Abstract
Pseudomonas putida converts benzoate to catechol using two enzymes that are encoded on the chromosome and whose expression is induced by benzoate. Benzoate also binds to the regulator XylS to induce expression of the TOL (toluene degradation) plasmid-encoded meta pathway operon for benzoate and methylbenzoate degradation. Finally, benzoate represses the ability of P. putida to transport 4-hydroxybenzoate (4-HBA) by preventing transcription of pcaK, the gene encoding the 4-HBA permease. Here we identified a gene, benR, as a regulator of benzoate, methylbenzoate, and 4-HBA degradation genes. A benR mutant isolated by random transposon mutagenesis was unable to grow on benzoate. The deduced amino acid sequence of BenR showed high similarity (62% identity) to the sequence of XylS, a member of the AraC family of regulators. An additional seven genes located adjacent to benR were inferred to be involved in benzoate degradation based on their deduced amino acid sequences. The benABC genes likely encode benzoate dioxygenase, and benD likely encodes 2-hydro-1,2-dihydroxybenzoate dehydrogenase. benK and benF were assigned functions as a benzoate permease and porin, respectively. The possible function of a final gene, benE, is not known. benR activated expression of a benA-lacZ reporter fusion in response to benzoate. It also activated expression of a meta cleavage operon promoter-lacZ fusion inserted in an E. coli chromosome. Third, benR was required for benzoate-mediated repression of pcaK-lacZ fusion expression. The benA promoter region contains a direct repeat sequence that matches the XylS binding site previously defined for the meta cleavage operon promoter. It is likely that BenR binds to the promoter region of chromosomal benzoate degradation genes and plasmid-encoded methylbenzoate degradation genes to activate gene expression in response to benzoate. The action of BenR in repressing 4-HBA uptake is probably indirect.
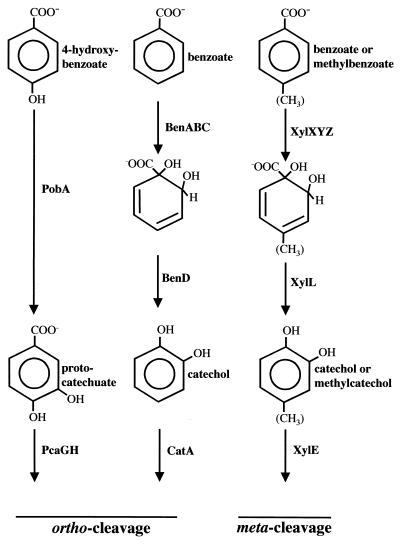
Pseudomonas putida converts a variety of environmental pollutants and plant phenolic compounds to a small number of structurally simple aromatic compounds that are the starting points for pathways of aromatic ring fission (20). Ring fission is termed ortho cleavage when it occurs between two adjacent hydroxyl groups and meta cleavage when it occurs adjacent to a single hydroxyl group. P. putida can degrade the aromatic acid benzoate, after converting it to catechol, by either a meta ring cleavage pathway or an ortho ring cleavage pathway (20). The aromatic acid 4-hydroxybenzoate (4-HBA) is degraded by an ortho ring cleavage pathway after conversion to protocatechuate (Fig. 1).
FIG. 1.
Initial steps for the ortho and meta cleavage pathways used by P. putida to degrade 4-HBA, benzoate, and methylbenzoates. The meta cleavage pathway is encoded by the TOL catabolic plasmid. The methyl group can be present in either the 3 or 4 position of the ring.
The TOL (toluene degradation) catabolic plasmid (pWW0) from P. putida carries genes for the degradation of toluene and xylenes (34). On the plasmid are found two operons involved in aromatic compound degradation. One of the operons encodes enzymes for the conversion of toluene and xylenes to benzoate and methylated benzoates (the upper pathway), and the other encodes enzymes that catalyze a meta ring cleavage and subsequent reactions leading to the formation of tricarboxylic acid cycle intermediates (the meta pathway). Transcription of the meta pathway genes is regulated by XylS, a protein also encoded on the TOL catabolic plasmid (34). P. putida will also convert benzoate to catechol using chromosomally encoded enzymes. The genes for these enzymes have not yet been sequenced or fully characterized (25). Catechol, but not methylated catechols, is then further degraded to trichloroacetic acid cycle intermediates by an ortho ring cleavage pathway (Fig. 1) (20).
Benzoate induces the synthesis of the TOL plasmid-encoded enzymes of the meta fission pathway (11, 22) as well as synthesis of the chromosomally encoded enzymes that convert benzoate to catechol. Additionally, a surprising recent finding is that benzoate represses the utilization of 4-HBA (40). When P. putida is given a mixture of benzoate and 4-HBA, it degrades benzoate in preference to 4-HBA. Presumably, this is a reflection of the fact that benzoate supports a slightly higher rate of growth than 4-HBA. Benzoate was found to depress the levels of 4-HBA hydroxylase and protocatechuate dioxygenase activity, as well as the level of 4-HBA transport in cells grown on benzoate plus 4-HBA. Benzoate represses 4-HBA transport by preventing transcription of pcaK, the gene encoding the 4-HBA permease (40).
Here we described a chromosomally encoded cluster of eight genes from P. putida that is involved in conversion of benzoate to catechol and that includes a new regulatory gene, benR. We determined that BenR activates expression of benzoate dioxygenase genes in response to benzoate. It is also necessary for benzoate-dependent repression of 4-HBA transport gene expression. In addition, we demonstrate the likeness of BenR to XylS by showing that BenR activates expression of the meta cleavage pathway operon of the TOL catabolic plasmid. BenR thus has roles as an activator of benzoate degradation via ortho ring fission, as an activator of benzoate and methylbenzoate degradation via meta ring fission, and in repression of 4-HBA degradation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The plasmids and bacterial strains used in this study are listed in Table 1. P. putida was grown at 30°C in basal mineral (BM) medium [25 mM KH2PO4, 25 mM Na2HPO4, 0.1% (NH4)2SO4, 1% Hutner mineral base (14) (final pH 6.8)] containing an appropriate carbon and energy source. Carbon sources were sterilized separately and added to final concentrations of 5 mM (benzoate or 4-HBA) and 10 mM (glucose or succinate). Plasmids were mobilized from E. coli DH5α into P. putida via a triparental mating system using E. coli HB101(pRK2013) as the mobilizing strain. The mating mixtures were incubated overnight on solid Luria-Bertani medium at 30°C. Unless specified otherwise, E. coli strains were grown at 37°C in Luria broth (LB) (7). The TOL plasmid (pWW0) was transferred from P. putida PaW1 to the P. putida benR mutant (strain 4157) by direct mating with selection on solid BM medium containing 5 mM 3-methylbenzoate plus kanamycin. Antibiotics were added to the following final concentrations (in micrograms per milliliter): ampicillin, 100; gentamicin, 5; kanamycin, 100; spectinomycin, 100; and tetracycline, 25. Solid media contained 1.5% agar.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | Carries T7 RNA polymerase under the control of lacUV5 promoter | 51 |
| CC118 Pm-lacZ | Tcr; CC118 with chromosomal mini-Tn5::Pm::lacZ insertion | 31 |
| DH5α | F− λ−recA1 Δ(lacZYA-argF)U169 hsdR17 thi-1 gyrA96 supE44 endA1 relA1 φ80dlacZΔM15 | GIBCO-BRL |
| HB101 | F− λ−recA1 Δ(lacZYA-argF)U169 hsdR17 thi-1 gyrA96 supE55 endA1 relA1 | 48 |
| S17-1λpir | thi pro hdsR hdsM+ recA, chromosomal insertion of RP4-2(Tc::Mu Km::Tn7) | 50 |
| P. putida strains | ||
| PRS2000 | Wild type | 44 |
| PRS4157 | Kmr; mini-Tn5Km insertion in benR | This work |
| Plasmids | ||
| pHRP309 | Gmr; IncQ, lacZ transcriptional fusion vector | 46 |
| pHRP311 | Gmr, Smr, Spr; negative control plasmid (Ω cassette from cohort vector inserted into pHRP309) | 46 |
| pHRP315 | Apr, Smr, Spr; cohort cloning vector for use with pHRP309 | 46 |
| pHRP317 | Kmr, Smr, Spr; cohort cloning vector for use with pHRP309 | 46 |
| pRK415 | Tcr; IncP1, mobilizable broad-host-range cloning vector | 29 |
| pRK2013 | Kmr; ori (ColE1), RP4 mobilization functions | 10 |
| pT7-7 | Apr; ori (ColE1), expression vector with T7 promoter | 51 |
| pUC19 | Apr; high-copy-number cloning vector | 55 |
| pUTminiTn5-Km | Apr, Kmr; delivery plasmid for mini-Tn5-Km | 8 |
| pCCH100 | Kmr, Smr, Spr; benA promoter region in pHRP317 | This work |
| pCCH101 | Gmr, Smr, Spr; benA-lacZ transcriptional fusion in pHRP309 | This work |
| pCCH106 | Apr; pT7-7 with the benR open reading frame inserted in the NdeI/PstI sites | This work |
| pCCH107 | Tcr; pRK415 with benR inserted in the EcoRI/PstI sites | This work |
| pCCH108 | Apr; 1.3-kb inverse-PCR-generated EcoRI-XmaI fragment cloned in pUC19 | This work |
| pCNN100 | Apr; 12.2-kb EcoRI PRS2000 genomic fragment in pUC19 | This work |
| pHNN211 | Apr, Smr, Spr; pcaK promoter region in pHRP315 | This work |
| pHNN216 | Gmr, Smr, Spr; pcaK-lacZ transcriptional fusion in pHRP309 | This work |
Apr, ampicillin resistance; Kmr, kanamycin resistance; Gmr, gentamicin resistance; Smr, streptomycin resistance; Spr, spectinomycin resistance; Tcr, tetracycline resistance.
Mutagenesis and screening for benzoate-nondegrading mutants.
P. putida cells were randomly mutagenized with the transposon mini-Tn5 by mating P. putida PRS2000 cells with E. coli S17-1λpir (pUTminiTn5-Km) at 30°C overnight on LB plates with a donor-to-recipient cell ratio of 1:5. Mating mixtures were suspended in BM and plated onto BM plates containing 5 mM benzoate, a low concentration of succinate (1 mM), and kanamycin. Small colonies were patched onto plates containing kanamycin and either benzoate, 4-HBA, or succinate as the sole carbon source to identify mutants that could not utilize benzoate as a sole carbon source.
Arbitrary PCR amplifications.
The region of DNA flanking the transposon insertion in strain PRS4157 was amplified by arbitrary PCR (3) using arbitrary primers as described elsewhere (45). During the first round of amplification, a primer specific to the 5′ end of the transposon and an arbitrary primer were used to amplify sequences flanking the upstream end of the inserted transposon. In a similar manner, a primer specific to the 3′ end and an arbitrary primer were used to amplify sequences flanking the downstream end of the transposon. Product from the first-round reactions was used as template in second-round reactions with primers annealing to the 5′ end of the arbitrary primers and the respective transposon-specific primers from the first-round reactions. PCR products were sequenced using the transposon-specific primers as sequencing primers.
Colony hybridization.
A 335-bp probe specific to the region flanking the transposon insertion in strain PRS4157 was labeled with [32P]dCTP using Ready To Go DNA labeling beads (−dCTP) and purified with a ProbeQuant G-50 Micro column (Pharmacia Biotech, Piscataway, N.J.). E. coli colonies carrying EcoRI-generated fragments of P. putida genomic DNA in pUC19 were screened by colony hybridization (48) to identify wild-type DNA corresponding to the region that was indicated by transposon mutagenesis to be required for benzoate degradation. The probe hybridized to an approximately 12-kb fragment of DNA that was designated pCNN100. A 1.3-kb EcoRI-XmaI fragment containing the portion of the benF reading frame not contained on pCNN100 was subsequently cloned by inverse PCR (41). This clone was designated pCCH108.
DNA sequencing and analysis.
DNA sequencing was performed by the University of Iowa DNA Sequencing Facility (Iowa City). Sequence assembly and analysis were done with GENE Inspector, version 1.0.1 (Textco Inc., West Lebanon, N.H.). The amino acid sequences of open reading frames were submitted to the National Center for Biotechnology Information (Bethesda, Md.) and analyzed using the BLASTp 2.0.9 algorithm (1). The sequence alignment was constructed using the CLUSTAL W multiple-sequence alignment program at the Baylor College of Medicine Human Genome Center (52), and the program BOXSHADE (version 3.21) was used to shade aligned sequences.
Cloning and DNA manipulations.
Standard protocols were used for cloning and transformations. Restriction digests and ligations were performed using standard techniques. Plasmid DNA was prepared by using a QIAprep Spin Miniprep kit, and DNA restriction fragments were isolated from agarose gels using the QIAquick gel extraction kit (Qiagen Inc., Santa Clarita, Calif.).
Total RNA was isolated from PRS2000 cells grown on either glucose or glucose plus benzoate (2.5 mM) using the SV total RNA isolation system as instructed by the manufacturer (Promega Corp., Madison, Wis.). The transcription start site of benA was determined by primer extension analysis using the Promega AMV-RT (Avian myeloblastosis virus reverse transcriptase) primer extension system. The primer was complementary to bases 45 to 28 of benA. Primer extension products were analyzed on a 6% polyacrylamide gel next to a sequence ladder generated with the same primer. The Access reverse transcription-PCR system (Promega) was used to determine the transcriptional organization of the benA, -B, and -C genes. In each case, a reverse transcriptase-free control was included to ensure that reaction mixtures did not contain contaminating DNA.
Reporter plasmids pHNN216 and pCCH101 were constructed using a two-step cloning procedure described previously (46). The promoter regions of benA and pcaK were amplified by PCR and then directionally inserted adjacent to a Ω Spr/Smr cassette in either pHRP315 or pHRP317. Fragments containing the ΩSpr/Smr cassette and promoter region were then inserted upstream of the promoterless lacZ gene of pHRP309 to create pCCH101 (benA-lacZ) or pHNN216 (pcaK-lacZ). The fusions of the promoter-containing fragments and lacZ were confirmed by sequencing.
A 1,189-bp PCR product containing the benR open reading frame and suspected promoter region was cloned into the EcoRI/PstI sites of pRK415 to generate pCCH107. A 989-bp PCR product containing only the benR open reading frame was cloned into the NdeI/PstI sites of pT7-7 downstream of the T7 promoter to construct the BenR expression plasmid, pCCH106.
β-Galactosidase assays.
β-Galactosidase activities of P. putida cells carrying benA-lacZ or pcaK-lacZ transcriptional fusion plasmids were assayed according to Miller (36). For analysis of P. putida strains carrying pCCH101, cultures were grown to an A660 of 0.1 with succinate as the sole carbon source, at which time compounds to be tested for the ability to induce gene expression were added to a final concentration of 1 mM. For analysis of P. putida strains carrying pHNN216, cultures were grown with the indicated concentrations of compounds. β-Galactosidase activities were determined for cells harvested at an A660 of 0.2. Six to eight independently grown cultures were assayed in triplicate, and the values were averaged. For analysis of gene expression in E. coli, cells were grown in LB at 30°C to an A660 of 0.25, at which time benzoate and catechol, if added, were added to a final concentration of 1 mM. E. coli cells were harvested at a final A660 of 0.5.
4-HBA uptake assays.
Cells (50 ml) grown in BM with either 4-HBA or a mixture of benzoate (2.5 mM) and 4-HBA (2.5 mM) as carbon sources were harvested at mid-logarithmic phase, washed in 25 ml of phosphate buffer (25 mM KH2PO4, 25 mM Na2HPO4), and resuspended in 2 ml of phosphate buffer. Resuspended cells (300 μl) were added to an equal volume of reaction mixture (25 mM KH2PO4, 25 mM Na2HPO4, 4.0 mM succinate, 4.0 mM glucose, 127 μM 14C-labeled 4-HBA) to start the assay. At timed intervals, 100-μl samples were removed, filtered through Nucleopore polycarbonate membranes (0.2-μm pore size; Costar Corp., Cambridge, Mass.), and washed with 1.8 ml of phosphate buffer. Accumulated substrate was determined by scintillation counting of the cells retained on the filters.
Protein determinations.
Whole cells were precipitated by addition of trichloroacetic acid to 5% and then boiled in 0.1 N NaOH for 10 min. Protein concentrations were determined using the Bio-Rad (Hercules, Calif.) protein assay, with bovine serum albumin as a standard.
Radiochemicals.
[14C]uniformly-ring-labeled 4-HBA (33 Ci/mmol) and [32P]dCTP were obtained from Amersham Corp. (Arlington Heights, Ill.).
Nucleotide accession number.
The nucleotide sequence has been assigned GenBank accession number AF218267.
RESULTS
Identification of P. putida genes involved in benzoate degradation.
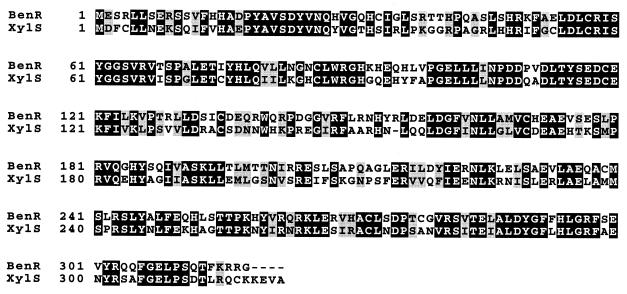
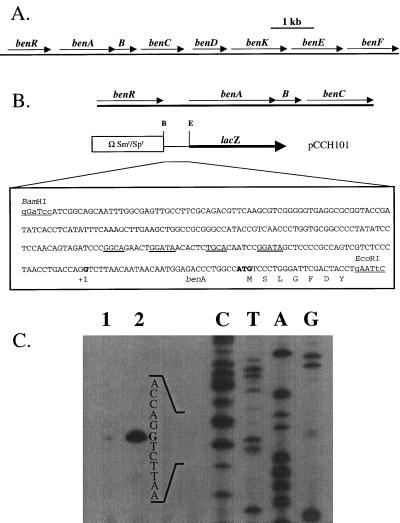
To identify genes involved in benzoate degradation, we isolated a transposon mutant, PRS4157, that was unable to utilize benzoate as a sole carbon source but that grew at wild-type rates on succinate and 4-HBA. Sequencing of the region of DNA flanking the transposon revealed that the transposon was inserted in a gene predicted to encode a regulator with high similarity to XylS, a TOL plasmid-encoded activator of benzoate and methylbenzoate degradation (Fig. 2) (17). We named this gene benR. A 13.3-kb segment of P. putida DNA was subsequently cloned and sequenced and was found to include eight genes that can be inferred to be involved in benzoate degradation based on their sequence similarity to known genes (Fig. 3; Table 2).
FIG. 2.
Amino acid sequence alignment of BenR and XylS proteins. Identical residues are outlined in black. Similar residues are shaded gray.
FIG. 3.
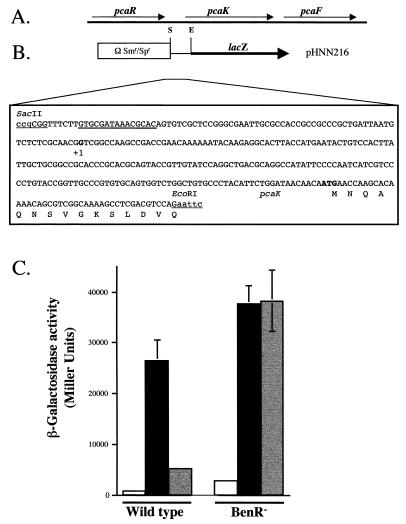
(A) Map of chromosomally encoded benzoate degradation genes from P. putida. (B) Map of the benA promoter region, diagram of the region incorporated to construct the benA-lacZ fusion plasmid, pCCH101, and nucleotide sequence of the benA promoter region. The putative BenR binding sites are heavily underlined. Introduced restriction sites are lightly underlined. The transcriptional and translational start sites are in bold. (C) Determination of the 5′ end of the benA transcript by primer extension. RNA was isolated from glucose-grown cells (lane 1) and glucose-benzoate-grown cells (lane 2) as described in the text. A sequence ladder was generated with the same primer (lanes C, T, A, and G). The first nucleotide in the transcript is shown in bold.
TABLE 2.
Benzoate degradation genes of P. putida
| Gene designation | Proposed function of gene product | Size of gene product in:
|
Most similar gene product (species) (% amino acid identity/% amino acid similarity) (accession no.) known function of gene product (reference) | |
|---|---|---|---|---|
| Residues | kDa | |||
| benR | Regulatory protein | 318 | 36.4 | XylS (P. putida) (59/73) (P07859) meta-cleavage benzoate degradation operon transcriptional activator (23) |
| EutR (Salmonella typhimurium) (27/44) (Q9ZFU7) ethanolamine degradation operon transcriptional activator (32) | ||||
| RhaS (S. typhimurium) (25/47) (P09377) l-rhamnose degradation operon transcriptional activator (53) | ||||
| ArgR (P. aeruginosa) (26/47) (AAC45653) arginine biosynthesis operon transcriptional activator (47) | ||||
| benA | Benzoate dioxygenase | 452 | 51.5 | XylX (P. putida) (73/86) (P23099) toluate 1,2-dioxygenase α subunit (18) |
| BenA (Acinetobacter sp.) (68/80) (P07769) benzoate 1,2-dioxygenase α subunit (38) | ||||
| CbdA (Burkholderia cepacia) (59/76) (CAA55681) 2-halobenzoate 1,2-dioxygenase α subunit (16) | ||||
| AntA (Acinetobacter sp.) (47/65) (AAC34813) anthranilate dioxygenase α-subunit (2) | ||||
| benB | Benzoate dioxygenase | 161 | 19.1 | XylY (P. putida) (76/89) (P23100) toluate 1,2-dioxygenase β subunit (18) |
| BenB (Acinetobacter sp.) (68/79) (P07770) benzoate 1,2-dioxygenase β subunit (38) | ||||
| CbdB (B. cepacia) (57/73) (CAA55682) 2-halobenzoate 1,2-dioxygenase β subunit (16) | ||||
| AntB (Acinetobacter sp.) (34/56) (AAC34814) anthranilate dioxygenase β subunit (2) | ||||
| benC | Ferredoxin reductase | 336 | 36.4 | XylZ (P. putida) (74/86) (P23101) methylcatechol 1,2-dioxygenase reductase component (18) |
| BenC (Acinetobacter sp.) (51/67) (S23479) benzoate 1,2-dioxygenase reductase component (37) | ||||
| CbdC (B. cepacia) (46/64) (CAA55683) 2-halobenzoate 1,2-dioxygenase reductase component (16) | ||||
| AntC (Acinetobacter sp.) (38/57) (AAC34815) anthranilate 1,2-dioxygenase reductase component (2) | ||||
| benD | cis-Diol dehydrogenase | 253 | 27.1 | XylL (P. putida) (78/86) (P23102) 2-hydro-1,2-dihydroxymethylbenzoate dehydrogenase (37) |
| BenD (Acinetobacter sp.) (58/71) (P07772) 2-hydro-1,2-dihydroxybenzoate dehydrogenase (37) | ||||
| CmtC (P. putida) (34/49) (AAB62287) 2,3-dihydroxy-2,3-dihydro-p-cumate dehydrogenase (9) | ||||
| TsaC (Comamonas testosteroni) (32/46) (AAB62287) p-sulfobenzyl alcohol dehydrogenase (27) | ||||
| benK | Benzoate transporter | 443 | 47.1 | BenK (Acinetobacter sp.) (41/58) (AAC46425) benzoate transporter (5) |
| PcaK (Acinetobacter sp.) (29/45) (Q43975) 4-hydroxybenzoate transporter (33) | ||||
| PcaK (P. putida) (29/44) (Q51955) 4-hydroxybenzoate transporter (19) | ||||
| VanK (Acinetobacter sp.) (24/41) (AAC27108) vanillate transporter (49) | ||||
| benE | Unknown | 399 | 41.3 | BenE (Acinetobacter sp.) (39/58) (P07775) membrane protein of unknown function |
| benF | Porin | 398 | 43.2 | PhaK (P. putida) (44/57) (AAC24339) porin-like phenylacetate-specific-channel-forming protein (43) |
| OprE (P. aeruginosa) (38/55) (S34969) anaerobically induced porin (54) | ||||
| OprD (P. aeruginosa) (37/51) (P32722) imipenem-specific-channel-forming protein (21) | ||||
| OprE3 (P. aeruginosa) (33/49) (BAA22267) porin-like outer membrane protein (42) | ||||
Downstream of benR are three genes, benA, -B, and -C, that have very high deduced amino acid sequence identity to the terminal oxygenase and reductase components of methylbenzoate and benzoate dioxygenases. BenD is similar to cis-diol dehydrogenases involved in the conversion of 2-hydro-1,2-dihydroxybenzoates (the products of benzoate dioxygenase reactions) to catechols (Fig. 1). Following benABCD are three genes that may encode proteins involved in benzoate uptake. The benK and benF genes almost certainly encode a benzoate transporter and a porin, respectively. The function of benE, predicted to encode a membrane protein, is unknown. Each ben gene has a guanine-plus-cytosine content of between 60 and 67%.
BenR activates the expression of benABC in response to benzoate.
The benR mutant (strain PRS4157) was able to grow on benzoate when a plasmid-borne copy of benR was supplied in trans on pCCH107. The complemented mutant grew on benzoate with a generation time of 2.4 h, compared to about 1.8 h for the wild-type strain. The sequence of the benR gene and the growth phenotype of the benR mutant suggested that the BenR protein is probably involved in regulating expression of the benA, benB, and benC genes, predicted to encode benzoate 1,2-dioxygenase. Reverse transcription-PCR amplification of the regions between benA and benB and between benB and benC showed that these three genes are cotranscribed in benzoate-grown cells (results not shown). Primer extension analysis indicated that the 5′ end of benA lies 30 bp upstream from its predicted translational start site (Fig. 3).
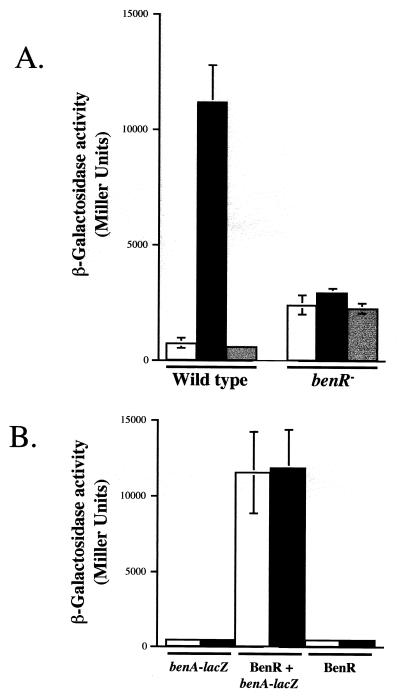
To test whether BenR regulates benA expression, we constructed a reporter plasmid that has the benA promoter fused to a promoterless lacZ gene (pCCH101). P. putida wild-type cells carrying the benA-lacZ fusion expressed β-galactosidase activity at levels that were 15-fold higher in cells grown on succinate in the presence of benzoate compared to succinate-grown cells (Fig. 4A). The presence of catechol, the product of cis-diol dehydrogenase, did not induce expression of the benA-lacZ fusion. Expression of the benA-lacZ fusion was not induced by benzoate in benR mutant cells. Overexpression of BenR from a T7 promoter (plasmid pCCH106) in E. coli BL21(DE3) cells carrying the benA-lacZ fusion plasmid resulted in a 25-fold increase in β-galactosidase expression over the levels seen in the absence of benR (Fig. 4B). This result shows that BenR directly activates the benA promoter. In this system, addition of benzoate did not influence the levels of β-galactosidase expression.
FIG. 4.
(A) β-Galactosidase activities of the benA-lacZ fusion (pCCH101) in wild-type (PRS2000) and benR mutant (PRS4157) cells. Cells were grown on succinate (white), succinate plus benzoate (black), or succinate plus catechol (gray) and assayed as described in Materials and Methods. (B) β-Galactosidase activities of E. coli cells harboring the benA-lacZ fusion (pCCH101) and/or the BenR expression construct (pCCH106) in the absence (white) or presence (black) of 1 mM benzoate. β-Galactosidase activity was assayed as described in Materials and Methods.
BenR activates expression of the TOL plasmid-encoded meta-cleavage pathway operon.
Pm, the promoter of the meta-cleavage operon from the TOL catabolic plasmid, is activated by the TOL plasmid-encoded regulator XylS when benzoate or methylbenzoates are present. A long-standing observation in studies of TOL plasmid gene regulation is that Pm can also be activated in the presence of benzoate by a chromosomally encoded regulator. The regulatory gene responsible for this activation was identified genetically in 1988 and given the name benR (6), but this gene was never sequenced. To determine whether the benR gene described here might be the regulatory gene that is responsible for XylS-independent activation of Pm, we expressed the BenR protein from pCCH107 in E. coli CC118 Pm-lacZ, a strain that has a Pm::lacZ fusion inserted in its chromosome. When BenR was present, 13,000 Miller units of β-galactosidase was expressed from Pm over an undetectable background. A slight increase in β-galactosidase production (17,000 Miller units) was seen when benzoate was included in the growth medium. The addition of catechol had no effect.
XylS responds to benzoate, but not 3-methylbenzoate, to modulate expression of benA.
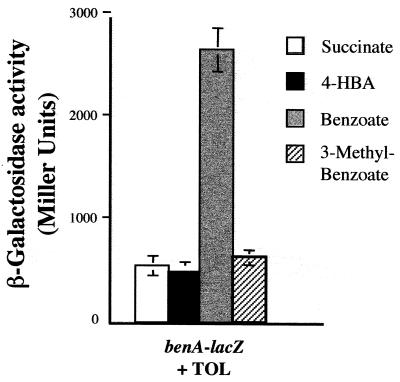
To determine whether XylS can restore benzoate-dependent regulation of the benA promoter in the absence of BenR, the TOL catabolic plasmid was introduced from P. putida PaW1 into the benR mutant (PRS4157) containing the benA-lacZ fusion plasmid (pCCH101). When the TOL plasmid was present, the observed level of benA-lacZ expression was fivefold higher when benzoate was included in the growth medium together with succinate than when cells were grown on succinate alone or on succinate plus 4-HBA or 3-methylbenzoate (Fig. 5). These results indicate that XylS, encoded on the TOL plasmid, can partially complement BenR function.
FIG. 5.
β-Galactosidase activities of benR mutant (PRS4157) cells carrying the benA-lacZ fusion plasmid (pCCH101) and the TOL plasmid. Cells were grown on succinate, succinate plus 4-hydroxybenzoate, succinate plus benzoate, or succinate plus 3-methylbenzoate, as indicated, and assayed as described in Materials and Methods.
BenR is required for benzoate-mediated repression of 4-HBA degradation.
When P. putida cells that are growing on 4-HBA as a sole carbon source are transferred to a medium that includes equal amounts of 4-HBA and benzoate, their rate of 4-HBA degradation decreases and they start to degrade benzoate at a high rate. Once all of the benzoate is depleted, rapid degradation of 4-HBA resumes (40). This preferred usage of benzoate over 4-HBA by P. putida can be partially explained by the observation that benzoate represses expression of pcaK, a gene that encodes a 4-HBA permease. To determine if BenR plays a role in this repression, we compared the levels of β-galactosidase produced by wild-type and benR mutant strains carrying a pcaK-lacZ transcriptional fusion (Fig. 6). As was previously demonstrated (40), wild-type P. putida cells grown on a mixture of benzoate and 4-HBA expressed β-galactosidase from the pcaK promoter to levels that were fivefold lower than those seen in cells grown on 4-HBA only. This repressive effect of benzoate on 4-HBA-induced pcaK expression was not seen when the pcaK-lacZ fusion was present in a benR mutant (Fig. 6). This indicates that BenR is involved in benzoate-mediated repression of pcaK expression. To determine whether BenR directly regulates pcaK, we examined the expression of the pcaK-lacZ fusion in E. coli BL21 cells in the presence and absence of BenR and benzoate. Low levels of pcaK-lacZ expression that were measured in E. coli were not influenced by BenR or benzoate (data not shown).
FIG. 6.
(A) Map of the chromosomally encoded pcaRKF cluster of 4-HBA degradation genes. (B) Map of the pcaK-lacZ fusion, pHNN216. The putative PcaR binding site is heavily underlined. Restriction sites introduced by cloning are lightly underlined. The transcriptional and translational start sites are in bold (36). (C) β-Galactosidase activities of the pcaK-lacZ fusion in wild-type (PRS2000) and benR mutant (PRS4157) cells. Cells were grown on succinate (white), 4-HBA (black), or 4-HBA plus benzoate (gray), and β-galactosidase activity was assayed as described in Materials and Methods.
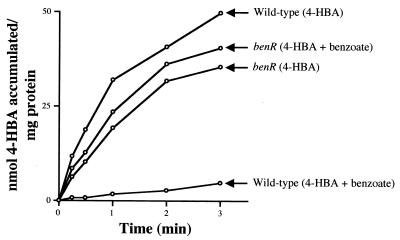
To show that the observed repressive effects of BenR on pcaK transcription have physiological significance, we measured rates of 4-HBA uptake in wild-type and benR mutant cells grown on 4-HBA only or on a mixture of 4-HBA and benzoate. When grown on 4-HBA alone, the wild type and the benR mutant accumulated 4-HBA intracellularly from the external medium at nearly identical rates of about 25 nmol per min per mg of protein. After growth on a mixture of benzoate and 4-HBA, wild-type cells accumulated 4-HBA very poorly at a 10-fold lower rate, whereas benR mutant cells took up 4-HBA from the medium at rates equivalent to those for cells grown on 4-HBA alone (Fig. 7).
FIG. 7.
Representative 4-HBA uptake assay of wild-type (PRS2000) and benR mutant (PRS4157) cells grown on 4-HBA or 4-HBA plus benzoate as indicated. Assays were performed as described in Materials and Methods.
DISCUSSION
A cluster of genes similar to the P. putida ben genes described here is also present in the gram-negative, nonmotile soil bacterium Acinetobacter sp. strain ADP1. This microbe has genes in the order benP benK benM benABDE (4, 26). Biochemical evidence indicates that the Acinetobacter benA, -B, and -C genes encode benzoate 1,2-dioxygenase and the benD gene encodes 2-hydro-1,2-dihydroxybenzoate dehydrogenase (39). The Acinetobacter benK gene encodes a benzoate transporter (5). We have assigned these functions to the homologous benABCD and -K genes from P. putida (Fig. 3). The benE genes of Acinetobacter and P. putida are homologous but do not resemble any known genes in the databases. benF from P. putida and benP from Acinetobacter each resemble porins in deduced amino acid sequence, although they do not resemble each other very much. The proposed porin function has yet to be demonstrated for either organism. A major difference between the two ben gene clusters is that they are controlled by members of two different families of regulatory proteins. Whereas expression of the Acinetobacter ben genes is controlled by BenM, a member of the LysR family of regulatory proteins (4), P. putida ben gene expression is regulated by BenR, an AraC/XylS family member.
BenR not only activates expression of the benABC genes in response to benzoate but also represses the 4-HBA-inducible expression of the 4-HBA transport protein, PcaK. Thus, BenR has the effect of shutting down the ability of cells to take up 4-HBA from their environment when benzoate is present. This provides a mechanism for the preferential degradation of benzoate by P. putida cells given a mixture of benzoate and 4-HBA. Regional regulation of aromatic compound degradation is not unique to P. putida; Acinetobacter sp. strain ADP1 also degrades benzoate in preference to 4-HBA (12). There is some evidence that BenM plays a role in mediating this preference (4).
Genes termed benR that regulated expression of chromosomal benABCD genes and that also activated Pm from the TOL plasmid were described for P. putida and P. aeruginosa some years ago (6, 25) but were not sequenced. Subsequently, Kessler et al. (30) concluded that very similar, if not identical, Pm sequence elements were recognized by the chromosomally encoded regulator BenR and the TOL plasmid-encoded regulator XylS. This prompted speculation that benR should be homologous to xylS; however, no hybridization between xylS and benR DNAs was detected in Southern hybridization experiments (25). This left open the possibility that two different types of regulators might be able to activate Pm. Results presented here show that the chromosomally encoded P. putida benR gene that activates Pm is, in fact, homologous to xylS.
Members of the AraC/XylS family of regulators are found widely distributed among bacteria. The family is characterized by a consensus sequence in the C-terminal 100 amino acids that includes two helix-turn-helix motifs that are proposed to mediate binding of the regulator to DNA (13). BenR has this consensus sequence. The activity of BenR and the benA promoter sequence have characteristics that match those of XylS activation of Pm transcription. The XylS protein is thought to bind to Pm as a dimer to a recognition sequence, TGCAN6GGNTA, that is repeated between nucleotides −70 and −56 and between nucleotides −49 and −35 (15). The benA promoter contains a direct repeat sequence between nucleotides −68 and −34 that matches the experimentally determined XylS binding site almost exactly (Fig. 3). In interacting with the downstream binding site that overlaps the −35 binding site for RNA polymerase, BenR may compete with RNA polymerase for binding to DNA. This is consistent with our observation of a threefold-higher basal level of β-galactosidase expression from the benA promoter under nonactivating conditions in the benR mutant compared to the wild type (Fig. 4A). Kaldalu et al. (28) have suggested that the N-terminal region of XylS interacts with its C-terminal domain to cause intramolecular repression of XylS function. On binding the benzoate effector, the N-terminal domain is proposed to undergo a change in conformation that then allows the C-terminal domain of XylS to function to allow initiation of transcription. The N-terminal regions of BenR and XylS share about 65% amino acid identity (Fig. 2), and BenR, like XylS, responds to benzoate as an effector. We found that BenR activates Pm expression to high levels in the absence of the benzoate effector when it is overexpressed in an E. coli background. A similar observation has been made with XylS (24, 28, 35). In the latter case, it has been proposed that a small amount of XylS is always present in a conformation that is active and able to stimulate transcription. When XylS is produced in large amounts, there is enough of the active form available to induce high levels of transcription in the absence of the effector (28, 35).
The effects of BenR in repressing transcription from the pcaK promoter may be indirect. We were unable to demonstrate an effect of BenR on pcaK expression in an E. coli background. Also, there are no detailed reports of an AraC/XylS family member responding to an effector molecule to mediate repression of gene transcription. Moreover, there is no recognizable XylS/BenR binding site in the pcaK promoter region. Clearly, much more study is required to determine the exact role of BenR in repressing pcaK transcription in response to benzoate.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant GM56665 from the National Institute of General Medical Sciences.
We thank Victor de Lorenzo for strains and for helpful discussions.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bundy B, Campbell A, Neidle E. Similarities between the antABC-encoded anthranilate dioxygenase and the benABC-encoded benzoate dioxygenase of Acinetobactersp. strain ADP1. J Bacteriol. 1998;180:4466–4474. doi: 10.1128/jb.180.17.4466-4474.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caetano-Anollés G. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 1993;3:85–94. doi: 10.1101/gr.3.2.85. [DOI] [PubMed] [Google Scholar]
- 4.Collier L S, Gaines G L I, Neidle E L. Regulation of benzoate degradation in Acinetobactersp. strain ADP1 by BenM, a LysR-type transcriptional activator. J Bacteriol. 1998;180:2493–2501. doi: 10.1128/jb.180.9.2493-2501.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collier L S, Nichols N N, Neidle E L. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobactersp. strain ADP1. J Bacteriol. 1997;179:5943–5946. doi: 10.1128/jb.179.18.5943-5946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuskey S M, Sprenkle A B. Benzoate-dependent induction from the OP2 operon-promoter region of the TOL plasmid pWW0 in the absence of known plasmid regulatory genes. J Bacteriol. 1988;170:3742–3746. doi: 10.1128/jb.170.8.3742-3746.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 8.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eaton R W. p-Cumate catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA carrying the cmtoperon. J Bacteriol. 1996;178:1351–1362. doi: 10.1128/jb.178.5.1351-1362.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franklin F C H, Bagdasarian M, Bagdasarian M M, Timmis K N. Molecular and functional analysis of the TOL plasmid pWW0 from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring metacleavage pathway. Proc Natl Acad Sci USA. 1981;78:7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaines G L I, Smith L, Neidle E L. Novel nuclear magnetic resonance spectroscopy methods demonstrate preferential carbon source utilization by Acinetobacter calcoaceticus. J Bacteriol. 1996;178:6833–6841. doi: 10.1128/jb.178.23.6833-6841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallegos M T, Schleif R, Bairoch A, Hofman K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. [Google Scholar]
- 15.González-Pérez M M, Ramos J L, Gallegos M T, Marqués S. Critical nucleotides in the upstream region of the XylS-dependent TOL meta-cleavage pathway operon promoter as deduced from analysis of mutants. J Biol Chem. 1999;274:2286–2290. doi: 10.1074/jbc.274.4.2286. [DOI] [PubMed] [Google Scholar]
- 16.Haak B, Fetzner S, Lingens F. Cloning, nucleotide sequence, and expression of the plasmid-encoded genes for the two-component 2-halobenzoate 1,2-dioxygenase from Pseudomonas cepacia2CBS. J Bacteriol. 1995;177:667–675. doi: 10.1128/jb.177.3.667-675.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harayama S, Rekik M. The metacleavage operon of TOL degradative plasmid pWW0 comprises 13 genes. Mol Gen Genet. 1990;221:113–120. doi: 10.1007/BF00280375. [DOI] [PubMed] [Google Scholar]
- 18.Harayama S, Rekik M, Bairoch A, Neidle E L, Ornston L N. Potential DNA slippage structures acquired during evolutionary divergence of Acinetobacter calcoaceticus chromosomal benABC and Pseudomonas putida TOL pWW0 plasmid xylXYZgenes encoding benzoate dioxygenases. J Bacteriol. 1991;173:7540–7548. doi: 10.1128/jb.173.23.7540-7548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harwood C S, Nichols N N, Kim M-K, Ditty J L, Parales R E. Identification of the pcaRKF gene cluster from Pseudomonas putida: involvement in chemotaxis, biodegradation, and transport of 4-hydroxybenzoate. J Bacteriol. 1994;176:6479–6488. doi: 10.1128/jb.176.21.6479-6488.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harwood C S, Parales R E. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 21.Huang H, Siehnel R J, Bellido F, Rawling E, Hancock R E. Analysis of two gene regions involved in the expression of the imipenem-specific, outer membrane porin protein OprD of Pseudomonas aeruginosa. FEMS Microbiol Lett. 1992;76:267–273. doi: 10.1016/0378-1097(92)90347-q. [DOI] [PubMed] [Google Scholar]
- 22.Inouye S, Nakazawa A, Nakazawa T. Molecular cloning of gene xylS of the TOL plasmid: evidence for positive regulation of the xylDEGF operon by xylS. J Bacteriol. 1981;148:413–418. doi: 10.1128/jb.148.2.413-418.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inouye S, Nakazawa A, Nakazawa T. Nucleotide sequence of the regulatory gene xylS on the Pseudomonas putidaTOL plasmid and identification of the protein product. Gene. 1986;44:235–242. doi: 10.1016/0378-1119(86)90187-3. [DOI] [PubMed] [Google Scholar]
- 24.Inouye S, Nakazawa A, Nakazawa T. Overproduction of the xylS gene product and activation of the xylDLEGFoperon of the TOL plasmid. J Bacteriol. 1987;169:3587–3592. doi: 10.1128/jb.169.8.3587-3592.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeffrey W H, Cuskey S M, Chapman P J, Resnick S, Olsen R H. Characterization of Pseudomonas putida mutants unable to catabolize benzoate: cloning and characterization of Pseudomonas genes involved in benzoate catabolism and isolation of a chromosomal DNA fragment able to substitute for xylSin activation of the TOL lower-pathway promoter. J Bacteriol. 1992;174:4986–4996. doi: 10.1128/jb.174.15.4986-4996.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones R M, Collier L S, Neidle E L, Williams P A. areABC genes determine the catabolism of aryl esters in Acinetobactersp. strain ADP1. J Bacteriol. 1999;181:4568–4575. doi: 10.1128/jb.181.15.4568-4575.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Junker F, Kiewitz R, Cook A M. Characterization of the p-toluenesulfonate operon tsaMBCD and tsaR in Comamonas testosteroniT-2. J Bacteriol. 1997;179:919–927. doi: 10.1128/jb.179.3.919-927.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaldalu N, Toots U, de Lorenzo V, Ustav M. Functional domains of the TOL plasmid transcription factor XylS. J Bacteriol. 2000;182:1118–1126. doi: 10.1128/jb.182.4.1118-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 30.Kessler B, Marqués S, Köhler T, Ramos J L, Timmis K N, de Lorenzo V. Cross talk between catabolic pathways in Pseudomonas putida: XylS-dependent and -independent activation of the TOL meta operon requires the same cis-acting sequences within the Pmpromoter. J Bacteriol. 1994;176:5578–5582. doi: 10.1128/jb.176.17.5578-5582.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kessler B, Timmis K N, de Lorenzo V. The organization of the Pmpromoter of the TOL plasmid reflects the structure of its cognate activator protein XylS. Mol Gen Genet. 1994;244:596–605. doi: 10.1007/BF00282749. [DOI] [PubMed] [Google Scholar]
- 32.Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimuriumencodes five homologues of carboxysome shell proteins. J Bacteriol. 1999;181:5317–5329. doi: 10.1128/jb.181.17.5317-5329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowalchuk G A, Hartnett G B, Benson A, Houghton J E, Ngai K-L, Ornston L N. Contrasting patterns of evolutionary divergence within the Acinetobacter calcoaceticus pcaoperon. Gene. 1994;146:23–30. doi: 10.1016/0378-1119(94)90829-x. [DOI] [PubMed] [Google Scholar]
- 34.Marques S, Ramos J L. Transcriptional control of the Pseudomonas putidaTOL plasmid catabolic pathways. Mol Microbiol. 1993;9:923–929. doi: 10.1111/j.1365-2958.1993.tb01222.x. [DOI] [PubMed] [Google Scholar]
- 35.Mermod N, Ramos J L, Bairoch A, Timmis K N. The xylS gene positive regulator of TOL plasmid pWW0: identification, sequence analysis and overproduction leading to constitutive expression of metacleavage operon. Mol Gen Genet. 1987;207:349–354. doi: 10.1007/BF00331600. [DOI] [PubMed] [Google Scholar]
- 36.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 37.Neidle E L, Hartnett C, Ornston L N, Bairoch A, Rekik M, Harayama S. Cis-diol dehydrogenases encoded by the TOL (pWW0) plasmid xylL gene and the Acinetobacter calcoaceticus chromosomal benDgene are members of the short-chain alcohol dehydrogenase superfamily. Eur J Biochem. 1992;204:113–120. doi: 10.1111/j.1432-1033.1992.tb16612.x. [DOI] [PubMed] [Google Scholar]
- 38.Neidle E L, Hartnett C, Ornston L N, Bairoch A, Rekik M, Harayama S. Nucleotide sequences of the Acinetobacter calcoaceticus benABCgenes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J Bacteriol. 1991;173:5385–5395. doi: 10.1128/jb.173.17.5385-5395.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neidle E L, Shapiro M K, Ornston L N. Cloning and expression in Escherichia coli of Acinetobacter calcoaceticusgenes for benzoate degradation. J Bacteriol. 1987;169:5496–5503. doi: 10.1128/jb.169.12.5496-5503.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nichols N N, Harwood C S. Repression of 4-hydroxybenzoate transport and degradation by benzoate: a new layer of regulatory control in the Pseudomonas putidaβ-ketoadipate pathway. J Bacteriol. 1995;177:7033–7040. doi: 10.1128/jb.177.24.7033-7040.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ochman H, Gerber A S, Hartl D L. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–623. doi: 10.1093/genetics/120.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto K, Gotoh N, Tsujimoto H, Yamada H, Yoshihara E, Nakae T, Nishino T. Molecular cloning and characterization of the oprQ gene coding for outer membrane protein OprE3 of Pseudomonas aeruginosa. Microbiol Immunol. 1999;43:297–301. doi: 10.1111/j.1348-0421.1999.tb02407.x. [DOI] [PubMed] [Google Scholar]
- 43.Olivera E R, Miñambres B, García B, Muñiz C, Moreno M A, Ferrández A, Díaz E, García J L, Luengo J M. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putidaU: the phenylacetyl-CoA catabolon. Proc Natl Acad Sci USA. 1998;95:6419–6424. doi: 10.1073/pnas.95.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ornston L N, Parke D. Properties of an inducible uptake system for β-ketoadipate in Pseudomonas putida. J Bacteriol. 1976;125:475–488. doi: 10.1128/jb.125.2.475-488.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Toole G A, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescensWCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 46.Parales R E, Harwood C S. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for Gram−bacteria. Gene. 1993;133:23–30. doi: 10.1016/0378-1119(93)90220-w. [DOI] [PubMed] [Google Scholar]
- 47.Park S M, Lu C D, Abdelal A T. Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosaPAO1. J Bacteriol. 1997;179:5300–5308. doi: 10.1128/jb.179.17.5300-5308.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 49.Segura A, Bünz P V, D'Argenio D A, Ornston L N. Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter. J Bacteriol. 1999;181:3494–3504. doi: 10.1128/jb.181.11.3494-3504.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivogenetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 51.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 52.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tobin J F, Schleif R F. Positive regulation of the Escherichia colil-rhamnose operon is mediated by the products of tandemly repeated regulatory genes. J Mol Biol. 1987;196:789–799. doi: 10.1016/0022-2836(87)90405-0. [DOI] [PubMed] [Google Scholar]
- 54.Yamano Y, Nishikawa T, Komatsu Y. Cloning and nucleotide sequence of anaerobically induced porin protein E1 (OprE) of Pseudomonas aeruginosaPAO1. Mol Microbiol. 1993;8:993–1004. doi: 10.1111/j.1365-2958.1993.tb01643.x. [DOI] [PubMed] [Google Scholar]
- 55.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]