Abstract
Cytolysin A (ClyA) is a pore-forming cytotoxic protein encoded by the clyA gene of Escherichia coli K-12. Genetic analysis suggested that clyA is silenced by the nucleoid protein H-NS. Purified H-NS protein showed preferential binding to clyA sequences in the promoter region, as evidenced by DNase I footprinting and gel mobility shift assays. Transcriptional derepression and activation of a chromosomal clyA::luxAB operon fusion were seen under conditions of H-NS deficiency and SlyA overproduction, respectively. In H-NS-deficient bacteria neither the absence nor the overproduction of SlyA affected the derepressed ClyA expression any further. Therefore, we suggest that overproduction of SlyA in hns+ E. coli derepresses clyA transcription by counteracting H-NS. The cyclic AMP receptor protein (CRP) was required for ClyA expression, and it interacted with a predicted, albeit suboptimal, CRP binding site in the clyA upstream region. Site-specific alterations of the CRP binding site to match the consensus resulted in substantially higher levels of ClyA expression, while alterations that were predicted to reduce CRP binding reduced ClyA expression. During anaerobic growth the fumarate and nitrate reduction regulator (FNR) was important for ClyA expression, and the clyA gene could be activated by overexpression of FNR. A major clyA transcript having its 5′ end (+1) located 72 bp upstream of the translational start codon and 61 bp downstream of the CRP-FNR binding site was detected in the absence of H-NS. The clyA promoter was characterized as a class I promoter that could be transcriptionally activated by CRP and/or FNR. According to DNA bending analyses, the clyA promoter region has high intrinsic curvature. We suggest that it represents a regulatory region which is particularly susceptible to H-NS silencing, and its features are discussed in relation to regulation of other silenced operons.
Bacteria having the ability to infect animals and humans are often capable of expressing virulence factors that can be of fundamental importance for the interactions that occur between the microorganism and the host. Molecular genetic analyes of different virulence determinants of enterobacteria encoding, e.g., cytotoxic substances, specific adhesins, and invasion proteins, have demonstrated that pathogenic isolates have complex gene systems that appear to be regulated in response to environmental growth conditions around the bacteria (36). Both enteropathogenic and uropathogenic isolates of Escherichia coli have become good model systems for this research. From analyses of genes controlling expression of fimbrial adhesins and invasiveness it was earlier shown that histone-like bacterial proteins are important for the regulation of virulence factors (20). The nucleoid-associated protein H-NS is known to influence the regulation of many genes in E. coli, and it appears that H-NS may cause silencing of many different operons (1). Even bacteria belonging to the normal flora presumably need to have their genes “tuned” to fit the environmental conditions within the host. Such regulation may be even more crucial for commensal organisms.
Cytolysin A (ClyA) is a 34-kDa cytolytic protein encoded by the clyA gene (also referred to as sheA and hlyE [13, 21]) located at 26.5 min on the E. coli K-12 chromosome. X-ray crystallography has shown that ClyA has unusual structural features and does not resemble any previously studied cytotoxin (59). We demonstrated recently that highly purified ClyA protein from E. coli K-12 causes lysis of mammalian cells by pore formation in a Ca2+-independent fashion (40) and apoptosis in murine-derived macrophage-like cells (30). It is interesting that the gene encoding this potentially toxic protein is found in E. coli K-12, which is considered to be nonpathogenic. In fact, it appears that most nonpathogenic strains of E. coli carry this gene and have the capacity to express cytotoxicity (39). Evidently, there is strict regulation of the clyA gene since it is phenotypically silent in E. coli K-12 under many tested laboratory conditions (39). The clyA gene is derepressed in H-NS-deficient E. coli strains (58; J. M. Gómez-Gómez, J. Blazquez, F. Baquero, and J. L. Martinez, Letter, Mol. Microbiol 19:909–910, 1996; Y. Mizunoe and B. E. Uhlin, Abstr. 34th Intersci. Conf. Antimicrob. Agents Chemother., p. 63, 1994), and strains overexpressing SlyA and MprA (13, 34, 35, 41). SlyA and MprA belong to a family of proteins thought to regulate diverse physiological processes in bacterial pathogens (57). A direct interaction between purified His-SlyAEC and the DNA upstream of the clyA-coding region was suggested from results obtained by band shift assays (41). In addition, ClyA expression is activated in E. coli K-12 by the expression of HlyX from Actinobacillus pleuropneumoniae (21). HlyX has 73% identical amino acid sequence compared with the oxygen-responsive transcriptional regulator, FNR, which binds to a putative FNR binding site in the clyA upstream region (21). Furthermore, it was recently shown that altered FNR proteins, similarly to HlyX, could activate the expression of clyA (43); i.e., minor alterations in a gene encoding a global regulator have a profound effect on the production of cytotoxic factors like ClyA. Because of the potential to express such a host-damaging product, the clyA gene represents a novel class of genes not previously characterized in commensal bacteria. In the present paper we present data from experiments aimed at elucidating features about the strict regulation of clyA.
MATERIALS AND METHODS
Bacterial strains and culture media.
The relevant genotypes of strains and plasmids used in this work are listed in Tables 1 and 2, respectively. The strains were grown in LB broth (4) with vigorous shaking or on LB broth solidified with 1.5% (wt/vol) agar. Blood agar plates were 5% horse erythrocytes solidified with 1% (wt/vol) Columbia agar base (Oxoid Ltd.). Antibiotic selection for pFZY1-derived plasmids was carried out using carbenicillin (25 μg · ml−1). In other cases the growth medium was supplemented with carbenicillin (50 μg · ml−1), kanamycin (50 μg · ml−1), chloramphenicol (10 μg · ml−1), or tetracycline (15 μg · ml−1). Anaerobic growth conditions were achieved by using the AnaeroGen compact atmosphere generation system of Oxoid Ltd., following the instructions of the manufacturer.
TABLE 1.
Bacterial strains used in this work
| Strain | Genotype or relevant characteristics | Reference or source |
|---|---|---|
| DH5α | endA1 hsdR17 (rK mK+) supE44 thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 [φ80(ΔlacZ)M15] | 23 |
| SY327(λpir) | Δ(lac-pro) argE(Am) rif nalA recA56 (λpir) | 38 |
| MC1061 | araD139 Δ(ara, leu) 7697 ΔlacX74 galU galK hsr hsm+ strA | 11 |
| BEU616 | MC1061 hns::cat (Cmr) | This work |
| M182 | Δ(lacIPOZY) X74 galK galU strA | 11 |
| M182 crp | M182 Δcrp | 9 |
| M182 fnr | M182 fnr::tet (Tcr) | 22 |
| M182 crp fnr | M182 Δcrp fnr::tet (Tcr) | 2 |
| JON31 | M182 hns::cat (Cmr) | This work |
| JON32 | M182 Δcrp hns::cat (Cmr) | This work |
| BEU701 | M182 fnr::tet hns::cat (Tcr Cmr) | This work |
| BEU705 | M182 Δcrp fnr::tet hns::cat (Tcr Cmr) | This work |
| MC4100 | araD139 Δ(lac)U169 strA thi | 10 |
| BSN26 | MC4100 trp::tet (Tcr) | 26 |
| BSN27 | MC4100 trp::tet Δhns (Tcr) | 26 |
| JON33 | BSN26 clyA::luxAB | This work |
| JON34 | BSN27 clyA::luxAB | This work |
| MWK6 | BSN27 ΔslyA | This work |
| MWK10 | MWK6 clyA::luxAB | This work |
| MWK11 | MC4100 with a consensus CRP binding site in the clyA promoter regiona | This work |
5′-AAATGTGATCTAGATCACATTT-3′
TABLE 2.
Plasmids used in this work
| Plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| pCH257 | Suicide vector, Cmr | 17 |
| pFZY1 | Mini-F transcriptional lacZ fusion vector, Cbr | 28 |
| pKO3 | Gene replacement vector, Cmr | 31 |
| pLG339 | Cloning vector, Tcr Kmr | 54 |
| pSL1180 | Cloning vector, Cbr | 7 |
| pYMZ62 | 3.5-kb subclone of the clyA locus in pUC18 | This laboratory |
| pYMZ80 | 1.6-kb subclone of the clyA locus in pUC18 | This laboratory |
| pYMZ81 | Same as plasmid pYMZ80, but with the 1.6-kb clyA locus in the opposite orientation | This laboratory |
| pYMZ83 | Transcriptional clyA::lacZ fusion in pFZY1 | This work |
| pMWK4 | clyA::luxAB in pCH257 | This work |
| pMWK9 | pYMZ81 with altered CRP binding site: 5′-TTGTTTGATATAGATCACATTT-3 | This work |
| pMWK10 | pYMZ81 with altered CRP binding site: 5′-AAATGTGATCTAGATCACATTT-3′ | This work |
| pMWK24 | pYMZ81 with altered CRP binding site: 5′-TTGTGTGATATTTATCATATTA-3′ | This work |
| pMWK28 | pYMZ81 with altered CRP binding site: 5′-TTGTTTAATATTTATCATATTA-3′ | This work |
| pMWK29 | pYMZ81 with altered CRP binding site: 5′-TTGTCTAATATTTATCATATTA-3′ | This work |
| pMWK31 | pYMZ81 with altered clyA −10 sequence: TATGAAT→CACGAAC | This work |
| pMWK32 | Subclone of the clyA locus from position −349 to −50 in pGEM-T Easy | This work |
| pMWK33 | Subclone of the clyA locus from position −250 to +50 in pGEM-T Easy | This work |
| pMWK34 | Subclone of the clyA locus from position −150 to +150 in pGEM-T Easy | This work |
| pMWK35 | Subclone of the clyA locus from position −50 to +250 in pGEM-T Easy | This work |
| pMWK36 | Subclone of the clyA locus from position +50 to +349 in pGEM-T Easy | This work |
| pMWK37 | Subclone of the clyA locus from position +150 to +449 in pGEM-T Easy | This work |
| pMWK45 | pJON78 with altered CRP binding site: 5′-AAATGTGATCTAGATCACATTT-3′ | This work |
| pJON22 | 2.1-kb subclone of the slyAEC locus in pACYC177 | This laboratory |
| pJON78 | 3.5-kb subclone of the clyA locus in pKO3 | This laboratory |
| pMOJ2 | slyA in-frame deletion in pKO3 | This work |
| pDW300 | crp gene in pLG339, formerly known as pLG339/CRP | 2, 60 |
| pGS24 | fnr gene in pBR322 | 50 |
| pGS215 | FNR-V208R;S212G;G216K | 53 |
| pGS297 | FNR-V208R;E209D | 52 |
Genetic techniques.
Standard procedures were used in all general molecular applications (46). Generalized bacteriophage P1 transduction was performed as described by Willets et al. (61). Sequencing oligonucleotides were made on an Applied Biosystems 394 synthesizer or obtained from DNA Technology, Aarhus, Denmark. Dideoxy sequencing was carried out using a T7 sequencing kit (Pharmacia Biotech) according to the specifications of the manufacturer, using pYMZ62 as the template. For general purposes DNA sequencing was performed using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA polymerase and an ABI PRISM 377 DNA sequencer. Site-specific alterations of DNA sequences were obtained by using the QuickChange site-directed mutagenesis kit of Stratagene, following the instructions of the manufacturer. The desired mutations were always placed in the middle of the primer with approximately 15 bases of correct sequence on each side.
Plasmid and strain constructions.
The construct pYMZ83 was made by ligation of a 0.4-kb EcoRI-BglII restriction fragment from pYMZ80 into EcoRI-BamHI-digested pFZY1, resulting in a clyA::lacZ transcriptional fusion having its 5′ end 290 bp upstream of the clyA start codon and its fusion junction 76 bp into the clyA coding sequence. The plasmid pMWK4 was constructed by ligating a blunt end-generated SmaI-BglII restriction fragment from pYMZ80, containing the DNA 290 bp upstream of and down to 76 bp within the clyA coding sequence, into EcoRV-digested pCH257 suicide vector, using E. coli SY327(λpir) as the host strain. The plasmid pMWK4 was integrated into the chromosome of MC1061 by a single recombination event between the 366-bp clyA sequence in our construct and the corresponding region of clyA on the chromosome. The resulting strain was designated MWK2. BSN26, BSN27, and MWK6 were transduced with P1 grown on MWK2 (clyA::luxAB), and transductants were isolated by selection for chloramphenicol resistance, resulting in the strains JON33, JON34, and MWK10, respectively. The plasmid pMWK24 was constructed by using the PCR-based strategy described above and the primers crp5 (5′-CATTAAACATTGTGTGATATTTATCATATT-3′) and crp6 (5′-AATATGATAAATATCACACAATGTTTAATG-3′), with pYMZ81 as the template. The same approach was utilized, with pYMZ81 as the template, to construct pMWK28 (primers crp7 [5′-CATTAAACATTGTTTAATATTTATCATATT-3′] and crp8 [5′-AATATGATAAATATTAAACAATGTTTAATG-3′], pMWK29 (primers crp9 [5′-TGACATTAAACATTGTCTAATATTTATCATATTAAT-3′] and crp10 [5′-ATTAATATGATAAATATTAGACAATGTTTAATGTCA-3′], pMWK31 (primers a-10 [5′-TCCCGCCCGGCTAACCACGAACTAGATGAAGTAAAA-3′] and b-10 [5′-TTTTACTTCATCTAGTTCGTGGTTAGCCGGGCGGGA-3′], and pMWK9 (primers crp1 [5′-CATTGTTTGATATAGATCACATTTATAGAAATAAAGAC-3′] and crp2 [5′-GTCTTTATTTCTATAAATGTGATCTATATCAAACAATG-3′]. The plasmid pMWK10 was constructed by the same method, with the primers crp3 (5′-GACATTAAACAAAATGTGATCTAGATCACATTTATAG-3′) and crp4 (5′-CTATAAATGTGATCTAGATCACATTTTGTTTAATGTC-3′) and pMWK9 as the template. A BbrPI-BglII promoter fragment containing the consensus cyclic AMP (cAMP) receptor protein (CRP) site (5′-AAATGTGATCTAGATCACATTT-3′) (see reference 16 and references therein) was cloned into the corresponding sites of the construct pJON78 to generate pMWK45. The derivative pJON78 is a 3.5-kb subclone of the clyA locus in the suicide donor plasmid pKO3. Using the derivative pMWK45, the CRP consensus site was introduced onto the chromosome of MC4100, as previously described (31), to generate the strain MWK11. Strain BEU616 (hns::cat) was constructed by transduction of MC1063 with P1 grown on a derivative of JC7623 carrying an hns::cat allele in the chromosome. JC7623 is recC22 recB21 sbcB15 sbc201 (24). MC1063 is MC1061 with trp::Tn10 (11). BEU701 and BEU705 were constructed by transduction of M182 fnr and M182 crp fnr, respectively, with P1 grown on BEU616. To construct a 429-bp in-frame deletion mutant of the slyA coding region, we used a PCR-based strategy (see above) and DH5α as the host strain: By using the primers SKO1 (5′-AATTATAAGGAGATGGAATTCGAATCGCCACTAGGT-3′), SKO2 (5′-ACCTAGTGGCGATTCGAATTCCATCTCCTTATAATT-3′), SKO3 (5′-ATTGAGTTACAGGCCGAATTCTGAAATGAAGGGGGC-3′), and SKO4 (5′-GCCCCCTTCATTTCAGAATTCGGCCTGTAACTCAAT-3′), and pJON22 as template, two new EcoRI restriction sites (underlined sequences) were introduced in the slyA gene, at bp 4 to 9 and bp 433 to 438, respectively. The resulting plasmid clone, designated pMWK11, was subsequently digested with EcoRI and religated to generate pMWK12. To facilitate cloning into pKO3, a 1.5-kb PstI fragment of pMWK12, encompassing the constructed in-frame deletion, was cloned into PstI-digested pSL1180 cloning vector, resulting in the plasmid pMWK13, which was used as an intermediate. A 1.5-kb PmlI-BamHI fragment of pMWK13, containing the slyA in-frame deletion, was cloned into SmaI-BamHI-digested pKO3, resulting in the plasmid pMOJ2. This construct, containing the slyA in-frame deletion, was introduced into BSN27 as previously described (31) to generate MWK6.
ClyA expression assays.
Lytic activity towards erythrocytes was scored by a clearance zone on blood agar plates after 16 to 17 h of incubation at 37°C or by quantification of the release of hemoglobin from erythrocytes as described below. Bacteria were grown to late logarithmic phase and diluted to 8.0 × 106 cells ml−1 in 1× phosphate-buffered saline. Fifty microliters of bacterial suspension was mixed with 50 μl of a horse erythrocyte suspension in 1× phosphate-buffered saline in a 96-well microtiter plate and incubated for 120 min at 37°C prior to determination of the release of hemoglobin, as described previously (47). β-Galactosidase activity was measured as previously described (36), with compensation for the number of plasmid-free cells (51). Luciferase assays were performed as described earlier (42).
Sodium dodecyl sulfate-PAGE and immunoblot analysis.
For determination of the ClyA and SlyA protein content in cells, bacterial samples grown at 37°C were harvested at late logarithmic phase, or from agar plates after 16 to 17 h of incubation, prior to analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) as previously described (29). Western immunoblotting was performed using an antiserum raised against ClyA as described previously (40) or using an antiserum raised against SlyA as described below at final dilutions of 1:1,000 and alkaline phosphatase-conjugated secondary antibody at a final dilution of 1:3,000. Immunoreactive bands were visualized using the enhanced chemiluminescence Western blotting detection system of Amersham Pharmacia Biotech, following the instructions of the manufacturer. Rabbit anti-SlyA antibodies were raised against His-SlyA which had been purified as described previously (41). An antiserum taken 8 weeks after the fourth injection was affinity purified as previously described (56).
RNA isolation and primer extension.
Total RNA was isolated from late logarithmic phase cultures, which had been grown in LB broth, using the hot-phenol method (63). Primer extension analysis was carried out as follows. Oligomers were 5′ end labeled using polynucleotide kinase and [γ-32P]ATP. A molar excess of the primer was annealed to 5 μg of total RNA in 8 μl of an annealing buffer (50 mM Tris-HCl [pH 8.3], 60 mM NaCl, 10 mM dithiothreitol [DTT]). Samples were heated for 5 min at 80°C and subsequently chilled on ice for 5 min. Eight microliters of extension mixture (25 mM Tris-HCl [pH 8.3], 30 mM NaCl, 15 mM MgCl2, 1.25 mM DTT, a 1 mM concentration of each deoxynucleoside triphosphate) was added together with 3 U of avian myeloblastosis virus reverse transcriptase, and samples were incubated at 42°C for 60 min. The oligonucleotides used were cct1 (5′-CCGTTTTATCTGCAACGATTTCAGTC-3′) and cct4 (5′-GGAGGCTGCCTGTGAATACTCCTGTTTAAAGCGACTTAAC-3′). The extension products were analyzed by electrophoresis on 6% polyacrylamide-urea gels.
DNA bending analysis.
Overlapping 300-bp DNA fragments of the clyA locus were generated by PCR using pYMZ62 as a template and the following oligonucleotide primers from the clyA locus (see Fig. 2): for fragment a, −349 (5′-GCGGAAAAGTCACAATTTCG-3′) and −50b (5′-TTAATATGATAAATATCAAA-3′); for fragment b, −250 (5′-CCAGCAGATCAATACTGATT-3′) and +50b (5′-ATAAATTGTAATGAAACTCC-3′); for fragment c, −150 (5′-ACGCTCATCCAGCAGAAATG-3′) and +150b (5′-AAGATCTAATGCTCCATCTG-3′); for fragment d, −50a (5′-ATAGAAATAAAGACATTGAC-3′) and +250 (5′-CGGAGGCTGCCTGTGAATAC-3′); for fragment e, +50a (5′-TATATTTAAAGAGGCGAATG-3′) and +349 (5′-ATTGCGTCGCAACACCACAC-3′); and for fragment f, +150a (5′-TTATAATAAATATCTCGATC-3′) and +449 (5′-TTCGTGATGCCGTCATCCAG-3′). The generated PCR fragments were cloned utilizing the pGEM-T Easy Vector System (Promega), as specified by the manufacturer, resulting in the constructs pMWK32 (position −349 to −50 [base pairs] relative to the clyA transcriptional start point), pMWK33 (−250 to +50), pMWK34 (−150 to +150), pMWK35 (−50 to +250), pMWK36 (+50 to +349), and pMWK37 (+150 to +449). These constructs were digested with EcoRI and used for DNA bending analysis.
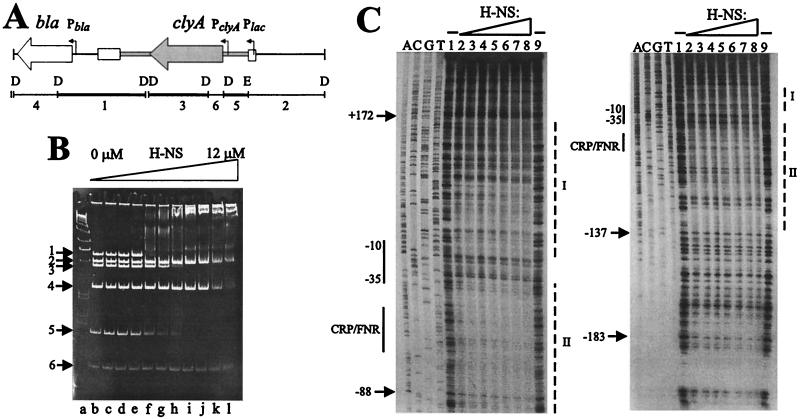
FIG. 2.
Binding of purified H-NS to clyA. (A) Schematic drawing of the plasmid pYMZ80. Relevant features, positions of the restriction endonuclease sites (D, DraI; E, EcoRI), and the extent of the resulting restriction endonuclease fragments used in gel shift assays with H-NS are indicated. Fragments bound preferentially by H-NS are indicated by thick black horizontal bars, and fragments showing no specific shift are indicated by thin black horizontal bars. Cloned chromosomal DNA encompassing the clyA locus is indicated in grey. (B) Gel shift assay of the clyA gene with purified H-NS protein. The different DraI and EcoRI restriction fragments generated from pYMZ80 are indicated within the figure. Lanes: a, DNA ladder; b, no protein; c, 0.8 μM H-NS; d, 1.7 μM; e, 2.5 μM; f, 3.4 μM; g, 4.2 μM; h, 5.0 μM; i, 6.7 μM; j, 8.4 μM, k, 10 μM; 1, 12 μM. (C) DNase I footprint assay of the clyA promoter region with H-NS. Addition of H-NS was as indicated at the top. Lanes 1 and 9 show samples without any H-NS added and lanes 2 to 8 show samples with increasing amounts (4.8, 5.6, 6.4, 7.2, 8.0, 8.8, and 9.6 μM) of H-NS added. The positions of the clyA promoter (−10 and −35 regions) and the CRP-FNR site are shown by solid lines along the left side. The two main regions of H-NS interaction (labelled I and II) are shown by dashed lines along the right.
The fragments were run on 6% polyacrylamide–bis-acrylamide gels (30:0.8, vol/vol) in a 90 mM TBE buffer (90 mM Tris, 90 mM boric acid, 2.5 mM disodium EDTA, pH 8.3) at 6 mA for 10 h at 5°C, room temperature, and 37°C, and subsequently stained with ethidium bromide. The migration lengths were measured and compared to a 1 Kb PLUS DNA Ladder molecular size standard (GibcoBRL). The ratio between the observed migration length (Mo) and the expected migration length (Me) was plotted against the center position of the DNA fragment.
Gel mobility shift assay.
Band shift assays were performed with DNA fragments from the plasmid pYMZ80 obtained by digestion with DraI and EcoRI. Purified H-NS protein was obtained from a strain carrying an expression plasmid (B. Sondén and B. E. Uhlin, unpublished data). DNA at a final concentration of approximately 36 nM was mixed with H-NS at a final concentration of 0.8 to 12 μM in buffer B (25 mM HEPES [pH 7.5], 0.1 mM EDTA, 5 mM DTT, 10% glycerol) in a total volume of 10 μl. The samples contained 100 ng of poly(dI-dC), and KCl was added to 50 mM. The reactions were incubated for 15 min at 26°C and then resolved by nondenaturing PAGE in a 6% gel using TBE running buffer. The gel was subsequently stained with ethidium bromide.
DNase I footprint analysis.
DNase I footprint analysis with CRP was carried out essentially as described previously (19). The DNA fragments were obtained by PCR of the strain MC1061, with the primers cct1 and umu1 (5′-AATATTTGTCGCTGC-3′) and the latter primer had been labeled with [γ-32P]dATP using T4 polynucleotide kinase. Reactions were carried out in a total volume of 50 μl. CRP was purified essentially as described previously (64), with the exception that CRP was precipitated with (NH4)2SO4 to obtain CRP free from cAMP instead of removing the cAMP by chromatography. Samples of CRP (final concentration, 4.7 to 38 nM) and/or RNA polymerase (22 to 87 nM) were added to approximately 10 ng of DNA in buffer B plus 50 mM KCl. When included, cAMP was added to a final concentration of 20 mM. Fifty nanograms of DNase I and MgCl2 to a final concentration of 5 mM were added to start the digestion. After 120 s (90 s for samples without protein) the reactions were stopped by the addition of 12 μl of stop mix (0.25 mM EDTA, 1.5 M NaCl, oyster glycogen [1.5 mg ml−1]). The samples were then phenol extracted, ethanol precipitated, and analyzed on 6% polyacrylamide-urea gels.
DNase I footprint analysis with H-NS was carried out essentially as described previously (19). The DNA fragments were obtained by PCR of the plasmid pYMZ62, with the primers p73 (5′-GAATGTCTTTCTGGGCGG-3′) and umu1, labeled with [γ-32P]dATP using T4 polynucleotide kinase. Reactions were carried out in a total volume of 50 μl. Samples of H-NS (final concentration, 4.8 to 9.6 μM) were added to approximately 30 ng of DNA in buffer B plus 50 mM KCl. Fifty nanograms of DNase I and MgCl2 at a final concentration of 5 mM was added to start the digestion. After 120 s (90 s for samples without protein) the reactions were stopped by the addition of 12 μl of stop mix (defined above). The samples were then phenol extracted, ethanol precipitated, and analyzed on 6% polyacrylamide-urea gels.
Computer projection of DNA curvature.
Projections of calculated DNA curvature were obtained by using the BEND program of the DNASTAR software package, which uses a wedge model to predict the helix trajectory. The dinucleotide bending angles used were according to published data (5).
RESULTS
Effects of H-NS deficiency and SlyA overproduction on clyA transcription as monitored by a chromosomal clyA::luxAB operon fusion.
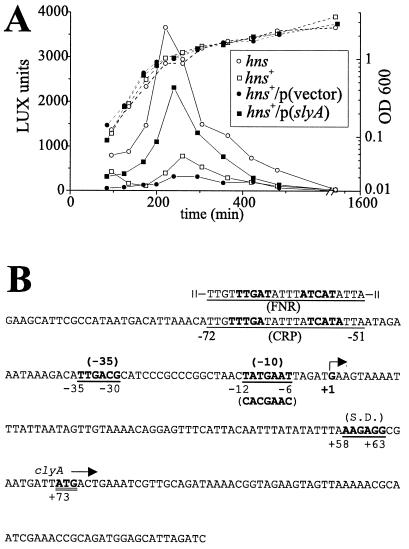
To determine at which level H-NS affects the expression of clyA, and to quantitatively monitor the transcription of clyA in hns mutant and SlyA-overproducing strains during different growth phases, we used the strains JON33 and JON34, which have a transcriptional clyA::luxAB fusion at the site of the clyA locus (see Materials and Methods). We investigated the transcription of clyA in isogenic hns wild-type and mutant strains (JON33 and JON34) by monitoring the expression of the chromosomal clyA::luxAB fusion throughout the growth cycle. As shown in Fig. 1, the luciferase activity of the hns strain JON34 peaked in late logarithmic phase and showed a more-than-fourfold increase in activity compared with the hns+ strain. In parallel we studied the activation of clyA by SlyA by assaying the expression of the chromosomal clyA::luxAB fusion throughout the growth cycle, using JON33 (hns+) as the host strain. As shown in Fig. 1, the luciferase activity of the SlyA-overproducing strain JON33/pJON22 peaked in late logarithmic phase and showed a more-than-fivefold increase in activity compared with the vector control strain JON33/pACYC177. Similar results were obtained when expression was monitored at the translational level using contact hemolysis assays with erythrocytes (see Materials and Methods; data not shown). These results were in accordance with the observation of a peak ClyA activity in samples taken from MprA- or SlyA-overproducing strains at late logarithmic phase (13, 34). Therefore, we conclude that the expression of clyA is mainly controlled at the transcriptional level and that H-NS is responsible for silencing the transcription of clyA. Furthermore, upon relief of this silencing (hns mutants and SlyA-overproducing strains), the highest expression of the chromosomal clyA::luxAB fusion occurred in the late logarithmic phase.
FIG. 1.
A. Effects of H-NS deficiency and SlyA overproduction on the transcription of clyA throughout the growth curve. (A) Expression of luciferase activity from a chromosomal clyA::luxAB fusion in the strains JON33 (hns+) (□), JON34 (hns) (○), JON33/pACYC177 (vector control) (●), and JON33/pJON22 p(SlyA) (■). The expressed luciferase activity was quantified by the luciferase assay (see Materials and Methods), and LUX units were displayed as millivolts/(milliliters × optical density at 600 nm). The growth curves are indicated with dotted lines. (B) DNA sequence of the clyA promoter region down to 76 bp into the clyA coding sequence, which is the position of the lux and lac fusion junctions. The positions of the transcriptional initiation point (+1) and putative regulatory elements, i.e., binding sites for CRP and FNR, Shine-Dalgarno (S.D.) sequence, and −10 and −35 boxes are shown. The mutational alterations in the plasmid pMWK31 are shown in parentheses below the −10 region sequence.
Site of initiation of derepressed clyA transcription and promoter analysis in hns mutant E. coli.
Analysis of clyA transcription by Northern blot hybridization suggested that it is a monocistronic operon (41). In order to further localize the clyA promoter active in the absence of H-NS, the clyA transcript was assayed by primer extension analysis. RNA was extracted from the hns strain, BEU616, that expresses phenotypically detectable levels of the ClyA protein. The clyA primer extension resulted in a distinct product that should represent one major transcript with the 5′ end 72 nucleotides upstream of the ATG translational start codon of the clyA structural gene (data not shown). Therefore, we concluded that the observed clyA transcriptional start point (+1) in the hns mutant strain was the same as in strains in which the clyA gene was activated by the cloned slyA locus (34). To functionally assess the predicted −10 promoter box (TATGAAT) (Fig. 1B), we introduced site-specific alterations in the clyA upstream sequence, using a PCR-based strategy (see Materials and Methods) and the plasmid pYMZ81 as the template. This plasmid contains the clyA sequence cloned in the opposite orientation to the promoter of the vector, the gene thus being controlled by its native promoter region only. The resulting construct (plasmid pMWK31), with the −10 sequence changed to (CACGAAC), had lost its promoter activity according to the in vivo tests. As shown below, DH5α harboring pMWK31 showed a lack of expression of ClyA protein and cytolytic activity compared with DH5α/pYMZ81. Thus, the predicted −10 promoter box is important for clyA expression, and we conclude that this analysis localised the promoter sequences.
H-NS shows preferential interaction in vitro with clyA sequences.
To examine whether there is a direct interaction between H-NS and the clyA locus, electrophoretic mobility shift assays were performed as described in Materials and Methods, using purified H-NS protein and clyA DNA. An initial indication of preferential binding of H-NS to clyA DNA (fragments 3 and 5) was observed using EcoRI-DraI-digested pYMZ80 as the target DNA (Fig. 2B). It was also observed that one of the vector DNA fragments in this experiment (fragment 1) shifted in the presence of H-NS, which is consistent with previous findings of H-NS interaction with the plasmid carried bla promoter region (33, 65). DNase I footprinting assays showed that H-NS interacted preferentially with two regions of the clyA promoter region (Fig. 2C). The H-NS protein interacted both in the downstream region of the promoter (designated I in Fig. 2C) and in the upstream region (designated II in Fig. 2C). These findings support a model where H-NS directly interacts with, and negatively affects, clyA transcription.
SlyA is not required for clyA expression in hns mutant E. coli.
Since SlyA, when overexpressed, activates the expression of ClyA, we wanted to investigate the requirement of SlyA for derepression of clyA in the absence of H-NS. We introduced (see Materials and Methods) a slyA in-frame deletion into the slyA locus of the hns strain BSN27, resulting in the strain MWK6, and a clyA::luxAB fusion into the clyA locus of MWK6, resulting in the strain MWK10. We found that there was no significant difference in luciferase activity throughout the growth curve in strain JON34 compared with MWK10 (data not shown), indicating that SlyA is not required for clyA expression in H-NS mutant strains. We also found that BSN27 and MWK6 showed similar ClyA activity on blood agar (Table 3) and quantified lytic activity using the erythrocyte assay (data not shown). This was in accordance with overproduction of SlyA in the hns clyA::luxAB strain JON34/pJON22 (data not shown), in the hns strain JON31/pJON22, and in the hns slyA strain MWK6/pJON22 (Table 3), which did not result in an additive effect on ClyA expression compared with the control strains JON34/pACYC177, JON31/pACYC177, and MWK6/pACYC177. We therefore concluded that SlyA was not essential for clyA expression when H-NS was absent.
TABLE 3.
Expression of ClyA as evidenced by lysisa of erythrocytes in agar
| E. coli strain | Relevant characteristic(s) | ClyA expression under the following conditions
|
|
|---|---|---|---|
| Aerobic | Anaerobic | ||
| BSN26 | Wild type | − | − |
| BSN27 | hns | ++ | ++ |
| MWK6 | hns slyA | ++ | ++ |
| JON31 | hns | ++ | ++ |
| JON32 | hns crp | (+) | (+) |
| BEU701 | hns fnr | ++ | ++ |
| BEU705 | hns crp fnr | (+) | (+) |
| JON32/pDW300 | hns crp/p(CRP) | ++ | ++ |
| JON32/pLG339 | hns crp | (+) | (+) |
| BEU701/pDW300 | hns fnr/p(CRP) | ++ | ++ |
| BEU701/pLG339 | hns fnr | ++ | ++ |
| BEU701/pGS24 | hns fnr/p(FNR) | ++ | ++ |
| BEU701/pGS215 | hns fnr/p(FNR-V208R;S212G;G216K) | ++ | ++ |
| BEU701/pGS297 | hns fnr/p(FNR-V208R;E209D) | ++ | ++ |
| BEU701/pBR322 | hns fnr | ++ | ++ |
| BEU705/pDW300 | hns crp fnr/p(CRP) | ++ | ++ |
| BEU705/pLG339 | hns crp fnr | (+) | (+) |
| BEU705/pGS24 | hns crp fnr/p(FNR) | (+) | + |
| BEU705/pGS215 | hns crp fnr/p(FNR-V208R;S212G;G216K) | (+) | + |
| BEU705/pGS297 | hns crp fnr/p(FNR-V208R;E209D) | (+) | + |
| BEU705/pBR322 | hns crp fnr | (+) | (+) |
| M182/pJON22 | Wild type/p(SlyA) | ++ | ++ |
| M182/pACYC177 | Wild type | − | − |
| M182 crp/pJON22 | crp/p(SlyA) | (+) | (+) |
| M182 crp/pACYC177 | crp | − | − |
| JON31/pJON22 | hns/p(SlyA) | ++ | ++ |
| JON31/pACYC177 | hns | ++ | ++ |
| MWK6/pJON22 | hns slyA/p(SlyA) | ++ | ++ |
| MWK6/pACYC177 | hns slyA | ++ | ++ |
Lysis of erythrocytes was scored on blood agar plates as ++, lysis around individual colonies; +, lysis beyond the edge of the bacterial cell mass only; (+), weak lysis in the center of the bacterial cell mass only; or −, no lysis.
The clyA promoter is dependent on CRP for efficient expression.
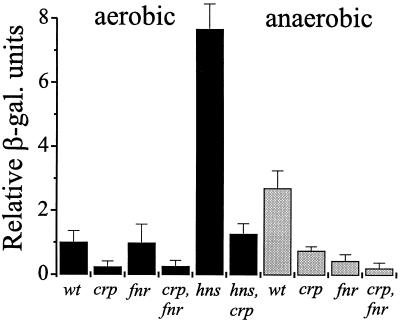
Analysis of the clyA promoter region revealed a potential CRP binding site (5′-TTGTTTGATATTTATCATATTA-3′) that matched the consensus in 13 out of 22 bases. The CRP binding site partly overlapped with a previously identified FNR binding site (21). To investigate the requirement of CRP for the transcription of clyA, a cat (Cmr) gene block was introduced into the hns locus of the strains M182 and M182 crp, resulting in the strains JON31 and JON32, respectively (see Materials and Methods). The low-copy (one to two copies per chromosome) clyA::lacZ reporter system pYMZ83 (based on the mini-F vector pFZY1) was used in these strains to study the level of clyA expression (Fig. 3). JON31 showed a more-than-sixfold-greater expression of β-galactosidase activity than JON32, which expressed the same low levels as the crp+ hns+ strains (even lower β-galactosidase activity was observed in hns+ crp strains). This was consistent with a much reduced cellular level of ClyA protein in the hns crp double mutant strain, as evidenced by Western immunoblotting (data not shown) and a substantially reduced lysis of erythrocytes in agar (Table 3). In addition, the results with the clyA::lacZ fusion indicated that the regulatory DNA sequences required for control of clyA transcription are present within the region spanning from 290 bp upstream of the clyA coding sequence to 76 bp into the clyA structural gene (the operon fusion junction). The reduced ClyA activity of JON32 could be restored by the reintroduction of CRP on a plasmid (pDW300). Thus, we concluded that CRP is required for derepression of clyA in hns strains. To investigate whether CRP is also required for the SlyA-mediated relief of H-NS silencing, we introduced the plasmid pJON22 (encoding SlyA) into the crp mutant and wildtype E. coli strains, M182 crp and M182, respectively. As shown in Table 3, overexpression of SlyA resulted in a strong cytolytic phenotype in M182, but only a weak cytolytic phenotype in M182 crp. This suggested that CRP is important for activation of ClyA expression by SlyA. The absence of CRP did not affect the level of SlyA protein, which was similar in M182/pJON22 and M182 crp/pJON22 as evidenced by western immunoblotting (data not shown), using an antiserum raised against SlyA (see Materials and Methods). Therefore, we concluded that the clyA promoter is dependent on CRP for efficient clyA expression.
FIG. 3.
CRP-dependent transcription of clyA. Shown is the quantification of clyA transcription from a clyA::lacZ reporter system on the plasmid pYMZ83 in the following strains grown under aerobic (solid black bars) and anaerobic (grey bars) conditions for 16 to 17 h at 37°C on LB agar: wt, M182; crp, M182 crp; fnr, M182 fnr; crp fnr, M182 crp fnr; hns, JON31; and hns crp, JON32. β-Galactosidase (β-gal.) activity was measured as described in Materials and Methods. A relative β-galactosidase activity of 1.0 equals the activity of the wild-type strain, M182/pYMZ83, under aerobic growth conditions (350 Miller units). Error bars indicate standard errors of the means from three separate experiments.
CRP interaction at the clyA promoter in vitro.
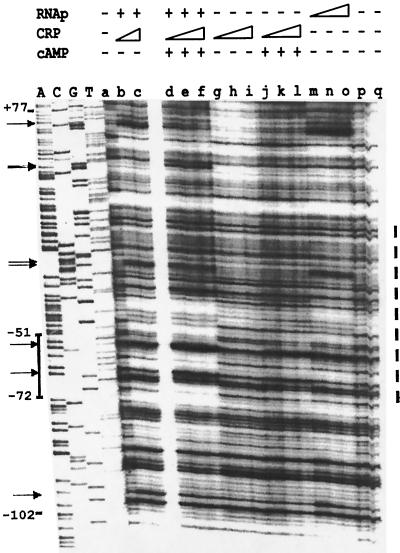
The involvement of CRP in the regulation of clyA gene expression and the presence of a potential CRP binding site in the clyA upstream region suggested a direct interaction of CRP with the clyA promoter. To investigate whether CRP could directly bind to the clyA promoter region, gel mobility shift assays and DNase I footprint analysis with purified CRP and the clyA DNA were carried out as described in Materials and Methods. A weak interaction between CRP and the clyA promoter was indicated by results from gel shift assays (data not shown). In the footprint analysis weakly footprinted regions were obtained only when CRP and RNA polymerase were both present. A region of protection from position −53 to −72, which encompasses the putative CRP binding site, was caused by CRP in the presence of cAMP and RNA polymerase (Fig. 4). Apparent hypersensitivity at positions −55 and −64 was caused by CRP in the presence of cAMP and RNA polymerase and by RNA polymerase at positions −21 and −22, which is similar to the hypersensitive sites −24, −25, and −54, noted in footprint analysis with FNR, HlyX, and RNA polymerase (21). Additional hypersensitive sites were found at position −98 with RNA polymerase (diminished by the addition of CRP and cAMP), at positions +35 and +36 with RNA polymerase plus CRP and cAMP, and at position +63 with RNA polymerase (diminished by the addition of CRP and cAMP). Evidently CRP interacted in a cAMP-dependent manner with the postulated suboptimal binding site. Based on the above observations we suggest a direct role for CRP in the positive regulation of clyA expression.
FIG. 4.
Binding of purified CRP to clyA. DNase I footprint assay of the clyA promoter region with CRP and RNA polymerase. The additions of CRP, RNA polymerase (RNAp), and cAMP were as indicated at the top. The extent of the DNA fragment used is shown by the indicated positions (base pairs). The position of the putative CRP binding site is shown by a solid line along the left side. The approximate region of interaction with RNA polymerase is shown by a dashed line along the right. Hypersensitive sites are indicated with arrows. When included, cAMP was added at a concentration of 20 mM. Lanes: a, no protein; b, CRP (4.7 nM) plus RNA polymerase (0.5 U); c, CRP (9.4 nM) plus RNA polymerase (22 nM); d, CRP (4.7 nM) plus cAMP plus RNA polymerase (22 nM); e, CRP (9.4 nM) plus cAMP plus RNA polymerase (22 nM); f, CRP (19 nM) plus cAMP plus RNA polymerase (22 nM); g, CRP (9.4 nM); h, CRP (19 nM); i, CRP (38 nM); j, CRP (9.4 nM) plus cAMP; k, CRP (19 nM) plus cAMP; 1, CRP (38 nM) plus cAMP; m, RNA polymerase (22 nM); n, RNA polymerase (44 nM); o, RNA polymerase (87 nM); p, no protein; q, no protein.
An altered CRP site in the clyA promoter results in altered expression of ClyA protein.
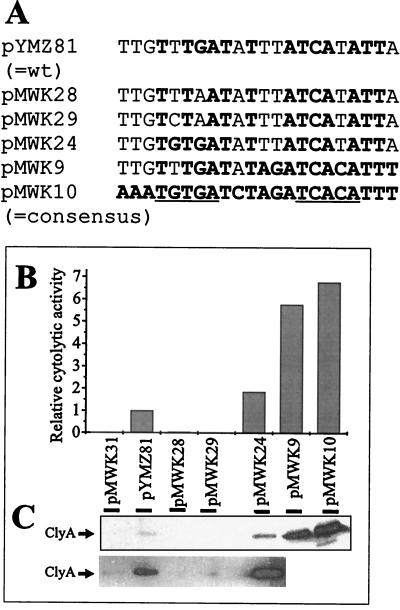
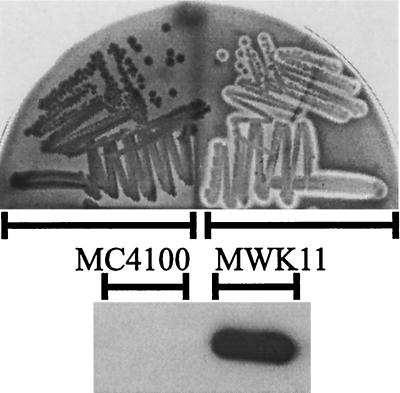
Since the potential CRP binding site in the clyA upstream region shows only partial homology (13 out of 22 bases) with the proposed consensus sequence, (5′AAATGTGATCTAGATCACATTT-3′) (16), we wanted to investigate whether the sequence features of this site are relevant for the regulation of ClyA expression. We therefore introduced site-specific changes in the clyA upstream sequence of the plasmid pYMZ81 (see Materials and Methods) (Fig. 5A). Site-specific alterations in the upstream pentamer of the potential CRP site resulted in the plasmid clones pMWK24 (TGTGA), pMWK28 (TTTAA), and pMWK29 (TCTAA). In addition, we substituted four positions in the predicted CRP binding site, resulting in the plasmid pMWK9, having an altered CRP site (5′-TTGTTTGATATAGATCACATTT-3′) which matches the consensus in 17 out of 22 bases. The construct pMWK10 contains an altered CRP site (5′-AAATGTGATCTAGATCACATTT-3′) that perfectly matches the consensus. We subsequently quantified the cytolytic activity of different E. coli strains carrying these constructs by using the erythrocyte assay (Fig. 5B) and by monitoring the cellular levels of ClyA protein with Western immunoblotting (Fig. 5C). Compared with DH5α/pYMZ81, substantial decreases in cellular ClyA protein and cytolytic activity were exhibited by DH5α/pMWK28 and DH5α/pMWK29. This is consistent with previous findings (25) which suggested that alterations in the upstream pentamer (TGTGA) at position two (G→C) and at position four (G→A) abolish CRP binding. DH5α/pMWK24, which has an improved CRP site (T→G) at position two in the upstream pentamer, showed an increased cellular level of ClyA protein and a stronger cytolytic activity than DH5α/pYMZ81. An even greater increase in cellular ClyA protein and cytolytic activity was exhibited by DH5α carrying pMWK9 or pMWK10. That the clyA expression was CRP-dependent in these cases was confirmed by tests with a crp mutant strain. Only a low, barely detectable level of cellular ClyA protein and cytolytic activity in the crp strain M182 crp carrying pMWK9, pMWK10, and pYMZ81 was observed (data not shown). We also constructed a strain with the CRP consensus DNA binding site in the clyA promoter region on the chromosome of the hns+ strain MC4100. The resulting derivative (strain MWK11) showed a strong hemolytic phenotype on blood agar plates, and there was a high level of ClyA protein in the cells detected by Western blot analysis (Fig. 6). These findings support a model where CRP is involved in expression of ClyA. We concluded that the sequence features of the CRP binding site are important for the positive role of CRP in the regulation of clyA expression.
FIG. 5.
Effect of site-specific alterations in the CRP DNA site and −10 sequence of the clyA promoter on clyA expression from various plasmids in DH5α. The construct pMWK31 contains an altered −10 promoter box (TATGAAT→CACGAAC). The strains were grown to late logarithmic phase and treated as described in Materials and Methods. (A) Sequences of the CRP binding sites in wild-type and mutant clyA clones. The consensus CRP binding site (see reference 16 and references therein) and positions showing identity to the consensus are shown in boldface type. Pentamers referred to in the text are underlined. (B) Cytolytic activity of the different strains towards erythrocytes. The cytolytic activity was measured as described in Materials and Methods, and the activity of the strain DH5α/pYMZ81 was arbitrarily set to 1.0. (C) Determination of ClyA protein content in the different strains by Western immunoblotting using a ClyA-specific antiserum (see Materials and Methods). Strains were grown in LB broth to late logarithmic phase. Approximately 107 bacteria were used for the extract loaded in each lane. The lower panel shows a prolonged exposure, and the ClyA reactive band is indicated with an arrow.
FIG. 6.
Effect of alterations in the CRP binding site in the clyA promoter region on the chromosome to match the consensus sequence (5′-AAATGTGATCTAGATCACATTT-3′). (Upper panel) MC4100 and MWK11 on a blood agar plate after incubation at 37°C for 17 h. (Lower panel) Detection of ClyA protein content in MC4100 and MWK11 by Western immunoblotting using a ClyA-specific antiserum (see Materials and Methods). Strains were grown in LB broth to late logarithmic phase. Approximately 107 bacteria were used for the extract loaded in each lane.
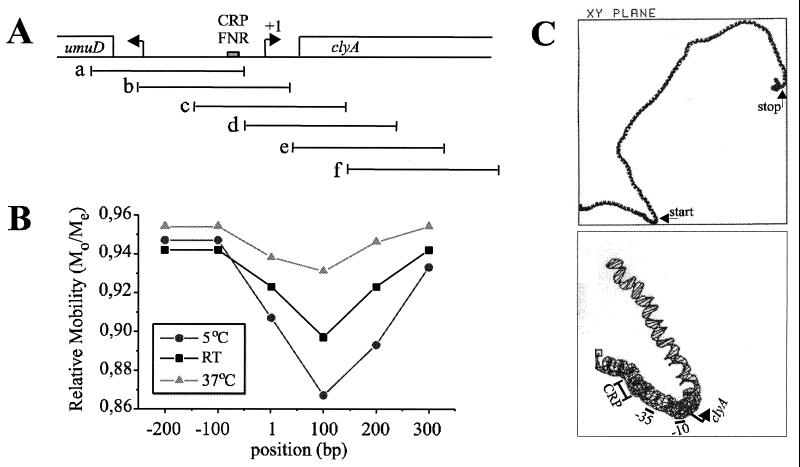
The interaction by CRP with typical binding sites may cause local bending of the DNA, which may affect the curvature properties of a nearby promoter. We noted that the clyA promoter region is rich in A-T base pairs (73.1% for the 186-bp region upstream of the start codon) and the region shows features typical of curved DNA. We studied the potential DNA bending properties by using overlapping DNA fragments in a gel migration analysis and by computer projection (see Materials and Methods). The results confirmed that, in particular, the region containing the 5′ end of clyA may be intrinsically curved (Fig. 7).
FIG. 7.
(A) Schematic drawing of the umuD clyA intercistronic region. The DNA site for CRP-FNR is shown as a grey box, and transcriptional start points for clyA and umuD are indicated by horizontal arrows. The extents of the overlapping DNA fragments generated by PCR, which were used in DNA bending analysis experiments, are shown as horizontal lines. (B) Mapping of the center of the sequence directed bend. Mos as distances from the top of the gel were divided by the Me for each fragment. The Mo/Me value was plotted against the center position (in base pairs) of the DNA fragment. RT, room temperature. (C) The upper panel shows a computer projection analysis of 1,402 bp from the clyA DNA region, which includes the 912-bp clyA coding sequence with 372 bp upstream of the clyA start codon and 118 bp downstream of the clyA stop codon. The positions of the translational start and stop codons of clyA are indicated by arrows. The lower panel shows an enlargement of the predicted curvature pattern in the clyA promoter region from position 200 to 400. The positions of the predicted CRP binding site (−72 to −51), the −35 sequence (−35 to −30), the −10 sequence (−12 to −6), and the transcriptional start point (+1) are indicated.
Involvement of both CRP and FNR in regulation of clyA expression during anaerobic growth conditions.
It was previously reported that overproduction of the FNR homolog HlyX of A. pleuropneumoniae in anaerobically grown E. coli K-12 results in binding to the FNR site and activation of clyA expression, while overproduction of the E. coli FNR protein results in less efficient activation (21). The requirement of CRP and FNR for anaerobic clyA expression was subsequently investigated. The low-copy clyA::lacZ reporter system pYMZ83 was used in different strains to study the level of clyA expression (Fig. 3), and a cat (Cmr) gene block was introduced into the hns locus of the strains M182 fnr and M182 crp fnr, resulting in the strains BEU701 and BEU705, respectively (see Materials and Methods). During anaerobic growth a clearly reduced β-galactosidase activity was observed in both fnr and crp strains (most reduced in the crp fnr double mutant strain) (Fig. 3), suggesting an involvement of both CRP and FNR in the transcriptional regulation of clyA expression. As evidenced by the lysis of erythrocytes in agar (Table 3), the absence of CRP, but not of FNR, reduced the level of clyA derepression in anaerobically grown hns strains. The attenuated lytic activity of BEU705 could be restored both with and without the presence of oxygen by the introduction of CRP on a plasmid (pDW300), and to some extent, under anaerobic conditions only, by the introduction of FNR (plasmid pGS24) or altered FNR proteins having CRP binding specificities (plasmids pGS215 and pGS297). We concluded that the clyA promoter is dependent on CRP also during anoxic growth conditions and that FNR to some extent can complement the requirement for CRP during anaerobic growth only.
DISCUSSION
Silencing of clyA by H-NS and relief of silencing by SlyA.
The clyA locus in the E. coli K-12 chromosome does not seem to be expressed under most laboratory growth conditions, and our present evidence established that the gene is subject to silencing by H-NS. The slyA and mprA genes have been shown to activate the expression of clyA when present in multiple copies (13, 34, 35, 41). In the present work we demonstrate that the major clyA transcript in the hns strain BEU616 has a 5′ end (+1) located 72 nucleotides upstream of the start of the clyA coding sequence. Thus, upon relief of the H-NS silencing, either in hns mutants (this work), or by overproduction of SlyA (34), the same promoter appeared to be active. Site-specific alterations of the putative −10 clyA promoter box (TATGAAT→CACGAAC) resulted in a significantly decreased expression of ClyA, establishing that this particular promoter is crucial for ClyA expression.
When H-NS acts as a silencer or repressor it binds to AT-rich, curved sequences and thereby blocks transcription of the gene in question (6). The DNA of the clyA promoter (the 186 bp immediately upstream of the clyA start codon) is notably A-T rich (73.1%). Computer bend predictions of the clyA locus suggested sharp bends both in the promoter and in the structural gene, and DNA bending analysis of the clyA promoter showed that it contains intrinsic curvature (Fig. 7). By studying the interaction with purified H-NS and the clyA gene in vitro using electrophoretic mobility shift and DNase I footprint assays (Fig. 2), it could be concluded that H-NS binds preferentially to DNA fragments upstream and downstream of the clyA transcriptional start point (+1). The protection of the clyA −10 and −35 regions was less pronounced than for surrounding sequences, something that has also been seen with the promoter for the proU operon, encoding a glycine betaine transport system (33). Hence, it appears that the very low level of clyA expression in E. coli K-12 strains is due to a direct interaction of H-NS with the clyA locus. By monitoring the expression from a chromosomal clyA::luxAB fusion, we observed that the highest clyA transcription coincided with the late logarithmic phase in both H-NS mutants and SlyA-overproducing strains (Fig. 1), which was consistent with previous observations with ClyA activity in MprA and SlyA-overproducing strains (13, 34). SlyA was not essential for clyA expression in an hns strain background, since the hns slyA strain MWK6 and the hns slyA clyA::luxAB strain MWK10 did not show a reduced ClyA expression. In addition, the overexpression of SlyA in the hns strain JON31/pJON22, the hns clyA::luxAB strain JON34/pJON22, and the hns slyA strain MWK6/pJON22, did not result in a further elevation of ClyA expression. Based on our findings, demonstrating no absolute requirement of SlyA for ClyA expression, but rather a copy number effect, we suggest that SlyA may not be involved specifically in the natural regulation of clyA. The observed regulatory effects on clyA with SlyA (and likely also MprA) may well be of a more general nature, e.g., competing with H-NS binding at the clyA locus. It has been suggested that SlyA-related proteins play key roles in the global regulation of diverse aspects of bacterial physiology (57). It has also been implied that rather than being a classical transcriptional activator, MprA may act like some histone-like E. coli proteins, modulating the transcription of specific promoters by locally altering DNA topology (14). In Salmonella, SlyA was demonstrated to regulate the expression of multiple proteins during stationary phase and during infection of macrophages (8), but the role of SlyA in E. coli is not yet understood. When present in multiple copies in E. coli K-12, the cloned slyA locus affected the expression of more than 50 proteins according to analyses using two-dimensional PAGE (39). This indicates that SlyA may not be specifically linked with the regulation of the H-NS-silenced clyA locus.
The clyA promoter is dependent on CRP for efficient expression.
In addition to the strict control exerted by H-NS, the clyA locus appeared to be controlled by the global regulatory protein CRP. According to our data CRP is required for efficient ClyA expression. A much reduced transcription of clyA in hns crp double mutants compared with hns mutants was evident by using a transcriptional clyA::lacZ fusion, and in line with these findings a substantial decrease in cellular ClyA protein and cytolytic activity was observed. The relief of H-NS silencing by SlyA was also much less efficient in the absence of CRP, since only a very weak cytolytic activity could be detected in CRP-deficient strains overexpressing SlyA (Table 3). Results from DNase I footprint experiments were consistent with the idea that the role of CRP in ClyA expression is to directly interact with the clyA promoter region (Fig. 4). Further evidence supporting the model that CRP is involved in the expression of ClyA was obtained by altering the sequence of the potential CRP binding site in the clyA upstream region, both located on the plasmid and on the chromosome (Fig. 5 and 6). By altering the DNA site for CRP to reduce its similarity to the consensus, ClyA expression was significantly lowered. In contrast, the altered CRP binding site that matched the consensus more closely resulted in substantially increased ClyA expression in crp+ but not crp strains. These findings supported a model in which the clyA promoter is dependent on CRP for efficient expression, and where the predicted DNA site for CRP is important for this regulation.
Anaerobic regulation of clyA involves both CRP and FNR.
Results from experiments using a low-copy plasmid-borne clyA promoter-lacZ fusion were consistent with the idea that both CRP and FNR are involved in the transcriptional regulation of clyA under anaerobic growth conditions (Fig. 3). Our findings suggest that FNR and CRP bind to the same sequence in the clyA promoter. There are other examples of binding of FNR and CRP to the same site (48). Evidently, CRP, and not FNR, was required for the expression of ClyA in hns mutants under anoxic conditions, although it appeared that FNR could partly complement CRP. We also observed that the clyA-lacZ fusion in the wild-type strain (M182) was expressed at a higher level (more than twofold) during anaerobic growth, suggesting that the clyA locus may be less repressed in the absence than in the presence of oxygen. It was shown previously that when expressed in anaerobically grown E. coli K-12, the FNR homolog HlyX of A. pleuropneumoniae and, although much less effective, FNR, are able to activate ClyA expression, presumably by binding to the FNR binding site in the clyA upstream region (21).
The clyA promoter: an H-NS silenced class I promoter.
Unlike the situation in eukaryotes, where gene expression is thought to be generally repressed by packaging of the DNA into nucleosomes, the DNA of prokaryotes is generally considered to be available for transcription at all times (55). There are, however, certain prokaryotic gene loci that are apparently not expressed under tested growth conditions. Such loci are referred to as cryptic, and some of them are efficiently silenced by the nucleoid-associated protein H-NS (3, 62). The clyA locus is an interesting new example of H-NS-silenced operons. The locus has some features in common with the cryptic β-glucoside (bgl) operon of E. coli, which is thought to be kept in a silenced state by a repressing nucleoprotein complex consisting of H-NS and other cellular factors. The complex renders the bgl promoter inaccessible to RNA polymerase and CRP (49). Silencing of the bgl operon is relieved by various mutations, including (i) mutations in hns (initially termed bglY) (12, 20) and in genes encoding the subunits of DNA gyrase (15), (ii) integration of insertion elements in cis to the promoter (44), and (iii) deletion of either one of the silencer sequences (32). These mutations may all, directly or indirectly, affect the locked conformation of the upstream region so that they allow more-efficient transcription at the bgl promoter. In addition, point mutations that improve the CRP binding site within the bgl promoter, resulting in CRP binding with higher affinity, cause activated bgl transcription (45). It appears that clyA, similar to the bgl operon, has a weak promoter and no classical operator site. The presence of a −10 promoter box (TATGAAT) centered at −9, a −35 sequence (TTGACG) centered at −32.5, and binding sites for CRP and FNR centered at −61.5 suggested that this clyA promoter is a class I promoter that could be transcriptionally activated by CRP or FNR. Another case of an H-NS silenced operon in which CRP has a positive role is the pap fimbrial adhesin determinant found in uropathogenic E. coli (19). However, transcription of pap is independent of CRP activation in the E. coli K-12 mutant lacking H-NS (18). That finding led to the suggestion of a new role for CRP: it can mediate its positive regulatory function by alleviating transcriptional silencing. In contrast to the situation found in the clyA locus, the binding site for CRP in pap is located relatively far from the promoters and the protein-DNA interaction there is rather clearly shown by in vitro footprint analysis (19). It is possible that CRP may also alleviate the action of H-NS in the case of clyA, e.g., by altering the local DNA conformation and/or by interfering with its DNA binding. An indication of such a role was obtained when the CRP site on the chromosome was altered to perfectly match the consensus, resulting in derepression of the clyA gene. However, the results were also consistent with the suggestion that CRP directly interacted with the RNA polymerase. The genetic evidence suggested a positive role for CRP both in the absence of H-NS and during SlyA overproduction. The suboptimal design of its binding site in the clyA DNA evidently did not allow for any efficient CRP-mediated alleviation of H-NS silencing, but there was a need for additional factors. The tight control of clyA transcription in wild-type E. coli during laboratory cultivation is intriguing, and it remains to be seen if there are different pathways for induction of its expression. For example, it will be of interest to consider whether or not CRP may act in direct cooperation with other factors under some conditions. It is also possible that expression of ClyA could be initiated at some stage during an infection process, as has been shown for the bgl operon (27). Considering the potent cytotoxic properties of this cytolysin, it appears reasonable that it would be strictly regulated, especially in nonpathogenic strains.
ACKNOWLEDGMENTS
We thank Monica Persson for skillful technical assistance and J. R. Guest for kindly providing fnr plasmids.
This work was supported by grants from the Swedish Natural Science Research Council, the Swedish Medical Research Council, and the Göran Gustafsson Foundation for Research in Natural Sciences and Medicine.
REFERENCES
- 1.Atlung T, Ingmer H. H-NS: a modulator of environmentally regulated gene expression. Mol Microbiol. 1997;24:7–17. doi: 10.1046/j.1365-2958.1997.3151679.x. [DOI] [PubMed] [Google Scholar]
- 2.Bell A I, Gaston K L, Cole J A, Busby S J. Cloning of binding sequences for the Escherichia coli transcription activators, FNR and CRP: location of bases involved in discrimination between FNR and CRP. Nucleic Acids Res. 1989;17:3865–3874. doi: 10.1093/nar/17.10.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender R A. Variations on a theme by Escherichia. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular Biology. Washington, D.C.: ASM Press; 1996. pp. 4–9. [Google Scholar]
- 4.Bertani J. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolshoy A, McNamara P, Harrington R E, Trifonov E N. Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc Natl Acad Sci USA. 1991;88:2312–2316. doi: 10.1073/pnas.88.6.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bracco L, Kotlarz D, Kolb A, Diekmann S, Buc H. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989;8:4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brosius J. Superpolylinkers in cloning and expression vectors. DNA. 1989;8:759–777. doi: 10.1089/dna.1989.8.759. [DOI] [PubMed] [Google Scholar]
- 8.Buchmeier N, Bossie S, Chen C Y, Fang F C, Guiney D G, Libby S J. SlyA, a transcriptional regulator of Salmonella typhimurium, is required for resistance to oxidative stress and is expressed in the intracellular environment of macrophages. Infect Immun. 1997;65:3725–3730. doi: 10.1128/iai.65.9.3725-3730.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busby S, Kotlarz D, Buc H. Deletion mutagenesis of the Escherichia coli galactose operon promoter region. J Mol Biol. 1983;167:259–274. doi: 10.1016/s0022-2836(83)80335-0. [DOI] [PubMed] [Google Scholar]
- 10.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 11.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 12.Defez R, De Felice M. Cryptic operon for beta-glucoside metabolism in Escherichia coli K12: genetic evidence for a regulatory protein. Genetics. 1981;97:11–25. doi: 10.1093/genetics/97.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.del Castillo F J, Leal S C, Moreno F, del Castillo I. The Escherichia coli K-12 sheA gene encodes a 34-kDa secreted hemolysin. Mol Microbiol. 1997;25:107–115. doi: 10.1046/j.1365-2958.1997.4391813.x. [DOI] [PubMed] [Google Scholar]
- 14.del Castillo I, Gonzalez-Pastor J E, San Millan J L, Moreno F. Nucleotide sequence of the Escherichia coli regulatory gene mprA and construction and characterization of mprA-deficient mutants. J Bacteriol. 1991;173:3924–3929. doi: 10.1128/jb.173.12.3924-3929.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiNardo S, Voelkel K A, Sternglanz R, Reynolds A E, Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 16.Ebright R H. Transcription activation at class I CAP-dependent promoters. Mol Microbiol. 1993;8:797–802. doi: 10.1111/j.1365-2958.1993.tb01626.x. [DOI] [PubMed] [Google Scholar]
- 17.Forsberg A J, Pavitt G D, Higgins C F. Use of transcriptional fusions to monitor gene expression: a cautionary tale. J Bacteriol. 1994;176:2128–2132. doi: 10.1128/jb.176.7.2128-2132.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forsman K, Sondén B, Göransson M, Uhlin B E. Antirepression function in Escherichia coli for the cAMP-cAMP receptor protein transcriptional activator. Proc Natl Acad Sci USA. 1992;89:9880–9884. doi: 10.1073/pnas.89.20.9880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Göransson M, Forsman K, Nilsson P, Uhlin B E. Upstream activating sequences that are shared by two divergently transcribed operons mediate cAMP-CRP regulation of pilus-adhesin in Escherichia coli. Mol Microbiol. 1989;3:1557–1565. doi: 10.1111/j.1365-2958.1989.tb00141.x. [DOI] [PubMed] [Google Scholar]
- 20.Göransson M, Sondén B, Nilsson P, Dagberg B, Forsman K, Emanuelsson K, Uhlin B E. Transcriptional silencing and thermoregulation of gene expression in Escherichia coli. Nature. 1990;344:682–685. doi: 10.1038/344682a0. [DOI] [PubMed] [Google Scholar]
- 21.Green J, Baldwin M L. The molecular basis for the differential regulation of the hlyE-encoded haemolysin of Escherichia coli by FNR and HlyX lies in the improved activating region 1 contact of HlyX. Microbiology. 1997;143:3785–3793. doi: 10.1099/00221287-143-12-3785. [DOI] [PubMed] [Google Scholar]
- 22.Griffiths L, Cole J A. Lack of redox control of the anaerobically-induced nirB+ gene of Escherichia coli K-12. Arch Microbiol. 1987;147:364–369. doi: 10.1007/BF00406134. [DOI] [PubMed] [Google Scholar]
- 23.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 24.Horii Z, Clark A J. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol. 1973;80:327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- 25.Jansen C, Gronenborn A M, Clore G M. The binding of the cyclic AMP receptor protein to synthetic DNA sites containing permutations in the consensus sequence TGTGA. Biochem J. 1987;246:227–232. doi: 10.1042/bj2460227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson J, Dagberg B, Richet E, Uhlin B E. H-NS and StpA proteins stimulate expression of the maltose regulon in Escherichia coli. J Bacteriol. 1998;180:6117–6125. doi: 10.1128/jb.180.23.6117-6125.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan M A, Isaacson R E. In vivo expression of the beta-glucoside (bgl) operon of Escherichia coli occurs in mouse liver. J Bacteriol. 1998;180:4746–4749. doi: 10.1128/jb.180.17.4746-4749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koop A H, Hartley M E, Bourgeois S. A low-copy-number vector utilizing beta-galactosidase for the analysis of gene control elements. Gene. 1987;52:245–256. doi: 10.1016/0378-1119(87)90051-5. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Lai X H, Arencibia I, Johansson A, Wai S N, Oscarsson J, Kalfas S, Sundqvist K G, Mizunoe Y, Sjöstedt A, Uhlin B E. Cytocidal and apoptotic effects of the ClyA protein from Escherichia coli on primary and cultured monocytes and macrophages. Infect Immun. 2000;68:4363–4367. doi: 10.1128/iai.68.7.4363-4367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Link A J, Phillips D, Church G M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopilato J, Wright A. Mechanisms of activation of the cryptic bgl operon of Escherichia coli K-12. In: Drlica K, Riley M, editors. The bacterial chromosome. Washington, D.C.: American Society for Microbiology; 1990. pp. 435–444. [Google Scholar]
- 33.Lucht J M, Dersch P, Kempf B, Bremer E. Interactions of the nucleoid-associated DNA-binding protein H-NS with the regulatory region of the osmotically controlled proU operon of Escherichia coli. J Biol Chem. 1994;269:6578–6586. [PubMed] [Google Scholar]
- 34.Ludwig A, Bauer S, Benz R, Bergmann B, Goebel W. Analysis of the SlyA-controlled expression, subcellular localization and pore-forming activity of a 34 kDa haemolysin (ClyA) from Escherichia coli K-12. Mol Microbiol. 1999;31:557–567. doi: 10.1046/j.1365-2958.1999.01196.x. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig A, Tengel C, Bauer S, Bubert A, Benz R, Mollenkopf H J, Goebel W. SlyA, a regulatory protein from Salmonella typhimurium, induces a haemolytic and pore-forming protein in Escherichia coli. Mol Gen Genet. 1995;249:474–486. doi: 10.1007/BF00290573. [DOI] [PubMed] [Google Scholar]
- 36.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 38.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oscarsson J. Ph.D. thesis. Umeå University Medical Dissertations; 1999. , New Series no. 628, Umeå, Sweden. [Google Scholar]
- 40.Oscarsson J, Mizunoe Y, Li L, Lai X H, Wieslander Å, Uhlin B E. Molecular analysis of the cytolytic protein ClyA (SheA) from Escherichia coli. Mol Microbiol. 1999;32:1226–1238. doi: 10.1046/j.1365-2958.1999.01435.x. [DOI] [PubMed] [Google Scholar]
- 41.Oscarsson J, Mizunoe Y, Uhlin B E, Haydon D J. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol Microbiol. 1996;20:191–199. doi: 10.1111/j.1365-2958.1996.tb02500.x. [DOI] [PubMed] [Google Scholar]
- 42.Owen-Hughes T A, Pavitt G D, Santos D S, Sidebotham J M, Hulton C S, Hinton J C, Higgins C F. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992;71:255–265. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- 43.Ralph E T, Guest J R, Green J. Altering the anaerobic transcription factor FNR confers a hemolytic phenotype on Escherichia coli K12. Proc Natl Acad Sci USA. 1998;95:10449–10452. doi: 10.1073/pnas.95.18.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynolds A E, Felton J, Wright A. Insertion of DNA activates the cryptic bgl operon in E. coli K12. Nature. 1981;293:625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- 45.Reynolds A E, Mahadevan S, LeGrice S F, Wright A. Enhancement of bacterial gene expression by insertion elements or by mutation in a CAP-cAMP binding site. J Mol Biol. 1986;191:85–95. doi: 10.1016/0022-2836(86)90424-9. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Sansonetti P J, Ryter A, Clerc P, Maurelli A T, Mounier J. Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect Immun. 1986;51:461–469. doi: 10.1128/iai.51.2.461-469.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sawers G, Kaiser M, Sirko A, Freundlich M. Transcriptional activation by FNR and CRP: reciprocity of binding-site recognition. Mol Microbiol. 1997;23:835–845. doi: 10.1046/j.1365-2958.1997.2811637.x. [DOI] [PubMed] [Google Scholar]
- 49.Schnetz K, Wang J C. Silencing of the Escherichia coli bgl promoter: effects of template supercoiling and cell extracts on promoter activity in vitro. Nucleic Acids Res. 1996;24:2422–2428. doi: 10.1093/nar/24.12.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaw D J, Guest J R. Amplification and product identification of the fnr gene of Escherichia coli. J Gen Microbiol. 1982;128:2221–2228. doi: 10.1099/00221287-128-10-2221. [DOI] [PubMed] [Google Scholar]
- 51.Sondén B, Uhlin B E. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 1996;15:4970–4980. [PMC free article] [PubMed] [Google Scholar]
- 52.Spiro S, Gaston K L, Bell A I, Roberts R E, Busby S J, Guest J R. Interconversion of the DNA-binding specificities of two related transcription regulators, CRP and FNR. Mol Microbiol. 1990;4:1831–1838. doi: 10.1111/j.1365-2958.1990.tb02031.x. [DOI] [PubMed] [Google Scholar]
- 53.Spiro S, Guest J R. Activation of the lac operon of Escherichia coli by a mutant FNR protein. Mol Microbiol. 1987;1:53–58. doi: 10.1111/j.1365-2958.1987.tb00526.x. [DOI] [PubMed] [Google Scholar]
- 54.Stoker N G, Fairweather N F, Spratt B G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982;18:335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- 55.Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 56.Taraseviciene L, Naureckiene S, Uhlin B E. Immunoaffinity purification of the Escherichia coli rne gene product. Evidence that the rne gene encodes the processing endoribonuclease RNase E. J Biol Chem. 1994;269:12167–12172. [PubMed] [Google Scholar]
- 57.Thomson N R, Cox A, Bycroft B W, Stewart G S, Williams P, Salmond G P. The Rap and Hor proteins of Erwinia, Serratia and Yersinia: a novel subgroup in a growing superfamily of proteins regulating diverse physiological processes in bacterial pathogens. Mol Microbiol. 1997;26:531–544. doi: 10.1046/j.1365-2958.1997.5981976.x. [DOI] [PubMed] [Google Scholar]
- 58.Uhlin B E, Mizunoe Y. Expression of a novel contact-hemolytic activity by E. coli. J Cell Biochem Suppl. 1994;18A:71. [Google Scholar]
- 59.Wallace A J, Stillman T J, Atkins A, Jamieson S J, Bullough P A, Green J, Artymiuk P J. E. coli hemolysin E (HlyE, ClyA, SheA): X-ray crystal structure of the toxin and observation of membrane pores by electron microscopy. Cell. 2000;100:265–276. doi: 10.1016/s0092-8674(00)81564-0. [DOI] [PubMed] [Google Scholar]
- 60.West D, Williams R, Rhodius V, Bell A, Sharma N, Zou C, Fujita N, Ishihama A, Busby S. Interactions between the Escherichia coli cyclic AMP receptor protein and RNA polymerase at class II promoters. Mol Microbiol. 1993;10:789–797. doi: 10.1111/j.1365-2958.1993.tb00949.x. [DOI] [PubMed] [Google Scholar]
- 61.Willets N S, Clark A J, Low B. Genetic locations of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol. 1969;97:244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams R M, Rimsky S. Molecular aspects of the E. coli nucleoid protein, H-NS: a central controller of gene regulatory networks. FEMS Microbiol Lett. 1997;156:175–185. doi: 10.1111/j.1574-6968.1997.tb12724.x. [DOI] [PubMed] [Google Scholar]
- 63.von Gabain A, Belasco J G, Schottel J L, Chang A C, Cohen S N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci USA. 1983;80:653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, Gunasekera A, Ebright Y W, Ebright R H. Derivatives of CAP having no solvent-accessible cysteine residues, or having a unique solvent-accessible cysteine residue at amino acid 2 of the helix-turn-helix motif. J Biomol Struct Dyn. 1991;9:463–473. doi: 10.1080/07391102.1991.10507929. [DOI] [PubMed] [Google Scholar]
- 65.Zuber F, Kotlarz D, Rimsky S, Buc H. Modulated expression of promoters containing upstream curved DNA sequences by the Escherichia coli nucleoid protein H-NS. Mol Microbiol. 1994;12:231–240. doi: 10.1111/j.1365-2958.1994.tb01012.x. [DOI] [PubMed] [Google Scholar]