Abstract
Anorexia nervosa (AN), a psychiatric condition defined by low body weight for age and height, is associated with numerous dermatological conditions. Yet, clinical observations report that patients with AN do not suffer from infectious skin diseases like those associated with primary malnutrition. Cell-mediated immunity appears to be amplified in AN; however, this proinflammatory state does not sufficiently explain the lower incidence of infections. Antimicrobial peptides (AMPs) are important components of the innate immune system protecting from pathogens and shaping the microbiota. In Drosophila melanogaster starvation precedes increased AMP gene expression. Here, we analyzed skin microbiota in patients with AN and age-matched, healthy-weight controls and investigated the influence of weight gain on microbial community structure. We then correlated features of the skin microbial community with psoriasin and RNase 7, two highly abundant AMPs in human skin, to clarify whether an association between AMPs and skin microbiota exists and whether such a relationship might contribute to the resistance to cutaneous infections observed in AN. We find significant statistical correlations between Shannon diversity and the highly abundant skin AMP psoriasin and bacterial load, respectively. Moreover, we reveal psoriasin significantly associates with Abiotrophia, an indicator for the healthy-weight control group. Additionally, we observe a significant correlation between an individual’s body mass index and Lactobacillus, a microbial indicator of health. Future investigation may help clarify physiological mechanisms that link nutritional intake with skin physiology.
Subject terms: Malnutrition, Microbiome
Introduction
Anorexia nervosa (AN) is a psychiatric condition typically affecting females with an estimated lifetime prevalence between 0.5 and 2.0% and a peak age in onset between 13 and 18 years of age1. The hallmark feature of AN is low body weight for age and height, usually achieved via extreme caloric restriction. AN is complicated by malnutrition that can lead to life-threatening medical consequences as a result of multiple organ failure and immune system dysfunction2,3.
Starvation, malnutrition, altered dietary patterns, and single-nutrient deficiencies can all cause impaired immune functioning that can lead to chronic inflammation and recurrent infections4,5. Indeed, malnourished children chiefly die from “common infections”4,6. Obese individuals, who often have micronutrient deficiencies, experience more frequent and more severe infections4. Paradoxically, clinical observations of patients with AN report an absence of infections as well as delayed or reduced physiological responses to infection7–11. Moreover, AN associated dermatological changes include xerosis (dry skin), increased acne, slower wound healing, generalized pruritis, and seborrheic dermatitis, but an increased risk of skin infections has not been reported12. This is in striking contrast to an increased risk of skin infections associated with primary malnutrition typically seen in developing nations13,14. Cell-mediated immunity appears to be amplified in AN; however, this proinflammatory state does not sufficiently explain the lower incidence of infections2,11,15,16.
Antimicrobial peptides (AMPs) are evolutionarily conserved effector molecules of the innate immune system with broad-spectrum antimicrobial activities17. Psoriasin and RNase 7 are the most abundant AMPs found on human skin that serve immunomodulatory roles in skin immunity through the induction of cytokines and chemokines18,19. In the chronic skin inflammatory disease psoriasis, keratinocytes proliferate in response to inflammatory cytokines, which in turn increases the synthesis of AMPs, including psoriasin, and contributes to the recruitment of T cell subsets and other immune effector cells into the skin20,21. RNase 7 is induced by proinflammatory cytokines and a wide spectrum of potential pathogenic microorganisms such as Staphylococcus aureus and Candida albicans19,22. Similar to psoriasin, RNase 7 is upregulated in psoriasis and atopic dermatitis23,24. It is thus intriguing that the expression of AMP genes are also induced by starvation in Drosophila melanogaster (common fruit fly) in the absence of infection and independent of the pathogen-response pathway25–27. It is possible that this mechanism evolved to ensure innate immune activity during periods of energy deprivation.
Previously, to evaluate whether weight status may also affect AMP expression in human skin, we analyzed the concentrations of the AMPs psoriasin and RNase 7 on the skin surface of patients with AN before and after weight gain. Surprisingly, we found AMP concentrations did not decrease with weight gain, but rather an association of weight gain with increasing AMP concentrations was observed28. While a link between AN and skin immune function has yet to be elucidated, we hypothesize here that changes in the skin microbial profile of patients with AN might contribute to the absence of skin infections observed in this population.
In this study, we conducted an analysis of skin microbiota based on 16S rRNA gene amplicon sequencing in female patients with AN before and after undergoing an inpatient treatment program to gain weight and compared to age-matched healthy-weight controls. To test for possible relationships between AMP concentrations, bacterial load, or body mass index (BMI) and skin microbiota, and to gain insight into whether such relationships might contribute to the resistance to dermatological infections observed in AN, we analyzed skin microbial profiles in conjunction with these measures. We observe increasing levels of bacterial load with weight gain in patients with AN, which is significant at the inner elbow sampling location. In a collective analysis of the sampled body sites, we reveal a significant correlation between psoriasin concentrations and the healthy-weight control group indicator taxon Abiotrophia. In a similar analysis, we further find Shannon diversity significantly negatively correlates with psoriasin concentrations as well as total bacterial load. Psoriasin concentrations also significantly correlate with bacterial loads at the forehead sampling site across study groups. Finally, we observe a significant correlation between an individual’s BMI and Lactobacillus, a significant microbial indicator of health.
Results
Study participants and skin sampling
Thirty-three females diagnosed with AN receiving inpatient medical care and thirty-three healthy-weight age-matched female control subjects from Germany were recruited for this study (see “Methods” for inclusion and exclusion details). One patient with AN withdrew from the study prior to the second sampling point. Patient metadata analyzed in this study are summarized in Table 1; complete study metadata are provided online in Supplementary Table S1. In patients with AN, the mean BMI was 12.56 kg/m2 (SD 1.70) before weight gain and 14.54 kg/m2 (SD 1.70) after weight gain, with a mean weight gain of 5.7 kg (SD 1.50) corresponding to an increase in BMI of 2.0 (SD 0.50) points. The mean BMI of the healthy-weight control group was 22.10 kg/m2 (SD 1.73). All patients with AN had been diagnosed with a severe and life-threatening stage of AN, according to DSM-5 criteria, as represented by a BMI of 15.0 kg/m2 or less. Body mass index standard deviation scores (BMI SDS) in regard to age (years) is provided for the anorexia study cohort in Supplementary Table S2.
Table 1.
Summarized metadata.
| AN (n = 33) | HC subjects (n = 33) | |
|---|---|---|
| Age at time of first measurement | ||
| Mean (SD) | 25.80 years (9.90) | 26.00 years (9.90) |
| Median | 23.00 | 22.50 |
| Range | 17–54 | 16–57 |
| Weight (kg) first measurement, before weight gain | ||
| Mean (SD) | 35.40 (5.2) | 61.55 (6.18) |
| Median | 36.90 | 61.00 |
| Range | 23.40–45.50 | 49–80 |
| BMI first measurement, before weight gain | ||
| Mean (SD) | 12.56 (1.70) | 22.10 (1.73) |
| Median | 12.87 | 22.28 |
| Range | 9.22–14.8 | 18.69–25.84 |
| AN (n = 32) | HC subjects (n = 33) | |
| Weight (kg) second measurement, after weight gain | ||
| Mean (SD) | 40.30 (5.20) | – |
| Median | 42.40 | – |
| Range | 29–47 | – |
| BMI second measurement, after weight gain | ||
| Mean (SD) | 14.54 (1.72) | – |
| Median | 14.81 | – |
| Range | 10.73–17.55 | – |
AN patients with anorexia nervosa, HC healthy weight controls.
To obtain skin bacterial profiles and bacterial load estimates, we extracted DNA from material derived from a skin-rinsing protocol that concurrently collected the antimicrobial peptides psoriasin and RNase 7 from three standardized body sites representing sebaceous, moist, and dry cutaneous zones (Fig. 1). Sampling sites were 1.77 cm2 in size and included the forehead (sebaceous), the antecubital fossa (referred to as inner elbow in this study; moist), and the ventral side of the lower forearm (dry). Patients were positioned accordingly to facilitate sampling procedures, e.g., were placed supine for forehead sampling.
Figure 1.
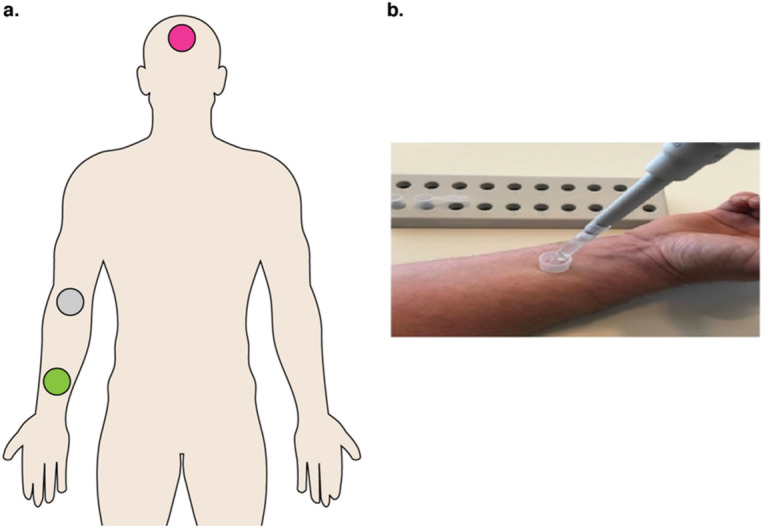
Overview of sampling procedure used in this study. (a) Standardized sampling locations for healthy-weight controls and patients with AN included the forehead, a sebaceous zone (pink), the antecubital fossa/ inner elbow, a moist zone (grey), and the ventral side of the lower arm, a dry zone (green). Patients were positioned accordingly to facilitate sampling procedures, e.g., supine for forehead sampling. Illustration by B. Hermes. (b) Sampling procedure of the distal (lower) forearm. Image photographed by Bendix et al.28.
For the subsequent analyses, we defined three study subject groupings: (1) healthy-weight controls (HC) defined by a BMI ranging between 18.5 and 25.0 kg/m2, (2) patients with AN prior to gain weight (hereafter referred to as AN before weight gain), and (3) patients with AN after undergoing an inpatient protocol to gain weight and after having gained at least 2 kg of body cell mass (hereafter referred to as AN after weight gain). The second sampling point for patients with AN was defined such that sufficient weight gain could be readily achieved by patients while under-going inpatient care (see “Methods” section). We expected an effect on biological parameters after a weight gain of one BMI unit. We analyzed these three groups according to total bacterial load derived from digital droplet PCR (ddPCR), the relative abundance of major taxa and diversity patterns identified in the 16S rRNA gene analysis, and our previously published concentrations of psoriasin and RNase 728. A summary of the mean and median concentrations of psoriasin and RNase 7 is provided in Supplementary Table S3.
Bacterial load
Due to the low microbial biomass of the skin environment and the associated technical challenges29, and the reasonable expectation that AMPs and/or AN disease status could influence bacterial load, we initially measured the total bacterial load of each sample using ddPCR to obtain a precise quantification of target DNA copies, as described by Sze and colleagues30 (see “Methods” section) (Supplementary Table S4). Digital droplet PCR is a method whereby a sample is fractionated into tens of thousands of individual droplets using a water–oil emulsion; PCR is then carried out within each droplet thereby providing reliable, absolute quantification of the target molecule, reducing PCR bias, and increasing signal-to-noise ratios, especially in low biomass samples such as skin30–35. We assessed the distribution of bacterial load between groups at individual sampling locations (Fig. 2). We observe an overall trend of increasing bacterial load with weight gain in patients with AN. However, we largely find that differences in bacterial load between groups are not significant, except for at the inner elbow (antecubital fossa). Here, differences in load between HC and patients with AN after weight gain reach statistical significance but differences are not significant between patients with AN before and after weight gain (Wilcoxon; p = 0.009; p = 0.053, respectively; Fig. 2). We additionally find that bacterial load significantly correlates with psoriasin concentrations at the forehead (Spearman; rs = 0.281, p = 0.019), but not at the inner elbow or lower arm (Supplementary Table S4). Bacterial load did not significantly correlate with RNase 7 concentrations or BMI.
Figure 2.
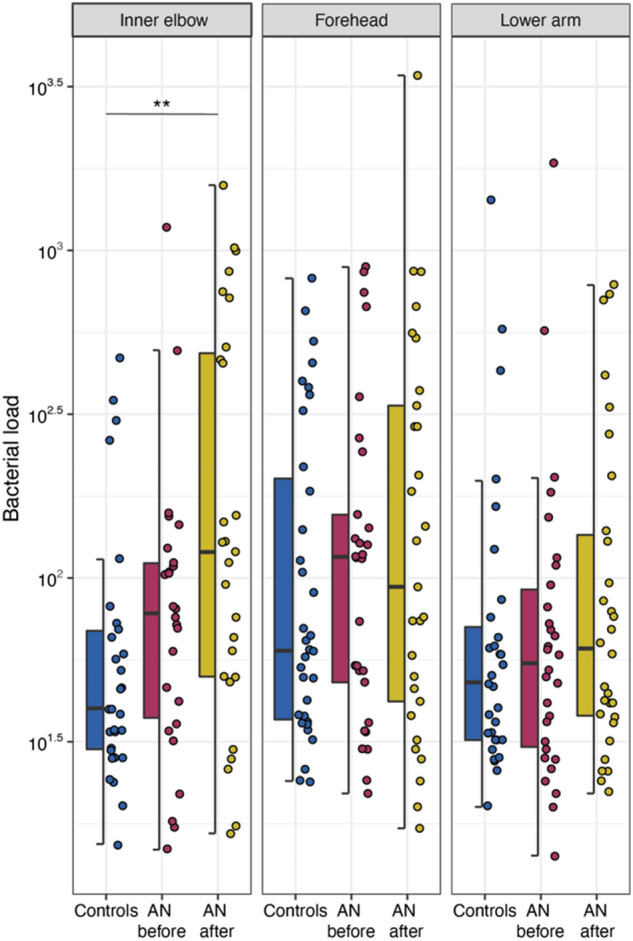
Box plots of total bacterial load, as measured by ddPCR, for study groups at individual sampling locations. AN = anorexia nervosa. Wilcoxon test (see “Methods” section); p-values: *< 0.05; **< 0.01; ***< 0.001. p-values were adjusted for multiple testing according to Benjamini and Hochberg94. Line indicates the median concentration; box shows the interquartile range (IQR), and the whiskers are 1.5 × IQR. Blue represents healthy-weight controls, red represents patients with AN before weight gain, and gold represents patients with AN after weight gain.
Next, we used ddPCR measurements to aid the assessment of potential contamination (see “Methods” section for a detailed description). Briefly, total bacterial load was used as a proxy for input bacterial DNA concentrations for the ‘frequency’ method within the R package ‘decontam’ (version 1.8.0; see “Methods” section)36,37. To verify amplicon sequence variants (ASVs) classified as contaminants (n = 154), we visualized five randomly selected contaminants in frequency plots to examine the distribution of the ASV with respect to bacterial loads. We find that the contaminant ASVs follow an expected pattern in which frequency is inversely proportional to bacterial load, as contaminating DNA will account for a larger fraction of this load in samples with low biomass36. We subsequently utilized the ‘prevalence’ method within ‘decontam’ (version 1.8.0), in which the prevalence (presence /absence) of ASVs in samples is compared to that in negative controls, to identify additional contaminants36. An additional 70 ASVs were identified as contaminants and removed from the dataset. Finally, following the recommendations of Weyrich et al.38, any ASV belonging to families Halomonadaceae (n = 0) or Shewanellaceae (n = 14) were removed. In total, we analyzed more than 400,000 sequences, with a normalized coverage of 1000 sequences per sample (see “Methods” section).
Overview of skin microbiota in patients with AN and healthy-weight controls
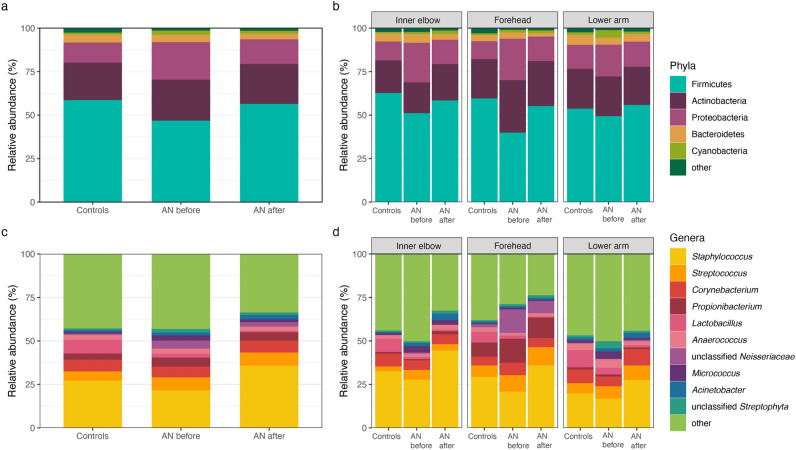
We first analyzed community composition at the phylum and genus levels. The dominant phyla include Firmicutes, Actinobacteria, Proteobacteria, Bacteroidetes, and the dominant genera include Staphylococcus, Streptococcus, Propionibacterium, Corynebacterium, Anaerococcus, and Lactobacillus, whose relative proportions are shown in Fig. 3. Comparisons of relative abundances between groups at the phylum level revealed significant differences in Proteobacteria between patients with AN before and after weight gain and between HC and patients with AN before weight gain (Wilcoxon; p = 0.005, p = 0.014, respectively). Additionally, we find significant differences in Firmicutes abundance between HC and patients with AN before weight gain (Wilcoxon; p = 0.003). At the genus-level, there are significant differences in the relative abundance of Lactobacillus between HC and AN before weight gain, as well as between HC and AN after weight gain groups (Wilcoxon; p = 4.23e−07, 5.12e−08, respectively). Other significant differences between groups include Staphylococcus for AN before compared to HC (Wilcoxon; p = 0.021) and AN before compared to AN after (Wilcoxon; p = 0.005), unclassified Neisseriaceae for both AN before and AN after weight gain compared to HC (Wilcoxon; p = 0.003, p = 0.026, respectively) and unclassified Streptophyta for AN after weight gain compared to HC (Wilcoxon; p = 0.049). Supplementary Table S5 provides a summary of statistical analyses comparing the most abundant phyla and genera between groups.
Figure 3.
Overview of dominant taxa at sampling sites. (a) Bar plot of relative abundances for the most abundant phyla, and (b) at sampling sites (inner elbow, forehead, and lower arm). (c) Bar plot of relative abundances for the most abundant genera, and (d) at sampling sites (inner elbow, forehead, and lower arm). AN anorexia nervosa, Controls healthy-weight controls, AN before patients with AN before weight gain treatment, AN after patients with AN after weight gain treatment.
Diversity indices
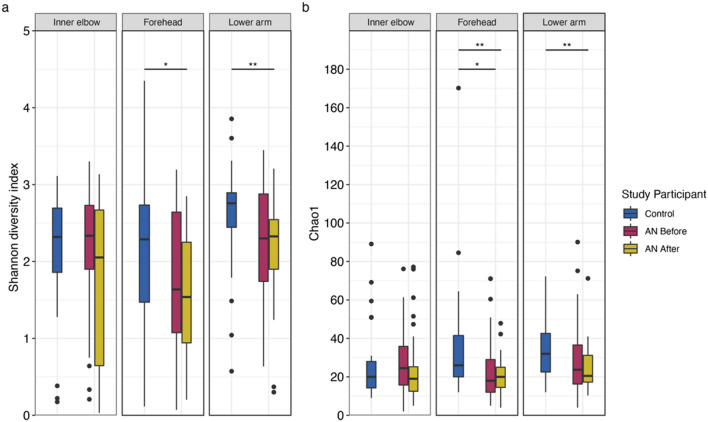
Next, we assessed alpha diversity at the ASV-level to investigate potential effects of AN on skin microbiota. Shannon diversity measures both the richness (number of different species) and evenness (how the species are distributed relative to one another) of the bacterial community, whereas the Chao1 index reflects expected species richness. Surprisingly, we show alpha diversity tends to decrease in patients with AN after weight gain therapy and find significant differences in both community richness and evenness in these patients compared to HC. Specifically, we find a significant difference in Shannon diversity at the forehead between HC and AN after weight gain (Wilcoxon; p = 0.018; Fig. 4a). We also find a significant difference in Shannon diversity at the lower forearm between HC and AN after weight gain (Wilcoxon; p = 0.005; Fig. 4a). For Chao1 diversity, we find a significant difference at the forehead between AN before weight gain and HC and between AN after weight gain and HC (Wilcoxon; p = 0.019; p = 0.007, respectively; Fig. 4b). As with Shannon diversity, there is a significant difference in Chao1 diversity at the lower forearm between HC and AN after weight gain (Wilcoxon; p = 0.009; Fig. 4b). Supplementary Table S6 provides summary statistics for group comparisons.
Figure 4.
Alpha diversity indices for healthy-weight controls and patients with anorexia nervosa (AN), by weight gain status (before and after), at each sampling location. (a) Shannon diversity index. (b) Chao1 diversity index. Wilcoxon test (see “Methods” section); p-values: *< 0.05; **< 0.01; ***< 0.001. p-values were adjusted for multiple testing according to Benjamini and Hochberg94. Line indicates the median concentration; box shows the interquartile range (IQR), and the whiskers are 1.5 × IQR. Blue represents healthy-weight controls, red represents patients with AN before weight gain, and gold represents patients with AN after weight gain.
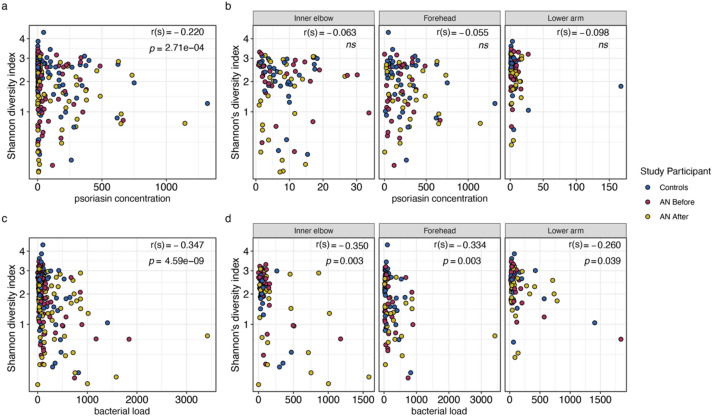
To further explore the trend of decreasing alpha diversity after weight gain in patients with AN, we calculated Spearman correlations for Shannon and Chao1 diversity measures with AMP concentrations, total bacterial load, and BMI (Supplementary Table S7). We find that Shannon diversity is significantly negatively correlated with psoriasin concentrations (Spearman; rho = − 0.220, p = 2.71e−04; Fig. 5a), but not within individual sampling sites (Fig. 5b). We find Shannon diversity also significantly negatively correlates with total bacterial load (Spearman; rho = − 0.347, p = 4.59e−09; Fig. 5c), and moreover, this significant relationship is preserved at the inner elbow (Spearman; rho = − 0.350, p = 0.003), the forehead (Spearman; rho = − 0.334, p = 0.003), and the lower arm (Spearman; rho = − 0.260, p = 0.039; Fig. 5d). Shannon diversity does not significantly covary with RNase 7 concentrations or BMI. Chao 1 index does not significantly correlate with AMP concentrations, total bacterial load, or BMI.
Figure 5.
Spearman correlations between Shannon diversity index and (a) psoriasin concentrations, (b) psoriasin concentrations at individual sampling locations, (c) total bacterial load, and (d) total bacterial load at individual sampling locations. r(s) = spearman’s Rho. AN = anorexia nervosa. ns = not significant. Blue represents healthy-weight controls, red represents patients with AN before weight gain, and gold represents patients with AN after weight gain. p-values were adjusted for multiple testing according to Benjamini and Hochberg94.
To assess overall community compositional differences between groups, we next performed beta diversity analyses. We find no distinguishable separation of study groups based on the Bray–Curtis dissimilarity index (based on abundance) or with the Jaccard index (based on presence/absence), suggesting similar microbial communities amongst the groups (Supplementary Fig. S1). A constrained analysis of principal coordinates of the Bray–Curtis distance (‘capscale’39) with respect to treatment status (i.e., HC, AN before, and AN after) reveals significant differences between the study groups, but treatment status explains only about 1% of the variation between groups (anova.cca; p = 1.00e−04; Supplementary Fig. S2).
Indicator species
To reveal potentially important individual taxa, we conducted indicator species analyses (‘indicspecies’40) at the ASV- and genus-level on a microbiota core defined by a prevalence threshold, whereby a taxon must be present in at least 5% of samples for inclusion in the analysis (see “Methods” section).
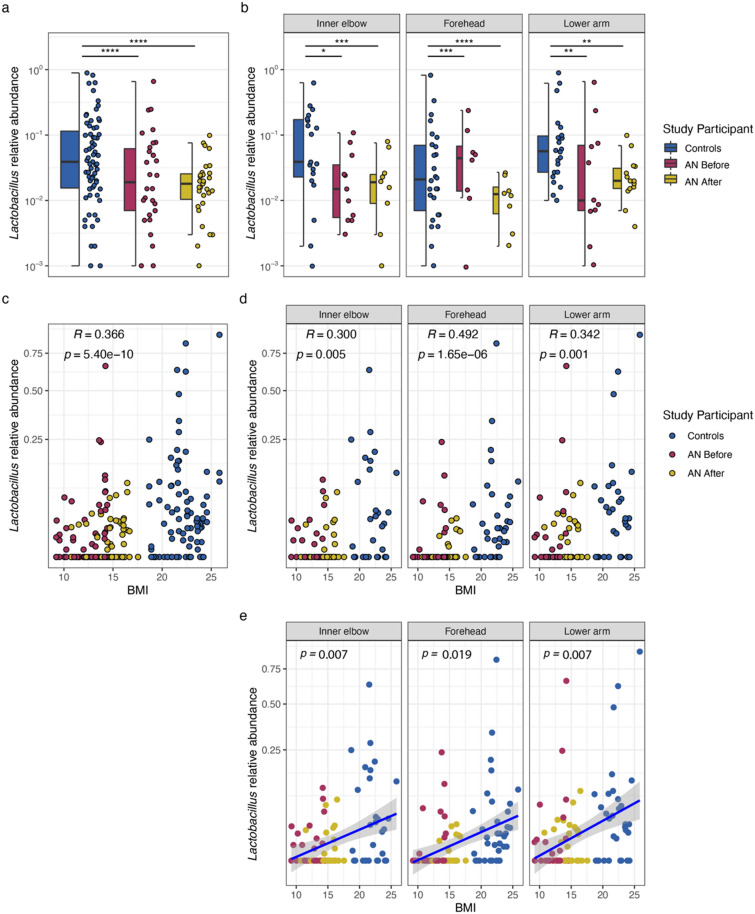
At the genus-level, Lactobacillus, Clostridium sensu stricto, and Abiotrophia associate with the HC group (Table 2). Accordingly, there is a statistically significant difference in the relative abundance of Lactobacillus in HC compared to both AN before and AN after weight gain groups (Wilcoxon; p = 4.23e−07, p = 5.12e−098, respectively; Fig. 6a; Supplementary Table S5). These significant differences are maintained at individual sampling locations (Fig. 6b; Supplementary Table S5). Further, we identify unclassified Neisseriaceae as a significant indicator for both the AN before and AN after groups. Summary statistics for differences in indicator genera between groups, and between groups at individual sampling locations, are presented in Supplementary Table S5.
Table 2.
Indicators at genus- and ASV-level with RDP SeqMatch results.
| Stat | p-value | adj. p-value | RDP SeqMatch result | S_ab score | |
|---|---|---|---|---|---|
| Healthy controls | |||||
| Lactobacillus | 0.706 | 2.00E−05 | 0.002 | Lactobacillus crispatus | 1.00 |
| ASV_29 | 0.706 | 4.00E−05 | 0.005 | ||
| Abiotrophia | 0.404 | 9.00E−04 | 0.030 | Abiotrophia defectiva | 1.00 |
| ASV_160 | 0.404 | 0.001 | 0.028 | ||
| Clostridium sensu stricto | 0.374 | 0.001 | 0.030 | Clostridium spp. | 1.00 |
| ASV_744 | 0.374 | 0.001 | 0.028 | ||
| Patients with AN | |||||
| unclassified Neisseriaceae | 0.502 | 0.002 | 0.047 | NA | NA |
| ASV_13 | 0.502 | 0.003 | 0.049 | ||
Indicator species analysis applied using ‘indicspecies’ (version 1.7.9) with “r.g.” function and 99,999 permutations on a microbial core defined by ASVs classified to the genus-level that are present in at least 5% of all samples (see “Methods” section). p-values were adjusted for multiple testing according to Benjamini and Hochberg94.
Representative 16S rRNA gene sequences were queried via Ribosomal Database Project SeqMatch (version 3).
S_ab scores represent the number of unique 7-base oligomers shared between the query sequence and a given RDP sequence for both type- and non-type strains. The higher the S-ab score, the better the match.
ASV amplicon sequent variant, RDP ribosomal database project, AN anorexia nervosa.
Figure 6.
Box plots of relative abundances, Spearman correlations, and single linear regressions for Lactobacillus, an indicator genus. (a) Box plot of Lactobacillus relative abundances for healthy-weight controls and patients with AN by weight gain group (before and after) and, (b) faceted by sampling location. Wilcoxon test (see “Methods” section); p-values: *< 0.05; **< 0.01; ***< 0.001. Line indicates the median concentration; box shows the interquartile range (IQR), and the whiskers are 1.5 × IQR. (c) Spearman correlation between indicator Lactobacillus and BMI, and at (d) individual sampling locations (e) Single linear regression model between BMI and relative abundance of Lactobacillus at the inner elbow (R2adj = 0.085; p = 0.007), the forehead (R2adj = 0.048; p = 0.019), and at the lower arm (R2adj = 0.076; p = 0.007). r(s) = spearman’s Rho. AN anorexia nervosa, BMI body mass index. Blue represents healthy-weight controls, red represents patients with AN before weight gain, and gold represents patients with AN after weight gain. p-values were adjusted for multiple testing according to Benjamini and Hochberg94).
Subsequently, because the ‘decontam’ procedure was performed on the level of ASVs, we conducted an additional screen to evaluate whether indicator genera could represent contaminants in our dataset, based on the expectation of negative correlations between bacterial load and contaminating taxa36. We thus accordingly calculated Spearman correlations between the relative abundance of indicator genera and bacterial loads. We find no significant correlations between total bacterial load and Lactobacillus, Abiotrophia, Clostridium sensu stricto, or unclassified Neisseriaceae (see “Methods” section; Supplementary Table S8). However, our analysis finds Jeotgalicoccus, an additional indicator genus for the HC group, to negatively correlate with total bacterial load (Spearman; rs = − 0.129, p = 0.034). This association is not significant at individual sampling locations. Nevertheless, following the logic that contaminant sequences are expected to negatively covary with bacterial loads, Jeotgalicoccus was excluded from additional analyses and not reported in Table 2.
At the ASV level, we find three significant indicators for the HC group, and one for both the patients with AN before and after weight gain (Table 2). To refine the taxonomic classification of indicator ASVs, we queried representative sequences using RDP SeqMatch (see “Methods” section; Supplementary Table S9). Indicator ASV_29 is a close match to Lactobacillus crispatus (S_ab score = 1.00). Lactobacilli spp. are well-known human commensals, with previous studies reporting Lactobacilli spp. in the gut, vagina, mouth, on skin, and in breastmilk41–43. A query of indicator ASV_160 reveals a close match to Abiotrophia defectiva (S_ab score = 1.00). Previous studies identified Abiotrophia spp., from the family Lactobacillales, in the oral and upper respiratory flora44. Clostridium sensu stricto was identified in the human gut microbiome in the context of chronic disease45 and was previously classified as a human-associated microbe with pathogenic capabilities46.
To evaluate potential associations between the relative abundance of indicator genera with AMP concentrations and BMI, we calculated Spearman correlations (see “Methods” section; Supplementary Table S8). We find that Abiotrophia significantly positively associates with psoriasin concentrations (Spearman; rs = 0.174, p = 0.004; Supplementary Fig. S3). However, at individual sampling locations, i.e., inner elbow, forehead, and lower arm, these correlations are not significant (Supplementary Fig. S3). Abiotrophia does not significantly correlate with RNase 7 concentrations or BMI. We find that Lactobacillus does not correlate with psoriasin or RNase 7 concentrations. However, we find significant associations between BMI and Lactobacillus (Spearman; rs = 0.366, p = 5.42e−10; Fig. 6c); further, this significant association is maintained at the inner elbow (Spearman; rs = 0.300, p = 0.005), the forehead (Spearman; rs = 0.492, p = 1.65e−06), and lower arm (Spearman; rs = 0.342, p = 0.001; Fig. 6d). Since Lactobacillus is an abundant taxon, we selected it to conduct single linear regression modeling to assess whether BMI predicts the relative abundance of Lactobacillus. We find BMI to be a weak, but significant predictor of Lactobacillus relative abundance at the inner elbow (R2adj = 0.085; p = 0.007), the forehead (R2adj = 0.048; p = 0.019), and at the lower arm (R2adj = 0.076; p = 0.007; Fig. 6e).
Discussion
Our study is the first to characterize the skin microbiota in female patients with AN. We conducted a 16S rRNA gene-based analysis in patients with AN before and after weight gain and with age-matched, healthy-weight controls and then correlated these findings with the concentrations of two highly abundant skin AMPs, psoriasin and RNase 7, and with BMI.
Notably, we find that the concentration of the AMP psoriasin weakly, but significantly, correlates with the indicator genus Abiotrophia for HC. However, at individual sampling locations, this genus does not significantly correlate with psoriasin concentrations, possibly due to low frequencies and low relative abundances.
Recently, Abiotrophia was found to be positively associated with the severity of psoriasis, a mixed autoimmune and autoinflammatory skin disorder marked by elevated psoriasin concentrations47. In this regard, our findings that Abiotrophia significantly positively associates with psoriasin, an established biomarker for psoriasis, supports evidence that links Abiotrophia and psoriasis disease severity. In our study, the relative abundance of Abiotrophia represents less than 1% of the total abundance of skin microbiota. This is fitting, as our study population did not exhibit signs of inflammatory skin disease or psoriasis, and thus, we would not expect this taxon to be a dominant genus. Still, it is interesting that Abiotrophia is an indicator for HC, but not for patients with AN, where one might expect inflammation to occur alongside AN-associated skin changes. The role of cell-mediated immunity in AN is controversial, with several studies reporting an increase in T-cell proliferation and inflammatory cytokine production, including interleukin-1, interleukin-6, and tumor-necrosis factor, when compared to healthy controls or to innate immunity responses in primary malnutrition, where immune function is suppressed 2,11,15,16. However, an earlier investigation conducted by Omodei and colleagues48 found that immune cell populations and the cytokines they produce are reduced in AN, but display greater antioxidant, stress resistance, and anti-inflammatory profiles compared to controls. It is possible that the AN population included in our study exhibits an augmented anti-inflammatory profile, thereby clarifying the relatively reduced levels of microbial taxa associated with inflammatory skin disease, such as Abiotrophia, observed in these subjects.
We also find the Shannon diversity index, which reflects both species richness and species evenness, to significantly negatively associate with psoriasin concentrations and with total bacterial load. Furthermore, we show that both Shannon and Chao1 diversity decreases in patients with AN after weight gain compared to baseline weight and compared to HC, and that there are significant differences in alpha diversity between HC and AN after weight gain.
These results are interesting in the context of our previous work, involving the same study cohort, in which we found that AMP concentrations, and psoriasin in particular, tended to increase in patients with AN after weight gain28. The relationship between the antimicrobial peptide psoriasin and skin microbiota is still unclear. Previous surveys of patients with psoriasis report higher alpha diversity, but with lower stability, compared to healthy skin49. However, other studies report decreased taxonomic diversity in psoriatic skin compared to healthy skin50. These data are notable as increased psoriasin expression is a well-established feature of psoriasis51. We propose that the observation of increasing psoriasin concentrations with weight gain in severely underweight patients may in turn reduce taxonomic diversity on the skin, perhaps through targeted bactericidal activity28,52. It is possible that our findings capture a shift in alpha diversity in low weight patients in response to rising psoriasin levels.
Additionally, we find that BMI significantly correlates with Lactobacillus, another indicator genus for HC. Since Lactobacillus represents a dominant genus in our study, we assessed the possibility that Lactobacillus is a contaminant. We visualized the distribution of Lactobacillus across bacterial loads obtained from ddPCR for the most abundant Lactobacillus ASVs in our dataset. We find that these Lactobacillus ASVs do not follow a pattern of contamination, whereby their frequency would be inversely proportional to input bacterial load. Rather, the frequency of Lactobacillus ASVs are independent of the input ddPCR load data. These findings are consistent with Lactobacillus representing a true biological signal.
The finding of Lactobacillus, and L. crispatus in particular, as an indicator of healthy-weight is congruent with previous studies demonstrating the potential role of Lactobacilli spp. as probiotics for improving skin health and barrier function53,54, anti-aging effects55, and balancing the gut microbial population, thereby preventing inflammatory disease and even cancer at different sites in the body, most likely through the production anti-inflammatory metabolites such as short chain fatty acids56. Moreover, this finding is supported by data showing that Lactobacillus colonizes healthy human skin57–61, including the inner elbow62, forehead63, and scalp64. Interestingly, one study exploring the effects of age on the structure of the skin microbiome found Lactobacillus to be present on the skin of participants aged 20–30 years, but not on those aged 50–60 years65. This finding is particularly interesting given that the mean age of our study population is 25 years for patients with AN and 26 years for HC. However, another survey of the skin microbiome in relation to age and photodamage found increasing age is associated with an increase in Lactobacillus61. Li et al.61 hypothesize that Lactobacillus spp. may increase in response to skin damage (e.g., from UV radiation) that accumulates with age, which may reduce inflammation and improve skin barrier integrity. Moreover, the authors speculate that Lactobacillus, Staphylococcus, and Propionibacterium (Cutibacterium) might act synergistically in skin immunity functions to protect and repair skin from photodamage and inflammation61.
In our study, it is possible that the skin microbiome is responding to inflammation and AN-associated skin changes in patients with AN as nutritional status improves. Although not significant, we find Lactobacillus to increase in patients with AN with weight gain at the inner elbow and lower arm. Conversely, Lactobacillus decreases at the forehead, but here, we also find increasing total bacterial load, which significantly correlates with increasing psoriasin concentrations, and we further observe a non-significant increase in Staphylococcus at the forehead (Supplementary Fig. S3, Supplementary Table S5). The synergistic actions of Lactobacillus, Propionibacterium, and Staphylococcus to mitigate inflammation and repair skin integrity could perhaps explain why we did not observe significant differences between patients with AN before and after weight gain when analyzing these taxa individually. Indeed, when we visualized the sums of the relative abundances for Lactobacillus, Propionibacterium, and Staphylococcus, we observe a significant difference in relative abundance between patients with AN before and after weight gain and between HC and patients before weight gain. Notably, the sum of these relative abundances in patients with AN after weight gain is not significantly different from HC (Supplementary Fig S4).
Lastly, our findings contribute to the growing body of evidence demonstrating that BMI significantly associates with skin microbial diversity. In a study of underweight (BMI 15–18.5), normal-weight (BMI 18.5–25.0), overweight (BMI 25–30), and obese individuals (BMI 30.0–45.0), Brandwein et al.66 found that skin microbial diversity was significantly associated with BMI. Specifically, the authors reported a significant difference in skin microbial diversity between underweight and overweight/ obese individuals and between underweight and normal-weight individuals, but not between normal-weight and overweight/ obese individuals. While limited in terms of sample size and weight categories (underweight and normal-weight only), our work supports the finding that features of the human skin microbiome covary with BMI.
Our study has some limitations. The range of diverse microenvironments (i.e., dry, moist, sebaceous) encompassing human skin as well as the need to consider bacteria living on the skin’s surface and those residing within its deeper layers introduce challenges67–72. Our sampling strategy included a skin washing method that, to our knowledge, has not been implemented in other skin microbiome surveys. This method is advantageous in that it allows for simultaneous collection of AMPs. A potential downside to this method is that the washing solution likely collects only superficial microbes that can be readily flushed off the skin using the rinsing solution. It is therefore possible that our findings are not necessarily comparable to other surveys of the skin microbiome in which methods such as skin scraping or swabbing, that can perhaps collect greater numbers of bacteria and bacteria at various depths, were utilized. This is perhaps evident in our bacterial load findings. Here, we believe the zero bacterial load scores for some samples represents values below the detection limit of our ddPCR method for particularly low biomass skin microbiota samples. Additionally, we find Propionibacterium at the oily forehead location in relative abundances less than that typically found in other skin microbiome surveys73. This finding may be a result of the rinsing solution not efficiently washing off Propionibacterium, which are known to adhere to free fatty acids on the skin73. Additionally, it is possible that other bacteria, such as Lactobacillus, are more readily washed off the skin’s surface and thus may be overrepresented in our results.
Our unique study population is also likely to influence the skin microbial profiles reported here. Our study included young women, including women under 18 years of age, with severe and life-threatening anorexia nervosa. Previous surveys found that pre-pubescent children often harbor low levels or no Propionibacterium on the skin74,75. A common side effect of malnutrition in severe and life-threatening anorexia nervosa is pubertal delay76. Thus, it is possible that the relative abundances of Propionibacterium in our study reflect those reported in surveys of pre-pubescent children. Moreover, a survey comparing skin microbiota profiles of hands between men and women found that taxa from Lactobacillaceae are more abundant on the hands of women77. The abundance of Lactobacillus reported in our study might reflect a larger phenomenon in which young women harbor greater abundances of these commensal bacteria on the skin compared to men. Future studies, especially those comparing the skin washing method with other established methods in the field, are necessary to verify these hypotheses.
Further, the timing of the second sampling point for AMP and skin microbiota collection may have occurred too soon to sufficiently capture additional meaningful changes in the composition of the skin microbiota. The skin microbiome is remarkably stable at the strain level, despite an ever-changing environment, and the composition of the skin microbiome is largely shaped by host physiology78. Given that the patients with AN were still significantly under-weight at the second sampling timepoint, with a mean BMI of 14.54 kg/m2, it is possible that any immune dysregulation affecting microbial composition at the first timepoint was still present after weight gain. Interestingly, Gibson et al.2 speculate that the proinflammatory state in AN is perhaps a primary immunity defect that contributes to the pathogenesis of AN. If immune dysregulation in AN is not necessarily secondary to malnutrition, then it stands to reason improvements in weight and nutrition status in patients with AN would not necessarily affect skin immune processes, and therefore may not lead to substantial changes in skin microbiota. Moreover, it is also feasible that the modest weight gain (at least 2 kg) in patients with AN between sampling timepoints one and two was not enough to alter skin physiology in other ways (e.g., increase sebum production), and therefore not enough to significantly alter microbial community structure. Finally, it is possible that the effect of starvation on AMP levels observed in Drosophila is not readily translated to humans27. Nutritional status and dietary intake affect human physiological and biochemical processes, yet little is known about the effect of nutrition on human skin physiology79,80.
In conclusion, this study explored whether changes in the skin microbial profile of patients with AN might contribute to the absence of skin infections observed in this population. Within the anorexia nervosa cohort, we do not observe an association between skin microbiota features and antimicrobial peptide concentrations. Moreover, we are unable to identify indicator species or other features of the skin microbiota in patients with AN that might provide an explanation for why this population seems to experience less frequent and less severe infections compared to other undernourished populations4,5,7–11. We do, however, find significant statistical correlations between the highly abundant skin AMP psoriasin and features of the skin microbiome of healthy-weight controls. Further investigation of the relationship between psoriasin and skin microbiota in the context of both healthy and diseased states is warranted.
Finally, we reveal a significant statistical correlation between an individual’s BMI and Lactobacillus, a significant microbial indicator of health, at all sampling locations, affirming a previously identified association between BMI and skin microbial diversity65. Further studies examining the relationship between caloric and nutritional intake and skin microbiota in the context of eating disorders may help clarify the physiological mechanisms that link nutritional intake with skin physiology.
Methods
Study subjects
The study was approved by the ethics committee of Hannover Medical School (3209-2016) and was conducted following the Declaration of Helsinki and in accordance with relevant guidelines and regulations. All participants or legal guardians provided written informed consent prior to study inclusion.
Thirty-three female patients diagnosed with AN according to DSM-5 criteria81, and with a body mass index (BMI) of 15 kg/m2 or below were recruited from two inpatient eating disorder facilities in Germany (Klinik Lüneburger Heide and Hannover Medical School). The DSM-5 defines AN by (a) a restriction of energy intake leading to a significant low body weight, (b) an intense fear of gaining weight or becoming fat, and (c) an unduly influence of body weight or shape on self-worth. Patients with AN were investigated shortly after hospitalization, prior to undergoing an inpatient treatment program to gain weight, and again after having achieved an increase in body cell mass of 2 kg or more. The second skin sampling time point was strategically defined. Full weight restoration in adult patients takes years to achieve and is not usually accomplished during inpatient care. Adult patients are typically discharged from inpatient care when they are still underweight, but after some degree of weight gain or symptom reduction has been achieved. Outpatient treatment usually follows discharge. Further, even modest weight gain, e.g., 2 kg, can be a therapeutic challenge in low weight patient populations. Moreover, we expected an effect on biological parameters after a modest weight gain of one BMI unit. Thus, an increase in body cell mass of 2 kg or more, which was deemed to be both therapeutically appropriate and achievable within a reasonable timeframe while still undergoing inpatient therapy, was defined as the second sampling point. One patient with AN withdrew from the study prior to the second sampling point. Randomly selected healthy-weight control subjects, defined by a BMI between 18.5 and 25.0 kg/m2, included thirty-three age-matched females without a psychiatric history and free of current mental disorders. Controls were investigated at one time point. Inclusion criteria for all subjects included a minimum age of 16 years, non-smoking status, and to be visually free from skin disorders. All subjects were free from inflammatory disease and immunosuppressive drugs. Final sample sizes included 287 skin rinsing samples from 32 patients with AN and 33 control subjects (Table 1; Supplementary Table S1).
Subjects underwent a clinical interview to gather socio-demographic information and medical history. Subjects were weighed using a standardized scale. BMI was calculated using height and weight data. Bioelectrical impedance analysis (BIACORPUS RX 4004 M, Medical Healthcare GmbH, Karlsruhe, Germany) was used to verify an increase in body cell mass of at least 2 kg prior to the second sampling point for the patients with AN.
Sampling procedures
A standardized sampling procedure was implemented by Bendix et al.28, whereby sampling was conducted in the same location by one investigator at the same time to minimize putative influences of the circadian rhythm82. Three standardized body sites measuring 1.77 cm2 in size and comprising diverse skin microenvironments (i.e., sebaceous, moist, and dry body regions) were selected: forehead (sebaceous), inner elbow (moist), and ventral side of the lower forearm (dry; Fig. 1). All subjects avoided cosmetics, lotions, and other topical products the morning of testing. Subjects abstained from physical exercise the morning of sampling days, as exercise may increase AMP expression83. Subjects were placed in appropriate positions to collect samples from the various locations. For example, forehead sampling could be carried out with the subject placed in a supine position. Negative sampling controls including ambient air, room controls, and/or negative extraction controls were included for each sampling batch. Ambient air controls containing aliquots of the rinsing buffer solution used at the study site were opened and closed quickly, and then processed as samples. Room controls containing aliquots of the rinsing buffer solution used at the study site were left open for the duration of the rinsing procedure before being processed.
AMPs investigated in this study included psoriasin and RNase 7, which represent the two most abundant AMPs on the surface of human skin84,85. AMP data analyzed in this study was previously reported by Bendix et al.28. AMP sampling was conducted using a skin rinsing method, previously described by Bendix et al.28, Gläser et al.85, and Wittersheim et al.86. Briefly, standardized skin sites were washed by pipet with 1 ml of a rinsing buffer solution (10 mM sodium phosphate buffer, pH 7.4 containing 0.1% Triton X-100) using a sterile, DNA-free plastic ring. The buffer solution was collected, centrifuged (10 min, 10.000×g), and diluted 1:10 with 10% (w/v) bovine serum albumin. Samples were stored at −80 °C until further processing. Quantitative determination of the AMPs was measured by ELISA, as previously described by Gläser et al.87 and Bendix et al.28. A monoclonal antibody derived from hybridoma mouse cells was used for the psoriasin ELISA85. A polyclonal antibody derived from goat was used for the RNase 7 ELISA84. ELISA was performed twice for each sample to ensure reliability. A mean value was calculated from the two sampling measurements and subsequently used in downstream analyses.
Bacteria were collected concurrently with antimicrobial peptides during the skin rinsing protocol and harvested by centrifugation (10 min, 10.000×g). Bacterial genomic DNA was extracted from the resulting pellet formed during the centrifugation step of the skin rinsing procedure using the Ultra-Deep Microbiome Prep extraction kit according to the supplier’s protocol. Samples were extracted in batches corresponding to collection dates and library preparation and subsequent sequencing (see below) was completed in two batches (Supplementary Table S1).
Bacterial load assessment and 16S rRNA gene sequencing
We adapted ddPCR to measure bacterial loads by targeting the V2 hypervariable region of the 16S rRNA gene, as described by Sze and colleagues30. The 20µL ddPCR master mix was prepared according to the manufacturer’s protocol with a final primer concentration of 120 nM and with 10 ng of nucleic acid template. PCR was performed on Bio-Rad C1000 Touch Thermal Cycler with following conditions: 95 °C for 5 min, 40 cycles at 95 °C for 15 s and 60 °C for 1 min, 4 °C for 5 min, 90 °C for 5 min, and incubation at 10 °C. Final products were transferred to QX200™ Droplet Reader and quantified as gene copies (per 20µL) using Bio-Rad QuantaSoft (v.1.7.4.0917).
16S rRNA amplicon library preparation and sequencing were performed as described in Belheouane et al.88 Briefly, hypervariable regions V1-V2 of the bacterial 16S rRNA gene were amplified and sequencing was performed using the dual-index sequencing strategy for amplicon sequencing on the Illumina MiSeq platform89. Negative controls were included in library preparation and sequencing batches. After PCR amplification, the final number of negative controls that were included for sequencing included: ambient air (n = 5), room controls (n = 5), and negative extraction controls with and without rinsing buffer (n = 16, respectively). All controls were processed alongside samples. ZymoBIOMICS Microbial Community Standard cells (Zymo Research) were used as extraction and sequencing controls to assess contamination36.
Data processing and 16S taxonomic classification
Data processing and statistical analyses were performed in R (version 4.0.5). Processing and taxonomic classification of 16S rRNA gene sequence data was performed as previously described88. Sequences were processed using DADA2 (version 1.16.0), resulting in ASV abundance tables90. Taxonomic assignment of ASVs was completed in ‘DADA2’ with the Bayesian classifier using the NR Silva database training set, version 13891.
Contamination assessment
As the skin harbors relatively low microbial biomass, the risk of contamination during skin sampling and sample processing is substantial and any contamination introduced during these steps can radically affect data interpretation, as contaminants tend to be preferentially amplified and sequenced over true biological signals within the sample 29,36,92. Here, we present a detailed description of steps taken throughout this study to mitigate and assess the contribution of contamination.
ddPCR load measurements were used to assess contamination in our dataset via the ‘frequency’ method within the R package ‘decontam’ (version 1.8.0) in conjunction with ‘phyloseq’ (version 1.32.0)36,37,93. The strict probability threshold of 0.1 was used and the batch feature within ‘decontam’ was utilized to analyze samples according to extraction batch to account for any batch effects and differences in contamination between batches. This resulted in the identification and subsequent removal of 154 ASVs labelled as likely contaminants. To verify ASVs classified as contaminants by the ‘frequency’ method within ‘decontam’, we visualized five randomly selected contaminant ASVs in frequency plots to view the distribution of the ASV with respect to total bacterial loads obtained from ddPCR. We find that the ASVs identified as contaminants follow the expected pattern in which frequency is inversely proportional to input ddPCR load, as contaminating DNA will account for a larger fraction of the ddPCR load in samples with low biomass.
Next, we utilized the ‘prevalence’ method within ‘decontam’, in which the prevalence (absence/ presence) of sequence features in samples is compared to the prevalence in negative controls to identify contaminants. The threshold parameter was set to the strict probability threshold of 0.5 and the batch function was utilized to analyze samples according to extraction batch and to account for differences in contamination between batches. An additional 70 ASVs were identified as likely contaminants and subsequently removed from the dataset. Finally, following recommendations of Weyrich et al.38, any ASV belonging to families Halomonadaceae (n = 0) and Shewanellaceae (n = 14) were removed, as these bacteria represent common contaminants in low biomass samples.
To normalize sequencing coverage after the ‘decontam’ filtering procedure, we calculated rarefaction curves to determine the sampling threshold; random sub-sampling to 1000 sequences per sample was performed. Twenty-three samples did not meet the 1000 sequences coverage threshold and were excluded from the analysis. Additionally, 761 ASVs were removed because they were no longer present in any sample after random sub-sampling.
Ecological and statistical analyses
Statistical significance was defined at 0.05 and below for all analyses. Alpha diversity was measured using Shannon and Chao1 indices with ‘vegan’ (version 2.5-6.) As data were not normally distributed, all three groups were compared against each other using paired (AN before versus AN after) and unpaired (AN before versus HC, AN after versus HC) Wilcoxon rank sum tests. Spearman correlations were performed to assess the relationship between alpha diversity and BMI. Overall differences between groups (beta diversity) were assessed using the Bray–Curtis dissimilarity index in Phyloseq; the ‘vegan’ package was used to conduct a constrained analysis of principal coordinates (‘capscale’), a hypothesis-driven ordination that limits the separation of the communities based on the variable tested, for which the ‘anova.cca’ function was applied to assess significance.
Between group relative abundances at the phylum- and genus-level were calculated in ‘phyloseq’ and compared using Wilcoxon rank sum tests. Spearman correlations were performed to assess the relationship between the relative abundances of individual taxa and BMI or concentrations of psoriasin or RNase 7 at individual sampling locations (i.e., forehead, inner elbow, lower arm). Correction for multiple testing was performed according to Benjamini and Hochberg method94.
Indicator species analysis was applied using ‘indicspecies’ (version 1.7.9) with the ‘r.g.’ function95 and 99,999 permutations on a microbial core defined by ASVs classified to the genus-level that are present in at least 5% of all samples. Significant indicator ASVs were selected after correction of p-values for multiple testing using the Benjamini and Hochberg method94. We additionally calculated Spearman correlations between significant indicator taxa at the genus-level and bacterial loads. In cases of contamination, contaminant features are usually inversely proportional to input DNA concentration, as contaminants tend to be preferentially amplified and sequenced over true biological signals within the sample29,36,92. We find no significant negative correlations between bacterial loads and indicator genera reported (see Results), suggesting that these indicator taxa likely represent true biological signal.
Representative 16S rRNA gene sequences were queried via Ribosomal Database Project SeqMatch (version 3)96,97. Results represent classification based on the RDP match score (S_ab), which is the number of unique 7-base oligomers shared between the query sequence and a given RDP sequence for both type- and non-type strains.
Supplementary Information
Acknowledgements
We would like to thank Katja Cloppenborg-Schmdit, Cornelia Wilgus, Heilwig Hinrichs, and Yasmin Claussen for excellent technical assistance. Thank you to Dr. Malte C. Rühlemann and Dr. Shauni Doms for statistical and analytical expertise. Fig. 1a was drawn by Britt M. Hermes and Fig. 1b was photographed by M. Bendix.
Abbreviations
- AN
Anorexia nervosa
- AMP
Antimicrobial peptide
- BMI
Body mass index
- ASV
Amplicon-sequence variants
Author contributions
Conceptualization: B.M.H., J.H., M.Z., G.T., J.F.B., F.R.; Data curation: B.M.H. (16S rRNA gene data); M.B. (AMP data); C.J. (ddPCR data); M.Z. (BMI-SDS); Study (experiment) conduct: M.B., M.Z.; Formal analysis: B.M.H.; Funding acquisition: M.Z., M.B., J.H., J.F.B.; Investigation: B.M.H.; Methodology: B.M.H., J.H., J.F.B.; Project administration: B.M.H., J.H., J.F.B.; Resources: B.M.H., J.H., J.F.B.; Software: B.M.H.; Supervision: J.H., J.F.B.; Validation: B.M.H., J.F.B.; Visualization: B.M.H.; Writing–Original draft preparation: B.M.H.; Writing–Review and editing: all authors.
Funding
Open Access funding enabled and organized by Projekt DEAL. This project was funded by the German Centre for Infection Research (DZIF) of the structured doctoral program (StrucMed) at Hannover Medical School (grant to M.B.). Funding was also provided by the German Science Foundation (DFG) grant RTG 1743 “Genes, Environment and Inflammation”, the DFG Cluster of Excellence 2167 “Precision Medicine in Chronic Inflammation (PMI)” (grant no. EXC2167 to J.F.B.), and the European Union and its program: ERA-NET (European Research Area Networks) Neuron, Grant/Award Number: 01EW1906A; European Union's Framework Programme for Research and Innovation Horizon 2020; German Ministry for Education and Research (BMBF).
Data availability
The data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB47175 (https://www.ebi.ac.uk/ena/browser/view/PRJEB47175).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Jürgen Harder and John F. Baines.
Contributor Information
Britt M. Hermes, Email: hermes@evolbio.mpg.de
Jürgen Harder, Email: jharder@dermatology.uni-kiel.de.
John F. Baines, Email: baines@evolbio.mpg.de
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-19676-6.
References
- 1.Campbell K, Peebles R. Eating disorders in children and adolescents: State of the art review. Pediatrics. 2014;134:582–592. doi: 10.1542/peds.2014-0194. [DOI] [PubMed] [Google Scholar]
- 2.Gibson D, Mehler PS. Anorexia nervosa and the immune system—A narrative review. J. Clin. Med. 2019;8:191. doi: 10.3390/jcm8111915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westmoreland P, Krantz MJ, Mehler PS. Medical complications of anorexia nervosa and bulimia. Am. J. Med. 2016;129:30–37. doi: 10.1016/j.amjmed.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 4.Bourke CD, Berkley JA, Prendergast AJ. Immune dysfunction as a cause and consequence of malnutrition. Trends Immunol. 2016;37:386–398. doi: 10.1016/j.it.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calder PC, Jackson AA. Undernutrition, infection and immune function. Nutr. Res. Rev. 2000;13:3–29. doi: 10.1079/095442200108728981. [DOI] [PubMed] [Google Scholar]
- 6.Rytter MJH, Kolte L, Briend A, Friis H, Christensen VB. The immune system in children with malnutrition—A systematic review. PLoS ONE. 2014;9:e105017. doi: 10.1371/journal.pone.0105017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowers TK, Eckert E. Leukopenia in anorexia nervosa. Lack of increased risk of infection. Arch. Intern. Med. 1978;138:1520–1523. doi: 10.1001/archinte.1978.03630350050015. [DOI] [PubMed] [Google Scholar]
- 8.Brown RF, Bartrop R, Birmingham CL. Immunological disturbance and infectious disease in anorexia nervosa: A review. Acta Neuropsychiatr. 2008;20:117–128. doi: 10.1111/j.1601-5215.2008.00286.x. [DOI] [PubMed] [Google Scholar]
- 9.Brown RF, Bartrop R, Beumont P, Birmingham CL. Bacterial infections in anorexia nervosa: Delayed recognition increases complications. Int. J. Eat Disord. 2005;37:261–265. doi: 10.1002/eat.20135. [DOI] [PubMed] [Google Scholar]
- 10.Nova E, Marcos A. Immunocompetence to assess nutritional status in eating disorders. Expert Rev. Clin. Immunol. 2006;2:433–444. doi: 10.1586/1744666X.2.3.433. [DOI] [PubMed] [Google Scholar]
- 11.Słotwiński SM, Słotwiński R. Immune disorders in anorexia. Cent. Eur. J. Immunol. 2017;42:294–300. doi: 10.5114/ceji.2017.70973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strumìa R, Varotti E, Manzato E, Gualandi M. Skin signs in anorexia nervosa. Dermatology. 2001;203:314–317. doi: 10.1159/000051779. [DOI] [PubMed] [Google Scholar]
- 13.Schaible UE, Kaufmann SHE. Malnutrition and Infection: Complex Mechanisms and Global Impacts. PLoS Med. 2007;4:e115. doi: 10.1371/journal.pmed.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heilskov S, et al. Dermatosis in children with oedematous malnutrition (Kwashiorkor): A review of the literature. J. Eur. Acad. Dermatol. Venereol. 2014;28:995–1001. doi: 10.1111/jdv.12452. [DOI] [PubMed] [Google Scholar]
- 15.Dalton B, et al. A meta-analysis of cytokine concentrations in eating disorders. J. Psychiatr. Res. 2018;103:252–264. doi: 10.1016/j.jpsychires.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Dalton B, et al. Inflammatory markers in anorexia nervosa: An exploratory study. Nutrients. 2018;10:1573. doi: 10.3390/nu10111573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasupuleti M, Schmidtchen A, Malmsten M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2012;32:143–171. doi: 10.3109/07388551.2011.594423. [DOI] [PubMed] [Google Scholar]
- 18.Chessa C, et al. Antiviral and immunomodulatory properties of antimicrobial peptides produced by human keratinocytes. Front. Microbiol. 2020;11:1155. doi: 10.3389/fmicb.2020.01155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rademacher F, et al. The antimicrobial and immunomodulatory function of RNase 7 in skin. Front. Immunol. 2019;10:2553. doi: 10.3389/fimmu.2019.02553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowes MA, Suárez-Fariñas M, Krueger JG. Immunology of psoriasis. Annu. Rev. Immunol. 2014;32:227–255. doi: 10.1146/annurev-immunol-032713-120225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson PH, Leygue ER, Murphy LC. Psoriasin (S100A7) Int. J. Biochem. Cell Biol. 1998;30:567–571. doi: 10.1016/S1357-2725(97)00066-6. [DOI] [PubMed] [Google Scholar]
- 22.Rademacher F, Simanski M, Harder J. RNase 7 in cutaneous defense. Int. J. Mol. Sci. 2016;17:560. doi: 10.3390/ijms17040560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harder J, et al. Enhanced expression and secretion of antimicrobial peptides in atopic dermatitis and after superficial skin injury. J. Investig. Dermatol. 2010;130:1355–1364. doi: 10.1038/jid.2009.432. [DOI] [PubMed] [Google Scholar]
- 24.Gambichler T, et al. Differential mRNA expression of antimicrobial peptides and proteins in atopic dermatitis as compared to psoriasis vulgaris and healthy skin. Int. Arch. Allergy Immunol. 2008;147:17–24. doi: 10.1159/000128582. [DOI] [PubMed] [Google Scholar]
- 25.Becker T, et al. FOXO-dependent regulation of innate immune homeostasis. Nature. 2010;463:369–373. doi: 10.1038/nature08698. [DOI] [PubMed] [Google Scholar]
- 26.Wu J, Randle KE, Wu LP. ird1 is a Vps15 homologue important for antibacterial immune responses in Drosophila. Cell. Microbiol. 2007;9:1073–1085. doi: 10.1111/j.1462-5822.2006.00853.x. [DOI] [PubMed] [Google Scholar]
- 27.Zinke I, Schütz CS, Katzenberger JD, Bauer M, Pankratz MJ. Nutrient control of gene expression in Drosophila: Microarray analysis of starvation and sugar-dependent response. EMBO J. 2002;21:6162–6173. doi: 10.1093/emboj/cdf600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bendix M-C, et al. Antimicrobial peptides in patients with anorexia nervosa: Comparison with healthy controls and the impact of weight gain. Sci. Rep. 2020;10:1–8. doi: 10.1038/s41598-020-79302-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisenhofer R, et al. Contamination in low microbial biomass microbiome studies: Issues and recommendations. Trends Microbiol. 2019;27:105–117. doi: 10.1016/j.tim.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 30.Sze MA, Abbasi M, Hogg JC, Sin DD. A comparison between droplet digital and quantitative PCR in the analysis of bacterial 16s load in lung tissue samples from control and COPD GOLD 2. PLoS ONE. 2014;9:e110351. doi: 10.1371/journal.pone.0110351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abellan-Schneyder I, Schusser AJ, Neuhaus K. ddPCR allows 16S rRNA gene amplicon sequencing of very small DNA amounts from low-biomass samples. BMC Microbiol. 2021;21:349. doi: 10.1186/s12866-021-02391-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quan P-L, Sauzade M, Brouzes E. dPCR: A technology review. Sensors (Basel) 2018;18:1271. doi: 10.3390/s18041271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogelstein B, Kinzler KW. Digital PCR. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gobert G, et al. Droplet digital PCR improves absolute quantification of viable lactic acid bacteria in faecal samples. J. Microbiol. Methods. 2018;148:64–73. doi: 10.1016/j.mimet.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 35.Maheshwari Y, Selvaraj V, Hajeri S, Yokomi R. Application of droplet digital PCR for quantitative detection of Spiroplasma citri in comparison with real time PCR. PLoS ONE. 2017;12:e0184751. doi: 10.1371/journal.pone.0184751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karstens L, et al. Controlling for contaminants in low-biomass 16S rRNA gene sequencing experiments. mSystems. 2019;4:e00290-19. doi: 10.1128/mSystems.00290-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis NM, Proctor DM, Holmes SP, Relman DA, Callahan BJ. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6:226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weyrich LS, et al. Laboratory contamination over time during low-biomass sample analysis. Mol. Ecol. Resour. 2019;19:982–996. doi: 10.1111/1755-0998.13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson MJ, Willis TJ. Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology. 2003;84:511–525. doi: 10.1890/0012-9658(2003)084[0511:CAOPCA]2.0.CO;2. [DOI] [Google Scholar]
- 40.De Caceres M, Legendre P. Associations between species and groups of sites: Indices and statistical inference. Ecology. 2009;90:3566–3574. doi: 10.1890/08-1823.1. [DOI] [PubMed] [Google Scholar]
- 41.Mu Q, Tavella VJ, Luo XM. Role of Lactobacillus reuteri in human health and diseases. Front. Microbiol. 2018;9:757. doi: 10.3389/fmicb.2018.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delanghe L, et al. The role of lactobacilli in inhibiting skin pathogens. Biochem. Soc. Trans. 2021;49:617–627. doi: 10.1042/BST20200329. [DOI] [PubMed] [Google Scholar]
- 43.Kim J-H, et al. Taxonomic profiling of skin microbiome and correlation with clinical skin parameters in healthy Koreans. Sci. Rep. 2021;11:16269. doi: 10.1038/s41598-021-95734-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanamoto T, Terakubo S, Nakashima H. Antimicrobial susceptibilities of oral isolates of Abiotrophia and Granulicatella according to the consensus guidelines for fastidious bacteria. Medicines (Basel) 2018;5:129. doi: 10.3390/medicines5040129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Si J, et al. Gut microbiome signatures distinguish type 2 diabetes mellitus from non-alcoholic fatty liver disease. Comput. Struct. Biotechnol. J. 2021;19:5920–5930. doi: 10.1016/j.csbj.2021.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rath S, Rud T, Karch A, Pieper DH, Vital M. Pathogenic functions of host microbiota. Microbiome. 2018;6:174. doi: 10.1186/s40168-018-0542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Assarsson M, Söderman J, Dienus O, Seifert O. Significant differences in the bacterial microbiome of the pharynx and skin in patients with psoriasis compared with healthy controls. Acta Derm Venereol. 2020;100:adv00273. doi: 10.2340/00015555-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Omodei D, et al. Immune-metabolic profiling of anorexic patients reveals an anti-oxidant and anti-inflammatory phenotype. Metabolism. 2015;64:396–405. doi: 10.1016/j.metabol.2014.10.025. [DOI] [PubMed] [Google Scholar]
- 49.Chang H-W, et al. Alteration of the cutaneous microbiome in psoriasis and potential role in Th17 polarization. Microbiome. 2018;6:154. doi: 10.1186/s40168-018-0533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alekseyenko AV, et al. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome. 2013;1:31. doi: 10.1186/2049-2618-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gläser R, Köten B, Wittersheim M, Harder J. Psoriasin: Key molecule of the cutaneous barrier? JDDG J. Deutsch. Dermatol. Ges. 2011;9:897–902. doi: 10.1111/j.1610-0387.2011.07683.x. [DOI] [PubMed] [Google Scholar]
- 52.Gläser R, et al. The antimicrobial protein psoriasin (S100A7) Is upregulated in atopic dermatitis and after experimental skin barrier disruption. J. Investig. Dermatol. 2009;129:641–649. doi: 10.1038/jid.2008.268. [DOI] [PubMed] [Google Scholar]
- 53.Nam B, et al. Regulatory effects of Lactobacillus plantarum HY7714 on skin health by improving intestinal condition. PLoS ONE. 2020;15:e0231268. doi: 10.1371/journal.pone.0231268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jung Y-O, et al. Lysates of a probiotic, Lactobacillus rhamnosus, can improve skin barrier function in a reconstructed human epidermis model. Int J Mol Sci. 2019;20:428. doi: 10.3390/ijms20020428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee DE, et al. Clinical evidence of effects of lactobacillus plantarum HY7714 on skin aging: A randomized, double blind, placebo-controlled study. J. Microbiol. Biotechnol. 2015;25:2160–2168. doi: 10.4014/jmb.1509.09021. [DOI] [PubMed] [Google Scholar]
- 56.Markowiak-Kopeć P, Śliżewska K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients. 2020;12:E1107. doi: 10.3390/nu12041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morvan P-Y, Vallee R, Py M. Evaluation of the effects of stressful life on human skin microbiota. Appl. Microbiol. Open Access. 2018;4:8. [Google Scholar]
- 58.Byeon J, et al. Insights into the skin microbiome of sickle cell disease leg ulcers. Wound Repair. Regen. 2021;29:801–809. doi: 10.1111/wrr.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukherjee S, et al. Sebum and hydration levels in specific regions of human face significantly predict the nature and diversity of facial skin microbiome. Sci. Rep. 2016;6:36062. doi: 10.1038/srep36062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Łoś-Rycharska E, et al. A combined analysis of gut and skin microbiota in infants with food allergy and atopic dermatitis: A pilot study. Nutrients. 2021;13:1682. doi: 10.3390/nu13051682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Z, et al. New insights into the skin microbial communities and skin aging. Front. Microbiol. 2020;11:565549. doi: 10.3389/fmicb.2020.565549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kates AE, Zimbric ML, Mitchell K, Skarlupka J, Safdar N. The impact of chlorhexidine gluconate on the skin microbiota of children and adults: A pilot study. Am. J. Infect. Control. 2019;47:1014–1016. doi: 10.1016/j.ajic.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park H, et al. Pilot study on the forehead skin microbiome and short chain fatty acids depending on the SC functional index in Korean cohorts. Microorganisms. 2021;9:2216. doi: 10.3390/microorganisms9112216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang L, et al. Amplicon-based sequencing and co-occurence network analysis reveals notable differences of microbial community structure in healthy and dandruff scalps. BMC Genom. 2022;23:312. doi: 10.1186/s12864-022-08534-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim H-J, et al. Segregation of age-related skin microbiome characteristics by functionality. Sci. Rep. 2019;9:16748. doi: 10.1038/s41598-019-53266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brandwein M, Katz I, Katz A, Kohen R. Beyond the gut: Skin microbiome compositional changes are associated with BMI. Hum. Microbiome J. 2019;13:100063. doi: 10.1016/j.humic.2019.100063. [DOI] [Google Scholar]
- 67.Grice EA, et al. A diversity profile of the human skin microbiota. Genome Res. 2008;18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim D. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5:14. doi: 10.1186/s40168-017-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kong HH. Details matter: Designing skin microbiome studies. J. Investig. Dermatol. 2016;136:900–902. doi: 10.1016/j.jid.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kong HH, et al. Performing skin microbiome research: A method to the madness. J. Investig. Dermatol. 2017;137:561–568. doi: 10.1016/j.jid.2016.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meisel JS, et al. Skin microbiome surveys are strongly influenced by experimental design. J. Investig. Dermatol. 2016;136:947–956. doi: 10.1016/j.jid.2016.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakatsuji T, et al. The microbiome extends to subepidermal compartments of normal skin. Nat. Commun. 2013;4:1431. doi: 10.1038/ncomms2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grice EA, Segre JA. The skin microbiome. Nat. Rev. Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mourelatos K, Eady EA, Cunliffe WJ, Clark SM, Cove JH. Temporal changes in sebum excretion and propionibacterial colonization in preadolescent children with and without acne. Br. J. Dermatol. 2007;156:22–31. doi: 10.1111/j.1365-2133.2006.07517.x. [DOI] [PubMed] [Google Scholar]
- 75.Leyden JJ, McGinley KJ, Mills OH, Kligman AM. Propionibacterium levels in patients with and without acne vulgaris. J. Investig. Dermatol. 1975;65:382–384. doi: 10.1111/1523-1747.ep12607634. [DOI] [PubMed] [Google Scholar]
- 76.Neale J, Pais SMA, Nicholls D, Chapman S, Hudson LD. What are the effects of restrictive eating disorders on growth and puberty and are effects permanent? A systematic review and meta-analysis. J. Adolesc. Health. 2020;66:144–156. doi: 10.1016/j.jadohealth.2019.08.032. [DOI] [PubMed] [Google Scholar]
- 77.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl. Acad. Sci. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oh J, Byrd AL, Park M, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell. 2016;165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cao C, Xiao Z, Wu Y, Ge C. Diet and skin aging-from the perspective of food nutrition. Nutrients. 2020;12:E870. doi: 10.3390/nu12030870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liakou AI, Theodorakis MJ, Melnik BC, Pappas A, Zouboulis CC. Nutritional clinical studies in dermatology. J. Drugs Dermatol. 2013;12:1104–1109. [PubMed] [Google Scholar]
- 81.Attia E, et al. Feeding and eating disorders in DSM-5. AJP. 2013;170:1237–1239. doi: 10.1176/appi.ajp.2013.13030326. [DOI] [PubMed] [Google Scholar]
- 82.Bilska B, et al. Expression of antimicrobial peptide genes oscillates along day/night rhythm protecting mice skin from bacteria. Exp. Dermatol. 2021;30:1418–1427. doi: 10.1111/exd.14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eda N, Shimizu K, Suzuki S, Lee E, Akama T. Effects of high-intensity endurance exercise on epidermal barriers against microbial invasion. J. Sports Sci. Med. 2013;12:44–51. [PMC free article] [PubMed] [Google Scholar]
- 84.Köten B, et al. RNase 7 contributes to the cutaneous defense against Enterococcus faecium. PLoS ONE. 2009;4:e6424. doi: 10.1371/journal.pone.0006424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gläser R, et al. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat. Immunol. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 86.Wittersheim M, et al. Differential expression and in vivo secretion of the antimicrobial peptides psoriasin (S100A7), RNase 7, human beta-defensin-2 and -3 in healthy human skin. Exp. Dermatol. 2013;22:364–366. doi: 10.1111/exd.12133. [DOI] [PubMed] [Google Scholar]
- 87.Gläser R, et al. UV-B radiation induces the expression of antimicrobial peptides in human keratinocytes in vitro and in vivo. J. Allergy Clin. Immunol. 2009;123:1117–1123. doi: 10.1016/j.jaci.2009.01.043. [DOI] [PubMed] [Google Scholar]
- 88.Belheouane M, et al. Assessing similarities and disparities in the skin microbiota between wild and laboratory populations of house mice. ISME J. 2020 doi: 10.1038/s41396-020-0690-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Salter SJ, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013 doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- 95.Cáceres MD, Legendre P. Associations between species and groups of sites: Indices and statistical inference. Ecology. 2009;90:3566–3574. doi: 10.1890/08-1823.1. [DOI] [PubMed] [Google Scholar]
- 96.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cole JR, et al. The ribosomal database project (RDP-II): Sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB47175 (https://www.ebi.ac.uk/ena/browser/view/PRJEB47175).