of 0.5% pullulan (a), 0.5% starch and 0.2% amylose (b), and 0.5% cyclodextrins (c). Samples were incubated at 80°C, and at different time intervals aliquots were withdrawn and analyzed on an Aminex HPX 42-A. The products formed after pullulan hydrolysis were incubated with commercial α-glucosidase and then analyzed by HPLC (a). Dp, degree of polymerization (Dp1, glucose; Dp2, maltose; etc.).
Incubation of soluble starch, amylose (Fig. 5b), maltoheptaose, maltohexaose, maltopentaose, maltotetraose, or cyclodextrins (Fig. 5c) with pullulanase for 16 h resulted in the formation of maltose and maltotriose. Maltotetraose is the smallest maltooligosaccharide which can be hydrolyzed by rapuDm. These results indicate that the enzyme acts on pullulan as a true pullulanase and has an endo-acting α-amylase activity. Since the known thermoactive type II pullulanases from archaea are not active on cyclodextrins and are inhibited by these cyclic compounds, the enzyme from D. mucosus should be considered a new archaeal pullulanase with a wider substrate specificity.
Kinetic studies.
The Km of the enzyme with pullulan and starch as the substrate was 0.25 and 5.88 M, respectively. Since the enzyme displayed dual activity with respect to glycosidic bond cleavage (α-1,4 and α-1,6), kinetic analysis of the purified enzyme in a system which contained both pullulan and amylose as competing substrates was performed. This approach would help us to estimate whether one or two different active sites were involved. The competition kinetic studies with mixed substrates were done with a pullulan concentration of between 0.1 and 0.5 M (the amylose concentration was kept constant at 0.1 M). The apparent maximum velocity for the cleavage of amylose (Va) and that for the cleavage of pullulan (Vp) was 102 and 170 μg (as glucose) per min per ml, respectively. The apparent maximum velocity (V) for the mixture of substrates (amylose-pullulan, 1:1) was estimated to be 175 μg (as glucose) per min per ml. Since the value of V is not larger than the sum of Va + Vp due to the competition of the substrates, this result indicates that the pullulanase from D. mucosus has putatively one single active site responsible for the cleavage of α-1,4 and α-1,6 glycosidic linkages.
Effect of metal ions and other reagents.
Divalent cations such as Zn2+, Cu2+, and Fe2+ inhibited enzyme activity, while Ca2+ and Mn2+ had no effect. This is in contrast to reported pullulanases, for which Ca2+ ions are required for full activity. EDTA was slightly inhibitory, suggesting that this chemical did not chelate a possible divalent cation(s) required for the activity of the thermostable pullulanase. Strong inhibition by N-bromosuccinimide (NBS) was observed at a low concentration, suggesting the crucial involvement of tryptophan residues as well as histidine residues at the active site. The activity of the pullulanase was not inhibited by α-, β-, and γ-cyclodextrins, which are generally known as possible competitive inhibitors of archaeal pullulanases (Table 3). In fact, they are enzyme substrates yielding maltotriose and maltose as products after prolonged incubation.
TABLE 3.
Influence of different reagents on pullulanase activitya
| Reagent | Concn | Pullulanase activity (% of control) |
|---|---|---|
| None | 100 | |
| Ammonium sulfate | 0.5 M | 36 |
| EDTA | 5 mM | 63 |
| SDS | 1 mM | 0 |
| Triton X-100 | 0.1% | 100 |
| 1% | 100 | |
| Urea | 4 M | 81 |
| 6 M | 63 | |
| Guanidine HCl | 0.4 M | 63 |
| 1 M | 0 | |
| Iodoacetamide | 10 mM | 100 |
| 20 mM | 95 | |
| MnCl2, MgCl2, CaCl2 | 5 mM | 100 |
| NiCl2 | 5 mM | 92 |
| FeSO4 | 5 mM | 60 |
| ZnCl2 | 5 mM | 51 |
| CuCl2 | 5 mM | 0 |
| N-Bromosuccinimide | 0.01% | 0 |
| α-Cyclodextrin | 0.1% | 120 |
| β-Cyclodextrin | 0.1% | 120 |
| γ-Cyclodextrin | 0.1% | 120 |
Purified pullulanase was extensively dialyzed against 50 mM sodium acetate buffer (pH 5.0). Samples (2 μg) were preincubated with the above reagents and ions at 60°C for 15 min (200 μl final volume). Aliquots (10 μl) were tested for pullulanase activity by incubating the samples with 0.5% pullulan in 50 mM sodium acetate buffer (pH 5.0) at 85°C for 20 min. The values shown are the percentages of the activity without additives.
Sequence comparisons of apuA with other pullulanases.
The amino acid sequence of the apuA product demonstrated highest amino acid homology (45.4%) to the pullulanase from the archaeon T. aggregans DSM10597 (EMBLNEW AJ251532). apuA from D. mucosus showed no homology to the pullulanase type II (amylopullulanase) genes of two other archaea, P. furiosus and T. hydrothermalis. On comparison with the sequence databases, ApuA showed up to 29% pairwise amino acid identity with pullulan hydrolase type I (neopullulanases), cyclomaltodextrinases, and maltogenic amylases. Sequence pair distances between all of the pullulytic and amylolytic enzymes are presented in Table 4. BLASTP alignment revealed low overall homology with sequences in the database, but areas of high amino acid conservation could be seen between the D. mucosus sequence and neopullulanases and cyclomaltodextrin hydrolases. The four regions conserved among amylolytic enzymes (31) could be identified in the pullulanase sequence (Table 5). These could not be identified in the T. hydrothermalis or P. furiosus pullulanase sequences. In contrast to other pullulanase sequences, ApuA is only 686 amino acids in length, making it one of the smallest pullulanases found to date.
TABLE 4.
Pairwise similarity between pullulanase amino acid sequencesa
| Pullulanase sequenceb | Similarity (%) with pullulanase sequence:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| 1. R08255 | 100 | 99.7 | 85.9 | 69.2 | 56.9 | 58.1 | 56.5 | 49.0 | 52.9 | 43.8 | 46.0 | 25.0 | 26.9 |
| 2. P38940 | 100 | 86.2 | 69.4 | 56.9 | 58.1 | 56.7 | 49.0 | 52.9 | 43.8 | 46.2 | 25.0 | 27.0 | |
| 3. O69007 | 100 | 69.2 | 56.1 | 57.9 | 57.0 | 49.7 | 53.2 | 42.7 | 46.3 | 24.8 | 26.9 | ||
| 4. Q45490 | 100 | 60.2 | 62.7 | 58.4 | 48.3 | 54.5 | 42.6 | 44.4 | 22.7 | 24.9 | |||
| 5. Q57482 | 100 | 59.0 | 55.2 | 47.0 | 49.8 | 39.1 | 42.2 | 26.2 | 25.2 | ||||
| 6. O82982 | 100 | 53.1 | 45.5 | 50.7 | 41.0 | 41.8 | 26.1 | 25.7 | |||||
| 7. P32818 | 100 | 46.9 | 48.6 | 38.8 | 42.5 | 25.6 | 28.7 | ||||||
| 8. Q08341 | 100 | 56.6 | 40.7 | 41.6 | 25.7 | 26.7 | |||||||
| 9. Q59226 | 100 | 43.9 | 45.0 | 22.2 | 23.7 | ||||||||
| 10. Q08571 | 100 | 44.4 | 23.2 | 26.7 | |||||||||
| 11. P29964 | 100 | 25.8 | 27.9 | ||||||||||
| 12. AJ251532 | 100 | 45.4 | |||||||||||
| 13. D. mucosus | 100 | ||||||||||||
Sequence pair distances were determined using Clustal with PAM250 residue weight table.
The sequences are as follows; R08255, neopullulanase from Lactobacillus bifidus (SHOKUHIN SANGYO KOS. 1990. WPI; 1990-380803/51); P38940, neopullulanase from B. stearothermophilus (28); O69007, maltogenic amylase from Thermus sp. (26); Q45490, B. stearothermophilus ET1 (9); Q57482, neopullulanase from Bacillus sp. (19); O82982, cyclomaltodextrinase from an alkaliphilic Bacillus; P32818, maltogenic α-amylase from Bacillus acidopullulyticus (23); Q08341, cyclomaltodextrinase from Bacillus sphaericus (35); Q59226, cyclomaltodextrinase from Bacillus sp. strain I-5 (25); Q08751, neopullulanase from Thermoactinomyces vugaris (45); P29964, cyclomaltodextrin hydrolase from Thermoanaerobacter ethanolicus (37); AJ251532, pullulanase from T. aggregans (33); D. mucosus, this study.
TABLE 5.
Regions conserved among pullulanases, cyclomaltodextrinases, and amylasesa
| Enzyme source | Starting positions and amino acid sequences of conserved regions
|
||||||
|---|---|---|---|---|---|---|---|
| Pos. 160–170 | I | II | III | Pos. 402–414 | IV | Pos. 498–512 | |
| D. mucosus | YQIFPDRFYNG | DFVPDH | DGLRIDTPLD | YIVGEIWDY | WLRGNAFDSLMNY | GSHDTSRVLT | GMPVVFQGDERGITG |
| AJ251532 | YQIFPDRFYNG | DFVPNH | DGIRIDAPQE | YIVGEIWEL | WVQGNMFDSLMNY | SSHDTSRVLT | GMPVTFQGDERGLLG |
| P29964 | YQIFPERFNNG | DAVFNH | DGWRLDVANE | IIVGEVWHD | WLRGDQFDSVMNY | GSHDTERFLT | GIPYIYYGDEVGMVG |
| P32818 | YQIFPERFANG | DAVFNH | DGWRLDVANE | YILGEIWHD | WLRGDQFDAVMNY | GSHDTPRILT | GTPCIYYGDEIGLTG |
| P38940 | YQIFPERFANG | DAVFNH | DGWRLDVANE | YILGEIWHD | WLRGDQFDAVMNY | GSHDTSRILT | GSPCIYYGDEIGMTG |
| Q08751 | YQIFPERFANG | DAVFNH | DGWRLDVANE | LIVGEIWHD | WLMGDQFDSVMNY | DSHDTERFLT | GTPLIYYGDEIGMAG |
| Q57482 | YQIFPERFPNG | DAVFNH | DGWRLDVANE | YILGEVWHD | WLQGDQFDAVMNY | GSHDTPRILT | GTPCIYYGDEIGLTG |
| Q45490 | YQIFPERFANG | DAVFNH | DGWRLDVANE | YILGEIWHD | WLRGDQFDAVMNY | GSHDTPRLLT | GTPCIYYGDEIGITG |
| Conserved | ∗∗∗∗∗ ∗∗ ∗∗ | ∗ | ∗∗ | ∗∗∗ | ∗ ∗ ∗∗ ∗∗∗ | ∗ ∗ ∗ ∗∗ | ∗ ∗ ∗∗∗ ∗ ∗ |
The underlined amino acids are conserved among all types of amylolytic enzymes, while those highlighted with an asterisk are conserved among all the enzymes listed here. Regions I, II, III, and IV have been previously defined. The numbering refers to the amino acid position (Pos.) in the D. mucosus ApuA sequence. The sequences are as follows: D. mucosus, this study; AJ251532, pullulanase from T. aggregans (33); P29964, cyclomaltodextrin hydrolase from T. ethanolicus (37); P32818, maltogenic α-amylase from B. acidopullulyticus (23); P38940, neopullulanase from B. stearothermophilus (28); Q08751, neopullulanase from T. vulgaris (45); Q57482, neopullulanase from Bacillus sp. (19); Q45490, B. stearothermophilus ET1 (9).
DISCUSSION
In this paper, it has been demonstrated, for the first time to our knowledge, that an archaeal gene encoding an extremely thermostable enzyme can be successfully expressed in B. subtilis and the recombinant enzyme can be purified from the culture fluid and characterized. The expression of the pullulanase in B. subtilis was relatively low at 15 mg/liter, but the protein was present in the supernatant at a level which allowed purification. The difficulties with high-level expression of proteins from archaea in mesophilic hosts may be due to many reasons, such as mRNA stability, codon bias, protein folding, and secretion. The pullulanase of D. mucosus has a typical archaeal codon bias. In this case, nine rare codons (for E. coli expression) are present in the first 22 amino acids.
The recombinant active protein has an apparent molecular mass of 66 kDa, as measured on SDS-PAGE, which was the same as that for the native enzyme isolated from D. mucosus. These results suggest that the recombinant pullulanase is correctly expressed in B. subtilis and secreted. Both wild-type and recombinant pullulanases could migrate faster in SDS-PAGE than expected from their predicted size (74 kDa). Variable apparent molecular masses can generally be attributed to the interactions of the enzyme with the gel filtration matrices or to retention of the globular structure. This has been observed for the amylases from Streptococcus bovis (16) and Clostridium acetobutylicum (36) and the pullulanase from P. woesei (39) and P. furiosus (14). On a native gel, the enzyme has a molecular mass of 110 kDa, which indicates that the enzyme is a dimer. The recombinant pullulanase type I from Fervidobacterium pennivorans Ven5 is a dimeric enzyme (4), whereas the pullulanases described so far are monomeric. In addition, the predicted mass of 74 kDa represents one of the lowest among the pullulanases isolated to date. In fact, only the pullulanase type I from Bacillus flavocaldarius KP1228 (54 kDa) (22) is smaller. Comparing the relative rates of hydrolysis with different substrates showed that the archaeal enzyme preferentially cleaves pullulan and amylose rather than amylopectin or glycogen. Since rapuDm attacks pullulan (producing maltotriose) and other polysaccharides such as starch and amylose, it has been classified as a pullulanase type II or amylopullulanase. Interestingly, rapuDm possesses wide substrate specificity, because it is also able to cleave cyclodextrins, which are usually known as competitive inhibitors of pullulanases. Among the archaeal pullulanases, only that from T. aggregans has been reported to hydrolyze cyclodextrins (33). The significant sequence homology with bacterial cyclodextrinases, the small size of the enzyme, and the hydrolytic action on cyclodextrins led us to conclude that this enzyme, although classified as a pullulanase type II (based on its preference for pullulan), is closely related to cyclodextrinases.
Recent investigations allow a clear distinction between bifunctional type II pullulanases which can be divided into two groups: those enzymes that hydrolyze both α-1,4 and α-1,6 glycosidic bonds at the same active site, and those that catalyze the two activities at different active sites (2, 24). Although more detailed studies are required to determine the number of the active sites present in a protein, our results are indicative of competition between pullulan and amylose, suggesting the presence of a single active site capable of hydrolysis of both types of linkages. In addition, linear sequence alignment with the cyclodextrinase of Thermoanaerobacter ethanolicus (formerly Clostridium thermohydrosulfuricum 39E) (37), which possesses one catalytic site, showed clearly that the three catalytic residues, Asp-325, Glu-354, and Asp-421, were absolutely conserved in the D. mucosus pullulanase sequence at positions Asp-363, Glu-394, and Asp-461.
The optimal pH for enzyme activity (pH 5.0) was within the range of pHs reported for most bacterial and archaeal amylases and pullulanases (4, 7, 8) and is in agreement with the acidic catalysis mechanism proposed for these enzymes (26, 41). The optimum temperature for activity was 85°C, reflecting the growth temperature of D. mucosus, which is 85°C. This novel thermostable pullulanase with activity at low pH (60% relative activity at pH 4.0) is an interesting candidate for application in the starch industry. Future investigations will focus on overexpression of this enzyme in the mesophilic industrial host B. subtilis and studies on structure-function relationships.
ACKNOWLEDGMENT
Financial support by the European Union (Generic EU-Project Extremophiles as Cell Factories) is greatly appreciated.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D L. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ara K, Igarashi K, Saeki K, Ito S. An alkaline amylopullulanase from alkalophilic Bacillus sp. KSM-1378: kinetic evidence for two independent active sites for the α-1,4 and α-1,6 hydrolytic reactions. Biosci Biotechnol Biochem. 1995;59:662–666. [Google Scholar]
- 3.Bernfeld P. Amylases α and β. Methods Enzymol. 1955;I:149–155. [Google Scholar]
- 4.Bertoldo C, Duffner F, Jørgensen P L, Antranikian G. Pullulanase type I from Fervidobacterium pennavorans Ven5: cloning, sequencing, and expression of the gene and biochemical characterization of the recombinant enzyme. Appl Environ Microbiol. 1999;65:2084–2091. doi: 10.1128/aem.65.5.2084-2091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blum H, Beier H, Gross H J. Improved silver staining of plants, proteins, RNA, DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 6.Bradford M M. A rapid sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Brown S H, Kelly R M. Characterization of amylolytic enzymes having both α-1,4 and α-1,6 hydrolytic activity from the thermophilic archaea Pyrococcus furiosus and Thermococcus litoralis. Appl Environ Microbiol. 1993;59:2614–2621. doi: 10.1128/aem.59.8.2614-2621.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canganella F, Andrade C, Antranikian G. Characterisation of amylolytic and pullulytic enzymes from thermophilic archaea and from a new Fervidobacterium species. Appl Microbiol Biotechnol. 1994;42:239–245. [Google Scholar]
- 9.Cha H J, Yoon H G, Kim Y W, Lee H S, Kim J W, Kweon K S, Oh B H, Park K H. Molecular and enzymatic characterization of a maltogenic amylase that hydrolyzes and transglycosylates acarbose. Eur J Biochem. 1998;253:251–262. doi: 10.1046/j.1432-1327.1998.2530251.x. [DOI] [PubMed] [Google Scholar]
- 10.Chung H, Yoon S, Lee M, Kim M, Kweon K, Lee I, Kim J, Oh B, Lee H-S. Characterization of a thermostable cyclodextrin glucanotransferase isolated from Bacillus stearothermophilus ET1 J. Agric Food Chem. 1998;3:952–959. [Google Scholar]
- 11.Crabb W D, Mitchinson C. Enzymes involved in the processing of starch to sugars. Trends Biotechnol. 1997;15:349–352. [Google Scholar]
- 12.Diderichsen B, Christiansen L. Cloning of a maltogenic α-amylase from Bacillus stearothermophilus. FEMS Microbiol Lett. 1988;56:53–60. [Google Scholar]
- 13.Diderichsen B, Wedsted U, Hedegaard L, Jensen B R, Sjøholm C. Cloning of aldB, which encodes α-acetolactate decarboxylase, an exoenzyme from Bacillus brevis. J Bacteriol. 1990;172:4315–4321. doi: 10.1128/jb.172.8.4315-4321.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong G, Vieille C, Zeikus J G. Cloning, sequencing, and expression of the gene encoding amylopullulanase from Pyrococcus furiosus and biochemical characterization of the recombinant enzyme. Appl Environ Microbiol. 1997;63:3577–3584. doi: 10.1128/aem.63.9.3577-3584.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erra-Pujada M, Debeire P, Duchiron F, O'Donohue M J. The type II pullulanase of Thermococcus hydrothermalis: molecular characterization of the gene and expression of the catalytic domain. J Bacteriol. 1999;181:3282–3287. doi: 10.1128/jb.181.10.3284-3287.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freer S. Purification and characterization of an extracellular α-amylase from Streptococcus bovis YB. Appl Environ Microbiol. 1993;59:1398–1402. doi: 10.1128/aem.59.5.1398-1402.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furegon L, Curioni A, Peruffo D B A. Direct detection of pullulanase activity in electrophoretic polyacrylamide gels. Anal Biochem. 1994;221:200–201. doi: 10.1006/abio.1994.1397. [DOI] [PubMed] [Google Scholar]
- 18.Gantelet H, Duchiron F. Purification and properties of a thermoactive and thermostable pullulanase from Thermococcus hydrothermalis, a hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Appl Microbiol Biotechnol. 1998;49:770–777. [Google Scholar]
- 19.Igarashi K, Ara K, Saeki K, Ozaki K, Kawai S, Ito S. Nucleotide sequence of the gene that encodes a neopullulanase from an alkalophilic Bacillus. Biosci Biotechnol Biochem. 1992;56:514–516. doi: 10.1271/bbb.56.514. [DOI] [PubMed] [Google Scholar]
- 20.Jørgensen P L, Skov K W, Diderichsen B. Cloning, sequence, and expression of a lipase gene from Pseudomonas cepatia: lipase production in heterologous hosts requires two Pseudomonas genes. J Bacteriol. 1991;173:559–567. doi: 10.1128/jb.173.2.559-567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jørgensen S, Vorgias C E, Antranikian G. Cloning, sequencing and expression of an extracellular α-amylase from the hyperthermophilic archaeon Pyrococcus furiosus in Escherichia coli and Bacillus subtilis. J Biol Chem. 1997;272:16335–16342. doi: 10.1074/jbc.272.26.16335. [DOI] [PubMed] [Google Scholar]
- 22.Kashiwabara S, Ogawa S, Myyoshi N, Oda M, Suzuki Y. Three domains comprised in thermostable molecular weight 54,000 pullulanase of type I from Bacillus flavocaldarius KP 1228. Biosci Biotechnol Biochem. 1999;63:1736–1748. doi: 10.1271/bbb.63.1736. [DOI] [PubMed] [Google Scholar]
- 23.Kelly A P, Diderichsen B, Jorgensen S, McConnell D J. Molecular genetic analysis of the pullulanase B gene of Bacillus acidopullulyticus. FEMS Microbiol Lett. 1994;115:97–106. doi: 10.1111/j.1574-6968.1994.tb06621.x. [DOI] [PubMed] [Google Scholar]
- 24.Kim C H, Kim Y S. Substrate specificity and detailed characterization of a bifunctional amylase-pullulanase enzyme from Bacillus circulans F-2 having two different active sites on the same polypeptide. Eur J Biochem. 1995;227:687–693. doi: 10.1111/j.1432-1033.1995.tb20189.x. [DOI] [PubMed] [Google Scholar]
- 25.Kim T J, Shin J H, Oh J H, Kim M J, Lee S B, Ryu S, Kwon K, Kim J W, Choi E H, Robyt J F, Park K H. Analysis of the gene encoding cyclomaltodextrinase from alkalophilic Bacillus sp. I-5 and characterization of enzymatic properties. Arch Biochem Biophys. 1998;353:221–227. doi: 10.1006/abbi.1998.0639. [DOI] [PubMed] [Google Scholar]
- 26.Kim T J, Kim M, Kim B, Kim J, Cheong T, Kim J, Park K. Modes of action of acarbose hydrolysis and transglycosylation catalyzed by a thermostable maltogenic amylase, the gene for which was cloned from a Thermus strain. Appl Environ Microbiol. 1999;65:1644–1651. doi: 10.1128/aem.65.4.1644-1651.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koch R, Zablowski P, Spreinat A, Antranikian G. Extremely thermostable amylolytic enzyme from the archaebacterium Pyrococcus furiosus. FEMS Microbiol Lett. 1990;71:21–26. [Google Scholar]
- 28.Kuriki T, Park J H, Imanaka T. Characteristics of thermostable pullulanases from Bacillus stearothermophilus and the nucleotide sequence of the gene. J Ferment Bioeng. 1989;69:204–210. [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Matzke J, Herrmann A, Schneider E, Bakker E P. Gene cloning, nucleotide sequence and biochemical properties of a cytoplasmic cyclomaltodextrinase (neopullulanase) from Alicyclobacillus acidocaldarius: reclassification of a group of enzymes. FEMS Microbiol Lett. 2000;183:55–61. doi: 10.1111/j.1574-6968.2000.tb08933.x. [DOI] [PubMed] [Google Scholar]
- 31.Nakajima R, Imanaka T, Aiba S. Comparison of amino acid sequences of eleven different α-amylases. Appl Microbiol Biotechnol. 1986;23:355–360. [Google Scholar]
- 32.Niehaus F, Bertoldo C, Kähler M, Antranikian G. Extremophiles as a source of novel enzymes for industrial application. Appl Microbiol Biotechnol. 1999;51:711–729. doi: 10.1007/s002530051456. [DOI] [PubMed] [Google Scholar]
- 33.Niehaus F, Peters A, Groudieva T, Antranikian G. Cloning, expression and biochemical characterization of a unique thermostable pullulan-hydrolyzing enzyme from the hyperthermophilic archaeon Thermococcus aggregans. FEMS Microbiol Lett. 2000;19:223–229. doi: 10.1111/j.1574-6968.2000.tb09290.x. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Oguma T, Matsuyama A, Kikuchi M, Nakano E. Cloning and sequence analysis of the cyclomaltodextrinase gene from Bacillus sphaericus and expression in Escherichia coli cells. Appl Microbiol Biotechnol. 1993;39:197–203. doi: 10.1007/BF00228606. [DOI] [PubMed] [Google Scholar]
- 36.Paquet V, Croux C, Goma G, Soucaille P. Purification and characterization of the extracellular α-amylase from Clostridium acetobutylicum ATCC824. Appl Environ Microbiol. 1991;57:212–218. doi: 10.1128/aem.57.1.212-218.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Podkovyrov S M, Zeikus J G. Structure of the gene encoding cyclomaltodextrinase from Clostridium thermohydrosulfuricum 39E and characterization of the enzyme purified from Escherichia coli. J Bacteriol. 1992;171:5400–5405. doi: 10.1128/jb.174.16.5400-5405.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramakrishnan V, Adams M W W. Preparation of genomic DNA from sulfur-dependent hyperthermophilic archaea. In: Robb F T, Place A R, editors. Archaea—a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 95–96. [Google Scholar]
- 39.Rüdiger A, Jorgensen P L, Antranikian G. Isolation and characterization of a heat-stable pullulanase from the hyperthermophilic archaeon Pyrococcus woesei after cloning and expression of its gene in Escherichia coli. Appl Environ Microbiol. 1995;61:567–575. doi: 10.1128/aem.61.2.567-575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Schwermann B, Pfau K, Liliensiek B, Schleyer M, Fischer T, Bakker E P. Purification, properties and structural aspects of a thermoacidophilic α-amylase from Alicyclobacillus acidocaldarius ATCC 27009: insight into acidostability of proteins. Eur J Biochem. 1994;226:981–991. doi: 10.1111/j.1432-1033.1994.00981.x. [DOI] [PubMed] [Google Scholar]
- 42.Spreinat A, Antranikian G. Purification and properties of a thermostable pullulanase from Clostridium thermosulfurogenes EM1 which hydrolyses both α-1,6 and α-1,4-glycosidic linkages. Appl Microbiol Biotechnol. 1990;33:511–518. [Google Scholar]
- 43.Terada Y, Kazutoshi F, Takaha T, Okada S. Thermus aquaticus ATCC 33923 amylomaltase gene cloning and expression and enzyme characterization: production of cycloamylose. Appl Environ Microbiol. 1999;65:910–915. doi: 10.1128/aem.65.3.910-915.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tonozuka T, Ohtsuka M, Mogi S, Sakai H, Ohta T, Sakano Y. A neopullulanase-type α-amylase gene from Thermoactinomyces vulgaris. Biosci Biotechnol Biochem. 1993;57:395–401. doi: 10.1271/bbb.57.395. [DOI] [PubMed] [Google Scholar]